Abstract
4-(Methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) is the most abundant and carcinogenic tobacco-specific nitrosamine in tobacco and tobacco smoke. The major metabolic pathway for NNK is carbonyl reduction to form the (R) and (S) enantiomers of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL) which, like NNK, is a potent lung carcinogen. The goal of this study was to characterize NNAL enantiomer formation in human lung and identify the enzymes responsible for this activity. While (S)-NNAL was the major enantiomer of NNAL formed in incubations with NNK in lung cytosolic fractions, (R)-NNAL comprised ~60 and ~95% of the total NNAL formed in lung whole cell lysates and microsomes, respectively. In studies examining the role of individual recombinant cytosolic reductase enzymes in lung NNAL enantiomer formation, AKR1C1, AKR1C2, AKR1C3, AKR1C4 and CBR1 all exhibited (S)-NNAL-formation activity. To identify the microsomal enzymes responsible for (R)-NNAL formation, 28 microsomal reductase enzymes were screened for expression by real-time PCR in normal human lung. HSD17β6, HSD17β12, KDSR, NSDHL, RDH10, RDH11 and SDR16C5 were all expressed at levels ≥HSD11β1, the only previously reported microsomal reductase enzyme with NNK-reducing activity, with HSD17β12 the most highly expressed. Of these lung-expressing enzymes, only HSD17β12 exhibited activity against NNK, forming primarily (>95%) (R)-NNAL, a pattern consistent with that observed in lung microsomes. siRNA knock-down of HSD17β12 resulted in significant decreases in (R)-NNAL-formation activity in HEK293 cells. These data suggest that both cytosolic and microsomal enzymes are active against NNK and that HSD17β12 is the major active microsomal reductase that contributes to (R)-NNAL formation in human lung.
The results of these studies demonstrate that NNK, one of the most abundant and potent carcinogens in tobacco and tobacco smoke, is metabolized in the lung primarily to the (R) enantiomer of NNAL, an activity manifested primarily by HSD17β12.
Introduction
Lung cancer is the leading cause of cancer death in the USA and worldwide, causing more deaths than that of the next three most common cancers combined (colon, breast and prostate) (1). Smoking is the major risk factor for lung cancer, contributing to over 80% of lung cancer deaths, with an estimated 155870 Americans expected to die from lung cancer in 2017, accounting for ~27% of all cancer deaths (1).
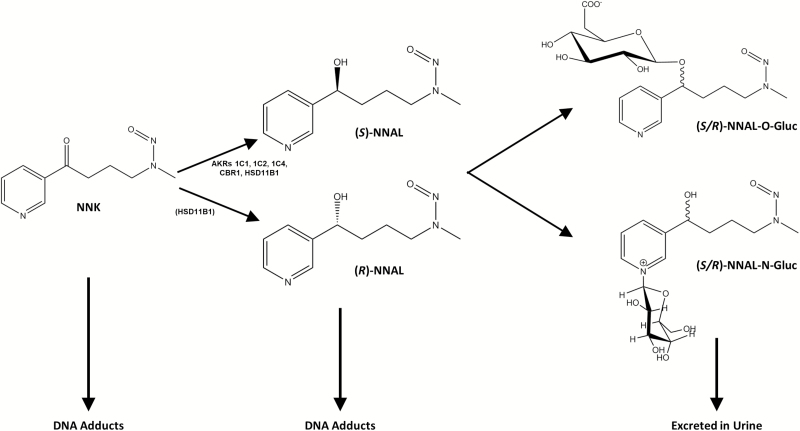
Among the carcinogens that have been found to induce lung tumors in laboratory animals, NNK is one of the most abundant (20–310 ng/cigarette) and carcinogenic, inducing mainly lung tumors in rodents regardless of the route of administration (2–4). The major metabolic pathway for NNK is reduction of its carbonyl group to form NNAL (Figure 1). Both NNK and NNAL are metabolically activated by cytochrome P450 (CYP450) enzymes via α-hydroxylation (α-methylhydroxylation or α-methylenehydroxylation) to form highly reactive metabolites that can methylate or alkylate DNA to form DNA adducts (5,6). Carbonyl reduction of NNK reduces the ketone moiety to a functional hydroxyl group that is readily glucuronidated and detoxified by phase II UDP-glucuronosyltransferases (UGTs) (5,7–17). NNAL and NNAL glucuronides (NNAL-Gluc), but not NNK, are detected in significant quantities in the urine of smokers (18).
Figure 1.
Schematic of major NNK and NNAL metabolic pathways. Enzymes known to be active in NNAL formation are shown; the lower relative (R)-NNAL forming activity of HSD11β1 is indicated by its presence in parenthesis.
NNAL formation is variable in smokers, with between 39 and 100% of the NNK dose from smokers estimated to be reduced to NNAL (18). In isolated human lung cells, Smith et al. (19) observed large interindividual differences in the amount of NNAL formed, a pattern also observed in human liver microsomes in vitro (9–11,20,21). In addition, the levels of total urinary NNAL (NNAL + NNAL-Gluc) were shown previously to be a predictor of lung cancer risk (22). This suggests that enzymes involved in NNAL formation may contribute to individual susceptibility to NNK-induced lung cancer.
NNAL is formed as (R)- and (S)-NNAL enantiomers. (S)-NNAL may be a better substrate for α-hydroxylation and better able to convert back to NNK as compared to (R)-NNAL (8,23). (S)-NNAL has also been shown to form more DNA adducts and to be more tumorigenic in rodent models than (R)-NNAL (8,24). However, (S)-NNAL has been shown to be more efficiently detoxified by glucuronidation in monkeys and likely in humans (23). Therefore, the relative contribution of each enantiomer to the carcinogenicity of NNK is still unknown.
Six known enzymes within the steroid dehydrogenase reductase (SDR) superfamily have been associated with NNK metabolism to form NNAL: 11β-hydroxysteroid dehydrogenase type 1 (HSD11β1), carbonyl reductase type 1 (CBR1) and aldo-keto reductases (AKRs) 1C1, 1C2, 1C4 and 1B10 (25–27). In these studies, five (HSD11β1, CBR1, AKR1C1, AKR1C2 and AKR1C4) were characterized by enzyme purification from human tissue fractions. Of the six SDR enzymes with NNAL-formation activity, only HSD11β1 is microsomal, but all form primarily (S)-NNAL (65% for HSD11β1, >90% for the five cytosolic enzymes). The pattern observed for CBR1 and the AKRs is consistent with that produced in vitro with human lung cytosolic fractions where (S)-NNAL was shown previously to comprise >80% of the total NNAL formed (26). Interestingly, (R)-NNAL comprises >65% of the total NNAL formed in microsomal fractions from all human tissues yet examined including >90% in human lung (26), a pattern not consistent with the preferential (S)-NNAL-formation activity observed for HSD11β1, which is the only NNK-reducing microsomal enzyme identified to date (26). This suggests that there exists at least one or more unidentified microsomal enzymes with (R)-NNAL-forming activity in human tissues.
The goal of this study was to better characterize lung NNAL formation and identify relevant NNAL-forming enzymes in lung. A major finding of this study is that HSD17β12 is the major enzyme responsible for the formation of (R)-NNAL in human lung.
