Abstract
Lysosomes play a central role in cellular homeostasis by regulating the cellular degradative machinery. Because aberrant lysosomal function has been associated with multiple lysosomal storage and neurodegenerative disorders, enhancement of lysosomal clearance has emerged as an attractive therapeutic strategy. Transcription factor EB (TFEB) is known as a master regulator of lysosomal biogenesis and, here, we reveal that aspirin, one of the most widely used medications in the world, upregulates TFEB and increases lysosomal biogenesis in brain cells. Interestingly, aspirin induced the activation of peroxisome proliferator-activated receptor alpha (PPARα) and stimulated the transcription of Tfeb via PPARα. Finally, oral administration of low-dose aspirin decreased amyloid plaque pathology in both male and female 5X familial Alzheimer's disease (5XFAD) mice in a PPARα-dependent fashion. This study reveals a new function of aspirin in stimulating lysosomal biogenesis via PPARα and suggests that low-dose aspirin may be used in lowering storage materials in Alzheimer's disease and lysosomal storage disorders.
SIGNIFICANCE STATEMENT Developing drugs for the reduction of amyloid β containing senile plaques, one of the pathological hallmarks of Alzheimer's disease (AD), is an important area of research. Aspirin, one of the most widely used medications in the world, activates peroxisome proliferator-activated receptor alpha (PPARα) to upregulate transcription factor EB and increase lysosomal biogenesis in brain cells. Accordingly, low-dose aspirin decreases cerebral plaque load in a mouse model of Alzheimer's disease via PPARα. These results reveal a new mode of action of aspirin that may be beneficial for AD and lysosomal storage disorders.
Keywords: Alzheimer's disease, amyloid plaques, aspirin, lysosomal biogenesis, PPARα, TFEB
Introduction
Alzheimer's disease (AD), which is clinically characterized by progressive cognitive impairment, is the most common neurodegenerative disorder. At present, there is no effective treatment for preventing or halting the disease (Goedert and Spillantini, 2006; Querfurth and LaFerla, 2010; Huang and Mucke, 2012). The major defining neuropathological features of AD are deposition of extracellular senile plaques composed of toxic amyloid beta (Aβ) aggregates and the formation of intracellular neurofibrillary tangles originated from hyperphosphorylation of the microtubule-associated protein tau (Querfurth and LaFerla, 2010; Yoon and Kim, 2016). The early onset familial forms of AD have genetic origins characterized by mutations in the gene encoding the amyloid precursor protein (APP), a neuronal transmembrane protein, and the presenilins PS1 and PS2, the catalytic subunit of the gamma secretase complex (Whyte et al., 2017). Although the familial forms of AD are rare, the major AD occurrences are sporadic in nature with etiology that remains elusive. Mounting evidence has identified impaired Aβ clearance as the underlying mechanism in patients with sporadic AD (Mawuenyega et al., 2010). Therefore, stimulation of the cellular degradative machinery for the efficient removal of Aβ has emerged as a growing field in AD research.
Attempts to enhance the lysosomal function and restore normal autophagy by modulating transcription factor EB (TFEB), the essential regulator of the lysosome system, have generated promising therapeutic results in rescuing the amyloid pathogenesis in AD. Enhancing lysosomal function with TFEB leads to increased lysosomal degradation of holo-APP in the neurons and thus reduces the amyloidogenic processing of APP and Aβ generation (Xiao et al., 2015). Recent studies have demonstrated TFEB overexpression can alleviate AD pathology by regulation of the autophagy–lysosome pathway (Zhang and Zhao, 2015). Targeted TFEB expression in astrocytes mediated by viral gene transfer promotes attenuation of the amyloid pathology by enhancing lysosomal biogenesis and facilitating Aβ uptake and lysosomal degradation by astrocytes (Xiao et al., 2014). Given the capability of astrocytic lysosomal stimulation in mitigating Aβ pathology, we explored the role of drug-mediated lysosomal biogenesis in astrocytes and its therapeutic effect in the 5X familial Alzheimer's disease (5XFAD) model of AD.
Acetylsalicylic acid, commonly known as aspirin, is one of the most frequently used pharmaceutics in medical practice and is available over the counter (Green, 2001). As a member of the nonsteroidal anti-inflammatory drug group, aspirin is known to exert its anti-inflammatory effects by inhibiting cycloxygenases and thereby suppressing the generation of proinflammatory molecules such as prostaglandins (Vane, 1971; Vane and Botting, 2003). Other than its extensive use as an analgesic and antipyretic, aspirin has also been demonstrated to have beneficial effects for atherosclerosis, cardiovascular disease, and several cancers (Moyad, 2001; Dai and Ge, 2012; Rothwell et al., 2012; Berk et al., 2013). Earlier studies have explored the neuroprotective effect of aspirin under different disease conditions. Aspirin was shown to have protective effects in an animal model of Parkinson's disease independent of its cycloxygenase inhibitory properties (Aubin et al., 1998). The memory-enhancing effects of aspirin was observed in an AlCl3-induced mouse model of neurotoxicity (Rizwan et al., 2016). In AD, epidemiological studies have shown that high-dose aspirin users exhibit lower prevalence of AD and better maintenance of cognitive functions (Nilsson et al., 2003). Another study investigating the role of low-dose aspirin treatment revealed promising effects (Kern et al., 2012). However, although epidemiological results point toward a potential role of aspirin in AD prevention and treatment, its therapeutic potential and the underlying molecular mechanism need further investigation. In this study, we tested the effect of aspirin in an AD mouse model. Previously, we found that activation of peroxisome proliferator-activated receptor alpha (PPARα) by its agonists can induce lysosomal biogenesis (Ghosh et al., 2015). Here, we reveal that aspirin induces lysosomal biogenesis in cultured mouse brain cells via activation of PPARα. We also describe a role for PPARα in aspirin-mediated attenuation of the amyloid plaque burden in the 5XFAD mouse model.
Materials and Methods
Reagents.
Cell culture materials (DMEM/F-12, l-glutamine, Hank's balanced salt solution, 0.05% trypsin, and antibiotic–antimycotic) were from Mediatech. FBS was from Atlas Biologicals. Aspirin and all molecular biology-grade chemicals were obtained from Sigma-Aldrich. Primary antibodies, their sources, and the concentrations used are listed in Table 1. Alexa Fluor antibodies used in immunostaining were from Jackson ImmunoResearch Laboratories and IR-dye-labeled reagents used for immunoblotting were from Li-Cor Biosciences.
Table 1.
List of antibodies used in this study
| Target and antibody | Source | Catalog no. | Application/dilution |
|---|---|---|---|
| LAMP2 | Millipore | MABC40 | WB/1:1000 |
| ICC/1:500 | |||
| IHC/1:250 | |||
| TPP1 | Abcam | ab96498 | WB/1:1000 |
| GFAP | DAKO | IS524 | ICC/1:1000 |
| IHC/1:1000 | |||
| Cathepsin D | Santa Cruz Biotechnology | sc-6487 | ICC/1:500 |
| TFEB | Abcam | ab2636 | WB/1:500 |
| ICC/1:500 | |||
| IHC/1:500 | |||
| MAP2 | Millipore | AB5622 | ICC/1:750 |
| PPARα | Santa Cruz Biotechnology | sc-398394 | ICC/1:500 |
| IHC/1:250 | |||
| ChIP/1:100 | |||
| PPARβ | Santa Cruz Biotechnology | sc-7197 | ChIP/1:100 |
| PPARγ | Santa Cruz Biotechnology | sc-7273 | ChIP/1:100 |
| CBP | Santa Cruz Biotechnology | sc-369 | ChIP/1:100 |
| RNA Pol | Millipore | 05–623 | ChIP/1:200 |
| β-Amyloid | |||
| Mouse monoclonal (6E10) | BioLegend | 803001 | WB/1:1000 |
| IHC/1:500 | |||
| Mouse monoclonal (4G8) | Anaspec | SIG-39220-100 | WB/1:1000 |
| NeuN | Invitrogen | 702022 | IHC/1:1000 |
| β-Tubulin | Santa Cruz Biotechnology | sc-9104 | WB/1:1500 |
| β-Actin | Abcam | ab6276 | WB/1:10000 |
WB, Western blot; ICC, immunocytochemistry; IHC, immunohistochemistry.
Isolation of primary mouse astroglia.
Astroglia were isolated from 7- to 9-d-old mouse pups as described previously (Brahmachari et al., 2006; Ghosh and Pahan, 2012; Khasnavis et al., 2012). Briefly, on day 9, the mixed glial cultures were subjected to shaking at 240 rpm for 2 h at 37°C on a rotary shaker to remove microglia, followed by another round of shaking on day 11 at 190 rpm for 18 h to remove oligodendroglia and residual microglia. The attached cells were washed and seeded onto new plates for further studies. Approximately 98% of this preparation was found to be positive for GFAP, a marker of astrocytes.
LysoTracker staining.
It was performed as described previously (Ghosh et al., 2015; Chandra et al., 2018). Briefly, after treatments, cells were incubated with 75 nm LysoTracker Red DND99 (Life Technologies) for 45 min. Cells were then washed thoroughly with filtered PBS, mounted on glass slides, and viewed under a BX41 fluorescence microscope.
Electron microscopy and counting of lysosomes.
After treatment, cells were fixed with a paraformaldehyde (2%) and glutaraldehyde (2.5%) mixture. After primary fixation, samples were prepared in the electron microscopy core facility of the University of Illinois at Chicago Research Resources Center. To stabilize cell components, samples were treated with 1% osmium tetroxide in phosphate buffer. Next, the samples were dehydrated through an increasing concentration of ethanol, passed through propylene oxide, and then infiltrated and embedded in a liquid resin. The resin block was then sectioned by ultramicrotomy with 50–70 nm thickness and collected on metal mesh grids, which were stained with electron-dense stains before observation in the TEM (JEOL JEM-1220). Electron microscopy images of primary astrocytes were analyzed for the presence of autophagic vesicles and lysosomes using the cell counter program in ImageJ. Data are shown as the mean number of vesicles ± SEM per cell. At least 20 images per condition were quantified for the analysis.
RT-PCR analysis.
RT-PCR was performed using the ABI-Prism7700 sequence detection system (Applied Biosystems) as described previously (Corbett et al., 2012; Ghosh and Pahan, 2012; Khasnavis et al., 2012). The mRNA expressions of respective genes were normalized to the level of GAPDH mRNA.
Immunoblotting.
Western blotting was conducted as described previously (Corbett et al., 2012; Ghosh and Pahan, 2012). Briefly, cells were scraped in PBS, transferred to microfuge tubes, and spun into pellets, followed by lysis in RIPA buffer or CHAPS buffer. The supernatant was collected and analyzed for protein concentration via the Bradford method (Bio-Rad). SDS sample buffer was added to 40–60 μg of total protein and boiled for 5 min. Denatured samples were electrophoresed on NuPAGE Novex 4–12% Bis-Tris gels (Invitrogen) and proteins transferred onto a nitrocellulose membrane (Bio-Rad) using the Thermo-Pierce Fast Semi-Dry Blotter. The membrane was then washed for 15 min in TBS plus Tween 20 (TBST) and blocked for 1 h in TBST containing BSA. Next, membranes were incubated overnight at 4°C under shaking conditions with primary antibodies. The next day, membranes were washed in TBST for 1 h, incubated in secondary antibodies for 1 h at room temperature, washed for an additional 1 h, and visualized under the Odyssey Infrared Imaging System (Li-COR).
Densitometric analysis.
Protein blots were analyzed using ImageJ and bands were normalized to their respective β-actin loading controls. Data are representative of the average fold change with respect to control for three independent experiments.
Immunofluorescence analysis.
Immunofluorescence analysis was performed as described previously (Brahmachari et al., 2009; Khasnavis and Pahan, 2012). Briefly, coverslips containing 100–200 cells/mm2 were fixed with 4% paraformaldehyde, followed by treatment with cold ethanol and two rinses in PBS. Samples were blocked with 3% BSA in PBS-Tween-20 (PBST) for 30 min and incubated in PBST containing 1% BSA and primary antibodies. After three washes in PBST (15 min each), slides were further incubated with Cy2 (Jackson ImmunoResearch Laboratories). For negative controls, a set of culture slides was incubated under similar conditions without the primary antibodies. The samples were mounted and observed under an Olympus IX81 fluorescence microscope.
Lysosomal enzyme [tri-peptidyl-peptidase 1 (TPP1) and Cathepsin B and D] assays.
