Abstract
Most strategies for life detection rely upon finding features known to be associated with terran life, such as particular classes of molecules. But life may be vastly different on other planets and moons, particularly as we expand our efforts to explore ocean worlds like Europa and Enceladus. We propose a new concept for life detection that harnesses the power of DNA sequencing to yield intricate informatics fingerprints, even for life that is not nucleic acid–based. The concept is based on the fact that folded nucleic acid structures (aptamers) have been shown to be capable of binding a wide variety of compounds, whether inorganic, organic, or polymeric, and irrespective of being from a biotic or abiotic source. Each nucleic acid sequence can be thought of as a code, and a combination of codes as a “fingerprint.” Over multiple analytes, the “fingerprint” of a non-terran sample can be analyzed by chemometric protocols to provide a classifier of molecular patterns and complexity. Ultimately the chemometric fingerprints of living systems, which may differ significantly from nonliving systems, could provide an empirical, agnostic means of detecting life. Because nucleic acids are exponentially amplified by the polymerase chain reaction, even very small input signals could be translated into a robust readable output. The derived sequences could be identified by a small, portable sequencing device or by capture and optical imaging on a DNA microarray. Without presupposing any particular molecular framework, this agnostic approach to life detection could be used from Mars to the far reaches of the Solar System, all within the framework of an instrument drawing little heat and power.
Key Words: : Agnostic biosignatures, Astrobiology, Chemometrics, DNA sequencing, Life detection, Proximity ligation assay
1. Introduction
An entirely new set of missions with astrobiological relevance is now coming into the realm of possibility: the idea of visiting the icy moons of Jupiter and Saturn. These ocean worlds are an order of magnitude farther away than our near neighbor Mars, but the development of larger rockets such as the Space Launch System (SLS), advanced power systems, and solar electric propulsion technologies are paving the way for their exploration (Klaus et al., 2013). Seeking life there will be challenging, and the possibilities for that life are diverse. These distant moons are much more foreign than Mars. For instance, Europa, orbiting Jupiter, and Enceladus, orbiting Saturn, hold their seas beneath icy crusts (Nimmo and Pappalardo, 2016). There are geysers on both worlds, which mysteriously fluctuate, and clear evidence of tidal warming and geologic activity (Kattenhorn and Prockter, 2014; Kite and Rubin, 2016; Lorenz, 2016; Nimmo and Pappalardo, 2016; Wyrick et al., 2016).
As we design instrumentation for future missions to these worlds, we can, should, and will apply traditional biosignature approaches: looking for isotopic signatures, particular classes of molecules, evidence of enantiomeric excess, non-equilibrium distribution of compounds, and patterning within the molecular weights of fatty acids or other lipids. Taking along a nucleic acid sequencing device has also been proposed, which could be utilized to monitor and identify potential contamination brought from Earth, or to look for nucleic acids based on a shared ancestry hypothesis (Carr et al., 2016; Mojarro et al., 2016; Bywaters et al., 2017). Rapid advances in miniaturization have already led to a small stand-alone version of the technology, Oxford Nanopore's MinION, which weighs only 85 g (Loman and Watson, 2015). Sequencing, however, may potentially have a broader and more useful application than targeting shared ancestry—which is arguably less tenable as we proceed deeper into the Solar System—or monitoring for contamination. Sequencing could also be used as a tool for detecting patterns of binding chemistry associated with non-terran biosignatures.
Here, we describe a chemometric approach to life detection that is particularly well suited for detecting agnostic biosignatures in low-biomass samples. First, we describe concepts for sample delivery and describe a method—the proximity ligation assay—that enables binding chemistry to be assayed in solution in liquid samples, such as a plume from Enceladus. Next, we outline possibilities for signal detection as well as statistical approaches to data analysis.
2. Chemometrics for Life Detection: The Concept
Patterns of binding to nucleic acids, independent of their biological function, can be used to probe and report on any chemical environment, opening up a new way to detect biosignatures. This concept builds on the fact that oligonucleotides naturally form secondary and tertiary structures that can have affinity and specificity for a variety of molecules, from biologicals such as peptides and proteins (Sun and Zu, 2015) to a wide variety of small organic molecules (Stoltenburg et al., 2007; Kim and Gu, 2013) to inorganics such as mineral surfaces (Cleaves et al., 2011) and individual metals (Ye et al., 2012). DNA sequences as short as 15 nucleotides in length (but more commonly 30–80 nucleotides in length) can form complex structures that, like antibodies, will bind to analytes, even in a complex mixture (see Fig. 1, which shows oligonucleotide binding to a cell surface protein). The mapping of diverse nucleic acid sequences and shapes to many different analytes forms the basis for why our chemometric analysis will be relevant to a wide variety of environments.
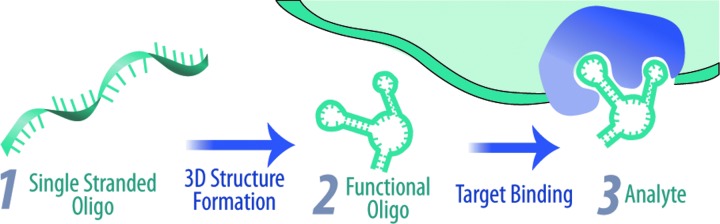
FIG. 1.
Primary, secondary, and tertiary structure contribute to single-stranded DNA sequences that fold into functional aptamers that can bind to a variety of analytes.