Materials and methods
Analysis of reductase enzyme expression in human lung
An initial screening of the HUGO Genome Nomenclature Committee (HNGC) database [(28); www.genenames.org; retrieved July, 2015] was performed to identify members of the SDR superfamily that were previously found to be expressed in lung. Subcellular location for each of the SDR enzymes were then characterized using the UniProt database [(29); www.uniprot.org]. Enzymes known to be expressed in human lung and to reside in the endoplasmic reticulum (i.e. within the microsomal fraction of a cell) were selected for expression analysis.
Real-time polymerase chain reaction (PCR) was performed for the identified microsomal SDRs along with known NNK-reducing cytosolic enzymes (CBR1, and AKRs 1C1, 1C2, 1C4 and 1B10) as well as AKR1C3 using the TaqMan Gene Expression Assay kit (Applied Biosystems, Foster City, CA) as per the manufacturer’s protocol (see Supplementary Table 1, available at Carcinogenesis Online). Real-time PCR was performed in incubations (20 µl total volume) containing 50 ng of complementary DNA (cDNA) generated from (i) pooled human lung RNA from five adults (purchased from BioChain, Newark, CA), or (ii) five lung specimens from five separate individuals [all were Caucasian postmortem non-cancer subjects: 1 female, 4 males; obtained from Banner Sun Health Research Institute (BSHRI); Sun City, AZ] in independent reactions using the SuperScript VILO cDNA Synthesis kit (Life Technologies, Grand Island, NY). GAPDH was used as the ‘housekeeping’ gene for normalizing expression, with each sample being analyzed in quadruplicate. Enzyme expression was determined using the microsomal HSD11β1 enzyme as the referent. Microsomal enzymes with lung expression levels >1.0-fold that observed for HSD11β1 (HSD17β6, HSD17β12, KDSR, NSDHL, RDH10, RDH11 and SDR16C5) were then selected to be cloned as stable over-expressing cell lines. Expression was calculated using the delta–delta-Ct method (30).
Cloning and over-expression of microsomal reductases
Amplification of microsomal SDR cDNAs was performed after an initial reverse transcriptase reaction using the SuperScript II cDNA Synthesis kit (Life Technologies) with 2 µg of total RNA from pooled normal human lung RNA from five adults (purchased from BioChain). Total RNA was purified from the BSHRI specimens using standard protocols. PCR amplification was subsequently performed using Pfx Polymerase (Life Technologies) in a GeneAmp 9700 thermocycler (Applied Biosystems), with an initial denaturing temperature of 94°C for 2 min, 35 cycles of 94°C for 30 s, 30 s at the specific annealing temperature (see Supplementary Table 1, available at Carcinogenesis Online), and 68°C for 75 s, followed by a final cycle of 68°C for 10 min. PCR products were gel-purified using the GeneJet Gel Purification kit (Thermo Scientific, Waltham, MA), sequenced by GeneWiz (Cambridge, MA) and compared with the reference sequence in GenBank (see Supplementary Table 1, available at Carcinogenesis Online). Primers utilized for PCR amplification are listed in Supplementary Table 1, available at Carcinogenesis Online. cDNAs for verified wild-type SDRs were individually cloned into the pcDNA3.1/V5-His-TOPO vector (Life Technologies) using standard protocols using One Shot TOP10 competent Escherichia coli (Life Technologies). SDR-containing plasmids were purified using the GeneJet Plasmid MiniPrep kit (Thermo Scientific) and sequenced by GeneWiz using two vector primers (T7 and BGH) to ensure proper incorporation of the gene and removal of the stop codon for generation of the V5-His tag at the 3′-end of each cDNA. After large-scale plasmid preparations, verified SDR-containing pcDNA3.1/V5-His-TOPO vectors were transfected into the HEK293 cell line using a standard lipofectamine protocol. These vectors were also transfected and over-expressed in the MCF-7 cell line using a similar protocol.
HEK293, HEK293T, MCF-7 and IMR-90 cells were obtained between 2014 and 2017 from American Type Culture Collection (ATCC, Manassas, VA). NHF cells were a kind gift from Dr Jiyue Zhu (College of Pharmacy, Washington State University, Spokane, WA) (31). HEK293, HEK293T and MCF-7 cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; Life Technologies), IMR-90 cells were cultured in Eagle’s Minimum Essential Medium (EMEM; Life Technologies), and NHF cells were cultured in mammary epithelial cell growth medium (MEGM; Lonza, Basel, Switzerland), all with 10% fetal bovine serum and 1% penicillin–streptomycin (Life Technologies) supplemented in their respective media. The major cell lines used in this study, HEK293 and MCF-7, were authenticated by ATCC in 2017 using short-tandem repeat polymorphisms analysis. For cloning of microsomal enzymes, HEK293 and MCF-7 cells were grown in DMEM supplemented with 10% fetal bovine serum and passaged at 24 h post-transfection and subsequently grown in geneticin (700 µg/ml medium; Life Technologies) for the selection of geneticin-resistant cells containing the SDR-pcDNA3.1/V5-His-TOPO vector in a humidified incubator at 37°C in an atmosphere of 5% CO2, with the selection medium changed every 2–3 days. Cell homogenates, cytosolic and microsomal fractions were prepared through differential centrifugation as previously described (13,32). All microsomal samples were washed once with phosphate-buffered saline. Total protein concentrations in each homogenate and cellular fraction were determined using the Pierce BCA protein assay (Thermo Scientific) as per the manufacturer’s protocol.
Cell culture and transfection with siRNA
Control siRNA (sc-37007, Santa Cruz, Dallas, TX) was diluted in 1 ml serum-free DMEM to 10 nM while human HSD17β12 siRNA (ID: 108811, Ambion; Thermo Scientific) was diluted in 1 ml serum-free DMEM to different concentrations (0.1, 0.5, 1, 5 and 10 nM). Forty microlitres of HiPerFect transfection reagent (Qiagen, Hilden, Germany) was added to diluted siRNA. The mixture was incubated for 10 min at room temperature after vortexing.
Twenty-four hours prior to siRNA treatment, HEK293 cells were plated in 10 cm culture dishes. For siRNA treatment, the culture medium was removed and replaced with 9 ml of fresh serum-free DMEM. siRNA–HiPerFect complexes were added dropwise to cells, and plates was swirled gently and then incubated under normal growth conditions, with cells harvested at 24 or 72 h after transfection. HSD17β12 expression was measured by real-time PCR, using the methods described above. HSD17β12 knock-down effects were confirmed in reductase activity assays with 1 mM NNK, using liquid chromatography-mass spectrometry (LC-MS) as described below. All transfections and analyses were performed in triplicate.
Cytosolic protein induction and purification
Polyhistidine-tagged recombinant cytosolic SDRs (AKR1C1, AKR1C2, AKR1C3, AKR1C4 and CBR1) were prepared and quantified as described previously (33). Briefly, chemically competent BL21 E. coli (Thermo Scientific) were transformed with AKR/CBR1-containing expression vectors and induced by the addition of 0.5 mmol/l β-d-1-thiogalactopyranoside (Sigma-Aldrich, St, Louis, MO). Polyhistidine-tagged recombinant protein was affinity purified using a Ni-NTA resin column (Thermo Scientific), washed and ultimately eluted with increasing concentrations of imidazole (Sigma-Aldrich). Dialysis was performed against phosphate-buffered saline to ensure removal of imidazole prior to use in pharmacokinetic assays. The purity and quantity of each cytosolic protein was determined by silver staining of sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) as described previously (33). As indicated previously (33), the relative expression of the purified cytosolic proteins were 1.2 (AKR1C1), 1.0 (AKR1C3), 1.7 (AKR1C4) and 1.5 (CBR1) relative to that observed for AKR1C2 (set to 1.0).
