Abstract
Humans possess three genetic isoforms of apolipoprotein E (ApoE)—ApoE2, ApoE3, and ApoE4—that confer differential risk for Alzheimer's disease (AD); however, the underlying mechanisms are poorly understood. This study sought to investigate the impact of human ApoE isoforms on brain energy metabolism, an area significantly perturbed in preclinical AD. A TaqMan custom array was performed to examine the expression of a total of 43 genes involved in glucose and ketone body transport and metabolism, focusing on pathways leading to the generation of acetyl-CoA, in human ApoE gene-targeted replacement female mice. Consistent with our previous findings, brains expressing ApoE2 exhibited the most robust profile, whereas brains expressing ApoE4 displayed the most deficient profile on the uptake and metabolism of glucose, the primary fuel for the brain. Specifically, the three ApoE brains differed significantly in facilitated glucose transporters, which mediate the entry of glucose into neurons, and hexokinases, which act as the “gateway enzyme” in glucose metabolism. Interestingly, on the uptake and metabolism of ketone bodies, the secondary energy source for the brain, ApoE2 and ApoE4 brains showed a similar level of robustness, whereas ApoE3 brains presented a relatively deficient profile. Further, ingenuity pathway analysis indicated that the PPAR-γ/PGC-1α signaling pathway could be activated in the ApoE2 brain and inhibited in the ApoE4 brain. Notably, PGC-1α overexpression ameliorated ApoE4-induced deficits in glycolysis and mitochondrial respiration. Overall, our data provide additional evidence that human ApoE isoforms differentially modulate brain bioenergetic metabolism, which could serve as a potential mechanism contributing to their discrete risk impact in AD.
SIGNIFICANCE STATEMENT We uncovered hexokinase as a key cytosolic point in the glucose metabolism that is differentially modulated by the three ApoE genotypes. The differences in hexokinase expression and activity exhibited in the three ApoE brains may underlie their distinct impact on brain glucose utilization and further susceptibility to AD. Therefore, a therapeutic approach that could circumvent the deficiencies in the cytosolic metabolism of glucose by providing glucose metabolizing intermediates, e.g., pyruvate, may hold benefits for ApoE4 carriers, who are at high risk for AD. The bioenergetic robustness may translate into enhanced synaptic activity and, ultimately, reduces the risk of developing AD and/or delays the onset of clinical manifestation.
Keywords: Alzheimer's disease, apolipoprotein E, energy metabolism, glucose, glycolysis, ketone body
Introduction
Alzheimer's disease (AD) currently affects 35 million people worldwide, including 5.4 million Americans. This number is predicted to nearly triple by 2050, projected to 13.8 million (Alzheimer's Association, 2016). Despite the extensive research effort put forth over the last 2 decades, the cause of AD remains elusive, and no effective treatment is currently available. The growing number of AD cases, along with continuous failures in AD drug development (Cummings et al., 2014), underscores the urgent need for advancement in the area of AD therapy. To that end, a better understanding of the mechanisms that counteract AD risk could potentially lead to new strategies aimed at AD prevention and early intervention.
The genetic polymorphism of human apolipoprotein E (ApoE) has been shown to confer differential susceptibility to late-onset AD, which accounts for the majority of the AD cases (Farrer et al., 1997). The three major isoforms, ApoE2, ApoE3, and ApoE4, are the products of three alleles at a single gene locus on the long arm of chromosome 19. The most common isoform, ApoE3, is present in ∼75% of the population and is believed to be risk-neutral. ApoE2 is rare, occurring in 5% of the population, and it shows protection against AD. In contrast, ApoE4 exists in 20% of the population; however it comprises nearly 65% of AD cases (Kim et al., 2009; Liu et al., 2013). Clinically, ApoE4 carriers exhibit accelerated rates of cognitive decline related to aging and AD (Craft et al., 1998; Martins et al., 2005; Caselli et al., 2009, 2011). On the molecular level, ApoE4 has been associated with aggravated AD neuropathology, enhanced neuroinflammation, deregulated lipid metabolism, mitochondrial dysfunction, and impaired synaptic plasticity (Nagy et al., 1995; Chang et al., 2005; Rapp et al., 2006; Klein et al., 2010; Rodriguez et al., 2014).
Previous studies have primarily focused on identifying AD risk mechanisms conferred by ApoE4, and relatively few studies have explored the role of ApoE2 in relation to AD. ApoE2 has been found to slow AD progression and be positively associated with cognitive functions during aging (Nagy et al., 1995; Craft et al., 1998; Martins et al., 2005; Shinohara et al., 2016). Results from preclinical studies are in line with the clinical findings. Forced expression of ApoE2 attenuated the progressive loss of synaptic integrity induced by ApoE4 (Klein et al., 2010). Gene delivery of ApoE2 markedly reduced brain amyloid pathology in an AD mouse model (Zhao et al., 2016). Recently, a nonbiased proteomics study from our laboratory revealed that the ApoE2 brain possessed the most robust regulatory mechanisms for synaptic transmission (Woody et al., 2016). Collectively, these studies suggest that ApoE2 brains are subjected to neuroprotective mechanisms; the uncovering of which may lead to novel AD prevention and risk reduction strategies.
It has been proposed that protection against AD should greatly precede AD onset. Indeed, the preclinical development of AD could last 10–20 years before its clinical manifestation. One of the major changes that typically occurs in the preclinical AD brain is the perturbed brain energy metabolism. Cognitively normal individuals at risk for AD, including those carrying ApoE4, showed reduced cerebral metabolism of glucose (Reiman et al., 2004, 2005). Treatment with a ketogenic agent improved cognitive functions in ApoE3 carriers but not ApoE4 carriers (Reger et al., 2004; Henderson et al., 2009), suggesting that ApoE genetic status affects brain ketone body utilization. Further, a pathway-focused gene profiling study revealed that mouse brains expressing ApoE2 exhibited the most robust profile in insulin-like growth factor 1 signaling and glucose transport (Keeney et al., 2015). Together, these findings suggest that human ApoE genetic status influences brain energy metabolism; however, the underlying mechanisms remain unclear. In the present study, we examined the pathways involved in the transport and metabolism of glucose and ketone bodies, the primary and secondary energy source for the brain, in humanized ApoE mouse and cell models.
Materials and Methods
Animals.
All the animal work presented has been approved by the Institutional Animal Care and Use Committee of the University of Kansas and followed NIH guidelines for the care and use of laboratory animals. Six-month-old human ApoE2, ApoE3, and ApoE4 gene-targeted replacement (hApoE2-TR, hApoE3-TR, and hApoE4-TR) female mice were obtained from Taconic Biosciences. These mice express levels of human ApoE protein similar to human tissue in the brain regions that are most affected by AD. Additionally, immunohistochemistry analyses revealed that human ApoE protein is predominantly expressed in the glia, which recapitulates the pattern of ApoE expression in the non-demented human brains (Sullivan et al., 2004). Therefore, utilization of this animal model would allow for direct measurement and comparison of human ApoE isoform-specific effects in vivo.
Cell culture.
Mouse neuroblastoma cell line Neuro-2a (N2a) was purchased from American Type Culture Collection (CCL-131). N2a cells were grown in DMEM High Glucose (Invitrogen, 11965092) supplemented with 10% fetal bovine serum (FBS; Invitrogen, 16000044). Cells were subcultivated every 3–4 d and maintained at 37°C in a humidified atmosphere with 5% CO2. A new vial of frozen cell stock was recovered when the passage number exceeded 21.
Induction of N2a cell differentiation.
N2a cells were plated at a density of 1 × 105/ml in complete culture medium. Twenty-four hours later, cells were treated with 20 μm retinoic acid (RA; Sigma-Aldrich) in DMEM supplemented with 2% FBS. The medium containing RA was refreshed every 48 h. Cells were subjected to morphological and biochemical analyses after 96 h incubation with RA. Differentiated N2a cells were defined by neuron-like morphology and the enhanced protein expressions of neuronal markers such as NeuN, synaptophysin, and PSD95.
Plasmids and transfection.
Human ApoE2, ApoE3, and ApoE4 cDNA clones expressed in the mammalian vector pCMV6 with C-terminal Myc-DDK Tag were procured from OriGene Technologies. ApoE3 cDNA clone (NM_000041) was directly purchased from OriGene. ApoE2 and ApoE4 cDNA clones were generated by genetic modification of ApoE3 cDNA sequence. Specifically, ApoE2 cDNA clone was obtained by mutation on nucleotide position 388 T to C, resulting in a Cysteine to Arginine substitution in codon 112. ApoE4 cDNA clone was obtained by mutation on nucleotide position 526 C to T, which resulted in an arginine to cysteine substitution in codon 158. peroxisome proliferator-activated receptor-gamma coactivator 1 α (PGC-1α; NM_008904) mouse cDNA clone was also purchased from OriGene. For all the experiments, empty pCMV6 vector was used as control plasmid. For plasmid transfection, 2 μg human ApoE2, ApoE3, and ApoE4 cDNA were introduced to undifferentiated or differentiated N2a cells using lipofectamine 3000 transfection reagent (Invitrogen). For undifferentiated N2a cells, treatment or assays were performed 48 h post-transfection. Differentiated N2a cells were evaluated 96 h after transfection. ApoE4-expressing cells were transfected with 500 ng mouse PGC-1α. Cells were incubated with plasmid DNA/transfection reagent complex for 48 h before protein expression analysis and further experiments.
Cell lines stably expressing human ApoE isoforms.
N2a cells stably expressing human ApoE isoforms were generated as described below. Briefly, a kill curve titration was performed on N2a cells to determine the optimal concentration of geneticin (G418, Invitrogen). Cells were then transfected with 2 μg human ApoE2, ApoE3, and ApoE4 cDNA using lipofectamine 3000 according to the manufacturer's instructions. Forty-eight hours after transfection, the medium containing transfection complexes was replaced with complete N2a culture medium containing 600 μg/ml G418. Cells stably transfected were selected for 10 d, then dissociated from the plates and subjected to serial dilution in 96-well plates. Wells containing single cells were marked and colonies formed in those wells were considered to be monoclonal. Cells were maintained in a humidified 37°C, 5% CO2 environment and G418 were replenished every 3–4 d. An individual colony in each well was transferred to one well in 24-well plates for further expansion. Cell lines stably expressing ApoE at similar levels, as determined by immunoblotting analysis, were used in the study.
Western immunoblotting.
Transfected cells were washed twice by cold Dulbecco's PBS and lysed with Neuronal Protein Extraction Reagent (N-PER, Thermo Scientific, 87792) supplemented with protease and phosphatase inhibitors (Thermo Scientific, A32965). Cortical tissue homogenates were prepared from cortices of 6-month-old female hApoE-TR mice using Tissue Protein Extraction Reagent (T-PER, Thermo Scientific, 78510) containing protease and phosphatase inhibitors. Protein concentrations of the cell extracts and tissue homogenates were determined by BCA protein assay kit (Thermo Scientific, 23225). Immunoblotting analyses were performed as previously published (Woody et al., 2016). The following primary antibodies were used: anti-NeuN (1:1000; Cell Signaling Technology, D3S3I), anti-synaptophysin (1:1000; Cell Signaling Technology, 5461S), anti-PSD95 (1:1000; Cell Signaling Technology, 3450S), anti-apolipoprotein E (1:5000; Millipore, 178479), anti-hexokinase I (1:4000; Cell Signaling Technology, C35C4), anti-hexokinase II (1:2000; Cell Signaling Technology, C64G5), anti-PGC-1α (1:1000; Millipore Sigma, ST1201), anti-β-actin (1:5000; Thermo Scientific, PA5-16914), and anti-β-tubulin (1:5000; Thermo Scientific, MA5-11732). Secondary antibodies were species-specific horseradish peroxidase-conjugated immunoglobulins purchased from Thermo Scientific. Protein bands were visualized using chemiluminescence (Bio-Rad) on C-Digit Blot Scanner (LI-COR). Band densities were quantified using Image Studio v4.0 image digitizing software with signal normalized to loading control proteins.
