Abstract
Emerging single-molecule sequencing technologies from Pacific Biosciences and Oxford Nanopore have revived interest in long-read mapping algorithms. Alignment-based seed-and-extend methods demonstrate good accuracy, but face limited scalability, while faster alignment-free methods typically trade decreased precision for efficiency. In this article, we combine a fast approximate read mapping algorithm based on minimizers with a novel MinHash identity estimation technique to achieve both scalability and precision. In contrast to prior methods, we develop a mathematical framework that defines the types of mapping targets we uncover, establish probabilistic estimates of p-value and sensitivity, and demonstrate tolerance for alignment error rates up to 20%. With this framework, our algorithm automatically adapts to different minimum length and identity requirements and provides both positional and identity estimates for each mapping reported. For mapping human PacBio reads to the hg38 reference, our method is 290 × faster than Burrows–Wheeler Aligner-MEM with a lower memory footprint and recall rate of 96%. We further demonstrate the scalability of our method by mapping noisy PacBio reads (each ≥5 kbp in length) to the complete NCBI RefSeq database containing 838 Gbp of sequence and >60,000 genomes.
Keywords: : Jaccard, long-read mapping, MinHash, minimizers, sketching, winnowing
1. Introduction
Mapping reads generated by high-throughput DNA sequencers to reference genomes is a fundamental and widely studied problem (Li and Homer, 2010; Ruffalo et al., 2011). The problem is particularly well studied for short read sequences, for which effective mapping algorithms and widely used software such as Burrows–Wheeler Aligner (BWA; Li and Durbin, 2009) and Bowtie (Langmead and Salzberg, 2012) have been developed. The increasing popularity of single-molecule sequencers from Pacific Biosciences and Oxford Nanopore, and their continually improving read lengths (10 kbp and up), is generating renewed interest in long-read mapping algorithms. However, the benefit of long-read lengths is currently accompanied by much higher error rates (up to 15%–20%). Despite their high error rates, long reads have proved advantageous in many applications, including de novo genome assembly (Chin et al., 2013; Koren et al., 2013) and real-time pathogen identification (Ashton et al., 2015; Quick et al., 2016).
Sequence data from nanopore devices are available just minutes after introducing the sample. This can enable real-time genomic analysis when coupled with fast computational methods that can map the data stream against large reference databases. However, mapping raw sequences continues to be a bottleneck for many applications. The problem is only going to worsen as Oxford Nanopore's PromethION is projected to generate multiple terabases of sequence per day. In parallel, reference databases are continually growing in size, with the nonredundant NCBI RefSeq database close to exceeding a terabase in size. The high error rate of raw single-molecule sequences further adds to the computational complexity.
Read mapping problems can be solved exactly by designing appropriate variants of the Smith–Waterman (SW) alignment algorithm (Smith and Waterman, 1981); however, it is computationally prohibitive when mapping reads from a high-throughput sequencer to large reference genomes. Seed-and-extend mapping heuristics address this limitation for both long and short reads by limiting the search to locations, where exact short word or maximal common substring matches occur before executing an alignment algorithm at these locations (Altschul et al., 1997; Delcher et al., 2002; Chaisson and Tesler, 2012). Accurate alignment-based long-read mappers include basic local alignment with successive refinement (BLASR; Chaisson and Tesler, 2012) and BWA-MEM (Li, 2013). However, repetitive seeds that do not translate to correct mappings combined with high sequencing error rates limit their scalability. Additionally, alignment-based mapping algorithms preserve the complete reference sequence in the index, and hence, cannot scale to terabase scale reference databases. Many genomics applications do not require detailed base-to-base alignment information, and instead use only the alignment boundary and identity summaries. Such applications include depth-of-coverage analysis, metagenomic read assignment, structural variant detection, and selective sequencing (Loose et al., 2016). Efficient algorithms for these problems, combined with nanopore sequencing, could enable the real-time genomic analysis of patients, pathogens, cancers, and microbiomes.
One class of algorithms for fast, approximate mapping relies on ideas originally developed for finding similarities between web documents. Broder (1997) proved that an unbiased estimate of the Jaccard similarity coefficient between two sets can be computed efficiently using a subset of hashed elements called a sketch. Schleimer et al. (2003) proposed the winnowing algorithm, which picks a minimum hashed item (also known as a minimizer; Roberts et al., 2004) from each consecutive window of text as a means to more quickly estimate local similarity between web documents. These ideas have been used to develop new mapping and assembly algorithms for long reads, such as the MinHash Alignment Process (Berlin et al., 2015), minimap (Li, 2016), and BALAUR (Popic and Batzoglou, 2016). To date, the effectiveness of these approaches has only been demonstrated empirically.
In this article, we propose a fast approximate algorithm for mapping long reads that scales to large reference databases with sufficient theoretical guarantees and practical validation on the quality of results reported. We propose a problem formulation that mathematically characterizes desired mapping targets by linking the Jaccard coefficient between the k-mer spectra of the read and its mapping region to a sequence error rate assuming a Poisson error model. We then provide an efficient algorithm to estimate the Jaccard coefficient through a combination of MinHash and winnowing techniques that characterizes and guarantees the types of mapping regions we find. On the quality side, we provide probabilistic bounds on sensitivity. We present techniques for choosing algorithmic parameters as a function of error rate and sequence lengths that guarantees the desired statistical significance. The theory is validated using PacBio and MinION datasets, and we demonstrate the scalability of our approach by mapping PacBio metagenomic reads to the entire RefSeq database. The speed and space efficiency of our algorithm enables real-time mapping, and compared with minimap, our method maintains high sensitivity with better precision for large, repetitive genomes. The implementation is available at github.com/MarBL/MashMap (v1.0).
2. Preliminaries
2.1. Read error model
We assume errors occur independently at the read positions, and use a Poisson error model as in previous works (Fan et al., 2015; Ondov et al., 2016). A binomial model would also be appropriate, but is not discussed in this study for brevity. Let 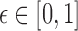 be the per-base error rate. The expected number of errors in a k-mer is
be the per-base error rate. The expected number of errors in a k-mer is 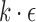 , and the probability of no errors within each k-mer, assumed independent, is
, and the probability of no errors within each k-mer, assumed independent, is 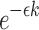 . We assume the statement is valid irrespective of error type.
. We assume the statement is valid irrespective of error type.
2.2. Jaccard similarity
Assuming 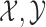 are the sets of k-mers in sequences X and Y, respectively, their Jaccard similarity is defined as
are the sets of k-mers in sequences X and Y, respectively, their Jaccard similarity is defined as 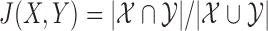 . The Poisson error model is used to compute the relationship between Jaccard similarity and alignment error rate (Ondov et al., 2016). We approximate the length of a read alignment to be the read length. Let A be a read derived from Bi, where Bi denotes the length
. The Poisson error model is used to compute the relationship between Jaccard similarity and alignment error rate (Ondov et al., 2016). We approximate the length of a read alignment to be the read length. Let A be a read derived from Bi, where Bi denotes the length  substring of reference B starting at position i. If c and n denote the number of error-free and total k-mers in A, respectively, then the expected value of c/n, termed k-mer survival probability, is
substring of reference B starting at position i. If c and n denote the number of error-free and total k-mers in A, respectively, then the expected value of c/n, termed k-mer survival probability, is 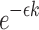 . This equation assumes k is large enough such that k-mers in A or Bi are unique, because
. This equation assumes k is large enough such that k-mers in A or Bi are unique, because 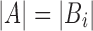 ,
, 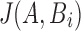 , abbreviated as J, equals
, abbreviated as J, equals 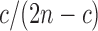 . Using the two equations, we derive the following functions
. Using the two equations, we derive the following functions 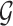 and
and 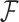 to estimate J and
to estimate J and 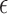 :
:
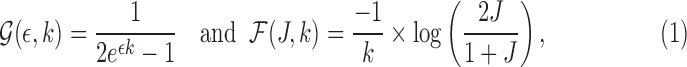 |
where 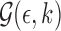 serves as an estimate of the Jaccard similarity given an error rate and
serves as an estimate of the Jaccard similarity given an error rate and 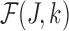 estimates the converse.
estimates the converse. 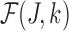 can be shown as the maximum likelihood estimator (MLE) for error rate (proof in Section 10). Note
can be shown as the maximum likelihood estimator (MLE) for error rate (proof in Section 10). Note 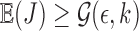 (using Jensen's inequality).