Western blot analysis of microsomal proteins
The level of expressed microsomal SDR protein in each over-expressed cell line was determined by western blot analysis using a monoclonal V5-HRP antibody (Invitrogen; Thermo Fisher) at a 1:5000 dilution as recommended by the manufacturer. Microsomal protein (0.7–30 µg) from each SDR-containing cell line was loaded into each lane. The monoclonal Calnexin-HRP antibody (Sigma-Aldrich) at a 1:5000 dilution was used as a loading control. The intensity of V5 signal was measured using Image J (National Institutes of Health, Bethesda, MD). The relative V5-protein expression of each SDR was calculated as the mean of three independent experiments and relative to the V5 signal for the HSD11β1-over-expressing cell line. All activity assays were normalized based on the relative V5-protein expression within each respective SDR-over-expressing cell line, with the V5-protein expression equal to 1.9, 0.9, 1.3, 4.3, 0.4, 0.4 and 0.2 for cell lines over-expressing HSD11β1, HSD17β6, HSD17β12, KDSR, NSDHL, RDH10, RDH11 and SDR16C5, respectively, as compared to that observed for HSD11β1 (set to 1.0).
Human tissue fractionation
Normal human lung tissue samples from postmortem non-cancer subjects were purchased from BSHRI. The three lung samples were all from Caucasian former smokers (2 males, 1 female) aged ≥79 years who had stopped smoking >10 years before expiration. Normal human liver specimens were provided by the Tissue Procurement Facility at the H. Lee Moffitt Cancer Center (Tampa, FL) from individuals undergoing surgery for hepatocellular carcinoma as previously described (11). Lung specimens were, on average, isolated and frozen at −70°C within 3 h postmortem; liver specimens were frozen at −70°C within 2 h after surgery. All protocols involving the collection and analysis of tissue specimens from these tissue banks were approved by the institutional review boards at their respective institutions and were in accordance with assurances filed and approved by the US Department of Health and Human Services. Tissue homogenization was performed with a Qiagen TissueLyser II in 2 ml tubes with a 5-mm bead at 22 Hz for 2 min. Preparation of cytosolic and microsomal fractions was performed as described previously (32). All microsomes were washed once with Tris-buffered saline (pH 7.4).
NNK-reduction assays
The NNK-reduction assay used for the present studies was adapted from a previous method (26). Briefly, 1 µg of purified recombinant cytosolic proteins (AKR1C1, AKR1C2, AKR1C3, AKR1C4 or CBR1), microsomes from SDR-over-expressing HEK293 cells or cellular fractions from lung or liver tissue (homogenate, cytosol or microsomes) were incubated at 37°C for 60 min with NNK (0.15–20 mM) in a total volume of 20 μl containing 100 mM KPO4 buffer (pH 7.5), and NADPH-regenerating system (1.3 mM NADP+, 3.3 mM glucose-6-phosphate, 0.4 U/ml glucose-6-phosphate dehydrogenase and 3.3 mM MgCl2; Corning, New York, NY). Incubation time and enzyme protein concentrations were chosen so that reaction rates were linear. Reactions were terminated by the addition of 20 μl ice-cold methanol with 5 µl of 0.4 ppm D4-NNAL added as an internal standard, the precipitate was removed by centrifugation (10 min, 4°C, 16100×g), and the supernatant was saved for LC-MS analysis. All reagents used for NNK-reduction assays were ACS grade or higher and obtained from Sigma-Aldrich (St. Louis, MO).
The parent HEK293 cell line exhibits low levels of endogenous NNK-reduction activity to form NNAL. To account for this, equal amounts of HEK293 microsomal protein were incubated with equivalent amounts of substrate for each experiment. The amount of NNAL produced by HEK293 microsomes was then subtracted from the amount produced by SDR-over-expressing cell microsomes for each concentration of NNK examined.
LC-MS/MS analysis
All reagents used for LC-MS/MS were obtained from Sigma-Aldrich. LC separation and detection of (S)- and (R)-NNAL was achieved by loading 2.0 µl NNK-reduction assay supernatants on an Acquity Class I ultrapressure LC equipped with an autosampler (model FTN) and a Zspray electrospray ionization interface (Waters, Milford, MA). LC separation was achieved using a 150 × 2 mm Amylose-2 HPLC chiral column (Phenomenex, Torrance, CA) in series with a 0.2-μm Waters assay frit filter (2.1 mm, Waters) using a flow rate of 0.2 ml/min and a linear gradient from 50% buffer A (5 mM ammonium acetate): 50% buffer B (methanol:isopropanol, 3:1) to 100% buffer B in 20 min. The optimized MS conditions were as follows: positive daughter scan and multiple reaction monitoring mode, capillary voltage 0.6 kV, cone voltage 15 V, collision voltage 10 V, source temperature 130°C and desolvation temperature 500°C. Nitrogen was used as the desolvation and cone gas with the flow rate at 800 and 50 l/h, respectively. Ultra-pure argon was used as the collision gas at flow rate of 0.1 l/h. The dwell time for each ion was 0.5 s. The amount of (S)- and (R)-NNAL formed was determined on the basis of the ratio of NNAL to d4-NNAL after calculating the area under the curve for the NNAL and d4-NNAL peaks and quantified by comparing this ratio with a standard curve using known amounts of NNAL and d4-NNAL added to each reaction (0.1–50 µM and 0.4 ppm, respectively). All data were quantified using the MassLynx™ NT 4.1 software within the TargetLynx™ program (Waters Corp., Milford, MA). Three independent experiments were performed to determine kinetic constants for each active enzyme.
For the detection of NNAL enantiomers, the mass spectrometer was operated in the multiple reaction monitoring mode; the concentrations of both NNAL enantiomers were determined simultaneously. The ion-related parameters for NNAL were a MS transition of 210.124>180.124, with a cone voltage and collision energy at 15 and 10 V, respectively. The MS transitions and LC retention times for each enantiomer were compared to the internal d4-NNAL standard (Toronto Research Chemicals, Toronto, Ontario, Canada).
Statistical analysis
Michaelis–Menten kinetic constants were determined using Prism Version 5 software (La Jolla, CA) and verified using R Version 3.4 (Vienna, Austria). All statistical analyses were two-sided and considered significant if P < 0.05 for all tests.