ATP levels.
ATP contents were determined using CellTiter-Glo Luminescent Cell Viability Assay (Promega) according to the manufacturer's instructions. ATP levels were normalized to the protein content in each sample.
ApoE gene expression.
The gene expression of ApoE after transfection was measured by real-time PCR as previously described (Calero et al., 2009). Genomic DNA was extracted from N2a cells transfected with human ApoE2, ApoE3, ApoE4 cDNA, and mock vector by PureLink Genomic DNA Mini Kit (Invitrogen) following manufacturer's instructions. The primers were synthesized and purchased from Integrated DNA Technologies. The sequences of the primers were as follows: 5′-CGGACATGGAGGACGTGT-3′ (Forward) and 5′-CTGGTACACTGCCAGGCA-3′ (Reverse) for ApoE2, 5′-CGGACATGGAGGACGTGT-3′ (Forward) and 5′-CTGGTACACTGCCAGGCG-3′ (Reverse) for ApoE3, 5′-CGGACATGGAGGACGTGC-3′ (Forward) and 5′-CTGGTACACTGCCAGGCG-3′ (Reverse) for ApoE4. DNA amplification was performed on 100 ng of genomic DNA mixed with 1× Power SYBR Green PCR Master Mix (Applied Biosystems) and 0.3 μm of each primer under the following protocol: Stage 1: AmpErase UNG activation at 50°C for 2 min; Stage 2: AmpliTaq Gold DNA Polymerase activation at 95°C for 10 min; Stage 3: denature at 95°C for 15 s followed by annealing/extension at 60°C for 1 min, for 40 cycles. Negative control contained the same reaction mixture without DNA. All the reactions were run in quadruplet. The fluorescence was detected on QuantStudio 7 Flex Real-Time PCR System (fast 96-well format) and data were calculated by the comparative Ct (ΔCt) method.
TaqMan gene array profiling.
TaqMan gene expression array cards (96-well plate format, which enables analysis of 2 samples against 48 genes) were custom manufactured at Applied Biosystems. A list of 43 genes and 5 candidate control genes examined in the array can be found in Fig. 1-1. Total RNA was extracted from cortex tissues of 6-month-old female hApoE-TR mice using PureLink RNA Mini Kit (Invitrogen). RNA quantity was determined using Qubit Assay Kits (Invitrogen) and RNA quality was analyzed using Agilent RNA 6000 Nano Kit on Agilent 2100 Bioanalyzer system. RNA samples with RNA integrity number greater or equal to 8 were converted to cDNA using the High Capacity RNA-to-cDNA Master Mix (Applied Biosystems) on a T100 thermocycler (Bio-Rad). Taqman qRT-PCR was performed on QuantStudio 7 Flex Real-Time PCR System (Applied Biosystems) under the following thermal cycling conditions: AmpErase UNG activation at 50°C for 2 min; AmpliTaq Gold DNA Polymerase activation at 95°C for 10 min followed by 40 cycles of denaturation at 95°C for 15 s and annealing/extension at 60°C for 1 min. Each reaction mixture contained 10 ng cDNA samples mixed with 1× TaqMan Universal PCR Master Mix.
Data were analyzed using the PCR Array Data Analysis Software (Qiagen). Relative gene expression levels or fold-change (FC) relative to the comparison group were calculated by the comparative Ct (ΔΔCt) method, with Ct denoting threshold cycle. Normalization factor calculation based on the geometric mean of multiple control genes was performed for selection of the endogenous control genes (Vandesompele et al., 2002). Samples collected from four animals per ApoE genotype group were included in the analysis. For each sample, ΔCt was calculated as the difference in average Ct of the target gene and the endogenous control gene. For each ApoE genotype, mean 2−ΔCt was calculated as the arithmetic mean of 2−ΔCt of the four samples in the group. FC was then calculated as mean 2−ΔCt (ApoE genotype group) divided by mean 2−ΔCt (comparison group). The 2−ΔCt values for each target gene between two ApoE genotype groups were statistically compared using Student's t test; p < 0.05 was considered statistically significant. The p values were adjusted by Bonferroni correction. The heat map graphically demonstrated the hierarchical clustering of gene expression data. Distances between samples and target genes were calculated based on ΔCt values using Pearson's correlations. The volcano plot displayed FC in the x-axis versus p values in the y-axis, which enabled the overview of gene-fold regulation and identification of genes with fold-change to the statistical significance level.
IPA network analysis.
Data containing gene identifiers and corresponding FC relative to the comparison group (ApoE3) were uploaded to IPA (ingenuity pathway analysis). IPA upstream regulator analysis identifies transcriptional regulators of the genes in the dataset, which could explain the observed gene expression changes. The analysis predicts likely relevant upstream molecules by assessing how many known downstream targets are present. The prediction is based on the information stored in the Ingenuity Knowledge Base and literature references. A p value was calculated to evaluate whether there is a significant overlap between the set of genes examined and genes regulated by a specific transcriptional factor. A z-score was calculated to predict the state of the upstream regulator: activating (z-score ≥2) or inhibiting (z-score ≤−2).
Hexokinase activity.
Hexokinase activity assay was performed as previously described (Ding et al., 2013). The assay is based upon the reduction of nicotinamide adenine dinucleotide (NAD+) through a coupled reaction with glucose-6-phosphate dehydrogenase and is determined spectrophotometrically by measuring the increase in absorbance at 340 nm. One hundred and fifty microliter reaction mixes containing 13.3 mm MgCl2, 0.112 m glucose, 0.55 mm adenosine 5′ triphosphate, 0.227 mm NAD+, and 1 IU/ml glucose-6-phosphate dehydrogenase in 0.05 m Tris-HCl buffer, pH 8.0, was loaded in each well of 96-well microplate. The plate was incubated at room temperature for 15 min to achieve temperature equilibration. Twenty microgram whole-cell lysates were added to start the reaction. Hexokinase solution prepared by dissolving hexokinase (Sigma-Aldrich) in 0.05 m Tris-HCl buffer, pH 8.0, served as positive control. Assay buffer without sample served as negative control. Absorbance (A) at 340 nm was recorded every 1 min for 30 min at 30°C. ΔA/min from initial linear portion of the curve was used to calculate the enzyme activity.
Mitochondrial respiration and glycolysis.
The XF96 Extracellular Flux Analyzer (Seahorse Bioscience), which measures oxygen consumption rate (OCR) and extracellular acidification rate (ECAR), was used to examine the ApoE isoform-specific effects on mitochondrial respiration and glycolysis in N2a cells. Transfected cells were seeded at a density of 3.1 × 105 cells/cm2 in the 96-well culture plate provided with the flux pack (Seahorse Bioscience). Mito Stress Test and the Glycolysis Stress Test were performed according to the manufacturer's instructions.
For Mito Stress Test, cell growth medium was replaced with prewarmed assay medium (XF base medium supplemented with 5.5 mm glucose, 2 mm glutamine, and 1 mm sodium pyruvate, pH 7.4) and incubated in 37°C incubator without CO2 for 1 h before the test. OCR was measured under basal condition followed by sequential injection of the compounds: ATP synthase inhibitor oligomycin (2 μg/ml), electron transport chain (ETC) accelerator FCCP (1 μm), and a mix of mitochondrial ETC complex inhibitors rotenone (1 μm) and antimycin A (1 μm) to measure ATP-linked respiration, maximal respiration, and non-mitochondrial respiration, respectively.
For Glycolysis Stress Test, cell growth medium was replaced with prewarmed assay medium (XF base medium containing 2 mm glutamine, pH 7.4) and incubated in 37°C incubator without CO2 for 1 h before the assay. ECAR was measured under basal condition followed by serial addition of compounds. The initial injection was a saturating concentration of glucose (10 mm), which provided the measurement of glycolysis rate under basal conditions. The addition of oligomycin (2 μg/ml) inhibited ATP generation and thus shunted the energy production to glycolysis, with the subsequent increase in ECAR revealing the maximum glycolytic capacity of the cells. The final injection, 2-deoxy-d-glucose (2-DG; 50 mm), terminated the glycolysis, as the phosphorylated product of 2-DG cannot be metabolized by phosphoglucose isomerase.
To examine the impact of human ApoE isoforms on cells' ability of using ketone bodies, OCR was measured in N2a-ApoE cells receiving sequential injections of the substrate, which was vehicle control or β-hydroxybutyrate (BHB; 5 mm), oligomycin (5 μg/ml), and FCCP (1 μm). OCR and ECAR values were normalized to the total protein content in each well. Data were analyzed using Seahorse XF Cell Mito Stress Test Report Generator.
Lactate production assay.
Lactate fluorometric assay kit (K607-100; BioVision) was used to determine lactate levels according to the manufacturer's instructions. Briefly, differentiated N2a cells transfected with ApoE isoforms were incubated in DMEM without FBS or glucose for 1 h. The medium was refreshed and cells were incubated in 37°C incubator without CO2 for another 1 h. The assay was started by adding 10 mm glucose to each well of transfected cells followed by incubation in 37°C, 5% CO2 for 10 min. 10 μl medium was collected from each sample and incubated with reaction mix for 30 min at room temperature. The fluorescence was detected at ex/em 540/35 nm using a plate reader (BioTek). Lactate levels were normalized to protein content in each sample.
Statistical analysis.
Each experiment was performed independently at least three times. Data are presented as mean ± SEM. All statistical analyses in the present study were performed using GraphPad Prism 6. One-way ANOVA with Bonferroni post hoc test were performed to compare mean values among the three ApoE genotype groups. A p value <0.05 was considered statistically significant and the degree of significance was represented by the number of asterisk and/or pound sign.