Cultured mouse primary astrocytes were treated with different doses (2, 5, and 10 μm) of aspirin for 24 h, after which the cells were pelleted down. For the TPP1 assay, the cell pellet was homogenized in a buffer containing 0.15 m NaCl and Triton X-100. The supernatant was incubated with 200 μm of the substrate Ala-Ala-Phe 7-amido-4-methylcoumarin (Sigma-Aldrich) in a 100 μl reaction at pH 4. The plate was measured at excitation/emission of 360 nm/460 nm in a Victor X2 microplate reader (Perkin-Elmer). Data are shown as the relative fluorescence unit (RFU) fold change of the treated groups with respect to the control at 20 min. For cathepsin B and D assays, the cells were lysed in a homogenization buffer (pH 5.5; 2.5 mm EDTA, Triton X-100, 2.5 mm DTT). For Cathepsin B, the supernatant was incubated with 100 μm of the substrate Z-Arg-Arg-7-amido-4-methylcoumarin hydrochloride at pH 6. The plate was measured at excitation/emission of 355/460 nm. For Cathepsin D, the supernatant was incubated at pH 4 with the 10 μm of the substrate 7-methoxycoumarin-4-acetyl-Gly-Lys-Pro-Ile-Leu-Phe-Phe-Arg-Leu-Lys(DNP)-d-Arg-amide (Enzo Life Sciences). The plate was measured for every 10 min for 90 min at excitation/emission of 320/420 nm. Data are shown as the RFU fold change of the treated groups compared with the control at 30 min.
Electrophoretic mobility shift assay (EMSA).
The DNA-binding activity of PPARα was analyzed by nonradioactive EMSA as described previously (Corbett et al., 2012; Ghosh and Pahan, 2012) using the peroxisomal proliferator-response element (PPRE) of the Tfeb promoter. After treatment, cells were washed with PBS, scraped into 1.5 ml tube,s and centrifuged in 4°C for 5 min at 500 rpm. The supernatant was aspirated and the pellet was resuspended in a membrane lysis buffer composed of HEPES, pH 8.0, MgCl2, KCl, DTT, and protease/phosphatase inhibitors (Sigma-Aldrich), vortexed, and centrifuged in 4°C at 720 × g for 5 min. Again, the supernatant was aspirated and the pellet was resuspended in a high-salt nuclear envelope lysis buffer composed of HEPES, pH 8.0, MgCl2, glycerol, NaCl, EDTA, DTT, and protease/phosphatase inhibitors, rotated vigorously in 4°C for 15 min, and centrifuged in 4°C at 13,000 rpm for 15 min. The resultant supernatant was complexed with a mixture of binding buffer (Tris-HCl, KCl, EDTA, DTT, 10× TGE, glycerol, and Triton X-100), custom-designed fluorescent PPRE-specific probe (Li-Cor Biosciences), and salmon sperm DNA (Invitrogen) for 15 min at room temperature and electrophoresed on custom-cast 6% polyacrylamide-TGE gels in 1× TGE for 2 h. The shift was visualized under the Odyssey Infrared Imaging System (Li-COR).
Construction of mouse Tfeb promoter-driven reporter construct.
The construct was made as described previously (Ghosh et al., 2015).
Cloning of Tfeb promoter and site-directed mutagenesis.
Cloning and mutagenesis were performed as described previously (Ghosh et al., 2015).
Assay of Tfeb promoter-driven reporter activity.
Cells plated at 50–60% confluence in 12-well plates were cotransfected with 0.25 μg of pTFEB(WT)-Luc, pTFEB(Mu)-Luc and using Lipofectamine Plus (Life Technologies). After 24 h of transfection, cells were stimulated with different agents under serum-free conditions for 6 h. Firefly luciferase activities were analyzed in cell extracts using the Luciferase Assay System kit (Promega) in a TD-20/20 Luminometer (Turner Designs) as described previously (Jana et al., 2007; Jana and Pahan, 2012; Ghosh et al., 2015).
Assay of transcriptional activities.
Cells plated at 70–80% confluence in 12-well plates were cotransfected with 0.25 μg of PPRE-Luc (an PPAR-dependent reporter construct) and 12.5 ng of pRL-TK using LipofectAMINE Plus (Corbett et al., 2012; Ghosh and Pahan, 2012). After 24 h of transfection, cells were treated with aspirin for 4 h, followed by measuring firefly and Renilla luciferase activities.
ChIP.
Recruitment of PPARα to the Tfeb gene promoter was determined using the EZ ChIP kit from Millipore as described previously (Corbett et al., 2012; Ghosh and Pahan, 2012; Ghosh et al., 2012). Briefly, 1 × 106 astrocytes were treated with aspirin and, after 1 h of stimulation, fixed by adding formaldehyde (1% final concentration, and cross-linked adducts were resuspended and sonicated. ChIP was performed on the cell lysate by overnight incubation at 4°C with 2 μg of Abs against PPARα, PPARβ, PPARγ, CBP, and RNA polymerase, followed by overnight incubation with protein G-agarose (Santa Cruz Biotechnology). The beads were washed and incubated with elution buffer. To reverse the cross-linking and purify the DNA, precipitates were incubated in a 65°C incubator overnight and digested with proteinase K. DNA samples were then purified and precipitated and the precipitates were washed with 75% ethanol, air-dried, and resuspended in Tris-EDTA buffer. The following primers were used to amplify the PPRE-containing fragment of the mouse Tfeb promoter: sense: 5′-GAA CAT TCC AGG TGG AGG CA-3′, antisense: 5′-CCC CCA ACA CAT GCT TCT CT-3′. PCR products were electrophoresed on 2% agarose gels.
Amyloid β uptake and degradation assay.
Mouse primary astrocytes were cultured in 96-well plates (Thermo Fisher Scientific) and treated with 5 μm aspirin for 24 h. Next, the cells were incubated in medium containing 500 nm oligomeric FAM-tagged Aβ(1–42) (Anaspec) for 2 or 4 h. For the degradation assay, after 4 h of FAM-Aβ incubation, the cells were allowed to grow in Aβ-free medium for an additional 6 h and fluorescence was measured. Next, the cells were washed in regular medium and the extracellular signal was quenched using Trypan blue solution. After another media wash, the fluorescence was read at excitation/emission of 485/535 in Perkin-Elmer Victor X2. Next, the cells were incubated with Hoechst and the fluorescence was measured at excitation/emission of 360/465 nm. FAM-Aβ fluorescence was normalized with Hoechst. The data are shown as the fold change of the aspirin-treated cells with respect to the controls.
Aβ uptake and degradation immunocytochemistry.
Mouse primary astrocytes, grown on coverslips, were treated with 5 μm aspirin for 24 h, followed by incubation with 500 nm Hilyte Fluor 647-tagged Aβ(1–42) (Anaspec) for 4 h. For monitoring the degradation, the cells were washed in normal Aβ-free medium for an additional 6 h. Next, the cells were fixed using chilled methanol. DAPI was used for staining nuclei. Finally, the coverslips were mounted using Fluoromount (Sigma-Aldrich).
Animals and aspirin treatment.
B6SJL-Tg(APPSwFlLon,PSEN1*M146L*L286V)6799Vas/J transgenic (5XFAD) mice were from The Jackson Laboratory. Animals were maintained and experiments were conducted in accordance with National Institutes of Health guidelines and were approved by the Rush University Medical Center Institutional Animal Care and Use Committee. Six- to seven-month-old male and female 5XFAD mice were treated with low-dose (2 mg/kg body weight/d) aspirin (solubilized in 0.1% methyl cellulose) via gavage for 30 d.
Immunohistochemistry.
Mice were anesthetized with ketamine–xylazine injectables and perfused with PBS, followed by dissection of half of the brain from each mouse for biochemical assays and half for immunohistochemistry (Ghosh et al., 2007; Khasnavis et al., 2014). Briefly, half brains were incubated in 4% paraformaldehyde (w/v), followed by 30% sucrose overnight at 4°C. Brains were then embedded in optimal cutting temperature medium (Tissue Tech) at −80°C and processed for conventional cryosectioning. Frozen sections (30 μm) were treated with cold ethanol (−20°C), followed by two rinses in PBS, blocking with 2% BSA in PBST, and double-labeling with two primary antibodies. After three washes in PBST, sections were further incubated with Cy2 and Cy5 (Jackson ImmunoResearch Laboratories). The samples were mounted and observed under an Olympus IX81 fluorescence microscope. Counting analysis was performed using ImageJ software.
Experimental design and statistical analysis.
Statistical analyses were performed with Student's t test (for two-group comparisons) and one-way ANOVA followed by Tukey's multiple-comparison tests as appropriate (for multiple groups comparison) using Prism 7 (GraphPad Software). Data are represented as mean ± SD or mean ± SEM as stated in figure legends. p < 0.05 was considered statistically significant. Sample sizes were similar to those described previously (Corbett et al., 2015; Rangasamy et al., 2015) and are provided in the figure legends. No statistical method was used to predetermine sample size.
Results
Induction of lysosomal biogenesis by aspirin in mouse primary astrocytes
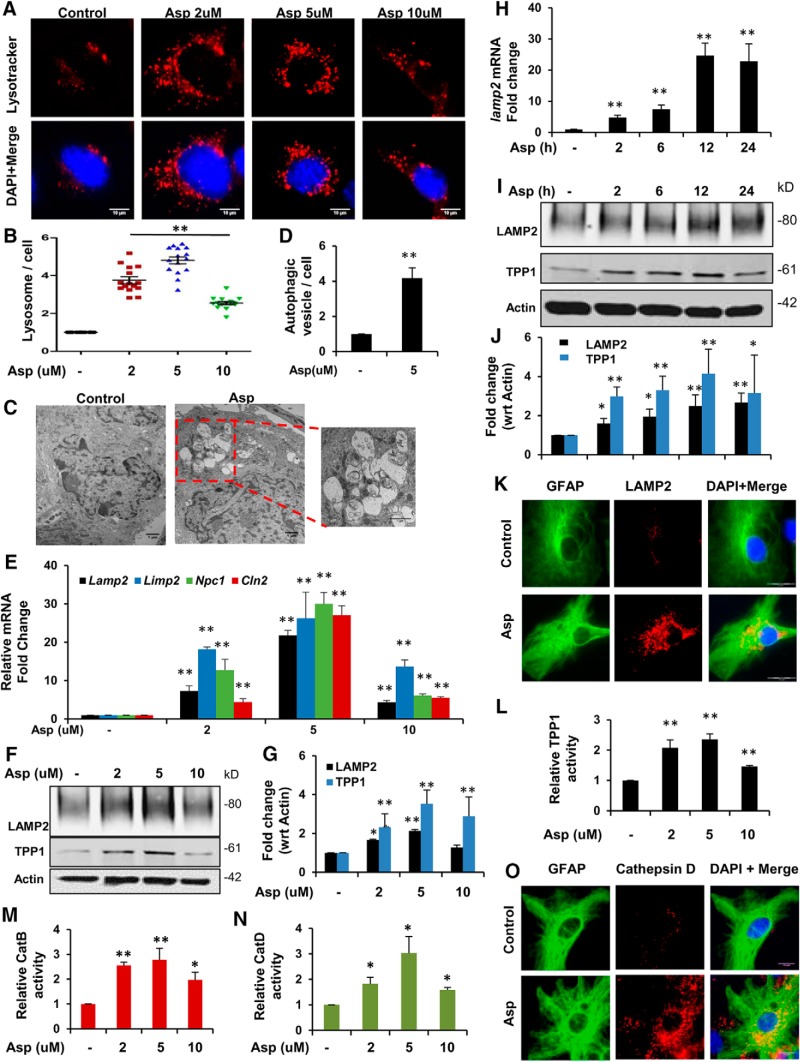
First, we investigated whether aspirin could induce lysosomal biogenesis in mouse primary astrocytes. To determine the lysosome content of the cell, astrocytes were treated with different doses of aspirin and further stained with LysoTracker Red, which selectively stains the acidic lysosomal organelles. We observed that aspirin treatment was able to significantly increase the lysosomal abundance, with 5 μm showing the highest effect (Fig. 1A,B). Electron microscopy study further confirmed the presence of increased number of lysosomes and different stages of autophagic vesicles in aspirin-treated astrocytes (Fig. 1C,D) compared with the control, indicating that aspirin was able to promote lysosomal biogenesis and autophagy in these cells. Next, we investigated the effect of aspirin on different lysosomal markers by quantitative RT-PCR and observed elevated mRNA expression of genes encoding lysosomal membrane proteins such as LAMP2, LIMP2, NPC1, as well as the lysosomal enzyme TPP1-encoding gene CLN2 (Fig. 1E). These results were further supported by Western blot analysis for LAMP2 and TPP1, which showed a similar pattern of increase in the expression of these proteins (Fig. 1F,G). Aspirin treatment was also found to augment the expression of LAMP2 mRNA in a time-dependent manner (Fig. 1H). The time point analysis showed an increase in the protein expression levels of both LAMP2 and TPP1 (Fig. 1I, J). Furthermore, immunofluorescence analysis showed a distinct increase in the expression of LAMP2 in primary astrocytes (Fig. 1K). Next, we tested whether aspirin could augment the activity of the lysosomal enzyme TPP1, dysfunction of which causes a lysosomal disorder known as late-infantile neuronal ceroid lipofuscinosis. Data showed that different doses of aspirin markedly upregulated TPP1 activity in primary astrocytes, suggesting that aspirin could increase the functionality of lysosomal enzyme (Fig. 1L). We further investigated the activity of Cathepsin B, a cysteine protease, and Cathepsin D, an aspartyl protease important for lysosomal proteolysis. Different doses of aspirin treatment remarkably upregulated the activity of Cathepsin B and D in primary astrocytes (Fig. 1M,N). Next, we analyzed the expression of Cathepsin D in astrocytes by double labeling Cathepsin D and GFAP. We observed a profound increase in the level of Cathepsin D after 5 μm aspirin treatment (Fig. 1O). Together, these results indicate that aspirin can stimulate lysosomal biogenesis and augment lysosomal functionality in astrocytes.