Results
NNAL-formation activity in normal human lung and liver tissue fractions
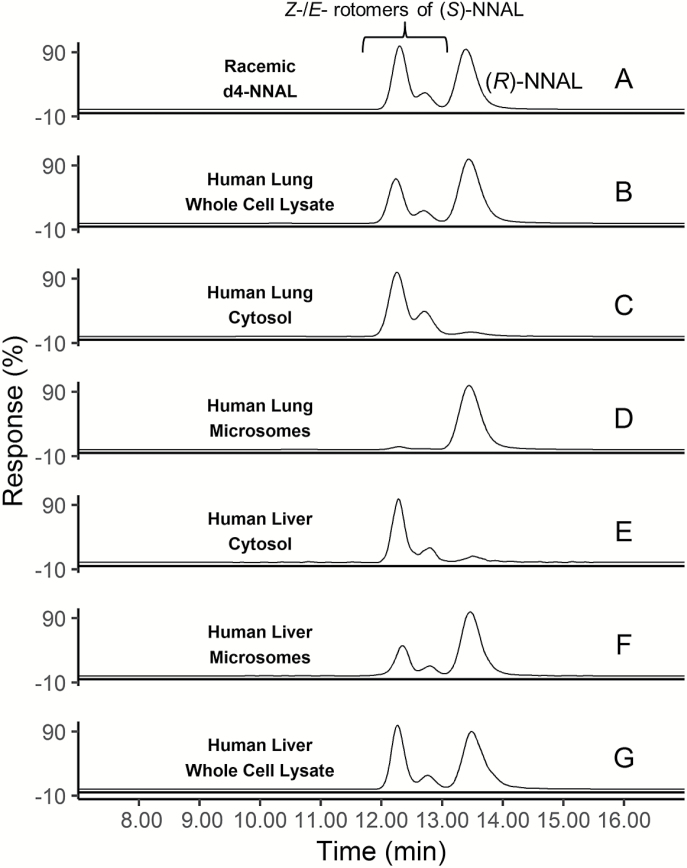
NNAL stereoisomers were separated and quantified by LC-MS/MS using a chiral HPLC column, with (S)- and (R)-NNAL peaks identified on the MS system with a positive multiple reaction monitoring mode at m/z 210→93 and confirmed using racemic d4-NNAL as the internal standard at m/z 214→97 (Figure 2A). The first two peaks were assigned as the Z- and E- rotomers of (S)-NNAL in accordance with previous studies (23,26,34), eluting at 12.25 and 12.70 min; the Z- and E-rotomers of (R)-NNAL were not separated in the LC-MS/MS method utilized in this study, which eluted as one peak at 13.38 min. As shown in representative traces of NNAL formation for human lung tissue fractions, whole cell lysates from human lung formed ~60% (R)-NNAL (Figure 2B). While the cytosolic fraction of lung tissue formed primarily (S)-NNAL (~90%, Figure 2C), the microsomal fraction formed primarily (R)-NNAL (~95%, Figure 2D). By comparison, whole cell lysates from human liver formed primarily (S)-NNAL (65%; Figure 2E). While a similar pattern was observed for the cytosolic and microsomal fractions from human liver as compared to human lung specimens (Figure 2E and F, respectively), the percentage of (R)-NNAL formed was slightly reduced in human liver microsomes (75%).
Figure 2.
Representative LC-MS traces of NNAL enantiomers formed in human lung and liver tissue fractions after incubating with 1 mM NNK. (A) Racemic d4-NNAL; (B) lung whole cell lysate; (C) lung cytosol; (D) lung microsomes; (E) liver whole cell lysate; (F) liver cytosol; (G) liver microsomes. [Z-(S)]-NNAL retention time 12.25; [E-(S)]-NNAL retention time = 12.70; [E-/Z-(R)]-NNAL retention time = 13.38 min).
SDR expression in human lung
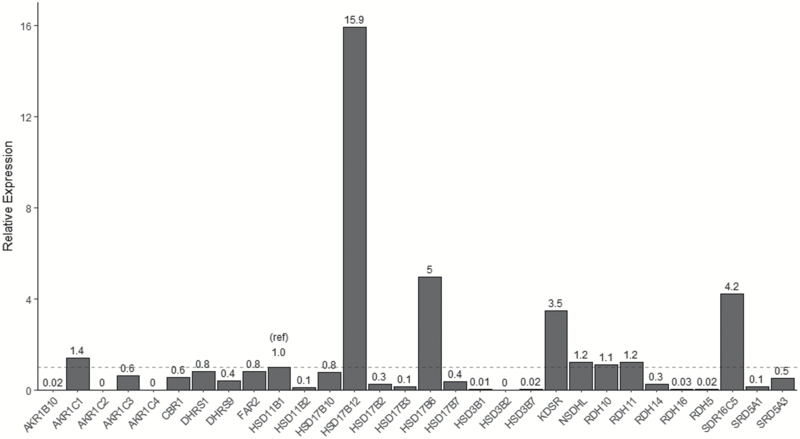
As described in the Methods, potential NNK-reducing microsomal enzymes expressed in lung were identified in silico; a total of 24 SDR enzymes previously suggested to be expressed in human lung and known to reside in the endoplasmic reticulum (i.e. microsomal fraction) were selected for a quantitative expression analysis in a sample of pooled lung RNA from five adults. These 24 microsomal enzymes, the five cytosolic enzymes previously screened for NNAL-formation activity (AKRs 1B10, 1C1, 1C2 and 1C4, and CBR1), and AKR1C3, which is the only other member of the AKR1C subfamily that was not previously examined for NNAL-formation activity, were all screened for expression in human lung. The microsomal enzymes RDH10, RDH11, NSDHL, KDSR, SDR16C5, HSD17β6 and HSD17β12 were expressed at levels that were 1.1-, 1.2-, 1.2-, 3.5-, 4.2-, 5.0- and 15.9-fold, respectively, compared to that observed for HSD11β1 in a pooled human lung RNA (Figure 3). The high level of expression of HSD17β12 relative to HSD11β1 was confirmed in total RNA purified from five lung specimens from individual subjects examined in independent reactions, with HSD17β12 exhibiting, on average, a 9.1-fold higher level of expression than HSD11β1 in these independent lung samples. While HSD11β1 was previously suggested not to play an important role in human lung microsomal NNK reductase activity (26), expression was observed for this enzyme in our pooled lung RNA sample. DHRS1, DHRS9 and FAR2, and hydroxysteroid dehydrogenases (HSDs) 11β2, 17β10, 17β2, 17β3, 17β7, 3β1 and 3β7, and RDH5, RDH14, RDH16, SDR5A1 and SDR5A3 were the other microsomal enzymes expressed in lung, but were at levels lower than that observed for HSD11β1; HSD3β2 was the only microsomal SDR that exhibited no detectable levels of expression in human lung. The cytosolic reductases, AKR1B10, AKR1C1, AKR1C3 and CBR1 exhibited expression in human lung at levels that were 0.02-, 1.4-, 0.6- and 0.6-fold that observed for HSD11β1, respectively; AKR1C2 and AKR1C4 were not detected.
Figure 3.
Expression of SDR enzymes in human lung (pooled RNA from five individuals). Values are relative to HSD11β1 (1.0 ref; dotted line), with GAPDH used as the housekeeping gene.