Results
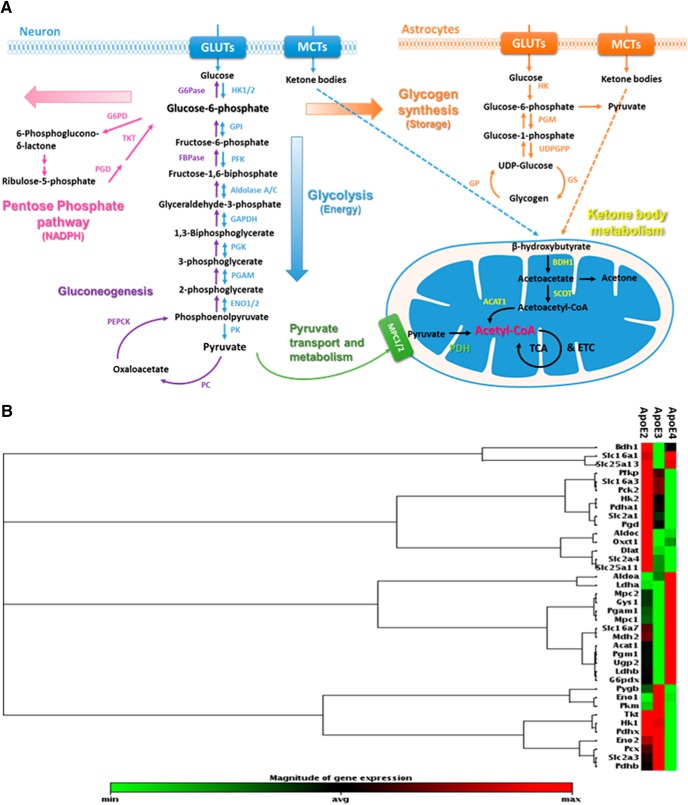
We sought to identify the energy substrate and specific metabolic pathways that are differentially affected by human ApoE isoforms. TaqMan gene expression profiling was conducted on cortical tissues collected from 6-month-old hApoE-TR female mice to evaluate the ApoE isoform-dependent differences in the molecular processes of brain uptake and metabolism of glucose and ketone bodies, two primary energy fuels for the brain. The diagram shown in Figure 1A illustrates the transporters and major metabolic pathways examined in the gene array analysis. A complete list of the 48 genes grouped according to the functional class can be found in Fig. 1-1. FC relative to expression levels in the comparison group (ApoE2 or ApoE3) with the corresponding p value are indicated in Fig. 1-2. The group heat map shown in Figure 1B revealed both distinct and consistent patterns among the three ApoE brains. Specifically, the gene expression profile associated with ApoE4 appeared most distinct from ApoE2 and ApoE3 brains; in comparison, ApoE2 and ApoE3 brains shared some similarities.
Figure 1.
Human ApoE isoforms differentially modulate brain energy metabolism. A, Schematic representation of brain energy substrate uptake and generation of acetyl-CoA. Hexokinase catalyzes the conversion of glucose to glucose-6-phosphate, a “branch-point” metabolite that has alternative metabolic fates. Acetyl-CoA is a critical point of convergence of glucose and ketone body metabolism. Brain glucose and ketone body uptake are strictly controlled by their respective transporters, GLUTs, and MCTs. The figure shows the key enzymes involved in the glucose and ketone body metabolic pathways. B, Gene expression profiles on energy substrate uptake and metabolism in the cortices of 6-month-old hApoE-TR female mice. The group heat map shows the expression of genes involved in glucose and ketone body transport and metabolism for all three ApoE genotypes. n = 4 per ApoE genotype. A complete list of the 48 genes grouped according to the functional class can be found in Figure 1-1. The relative expression (FC) with the corresponding p value are indicated in Figure 1-2.
List of genes examined in the brains of hApoE-TR mice. Download Figure 1-1, DOCX file (16.1KB, docx)
Relative gene expression in the brains of hApoE-TR mice. Download Figure 1-2, DOCX file (16.6KB, docx)
ApoE2-expressing brains possessed the most robust metabolic profile in glucose uptake and glycolysis
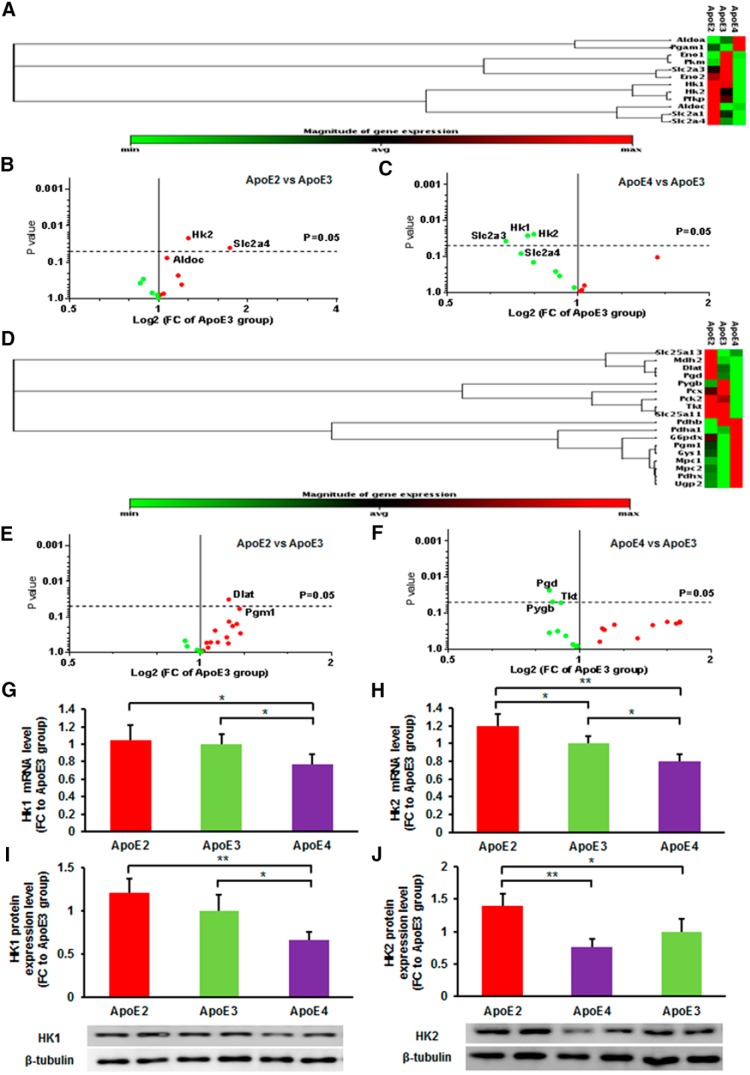
We first examined the gene expression profiles in brain uptake and metabolism of glucose, the primary fuel used by the brain. The volcano plots showed the FCs and p values for significant differences in ApoE2 brains (Fig. 2B) and ApoE4 brains (Fig. 2C) compared with ApoE3 brains. Of the 12 genes analyzed, two exhibited significantly higher levels in ApoE2 brains versus ApoE3 brains, including solute carrier family 2, member 4 (Slc2a4; FC = 2.05, p = 0.040) and hexokinase 2 (Hk2; FC = 1.19, p = 0.022). In contrast, a comparison between ApoE4 and ApoE3 brains yielded three genes that were significantly downregulated in ApoE4 brains: Slc2a3 (FC = 0.68, p = 0.037), Hk1 (FC = 0.76, p = 0.025), and Hk2 (FC = 0.79, p = 0.024).
Figure 2.
ApoE2-expressing brains exhibit the most robust profile on glucose uptake and glycolysis. A, The group heat map shows the expression of genes involved in glucose transport and glycolysis for all three ApoE genotypes. B, C, The volcano plots illustrate FCs (x-axis) and p values (y-axis) between (B) ApoE2 brains versus ApoE3 brains and (C) ApoE4 brains versus ApoE3 brains; significantly altered expression: Hk1/2, Slc2a3 (Glut3), Slc2a4 (Glut4). p < 0.05 was considered statistically significant with Student's t test. D, The heat map shows the expression of key genes involved in pyruvate metabolism, glycogen utilization, and pentose phosphate pathways for all three ApoE genotypes. E, F, The volcano plots show FCs (x-axis) and p values (y-axis) between (E) ApoE2 brains versus ApoE3 brains and (F) ApoE4 brains versus ApoE3 brains; significantly altered expression: Dlat and Pgd. p < 0.05 by Student's t test. n = 4 per group. G–I, Gene and protein expression levels of Hk1 were significantly lower in ApoE4 brains than in ApoE2 and ApoE3 brains. H–J, ApoE2 brains exhibited significantly higher levels of mRNA and protein expression of Hk2. Results were compared using one-way ANOVA with Bonferroni post hoc test. All of the analyses were performed in the cortical tissues of the same 6-month-old hApoE-TR mice. *p < 0.05, **p < 0.01. n = 4–5 per group.
Next, we evaluated the gene expression profiles in other glucose metabolic pathways, including pyruvate transport and metabolism, pentose phosphate pathway, and glycogen utilization. The volcano plot indicated that 14 of 18 genes we examined showed greater expression in ApoE2 brains than in ApoE3 brains (Fig. 2E). In comparison, 10 of 18 genes showed lower expression in ApoE4 brains than in ApoE3 brains (Fig. 2F). Genes that differed significantly included dihydrolipoamide S-acetyltransferase (Dlat; FC = 1.16, p = 0.033) and phosphogluconate dehydrogenase (Pgd; FC = 0.85, p = 0.021). Together, these data indicate that the ApoE2 brain possesses the most robust profile in glucose uptake and metabolism, particularly the glycolysis. In contrast, the ApoE4 brain bears the most deficient profile in glucose utilization.
We further validated the results of the gene array study using independent mRNA quantitation and immunoblot analyses. Among the genes that differed significantly between the ApoE genotype groups, we were particularly interested in Hk1/2 for two reasons: (1) the expression levels of both Hk1 and Hk2 were significantly higher in ApoE2 brains compared with ApoE4 brains; and (2) hexokinase is the “gateway” enzyme that catalyzes the initial step in glucose metabolism. Consistent with the results from the gene array study, protein expression of HK1 was significantly lower in ApoE4 brains than in ApoE2 (FC = 0.55, p = 0.001) and ApoE3 brains (FC = 0.66, p = 0.035; Fig. 2I). Similarly, ApoE2 brains exhibited significantly higher protein levels of HK2 than in both ApoE3 (FC = 1.40, p = 0.029) and ApoE4 brains (FC = 1.83, p = 0.002; Fig. 2J).