(using Jensen's inequality).
2.3. MinHash approximation
The MinHash algorithm is a fast and space-efficient approximation technique to compute an unbiased estimate of Jaccard similarity (Broder, 1997), without explicitly computing the underlying set intersection and union. Let s be a fixed parameter. Assuming universe U is the totally ordered set of all possible items, and 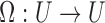 is a permutation chosen uniformly at random, Broder (1997) proved that
is a permutation chosen uniformly at random, Broder (1997) proved that 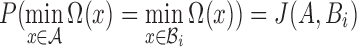 , and that
, and that
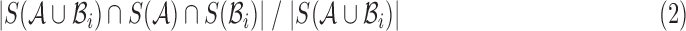 |
is an unbiased estimate of 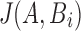 , where
, where 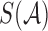 (called the sketch of
(called the sketch of 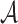 ) is the set of the smallest s hashed items in
) is the set of the smallest s hashed items in 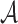 , that is,
, that is, 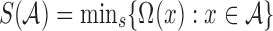 . Typically, the denominator
. Typically, the denominator 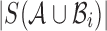 is referred as the MinHash sketch size and the numerator as the count of shared sketch elements. This estimate is unbiased provided
is referred as the MinHash sketch size and the numerator as the count of shared sketch elements. This estimate is unbiased provided 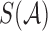 is a simple random sample of
is a simple random sample of 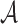 . Increasing the sketch size improves the accuracy of the estimate.
. Increasing the sketch size improves the accuracy of the estimate.
Assuming s is fixed and the true Jaccard similarity 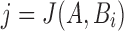 is known, the count of shared sketch elements between
is known, the count of shared sketch elements between 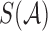 and
and 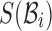 follows a hypergeometric distribution. Since s is much smaller than
follows a hypergeometric distribution. Since s is much smaller than 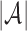 , it can be approximated by the binomial distribution.
, it can be approximated by the binomial distribution.
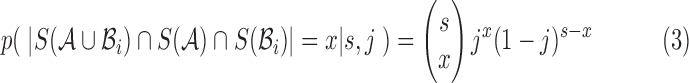 |
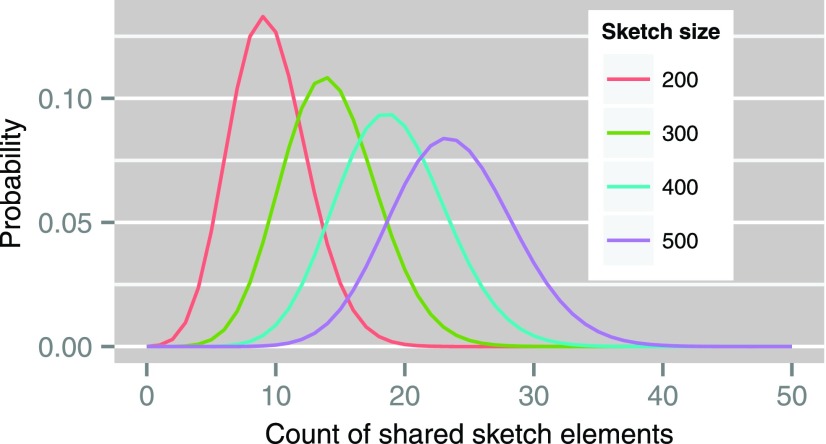
As an example, Figure 1 illustrates this distribution for a read with known Jaccard similarity 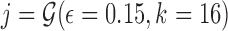 (using Eq. 1) and sketch size s varying from 200 to 500.
(using Eq. 1) and sketch size s varying from 200 to 500.
FIG. 1.
Probability distributions of count of shared sketch elements for a read with 15% alignment error (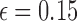 ) and k-mer size of 16, with varying sketch sizes. Estimated Jaccard similarity computed using Equation (1) is
) and k-mer size of 16, with varying sketch sizes. Estimated Jaccard similarity computed using Equation (1) is 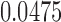 .
.
2.4. Winnowing
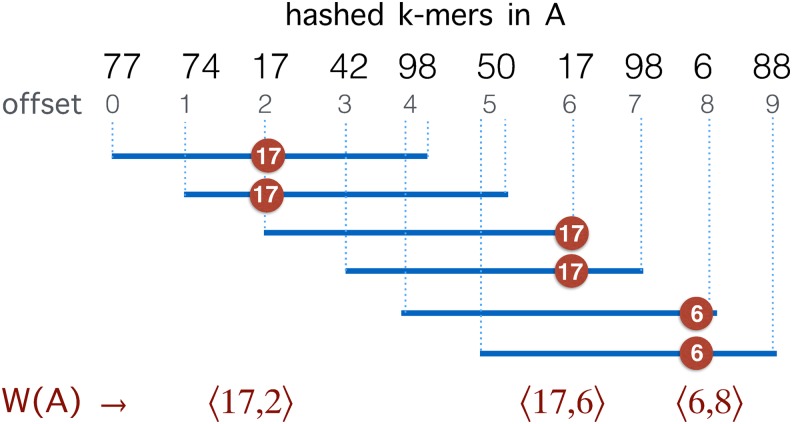
Winnowing is a local fingerprinting algorithm, proposed to measure similarity between documents by using a subset of hashed words (Schleimer et al., 2003). Unlike MinHash sketching, it bounds the maximum positional gap between any two consecutive selected hashes. It works by sampling the smallest hashed item in every consecutive fixed size sliding window (Fig. 2). Formal description of this algorithm in the context of genomic sequences follows.
FIG. 2.
Illustration of the winnowing method on a sequence of hashed k-mers in A.  represents the minimizers sampled from the sequence with window size
represents the minimizers sampled from the sequence with window size 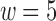 .
.
Let A0 denote the set of all k-mer tuples 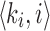 in sequence A, i denoting the k-mer position. Let w be the window size used for winnowing, and Kj be the set of w consecutive k-mer tuples starting at position j in A, that is,
in sequence A, i denoting the k-mer position. Let w be the window size used for winnowing, and Kj be the set of w consecutive k-mer tuples starting at position j in A, that is, 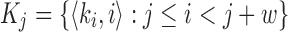 . Assume
. Assume 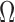 is a hash function defined as a random permutation. Then, the set of minimizers sampled by the winnowing algorithm in sequence A is
is a hash function defined as a random permutation. Then, the set of minimizers sampled by the winnowing algorithm in sequence A is 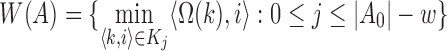 , where
, where
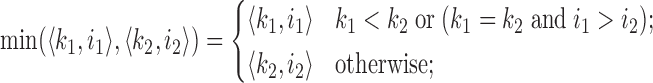 |
Schleimer et al. (2003) prove that the expected set count of minimizers selected from a random sequence A is 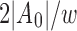 . Moreover,
. Moreover, 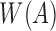 can be computed efficiently in
can be computed efficiently in 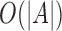 time and
time and 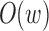 space using a double-ended queue, as sequence A is read in a streaming fashion (Smith, 2016).
space using a double-ended queue, as sequence A is read in a streaming fashion (Smith, 2016).