Figure 1.
Aspirin induces lysosomal biogenesis in mouse primary astrocytes. A, Mouse primary astrocytes from WT C57BL/6 pups were treated with different concentrations of aspirin (2, 5, and 10 μm) under serum-free conditions for 24 h, followed by staining with LysoTracker Red to selectively label the lysosomes and acidic organelles in live cells. DAPI was used to stain the nuclei. Scale bar 10 μm. B, Quantification of the number of LysoTracker-positive puncta per cell. Data are shown as mean ± SEM for fold change with respect to the untreated control for at least 15 images per group from three independent set of experiments quantified using ImgaeJ. C, Astrocytes treated with 5 μm aspirin were subjected to TEM to monitor authophagic and lysosomal vesicles. Scale bar, 1 μm. D, Quantitative analysis of autophagic vesicle per cell represented as mean ± SEM; 20 images per condition were quantified using ImageJ. E–G, Mouse primary astrocytes treated with different doses (2, 5, and 10 μm) of aspirin followed by mRNA expression analysis at 8 h for the lysosomal genes Lanp2, Limp2, Npc1, and Cln2 by quantitative RT-PCR (E); immunoblot analysis at 24 h for LAMP2 TPP1 (F); and densitometric analysis of LAMP2 TPP1 protein expression (G). Data are shown as fold change (mean ± SD) with respect to untreated controls. H–J, Astrocytes treated with 5 μm aspirin for different time points (2, 6, 12, and 24 h), followed by determining mRNA expression for Lamp2 by quantitative RT-PCR analysis (H) and protein levels for LAMP2 TPP1 by Western blot analysis (I). Graph represents fold change (mean ± SD) relative to untreated controls. K, Immunofluorescence analysis of astrocytes treated with 5 μm aspirin for 24 h, followed by double labeling with LAMP2 and the astrocytic marker GFAP. Scale bar, 20 μm. L, TPP1 enzyme activity in astrocytes treated with different doses of aspirin for 24 h. Data are shown as mean ± SEM for the RFU fold change with respect to control. M, N, Cathepsin B and Cathepsin D enzyme activity in primary astrocytes treated with 2, 5, and 10 μm aspirin for 24 h. Data are shown as mean ± SD for the RFU fold change with respect to untreated controls. O, Immunocytochemistry analyzing Cathepsin D levels in astrocytes treated with 5 μm aspirin for 24 h. All statistical analysis were performed using Student's t test. *p < 0.05, **p < 0.001.
Aspirin stimulates lysosomal biogenesis via upregulation of TFEB
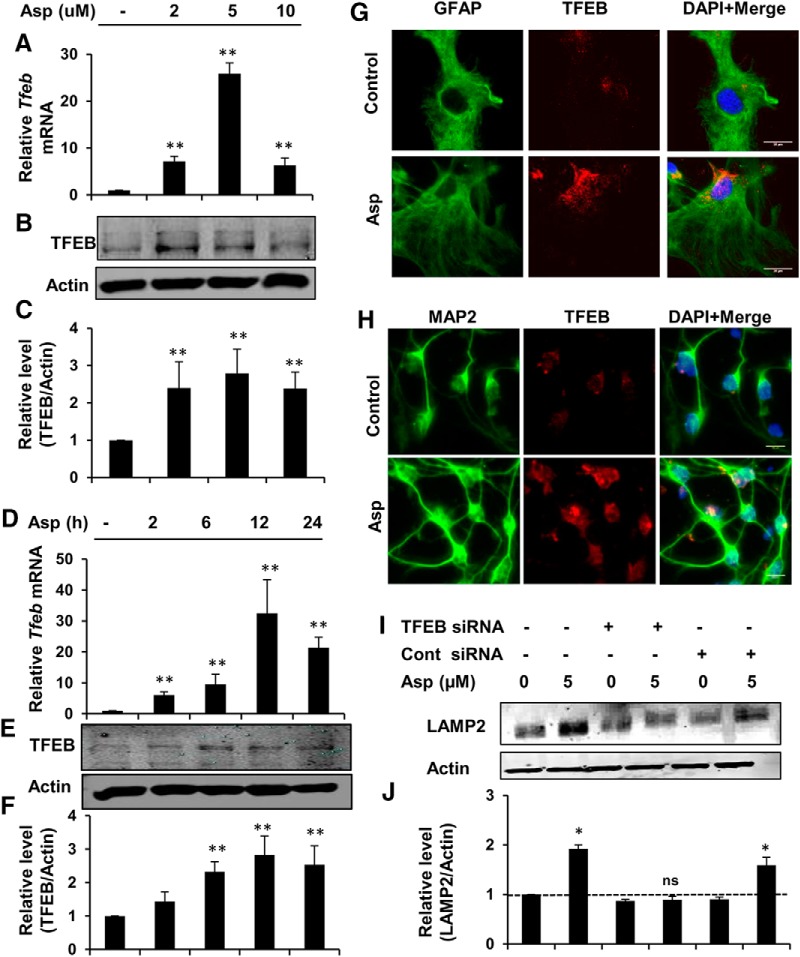
Next, we investigated whether aspirin treatment could augment the expression of TFEB, the master regulator of lysosomal biogenesis. Given that induction of TFEB transcriptionally upregulates other lysosomal genes and causes lysosomal biogenesis, we explored whether aspirin-mediated stimulation of lysosomal biogenesis requires TFEB. Mouse primary astrocytes were treated with different doses of aspirin and analyzed for TFEB expression. Expression of Tfeb mRNA increased in a dose-dependent manner after aspirin treatment, with the highest increase obtained with 5 μm aspirin (Fig. 2A). Western blot analysis also showed elevated levels of TFEB with different doses of aspirin (Fig. 2B,C). Furthermore, a time course study showed that aspirin increased the expression of Tfeb mRNA and TFEB protein with the peak increase seen at 12 h after aspirin treatment (Fig. 2D–F). Next, we performed immunocytochemistry in mouse primary astrocytes and cortical neuronal culture and observed that aspirin could markedly augment the overall expression of TFEB in these cells (Fig. 2G,H). In addition, TFEB immunoreactivity was more localized in and surrounding the nucleus in treated cells compared with the control group. This nuclear translocation of TFEB with treatment further indicated that TFEB was activating transcription. To further ascertain whether TFEB induction by aspirin is necessary for stimulating lysosomal biogenesis, we knocked down TFEB in astrocytes using Tfeb-specific siRNA and determined the expression of LAMP2 under different treatment conditions. Western blot analysis showed that the upregulation of LAMP2 by aspirin was abolished after TFEB silencing, whereas aspirin was still able to increase LAMP2 expression in scrambled siRNA-transfected cells (Fig. 2I,J). Collectively, these results demonstrate that aspirin induces lysosomal biogenesis via upregulating the central regulator of the lysosomal machinery, TFEB.
Figure 2.
Aspirin stimulates lysosomal biogenesis via upregulating TFEB. A–C, Mouse primary astrocytes were treated under serum-free conditions with different doses of aspirin, followed by monitoring of Tfeb mRNA expression at 8 h (A) and protein levels of TFEB at 24 h (B) of treatment and subsequent densitometric analysis of TFEB protein expression (C). Data are shown as fold change (mean ± SD) with respect to untreated control. D–F, Time point analysis for TFEB expression with 5 μm aspirin treatment by monitoring mRNA levels by quantitative RT-PCR (D), protein expression by immunoblot (E), and densitometry of TFEB protein levels (F). Data are shown as fold change (mean ± SD) with respect to untreated control. G, Mouse primary astrocytes treated with 5 μm aspirin for 24 h, followed by double labeling with TFEB and GFAP. Scale bar, 20 μm. H, Primary cortical neurons were treated under the same conditions and stained for TFEB and neuronal marker MAP2. Scale bar, 10 μm. I, J, Primary astrocytes were transfected with Tfeb siRNA or scrambled siRNA for 48 h and subsequently treated with 5 μm aspirin for 24 h, followed by Western blot (I) and densitometric (J) analysis for LAMP2 protein levels. All data are shown as mean ± SD. Statistical analysis was conducted using Student's t test; ns, nonsignificant; *p < 0.05, **p < 0.001.
Activation of PPARα by aspirin transcriptionally regulates TFEB expression
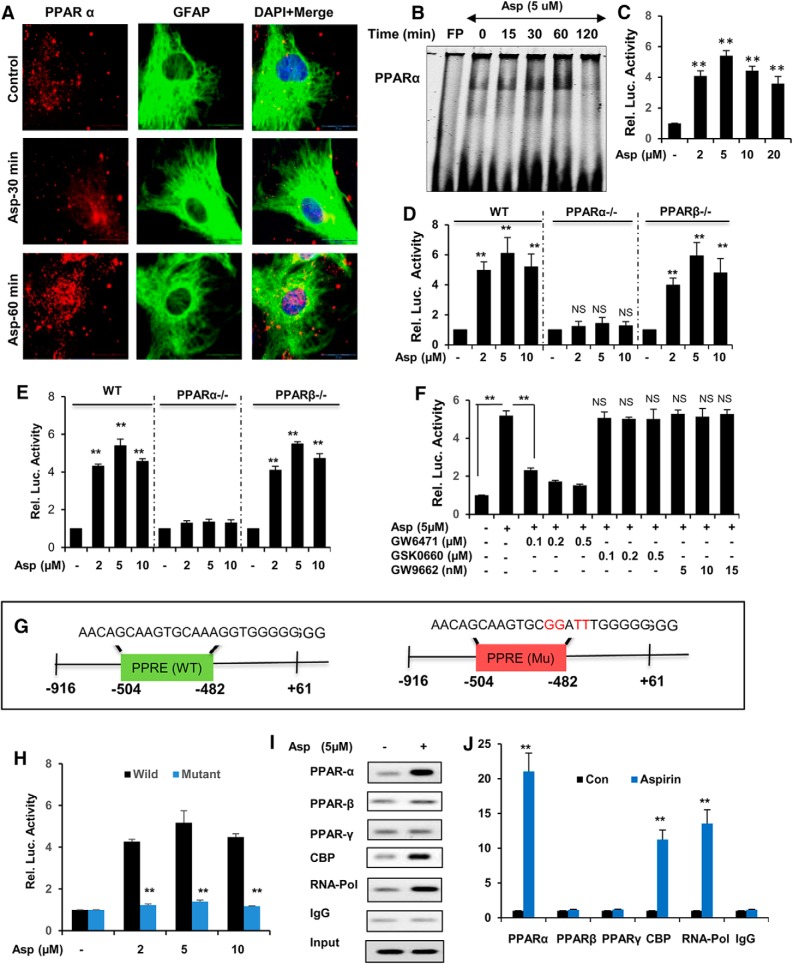
Given the evidence that aspirin can stimulate TFEB expression, we intended to characterize the upstream factors that can regulate TFEB. The presence of a PPRE site on the Tfeb promoter 480 bp upstream of the transcription start site prompted us to determine the activation status of PPARα after aspirin treatment. In a previous study, stimulation of PPARα by gemfibrozil resulted in lysosomal biogenesis in brain cells (Ghosh et al., 2015). Therefore, we hypothesized that activation of the PPRE on Tfeb promoter by aspirin could be the underlying mechanism by which aspirin mediated lysosomal biogenesis. Therefore, we monitored whether aspirin was capable of activating PPARα. First, we investigated whether aspirin treatment could induce nuclear translocation of PPARα. Mouse astrocytes were treated with 5 μm aspirin for different time points and analyzed by immunocytochemistry. Double labeling of PPARα and GFAP showed increased localization of PPARα in the nucleus and the surrounding perinuclear space, indicating activation of PPARα at 30 and 60 min after aspirin treatment (Fig. 3A). Further, we determined the DNA-binding activity of PPARα by EMSA. Wild-type astrocytes were treated with 5 μm aspirin for 15, 30, 60, and 120 min and a probe against the Tfeb gene PPRE was used for analyzing DNA-binding activity by PPARα. Data showed a shift with respect to the free probe and increased DNA binding of PPARα upon treatment with aspirin (Fig. 3B). Next, we tested whether activation of PPARα by aspirin could transcriptionally regulate Tfeb expression. WT astrocytes were transfected with pPPRE-luciferase construct for 24 h, followed by treatment with different doses of aspirin (2, 5, 10, and 20 μm) for 8 h. We observed a marked increase in the PPRE-driven luciferase activity in the aspirin-treated groups, with 5 μm showing the highest activity (Fig. 3C). Furthermore, astrocytes isolated from WT, Ppara-null, and Pparb-null mice were treated with aspirin and subjected to the pPPRE-luciferase assay. We observed a profound increase in the luciferase activity with aspirin treatment in WT and Pparb-null, but not Ppara-null, astrocytes, confirming that aspirin specifically activates PPARα (Fig. 3D). Next, we cloned the Tfeb promoter consisting of the PPRE site, denoted pTFEB(WT) (Fig. 3G), and observed that aspirin treatment remarkably induced TFEB-driven luciferase activity in WT and Pparb-null, but not Ppara-null, astrocytes, indicating the involvement of PPARα in activation of the Tfeb promoter (Fig. 3E). Moreover, we determined the effect of antagonists of PPARα, (GW6471), PPARβ (GSK0660), and PPARγ (GW9662) on TFEB promoter activation in WT astrocytes and found that only the PPARα antagonist GW6471 abolished aspirin-induced Tfeb promoter-driven luciferase activity, confirming that PPARα, but neither PPARβ nor PPARγ, is responsible for aspirin-stimulated activation of TFEB (Fig. 3F). In addition, when we performed the luciferase assay in WT astrocytes transfected with a construct containing mutated PPRE core sequence on the TFEB promoter, denoted pTFEB(Mu) (Fig. 3G), a drastic reduction in the mutated TFEB-driven luciferase activity was observed, further confirming the underlying role of PPARα in aspirin-mediated transcriptional induction of the TFEB promoter (Fig. 3H). Next, we performed ChIP followed by semiquantitative and RT-PCR to characterize the TFEB promoter DNA-binding complex. We observed that aspirin treatment markedly stimulated the recruitment of PPARα, CBP, and RNA polymerase to the PPRE site on the TFEB promoter (Fig. 3I,J). This result was specific because aspirin remained unable to induce the recruitment of either PPARβ or PPARγ to the Tfeb promoter (Fig. 3I,J). Together, our data indicated that aspirin-mediated PPARα activation transcriptionally regulates TFEB expression.