ApoE2-expressing cells exhibited the most potent glycolytic activity and mitochondrial respiration
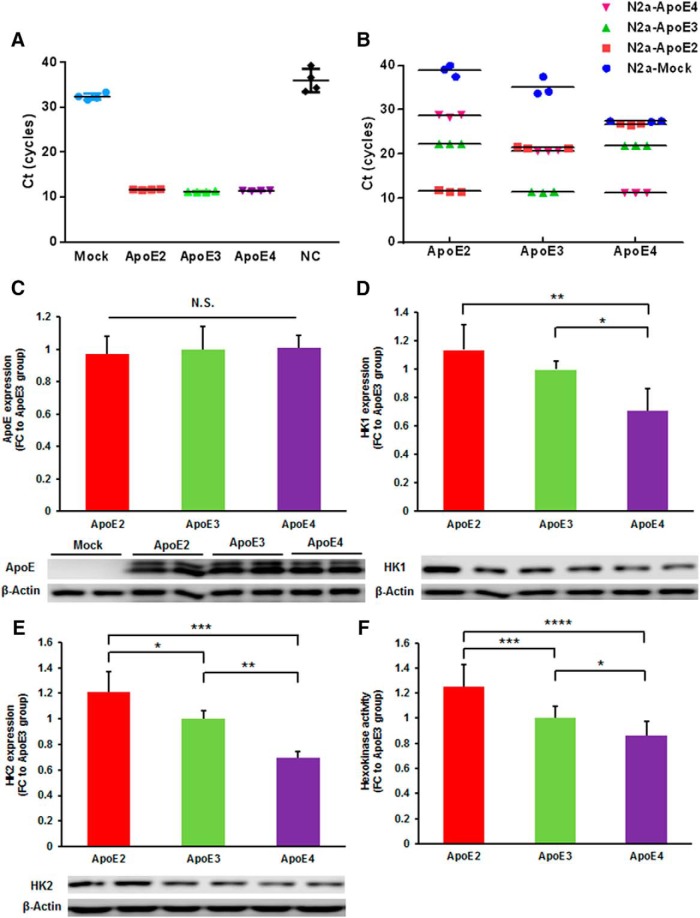
We used cell models expressing human ApoE2, ApoE3, and ApoE4 (N2a-ApoE2, N2a-ApoE3, and N2a-ApoE4) to further study how ApoE isoforms modulate glucose metabolism, focusing on hexokinase and glycolysis. Gene expression and protein levels of ApoE were comparable in N2a cells transfected with human ApoE2, ApoE3 and ApoE4 cDNA clones (Fig. 3A,C). The primers used for PCR amplification for each genotype were highly specific as the difference in Ct value between positive and negative was at least 11 cycles, suggesting that the specific amplifications are 2048-fold more efficient than the nonspecific ones (Fig. 3B). In line with our findings in the hApoE-TR mice, N2a cells transfected with ApoE2 exhibited significantly higher protein levels of Hk2 than cells transfected with ApoE3 (FC = 1.21, p = 0.022; Fig. 3E). Hk1 protein levels were also higher in ApoE2-transfected cells; however, the difference was not statistically significant (FC = 1.14, p = 0.205; Fig. 3D). In contrast, cells transfected with ApoE4 showed significantly lower protein expression of both Hk1 (FC = 0.71, p = 0.016) and Hk2 (FC = 0.69, p = 0.002) than those transfected with ApoE3 (Fig. 3D,E). Additionally, we examined hexokinase activity in the whole-cell lysates collected from transfected cells. Consistent with the protein expression data, N2a-ApoE2 cells showed significantly higher hexokinase activity (FC = 1.25, p = 0.0002), whereas N2a-ApoE4 cells exhibited significantly lower (FC = 0.86, p = 0.026) hexokinase activity than N2a-ApoE3 cells (Fig. 3F).
Figure 3.
Characterization of N2a cells expressing human ApoE isoforms. A, Gene expression levels of ApoE were determined in N2a-ApoE cells by real-time PCR. Amplification plots of three ApoE genotypes are shown. NC, Negative control. Four replicates each group. B, Genomic DNA extracted from N2a cells transfected with human ApoE cDNA or empty vector were amplified with specific primers for ApoE2, ApoE3, and ApoE4 genotyping. Three replicates per group. The difference in Ct values between positive and negative reactions across all genotypes was at least 10 cycles, which indicated the high specificity of the primers. C, ApoE protein levels were comparable among the three ApoE-transfected groups. D, E, Hexokinase expression was assessed in N2a-ApoE cells. Protein levels of Hk1 and Hk2 were significantly lower in ApoE4-transfected group than in ApoE2- and ApoE3-transfected groups. F, Hexokinase activities were determined in the cell lysates of N2a-ApoE cells. Hexokinase activity was the highest in N2a-ApoE2 cells, but the lowest in N2a-ApoE4 cells. Results were normalized to N2a-ApoE3 group. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 by one-way ANOVA. n = 4–5 per group.
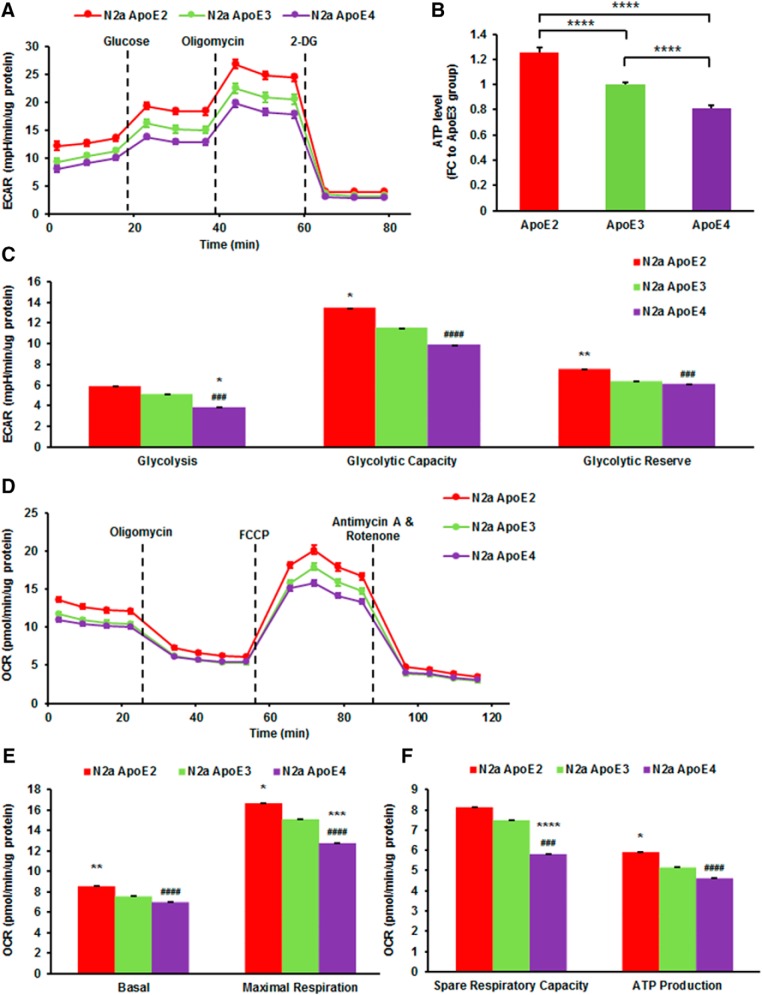
Because hexokinase is the “pacemaker” for glycolysis, and its expression was modulated by ApoE isoforms, we subsequently assessed glycolytic function in N2a-ApoE cells by glycolysis stress test. Kinetic graph displayed ECAR versus time before and after sequential injections of modulating compounds (Fig. 4A). Individual parameters for glycolysis were calculated and shown in Figure 4C. Basal glycolysis rate was significantly lower in ApoE4-expressing cells compared with ApoE2-expressing (FC = 0.65, p < 0.0001) and ApoE3-expressing cells (FC = 0.76, p = 0.017). Oligomycin-induced increase in ECAR was significantly larger, suggesting a higher maximum glycolytic capacity in ApoE2-expressing cells than in ApoE3-expressing cells (FC = 1.17, p = 0.012). ApoE2-expressing cells also had greater glycolytic reserve (FC = 1.19, p = 0.008). Compared with ApoE2-expressing cells, the glycolytic capacity was significantly lower in ApoE4-expressing cells (FC = 0.74, p < 0.0001). Similarly, the glycolytic reserve was markedly lower in ApoE4-expressing cells (FC = 0.95, p = 0.001).
Figure 4.
ApoE2-expressing cells are most robust in glycolytic function and mitochondrial respiration. A, B, Glycolytic stress tests were performed using the Seahorse XF96 Extracellular Flux Analyzer to measure the basal glycolysis, glycolytic capacity, and glycolytic reserve in N2a-ApoE cells. Results were compared using one-way ANOVA with Bonferroni post hoc test. n = 18–20 per group. *p < 0.05, **p < 0.01 vs N2a-ApoE3 group. ###p < 0.001, ####p < 0.0001 vs N2a-ApoE2 group by one-way ANOVA with Bonferroni post hoc test. C–E, Mitochondrial stress tests were performed using the Seahorse XF96 Extracellular Flux Analyzer to measure mitochondrial basal respiration, maximal respiration capacity and ATP-linked respiration in N2a-ApoE cells. n = 18–20 per group. F, ATP levels were determined in cell lysates of N2a-ApoE cells. n = 5 per group. Results were compared using one-way ANOVA with Bonferroni post hoc test. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 vs N2a-ApoE3 group. ###p < 0.001, ####p < 0.0001 vs N2a-ApoE2 group.
In addition to glycolysis, we also evaluated the impact of human ApoE isoforms on mitochondrial respiration by performing mito stress test on N2a-ApoE cells (Fig. 4D). The basal mitochondrial respiration rate was significantly higher in cells expressing ApoE2 than in cells expressing ApoE3 (FC = 1.13, p = 0.004; Fig. 4E). N2a-ApoE4 cells showed significantly lower basal respiration than N2a-ApoE2 cells (FC = 0.81, p < 0.0001). ApoE2-expressing cells demonstrated significantly higher maximal respiratory capacity than ApoE3-expressing cells (FC = 1.11, p = 0.021). In comparison, ApoE4-expressing cells exhibited significantly lower maximal respiratory capacity than ApoE3-expressing (FC = 0.85, p = 0.001) and ApoE2-expressing cells (FC = 0.76, p < 0.0001; Fig. 4E). Additionally, the spare respiratory capacity was the lowest in ApoE4-expressing cells (FC = 0.77, p = 0.0001 vs ApoE3, FC = 0.71, p < 0.0001 vs ApoE2; Fig. 4F), suggesting that cells expressing ApoE4 may produce less ATP in response to an increase in energy demand, which may render these cells vulnerable to cellular stress. Consistent with more robust glycolysis and mitochondrial respiration, ATP levels were significantly higher in N2a-ApoE2 cells than N2a-ApoE3 cells (FC = 1.25, p < 0.0001; Fig. 4B). By contrast, ATP levels were significantly lower in N2a-ApoE4 cells (FC = 0.81, p < 0.0001). Collectively, these data indicate that ApoE2-expressing cells are the most bioenergetically robust whereas ApoE4-expressing cells are relatively deficient in both glycolytic activity and mitochondrial respiration.