3. Problem Formulation
Given a read A and the maximum per-base error rate 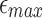 , our goal is to identify target positions in reference B, where A aligns with
, our goal is to identify target positions in reference B, where A aligns with 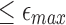 per-base error rate. This problem can be exactly solved in
per-base error rate. This problem can be exactly solved in 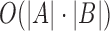 time by designing a suitable quadratic time alignment algorithm. When mapping to a large database of reference sequences, solving this problem exactly is computationally prohibitive. Hence, we define an approximate version of this problem using the Jaccard coefficient as a proxy for the alignment as follows: Let Bi denote the substring of size
time by designing a suitable quadratic time alignment algorithm. When mapping to a large database of reference sequences, solving this problem exactly is computationally prohibitive. Hence, we define an approximate version of this problem using the Jaccard coefficient as a proxy for the alignment as follows: Let Bi denote the substring of size  in B starting at position i (
in B starting at position i (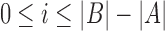 ). For a given k, we seek all mapping positions i in B such that
). For a given k, we seek all mapping positions i in B such that
 |
Note that if A aligns with Bi with per-base error rate 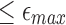 , then
, then 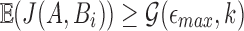 (using Eq. 1). As this equation applies only to the expected value of
(using Eq. 1). As this equation applies only to the expected value of 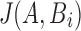 , we lower this threshold by
, we lower this threshold by 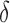 to account for variation in the estimate. The parameter
to account for variation in the estimate. The parameter 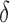 is defined as the margin of error in Jaccard estimation using a 90% confidence interval.
is defined as the margin of error in Jaccard estimation using a 90% confidence interval.
4. The Proposed Algorithm
Directly computing 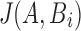 for all positions i is as asymptotically expensive as the alignment algorithm. The rationale for reformulating the problem in terms of Jaccard coefficients is that it enables the design of fast heuristic algorithms. We present an algorithm to estimate
for all positions i is as asymptotically expensive as the alignment algorithm. The rationale for reformulating the problem in terms of Jaccard coefficients is that it enables the design of fast heuristic algorithms. We present an algorithm to estimate 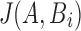 efficiently using a combination of MinHash and winnowing techniques. In addition, we compute an estimate of the alignment error rate
efficiently using a combination of MinHash and winnowing techniques. In addition, we compute an estimate of the alignment error rate 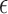 for each mapping reported. Our method relies on an indexing and search strategy we developed to prune the incorrect mapping positions efficiently.
for each mapping reported. Our method relies on an indexing and search strategy we developed to prune the incorrect mapping positions efficiently.
4.1. Definitions
Let 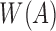 be the set of minimizers computed for read A using the winnowing method with window size w. We sketch
be the set of minimizers computed for read A using the winnowing method with window size w. We sketch 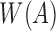 instead of sketching A itself. Assuming s is a fixed parameter, we define
instead of sketching A itself. Assuming s is a fixed parameter, we define 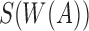 as the set of the s smallest hashed k-mers that were sampled using winnowing of A, that is,
as the set of the s smallest hashed k-mers that were sampled using winnowing of A, that is, 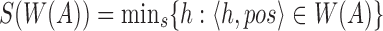 . To estimate
. To estimate 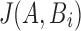 , we define winnowed-minhash estimate
, we define winnowed-minhash estimate 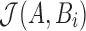 for
for 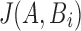 as
as
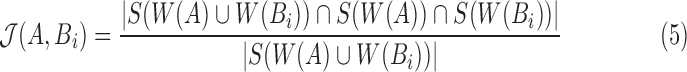 |
In contrast to the MinHash approximation (Eq. 2), our estimator 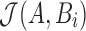 uses winnowing to reduce the sampling frame before picking the minimum hash values. Even though
uses winnowing to reduce the sampling frame before picking the minimum hash values. Even though 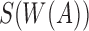 is no longer a simple random sample of the k-mers in A, we empirically show in Section 8.1 that the quality of the Jaccard estimation using
is no longer a simple random sample of the k-mers in A, we empirically show in Section 8.1 that the quality of the Jaccard estimation using 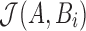 is as good as the MinHash estimation. We use
is as good as the MinHash estimation. We use 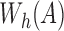 to denote the set of hashed k-mers in
to denote the set of hashed k-mers in 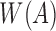 , that is,
, that is, 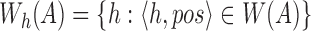 .
.
4.2. Indexing the reference
Retaining the minimizers 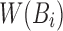 is sufficient for Jaccard similarity estimation
is sufficient for Jaccard similarity estimation 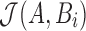 (Eq. 5). Since
(Eq. 5). Since 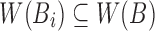 (Section 2), we compute
(Section 2), we compute 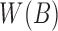 from the reference sequence B to be able to extract
from the reference sequence B to be able to extract 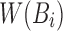 efficiently for any i. The set
efficiently for any i. The set 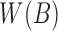 can be computed from B in a linear scan in
can be computed from B in a linear scan in 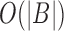 time. We store
time. We store 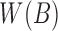 as an array
as an array 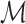 of tuples
of tuples 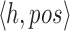 . When created, the set is naturally in ascending sorted order of the positions. Furthermore, to enable
. When created, the set is naturally in ascending sorted order of the positions. Furthermore, to enable 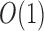 look-up of all the occurrences of a particular minimizer's hashed value h, we also replicate
look-up of all the occurrences of a particular minimizer's hashed value h, we also replicate 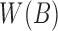 as a hash table
as a hash table 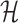 with h as the key and an array of its positions
with h as the key and an array of its positions 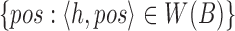 as the mapped value. The expected space requirements for
as the mapped value. The expected space requirements for 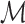 and
and 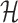 are
are 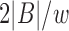 (Section 2). We postpone our discussion on how to compute an appropriate window size w to Section 5. Besides low memory requirements, a key advantage of this indexing strategy is that a new reference sequence can be incrementally added to the existing data structure in time linear to its length, which is not feasible for suffix array or Burrows–Wheeler transform-based indices, typically used in most mapping software.
(Section 2). We postpone our discussion on how to compute an appropriate window size w to Section 5. Besides low memory requirements, a key advantage of this indexing strategy is that a new reference sequence can be incrementally added to the existing data structure in time linear to its length, which is not feasible for suffix array or Burrows–Wheeler transform-based indices, typically used in most mapping software.
4.3. Searching the reference
The goal of the search phase is to identify for each read A, positions i such that 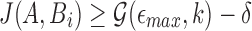 . We instead compute the winnowed-minhash estimate
. We instead compute the winnowed-minhash estimate 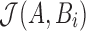 . Let
. Let 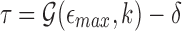 . To avoid directly evaluating
. To avoid directly evaluating 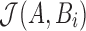 for each Bi, we state and prove the following Theorem 1:
for each Bi, we state and prove the following Theorem 1:
Theorem 1.
Assuming sketch size
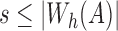 ,
,
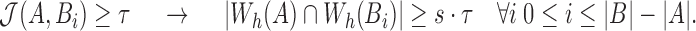
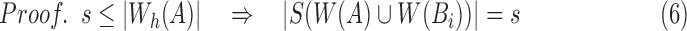 |
From Equation (5),
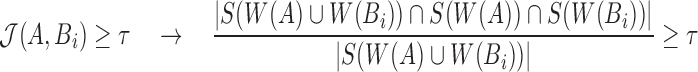
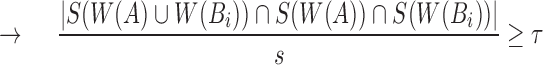 (using Eq. 6)
(using Eq. 6)
Note that 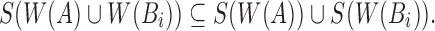 Therefore,
Therefore,
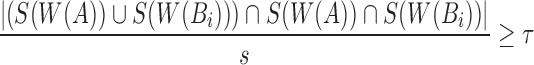
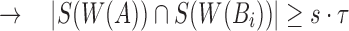
But, 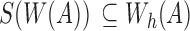 and
and 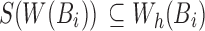
Therefore, 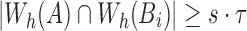 ■
■
We use the above condition as a filter and only consider positions in B which satisfy 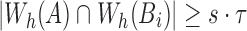 . To maximize effectiveness of the filter, we set the sketch size
. To maximize effectiveness of the filter, we set the sketch size 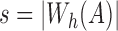 . The search proceeds in two successive stages. The first stage identifies candidate positions i using Theorem 1, and the second stage computes
. The search proceeds in two successive stages. The first stage identifies candidate positions i using Theorem 1, and the second stage computes 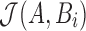 at each candidate position i. The position is retained as output if
at each candidate position i. The position is retained as output if 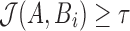 , and discarded otherwise.