Figure 3.
Activation of PPARα by aspirin transcriptionally modulates TFEB expression. A, Primary astrocytes were treated with 5 μm aspirin for 30 min and 60 min, followed by double labeling for PPARα and GFAP. Scale bar, 20 μm. DAPI was used for staining nuclei. B, DNA-binding activity of PPARα after 15, 30, 60, and 120 min treatment of astrocytes with 5 μm aspirin as demonstrated by EMSA. FP, Free probe. C, WT astrocytes were transfected with pPPRE-luciferase construct for 24 h, followed by treatment with different doses of aspirin (2, 5, and 10 μm) for 8 h and were examined by luciferase assay. WT, Ppara-null, and Pparb-null astrocytes transfected with pPPRE-luciferase (D) or pTFEB(WT)-luciferase (E) construct for 24 h were treated with 2, 5, and 10 μm aspirin for 8 h and analyzed by luciferase assay. F, WT mouse astrocytes transfected with pTFEB(WT)-luciferase for 24 h were pretreated with different doses of PPARα antagonist GW6471 (100, 200, and 300 nm), PPARβ antagonist GSK0660 (100, 200, and 300 nm), and PPARγ antagonist GW9662 (5, 10, and 15 nm) for 1 h, followed by 5 μm aspirin treatment and subsequently subjected to luciferase assay. G, WT astrocytes transfected with pTFEB(WT)-luciferase or pTFEB(mutant)-luciferase construct for 24 h were treated with 5 μm aspirin for 8 h and analyzed by luciferase assay. H, Primary astrocytes treated with 5 μm aspirin for 2 h were analyzed for the recruitment of PPARα, PPARβ, PPARγ, CBP, and RNA polymerase to the conserved PPRE site on Tfeb promoter by performing ChIP, followed by semiquantitative RT-PCR (I) and quantitative RT-PCR (J). All values were normalized with input and are represented as fold change with respect to untreated controls. Data are shown as mean ± SD. IP, Immunoprecipitation; CBP, CREB binding protein. Statistical analysis was conducted using Student's t test; **p < 0.001.
Aspirin induces lysosomal biogenesis via PPARα
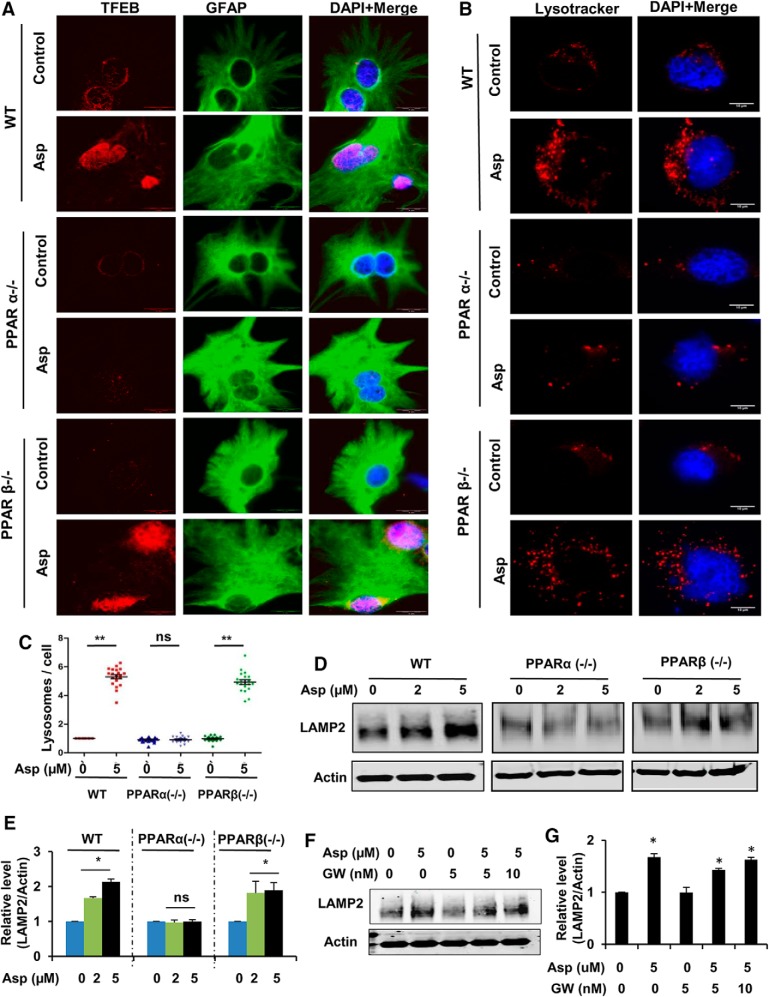
We next sought to determine whether PPARα is involved in aspirin-mediated induction of lysosomal biogenesis by conducting experiments in WT, Ppara-null, and Pparb-null astrocytes. Mouse primary astrocytes isolated from WT, Ppara-null, and Pparb-null mice were treated with aspirin and analyzed for different lysosomal markers. To establish this, first, we determined the expression of TFEB, the central transcription factor regulating lysosomal genes. Immunocytochemical analysis showed that aspirin treatment could distinctly upregulate TFEB expression in astrocytes isolated from WT and Pparb-null mice, but not Ppara-null mice. Moreover, higher levels of nuclear localized TFEB was observed in aspirin-treated astrocytes isolated from WT and Pparb-null mice, but not Ppara-null mice (Fig. 4A). These results suggest that TFEB stimulation by aspirin is specifically dependent on PPARα. Next, we tested whether aspirin-mediated lysosomal enrichment requires PPARα by performing the LysotrackerRed staining, followed by quantification of the total number of lysosomes per cell. Data showed that the overall lysosome content was significantly increased in WT and Pparb-null astrocytes after aspirin treatment; however, this effect was not observed in the case of aspirin-treated PPARα −/− astrocytes (Fig. 4B,C). These findings were further supported by Western blot analysis for LAMP2 showing a distinct increase with aspirin treatment in WT and Pparb-null astrocytes, but not in Ppara-null astrocytes (Fig. 4D,E). After establishing the involvement of PPARα in this process, we further investigated whether PPARγ plays any role in aspirin-mediated lysosomal induction. We pretreated WT astrocytes with the PPARγ-specific inhibitor GW9662, followed by treatment with aspirin, and determined the expression of LAMP2. Western blot analysis showed that, even in the presence of GW9662, aspirin was still able to upregulate LAMP2 levels, suggesting that PPARγ might not be required by aspirin for exerting its effect on the lysosomes (Fig. 4F,G). Together, these findings demonstrate that aspirin specifically requires PPARα for inducing lysosomal biogenesis.
Figure 4.
Induction of aspirin-mediated lysosomal biogenesis in WT and PPARβ−/− astrocytes, but not in PPARα−/− astrocytes. A–E, Primary astrocytes from WT, Ppara-null, and Pparb-null mice were treated with 5 μm aspirin for 24 h, followed by immunofluorescence staining for TFEB and GFAP (A; scale bar, 20 μm). Nuclei were stained using DAPI. B, Live cell staining using LysoTracker Red. Scale bar, 10 μm. C, Quantification of the lysosome number per cell using ImageJ. Data are shown as mean ± SEM of fold change relative to the untreated WT control. Twenty images per condition from three independent experiments were quantified. Immunoblot analysis for LAMP2 (D) and densitometric analysis of LAMP2 protein levels (E). Data are shown as mean ± SD for fold change with respect to untreated control in each group. F, G, WT astrocytes were pretreated with 5 and 10 nm PPARγ inhibitor GW9662 for 1 h, followed by 24 h treatment with 5 μm aspirin and analyzed for LAMP2 protein levels by Western blot (F) and densitometric analysis (G). Data are shown as mean ± SD for fold change normalized to the untreated control. All statistical analyses were done using Student's t test; *p < 0.05, **p < 0.001.
Aspirin enhances uptake and degradation of Aβ in primary astrocytes
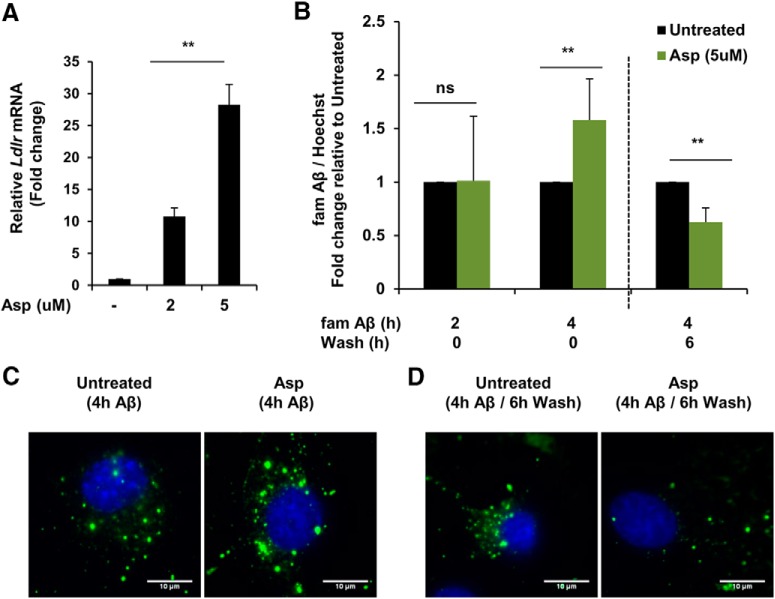
We further explored whether aspirin-mediated lysosomal biogenesis could enhance the cellular degradative capacity by uptake and effective lysosomal clearance of Aβ in astrocytes. Astrocytes can uptake Aβ via the cell surface low-density lipoprotein receptor (LDLR), followed by lysosomal degradation of Aβ (Wyss-Coray et al., 2003; Basak et al., 2012). In addition, PPARα activation has been reported to induce hepatic LDLR expression (Huang et al., 2008). Having demonstrated that aspirin activates PPARα, we determined the effect of aspirin on LDLR expression in primary astrocytes. Low doses of aspirin were observed to remarkably increase the mRNA expression of Ldlr (Fig. 5A). Because LDLR facilitates the internalization of extracellular Aβ, it is highly likely that enhanced LDLR expression would lead to a higher uptake of Aβ inside the cell. We performed a quantitative in vitro assay to study the effect of aspirin on uptake and degradation of FAM-tagged Aβ(1–42) in astrocytes. After 4 h of incubation with Aβ42-containing medium, aspirin treatment significantly enhanced the intracellular amount of Aβ42 compared with control, indicating that aspirin increases the uptake of Aβ by astrocytes (Fig. 5B). After the 4 h Aβ42 incubation, when cells were grown for an additional 6 h in Aβ-free medium (6 h wash), aspirin-treated astrocytes demonstrated a significantly lower level of intracellular Aβ than the control, suggesting that the internalized Aβ was effectively degraded (Fig. 5B). We further confirmed these observations by labeling astrocytes with HF-Aβ(1–42). Similar to the in vitro kinetic assay, immunocytochemistry showed that aspirin augmented the uptake of Aβ and the degradation of internalized Aβ compared with the control after 4 h of incubation with HF-Aβ42 and an additional 6 h wash, respectively (Fig. 5C,D). Collectively, these results demonstrate that aspirin can enhance astrocytic clearance of Aβ via increasing the uptake and lysosomal degradation of Aβ by astrocytes.