NNAL-formation activities of microsomal SDR enzymes
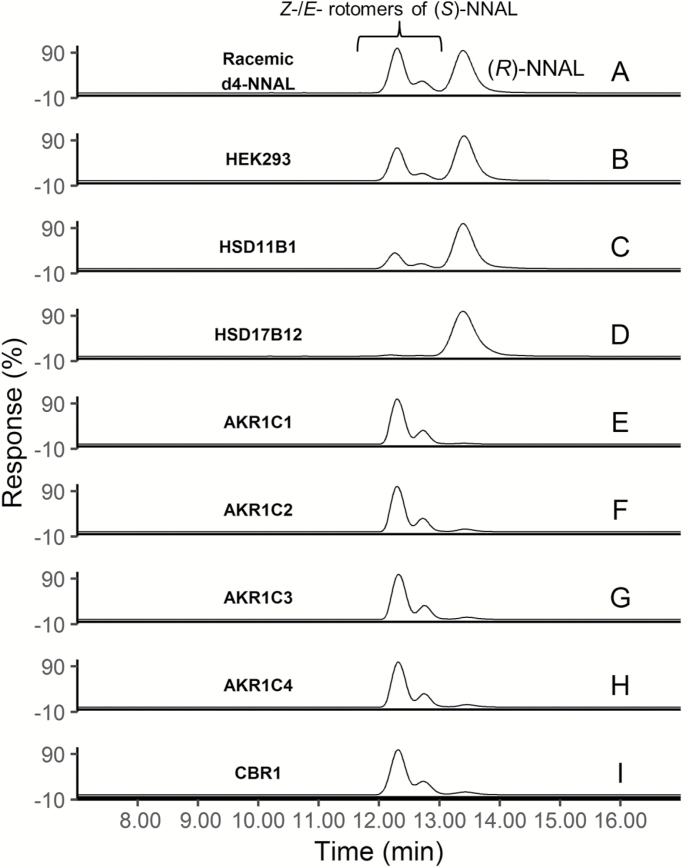
To develop an effective in vitro model for characterizing the microsomal enzymes responsible for NNAL formation, several cell lines including HEK293, HEK293T, MCF-7, IMR-90 and NHF were screened for endogenous reductase activity against NNK. Microsomes for all of the cell lines examined exhibited endogenous NNAL-formation activity, with HEK293 and MCF-7 cells exhibiting lower endogenous activities than IMR-90, NHF and HEK293T cells (results not shown). As shown for microsomes prepared from untransfected HEK293 cells, the (R) enantiomer was the major one formed in incubations with 1 mM NNK, with (R)-NNAL comprising 60% of the total NNAL (Figure 4B).
Figure 4.
Representative LC-MS traces of NNAL enantiomers from SDR over-expressing cell lines (C, D), and recombinant SDR proteins (E–I). (A) Racemic d4-NNAL; (B) HEK293 parent cell line; (C) HSD11β1-over-expressing cell microsomes; (D) HSD17β12-over-expressing cell microsomes; (E) recombinant AKR1C1; (F) recombinant AKR1C2; (G) recombinant AKR1C3; (H) recombinant AKR1C4; (I) recombinant CBR1. [Z-(S)]-NNAL retention time = 12.25; [E-(S)]-NNAL retention time = 12.70; [E-/Z-(R)]-NNAL retention time = 13.38 min.
HSD11β1 and the seven microsomal SDR enzymes determined by real-time PCR to be expressed in lung at levels similar to or greater than HSD11β1 (HSD17β6, HSD17β12, KDSR, NSDHL, RDH10, RDH11 and SDR16C5) were over-expressed in the HEK293 cell line as verified by western blot analysis (see Supplementary Figure 1, available at Carcinogenesis Online). Of the eight microsomal enzymes over-expressed in HEK293 cells, only HSD11β1 and HSD17β12 were active against NNK; HSD17β6, KDSR, NSDHL, RDH10, RDH11 and SDR16C5 were not active above background levels in the HEK293 cell line. HSD11β1 formed both (S)-NNAL (E- and Z- enantiomers eluting at 12.3–12.7 min) and (R)-NNAL (eluting at 13.4 min) as determined by LC-MS/MS (Figure 4C), with (R)-NNAL comprising 70% of the total NNAL formed. Interestingly, HSD17β12 formed almost exclusively (>90%) (R)-NNAL (Figure 4D). To confirm these data, both HSD11β1 and HSD17β12 were also over-expressed in the MCF-7 cell line; identical patterns of stereoisomer-specific formation of NNAL was observed for each enzyme in this cell line as compared to that observed in HEK293 cells (result not shown).
The kinetic constants (KM and Vmax) of NNK reduction were calculated for microsomes for HEK293 cells over-expressing each of the active recombinant enzyme (substrate concentrations ranging from 0.04 to 20 mM; Table 1), and values for HSD17β12 were normalized relative to tagged HSD11β1 expression). For (R)-NNAL formation, HSD11β1 exhibited a KM (1.1 mM) and Vmax (3.57 nmol/min/mg protein) that was similar to that observed for HSD17β12 (KM = 0.91 mM; Vmax = 6.42 nmol/min/mg protein), but resulted in a Vmax/KM that was 2.3-fold higher for HSD17β12 (as compared to HSD11β1). The KM values observed for the two enzymes for (R)-NNAL formation were similar to those observed in human lung microsomes (KM = 0.89 ± 0.3; Table 1). Consistent with the increased stereoselectivity for (R)-NNAL formation observed for HSD17β12 as compared with HSD11β1, the Vmax/KM was 5-fold higher for (R)-NNAL formation as compared to (S)-NNAL formation for HSD17β12, but was only 1.3-fold higher for HSD11β1.
Table 1.
Kinetic parameters of (S)- and (R)-NNAL formation by recombinant SDR enzymes and normal human lung cellular fractions
| (S)-NNAL | (R)-NNAL | |||||
|---|---|---|---|---|---|---|
| Enzyme |
K
M
(mM) |
V
max
(nmol/min/mg protein) |
V
max/KM (µl/min/mg protein) |
K
M
(mM) |
V
max
(nmol/min/mg protein) |
V
max/KM (µl/min/mg protein) |
| HSD11β1a | 0.80 ± 0.0 | 2.01 ± 0.5 | 2.45 ± 0.7 | 1.10 ± 0.1 | 3.57 ± 0.8 | 3.17 ± 0.7 |
| HSD17β12a | 0.45 ± 0.2 | 0.53 ± 0.1 | 1.43 ± 0.6 | 0.91 ± 1.4 | 6.42 ± 2.1 | 7.15 ± 5.2 |
| HEK293 | 1.28 ± 0.9 | 0.88 ± 0.4 | 0.64 ± 0.5 | 0.73 ± 0.1 | 1.15 ± 0.6 | 1.61 ± 1.0 |
| AKR1C1b | 1.12 ± 0.2 | 100 ± 16 | 89.9 ± 3.2 | 1.39 ± 0.1 | 1.83 ± 0.1 | 1.33 ± 0.1 |
| AKR1C2b | 1.63 ± 0.1 | 23.9 ± 0.8 | 14.7 ± 1.1 | 3.79 ± 0.9 | 1.28 ± 0.1 | 0.35 ± 0.1 |
| AKR1C3b | 1.15 ± 0.3 | 24.4 ± 0.7 | 21.9 ± 5.0 | 0.62 ± 0.4 | 1.20 ± 0.9 | 1.95 ± 0.6 |
| AKR1C4b | 0.80 ± 0.1 | 10.4 ± 0.2 | 12.5 ± 1.5 | 2.33 ± 0.4 | 0.85 ± 0.1 | 0.37 ± 0.1 |
| CBR1† | 0.70 ± 0.1 | 72.5 ± 6.6 | 106 ± 28 | 0.53 ± 0.1 | 0.42 ± 0.1 | 0.82 ± 0.2 |
| Lung microsomesc | 1.98 ± 0.8 | 0.86 ± 0.4 | 0.46 ± 0.4 | 0.89 ± 0.3 | 6.27 ± 3.9 | 7.05 ± 3.1 |
| Lung cytosolc | 1.75 ± 1.0 | 4.11 ± 3.6 | 2.35 ± 0.5 | 4.93 ± 4.0 | 1.03 ± 1.2 | 0.27 ± 0.9 |
aFor the microsomal enzymes HSD11β1 and HSD17β12, Vmax values were normalized based on the expression of HSD11β1 as described in the Materials and Methods.
bFor the cytosolic enzymes (AKRs 1C1, 1C2, 1C3 and 1C4, and CBR1), Vmax values were calculated based on ng of purified cytosolic enzyme added to the reaction as described in the Materials and Methods.
c V max values were normalized relative to total protein as described in the Materials and Methods.