Typically, in vitro selection, sometimes known as Systematic Evolution of Ligands by EXponential Enrichment (SELEX), is used to isolate and identify oligonucleotides that bind tightly to a very specific analyte (Keefe et al., 2010). In the early rounds of SELEX, it is frequently found that there are a very large number of nucleic acid sequences that have low affinity and low specificity for a given molecule, and that these sequences are cast aside during subsequent iterative rounds for high-affinity species. These early, less-fit species are precisely the ones we want. The important point for the method proposed herein is that in a random sequence pool there are multiple species that are already capable of interacting with a wide variety of compounds, and this unbiased wealth of nucleic acid receptors can potentially be captured and then directly amplified and sequenced. Instead of iterative rounds of selection, a single-step approach should recover diverse sets of sequences, a rich informatic landscape that can reflect even slight analyte signals or environmental differences and changes once properly analyzed. By accumulating large numbers of binding sequences that reflect different compounds in a mixture, statistical data analyses of oligonucleotide sequences and sequence counts enable patterns associated with increasing levels of complexity to be analyzed using chemometrics.
Chemometrics is a field that uses a wealth of statistical pattern-recognition protocols to analyze chemical data and has been used extensively to find patterns that classify specific chemical identities, as well as mixture consistency and complexity, in applications ranging from protein kinases to explosive materials (Wright et al., 2005; Hughes et al., 2008; Pai and Ellington, 2009; Umali and Anslyn, 2010; Stewart et al., 2011; Zamora-Olivares et al., 2014; Goodwin et al., 2015).
At the outset, we note a common misconception with using chemometric analyses with nucleic acid amplicons: the oligonucleotides in the libraries that are necessary to generate shapes capable of binding are longer than the binding sequences themselves, in much the same way that an enzyme is much longer than its active site residues. Thus, functional binding motifs can be short and well represented at multiple positions even within a much longer library (e.g., a given hexamer will be well represented in a library of a million 35-mers, while every possible 35-mer will not be well represented). That said, the short functional binding motifs are presented to the analyte by the folded structure of the longer oligonucleotide in much the same way that active site residues in an enzyme are presented to a substrate by the remainder of the folded enzyme.
Signatures resulting from the short functional motifs of nucleic acids involved in binding chemistry complement the astrobiology community's search for specific types of molecules of known biological origin (such as complex carotenoids or indigenous nucleic acids), but also enable an extension beyond terrestrial conceptions of what life may look like. This is because nucleic acids will bind all types of chemical structures in complex samples, from simple inorganics or minerals (e.g., Cleaves et al., 2011) to highly complicated biologicals, such as cell surfaces (e.g., Jayasena, 1999) (See Fig. 2). The method's power is that it is entirely agnostic; no prior knowledge of the surface attributes or about the 3D structures of the nucleic acids that do the binding is required. This contrasts with the only other known set of broadly binding receptors—antibodies—which by natural selection have already been heavily skewed toward the recognition of biologicals. In the end, the only necessary pieces of information are the selected nucleic acid sequences (short functional motifs, identified by comparative analyses), sequence counts, and a chemometric training set that differentiates between the various levels of chemical complexity.
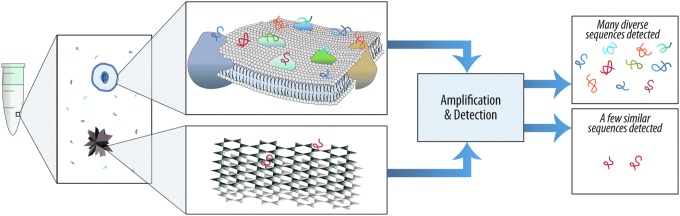
FIG. 2.
Nucleic acid binding can provide information that could reveal unknown life. After mixing a large, randomly generated library of folded oligonucleotides with a sample, many diverse oligonucleotides will bind to a complex surface like a cell membrane, whereas far fewer will bind to an inorganic crystalline substrate. Bound sequences can then be amplified and sequenced, revealing the diversity of binding sites within a sample.
3. Oligonucleotide Ligation and Amplification
This life-detection approach could be utilized on solid substrates or head gas, or samples in a liquid phase, such as a plume from Enceladus. Samples could be prepared for delivery in a variety of ways. For instance, suspended substrates in a liquid sample—particles in the form of mineral fines or other nonbiological source material and, potentially, cells or vesicles—could be size-sorted within a microfluidic device by using physical size-based filtration (Bhagat et al., 2010), hydrodynamic sorting (Woo et al., 2013), or combined separation of particles (Yoon et al., 2016). This would enable the downstream analysis of free-floating small inorganics or organics separately from larger substances, including those within the size range of microbial cells. Comparisons of the nucleic acid patterns between size-sorted compartments could then provide insight into the heterogeneity of samples, as different DNA sequences returned from different compartments would indicate different analytes.
Once samples are prepared, we propose that one of the best means of capturing sequences that associate with a given analyte or set of analytes is via a method called the proximity ligation assay (or PLA) (Fredriksson et al., 2002; Gustafsdottir et al., 2005). In PLA, two sequences that have some affinity for a given analyte are brought into proximity by binding and are then ligated. This ligation event produces an amplicon that can then be amplified and sequenced. In short, the binding sequences are “purified” not by sieving but by the fact that they create a small amount of an amplicon, which would otherwise not exist and not be capable of exponential amplification via the PCR; another way to look at this is that ligation is the only way that the PCR can achieve end-to-end amplification between two primer-binding sites. Because ligation in the absence of proximity is an extremely rare event, PLA has previously been shown to have the capability to identify analytes with almost single-molecule resolution (Söderberg et al., 2008; Ke et al., 2013), and thus also represents a means of garnering precise profiles of binding sequences. Importantly, because there are no cycles of selection (sieving), high-affinity aptamers are not the final product of the technique; rather, we capture the lower-affinity and lower-specificity sequences that are typically lost from SELEX, but which still likely contain a high information content regarding analytes and the environment in which the analytes are found.