Figure 5.
Aspirin enhances uptake and degradation of Aβ42 in mouse primary astrocytes. A, Primary astrocytes were treated with 5 μm aspirin for 4 h, followed by monitoring the mRNA expression of Ldlr by quantitative RT-PCR. Data are shown as fold change (mean ± SD) with respect to untreated controls. B, Treatment for 24 h followed by FAM-Aβ(1–42) uptake and degradation assay. Cells were incubated in medium containing 500 nm FAM-Aβ for 2 h, 4 h for monitoring the uptake. For degradation, the cells were grown in Aβ-free regular medium for an additional 6 h. Data are shown as fold change (mean ± SD) with respect to untreated control. C, Treatment for 24 h followed by 4 h incubation in medium containing HiLyte Fluor 647-Aβ for monitoring Aβ uptake. D, For degradation, the cells were washed in medium for an additional 6 h. DAPI was used to stain the nuclei. All statistical analyses were performed using Student's t test; **p < 0.001.
Stimulation of lysosomal biogenesis by aspirin in vivo in an 5XFAD model
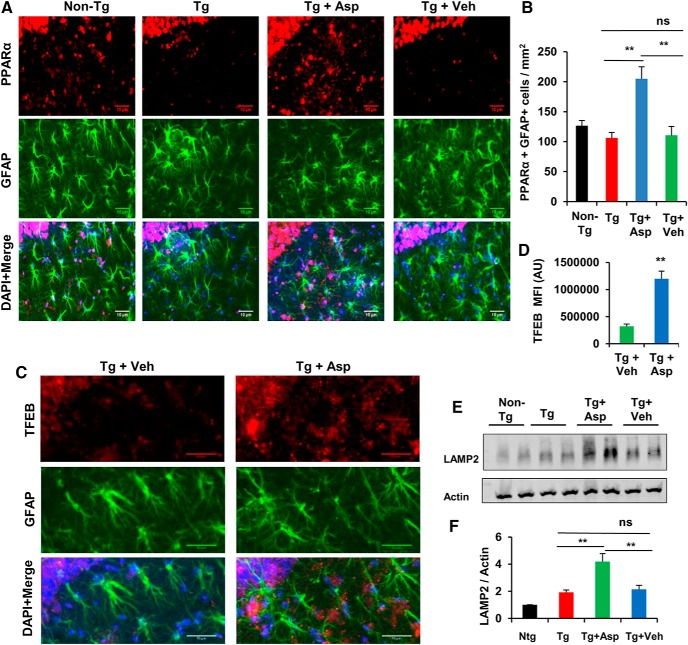
Next, we investigated whether aspirin treatment could stimulate lysosomal biogenesis in vivo in the hippocampus of the 5XFAD mouse model, which contains five familial AD-linked mutations in APP and PS1. Six- to 7-month-old 5XFAD transgenic (Tg) and age-matched nontransgenic (non-Tg) mice from the same background were used for the study. Mice received aspirin (2 mg/kg body weight/d) or vehicle (0.1% methylcellulose) treatment orally for 1 month. We first examined the astrocytic PPARα level by colabeling of hippocampal sections with the astrocytic marker GFAP and PPARα. Data showed that aspirin treatment remarkably increased the PPARα level in the hippocampus of Tg mice relative to either untreated or vehicle-treated Tg mice (Fig. 6A,B). Next, we determined the lysosomal abundance in these cohorts of mice. Immunohistochemistry of the hippocampal sections for TFEB and GFAP revealed significantly higher levels of TFEB in the aspirin-treated group compared with the vehicle group, indicating enhanced lysosomal biogenesis (Fig. 6C,D). Moreover, Western blot analysis of hippocampal homogenates also showed increased levels of LAMP2 in aspirin-treated mice (Fig. 6E,F). Together, our results illustrate that oral administration of aspirin augments the PPARα-mediated lysosomal biogenesis in 5XFAD mice.
Figure 6.
Stimulation of lysosomal biogenesis by aspirin in vivo in a 5XFAD mouse model. A, B, 5XFAD mice (6–7 months old, n = 6) were treated orally with aspirin (2 mg/ kg body weight/d) or vehicle (.1% methylcellulose) for 1 month, after which the level of PPARα in the hippocampus was monitored by immunostaining and quantification of astrocytic PPARα (PPARα+ GFAP+ cells). C, D, Lysosomal biogenesis was monitored by staining of hippocampal sections by TFEB, GFAP, and DAPI (C) and TFEB mean fluorescence intensity analysis using ImageJ (D). Three different regions from each of 12 different images for each group were quantified. E, F, LAMP2 levels in the hippocampal homogenates was analyzed by Western blotting (E) and densitometric analysis (F). Statistical analysis was performed using Student's t test or one-way ANOVA followed by Tukey's multiple-comparisons test; ns, not significant; **p < 0.001.
Aspirin reduces the amyloid burden in the hippocampus of 5XFAD mice
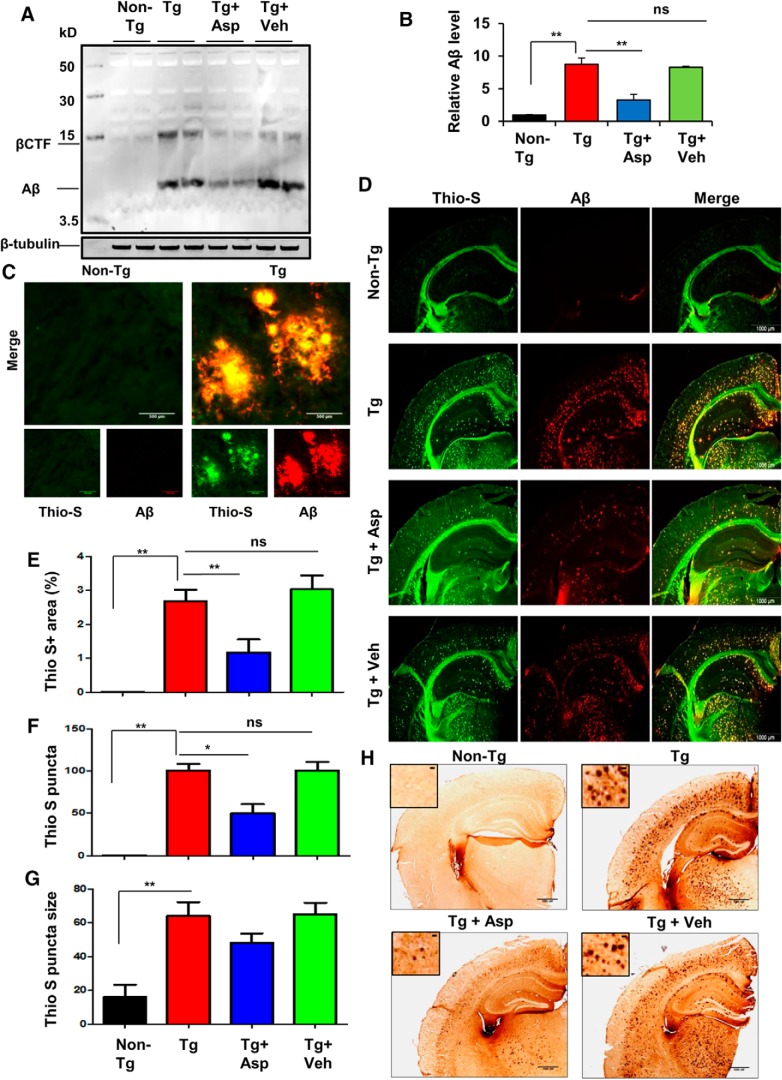
Having confirmed that aspirin causes enhancement of lysosomal biogenesis via stimulation of TFEB, we intended to evaluate the efficacy of this induction of the cellular clearance pathways to attenuate the amyloid plaque burden in the 5XFAD mouse model. Multiple reports have demonstrated the beneficial role of exogenously induced TFEB expression to enhance the lysosomal activity in AD models of amyloid pathology, confirming that activation of TFEB is an effective strategy to ameliorate amyloid plaque pathogenesis by facilitating APP and Aβ clearance (Xiao et al., 2014, 2015; Zhang and Zhao, 2015). We investigated whether oral administration of aspirin was capable of reducing the amyloid load in the hippocampus, the most affected brain region in AD, of Tg mice. Immunoblot analysis using the Aβ-specific antibody 4G8 showed a remarkable increase in the Aβ levels with a parallel upregulation of the β-CTF in the hippocampal homogenates from Tg mice compared with non-Tg mice. However, oral administration of aspirin significantly reduced the level of Aβ in the hippocampus of Tg mice, whereas the vehicle group did not exhibit such effects (Fig. 7A,B). Next, to determine the amyloid deposition in the brain, we performed double labeling of the hippocampal sections with thioflavin-S (thio-S), a classic amyloid-binding dye that detects the β-pleated sheet of the amyloid plaques and Aβ-specific monoclonal antibody 6E10. Parallel to the immunoblot results, there was marked abundance of thio-S positive and Aβ-immunoreactive plaques in the Tg mice (Fig. 7C,D). However, treatment of Tg mice with aspirin, but not vehicle, was capable of ameliorating the plaque load (Fig. 7D). Further quantitative analysis of thio-S staining revealed that aspirin treatment led to a significant reduction in the thio-S-positive fraction in the hippocampus, along with a lower number of thio-S puncta relative to the Tg and vehicle groups, demonstrating that aspirin was able to alleviate the amyloid pathology in the hippocampus (Fig. 7E,F). A reduction in the size of the plaques was observed in aspirin-treated Tg mice compared with vehicle-treated Tg mice, but this was nonsignificant (Fig. 7G). A similar pattern of results were seen with diaminobenzidine staining with Aβ 6E10 antibody, which showed robust amyloid deposition in Tg mice that was reduced by aspirin treatment (Fig. 7H). Collectively, these results demonstrate that aspirin treatment can substantially attenuate the burden of amyloid plaques in the 5XFAD model.
Figure 7.
Aspirin treatment reduces Aβ burden in the hippocampus of a 5XFAD mouse model of AD. A, Six- to 7-month-old 5XFAD mice (n = 6 per group) were treated orally with aspirin (2 mg/ kg body weight/d) or vehicle (0.1% methylcellulose) for 1 month and subsequently analyzed for Aβ levels in the hippocampal homogenates by immunoblotting using the 4G8 monoclonal antibody. β-Tubulin was used as the loading control. B, Densitometric analysis representing mean ± SEM for Aβ levels relative to non-Tg controls. C, Hippocampal sections demonstrating Aβ plaque pathology in transgenic 5XFAD mice in the CA1 region of hippocampus. D, Hhippocampal sections were double labeled using thio-S and Aβ 6E10 antibody. E–G, The thio-S-positive plaques in the hippocampus were further characterized for total area faction (thio-S positive area as a percentage of total hippocampal area; E), plaque count (F), and average plaque size (G). Data are shown as mean ± SEM. Quantification of thio-S-positive plaques were performed using ImageJ. H, Diaminobenzidine staining of hippocampal sections was performed using Aβ 6E10 antibody. In all cases, one-way ANOVA followed by Tukey's multiple-comparisons test was used for statistical analysis; *p < 0.05, **p < 0.001.