NNAL-formation activities of cytosolic SDR enzymes
Previous studies by Maser et al. (25) characterized the kinetics of several cytosolic SDR enzymes responsible for the conversion of NNK to NNAL using enzyme fractions isolated from human placenta. To better examine the specific activities of these enzymes against NNK, wild-type cytosolic reductases AKR1C1, AKR1C2, AKR1C3, AKR1C4 and CBR1 were cloned, histidine-tagged and purified over nickel columns to >90% purity. Similar to that described previously (4,35) for AKR1C1, AKR1C2, AKR1C4 and CBR1, all exhibited NNAL-formation activity in this study, forming primarily (S)-NNAL (≥93%; Figure 4E, F, H and I, respectively). Previous studies examining cytosolic NNAL formation had not included an assessment of AKR1C3 (4,35); the recombinant form of this enzyme examined in the present study was active, also forming primarily (94%) (S)-NNAL (Figure 4G).
Kinetic analysis suggested that AKR1C3 exhibited comparable or higher levels of activity against NNK than AKRs 1C2 and 1C4 (Table 1). Kinetic analysis further demonstrated that AKR1C1 and CBR1 appear to be the most active cytosolic reductases, with Vmax/KM’s that were 4.1- to 8.5-fold that observed for the other AKRs (Table 1), findings that were comparable to previous studies using placenta-fractionated cytosolic enzymes (25). The KM values observed for these SDRs (ranging from 0.70 mM for CBR1 to 1.63 mM for AKR1C2) were similar to those observed in three human lung cytosolic specimens (KM = 1.75 ± 1.0 mM; Table 1). While the KM values observed for these cytosolic SDRs were comparable for (S)- versus (R)-NNAL formation, the overall Vmax/KM’s ranged from 11- (AKR1C3) to 129- (CBR1) fold in favor of (S)-NNAL formation, due primarily to large differences in Vmax values.
siRNA knock-down of HSD17β12-induced NNAL-formation activity
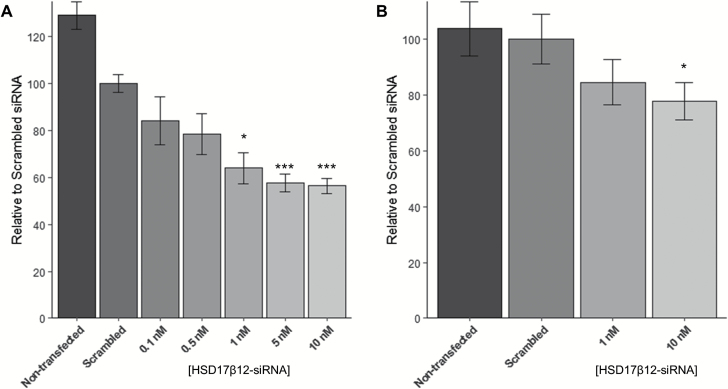
As described above, microsomes from several cell lines including HEK293 cells exhibited low levels of endogenous NNAL-formation activity, all forming almost exclusively (R)-NNAL. The Human Protein Atlas [www.proteinatlas.org (36)] was queried for cell lines regarding endogenous expression of HSD11β1 and HSD17β12; there were no cell lines without endogenous HSD17β12 expression. Since the only enzyme examined in this study that exhibited selectivity towards the (R)-NNAL enantiomer was HSD17β12, HEK293 cells were transfected with HSD17β12-siRNA to examine whether this endogenous activity was due at least in part to the endogenous expression of HSD17β12. As shown in Figure 5, HSD17β12 expression decreased in HEK293 cells in a dose-dependent manner, with maximal inhibition observed at 10 nM HSD17β12-siRNA as compared to HEK293 cells transfected with scrambled control (44% reduction, P = 0.001; Figure 5A). Cells were not viable at concentrations of >10 nM siRNA, suggesting that some level of HSD17β12 expression was necessary for cell growth. A corresponding 30% decrease in the levels of (R)-NNAL formation were observed in cells transfected with 10 nM HSD17β12-siRNA relative to cells transfected with scrambled siRNA (P = 0.048; Figure 5B).
Figure 5.
Results of siRNA knock-down of HSD17β12 in HEK293 cells. (A) RNA expression of HSD17β12 after administration of HSD17β12-siRNA; (B) NNAL formation from NNK in HEK293 cells after administration of HSD17β12-siRNA. Error bars represent standard error. *P < 0.05; ***P < 0.001.
Discussion
Reduction of the tobacco-specific nitrosamine NNK to its metabolite NNAL is a crucial step in the detoxification of this potent carcinogen. Consistent with previous studies (26), this study finds human lung tissue capable of forming both (S)-NNAL and (R)-NNAL from NNK, with the cytosolic fraction forming primarily (90%) (S)-NNAL and the microsomal fraction forming primarily (95%) (R)-NNAL. Interestingly, whole-cell lysates from human lung tissue (comprising both the cytosolic and microsomal fractions) make about 60% (R)-NNAL, which is higher than that observed for human liver (35%). These data suggest that while (S)-NNAL may be the primary enantiomer formed from NNK in human liver and is the major form targeted for whole-body excretion by glucuronidation (23,37), (R)-NNAL is the primary enantiomer formed in human lung, a major target tissue for tobacco smoke exposure.
This study is the first to identify and characterize the enantioselective reduction of NNK by recombinant microsomal enzymes. HSD11β1 was shown previously to exhibit NNAL-formation activity (38–40), but the possibility that a second microsomal enzyme capable of reducing NNK in a stereospecific manner had been previously hypothesized (26,38). Of the 24 SDR enzymes identified in silico to be microsomal and potentially expressed in human lung in this study, seven were expressed at levels similar to or higher than HSD11β1. Of these seven enzymes, only a single SDR, HSD17β12, exhibited activity against NNK. In addition, HSD17β12 formed almost exclusively (R)-NNAL, with a 19:1 ratio of (R)-NNAL:(S)-NNAL formation, higher than that observed (7:3) for HSD11β1 but was similar to that observed in human lung microsomes. This pattern of NNAL enantiomer stereoselectivity was also observed after kinetic analysis, with HSD17β12 exhibiting a relative CLint for (R)-NNAL formation that was 3.9-fold higher that observed for HSD11β1 (5-fold versus 1.3-fold). Combined with real-time PCR data demonstrating much higher expression for HSD17β12 versus HSD11β1 in lung (~16-fold), this suggests that HSD17β12 may be the principal enzyme involved in (R)-NNAL formation in the lung of smokers. A larger analysis of many tissue specimens will be necessary to better examine the association between gender, ethnicity/race or demographic factors (e.g. smoking status) on the relative expression of these enzymes. In addition, in a screening of The Human Protein Atlas (36), HSD17β12 was shown to be expressed at similar levels in human lung versus liver while HSD11β1 is expressed in liver at >20 times that observed in lung. This suggests that HSD11β1 has a more predominant role in NNAL formation in liver and is consistent with the higher level of (S)-NNAL formation observed in liver microsomes, a pattern also observed for HSD11β1. The fact that HSD17β12 and HSD11β1 were the only two microsomal SDRs with NNAL-formation activity in this study is consistent with the functional and structural overlap between carbonyl reductases such as the AKRs and HSDs, including the 11β-HSD and 17β-HSD subfamilies, as described previously (41).