, and discarded otherwise.
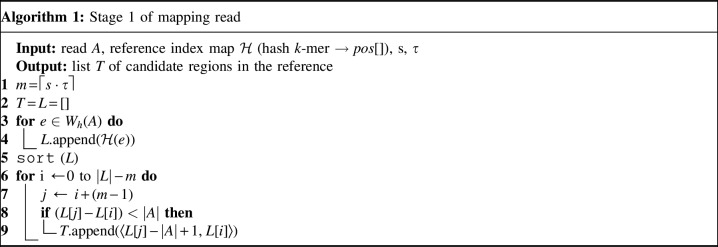
Stage 1:
Algorithm 1 outlines the first stage of our mapping procedure. It calculates all offset positions i in B such that 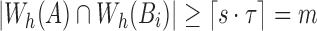 . The output list T is created in the form of one or more tuple ranges
. The output list T is created in the form of one or more tuple ranges 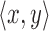 , implying that the criterion holds true for all Bi,
, implying that the criterion holds true for all Bi, 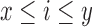 . We begin by computing the minimizer hashed values
. We begin by computing the minimizer hashed values 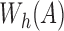 by winnowing the read A, and compute the positions of their occurrence in the reference (line 4). Accordingly,
by winnowing the read A, and compute the positions of their occurrence in the reference (line 4). Accordingly, 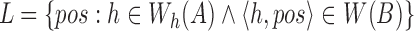 . Next, we sort the array L to process all the positions in ascending order. If Bi satisfies the filtering criterion, there should be at least m entries in L with values between
. Next, we sort the array L to process all the positions in ascending order. If Bi satisfies the filtering criterion, there should be at least m entries in L with values between 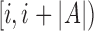 . It also implies that m consecutive entries should exist in L with positional difference between the first and
. It also implies that m consecutive entries should exist in L with positional difference between the first and 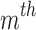 entry being
entry being 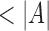 . This criterion is efficiently evaluated for all Bi using a linear scan on L (lines 6–9). If satisfied, we push the associated candidate range into T. To avoid reporting Bi more than once, we merge two consecutive overlapping tuple ranges into one.
. This criterion is efficiently evaluated for all Bi using a linear scan on L (lines 6–9). If satisfied, we push the associated candidate range into T. To avoid reporting Bi more than once, we merge two consecutive overlapping tuple ranges into one.
Stage 2: Evaluation of each tuple 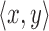 in the Stage 1 output array T requires computing
in the Stage 1 output array T requires computing 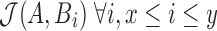 . Accordingly, we compute the minimum s unique sketch elements within
. Accordingly, we compute the minimum s unique sketch elements within 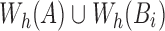 , and count the ones shared between A and Bi. We show the step-by-step procedure in Algorithm 2. We use
, and count the ones shared between A and Bi. We show the step-by-step procedure in Algorithm 2. We use 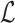 to contain the minimizer hashed values
to contain the minimizer hashed values 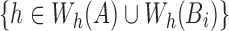 . To implement
. To implement 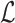 , we make use of the C++ ordered map data structure that supports logarithmic time insertion, deletion, and linear time iteration over unique ordered keys. We keep the hashed value as the map's key, and map it to 1 if it appears in both the reference and the read, and 0 otherwise. For each tuple
, we make use of the C++ ordered map data structure that supports logarithmic time insertion, deletion, and linear time iteration over unique ordered keys. We keep the hashed value as the map's key, and map it to 1 if it appears in both the reference and the read, and 0 otherwise. For each tuple 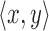 , we begin by saving the hashed values
, we begin by saving the hashed values 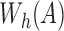 in read A into map
in read A into map 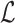 (lines 1 and 3). Two loops (lines 2 and 7) evaluate each tuple
(lines 1 and 3). Two loops (lines 2 and 7) evaluate each tuple 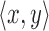 in T, and consider each
in T, and consider each 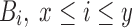 for Jaccard estimation
for Jaccard estimation 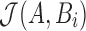 . The function getMinimizers gathers the reference minimizer hashes
. The function getMinimizers gathers the reference minimizer hashes 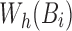 by sequentially iterating over
by sequentially iterating over 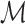 in the required position range and populating the minimizers associated with each Bi into the map
in the required position range and populating the minimizers associated with each Bi into the map 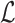 (lines 4, 8–9).
(lines 4, 8–9).
Note that a few incorrect corner minimizers 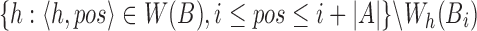 can appear in
can appear in 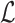 that were winnowed from windows overlapping with Bi. However, these can be discarded by recomputing the minimum of the first and last window of Bi. Finally, function solveJaccard computes
that were winnowed from windows overlapping with Bi. However, these can be discarded by recomputing the minimum of the first and last window of Bi. Finally, function solveJaccard computes 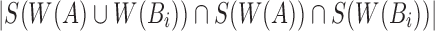 by iterating over s minimum unique sketch elements and counting the ones shared between A and Bi. If
by iterating over s minimum unique sketch elements and counting the ones shared between A and Bi. If 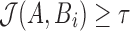 , then the position i and Jaccard estimate
, then the position i and Jaccard estimate 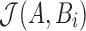 are saved into the output
are saved into the output 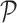 as pair
as pair 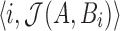 . The corresponding estimate of the alignment error rate
. The corresponding estimate of the alignment error rate 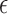 in this case, computed using Equation (1), would be
in this case, computed using Equation (1), would be 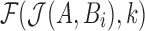 .
.
5. Selecting Window and Sketch Sizes
The sketch size for Jaccard similarity estimation is inversely proportional to the window size w (Section 4.3). A larger window size improves the runtime and space requirement during the search, but also negatively affects the statistical significance and accuracy of our estimate. To achieve the right balance, we analyze the p-value of a mapping location being reported under the null hypothesis that both query and reference sequences are random. For the subsequent analysis, we will assume the sketch size is s, the count of shared sketch elements is a discrete random variable Z, the k-mer size is k, the alphabet set is 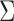 , and the read and reference sequence sizes are q and r, respectively.
, and the read and reference sequence sizes are q and r, respectively.
Location i is reported if 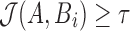 , that is, at least
, that is, at least 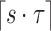 sketch elements are shared. Following Ondov et al. (2016), consider two random sequences of length q with k-mer sets X and Y, respectively. The probability of a random k-mer
sketch elements are shared. Following Ondov et al. (2016), consider two random sequences of length q with k-mer sets X and Y, respectively. The probability of a random k-mer 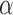 appearing in X or Y, assuming
appearing in X or Y, assuming 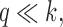 is
is 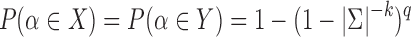 . Therefore, the expected Jaccard similarity
. Therefore, the expected Jaccard similarity 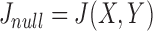 is given by
is given by
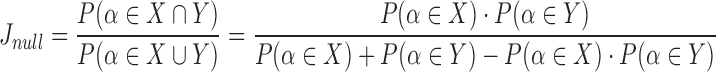 |
For sketch size s, the probability that x or more sketch elements are shared is 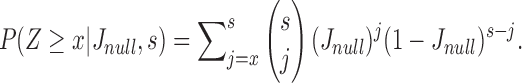 Using this equation, we compute the probability of a random sequence of length q mapping to at least one substring in a random reference sequence of size
Using this equation, we compute the probability of a random sequence of length q mapping to at least one substring in a random reference sequence of size 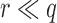 as
as 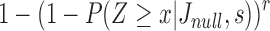 . For a minimum read length l0 and
. For a minimum read length l0 and 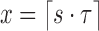 , we wish to ensure that this probability is kept below a user-specified threshold
, we wish to ensure that this probability is kept below a user-specified threshold 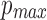 . As reported mapping locations i must satisfy
. As reported mapping locations i must satisfy 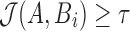 and
and 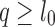 , a mapping with
, a mapping with 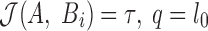 , in general, will have the highest probability of generating a random match.