Aspirin reduces intraneuronal amyloid β (Aβ) accumulation
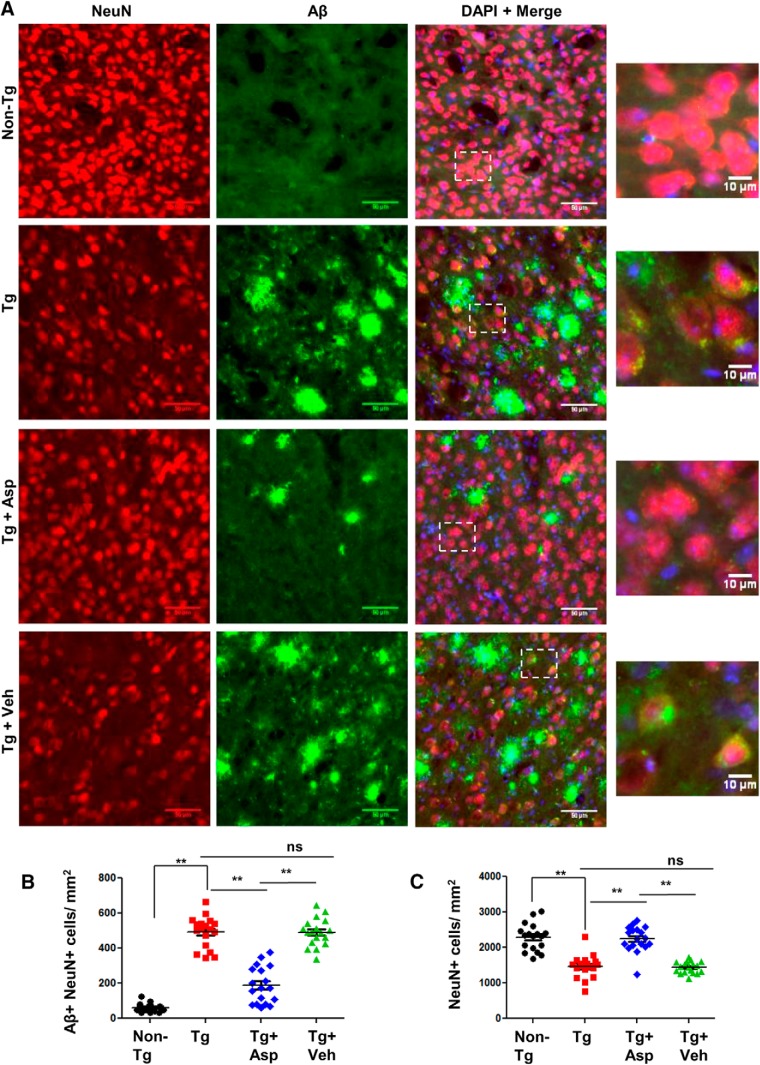
Next, we explored the effect of low-dose aspirin treatment on intraneuronal amyloid pathology in 5XFAD mice. In these mice, the five FAD mutations in APP and PS1 are expressed under the neuron-specific Thy-1 promoter and lead to accumulation of intraneuronal Aβ42, initiating plaque development (Oakley et al., 2006). A recent study has highlighted the beneficial role of neuronal-targeted TFEB in attenuating amyloid plaque pathology (Xiao et al., 2015). To study intraneuronal Aβ deposition, we performed double labeling of NeuN, a neuronal marker, and Aβ 6E10 in the cortex. As expected, 5XFAD Tg mice exhibited extensive neuronal Aβ accumulation compared with the non-Tg mice. Aspirin treatment remarkably reduced the intraneuronal level of Aβ, whereas no such effect was observed in the vehicle-treated mice (Fig. 8A,B). Along with Aβ accumulation, Tg mice also exhibit extensive neuronal loss in the cortex. We observed profound loss of cortical neurons in the Tg mice compared with the non-Tg mice, as demonstrated by quantification of the NeuN-positive cells. Importantly, aspirin-treated mice had marked improvement in the neuronal survival compared with the Tg and vehicle-treated groups (Fig. 8A,C). Together, these results suggest that aspirin treatment attenuates intraneuronal Aβ accumulation and enhances neuronal survival in 5XFAD mice.
Figure 8.
Aspirin reduces intraneuronal Aβ accumulation. 5XFAD mice (6–7 months old, n = 6) were treated orally with aspirin (2 mg/ kg body weight/d) or vehicle (.1% methylcellulose) for 1 month, after which the intraneuronal level of Aβ in the cortex was monitored by double labeling with NeuN and Aβ 6E10 (A), quantification of NeuN+ Aβ+ cells (B), and quantification of NeuN+ cells in the cortex by ImageJ (C). Eighteen images (20× magnification) from six different mice per group were quantified. One-way ANOVA followed by Tukey's multiple-comparisons test was used for statistical analysis; **p < 0.001.
Aspirin attenuates amyloid pathology in 5XFAD mice via PPARα
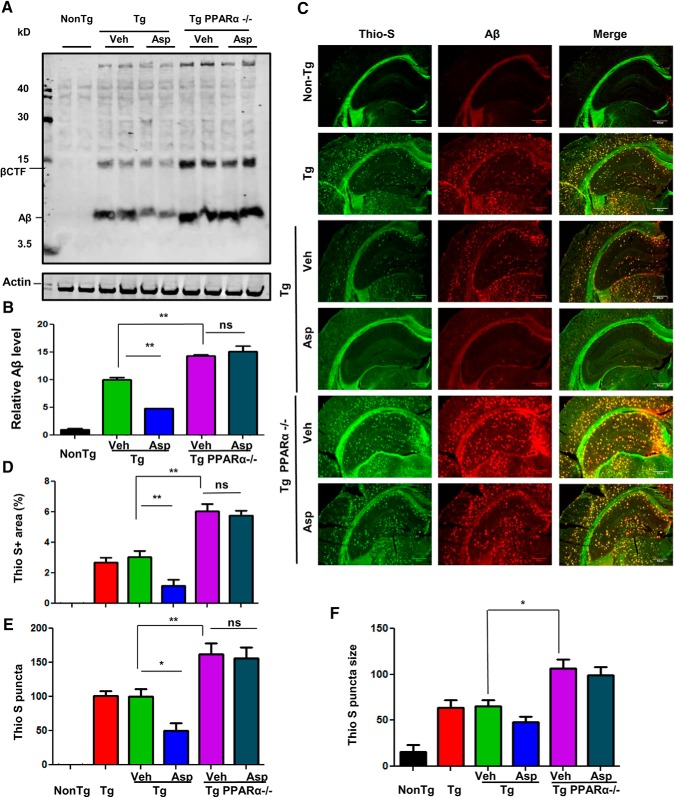
Given the observation that aspirin can activate PPARα and thus drive TFEB-mediated lysosomal enrichment, we explored our hypothesis that activation of PPARα by aspirin could be the underlying mechanism by which it exhibits the amyloid-lowering effects. We compared the amyloid pathology between Tg and Tg-Ppara-null mice, which harbors the five familial AD related mutations and are null for PPARα (Corbett et al., 2015). Immunoblot analysis from the hippocampal extracts demonstrated that Tg-Ppara-null mice receiving vehicle presented significantly higher levels of Aβ compared with Tg mice. However, in contrast to Tg mice, aspirin treatment remained unable to lower Aβ pathology in Tg-Ppara-null mice, suggesting that activation of PPARα by aspirin could be the underlying mechanism behind its amyloid-attenuating effects (Fig. 9A,B). It is noteworthy that Tg-Ppara-null mice exhibited remarkably more amyloid pathology relative to Tg mice, indicating that ablation of PPARα is a contributing factor driving more amyloidogenesis in these mice. In addition, the rise in Aβ levels in Tg-Ppara-null mice was accompanied by a parallel increase in the abundance of the β-CTF, another fragment generated by the amyloidogenic processing of APP, further supporting that PPARα could be implicated as a beneficial factor in relieving amyloidosis. Colabeling of hippocampal sections with thio-S and Aβ 6E10 showed a similar pattern of results, with significantly higher amyloid plaque deposition in Tg-Ppara-null mice, whereas aspirin treatment did not exhibit any improvement (Fig. 9C). Further analysis of the thio-S-positive plaques revealed a greater area fraction, higher count, and larger-sized plaques in the hippocampus of Tg-Ppara-null groups compared with the Tg groups (Fig. 9C–F). Together, these findings suggest that aspirin could mitigate amyloid pathology in a PPARα-dependent manner.
Figure 9.
Aspirin attenuates amyloid plaque pathology in 5XFAD mice via the PPARα pathway. A, Aspirin was administered orally in cohorts of 6- 7-month-old 5XFAD and 5XFAD/Ppara-null mice for 30 d, followed by analysis of the Aβ levels in the hippocampal extracts using Aβ 6E10 antibody. β-Actin was used as a loading control. B, Densitometric analysis (mean ± SEM) of Aβ levels with respect to nontransgenic control group. C, Colabeling of hippocampal sections with thio-S and Aβ 6E10 and characterization of thio-S-positive plaques for total area fraction (D), puncta count (E), and average plaque size (F) in the hippocampus. Statistical analysis was performed using one-way ANOVA followed by Tukey's multiple-comparisons test; *p < 0.05, **p < 0.001.
Discussion
Decreasing cerebral plaque load may have therapeutic importance in AD. Here, we demonstrate that low doses of aspirin, one of the most widely used medications in the world, upregulates lysosomal biogenesis and decreases amyloid plaque pathology in an animal model of AD. In cultured brain cells, aspirin increased the expression of different lysosome-specific molecules and upregulated lysosomal biogenesis. Similarly, after oral administration, low-dose aspirin also upregulated lysosomal markers in the hippocampus and decreased the load of amyloid plaques in 5XFAD mice. Therefore, our study suggests that aspirin may be a therapeutic agent for the treatment of AD.
Mechanisms by which lysosomal biogenesis occurs are becoming clear. Lysosomal adaptation under different physiological and pathological scenarios is dependent on a regulatory gene network known as CLEAR (coordinated lysosomal expression and regulation), with the master regulator TFEB at its core (Sardiello et al., 2009; Martini-Stoica et al., 2016). Here, we have shown that aspirin upregulates TFEB and stimulates lysosomal biogenesis via TFEB. After oral administration, aspirin also increased the level of TFEB in the hippocampus of 5XFAD mice. A recent study suggested that TFEB overexpression alleviates the disease progression by decreasing Aβ accumulation through the autophagy lysosome pathway (Zhang and Zhao, 2015). Furthermore, enhancing astrocytic lysosomal biogenesis by targeted TFEB expression through viral gene transfer was found to promote attenuation of amyloid pathogenesis by facilitating uptake and lysosomal degradation of Aβ in the astrocytes (Xiao et al., 2014). Similarly, in neurons, enhancing lysosomal function with TFEB accelerates lysosomal degradation of holo-APP and thus attenuates Aβ generation and plaque pathogenesis (Xiao et al., 2015). Therefore, upregulation of TFEB and lysosomal biogenesis by aspirin may have treatment prospects in AD.
How does aspirin upregulate TFEB? The presence of a PPRE site on the Tfeb promoter led us to explore the role of PPARα in aspirin-mediated TFEB upregulation. PPARs are ligand-inducible transcription factors belonging to the class of nuclear hormone receptors. Recently, we demonstrated that activation of PPARα by gemfibrozil leads to increased transcription of TFEB and stimulation in lysosomal biogenesis in brain cells (Ghosh et al., 2015). Several lines of evidence presented in this study also clearly suggest that aspirin upregulates TFEB and augments lysosomal biogenesis via PPARα. Numerous studies have highlighted the wide applicability and therapeutic potential of TFEB-mediated lysosomal modulation in lysosomal storage diseases (LSDs) such as Gaucher disease, Tay–Sachs disease (Song et al., 2013), Pompe disease (Spampanato et al., 2013), and cystinosis (Rega et al., 2016). A mechanistic connection between α-synuclein toxicity and impaired function of TFEB was reported and genetic activation of TFEB was shown to provide robust neuroprotection via α-synuclein clearance in dopaminergic neurons (Decressac et al., 2013). In another study, TFEB activation by induction of PGC-1α was able to promote htt turnover and clearance of protein aggregates in a mouse model of HD (Tsunemi et al., 2012). Therefore, the upregulation of TFEB and induction of lysosomal biogenesis by aspirin may have implications for the treatment of multiple LSDs and neurodegenerative disorders.
Mounting evidence suggest that astrocytes can efficiently uptake and degrade Aβ in lysosomes (Shaffer et al., 1995; Wyss-Coray et al., 2003; Nielsen et al., 2009). Therefore, enhancement of astrocytic Aβ clearance is an important therapeutic strategy for treatment of AD. The LDLR has been implicated in direct binding and internalization of Aβ by astrocytes (Basak et al., 2012). LDLR deficiency has been shown to reduce the glial response and enhance amyloid deposition in 5XFAD mice (Katsouri and Georgopoulos, 2011). Importantly, activation of the nuclear receptor PPARα by its agonist, fenofibrate, has been reported to induce hepatic LDLR expression (Huang et al., 2008). Here, we demonstrated that, along with inducing lysosomal biogenesis, aspirin at low doses can increase the mRNA expression of Ldlr and lead to enhanced uptake and degradation of extracellular Aβ by astrocytes.