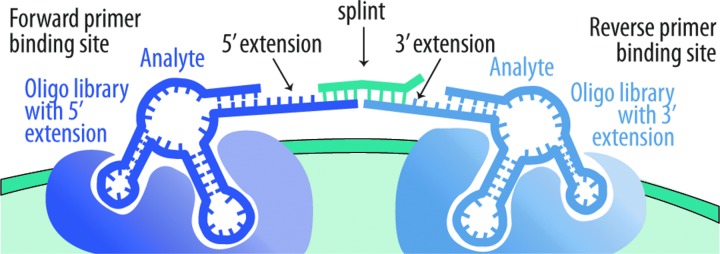
During PLA, two different DNA libraries—one library with a constant region at its 5' end and the other library with a constant region at its 3' end—are allowed to interact with a material or liquid. When two library members bind in proximity to one another on a material, or on an analyte in a liquid, a DNA ligase can covalently join them, creating a sequence that can be amplified by forward and reverse primers that bind to the 5' and 3' constant regions (see Fig. 3). In the absence of proximity, ligation between these two oligonucleotides in solution occurs at a vanishingly small rate, and thus only the bound, ligated species will be amplified, while any unbound and thus un-ligated species will not amplify. In essence, PLA is purifying binding species (and their associated functional submotifs) on the basis of ligation and amplification, rather than through sieving, which would be more cumbersome and would carry through nonbinding molecules at a higher rate, leading to a higher background that would confound downstream analyses.
FIG. 3.
For each library member, only one primer-binding site is present at the 35-mer terminus. For amplification to occur, two 35-mers must be ligated. Ligation can be carried out in the presence of a short binding DNA splint and a ligase, such as T4 DNA ligase, that utilizes a template to increase the efficiency of ligation (the ligation can also be carried out without a splint, using ligases that do not require templates).
One principal advantage of PLA is that the probability of a quaternary association (i.e., an analyte, two library members, and ligase) is exquisitely low, and thus only “real” analyte-dependent interactions should be cemented into the amplified and sequenced DNA; in practice, PLA shows almost no background, allowing for zeptomole detection of biomedically relevant analytes (Fredriksson et al., 2002). This further means that not only high-affinity binding species created from the juxtaposed, folded 35-mers can be captured, but also low-affinity or transient associations that could be cemented in place by the ligase. A single PLA reaction could therefore capture a range of sequences and structures with diverse binding properties, capable of targeting virtually any molecule, whether inorganic, organic, or polymeric. And, again, within each of the libraries that constitute the two halves of any given amplicon there will be many, much shorter, functional motifs available for binding and eventually for sequence analysis.
Each library could be ca. 35 random nucleotides in length, ultimately creating a 70-mer random sequence, flanked by two ca. 20-mer primer-binding sites, for an overall length of ca. 115 nucleotides (such oligonucleotides are synthetically accessible and readily amplified by PCR). The details of both libraries and PLA reactions that might prove most suitable for non-terran biosignature capture could be worked out in advance by using terran materials ranging from rocks to analytes in ionic liquids. Variables could include the use of a so-called “splint” that would improve the efficiencies of proximity ligation (see Fig. 3), or the elimination of an enzymatic ligase in favor of chemical ligation.
Following amplification of ligated species, direct DNA sequencing or hybridization of the entire mixture would then generate a “fingerprint.” The advantage of this approach is that no purification of the mixture would be required other than the amplification itself in the low-background PLA regime. The entire process would involve introduction of the library, ligation, amplification, and then analysis of the amplified species against a backdrop of many fewer non-ligated, non-amplified species.
4. Signal Detection
There are multiple possibilities for signal detection, including several commercial-off-the-shelf detection technologies. Examples include handheld nanopore sequencers as well as DNA microarrays, where, upon hybridization, data in the form of a pattern of fluorescence could be simply and quickly optically imaged.
The former is of particular relevance to Mars, where data transfer limitations are less stringent and detailed sequence information might also be utilized for testing a shared ancestry hypothesis (Mileikowsky et al., 2000; Kirschvink and Weiss, 2002). When coupled to a phylogenetic analysis, a sequencing capability could readily distinguish between modern microbial contaminants and novel, deeply branching DNA-based life-forms that could be similar to, but long isolated from, terrestrial life in terms of evolutionary distance.
One possible instrument choice is Oxford Nanopore Technologies' MinION Mk 1B handheld sequencer, which is extremely small (10.5 × 3.3 × 2.3 cm), lightweight, (85 g) and draws little power (<1 W). As nucleic acids pass through a small nanopore, characteristic changes in ionic current are used for base calling and directly sequencing the DNA; extremely long continuous read lengths (>100 kb) are possible (Mikheyev and Tin, 2014; Loman and Watson, 2015).