, in general, will have the highest probability of generating a random match.
Therefore, we compute the maximum value of w that satisfies the 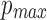 constraint for this instance. Sketch size s is set to
constraint for this instance. Sketch size s is set to 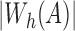 , which from Section 4.3 is expected to be
, which from Section 4.3 is expected to be 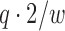 . Since x, s, and w have a circular dependency, we iteratively solve for w, starting from the maximum value l0, until the probability of a random mapping is
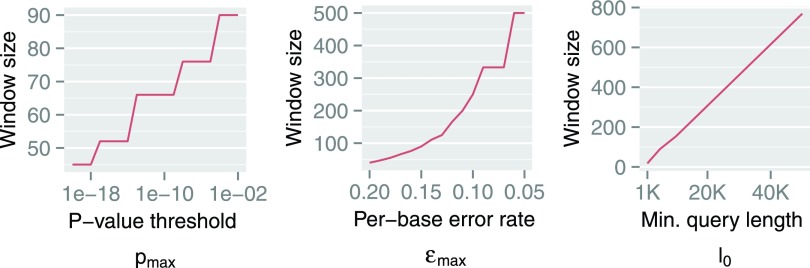
. Since x, s, and w have a circular dependency, we iteratively solve for w, starting from the maximum value l0, until the probability of a random mapping is 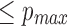 . Influence of different parameters on window size is shown in Figure 3. The window size w increases with increasing
. Influence of different parameters on window size is shown in Figure 3. The window size w increases with increasing 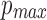 or l0, but has an inverse relationship with
or l0, but has an inverse relationship with 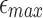 . These plots also highlight that as read length and error rate improve, our algorithm automatically adapts to a larger window size, greatly improving efficiency.
. These plots also highlight that as read length and error rate improve, our algorithm automatically adapts to a larger window size, greatly improving efficiency.
FIG. 3.
Illustration of how w varies with 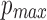 ,
, 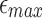 , and l0, respectively. The default values are set as
, and l0, respectively. The default values are set as 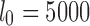 ,
, 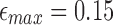 ,
, 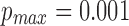 ,
, 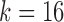 , and
, and 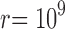 . Steps appear in the first two curves because Z is a discrete variable.
. Steps appear in the first two curves because Z is a discrete variable.
6. Proof of Sensitivity
We analyze the sensitivity exhibited by our algorithm in identifying correct mapping locations as a function of the read alignment error rate. Let i be a correct mapping location for read A. If 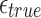 is the true error rate in aligning A with Bi, then
is the true error rate in aligning A with Bi, then 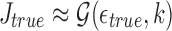 . Our algorithm reports this mapping location if the Jaccard estimate
. Our algorithm reports this mapping location if the Jaccard estimate 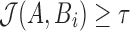 , that is, the count of shared sketch elements
, that is, the count of shared sketch elements 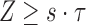 . The associated probability is given by
. The associated probability is given by 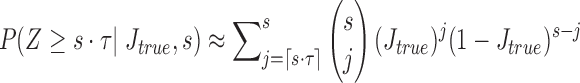 . We report the corresponding values in Table 1 while varying
. We report the corresponding values in Table 1 while varying 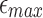 and
and 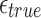 from 0.04 to 0.20 error rate, for two sketch sizes
from 0.04 to 0.20 error rate, for two sketch sizes 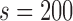 and 500, respectively. In an ideal scenario, a mapping should be reported only if
and 500, respectively. In an ideal scenario, a mapping should be reported only if 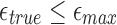 , that is, a perfect algorithm would have “1” in each of the entries at or above the diagonal, and “0” in all other positions. From the table, it is evident our algorithm achieves close to ideal sensitivity for alignment error rates up to 20%.
, that is, a perfect algorithm would have “1” in each of the entries at or above the diagonal, and “0” in all other positions. From the table, it is evident our algorithm achieves close to ideal sensitivity for alignment error rates up to 20%.
Table 1.
Probability of a Mapping Location Being Reported by Our Algorithm for Different Values of 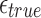 and
and 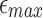
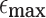 |
|||||
|---|---|---|---|---|---|
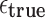 |
0.04 | 0.08 | 0.12 | 0.16 | 0.20 |
| 0.04 | 0.951 | 1 | 1 | 1 | 1 |
| 0.08 | 0 | 0.937 | 1 | 1 | 1 |
| 0.12 | 0 | 0.016 | 0.925 | 1 | 1 |
| 0.16 | 0 | 0 | 0.184 | 0.907 | 0.997 |
| 0.20 | 0 | 0 | 0.003 | 0.403 | 0.922 |
| 0.04 | 0.08 | 0.12 | 0.16 | 0.20 | |
|---|---|---|---|---|---|
| 0.04 | 0.939 | 1 | 1 | 1 | 1 |
| 0.08 | 0 | 0.949 | 1 | 1 | 1 |
| 0.12 | 0 | 0 | 0.937 | 1 | 1 |
| 0.16 | 0 | 0 | 0.013 | 0.904 | 1 |
| 0.20 | 0 | 0 | 0 | 0.104 | 0.896 |
True mapping locations correspond to 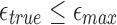 , that is, entries at or above the diagonal in the tables. Sketch sizes are set to 200 and 500 for the top and bottom tables, respectively. The k-mer size k is set to 16.
, that is, entries at or above the diagonal in the tables. Sketch sizes are set to 200 and 500 for the top and bottom tables, respectively. The k-mer size k is set to 16.
7. Other Implementation Details
7.1. Optimizing for variable read lengths
In contrast to cyclic short-read sequencing, single-molecule technologies can generate highly variable read lengths (e.g.,  –
–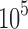 bases). Previously, we discussed how the window size w is determined using the minimum read length l0 in Section 5. From Figure 3c, notice that we can further reduce the sampling rate (i.e., use a larger window size) for reads longer than l0 while still satisfying the p-value constraint. However, to realize this, the sampling scheme for indexing the reference sequence B needs to be consistent with that of query. We propose the idea of multilevel winnowing to further optimize the runtime of our algorithm by choosing custom window size for each input read. Suppose
bases). Previously, we discussed how the window size w is determined using the minimum read length l0 in Section 5. From Figure 3c, notice that we can further reduce the sampling rate (i.e., use a larger window size) for reads longer than l0 while still satisfying the p-value constraint. However, to realize this, the sampling scheme for indexing the reference sequence B needs to be consistent with that of query. We propose the idea of multilevel winnowing to further optimize the runtime of our algorithm by choosing custom window size for each input read. Suppose 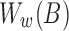 denotes the set of winnowed fingerprints in the reference computed using window size w, then
denotes the set of winnowed fingerprints in the reference computed using window size w, then 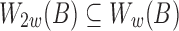 (Schleimer et al., 2003). We exploit this property to construct a multilevel reference index with multiple window sizes
(Schleimer et al., 2003). We exploit this property to construct a multilevel reference index with multiple window sizes 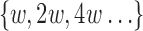 recursively. This optimization yields us faster mapping time per base pair for reads longer than l0 as we independently compute the window size for a given read length
recursively. This optimization yields us faster mapping time per base pair for reads longer than l0 as we independently compute the window size for a given read length 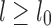 , and round it to the closest smaller reference window size
, and round it to the closest smaller reference window size 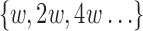 . The expected time and space complexity to index the reference using multiple levels is unaffected because the expected size of
. The expected time and space complexity to index the reference using multiple levels is unaffected because the expected size of 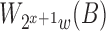 is half of
is half of 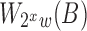 and
and 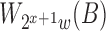 can be determined in linear time from
can be determined in linear time from 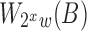 .
.