ApoE2 and ApoE4 brains displayed similar robustness in the metabolism of ketone bodies
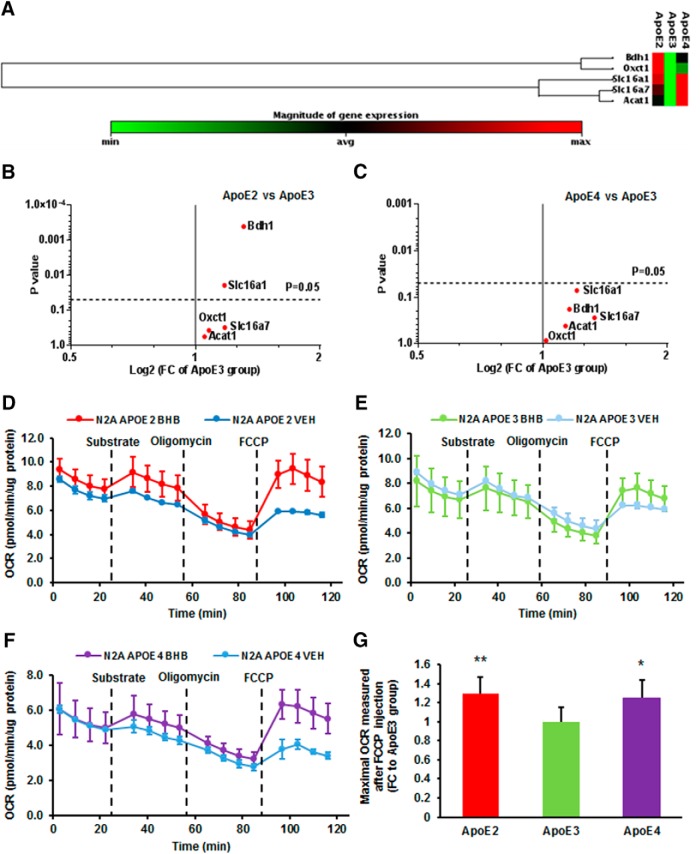
Brain ketone body uptake is mediated by the monocarboxylic acid transporters (MCTs; Pierre and Pellerin, 2005), and the metabolism of ketone bodies occurs exclusively in the mitochondria. Several clinical studies reported that ApoE4 carriers did not benefit from a ketogenic agent (Reger et al., 2004; Costantini et al., 2008), suggesting that individuals with ApoE4 may be deficient in the transporters that control the cellular uptake of ketone bodies or/and mitochondrial enzymes that convert the ketone bodies to their metabolites. Unexpectedly, both ApoE2 and ApoE4 brains demonstrated robust profiles in ketone body uptake and metabolism (Fig. 5A). More specifically, all five genes examined showed greater expression in ApoE2 (Fig. 5B) and ApoE4 brains (Fig. 5C) than in ApoE3 brains. Two genes exhibited significantly higher levels in ApoE2 brains than in ApoE3 brains, including 3-hydroxybutyrate dehydrogenase, type 1 (Bdh1; FC = 1.31, p = 0.0004) and solute carrier family 16, member 1 (Slc16a1; FC = 1.18, p = 0.019).
Figure 5.
ApoE2 and ApoE4 brains present similar level of robustness on ketone body uptake and metabolism. A, The heat map shows the expression of genes involved in brain ketone body transport and metabolism in the cortices of 6-month-old hApoE-TR mice. B, C, The volcano plots illustrate fold-changes and p values between (B) ApoE2 brains versus ApoE3 brains and (C) ApoE4 brains versus ApoE3 brains; Highlighted significantly altered expression: Bdh1 and Slc16a1. p < 0.05 was considered statistically significant with Student's t test. D–G, The impact of ApoE isoforms on ketone body utilization was evaluated in N2a cells expressing human ApoE isoforms. N2a-ApoE cells received sequential injections of substrate (BHB or vehicle) and modulating compounds, and OCR was measured by Seahorse XF96 Extracellular Flux Analyzer. The ketone body-metabolizing capacity of cells was represented by the maximum OCR measurement after FCCP injection. FCCP-induced OCR of cells treated with BHB was normalized to the vehicle-treated group. Results were compared using one-way ANOVA with Bonferroni post hoc test. n = 8 per group. *p < 0.05, **p < 0.01 vs N2a-ApoE3 group.
To further assess the effects of ApoE genotypes on cell capability to metabolize ketone bodies, we measured OCR in N2a-ApoE cells receiving sequential injections of substrate, BHB, or vehicle control, ATP synthase inhibitor oligomycin, and mitochondrial uncoupler FCCP. For all three ApoE genotypes, FCCP-induced response was significantly greater in the BHB-treated group than in the vehicle-treated group, suggesting that N2a cells are capable of metabolizing ketone bodies for energy supply (Fig. 5D–F). When normalized to their corresponding vehicle-treated groups, maximal OCR after FCCP injection was significantly higher in cells expressing ApoE2 and ApoE4 than in cells expressing ApoE3. The values of FCCP-induced maximal respiration rate in BHB-treated group divided by the mean of vehicle-treated group in each ApoE genotype were compared using one-way ANOVA with Bonferroni correction (Fig. 5G). In comparison with ApoE3-expressing cells, both ApoE2-expressing (FC = 1.30, p = 0.007) and ApoE4-expressing cells (FC = 1.25, p = 0.026) showed significantly higher maximal mitochondrial respiration upon BHB treatment. Collectively, our data indicate that cells expressing ApoE2 and ApoE4 are robust in metabolizing ketone bodies, whereas cells expressing ApoE3 are less efficient in ketone body utilization.
Upstream regulator PGC-1α was predicted to be activated in the ApoE2 brain
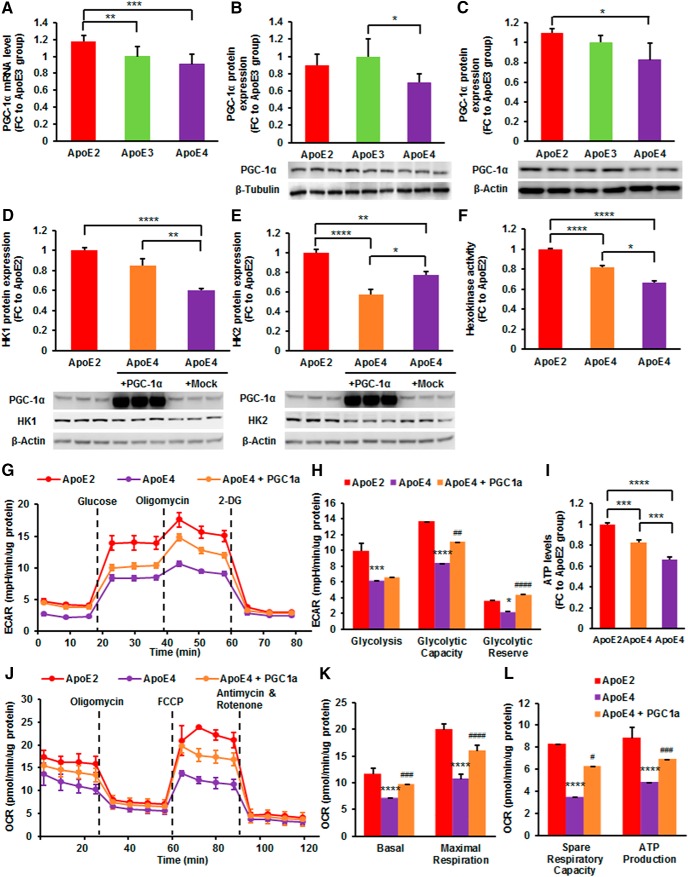
To further understand the biological relevance of the changes in gene expression, we conducted a regulatory pathway and molecular network analysis using the IPA. IPA predicted that Ppargc1a, which encodes PGC-1α, would be activated in the ApoE2 brain compared with the ApoE3 brain (z-score = 2.507, p value of overlap = 1.13E-12). Additionally, IPA predicted that peroxisome proliferator-activated receptor gamma (PPAR-γ) would be inhibited in the ApoE4 brain compared with the ApoE2 brain (z-score = −2.146, p value of overlap = 1.46E-09), which is consistent with our previous observations of lower PPAR-γ expression in the brains of ApoE4-TR mice in comparison with ApoE2-TR mice (Keeney et al., 2015). To validate the results from bioinformatic analysis, mRNA and protein levels of PGC-1α were determined in the cortices of hApoE-TR mice. In line with the prediction, ApoE2 brains expressed significantly higher levels of PGC-1α mRNA than in ApoE3 (FC = 1.13, p = 0.009) and ApoE4 brains (FC = 1.25, p = 0.0003; Fig. 6A). Protein level of PGC-1α was significantly lower in ApoE4 brains than in ApoE3 brains (FC = 0.70, p = 0.026; Fig. 6B). Consistent with tissue data, cells expressing ApoE4 showed markedly lower protein levels of PGC-1α than cells expressing ApoE2 (FC = 0.75, p = 0.005; Fig. 6C).
Figure 6.
PGC-1α overexpression ameliorates ApoE4-induced deficiencies in glycolysis and mitochondrial respiration. A, qRT-PCR was performed to examine the gene expression level of PGC-1α in the cortices of 6-month-old hApoE-TR mice. B, C, Protein expression of PGC-1α was determined in the cortices of the same hApoE-TR mice (B) and N2a-ApoE cells (C). Results were normalized to ApoE3 group. *p < 0.05, **p < 0.01, ***p < 0.001 by one-way ANOVA, n = 4–5 per group. D, E, Protein levels of Hk1 (D) and Hk2 (E) were measured in ApoE4-expressing cells transfected with vectors expressing mouse PGC-1α or mock control. PGC-1α overexpression increased Hk1 but decreased Hk2 in cells expressing ApoE4. F, Hexokinase activity was determined in the whole-cell lysates for the indicated groups. PGC-1α overexpression improved hexokinase activity in ApoE4-expresssing cells. *p < 0.05, ****p < 0.0001 by one-way ANOVA, n = 4–5 per group. G, ECAR was measured following sequential injections of glucose, oligomycin, and 2-DG in ApoE4-expressing cells transfected with mouse PGC-1α. H, Individual parameters for glycolysis, glycolytic capacity, and glycolytic reserve were calculated for the indicated groups. I, ATP levels were measured in ApoE4-expressing cells transfected with PGC-1α or empty vectors. Results were normalized to ApoE2 group. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 by one-way ANOVA with Bonferroni post hoc test. n = 6–7 per group. J, Mitochondrial OCR was measured in ApoE4-expressing cells transfected with mouse PGC-1α after sequential additions of oligomycin, FCCP, and antimycin A and rotenone as indicated. K, L, Quantification of individual parameters for basal respiration, maximal respiration, spare respiratory capacity, and ATP-linked respiration. Results were compared using one-way ANOVA with Bonferroni post hoc test. *p < 0.05, ***p < 0.001, ****p < 0.0001 vs ApoE2 group. #p < 0.05, ##p < 0.01, ###p < 0.001, ####p < 0.0001 vs ApoE4 group. n = 18–20 per group.
PGC-1α overexpression ameliorated bioenergetic deficits in N2a cells stably expressing ApoE4
Given that PGC-1α activation may underlie the bioenergetic robustness in ApoE2-expressing cells, we hypothesized that PGC-1α overexpression might be capable of ameliorating ApoE4-induced bioenergetic deficits. To test this hypothesis, N2a cells stably expressing ApoE4 were transfected with empty vector or vectors expressing mouse PGC-1α. Hexokinase protein expression, activities, as well as cellular bioenergetics were assessed in the transfected cells. Cells stably expressing ApoE2 served as positive control. Interestingly, we found that PGC-1α modulated hexokinase isozymes in opposite directions. Specifically, PGC-1α overexpression significantly increased Hk1 expression (FC = 1.41, p = 0.004) but decreased Hk2 expression (FC = 0.73, p = 0.012) in ApoE4-expressing cells (Fig. 6D,E). Additionally, PGC-1α overexpression significantly improved the enzymatic activity of hexokinase (FC = 1.23, p < 0.0001; Fig. 6F), likely attributed to enhanced Hk1 expression.