Another possibility for detection is a DNA microarray, which may better accommodate the technical limitations of an ocean worlds mission. Ligated amplicons would create large numbers of double-stranded DNAs, representative of the analytes in the mixture. These DNAs, once denatured, could serve as single-stranded inputs to a “molecular beacon array” (Tyagi and Kramer, 1996), hybridizing to potential complements. For instance, roughly a million hybridization sites could be patterned on a microarray that is the size of a microscope slide, each associated with one of the full set of every possible 410 10-mers. Perfect complementarity would not be necessary to activate a molecular beacon; that is, even though the library and the molecular beacon sequences would be somewhat random, there would be enough overlap to ensure activation of many of the positions in the DNA array, thereby transducing the analytes to sequences, and sequences to optical signals.
An electrochemical approach could also be utilized for the molecular beacons, such as short hairpins that terminate in an electrochemical reporter like methylene blue or ferrocene (Radi et al., 2006). If an amplicon were to bind to the loop portion of the molecular beacon, then the electrochemical reporter would move away from the surface of the chip, resulting in an electrochemical signal that could be read. Highly parallel electrochemical arrays could be multiplexed with a very simple electrical readout scheme, furthering the possibilities for autonomous data analysis.
5. Statistical Data Analysis
There are many statistical approaches that could be employed for chemical pattern recognition, as nucleic acid strands carry information inherent in their sequence. A sequence is analogous to a string of numbers, and sequence count is analogous to the number of times a particular string of numbers occurs. Thus, a predefined mathematical relationship for each DNA strand can be used as the input data for chemometric analysis. Examples include simple number count for all the sequences, or fold-change for the sequences (i.e., comparison between the biased and naïve libraries), or changes in repetitive patterns of smaller DNA sequences, such as pentamers, hexamers, and so on, across the entire nucleic acid pool. Irrespective of the manner in which the data are entered into the statistical protocol, the key is that each nucleic acid sequence is a code, and a combination of codes from a biased pool of sequences is a “fingerprint.”
Variations in nucleic acid strands, representing differences in chemical complexity of the target samples, can be analyzed to generate high-dimensionality chemometric score plots, similar to the example shown in Fig. 4. Principal component analysis (PCA), for instance, is a statistical method that merges data with many variables into a smaller number of axes that combine to concisely and graphically represent the overall trends and associations. Briefly, this method generates sample scores based on a simultaneous ordination of both rows and columns of a data matrix (McCune and Grace, 2002). PCA assigns correlation coefficients for all available data—a measure of the variance within the data set—and quantifies the amount to which any particular trait is forcing a trend. This is an unsupervised protocol, meaning that there is no prior knowledge of “like samples” that bias the mathematical analysis. In the context of discovering biosignatures, the goal would be to find patterns from the amplicons that are unlike anything that is recognized as possessing no life signs.
FIG. 4.
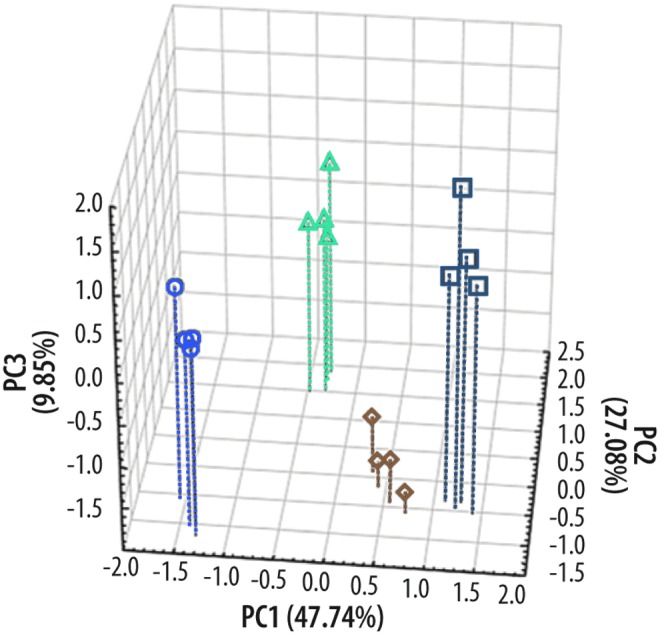
Initial data demonstrate the power of chemometrics. A small collection of aptamers and four human cell lines suggest unique patterns of response to the sequenced aptamer panel, with some aptamers only binding to a particular cell line and with many aptamers binding to multiple cell lines in characteristic amounts. This PCA score plot shows that four cell lines can be classified based solely on the variance in the fold change of the aptamer pool. PCA reveals, with no prior bias, that the pool of nucleic acids could pattern differences between very similar cell surface attributes, yet reproducibly reveal differences in repetitions of the cell samples. While the focus of these results is on cells (useful from the point of view of identifying cell-like objects), they are equally applicable to smaller analytes as well. Adapted from the work of Goodwin et al. (2015).
6. Alternative Statistical Methods
Another method, linear discriminant analysis, or LDA, is a supervised system that uses information in a learning set of labeled observations to construct a classification rule that will separate predefined classes to the maximum extent possible (Izenman, 2013). The resulting score plots can be considered a training space, where unknown samples can be classified as belonging, or not belonging, to one of the training sample sets. While the basis of biosignatures is still in question, it is expected that a certain level of chemical complexity would be one good sign of life (Cronin and Walker, 2016). LDA should always be accompanied by PCA, because the biased mathematics of LDA is so powerful that spurious differences between samples, such as subtle but systematic differences in experimental techniques (i.e., pipetting, plates, lamp intensities, etc.), can be derived as patterns, even when there may be no chemical differences between the samples (Worley and Powers, 2016).