7.2. Strand prediction
To account for the reads sequenced from the reverse strand relative to the reference genome, we compute and store only canonical k-mers, that is, the lexicographically smaller of the forward and reverse-complemented k-mer. For each k-mer tuple 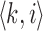 in
in 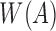 and
and 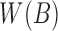 , we append a strand bit 1 if the forward k-mer is lexicographically smaller and −1 otherwise. While evaluating the read mappings in Stage 2, we compute the mapping strand of the read through a consensus vote among the shared sketches using sum of pairwise products of the strand bits.
, we append a strand bit 1 if the forward k-mer is lexicographically smaller and −1 otherwise. While evaluating the read mappings in Stage 2, we compute the mapping strand of the read through a consensus vote among the shared sketches using sum of pairwise products of the strand bits.
8. Experimental Results
8.1. Quality of Jaccard estimation
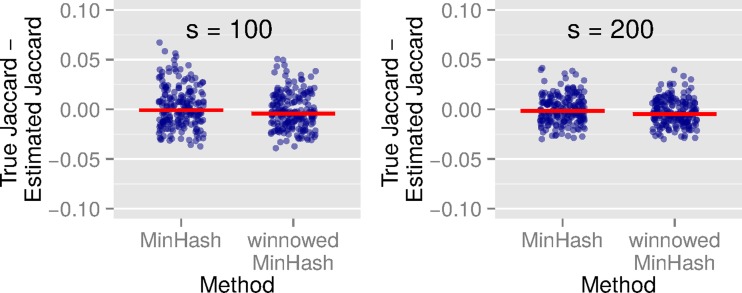
We first show that the accuracy of the winnowed-minhash estimator 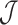 to estimate the Jaccard similarity is as good as the direct MinHash approximation, which is an unbiased statistical estimator. We construct a random sequence of length 5 kbp with each character having equal probability of being either A, C, G or T. We generate reads while introducing substitution errors at each position with probability 0.15. Note that both substitutions and indels have a similar effect of altering the k-mers containing them, and a uniform distribution of errors alters more k-mers than a clustering of errors. Figure 4 shows the estimation difference against the true Jaccard similarity using MinHash and our estimator for two different sketch sizes s = 100 and s = 200. Based on these results, we conclude that the bias in our estimation is practically negligible as the mean error by our method in estimating Jaccard similarity is <0.003 for both sketch sizes. Similar to MinHash approximation, we note that the magnitude of estimation error reduces with increasing sketch size.
to estimate the Jaccard similarity is as good as the direct MinHash approximation, which is an unbiased statistical estimator. We construct a random sequence of length 5 kbp with each character having equal probability of being either A, C, G or T. We generate reads while introducing substitution errors at each position with probability 0.15. Note that both substitutions and indels have a similar effect of altering the k-mers containing them, and a uniform distribution of errors alters more k-mers than a clustering of errors. Figure 4 shows the estimation difference against the true Jaccard similarity using MinHash and our estimator for two different sketch sizes s = 100 and s = 200. Based on these results, we conclude that the bias in our estimation is practically negligible as the mean error by our method in estimating Jaccard similarity is <0.003 for both sketch sizes. Similar to MinHash approximation, we note that the magnitude of estimation error reduces with increasing sketch size.
FIG. 4.
Jaccard similarity estimation using MinHash and winnowed-minhash estimator J (A, Bi) over simulated reads, with sketch sizes s = 100 and s = 200. Red bar indicates the average estimation difference over all reads.
8.2. Mapping MinION and PacBio reads
We refer the C++ implementation of our algorithm as mashmap and compare its run-time performance and memory usage against alignment-based long-read mappers BWA-MEM (v0.7.15-r114; Li, 2013), BLASR (vSMRTportal 2.3.0; Chaisson and Tesler, 2012), and minimap (v0.2; Li, 2016). We also perform a comparison of the approximate mapping targets generated by mashmap and minimap. Like mashmap, minimap uses winnowing to index the reference, but does not use the MinHash approximation to estimate Jaccard similarity or nucleotide identity. Instead, minimap seeks clusters of minimizer matches to identify regions of local similarity. Importantly, minimap approximates a local alignment process, which is useful for split-read mapping. However, because mashmap is currently designed to find complete read mappings, we only consider this case for the following comparisons.
8.2.1. Datasets and methodology
We evaluated the algorithms by mapping long-read datasets generated using single-molecule sequencers from Pacific Biosciences and Oxford Nanopore, and report single-threaded execution timings on an AMD Opteron 2376 CPU with 64 GB RAM. We use two datasets, labeled N1 and P1, respectively, both containing reads of length 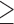 5 kbp. Dataset N1 is a random sample of 30,000 reads from the MinION (R9/1D) sequencing dataset of the Escherichia coli K12 genome (Loman, 2016). Dataset P1 contains 18,000 reads generated through a single SMRT (single molecule real-time) cell from PacBio's (P6/C4) sequencing of the CHM1 human genome (Chaisson et al., 2015). We map N1 to E. coli K12 (4.6 Mbp) and P1 to the human reference (3.2 Gbp). For mashmap, we use the following parameters:
5 kbp. Dataset N1 is a random sample of 30,000 reads from the MinION (R9/1D) sequencing dataset of the Escherichia coli K12 genome (Loman, 2016). Dataset P1 contains 18,000 reads generated through a single SMRT (single molecule real-time) cell from PacBio's (P6/C4) sequencing of the CHM1 human genome (Chaisson et al., 2015). We map N1 to E. coli K12 (4.6 Mbp) and P1 to the human reference (3.2 Gbp). For mashmap, we use the following parameters: 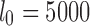 ,
, 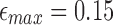 , and
, and 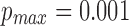 . When a read maps to multiple locations, mashmap only reports locations where mapping error rate is no >1% above the minimum of error rate over all such locations.
. When a read maps to multiple locations, mashmap only reports locations where mapping error rate is no >1% above the minimum of error rate over all such locations.
8.2.2. Run-time performance
Run-times for the index building and mapping stages, and memory used, for the four methods are compared in Table 2. As both BWA-MEM and BLASR are alignment-based methods, we expect their run-times to be significantly higher. Indeed, they take several hours in comparison to seconds (N1) or a few minutes (P1) taken by mashmap and minimap. The principal challenge is whether the latter methods can retain the quality obtainable through alignment-based methods. We note that mashmap has the lowest memory footprint for both datasets, and its run-time compares favorably with minimap. The ability to compute the sampling rate at run-time gives mashmap its edge in terms of memory usage.
Table 2.
Run-time and Memory Usage Comparison of Mashmap Against Minimap, Burrows–Wheeler Aligner-MEM and Basic Local Alignment with Successive Refinement for N1, P1 Datasets
| N1 (MinION-K12) | P1 (Pacbio-CHM1) | |||||
|---|---|---|---|---|---|---|
| Method | Index | Map | Memory (MB) | Index | Map | Memory (GB) |
| Mashmap | 0.5 seconds | 54 seconds | 17 | 5 minutes 52 seconds | 1 minutes 24 seconds | 3.7 |
| Minimap | 0.7 seconds | 37 seconds | 232 | 3 minutes 7 seconds | 1 minute 56 seconds | 6.8 |
| BWA-MEM | 2.6 seconds | 5 hours 39 minutes | 72 | 1 hours 19 minutes | 6 hours 46 minutes | 5.5 |
| BLASR | 1.3 seconds | 10 hours 17 minutes | 697 | 40 minutes 36 seconds | 20 hours 40 minutes | 17.6 |
BWA-MEM was executed with long-read mapping parameters-x pacbio/ont2d.
BWA, Burrows–Wheeler Aligner.
The best results are highlighted in bold.