In a screening of the The Human Protein Atlas (36), HSD17β12 was shown to be expressed in all cell lines queried. The fact that microsomes from all of the cell lines screened in this study exhibited primarily (>95%) (R)-NNAL-formation activity is consistent with the expression of HSD17β12 in these cell lines and its high stereoselective (R)-NNAL-formation activity. In this study, siRNA-induced knock-out of HSD17β12-dependent (R)-NNAL-formation activity in HEK293 cells is consistent with a role for HSD17β12 in (R)-NNAL formation in cells and human tissues.
A potential limitation of this study was that only SDRs with relatively high levels of expression in lung were examined for their reductase activity against NNK in vitro, with eight microsomal enzymes examined for activity. In addition to the eight tested enzymes, 10 others exhibited expression in lung by real-time PCR but at levels lower than that observed for HSD11β1. It is possible that one or more reductases with lower expression in lung could also exhibit significant NNAL-forming activity, a possibility missed in this study.
Recombinant AKRs 1C1, 1C2 and 1C4 as well as CBR1 were active against NNK to form primarily (S)-NNAL in this study, with AKR1C1 and CBR1 exhibiting the highest overall Vmax/KM of the four active cytosolic enzymes. The data from the present studies examining recombinant cytosolic SDRs for activity against NNK generally confirm the data from previous studies for cytosolic SDRs purified using fractionation techniques from human tissues (25,26,39,42,43). It is important to note that, unlike the analysis performed in this study, previous studies did not perform kinetic analysis of (R)- and (S)-NNAL formation independently, with previous studies instead analyzing racemic NNAL. Therefore, it makes any comparison between studies more difficult to interpret. However, since all previously identified cytosolic enzymes with NNAL-formation activity primarily form (S)-NNAL, one could make an approximate comparison between previous studies of cytosolic enzymes using racemic NNAL versus the present study using (S)-NNAL. The KM value for CBR1 was lower than those observed in previous studies (42), but the findings from the current study support previous studies which find that CBR1 is the primary cytosolic enzyme responsible for NNK reduction. A ~5-fold higher KM and a corresponding 4-fold higher Vmax/KM was observed for AKR1C2 in previous studies [comparing racemic NNAL versus (S)-NNAL], but the KM was exactly the same between studies for AKR1C4, while the Vmax/KM was nearly identical for both AKRs 1C1 and 1C4 (42). The KM and Vmax/KM data were also similar for the microsomal enzyme, HSD11β1, as well as for lung microsomes and lung cytosols between studies comparing racemic NNAL versus (S)-NNAL (35,39).
Although AKR1C3 was not included in previous studies (25,26,42,43), this enzyme was shown to be active against NNK in this study, exhibiting a Vmax/KM that was similar to or slightly higher than that observed for AKRs 1C2 and 1C4. Interestingly, no expression of AKRs 1C2 and 1C4 was observed in lung by real-time PCR in the present studies, suggesting that they are not important in NNAL formation in this organ. In addition, there was a low relative expression of cytosolic compared to microsomal enzymes in the present study. AKR1C1 was expressed at similar levels to HSD11β1, whereas AKR1B10, AKR1C3 and CBR1 were expressed at levels lower than HSD11β1 and there was little or no PCR amplification of AKR1C2 or AKR1C4 in human lung tissue. This information supports previous findings suggesting that lung microsomes formed NNAL more efficiently than lung cytosol (35) and supports the fact that whole cell lung lysates formed more (R)-NNAL (formed primarily by microsomal enzymes) than (S)-NNAL (formed primarily by cytosolic enzymes).
In summary, the results from this study indicate that the primary enzyme involved in (R)-NNAL formation in human lung tissue is the microsomal enzyme, HSD17β12. The stereo-selective pattern of (R)-NNAL formation by HSD17β12 is consistent with that observed in lung microsomes and with the high level of (R)-NNAL versus (S)-NNAL formation in whole cell lung lysates. Further, the present studies confirm the (S)-NNAL-formation activity of cytosolic enzymes including AKR1C1 and CBR1 and demonstrate that AKR1C3 also exhibits (S)-NNAL-formation activity. Further research is required to characterize functional variants in these enzymes that may affect NNK metabolism and overall risk for lung cancer.
Supplementary material
Supplementary materials can be found at Carcinogenesis online.
Funding
National Institutes of Health (Grant R01-ES025460 to P. Lazarus).
Acknowledgements
We would like to thank Gang Chen at Washington State University for helping to troubleshoot the LC-MS methodology used in this study, and Jiyue Zhu at Washington State University College of Pharmacy for providing NHF cells.
Conflict of Interest Statement: None declared.
Abbreviations
- AKR
aldo-keto reductase
- CBR
carbonyl reductase
- cDNA
complementary DNA
- DMEM
Dulbecco’s Modified Eagle’s Medium
- HSD
hydroxysteroid dehydrogenase
- LC-MS
liquid chromatography-mass spectrometry
- NNK
4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone
- PCR
polymerase chain reaction
- SDR
steroid dehydrogenase reductase
References
- 1. Siegel R.L., et al. (2017)Cancer statistics, 2017. CA. Cancer J. Clin., 67, 7–30. [DOI] [PubMed] [Google Scholar]
- 2. Akopyan G., et al. (2006)Understanding tobacco smoke carcinogen NNK and lung tumorigenesis. Int. J. Oncol., 29, 745–752. [PubMed] [Google Scholar]
- 3. Hecht S.S. (1999)Tobacco smoke carcinogens and lung cancer. J. Natl. Cancer Inst., 91, 1194–1210. [DOI] [PubMed] [Google Scholar]
- 4. Maser E. (2004)Significance of reductases in the detoxification of the tobacco-specific carcinogen NNK. Trends Pharmacol. Sci., 25, 235–237. [DOI] [PubMed] [Google Scholar]
- 5. Hecht S.S. (1998)Biochemistry, biology, and carcinogenicity of tobacco-specific N-nitrosamines. Chem. Res. Toxicol., 11, 559–603. [DOI] [PubMed] [Google Scholar]
- 6. Zhang S., et al. (2009)Analysis of pyridyloxobutyl and pyridylhydroxybutyl DNA adducts in extrahepatic tissues of F344 rats treated chronically with 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and enantiomers of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol. Chem. Res. Toxicol., 22, 926–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hecht S.S., et al. (1993)Metabolism of the tobacco-specific nitrosamine 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone in the patas monkey: pharmacokinetics and characterization of glucuronide metabolites. Carcinogenesis, 14, 229–236. [DOI] [PubMed] [Google Scholar]
- 8. Upadhyaya P., et al. (1999)Tumorigenicity and metabolism of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol enantiomers and metabolites in the A/J mouse. Carcinogenesis, 20, 1577–1582. [DOI] [PubMed] [Google Scholar]
- 9. Ren Q., et al. (1999)Glucuronidation of the lung carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL) by rat UDP-glucuronosyltransferase 2B1. Drug Metab. Dispos., 27, 1010–1016. [PubMed] [Google Scholar]
- 10. Ren Q., et al. (2000)O-Glucuronidation of the lung carcinogen 4-(methylnitrosamino)-1- (3-pyridyl)-1-butanol (NNAL) by human UDP-glucuronosyltransferases 2B7 and 1A9. Drug Metab. Dispos., 28, 1352–1360. [PubMed] [Google Scholar]