Both PCA and LDA generate loading plots containing what are referred to as loading vectors. In our proposal, each loading vector will be associated with a short oligonucleotide sequence. When overlaid with the score plots (known as a biplot), the loading vectors will identify which oligonucleotides contributed to the classification between samples, and the relative extent that each led to that classification. This is essential to the outcome, because as discussed herein the level of complexity in oligonucleotide sequences is expected to reflect the chemical complexity of the samples being interrogated.
Support vector machines, or SVM, is a significantly more advanced mathematical method. Unlike LDA, nonlinear correlations between variance in the data are found in the form of kernel functions—which compute the similarity of inputs—that are then transformed into linear representations (Steinwart and Christmann, 2008). This technique is often used in chemistry to find patterns in data that correlate with the concentrations of specific components in a background mixture of several chemicals of unknown identity and varying composition (Ivanciuc, 2007). It might be expected that the nucleic acids that interact with extraterrestrial samples would give patterns not predicted by any training sets of terran life. Thus, the complexity of the patterns would be unpredictable and not necessarily linear, thereby increasing the utility of an SVM approach.
Finally, artificial neural networks, or ANNs, could also be utilized. ANNs act to process data by mimicking animal neurons (Abraham, 2005). A series of mathematical functions (linear, polynomial, exponential, etc.) tie the original data set to “hidden layers” with different weights, whose results are then transformed again via other functions and weights to an outcome. As outcomes, we might imagine (1) no life signs, (2) minimal biosignatures, or (3) strong biosignatures. Because ANNs have been found to model almost any data set, they are prone to overtraining when small data sets are used for the number of connections employed in the training, validation, and test sets, meaning that while a model is derived that fits the data, it can have little to no predictive power to analyze unknowns. Hence, an ANN would always be the protocol of last resort and require significant testing to gain confidence in its validity.
7. Chemometric Training and Outlier Identification
As alluded to above, when using any of the chemometric protocols, the crux of discovering biosignatures is to identify a particular level of chemical complexity that gives confidence that life exists or has existed. Chemical complexity and anomalous patterns are our guiding principles for discovery, based upon the ideas put forth by Conrad and Nealson (2001), Scharf et al. (2015), Schulze-Makuch and Irwin (2008), and others. To determine a baseline level for chemical complexity or anomalous patterns, a reference state is required, which we can define as materials that are generally accepted to have no life-like characteristics, such as mineral surfaces and abiotic organics sourced from meteorites.
Because life is so ubiquitous on Earth, most naturally occurring solid substrates do not suffice. Given widespread microbial overprinting, it is challenging to find contexts as a reference that can be considered as no sign of life. Yet solid substrates will be found on any planet created by natural planetary events independent of life, and such substrates on Earth can be sterilized or synthesized in the lab. Similarly, prebiotic organic molecules are associated with comets and asteroids and prevalent throughout the Solar System (Anders, 1989). Abiotic organics extracted from meteorites (Cronin and Chang, 1993), as well as organics that are abiotically synthesized under hydrothermal conditions (McCollom and Seewald, 2007), could also provide starting reference points for comparison.
Chemometric scores could then be contrasted to materials where life is evident on Earth. For example, inorganic crystalline materials are of consistent and relatively uniform chemical composition, and thus the oligonucleotides that bind to these materials will have relatively consistent and repetitive sequences, giving a preponderance of the same, similar, or repetitive patterns. Hence, a loading plot will reveal that similar sequences are found to classify such materials. However, due to the chemical complexity of life as we understand it on Earth, one would expect a far larger variety of sequences to bind complex biotic surfaces. This would result in score plots whose loading vectors are consistent with a greater variety of the oligonucleotide amplicons (differing sequences and differing repeat units). Additional work is needed to determine when such a threshold is large enough to be confidently interpreted as a biosignature. This will only be determined with experience, time, and experimentation.
8. Implications
Enceladus has been added to the current list of New Frontier Program targets, and it is likely that a competed or directed Enceladus mission will be selected in the coming years. Formulation of a Europa surface lander mission to complement the Europa flyby (Clipper) mission is presently in progress, although exact dates for implementation of such a flagship mission remain uncertain. Likewise, tantalizing signatures of transient atmospheric methane on Mars may motivate robotic mission architects to target special regions on that planet that are off limits to astronauts with the intent to melt ice and search for extant life. It is thus imperative to broaden our techniques for life detection to include those that are based on recent dramatic advances in sequencing and to adapt these to the challenges of deep space exploration.
The approach described herein would enable the scientific community to distinguish samples with chemistries suggestive of biology with great sensitivity, that is, to “read” patterns of molecules arising, for example, from the vast amount of information stored on the surface of a primitive microbial cell. It is possible that the amount of biomass produced lithoautotrophically on ocean worlds like Europa and Enceladus may be extremely small compared to the biomass produced photosynthetically on Earth (McCollom, 1999). If a plume flyby were to only collect the equivalent of a few tens to a few hundreds of cells, it would require exquisite sensitivity within the suite of life-detection instruments. Although the analysis would likely be destructive, it is possible that only a tiny amount of sample would be required. By utilizing the power of PCR, this technique could be capable of amplifying the signal associated with an exceedingly small input.
The approach also provides a built-in way to monitor potential sources of contamination from Earth organisms, a key challenge for any life-detection approach. In the run-up to a mission launch, PLA could be carried out with fluids used to wash clean room surfaces, which would provide data about characteristic patterns that are associated with common clean room contaminants, such as Bacillus pumilus (Kempf et al., 2005), Bacillus atrophaeus (Probst et al., 2011), Acinetobacter johnsonii (Ghosh et al., 2010), or Tersicoccus phoenicis (Vaishampayan et al., 2013).