Emerging studies have pointed toward an aberrant autophagic function in the neurons of AD models. The principal defect is believed to be at the autolysosomal proteolysis stage, which ultimately leads to accumulation of autophagic vesicles in dystrophic neurites (Nixon et al., 2005; Nixon, 2007; Wolfe et al., 2013). Several reports have demonstrated that the impairment in lysosomal substrate degradation is due to PS1 mutations that disrupt the regular acidification of lysosomes (Lee et al., 2010, 2015; Wolfe et al., 2013; Coffey et al., 2014). PS1 holoprotein facilitates the effective N-glycosylation and lysosomal targeting of V0a1 subunit of the vacuolar-ATPase (v-ATPase) proton pump, which acidifies the lysosome. Therefore, enhanced expression of the V0a1 subunit might result in escape from ER quality control mechanisms and lead to a greater abundance of this protein in the lysosomes and thereby rectify the lysosomal acidification defect. Importantly, v-ATPase can be regulated by TFEB (Palmieri et al., 2011; Peña-Llopis et al., 2011; Settembre et al., 2011). Therefore, TFEB-mediated transcriptional upregulation of v-ATPase and thereby restoration of the acidic environment of the neuronal lysosomes, leading to improved lysosomal proteolysis, could be one of the plausible mechanisms underlying aspirin-mediated reduction in intraneuronal Aβ. Interestingly, aspirin has been reported to inhibit the activity of glycogen synthase kinase-3β (GSK-3β) (Ou et al., 2010), a serine/threonine kinase that has been widely regarded to be involved in numerous aspects of AD development and progression, including senile plaque pathology (Balaraman et al., 2006; Llorens-Martín et al., 2014). A GSK-3β inhibitor has been shown to restore acidification under PS1/2-deficient conditions (Avrahami et al., 2013). Inhibition of GSK-3β reacidified lysosomes, reduced amyloid deposition, and ameliorated cognitive impairment in the 5XFAD mouse (Avrahami and Eldar-Finkelman, 2013; Avrahami et al., 2013). Mounting evidence has highlighted the role of GSK-3β inhibition in improving several AD-related phenotypes in different models of AD (Koh et al., 2008; Serenó et al., 2009; Parr et al., 2012; Ly et al., 2013). Although we did not explore the effect of aspirin on GSK-3β inhibition in our study, it could be a relevant mechanism through which aspirin restores the neuronal acidification. Therefore, a combinatorial effect of TFEB-mediated enhancement of lysosome number and improved lysosomal acidification might facilitate the functionality and proteolysis of neuronal lysosomes and possibly underlie the attenuation of intraneuronal Aβ and improved neuronal survival by aspirin.
Neuroprotective effects of aspirin have been explored in multiple animal models of neurodegenerative disorders including AD. A recent study has reported memory enhancing effects of aspirin in an AlCl3-induced mouse model of neurotoxicity (Rizwan et al., 2016). AD is known to have a prominent inflammatory component that contributes to and aggravates the AD pathology (Heppner et al., 2015; Zhang and Jiang, 2015). Aspirin is also well established for its anti-inflammatory properties and as an inhibitor of NF-κB (Vane, 1971; Kopp and Ghosh, 1994). Aspirin-triggered lipoxin A4 (5S,6R,15Rtrihydroxy-7,9,13-trans-11-cis-eicosatetraenoic acid), an anti-inflammatory molecule generated after acetylation of cyclooxygenase-2, has been reported to reduce NF-κB activation and stimulate alternative activation of microglia leading to enhanced phagocytosis facilitating Aβ clearance and an improvement in cognitive functions in Tg2576 mice (Medeiros et al., 2013). Although we did not address the issue in our study, it is likely that aspirin may also regulate inflammation in 5XFAD mice.
Epidemiological studies have pointed toward a beneficial role of aspirin in AD. Studies by Nilsson et al. (2003) revealed that high-dose aspirin users exhibit lower prevalence of AD and better cognition. A systematic study by Wang et al. (2015) suggests that aspirin can significantly lower the risk of AD. A protective effect of aspirin in AD patients was reported by Etminan et al. (2003); however, it did not reach significance. Conversely, some studies have reported no effect of aspirin on the risk of cognitive decline in older individuals (Kelley et al., 2015; Veronese et al., 2017). One of the possible explanations why aspirin may not show much effect in AD patients or older individuals could be that the level of PPARα might be less in the brains of older AD and dementia patients. In our study, we observed that aspirin attenuates amyloid plaque pathology in AD mouse model in a PPARα-dependent manner. Therefore, in the absence of a basal level of PPARα, aspirin might not exhibit an optimal effect. Although several studies have indicated a beneficial role of aspirin in AD or dementia patients, the results are not conclusive and certainly need further exploration. Our study provides evidence that low-dose aspirin ameliorates amyloid pathology in a mouse model of AD via PPARα.
In summary, our study demonstrates that aspirin can enhance lysosomal biogenesis via PPARα-mediated upregulation of TFEB and may have therapeutic potential for LSDs. At present, there is no therapy available to prevent or halt the progression of AD. Pharmacological compounds targeting TFEB as a therapeutic strategy in AD are still understudied. Therefore, stimulation of lysosomal biogenesis and reduction of amyloid plaque pathology by low-dose aspirin holds promising therapeutic potential for the treatment of AD.
Footnotes
This work was supported by the Department of Veteran Affairs (Merit Award I01BX002174), the Alzheimer's Association (Zenith Fellows Award ZEN-17-438829), and the National Institutes of Health (Grant AG050431).
The authors declare no competing financial interests.
References
- Aubin N, Curet O, Deffois A, Carter C (1998) Aspirin and salicylate protect against MPTP-induced dopamine depletion in mice. J Neurochem 71:1635–1642. [DOI] [PubMed] [Google Scholar]
- Avrahami L, Eldar-Finkelman H (2013) GSK-3 and lysosomes meet in Alzheimer's disease. Commun Integr Biol 6:e25179. 10.4161/cib.25179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avrahami L, Farfara D, Shaham-Kol M, Vassar R, Frenkel D, Eldar-Finkelman H (2013) Inhibition of glycogen synthase kinase-3 ameliorates beta-amyloid pathology and restores lysosomal acidification and mammalian target of rapamycin activity in the alzheimer disease mouse model: in vivo and in vitro studies. J Biol Chem 288:1295–1306. 10.1074/jbc.M112.409250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaraman Y, Limaye AR, Levey AI, Srinivasan S (2006) Glycogen synthase kinase 3beta and Alzheimer's disease: pathophysiological and therapeutic significance. Cell Mol Life Sci 63:1226–1235. 10.1007/s00018-005-5597-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basak JM, Verghese PB, Yoon H, Kim J, Holtzman DM (2012) Low-density lipoprotein receptor represents an apolipoprotein E-independent pathway of abeta uptake and degradation by astrocytes. J Biol Chem 287:13959–13971. 10.1074/jbc.M111.288746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk M, Dean O, Drexhage H, McNeil JJ, Moylan S, O'Neil A, Davey CG, Sanna L, Maes M (2013) Aspirin: a review of its neurobiological properties and therapeutic potential for mental illness. BMC Med 11:74. 10.1186/1741-7015-11-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahmachari S, Fung YK, Pahan K (2006) Induction of glial fibrillary acidic protein expression in astrocytes by nitric oxide. J Neurosci 26:4930–4939. 10.1523/JNEUROSCI.5480-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahmachari S, Jana A, Pahan K (2009) Sodium benzoate, a metabolite of cinnamon and a food additive, reduces microglial and astroglial inflammatory responses. J Immunol 183:5917–5927. 10.4049/jimmunol.0803336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra G, Kundu M, Rangasamy SB, Dasarathy S, Ghosh S, Watson R, Pahan K (2018) Increase in mitochondrial biogenesis in neuronal cells by RNS60, a Physically-modified saline, via phosphatidylinositol 3-kinase-mediated upregulation of PGC1alpha. J Neuroimmune Pharmacol 13:143–162. 10.1007/s11481-017-9771-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffey EE, Beckel JM, Laties AM, Mitchell CH (2014) Lysosomal alkalization and dysfunction in human fibroblasts with the Alzheimer's disease-linked presenilin 1 A246E mutation can be reversed with cAMP. Neuroscience 263:111–124. 10.1016/j.neuroscience.2014.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett GT, Gonzalez FJ, Pahan K (2015) Activation of peroxisome proliferator-activated receptor alpha stimulates ADAM10-mediated proteolysis of APP. Proc Natl Acad Sci U S A 112:8445–8450. 10.1073/pnas.1504890112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett GT, Roy A, Pahan K (2012) Gemfibrozil, a lipid-lowering drug, upregulates IL-1 receptor antagonist in mouse cortical neurons: implications for neuronal self-defense. J Immunol 189:1002–1013. 10.4049/jimmunol.1102624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Y, Ge J (2012) Clinical use of aspirin in treatment and prevention of cardiovascular disease. Thrombosis 2012:245037. 10.1155/2012/245037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decressac M, Mattsson B, Weikop P, Lundblad M, Jakobsson J, Bjorklund A (2013) TFEB-mediated autophagy rescues midbrain dopamine neurons from alpha-synuclein toxicity. Proc Natl Acad Sci U S A 110:E1817–E1826. 10.1073/pnas.1305623110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etminan M, Gill S, Samii A (2003) Effect of non-steroidal anti-inflammatory drugs on risk of Alzheimer's disease: systematic review and meta-analysis of observational studies. BMJ 327:128. 10.1136/bmj.327.7407.128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A, Corbett GT, Gonzalez FJ, Pahan K (2012) Gemfibrozil and fenofibrate, FDA-approved lipid-lowering drugs, upregulate tripeptidyl-peptidase 1 in brain cells via peroxisome proliferator-activated receptor-a: implications for late infantile neuronal ceroid lipofuscinosis therapy. J Biol Chem 287:38922–38935. 10.1074/jbc.m112.365148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A, Jana M, Modi K, Gonzalez FJ, Sims KB, Berry-Kravis E, Pahan K (2015) Activation of peroxisome proliferator-activated receptor alpha induces lysosomal biogenesis in brain cells: implications for lysosomal storage disorders. J Biol Chem 290:10309–10324. 10.1074/jbc.M114.610659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A, Pahan K (2012) Gemfibrozil, a lipid-lowering drug, induces suppressor of cytokine signaling 3 in glial cells: implications for neurodegenerative disorders. J Biol Chem 287:27189–27203. 10.1074/jbc.M112.346932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A, Roy A, Liu X, Kordower JH, Mufson EJ, Hartley DM, Ghosh S, Mosley RL, Gendelman HE, Pahan K (2007) Selective inhibition of NF-kappaB activation prevents dopaminergic neuronal loss in a mouse model of Parkinson's disease. Proc Natl Acad Sci U S A 104:18754–18759. 10.1073/pnas.0704908104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedert M, Spillantini MG (2006) A century of Alzheimer's disease. Science 314:777–781. 10.1126/science.1132814 [DOI] [PubMed] [Google Scholar]
- Green GA. (2001) Understanding NSAIDs: from aspirin to COX-2. Clin Cornerstone 3:50–60. 10.1016/S1098-3597(01)90069-9 [DOI] [PubMed] [Google Scholar]
- Heppner FL, Ransohoff RM, Becher B (2015) Immune attack: the role of inflammation in Alzheimer disease. Nat Rev Neurosci 16:358–372. 10.1038/nrn3880 [DOI] [PubMed] [Google Scholar]