8.2.3. Quality of mapping
As there is no standard benchmark using real datasets, we assess sensitivity/recall using BWA-MEM's starting read mapping positions, and precision by computing SW alignments of the reported mappings (Table 3). Since both minimap and BWA-MEM also report split-read alignments, we post-filter their results to only keep alignments with ≥80% read coverage. Recall is measured against BWA-MEM alignments which satisfy the 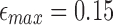 cutoff (≥80% identity). Because both minimap and mashmap estimate mapping positions, the reported mapping is assumed equivalent to BWA-MEM if the predicted starting position of a read is within ±50% of its length. Precision was directly validated using SW alignment (with scoring matrix:
cutoff (≥80% identity). Because both minimap and mashmap estimate mapping positions, the reported mapping is assumed equivalent to BWA-MEM if the predicted starting position of a read is within ±50% of its length. Precision was directly validated using SW alignment (with scoring matrix: 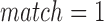 ,
, 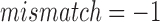 ,
, 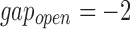 ,
, 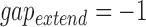 ). For minimap's and our results, we allow SW-identity ≥75% and query coverage ≥80%. Results in Table 3 show that both mashmap and minimap have close to ideal sensitivity/recall, demonstrating their ability to uncover the right target locations.
). For minimap's and our results, we allow SW-identity ≥75% and query coverage ≥80%. Results in Table 3 show that both mashmap and minimap have close to ideal sensitivity/recall, demonstrating their ability to uncover the right target locations.
Table 3.
Precision and Recall Statistics of Mashmap and Minimap Using Datasets N1 and P1
| Recall statistics | Precision statistics | ||||
|---|---|---|---|---|---|
| ID | Mashmap | Minimap | No. of BWA mappings | Mashmap | Minimap |
| N1 | 100% | 99.87% | 10,823 | 94.39% | 94.32% |
| P1 | 96.8% | 98.7% | 10,115 | 84.59% | 30.34% |
The best results are highlighted in bold.
Mashmap also achieves high precision, avoiding false positives on the repetitive human genome. Minimap's low precision on human is largely driven by false-positive mappings to repetitive sequence, which could potentially be resolved with alternative clustering parameters. Mashmap false positives are dominated by reported mappings with a SW query coverage <80% of the read length. It may be possible to avoid such mappings by considering the positional distribution of shared sketch elements during the second-stage filter, or by adopting a local alignment reporting strategy like minimap.
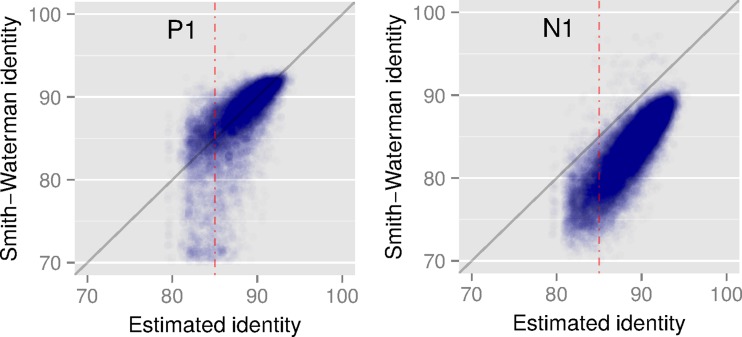
We compare our identity estimates (1–ε) × 100 against the SW alignment identities in Figure 5. For the PacBio reads, we observe that most of the points are aligned close to y = x. However, for the nanopore reads, our approach overestimates the identity. This is because PacBio sequencing produces mostly random errors, whereas current nanopore errors are more clustered and systematic (Laehnemann et al., 2016).
FIG. 5.
Correlation between Smith–Waterman identity and the identity estimated by mashmap using datasets P1 (PacBio) and N1 (MinION). Red dotted line corresponds to the error cutoff 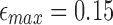 .
.
8.2.4. Performance gain from future improvements in long-read sequencing technologies
We discussed how mashmap adjusts its k-mer sampling rate for estimating the Jaccard similarity based on the provided error rate (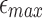 ) and minimum length (l0) cutoffs in Section 5. In this study, we show that improvement in read lengths and sequencing error rate can boost the performance of mashmap without affecting its output accuracy. For dataset P1, memory usage by mashmap drops significantly with decreasing per-base error rate threshold
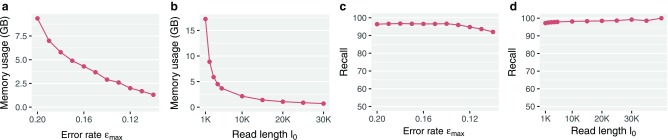
) and minimum length (l0) cutoffs in Section 5. In this study, we show that improvement in read lengths and sequencing error rate can boost the performance of mashmap without affecting its output accuracy. For dataset P1, memory usage by mashmap drops significantly with decreasing per-base error rate threshold 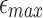 from 0.20 to 0.10 (Fig. 6a), or increasing minimum length threshold l0 from 5 kbp to 30 kbp (Fig. 6b). Note that the reduced k-mer sampling rate from reference and query sequences would also translate to faster mapping time. All this is achieved while maintaining high recall scores (>90%) against the BWA-MEM mappings that satisfy the input thresholds (Fig. 6c, d).
from 0.20 to 0.10 (Fig. 6a), or increasing minimum length threshold l0 from 5 kbp to 30 kbp (Fig. 6b). Note that the reduced k-mer sampling rate from reference and query sequences would also translate to faster mapping time. All this is achieved while maintaining high recall scores (>90%) against the BWA-MEM mappings that satisfy the input thresholds (Fig. 6c, d).
FIG. 6.
(a) Drop in memory usage of mashmap with varying values of maximum per-base error rate threshold 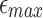 from
from 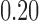 to
to 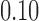 . Here l0 is fixed to 5 kbp. (b) Drop in memory usage of mashmap with varying values of read length cutoff l0 from 5 kbp to 40 kbp. Here
. Here l0 is fixed to 5 kbp. (b) Drop in memory usage of mashmap with varying values of read length cutoff l0 from 5 kbp to 40 kbp. Here 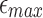 is fixed to 0.15. (c) Recall scores against BWA-MEM mappings, which satisfy input cutoffs with varying
is fixed to 0.15. (c) Recall scores against BWA-MEM mappings, which satisfy input cutoffs with varying 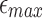 . These values are consistently above 90%. Note that recall score is relatively higher at
. These values are consistently above 90%. Note that recall score is relatively higher at 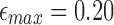 because a significant fraction of PacBio reads in dataset P1 have error rates much <20%. With decreasing
because a significant fraction of PacBio reads in dataset P1 have error rates much <20%. With decreasing 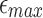 threshold, fraction of borderline cases increases. (d) Recall scores against BWA-MEM mappings with varying l0 parameter. These scores are consistently above 97%. BWA, Burrows–Wheeler Aligner.
threshold, fraction of borderline cases increases. (d) Recall scores against BWA-MEM mappings with varying l0 parameter. These scores are consistently above 97%. BWA, Burrows–Wheeler Aligner.
8.3. Mapping to RefSeq
We perform mapping of a publicly available PacBio read set consisting of 127,565 reads (each ≥5 kbp) sequenced from a mock microbial community containing 20 strains (Pacific Biosciences, 2014). To demonstrate the scalability of our algorithm, we map these reads against the complete NCBI RefSeq database (838 Gbp) containing sequences from 60,892 organisms. This experiment was executed using default parameters (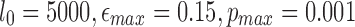 ) on an Intel Xeon CPU E7-8837 with 1 TB memory. BWA-MEM and minimap could not index the entire RefSeq database at once with this memory limitation. Mashmap took 29 CPU hours to index the reference and 16 CPU hours for mapping, with a peak memory usage of 660 GB. Note that the same index can be repeatedly used for mapping sequences, conferring our method the ability to process data in real-time. To check the accuracy of our results, we ran BWA-MEM against the 20 known genomes of the mock community. The recall of mashmap against BWA-MEM mappings ranged from 97.7% to 99.1% for all the 20 genomes in the mock community.