Proximity ligation assay is an exquisitely sensitive technique, capable of identifying even single molecules (Söderberg et al., 2008; Ke et al., 2013), and will likely detect even the very low bioburden levels required by planetary protection—2.78 × 105 bacterial endospores in the case of the Mars Science Laboratory (Benardini et al., 2014). Amplifiable oligonucleotides could even be painted or printed on spacecraft components to monitor for cross contamination, for instance from the avionics to the sample collection device. During or shortly after a mission, oligonucleotide libraries could be exposed to synthesized laboratory mixtures of new or unusual minerals and organics detected by other instruments aboard the spacecraft, testing to discern whether patterns match such abiotic materials.
9. Conclusion
The plans to explore ocean worlds in our own solar system motivate the development of techniques to search for evidence of biosignatures that are agnostic with respect to the characteristics of terrestrial biochemistry. On other planetary bodies, particularly those isolated from exchange with terrestrial material, life may not have originated along the same pathway as it did on Earth. Techniques such as DNA and RNA sequencing that precisely detect terrestrial biology, while highly definitive, may not be the optimal approach for life-detection missions designed to collect material from plumes of Enceladus or Europa, or from the cold icy surfaces of these moons. Yet with chemometrics, a sequencing approach to life detection could enable the science community not only to look for biosignatures associated with life similar to our own (i.e., nucleic acid–based life, based on DNA, RNA, and/or nonstandard bases), but also to recognize life that is vastly different from that of Earth by probing for indications of complex biochemistry that may be unimaginable within the confines of our current thinking.
Without presupposing any particular molecular framework, this chemometric life-detection approach could be used from Mars to the far reaches of the Solar System, all within the framework of a miniaturized chip that draws little heat and power. It would facilitate truly agnostic biosignature detection and enable us to fingerprint intricate patterns of binding chemistry.
Abbreviations Used
- ANNs
artificial neural networks
- LDA
linear discriminant analysis
- PCA
principal component analysis
- PLA
proximity ligation assay
- SELEX
Systematic Evolution of Ligands by EXponential Enrichment
- SVM
support vector machines
Acknowledgments
We would like to thank the NASA Agnostic Biosignatures Workshop, held in Washington, DC, in late 2016, for helping to spark this idea, as well as Brittany Klein for generating the graphics for the figures in this manuscript. E.V.A. thanks the Welch Regents Chair (F-0045) for continual financial support of his differential sensing projects. Similarly, A.D.E. thanks the Fraser Chair for continual financial support of nucleic acid chemistry research.
Author Disclosure Statement
No competing financial interests exist.
References
- Abraham A. (2005) Artificial neural networks. In Handbook of Measuring System Design, Part 8. Elements: B—Signal Conditioning, Wiley, Chichester, UK, doi: 10.1002/0471497398.mm421 [DOI] [Google Scholar]
- Anders E. (1989) Pre-biotic organic matter from comets and asteroids. Nature 342:255–257 [DOI] [PubMed] [Google Scholar]
- Benardini J.N., III, La Duc M.T., Beaudet R.A., and Koukol R. (2014) Implementing planetary protection measures on the Mars Science Laboratory. Astrobiology 14:27–32 [DOI] [PubMed] [Google Scholar]
- Bhagat A.A.S., Bow H., Hou H.W., Tan S.J., Han J., and Lim C.T. (2010) Microfluidics for cell separation. Med Biol Eng Comput 48:999–1014 [DOI] [PubMed] [Google Scholar]
- Bywaters K.B., Schmidt H., Vercoutere W., Deamer D., Hawkins A.R., Quinn R.C., Burton A.S., and McKay C.P. (2017) Development of solid-state nanopore technology for life detection [abstract 3437]. In Astrbiology Science Conference Abstracts, Lunar and Planetary Institute, Houston [Google Scholar]
- Carr C.E., Mojarro A., Tani J., Bhattaru S.A., Zuber M.T., Doebler R., Brown M., Herrington K., Talbot R., and Fuller CW. (2016) Advancing the search for extra-terrestrial genomes. In 2016 IEEE Aerospace Conference, IEEE, Piscataway, NJ, doi: 10.1109/AERO.2016.7500859 [DOI] [Google Scholar]
- Cleaves H.J., Crapster-Pregont E., Jonsson C.M., Jonsson C.L., Sverjensky D.A., and Hazen R.A. (2011) The adsorption of short single-stranded DNA oligomers to mineral surfaces. Chemosphere 83:1560–1567 [DOI] [PubMed] [Google Scholar]
- Conrad P.G. and Nealson K.H. (2001) A non-Earthcentric approach to life detection. Astrobiology 1:15–24 [DOI] [PubMed] [Google Scholar]
- Cronin J.R. and Chang S. (1993) Organic matter in meteorites: molecular and isotopic analyses of the Murchison meteorite. In The Chemistry of Life's Origins, Springer, Dordrecht, the Netherlands, pp 209–258 [Google Scholar]