- Huang Y, Mucke L (2012) Alzheimer mechanisms and therapeutic strategies. Cell 148:1204–1222. 10.1016/j.cell.2012.02.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Zhou X, Nicholson AC, Gotto AM Jr, Hajjar DP, Han J (2008) Activation of peroxisome proliferator-activated receptor-alpha in mice induces expression of the hepatic low-density lipoprotein receptor. Br J Pharmacol 155:596–605. 10.1038/bjp.2008.331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jana M, Pahan K (2012) Gemfibrozil, a lipid lowering drug, inhibits the activation of primary human microglia via peroxisome proliferator-activated receptor beta. Neurochem Res 37:1718–1729. 10.1007/s11064-012-0781-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jana M, Jana A, Liu X, Ghosh S, Pahan K (2007) Involvement of phosphatidylinositol 3-kinase-mediated up-regulation of I kappa B alpha in anti-inflammatory effect of gemfibrozil in microglia. J Immunol 179:4142–4152. 10.4049/jimmunol.179.6.4142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsouri L, Georgopoulos S (2011) Lack of LDL receptor enhances amyloid deposition and decreases glial response in an Alzheimer's disease mouse model. PLoS One 6:e21880. 10.1371/journal.pone.0021880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley BJ, McClure LA, Unverzagt FW, Kissela B, Kleindorfer D, Howard G, Wadley VG (2015) Regular aspirin use does not reduce risk of cognitive decline. J Am Geriatr Soc 63:390–392. 10.1111/jgs.13271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern S, Skoog I, Ostling S, Kern J, Borjesson-Hanson A (2012) Does low-dose acetylsalicylic acid prevent cognitive decline in women with high cardiovascular risk? A 5-year follow-up of a non-demented population-based cohort of Swedish elderly women. BMJ Open 2: pii: e001288. 10.1136/bmjopen-2012-001288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khasnavis S, Pahan K (2012) Sodium benzoate, a metabolite of cinnamon and a food additive, upregulates neuroprotective parkinson disease protein DJ-1 in astrocytes and neurons. J Neuroimmune Pharmacol 7:424–435. 10.1007/s11481-011-9286-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khasnavis S, Jana A, Roy A, Mazumder M, Bhushan B, Wood T, Ghosh S, Watson R, Pahan K (2012) Suppression of nuclear factor-kappaB activation and inflammation in microglia by physically modified saline. J Biol Chem 287:29529–29542. 10.1074/jbc.M111.338012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khasnavis S, Roy A, Ghosh S, Watson R, Pahan K (2014) Protection of dopaminergic neurons in a mouse model of Parkinson's disease by a physically-modified saline containing charge-stabilized nanobubbles. J Neuroimmune Pharmacol 9:218–232. 10.1007/s11481-013-9503-3 [DOI] [PubMed] [Google Scholar]
- Koh SH, Noh MY, Kim SH (2008) Amyloid-beta-induced neurotoxicity is reduced by inhibition of glycogen synthase kinase-3. Brain Res 1188:254–262. 10.1016/j.brainres.2007.10.064 [DOI] [PubMed] [Google Scholar]
- Kopp E, Ghosh S (1994) Inhibition of NF-kappa B by sodium salicylate and aspirin. Science 265:956–959. 10.1126/science.8052854 [DOI] [PubMed] [Google Scholar]
- Lee JH, Yu WH, Kumar A, Lee S, Mohan PS, Peterhoff CM, Wolfe DM, Martinez-Vicente M, Massey AC, Sovak G, Uchiyama Y, Westaway D, Cuervo AM, Nixon RA (2010) Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by alzheimer-related PS1 mutations. Cell 141:1146–1158. 10.1016/j.cell.2010.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, McBrayer MK, Wolfe DM, Haslett LJ, Kumar A, Sato Y, Lie PP, Mohan P, Coffey EE, Kompella U, Mitchell CH, Lloyd-Evans E, Nixon RA (2015) Presenilin 1 maintains lysosomal Ca(2+) homeostasis via TRPML1 by regulating vATPase-mediated lysosome acidification. Cell Rep 12:1430–1444. 10.1016/j.celrep.2015.07.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorens-Martín M, Jurado J, Hernández F, Avila J (2014) GSK-3beta, a pivotal kinase in Alzheimer disease. Front Mol Neurosci 7:46. 10.3389/fnmol.2014.00046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly PT, Wu Y, Zou H, Wang R, Zhou W, Kinoshita A, Zhang M, Yang Y, Cai F, Woodgett J, Song W (2013) Inhibition of GSK3beta-mediated BACE1 expression reduces alzheimer-associated phenotypes. J Clin Invest 123:224–235. 10.1172/JCI64516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini-Stoica H, Xu Y, Ballabio A, Zheng H (2016) The autophagy-lysosomal pathway in neurodegeneration: a TFEB perspective. Trends Neurosci 39:221–234. 10.1016/j.tins.2016.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mawuenyega KG, Sigurdson W, Ovod V, Munsell L, Kasten T, Morris JC, Yarasheski KE, Bateman RJ (2010) Decreased clearance of CNS beta-amyloid in Alzheimer's disease. Science 330:1774. 10.1126/science.1197623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros R, Kitazawa M, Passos GF, Baglietto-Vargas D, Cheng D, Cribbs DH, LaFerla FM (2013) Aspirin-triggered lipoxin A4 stimulates alternative activation of microglia and reduces Alzheimer disease-like pathology in mice. Am J Pathol 182:1780–1789. 10.1016/j.ajpath.2013.01.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyad MA. (2001) An introduction to aspirin, NSAids, and COX-2 inhibitors for the primary prevention of cardiovascular events and cancer and their potential preventive role in bladder carcinogenesis: part II. Semin Urol Oncol 19:306–316. [PubMed] [Google Scholar]
- Nielsen HM, Veerhuis R, Holmqvist B, Janciauskiene S (2009) Binding and uptake of A beta1–42 by primary human astrocytes in vitro. Glia 57:978–988. 10.1002/glia.20822 [DOI] [PubMed] [Google Scholar]
- Nilsson SE, Johansson B, Takkinen S, Berg S, Zarit S, McClearn G, Melander A (2003) Does aspirin protect against Alzheimer's dementia? A study in a swedish population-based sample aged > or =80 years. Eur J Clin Pharmacol 59:313–319. 10.1007/s00228-003-0618-y [DOI] [PubMed] [Google Scholar]
- Nixon RA. (2007) Autophagy, amyloidogenesis and alzheimer disease. J Cell Sci 120:4081–4091. 10.1242/jcs.019265 [DOI] [PubMed] [Google Scholar]
- Nixon RA, Wegiel J, Kumar A, Yu WH, Peterhoff C, Cataldo A, Cuervo AM (2005) Extensive involvement of autophagy in alzheimer disease: an immuno-electron microscopy study. J Neuropathol Exp Neurol 64:113–122. 10.1093/jnen/64.2.113 [DOI] [PubMed] [Google Scholar]
- Oakley H, Cole SL, Logan S, Maus E, Shao P, Craft J, Guillozet-Bongaarts A, Ohno M, Disterhoft J, Van Eldik L, Berry R, Vassar R (2006) Intraneuronal beta-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer's disease mutations: potential factors in amyloid plaque formation. J Neurosci 26:10129–10140. 10.1523/JNEUROSCI.1202-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou YQ, Zhu Wb, Li Y, Qiu PX, Huang YJ, Xie J, He SM, Zheng XK, Leng TD, Xu D, Yan GM (2010) Aspirin inhibits proliferation of gemcitabine-resistant human pancreatic cancer cells and augments gemcitabine-induced cytotoxicity. Acta Pharmacol Sin 31:73–80. 10.1038/aps.2009.172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri M, Impey S, Kang H, di Ronza A, Pelz C, Sardiello M, Ballabio A (2011) Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum Mol Genet 20:3852–3866. 10.1093/hmg/ddr306 [DOI] [PubMed] [Google Scholar]
- Parr C, Carzaniga R, Gentleman SM, Van Leuven F, Walter J, Sastre M (2012) Glycogen synthase kinase 3 inhibition promotes lysosomal biogenesis and autophagic degradation of the amyloid-beta precursor protein. Mol Cell Biol 32:4410–4418. 10.1128/MCB.00930-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña-Llopis S, Vega-Rubin-de-Celis S, Schwartz JC, Wolff NC, Tran TA, Zou L, Xie XJ, Corey DR, Brugarolas J (2011) Regulation of TFEB and V-ATPases by mTORC1. EMBO J 30:3242–3258. 10.1038/emboj.2011.257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querfurth HW, LaFerla FM (2010) Alzheimer's disease. N Engl J Med 362:329–344. 10.1056/NEJMra0909142 [DOI] [PubMed] [Google Scholar]
- Rangasamy SB, Corbett GT, Roy A, Modi KK, Bennett DA, Mufson EJ, Ghosh S, Pahan K (2015) Intranasal delivery of NEMO-binding domain peptide prevents memory loss in a mouse model of Alzheimer's disease. J Alzheimers Dis 47:385–402. 10.3233/JAD-150040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rega LR, Polishchuk E, Montefusco S, Napolitano G, Tozzi G, Zhang J, Bellomo F, Taranta A, Pastore A, Polishchuk R, Piemonte F, Medina DL, Catz SD, Ballabio A, Emma F (2016) Activation of the transcription factor EB rescues lysosomal abnormalities in cystinotic kidney cells. Kidney Int 89:862–873. 10.1016/j.kint.2015.12.045 [DOI] [PubMed] [Google Scholar]
- Rizwan S, Idrees A, Ashraf M, Ahmed T (2016) Memory-enhancing effect of aspirin is mediated through opioid system modulation in an AlCl3-induced neurotoxicity mouse model. Exp Ther Med 11:1961–1970. 10.3892/etm.2016.3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothwell PM, Wilson M, Price JF, Belch JF, Meade TW, Mehta Z (2012) Effect of daily aspirin on risk of cancer metastasis: a study of incident cancers during randomised controlled trials. Lancet 379:1591–1601. 10.1016/S0140-6736(12)60209-8 [DOI] [PubMed] [Google Scholar]
- Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS, Banfi S, Parenti G, Cattaneo E, Ballabio A (2009) A gene network regulating lysosomal biogenesis and function. Science 325:473–477. 10.1126/science.1174447 [DOI] [PubMed] [Google Scholar]
- Serenó L, Coma M, Rodríguez M, Sánchez-Ferrer P, Sánchez MB, Gich I, Agulló JM, Pérez M, Avila J, Guardia-Laguarta C, Clarimón J, Lleó A, Gómez-Isla T (2009) A novel GSK-3beta inhibitor reduces Alzheimer's pathology and rescues neuronal loss in vivo. Neurobiol Dis 35:359–367. 10.1016/j.nbd.2009.05.025 [DOI] [PubMed] [Google Scholar]
- Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, Sardiello M, Rubinsztein DC, Ballabio A (2011) TFEB links autophagy to lysosomal biogenesis. Science 332:1429–1433. 10.1126/science.1204592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer LM, Dority MD, Gupta-Bansal R, Frederickson RC, Younkin SG, Brunden KR (1995) Amyloid beta protein (A beta) removal by neuroglial cells in culture. Neurobiol Aging 16:737–745. 10.1016/0197-4580(95)00055-J [DOI] [PubMed] [Google Scholar]
- Song W, Wang F, Savini M, Ake A, di Ronza A, Sardiello M, Segatori L (2013) TFEB regulates lysosomal proteostasis. Hum Mol Genet 22:1994–2009. 10.1093/hmg/ddt052 [DOI] [PubMed] [Google Scholar]
- Spampanato C, Feeney E, Li L, Cardone M, Lim JA, Annunziata F, Zare H, Polishchuk R, Puertollano R, Parenti G, Ballabio A, Raben N (2013) Transcription factor EB (TFEB) is a new therapeutic target for pompe disease. EMBO Mol Med 5:691–706. 10.1002/emmm.201202176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunemi T, Ashe TD, Morrison BE, Soriano KR, Au J, Roque RA, Lazarowski ER, Damian VA, Masliah E, La Spada AR (2012) PGC-1alpha rescues Huntington's disease proteotoxicity by preventing oxidative stress and promoting TFEB function. Sci Transl Med 4:142ra97. 10.1126/scitranslmed.3003799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vane JR. (1971) Inhibition of prostaglandin synthesis as a mechanism of action for aspirin-like drugs. Nat New Biol 231:232–235. 10.1038/newbio231232a0 [DOI] [PubMed] [Google Scholar]
- Vane JR, Botting RM (2003) The mechanism of action of aspirin. Thromb Res 110:255–258. 10.1016/S0049-3848(03)00379-7 [DOI] [PubMed] [Google Scholar]
- Veronese N, Stubbs B, Maggi S, Thompson T, Schofield P, Muller C, Tseng PT, Lin PY, Carvalho AF, Solmi M (2017) Low-dose aspirin use and cognitive function in older age: a systematic review and meta-analysis. J Am Geriatr Soc 65:1763–1768. 10.1111/jgs.14883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Tan L, Wang HF, Tan CC, Meng XF, Wang C, Tang SW, Yu JT (2015) Anti-inflammatory drugs and risk of Alzheimer's disease: an updated systematic review and meta-analysis. J Alzheimers Dis 44:385–396. 10.3233/JAD-141506 [DOI] [PubMed] [Google Scholar]
- Whyte LS, Lau AA, Hemsley KM, Hopwood JJ, Sargeant TJ (2017) Endo-lysosomal and autophagic dysfunction: a driving factor in Alzheimer's disease? J Neurochem 140:703–717. 10.1111/jnc.13935 [DOI] [PubMed] [Google Scholar]
- Wolfe DM, Lee JH, Kumar A, Lee S, Orenstein SJ, Nixon RA (2013) Autophagy failure in Alzheimer's disease and the role of defective lysosomal acidification. Eur J Neurosci 37:1949–1961. 10.1111/ejn.12169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyss-Coray T, Loike JD, Brionne TC, Lu E, Anankov R, Yan F, Silverstein SC, Husemann J (2003) Adult mouse astrocytes degrade amyloid-beta in vitro and in situ. Nat Med 9:453–457. 10.1038/nm838 [DOI] [PubMed] [Google Scholar]
- Xiao Q, Yan P, Ma X, Liu H, Perez R, Zhu A, Gonzales E, Burchett JM, Schuler DR, Cirrito JR, Diwan A, Lee JM (2014) Enhancing astrocytic lysosome biogenesis facilitates abeta clearance and attenuates amyloid plaque pathogenesis. J Neurosci 34:9607–9620. 10.1523/JNEUROSCI.3788-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Q, Yan P, Ma X, Liu H, Perez R, Zhu A, Gonzales E, Tripoli DL, Czerniewski L, Ballabio A, Cirrito JR, Diwan A, Lee JM (2015) Neuronal-targeted TFEB accelerates lysosomal degradation of APP, Reducing Abeta Generation and Amyloid Plaque Pathogenesis. J Neurosci 35:12137–12151. 10.1523/JNEUROSCI.0705-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon SY, Kim DH (2016) Alzheimer's disease genes and autophagy. Brain Res 1649:201–209. 10.1016/j.brainres.2016.03.018 [DOI] [PubMed] [Google Scholar]
- Zhang F, Jiang L (2015) Neuroinflammation in Alzheimer's disease. Neuropsychiatr Dis Treat 11:243–256. 10.2147/ndt.s75546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YD, Zhao JJ (2015) TFEB participates in the abeta-induced pathogenesis of Alzheimer's disease by regulating the autophagy-lysosome pathway. DNA Cell Biol 34:661–668. 10.1089/dna.2014.2738 [DOI] [PubMed] [Google Scholar]