) on an Intel Xeon CPU E7-8837 with 1 TB memory. BWA-MEM and minimap could not index the entire RefSeq database at once with this memory limitation. Mashmap took 29 CPU hours to index the reference and 16 CPU hours for mapping, with a peak memory usage of 660 GB. Note that the same index can be repeatedly used for mapping sequences, conferring our method the ability to process data in real-time. To check the accuracy of our results, we ran BWA-MEM against the 20 known genomes of the mock community. The recall of mashmap against BWA-MEM mappings ranged from 97.7% to 99.1% for all the 20 genomes in the mock community.
9. Conclusions
We have presented a fast approximate algorithm for mapping long reads to large reference genomes. Instead of reporting base-level alignments, mashmap reports all reference intervals with sufficient Jaccard similarity compared with the k-mer spectrum of the read. In contrast to earlier techniques based on MinHash and winnowing, we provide a formal characterization of the mappings the algorithm is intended to uncover, and provide a provably good algorithm for computing them. In addition, we report an estimate of the alignment error rate tailored to each mapping under an assumed error model. Mashmap provides significant benefits in run-time, memory usage, and scalability, while achieving precision and recall similar to alignment-based methods. Future work aims to extend this method to split-read mapping, compressed reference databases, and additional error models. For example, the winnowed-minhash operation could be applied to paths within a de Bruijn graph to recover identity estimates and identify the database sequences most similar to a query sequence. Such approximate algorithms promise to help address the ever-increasing scale of genomic data.
10. Appendix
Below, we show that the estimator 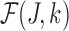 (defined in Section 2) for estimating the per-base error rate
(defined in Section 2) for estimating the per-base error rate 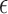 is a MLE.
is a MLE.
Claim: Under the assumed read error model in Section 2, and given Jaccard similarity J, 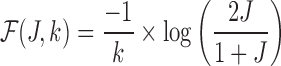 is an MLE for error rate
is an MLE for error rate 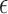 .
.
Proof. We follow the same notation as in Section 2. If c, n are the counts of conserved k-mers and total k-mers in a read, respectively, then 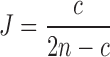 , or
, or 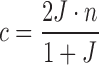 . Also, since
. Also, since 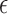 denotes the error rate, the probability of a k-mer being conserved (denoted by
denotes the error rate, the probability of a k-mer being conserved (denoted by 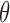 ) equals
) equals 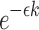 . Note that the fraction of the conserved k-mers follows a binomial distribution with parameters
. Note that the fraction of the conserved k-mers follows a binomial distribution with parameters 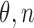 :
:
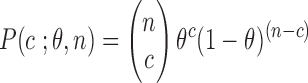 |
Therefore, likelihood function 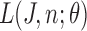 is given by:
is given by:
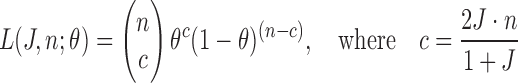 |
To compute 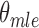 that maximizes likelihood L, we set
that maximizes likelihood L, we set 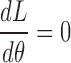 , therefore,
, therefore,
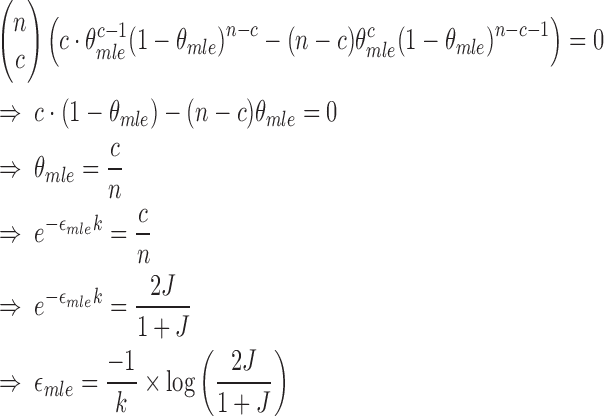 |
Acknowledgments
This research was supported in part by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health, and the U.S. National Science Foundation under IIS-1416259.
Author Disclosure Statement
No competing financial interests exist.
References
- Altschul S.F., Madden T.L., Schäffer A.A., et al. 1997. Gapped blast and psi-blast: A new generation of protein database search programs. Nucleic Acids Res. 250, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashton P.M., Nair S., Dallman T., et al. 2015. MinION nanopore sequencing identifies the position and structure of a bacterial antibiotic resistance island. Nat Biotechnol. 33, 296–300 [DOI] [PubMed] [Google Scholar]
- Berlin K., Koren S., Chin C.-S., et al. 2015. Assembling large genomes with single-molecule sequencing and locality-sensitive hashing. Nat Biotechnol. 33, 623–630 [DOI] [PubMed] [Google Scholar]
- Broder A.Z. 1997. On the resemblance and containment of documents. Proceedings of the Compression and Complexity of Sequences 1997, Salerno, Italy pp. 21–29 [Google Scholar]
- Chaisson M.J., and Tesler G. 2012. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): Application and theory. BMC Bioinformatics. 13, 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaisson M.J.P., Huddleston J., Dennis M.Y., et al. 2015. Resolving the complexity of the human genome using single-molecule sequencing. Nature. 517, 608–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin C.-S., Alexander D.H., Marks P., et al. 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods. 10, 563–569 [DOI] [PubMed] [Google Scholar]
- Delcher A.L., Phillippy A., Carlton J., and Salzberg S.L. 2002. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res. 30, 2478–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H., Ives A.R., Surget-Groba Y., and Cannon C.H. 2015. An assembly and alignment-free method of phylogeny reconstruction from next-generation sequencing data. BMC Genomics. 16, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S., Harhay G.P., Smith T.P.L., et al. 2013. Reducing assembly complexity of microbial genomes with single-molecule sequencing. Genome Biol. 14, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laehnemann D., Borkhardt A., and McHardy A.C. 2016. Denoising DNA deep sequencing data—high-throughput sequencing errors and their correction. Brief Bioinform. 17, 154–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., and Salzberg S.L. 2012. Fast gapped-read alignment with bowtie 2. Nat Methods. 9, 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:1303.3997
- Li H. 2016. Minimap and miniasm: Fast mapping and de novo assembly for noisy long sequences. Bioinformatics 32, 2103–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., and Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 25, 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., and Homer N. 2010. A survey of sequence alignment algorithms for next-generation sequencing. Brief Bioinform. 11, 473–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loman N.J. Nanopore r9 rapid run data release. 2016. Available at: https://goo.gl/UlHVtL Accessed September8, 2016
- Loose M., Malla S., and Stout M. 2016. Real time selective sequencing using nanopore technology. Nat Methods. 13, 751–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ondov B.D., Treangen T.J., Melsted P., et al. 2016. Mash: Fast genome and metagenome distance estimation using minhash. Genome Biol 17, 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacific Biosciences. Human microbiome mock community shotgun sequencing data. 2014. Available at: https://goo.gl/kjRcLb Accessed September8, 2016
- Popic V., and Batzoglou S. 2017. A hybrid cloud read aligner based on MinHash and kmer voting that preserves privacy. Nature Commun. 8, 15311 DOI: 10.1038/ncomms15311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quick J., Loman N.J., Duraffour S., et al. 2016. Real-time, portable genome sequencing for ebola surveillance. Nature. 530, 228–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts M., Hayes W., Hunt B.R., et al. 2004. Reducing storage requirements for biological sequence comparison. Bioinformatics. 20, 3363–3369 [DOI] [PubMed] [Google Scholar]
- Ruffalo M., LaFramboise T., and Koyutürk M. 2011. Comparative analysis of algorithms for next-generation sequencing read alignment. Bioinformatics. 27, 2790–2796 [DOI] [PubMed] [Google Scholar]
- Schleimer S., Wilkerson D.S., and Aiken A. 2003. Winnowing: Local algorithms for document fingerprinting. Proceedings of the 2003 ACM SIGMOD international conference on Management of data, San Diego, CA, pp. 76–85 [Google Scholar]
- Smith K.C. Sliding window minimum implementations. 2016. Available at: https://goo.gl/8RC54b Accessed September8, 2016
- Smith T.F., and Waterman M.S. 1981. Identification of common molecular subsequences. J Mol Biol. 147, 195–197 [DOI] [PubMed] [Google Scholar]