- Cronin L. and Walker S.I. (2016) Beyond prebiotic chemistry. Science 352:1174–1175 [DOI] [PubMed] [Google Scholar]
- Fredriksson S., Gullberg M., Jarvius J., Olsson C., Pietras K., Gústafsdóttir S.M., Östman A., and Landegren U. (2002) Protein detection using proximity-dependent DNA ligation assays. Nat Biotechnol 20:473–477 [DOI] [PubMed] [Google Scholar]
- Ghosh S., Osman S., Vaishampayan P., and Venkateswaran K. (2010) Recurrent isolation of extremotolerant bacteria from the clean room where Phoenix spacecraft components were assembled. Astrobiology 10:325–335 [DOI] [PubMed] [Google Scholar]
- Goodwin S., Gade A.M., Byrom M., Herrera B., Spears C., Anslyn E.V., and Ellington A.D. (2015) Next-generation sequencing as input for chemometrics in differential sensing routines. Angew Chem Int Ed Engl 127:6437–6440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsdottir S.M., Schallmeiner E., Fredriksson S., Gullberg M., Soderberg O., Jarvius M., Jarvius J., Howell M., and Landegren U. (2005) Proximity ligation assays for sensitive and specific protein analyses. Anal Biochem 345:2–9 [DOI] [PubMed] [Google Scholar]
- Hughes A.D., Glenn I.C., Patrick A.D., Ellington A., and Anslyn E.V. (2008) A pattern recognition based fluorescence quenching assay for the detection and identification of nitrated explosive analytes. Chemistry 14:1822–1827 [DOI] [PubMed] [Google Scholar]
- Ivanciuc O. (2007) Applications of support vector machines in chemistry. Reviews in Computational Chemistry 23, doi: 10.1002/9780470116449.ch6 [DOI] [Google Scholar]
- Izenman A.J. (2013) Linear discriminant analysis. In Modern Multivariate Statistical Techniques, Springer, New York, pp 237–280 [Google Scholar]
- Jayasena S.D. (1999) Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clin Chem 45:1628–1650 [PubMed] [Google Scholar]
- Kattenhorn S.A. and Prockter L.M. (2014) Evidence for subduction in the ice shell of Europa. Nat Geosci 7:762–767 [Google Scholar]
- Ke R., Nong R.Y., Fredriksson S., Landegren U., and Nilsson M. (2013) Improving precision of proximity ligation assay by amplified single molecule detection. PLoS One 8, doi: 10.1371/journal.pone.0069813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keefe A.D., Pai S., and Ellington A. (2010) Aptamers as therapeutics. Nat Rev Drug Discov 9:537–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempf M.J., Chen F., Kern R., and Venkateswaran K. (2005) Recurrent isolation of hydrogen peroxide-resistant spores of Bacillus pumilus from a spacecraft assembly facility. Astrobiology 5:391–405 [DOI] [PubMed] [Google Scholar]
- Kim Y.S. and Gu M.B. (2013) Advances in aptamer screening and small molecule aptasensors. In Biosensors Based on Aptamers and Enzymes, Springer, Berlin, pp 29–67 [DOI] [PubMed] [Google Scholar]
- Kirschvink J.L. and Weiss B.P. (2002) Mars, panspermia, and the origin of life: where did it all begin. Palaeontologia Electronica 4:8–15 [Google Scholar]
- Kite E.S. and Rubin A.M. (2016) Sustained eruptions on Enceladus explained by turbulent dissipation in tiger stripes. Proc Natl Acad Sci USA 113:3972–3975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaus K.K., Elsperman M., and Rogers F. (2013) Mission concepts enabled by solar electric propulsion and advanced modular power systems [id. 211.14]. In AAS/Division for Planetary Sciences Meeting Abstracts, American Astronomical Society, Washington, DC [Google Scholar]
- Loman N.J. and Watson M. (2015) Successful test launch for nanopore sequencing. Nat Methods 12:303–304 [DOI] [PubMed] [Google Scholar]
- Lorenz R.D. (2016) Europa ocean sampling by plume flythrough: astrobiological expectations. Icarus 267:217–219 [Google Scholar]
- McCollom T.M. (1999) Methanogenesis as a potential source of chemical energy for primary biomass production by autotrophic organisms in hydrothermal systems on Europa. J Geophys Res: Planets 104:30729–30742 [Google Scholar]
- McCollom T.M. and Seewald J.S. (2007) Abiotic synthesis of organic compounds in deep-sea hydrothermal environments. Chem Rev 107:382–401 [DOI] [PubMed] [Google Scholar]
- McCune B. and Grace J.B. (2002) Analysis of Ecological Communities. MjM Software Design, Gleneden Beach, OR, USA [Google Scholar]
- Mikheyev A.S. and Tin M.M. (2014) A first look at the Oxford Nanopore MinION sequencer. Mol Ecol Resour 14:1097–1102 [DOI] [PubMed] [Google Scholar]
- Mileikowsky C., Cucinotta F.A., Wilson J.W., Gladman B., Horneck G., Lindegren L., Melosh J., Rickman H., Valtonen M., and Zheng J.Q. (2000) Natural transfer of viable microbes in space: 1. From Mars to Earth and Earth to Mars. Icarus 145:391–427 [DOI] [PubMed] [Google Scholar]
- Mojarro A., Hachey J., Tani J., Smith A., Bhattaru S., Pontefract A., Doebler R., Brown M., Ruvkun G., and Zuber M. (2016) SETG: nucleic acid extraction and sequencing for in situ life detection on Mars [abstract 4095]. In 3rd International Workshop on Instrumentation for Planetary Missions, Lunar and Planetary Institute, Houston, LPI contribution 1980 [Google Scholar]