Although both hexokinase isoforms are expressed in a variety of cell types, cells that are sensitive to insulin, such as adult muscle cells, express primarily HK2, whereas cells that rely heavily on glycolysis for energy production, such as neurons, express predominantly Hk1 (Mandarino et al., 1995; Wilson, 2003). Additionally, it has been proposed that HK1 remains associated with mitochondria to promote glycolysis whereas HK2 dynamically translocates between cytosol and mitochondria where it directs the metabolic fate of glucose between anabolic (glycogen synthesis and pentose phosphate shunt) and catabolic (glycolysis) metabolism (Wilson, 2003; John et al., 2011). To examine the functional consequence of the differential regulation of hexokinase expression by PGC-1α, glycolytic function was evaluated in ApoE4-expressing cells transfected with PGC-1α. Sequential injections of glucose, oligomycin, and 2-DG measured basal glycolysis, glycolytic capacity, and nonglycolytic acidification (Fig. 6G). Glucose-induced increase in ECAR was significantly lower in ApoE4-expressing cells than in ApoE2-expressing cells (FC = 0.62, p = 0.0002), suggesting that ApoE4-expressing cells had a lower basal glycolytic rate (Fig. 6H). Unexpectedly, PGC-1α overexpression did not increase basal glycolysis in ApoE4-expressing cells. However, maximum glycolytic capacity was greatly improved by PGC-1α overexpression (FC = 1.33, p = 0.003). Further, PGC-1α overexpression completely reversed ApoE4-induced defects in glycolytic reserve capacity (FC = 1.99, p < 0.0001; Fig. 6H). These results indicate that the enhanced PGC-1α activity ameliorated ApoE4-associated deficits in glycolysis and the improvement in glycolytic function by PGC-1α is attributable in part to an increase in Hk1 expression.
Given that PGC-1α is the master regulator of mitochondrial biogenesis and oxidative phosphorylation, we also examined the effect of PGC-1α overexpression on mitochondrial respiration in cells stably expressing ApoE4. Higher levels of basal and maximal OCR following FCCP injection were observed in ApoE4-expressing cells transfected with PGC-1α than those transfected with mock control (Fig. 6J). Quantitative analysis confirmed that PGC-1α overexpression significantly improved basal (FC = 1.36, p = 0.0002) and maximal mitochondrial respiration (FC = 1.51, p < 0.0001) in ApoE4-expresssing cells (Fig. 6K). Similarly, PGC-1α overexpression resulted in a marked elevation in the spare respiratory capacity (FC = 1.80, p = 0.012) and ATP-linked respiration (FC = 1.44, p = 0.0003) in ApoE4-expressing cells (Fig. 6L). Further, total ATP levels were significantly elevated by PGC-1α overexpression in ApoE4-expressing cells (FC = 1.24, p = 0.0006), which may have resulted from improvements in glycolysis and mitochondrial respiration (Fig. 6I). Together, these results indicate that the enhanced PGC-1α activity counteracts the detrimental effects of ApoE4 in mitochondrial bioenergetics.
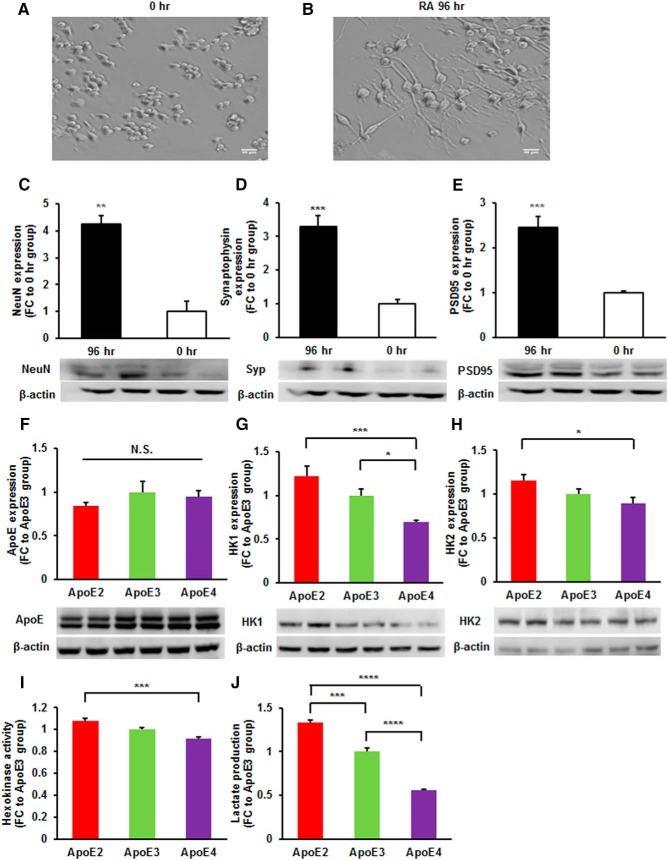
Differentiated N2a cells expressing ApoE2 exhibited the most potent hexokinase expression and glycolytic activity
To confirm the above findings of the effects of ApoE isoforms on hexokinase and glycolysis were not a result of potentially altered metabolic profile associated with neuroblastoma cells, we differentiated N2a cells with retinoid acid in combination with serum deprivation and performed further analyses using this more physiologically relevant cell model. The differentiated N2a cells displayed a typical neuronal phenotype as demonstrated by both morphological and biochemical properties. As shown in Figure 7B, the differentiated N2a cells contained one or more processes that were significantly longer than the diameter of the cell body. Additionally, protein expressions of NeuN, a widely used marker for mature neurons, and markers for synapses, including synaptophysin and PSD95, were significantly enhanced after 96 h incubation with RA (Fig. 7C–E), which further confirmed the differentiation. The differentiated N2a cells were then transfected with ApoE isoforms and hexokinase expression, enzyme activity, and glycolytic function were evaluated. Consistent with the observations in undifferentiated N2a cells, differentiated N2a cells expressing ApoE2 exhibited significantly higher protein expression of both isoforms of hexokinase, HK1 (FC = 1.76, p = 0.0002) and HK2 (FC = 1.29, p = 0.025), as well as hexokinase activity (FC = 1.18, p = 0.0002) compared with those expressing ApoE4 (Fig. 7G–I). Furthermore, the greatest production of lactate, an end product of glycolysis, was detected in ApoE2-expressing differentiated N2a cells (FC = 1.33 vs ApoE3 group, p = 0.0003), and the least production was detected in ApoE4-expressing differentiated cells (FC = 0.56 vs ApoE3 group, p < 0.0001; Fig. 7J). These results confirm our previous findings of modulations of hexokinase and glycolysis by ApoE isoforms in neuroblastoma cells also exist in postmitotic neurons.
Figure 7.
Differentiated N2a cells expressing ApoE2 displayed the most potent hexokinase expression and glycolytic activity. A, B, N2a cell morphology was evaluated by phase contrast imaging. The formation of neuronal processes were observed after RA incubation for 96 h. Scale bar = 40 μm. C–E, The differentiation of N2a cells was also confirmed by immunoblotting for markers of mature neurons. Expressions of NeuN, synaptophysin, and PSD95 were significantly increased in the differentiated N2a cells. n = 3–8 per group. F, ApoE expression levels were comparable in differentiated N2a cells transfected with ApoE isoforms. n = 6 per group. G, H, Protein expressions of Hk1 1 and Hk2 were significantly higher in neuron-like differentiated N2a cells expressing ApoE2. n = 7–13 per group. I, J, Differentiated N2a cells expressing ApoE2 exhibited the most potent hexokinase activity and glycolytic function. Results were normalized to ApoE3 group. n = 3–6 per group. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 by one-way ANOVA with Bonferroni post hoc test.
Discussion
Human ApoE isoforms have been shown to confer differential risk in late-onset AD, which comprises 95% of total AD cases. Previous work has primarily focused on the role of ApoE4 in AD pathogenesis, and few studies have explored the role of ApoE2 in relation to AD; yet, overall, the results of these studies suggest that ApoE2 is neuroprotective (Wu and Zhao, 2016). However, the underlying mechanisms by which ApoE2 confers neuroprotection remain largely unexplored. Results from the present study demonstrate that human ApoE isoforms differentially modulate brain uptake and cytosolic metabolism of the energy substrates, including glucose and ketone bodies (Fig. 8). Our data provide one potential mechanistic rationale for the neuroprotective effects associated with ApoE2 and the detrimental effects linked to ApoE4 in the aging and AD brain.
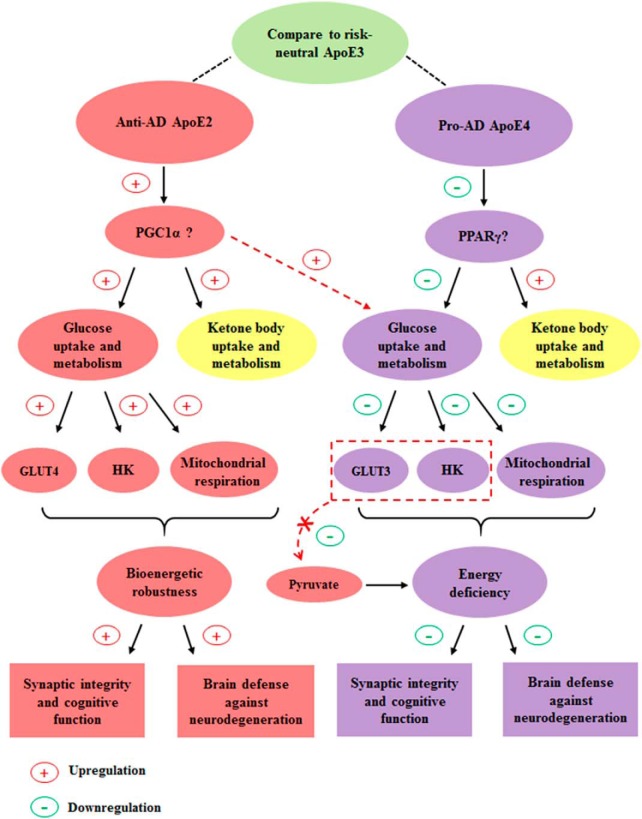
Figure 8.
Our working hypotheses and conclusions. The ApoE2 brain exhibits the most metabolically robust profile, whereas the ApoE4 brain presents the most deficient profile on the uptake and metabolism of glucose, the primary energy source for the brain. Particularly, these two ApoE genotypes are significantly different in the expression of GLUTs and hexokinases. On the uptake and metabolism of ketone bodies, the secondary fuel for the brain, both ApoE2 and ApoE4 brains show more robust profiles than the ApoE3 brain. PGC-1α may serve as an upstream master regulator of the bioenergetic robustness of ApoE2 whereas an inhibition of PPARγ signaling pathway may underlie the energy deficiency associated with ApoE4. In support of this notion, PGC-1α overexpression attenuated the bioenergetic deficits induced by ApoE4. A therapeutic approach that can bypass the deficiency in glucose uptake and glycolysis by providing glucose metabolizing intermediates, e.g., pyruvate, may hold the promise to reduce AD risk in ApoE4 carriers; however, a ketogenic strategy may provide a more meaningful benefit in ApoE3 than in ApoE4 carriers.