- 11. Wiener D., et al. (2004)Characterization of N-glucuronidation of the lung carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL) in human liver: importance of UDP-glucuronosyltransferase 1A4. Drug Metab. Dispos., 32, 72–79. [DOI] [PubMed] [Google Scholar]
- 12. Lazarus P., et al. (2005)Genotype-phenotype correlation between the polymorphic UGT2B17 gene deletion and NNAL glucuronidation activities in human liver microsomes. Pharmacogenet. Genomics, 15, 769–778. [DOI] [PubMed] [Google Scholar]
- 13. Dellinger R.W., et al. (2006)Importance of UDP-glucuronosyltransferase 1A10 (UGT1A10) in the detoxification of polycyclic aromatic hydrocarbons: decreased glucuronidative activity of the UGT1A10139Lys isoform. Drug Metab. Dispos., 34, 943–949. [DOI] [PubMed] [Google Scholar]
- 14. Chen G., et al. (2008)Glucuronidation of tobacco-specific nitrosamines by UGT2B10. Drug Metab. Dispos., 36, 824–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bushey R.T., et al. (2011)Characterization of UDP-glucuronosyltransferase 2A1 (UGT2A1) variants and their potential role in tobacco carcinogenesis. Pharmacogenet. Genomics, 21, 55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bushey R.T., et al. (2013)Importance of UDP-glucuronosyltransferases 2A2 and 2A3 in tobacco carcinogen metabolism. Drug Metab. Dispos., 41, 170–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kozlovich S., et al. (2015)Stereospecific metabolism of the tobacco-specific nitrosamine, NNAL. Chem. Res. Toxicol., 28, 2112–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Carmella S.G., et al. (1993)Metabolites of the tobacco-specific nitrosamine 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone in smokers’ urine. Cancer Res., 53, 721–724. [PubMed] [Google Scholar]
- 19. Smith G.B., et al. (1999)Biotransformation of the tobacco-specific carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) in freshly isolated human lung cells. Carcinogenesis, 20, 1809–1818. [DOI] [PubMed] [Google Scholar]
- 20. Muscat J.E., et al. (2009)Effects of menthol on tobacco smoke exposure, nicotine dependence, and NNAL glucuronidation. Cancer Epidemiol. Biomarkers Prev., 18, 35–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wiener D., et al. (2004)Correlation between UDP-glucuronosyltransferase genotypes and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone glucuronidation phenotype in human liver microsomes. Cancer Res., 64, 1190–1196. [DOI] [PubMed] [Google Scholar]
- 22. Yuan J.M., et al. (2009)Urinary levels of tobacco-specific nitrosamine metabolites in relation to lung cancer development in two prospective cohorts of cigarette smokers. Cancer Res., 69, 2990–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Upadhyaya P., et al. (2000)Formation and metabolism of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol enantiomers in vitro in mouse, rat and human tissues. Carcinogenesis, 21, 1233–1238. [PubMed] [Google Scholar]
- 24. Lao Y., et al. (2007)Formation and accumulation of pyridyloxobutyl DNA adducts in F344 rats chronically treated with 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and enantiomers of its metabolite, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol. Chem. Res. Toxicol., 20, 235–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Atalla A., et al. (2001)Characterization of enzymes participating in carbonyl reduction of 4-methylnitrosamino-1-(3-pyridyl)-1-butanone (NNK) in human placenta. Chem. Biol. Interact., 130-132, 737–748. [DOI] [PubMed] [Google Scholar]
- 26. Breyer-Pfaff U., et al. (2004)Enantioselectivity of carbonyl reduction of 4-methylnitrosamino-1-(3-pyridyl)-1-butanone by tissue fractions from human and rat and by enzymes isolated from human liver. Drug Metab. Dispos., 32, 915–922. [PubMed] [Google Scholar]
- 27. Martin H.J., et al. (2006)Purification and characterization of akr1b10 from human liver: role in carbonyl reduction of xenobiotics. Drug Metab. Dispos., 34, 464–470. [DOI] [PubMed] [Google Scholar]
- 28. Gray K.A., et al. (2015)Genenames.org: the HGNC resources in 2015. Nucleic Acids Res., 43(Database issue), D1079–D1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. The UniProt Consortium(2016)UniProt: the universal protein knowledgebase. Nucleic Acids Res, 45, D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Livak K.J., et al. (2001)Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- 31. Ma Y., et al. (2015)A combinatory strategy for detection of live CTCs using microfiltration and a new telomerase-selective adenovirus. Mol. Cancer Ther., 14, 835–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dellinger R.W., et al. (2007)Glucuronidation of PhIP and N-OH-PhIP by UDP-glucuronosyltransferase 1A10. Carcinogenesis, 28, 2412–2418. [DOI] [PubMed] [Google Scholar]
- 33. Platt A., et al. (2016)Impact of nonsynonymous single nucleotide polymorphisms on in-vitro metabolism of exemestane by hepatic cytosolic reductases. Pharmacogenet. Genomics, 26, 370–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hecht S.S., et al. (1997)Absolute configuration of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol formed metabolically from 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Carcinogenesis, 18, 1851–1854. [DOI] [PubMed] [Google Scholar]
- 35. Maser E., et al. (2000)Carbonyl reduction of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) by cytosolic enzymes in human liver and lung. Cancer Lett., 148, 135–144. [DOI] [PubMed] [Google Scholar]
- 36. Uhlen M., et al. (2015)Proteomics. Tissue-based map of the human proteome. Science, 347(6220)1260419. [DOI] [PubMed] [Google Scholar]
- 37. Carmella S.G., et al. (1999)Stereochemistry of metabolites of a tobacco-specific lung carcinogen in smokers’ urine. Cancer Res., 59, 3602–3605. [PubMed] [Google Scholar]
- 38. Finckh C., et al. (2001)Expression and NNK reducing activities of carbonyl reductase and 11beta-hydroxysteroid dehydrogenase type 1 in human lung. Chem. Biol. Interact., 130-132, 761–773. [DOI] [PubMed] [Google Scholar]
- 39. Maser E. (1998)11Beta-hydroxysteroid dehydrogenase responsible for carbonyl reduction of the tobacco-specific nitrosamine 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone in mouse lung microsomes. Cancer Res., 58, 2996–3003. [PubMed] [Google Scholar]
- 40. Soldan M., et al. (1999)Interindividual variability in the expression and NNK carbonyl reductase activity of 11beta-hydroxysteroid dehydrogenase 1 in human lung. Cancer Lett., 145, 49–56. [DOI] [PubMed] [Google Scholar]
- 41. Maser E. (1995)Xenobiotic carbonyl reduction and physiological steroid oxidoreduction. The pluripotency of several hydroxysteroid dehydrogenases. Biochem. Pharmacol., 49, 421–440. [DOI] [PubMed] [Google Scholar]
- 42. Atalla A., et al. (2000)Purification and characterization of oxidoreductases-catalyzing carbonyl reduction of the tobacco-specific nitrosamine 4-methylnitrosamino-1-(3-pyridyl)-1-butanone (NNK) in human liver cytosol. Xenobiotica, 30, 755–769. [DOI] [PubMed] [Google Scholar]
- 43. Breyer-Pfaff U., et al. (2000)High-affinity stereoselective reduction of the enantiomers of ketotifen and of ketonic nortriptyline metabolites by aldo-keto reductases from human liver. Biochem. Pharmacol., 59, 249–260. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.