- Nimmo F. and Pappalardo R. (2016) Ocean worlds in the outer Solar System. J Geophys Res: Planets 121:1378–1399 [Google Scholar]
- Pai S.S. and Ellington A.D. (2009) Using RNA aptamers and the proximity ligation assay for the detection of cell surface antigens. In Biosensors and Biodetection: Methods and Protocols: Electrochemical and Mechanical Detectors, Lateral Flow and Ligands for Biosensors, edited by Rasooly A. and Herold K.E., Springer, New York, pp 385–398 [DOI] [PubMed] [Google Scholar]
- Probst A., Facius R., Wirth R., Wolf M., and Moissl-Eichinger C. (2011) Recovery of Bacillus spore contaminants from rough surfaces: a challenge to space mission cleanliness control. Appl Environ Microbiol 77:1628–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radi A.-E., Acero Sánchez J.L., Baldrich E., and O'Sullivan CK. (2006) Reagentless, reusable, ultrasensitive electrochemical molecular beacon aptasensor. J Am Chem Soc 128:117–124 [DOI] [PubMed] [Google Scholar]
- Scharf C., Virgo N., Cleaves H.J., Aono M., Aubert-Kato N., Aydinoglu A., Barahona A., Barge L.M., Benner S.A., Biehl M., Brasser R., Butch C.J., Chandru K., Cronin L., Danielache S., Fischer J., Hernlund J., Hut P., Ikegami T., Kimura J., Kobayashi K., Mariscal C., McGlynn S., Menard B., Packard N., Pascal R., Pereto J., Rajamani S., Sinapayen L., Smith E., Switzer C., Takai K., Tian F., Ueno Y., Voytek M., Witkowski O., and Yabuta H. (2015) A strategy for origins of life research. Astrobiology 15:1031–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze-Makuch D. and Irwin L.N. (2008) Life in the Universe: Expectations and Constraints, Springer Science & Business Media, Berlin [Google Scholar]
- Söderberg O., Leuchowius K.-J., Gullberg M., Jarvius M., Weibrecht I., Larsson L.-G., and Landegren U. (2008) Characterizing proteins and their interactions in cells and tissues using the in situ proximity ligation assay. Methods 45:227–232 [DOI] [PubMed] [Google Scholar]
- Steinwart I. and Christmann A. (2008) Support Vector Machines, Springer Science & Business Media, Berlin [Google Scholar]
- Stewart S., Syrett A., Pothukuchy A., Bhadra S., Ellington A., and Anslyn E. (2011) Identifying protein variants with cross-reactive aptamer arrays. ChemBioChem 12:2021–2024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltenburg R., Reinemann C., and Strehlitz B. (2007) SELEX—a (r) evolutionary method to generate high-affinity nucleic acid ligands. Biomol Eng 24:381–403 [DOI] [PubMed] [Google Scholar]
- Sun H. and Zu Y. (2015) A highlight of recent advances in aptamer technology and its application. Molecules 20:11959–11980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi S. and Kramer F.R. (1996) Molecular beacons: probes that fluoresce upon hybridization. Nat Biotechnol 14:303–308 [DOI] [PubMed] [Google Scholar]
- Umali A.P. and Anslyn E.V. (2010) A general approach to differential sensing using synthetic molecular receptors. Curr Opin Chem Biol 14:685–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishampayan P., Moissl-Eichinger C., Pukall R., Schumann P., Spröer C., Augustus A., Roberts A.H., Namba G., Cisneros J., Salmassi T., and Venkateswaran K. (2013) Description of Tersicoccus phoenicis gen. nov., sp. nov. isolated from spacecraft assembly clean room environments. Int J Syst Evol Microbiol 63:2463–2471 [DOI] [PubMed] [Google Scholar]
- Woo Y., Heo Y., Shin K., and Yi G.-R. (2013) Hydrodynamic filtration in microfluidic channels as size-selection process for giant unilamellar vesicles. J Biomed Nanotechnol 9:610–614 [DOI] [PubMed] [Google Scholar]
- Worley B. and Powers R. (2016) PCA as a predictor of OPLS-DA model reliability. Curr Metabolomics 4:97–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright A.T., Griffin M.J., Zhong Z., McCleskey S.C., Anslyn E.V., and McDevitt J.T. (2005) Differential receptors create patterns that distinguish various proteins. Angew Chem Int Ed Engl 117:6533–6536 [DOI] [PubMed] [Google Scholar]
- Wyrick D., Teolis B., Bouquet A., Magee B., and Waite J. (2016) The effects of plumes and other geologic activity on Europa's exospheric structure and composition [abstract 2258]. In 47th Lunar and Planetary Science Conference Abstracts, Lunar and Planetary Institute, Houston [Google Scholar]
- Ye B.F., Zhao Y.J., Cheng Y., Li T.T., Xie Z.Y., Zhao X.W., and Gu Z.Z. (2012) Colorimetric photonic hydrogel aptasensor for the screening of heavy metal ions. Nanoscale 4:5998–6003 [DOI] [PubMed] [Google Scholar]
- Yoon Y., Kim S., Lee J., Choi J., Kim R.-K., Lee S.-J., Sul O., and Lee S.-B. (2016) Clogging-free microfluidics for continuous size-based separation of microparticles. Sci Rep 6, doi: 10.1038/srep26531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamora-Olivares D., Kaoud T.S., Jose J., Ellington A., Dalby K.N., and Anslyn E.V. (2014) Differential sensing of MAP kinases using SOX-Peptides. Angew Chem Int Ed Engl 126:14288–14292 [DOI] [PubMed] [Google Scholar]