Under normal physiological conditions, glucose is the sole substrate for energy production in the brain. Because neurons have a high energy requirement, their functionality depends heavily on glucose availability and utilization (Mosconi, 2013). The availability of glucose to cells is governed by the facilitated glucose transporters (GLUTs). Here we examined mRNA levels of major GLUTs in the brains of hApoE-TR mice, and detected no significant difference in GLUT1 expression among the three ApoE genotypes. However, significant reduction in glucose transport across the blood–brain barrier without altered mRNA or protein expression of GLUT1 has been reported in hApoE4-TR mice, suggesting that the function of GLUT1 may involve post-translational modification (Alata et al., 2015). In contrast to GLUT1, we found a significantly lower mRNA level of GLUT3 in ApoE4 brains than in ApoE3 brains. GLUT3 is the predominant isoform in the neuron populations (Nagamatsu et al., 1992), and protein expression of GLUT3 has been shown to decrease in parallel with reduced cerebral glucose metabolism in AD-vulnerable brain regions (Simpson et al., 1994). Therefore, lower GLUT3 expression could lead to insufficient energy supply and perturbation of neuronal function in the ApoE4 brain. In addition, the mRNA level of GLUT4 was significantly higher in ApoE2 brains compared with ApoE4 brains, which is consistent with our previous findings (Keeney et al., 2015). Further in-depth studies are required to determine the impact of ApoE isoforms on GLUTs activity.
Glucose metabolism includes glycolysis in the cytoplasm, the tricarboxylic acid cycle, and oxidative phosphorylation in the mitochondria. We found that the three ApoE genotypes differed significantly in the glycolysis, particularly hexokinase. Specifically, the ApoE2 and ApoE4 genotypes were associated with the most robust and deficient glycolytic function, which is likely attributable to their discrete impact on hexokinase expression. Despite the fact that ATP generated by glycolysis only accounts for a small percentage of total energy used in the brain, emerging evidence suggests that glycolysis plays a crucial role in maintaining synaptic functions. Several glycolytic enzymes have been found in the nerve terminals where they interact with the subunits of V-type H+-ATPase (V-ATPase), a proton pump that mediates the concentration of neurotransmitters into synaptic vesicles, which therefore is crucial for synaptic transmission (Lu et al., 2004). Chan et al. (2016) reported that the glycolysis flow controlled V-ATPase reassembly after the addition of glucose into glucose-deprived cells. Previously, we demonstrated that ApoE2 brains express significantly higher levels of the β subunit of V-ATPase (Atp6v1B2) compared with both ApoE3 and ApoE4 brains (Woody et al., 2016). Here, we show that ApoE2 brains also possess the most robust metabolic profile for glycolysis, which provides a direct source of ATP and protons for V-ATPase. Therefore, our data indicate that the ApoE isoform-mediated differential modulation of brain glucose metabolism could translate into a significant impact on neurotransmission through synaptic proteins with activities heavily relying on energy availability such as V-ATPase. The bioenergetic robustness may serve as an important mechanism underlying the cognition-favoring and anti-AD properties associated with ApoE2.
Apart from glucose, we also examined the impact of human ApoE isoforms on the metabolism of ketone bodies, the main alternative fuel for the brain under energy challenging conditions. Although clinical studies have shown that treatment with a ketogenic agent improved cognitive performance in AD patients, the therapeutic benefits appeared to be confined to ApoE4 noncarriers (Reger et al., 2004; Costantini et al., 2008; Henderson et al., 2009). This suggests that ApoE4 brains might be deficient in ketone body transport and/or key enzymes that metabolize ketone bodies. Intriguingly, our gene array data indicate a more robust metabolic profile for ketone body uptake and metabolism in both ApoE2 and ApoE4 brains compared with ApoE3 brains, suggesting that ApoE2 and ApoE4 brains are better equipped for ketone body utilization. In support of this notion, ApoE2- and ApoE4-expresssing cells metabolized ketone bodies more efficiently than ApoE3-expressing cells. Results from these studies provide an alternative explanation for the clinical observation that ketosis benefits ApoE3 carriers more than ApoE4 carriers. In the AD context, because the ApoE4 brain is associated with glucose hypometabolism, the robust ketone body metabolism could serve as an adaptive response to compensate for the bioenergetic deficits. We may speculate that the ApoE4 brain may possess an upper limit of ketone body utilization as a result of compensatory response. Consequently, the ApoE4 brain would not be able to further metabolize exogenously supplied ketone bodies for energy production and would thus not induce improved cognitive performance. However, robust ability in ketone body metabolism may be another important contributor to the intact cognition associated with ApoE2.
Although the mechanism underlying ApoE isoform-specific modulation in brain energy metabolism is not clear, the transcription cofactor PGC-1α may play a critical role. PGC-1α downregulation has been associated with extensive alterations in several metabolic pathways, particularly in oxidative phosphorylation and the mitochondrial electron transport chain (Cui et al., 2006; Taherzadeh-Fard et al., 2009). Additionally, gene expression analysis of PGC-1α null mouse brain revealed significant downregulation of genes associated with neuronal functions (Lin et al., 2004; Ma et al., 2010). These findings suggest a pivotal role of PGC-1α in maintaining neuronal functions by controlling mitochondrial oxidative metabolism and energy homeostasis. In this study, we found significantly higher mRNA levels of PGC-1α in ApoE2 brains than in ApoE3 brains. In contrast, PGC-1α expression was the lowest in ApoE4 brains. In addition to lower protein expression, the activity of PGC-1α may also be diminished. Theendakara et al. (2013) reported that forced expression of ApoE4 reduced mRNA and protein levels of sirtuin 1 (Sirt1), an enzyme that mediates NAD+-dependent deacetylation of PGC-1α, thus enhancing its transcriptional activity (Cantó and Auwerx, 2009). Therefore, lower expression of Sirt1 may result in reduced PGC-1α activation, which may underlie the bioenergetic deficits associated with ApoE4. Indeed, PGC-1α overexpression attenuated ApoE4-induced defects in glycolysis and mitochondrial respiration. To date, there is no report on Sirt1 expression and transcriptional regulation in the ApoE2 genotype. However, ApoE2 may modulate PGC-1α activity through a similar mechanism. Given that PGC-1α is a master regulator of mitochondrial oxidative metabolism, the higher expression and activity of PGC-1α may significantly contribute to the bioenergetic robustness associated with ApoE2. Further investigations are warranted to identify other upstream regulators and associated signaling pathways involved in ApoE-isoform-dependent role in glucose metabolism.
Our results also suggest the potential benefit of pyruvate supplement in ApoE4 carriers. Suh et al. (2005) reported that the overactivation of poly (ADP-ribose) polymerase-1 (PARP-1) resulted in the depletion of cytosolic NAD+ followed by inhibition of glycolysis, leading to sustained energy failure and ultimately cell death. As a direct substrate for mitochondrial metabolism, pyruvate can be oxidized in the absence of cytosolic NAD+; therefore, it can bypass the restrictions imposed by PARP-1 and thus restore energy deficiency (Zilberter et al., 2015). Consistent with this concept, administration of pyruvate significantly improved neuronal survival in the brains of rats subjected to insulin-induced hypoglycemia (Suh et al., 2005). Therefore, pyruvate may provide a more efficient energy source for the ApoE4 brain by circumventing a sustained impairment in neuronal glucose utilization due to defects in glucose uptake and glycolysis (Fig. 8). More work is necessary to examine the therapeutic benefits of pyruvate in aging and AD brains, particularly in those bearing the ApoE4 genotype.
We should point out that this study presented both technical strengths and limitations. One major concern was the utilization of a neuroblastoma cell line, which may have altered metabolic phenotype compared with postmitotic neurons. To address this concern, we examined and confirmed the effects of ApoE isoforms on hexokinase and glycolysis in differentiated N2a cells, a more physiologically relevant neuronal model. The advantage of using N2a cells was that the transfected cells expressed comparable human ApoE, which enabled the comparisons of the three ApoE isoforms in parallel and precluded compounding complexity resulting from the differences in ApoE expression levels as reported in the hApoE-TR mice (Riddell et al., 2008; Sullivan et al., 2011). Collectively, our findings from the combined utilization of the hApoE-expressing mouse as well as cell models provide complementary and consistent evidence that (a) human ApoE isoforms differentially modulate brain bioenergetic metabolism, and (b) the distinct functional properties of the ApoE isoforms rather than the differences in ApoE protein levels in the three isoform-expressing brains cause this differential modulation. Of the most significance, we pinpointed hexokinase as a key cytosolic point that the three ApoE isoforms differentially affect, which may contribute to their distinct impact on brain glucose utilization. The glycolytic robustness exhibited in ApoE2-expressing brains may translate into enhanced synaptic activity and ultimately the clinically proven cognition-favoring and anti-AD properties associated with this genotype.
Footnotes
This work was supported by Grants from the National Institutes of Health (R21AG055964, P20GM103418, P30GM110761) and the University of Kansas NFGRF, GRF, and start-up funds to L.Z.
The authors declare no competing financial interests.
References
- Alata W, Ye Y, St-Amour I, Vandal M, Calon F (2015) Human apolipoprotein E ε4 expression impairs cerebral vascularization and blood–brain barrier function in mice. J Cereb Blood Flow Metab 35:86–94. 10.1038/jcbfm.2014.172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alzheimer's Association (2016) 2016 Alzheimer's disease facts and figures. Alzheimers Dement 12:459–509. 10.1016/j.jalz.2016.03.001 [DOI] [PubMed] [Google Scholar]
- Calero O, Hortigüela R, Bullido MJ, Calero M (2009) Apolipoprotein E genotyping method by real time PCR, a fast and cost-effective alternative to the TaqMan® and FRET assays. J Neurosci Methods 183:238–240. 10.1016/j.jneumeth.2009.06.033 [DOI] [PubMed] [Google Scholar]
- Cantó C, Auwerx J (2009) PGC-1alpha, SIRT1 and AMPK, an energy sensing network that controls energy expenditure. Curr Opin Lipidol 20:98–105. 10.1097/MOL.0b013e328328d0a4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caselli RJ, Dueck AC, Osborne D, Sabbagh MN, Connor DJ, Ahern GL, Baxter LC, Rapcsak SZ, Shi J, Woodruff BK, Locke DE, Snyder CH, Alexander GE, Rademakers R, Reiman EM (2009) Longitudinal modeling of age-related memory decline and the APOE epsilon4 effect. New Engl J Med 361:255–263. 10.1056/NEJMoa0809437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caselli RJ, Dueck AC, Locke DE, Hoffman-Snyder CR, Woodruff BK, Rapcsak SZ, Reiman EM (2011) Longitudinal modeling of frontal cognition in APOE epsilon4 homozygotes, heterozygotes, and noncarriers. Neurology 76:1383–1388. 10.1212/WNL.0b013e3182167147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CY, Dominguez D, Parra KJ (2016) Regulation of vacuolar H+-ATPase (V-ATPase) reassembly by glycolysis flow in 6-phosphofructo-1-kinase (PFK-1)-deficient yeast cells. J Biol Chem 291:15820–15829. 10.1074/jbc.M116.717488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, ran Ma T, Miranda RD, Balestra ME, Mahley RW, Huang Y (2005) Lipid- and receptor-binding regions of apolipoprotein E4 fragments act in concert to cause mitochondrial dysfunction and neurotoxicity. Proc Natl Acad Sci U S A 102:18694–18699. 10.1073/pnas.0508254102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini LC, Barr LJ, Vogel JL, Henderson ST (2008) Hypometabolism as a therapeutic target in Alzheimer's disease. BMC neuroscience 9:S16. 10.1186/1471-2202-9-S2-S16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craft S, Teri L, Edland SD, Kukull WA, Schellenberg G, McCormick WC, Bowen JD, Larson EB (1998) Accelerated decline in apolipoprotein E-epsilon4 homozygotes with Alzheimer's disease. Neurology 51:149–153. 10.1212/WNL.51.1.149 [DOI] [PubMed] [Google Scholar]
- Cui L, Jeong H, Borovecki F, Parkhurst CN, Tanese N, Krainc D (2006) Transcriptional repression of PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell 127:59–69. 10.1016/j.cell.2006.09.015 [DOI] [PubMed] [Google Scholar]
- Cummings JL, Morstorf T, Zhong K (2014) Alzheimer's disease drug-development pipeline: few candidates, frequent failures. Alzheimers Res Ther 6:37. 10.1186/alzrt269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding F, Yao J, Rettberg JR, Chen S, Brinton RD (2013) Early decline in glucose transport and metabolism precedes shift to ketogenic system in female aging and Alzheimer's mouse brain: implication for bioenergetic intervention. PloS One 8:e79977. 10.1371/journal.pone.0079977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, Mayeux R, Myers RH, Pericak-Vance MA, Risch N, van Duijn CM (1997) Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA 278:1349–1356. 10.1001/jama.1997.03550160069041 [DOI] [PubMed] [Google Scholar]
- Henderson ST, Vogel JL, Barr LJ, Garvin F, Jones JJ, Costantini LC (2009) Study of the ketogenic agent AC-1202 in mild to moderate Alzheimer's disease: a randomized, double-blind, placebo-controlled, multicenter trial. Nutr Metab (Lond) 6:31. 10.1186/1743-7075-6-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John S, Weiss JN, Ribalet B (2011) Subcellular localization of hexokinases I and II directs the metabolic fate of glucose. PloS One 6:e17674. 10.1371/journal.pone.0017674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney JT, Ibrahimi S, Zhao L (2015) Human ApoE isoforms differentially modulate glucose and amyloid metabolic pathways in female brain: evidence of the mechanism of neuroprotection by ApoE2 and implications for Alzheimer's disease prevention and early intervention. J Alzheimers Dis 48:411–424. 10.3233/JAD-150348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Basak JM, Holtzman DM (2009) The role of apolipoprotein E in Alzheimer's disease. Neuron 63:287–303. 10.1016/j.neuron.2009.06.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein RC, Mace BE, Moore SD, Sullivan PM (2010) Progressive loss of synaptic integrity in human apolipoprotein E4 targeted replacement mice and attenuation by apolipoprotein E2. Neuroscience 171:1265–1272. 10.1016/j.neuroscience.2010.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Wu PH, Tarr PT, Lindenberg KS, St-Pierre J, Zhang CY, Mootha VK, Jäger S, Vianna CR, Reznick RM, Cui L, Manieri M, Donovan MX, Wu Z, Cooper MP, Fan MC, Rohas LM, Zavacki AM, Cinti S, Shulman GI, et al. (2004) Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell 119:121–135. 10.1016/j.cell.2004.09.013 [DOI] [PubMed] [Google Scholar]
- Liu CC, Kanekiyo T, Xu H, Bu G (2013) Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nat Rev Neurol 9:106–118. 10.1038/nrneurol.2012.263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M, Sautin YY, Holliday LS, Gluck SL (2004) The glycolytic enzyme aldolase mediates assembly, expression, and activity of vacuolar H+-ATPase. J Biol Chem 279:8732–8739. 10.1074/jbc.M303871200 [DOI] [PubMed] [Google Scholar]
- Ma D, Li S, Lucas EK, Cowell RM, Lin JD (2010) Neuronal inactivation of peroxisome proliferator-activated receptor gamma coactivator 1alpha (PGC-1alpha) protects mice from diet-induced obesity and leads to degenerative lesions. J Biol Chem 285:39087–39095. 10.1074/jbc.M110.151688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandarino LJ, Printz RL, Cusi KA, Kinchington P, O'Doherty RM, Osawa H, Sewell C, Consoli A, Granner DK, DeFronzo RA (1995) Regulation of hexokinase II and glycogen synthase mRNA, protein, and activity in human muscle. Am J Physiol 269:E701–E708. 10.1152/ajpendo.1995.269.4.E701 [DOI] [PubMed] [Google Scholar]
- Martins CA, Oulhaj A, de Jager CA, Williams JH (2005) APOE alleles predict the rate of cognitive decline in alzheimer disease: a nonlinear model. Neurology 65:1888–1893. 10.1212/01.wnl.0000188871.74093.12 [DOI] [PubMed] [Google Scholar]
- Mosconi L. (2013) Glucose metabolism in normal aging and Alzheimer's disease: methodological and physiological considerations for PET studies. Clin Transl Imaging 1:4. 10.1007/s40336-013-0026-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamatsu S, Kornhauser JM, Burant CF, Seino S, Mayo KE, Bell GI (1992) Glucose transporter expression in brain. cDNA sequence of mouse GLUT3, the brain facilitative glucose transporter isoform, and identification of sites of expression by in situ hybridization. J Biol Chem 267:467–472. [PubMed] [Google Scholar]
- Nagy Z, Esiri MM, Jobst KA, Johnston C, Litchfield S, Sim E, Smith AD (1995) Influence of the apolipoprotein E genotype on amyloid deposition and neurofibrillary tangle formation in Alzheimer's disease. Neuroscience 69:757–761. 10.1016/0306-4522(95)00331-C [DOI] [PubMed] [Google Scholar]
- Pierre K, Pellerin L (2005) Monocarboxylate transporters in the central nervous system: distribution, regulation and function. J Neurochem 94:1–14. 10.1111/j.1471-4159.2005.03168.x [DOI] [PubMed] [Google Scholar]
- Rapp A, Gmeiner B, Hüttinger M (2006) Implication of apoE isoforms in cholesterol metabolism by primary rat hippocampal neurons and astrocytes. Biochimie 88:473–483. 10.1016/j.biochi.2005.10.007 [DOI] [PubMed] [Google Scholar]
- Reger MA, Henderson ST, Hale C, Cholerton B, Baker LD, Watson GS, Hyde K, Chapman D, Craft S (2004) Effects of beta-hydroxybutyrate on cognition in memory-impaired adults. Neurobiol Aging 25:311–314. 10.1016/S0197-4580(03)00087-3 [DOI] [PubMed] [Google Scholar]
- Reiman EM, Chen K, Alexander GE, Caselli RJ, Bandy D, Osborne D, Saunders AM, Hardy J (2004) Functional brain abnormalities in young adults at genetic risk for late-onset Alzheimer's dementia. Proc Natl Acad Sci U S A 101:284–289. 10.1073/pnas.2635903100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiman EM, Chen K, Alexander GE, Caselli RJ, Bandy D, Osborne D, Saunders AM, Hardy J (2005) Correlations between apolipoprotein E epsilon4 gene dose and brain-imaging measurements of regional hypometabolism. Proc Natl Acad Sci U S A 102:8299–8302. 10.1073/pnas.0500579102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddell DR, Zhou H, Atchison K, Warwick HK, Atkinson PJ, Jefferson J, Xu L, Aschmies S, Kirksey Y, Hu Y, Wagner E, Parratt A, Xu J, Li Z, Zaleska MM, Jacobsen JS, Pangalos MN, Reinhart PH (2008) Impact of apolipoprotein E (ApoE) polymorphism on brain ApoE levels. J Neurosci 28:11445–11453. 10.1523/JNEUROSCI.1972-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez GA, Tai LM, LaDu MJ, Rebeck GW (2014) Human APOE4 increases microglia reactivity at aβ plaques in a mouse model of aβ deposition. J Neuroinflammation 11:111. 10.1186/1742-2094-11-111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara M, Kanekiyo T, Yang L, Linthicum D, Shinohara M, Fu Y, Price L, Frisch-Daiello JL, Han X, Fryer JD, Bu G (2016) APOE2 eases cognitive decline during aging: clinical and preclinical evaluations. Ann Neurol 79:758–774. 10.1002/ana.24628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson IA, Chundu KR, Davies-Hill T, Honer WG, Davies P (1994) Decreased concentrations of GLUT1 and GLUT3 glucose transporters in the brains of patients with Alzheimer's disease. Ann Neurol 35:546–551. 10.1002/ana.410350507 [DOI] [PubMed] [Google Scholar]
- Suh SW, Aoyama K, Matsumori Y, Liu J, Swanson RA (2005) Pyruvate administered after severe hypoglycemia reduces neuronal death and cognitive impairment. Diabetes 54:1452–1458. 10.2337/diabetes.54.5.1452 [DOI] [PubMed] [Google Scholar]
- Sullivan PM, Mace BE, Maeda N, Schmechel DE (2004) Marked regional differences of brain human apolipoprotein E expression in targeted replacement mice. Neuroscience 124:725–733. 10.1016/j.neuroscience.2003.10.011 [DOI] [PubMed] [Google Scholar]
- Sullivan PM, Han B, Liu F, Mace BE, Ervin JF, Wu S, Koger D, Paul S, Bales KR (2011) Reduced levels of human apoE4 protein in an animal model of cognitive impairment. Neurobiol Aging 32:791–801. 10.1016/j.neurobiolaging.2009.05.011 [DOI] [PubMed] [Google Scholar]
- Taherzadeh-Fard E, Saft C, Andrich J, Wieczorek S, Arning L (2009) PGC-1alpha as modifier of onset age in Huntington disease. Mol Neurodegener 4:10. 10.1186/1750-1326-4-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theendakara V, Patent A, Peters Libeu CA, Philpot B, Flores S, Descamps O, Poksay KS, Zhang Q, Cailing G, Hart M, John V, Rao RV, Bredesen DE (2013) Neuroprotective sirtuin ratio reversed by ApoE4. Proc Natl Acad Sci U S A 110:18303–18308. 10.1073/pnas.1314145110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JE. (2003) Isozymes of mammalian hexokinase: structure, subcellular localization and metabolic function. J Exp Biol 206:2049–2057. 10.1242/jeb.00241 [DOI] [PubMed] [Google Scholar]
- Woody SK, Zhou H, Ibrahimi S, Dong Y, Zhao L (2016) Human ApoE varepsilon2 promotes regulatory mechanisms of bioenergetic and synaptic function in female brain: a focus on V-type H+-ATPase. J Alzheimers Dis 53:1015–1031. 10.3233/JAD-160307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Zhao L (2016) ApoE2 and Alzheimer's disease: time to take a closer look. Neural Regener Res 11:412–413. 10.4103/1673-5374.179044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Gottesdiener AJ, Parmar M, Li M, Kaminsky SM, Chiuchiolo MJ, Sondhi D, Sullivan PM, Holtzman DM, Crystal RG, Paul SM (2016) Intracerebral adeno-associated virus gene delivery of apolipoprotein E2 markedly reduces brain amyloid pathology in Alzheimer's disease mouse models. Neurobiol Aging 44:159–172. 10.1016/j.neurobiolaging.2016.04.020 [DOI] [PubMed] [Google Scholar]
- Zilberter Y, Gubkina O, Ivanov AI (2015) A unique array of neuroprotective effects of pyruvate in neuropathology. Front Neurosci 9:17. 10.3389/fnins.2015.00017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of genes examined in the brains of hApoE-TR mice. Download Figure 1-1, DOCX file (16.1KB, docx)
Relative gene expression in the brains of hApoE-TR mice. Download Figure 1-2, DOCX file (16.6KB, docx)