Abstract
Members of the cytochrome P450 3A (CYP3A) subfamily of drug metabolizing enzymes exhibit developmental changes in expression in human liver characterized by a transition between CYP3A7 and CYP3A4 over the first few years of life. In contrast, the developmental expression of CYP3A5 is less well understood due to polymorphic expression of the enzyme in human tissues as a result of the prevalence of the CYP3A5*3 allele, which leads to alternative splicing. We further explored the expression of CYP3A5 and the impact of alternative splicing on the variability of CYP3A5 functional activity in a large bank of human prenatal liver samples (7 to 32 weeks of age postconception). The expression of normally spliced CYP3A5 mRNA in all human fetal liver samples varied 235-fold whereas CYP3A5 SV1 mRNA was only detected in fetal liver samples with at least one CYP3A5*3 allele. Formation of 1ʹ-OH midazolam (MDZ) varied 79-fold, and the ratio of 1ʹ-OH MDZ to 4-OH MDZ varied 8-fold and depended on the presence or absence of the CYP3A5*3 allele. Formation of 4-OH MDZ was significantly associated with 1ʹ-OH MDZ (r2 = 0.76, P < 0.0001) but varied (36-fold) independently of CYP3A5 genotype or expression. The substantial interindividual variability that remains even after stratification for CYP3A5 genotype suggests that factors such as environmental exposure and epigenetic alterations act in addition to genetic variation to contribute to the variability of CYP3A5 expression in human prenatal liver.
Introduction
The cytochrome P450 3A (CYP3A) subfamily in humans consists of four members, CYP3A4, CYP3A5, CYP3A7, and CYP3A43. In adults, it is qualitatively the most important subfamily of cytochrome P450 enzymes in terms of hepatic drug biotransformation. The biotransformation of more than 50% of medications used clinically is dependent, to some extent, on CYP3A4, the most abundant CYP3A isoform in the adult liver and intestine (Guengerich, 1999). CYP3A5 demonstrates distinct catalytic activity but overlapping substrate specificity relative to CYP3A4 (Williams et al., 2002). Although its contribution to overall hepatic CYP3A activity remains a matter of some debate (Hustert et al., 2001; Kuehl et al., 2001; Westlind-Johnsson et al., 2003), CYP3A5 is clearly an important driver of the metabolic clearance of tacrolimus and vincristine in individuals carrying the active CYP3A5*1 allele (Dennison et al., 2007; Birdwell et al., 2015).
CYP3A7 is widely accepted to be the major cytochrome P450 isoform expressed in human fetal liver (Lacroix et al., 1997; Stevens et al., 2003), as CYP3A4 does not appear to be expressed in fetal liver to any appreciable extent (Leeder et al., 2005; Shuster et al., 2014). However, data from several investigations indicate that CYP3A5 is expressed in a subset of fetal livers and may contribute to the metabolism of certain CYP3A substrates in a genotype-dependent manner. In previous studies, immunoreactive CYP3A5 protein was detected in approximately 50% of fetal liver microsomal samples (Hakkola et al., 2001; Stevens et al., 2003). Additionally, in a study of 54 human fetal livers, we observed that microsomal testosterone 2α- and 6β-hydroxylase activities were highly correlated (r2 = 0.859), and the correlation improved (r2 = 0.974) in fetal livers in which CYP3A5 activity was predicted to be minimal based on genotype (CYP3A5*3*3), indirectly implying the presence of functional CYP3A5 expression. In contrast, dehydroepiandrosterone (DHEA) 16α-hydroxylase activity was not affected by the CYP3A5 genotype at the concentration of substrate examined, which further suggests that fetal liver CYP3A5 may contribute to the biotransformation of some substrates but not others (Leeder et al., 2005). Given the controversy regarding the relative contribution of CYP3A5 to overall CYP3A activity (Hustert et al., 2001; Kuehl et al., 2001; Westlind-Johnsson et al., 2003), further investigation of the range of its contribution to CYP3A activity in fetal liver is warranted.
Historically, the high degree of structural similarity and overlapping substrate specificities between CYP3A4 and CYP3A5 have hampered attempts to accurately and reliably ascertain the relative contributions and activities of each isoform in the adult liver using available substrates and reagents (Stevens et al., 2003). CYP3A7 expression in fetal liver adds even more complexity for assigning CYP3A isoform-specific metabolism. One strategy that has proven useful is characterization of regiospecific patterns of metabolite formation from probe substrates. For example, Stevens et al. (2003) exploited differential patterns of DHEA 16α- and 7β-hydroxylation by CYP3A7 and CYP3A4 in conjunction with a nonlinear multivariate regression model to estimate CYP3A4 and CYP3A7 protein levels in prenatal and postnatal livers. We used published testosterone hydroxylation data generated using heterologously expressed human cytochrome P450 enzymes (Usmani et al., 2003) as the basis for selecting 2α- and 6β-hydroxylation activities for our previous analyses (Leeder et al., 2005). Similarly, CYP3A5 preferentially catalyzes the formation of 1ʹ-hydroxymidazolam (1ʹ-OH MDZ) versus 4-hydroxymidazolam (4-OH MDZ). The ratio of 1ʹ-OH MDZ to 4-OH MDZ depends on the substrate concentration and the CYP3A isoform present, with the 1ʹ-OH MDZ/4-OH MDZ ratio typically 2-fold greater for CYP3A5 than for CYP3A4 (Gorski et al., 1994; Williams et al., 2002). Furthermore, the 1ʹ-OH MDZ/4-OH MDZ ratio was lower in liver microsomes from adults that were homozygous CYP3A5*3*3 versus individuals with at least one CYP3A5*1 allele (5.9 versus 8.1, respectively) (Kuehl et al., 2001). In contrast, CYP3A7 preferentially catalyzes the formation of 4-OH MDZ (Williams et al., 2002). Therefore, the metabolic ratio of midazolam (MDZ) is considered useful for inferring the relative amounts of CYP3A enzymes present in a tissue of interest.
Our investigation characterized CYP3A5 mRNA and protein expression and functional CYP3A5 capacity in a panel of human fetal liver samples and elucidated the mechanisms responsible for the variability observed. MDZ was selected as the most appropriate phenotyping probe for this in vitro investigation, given that 1) CYP3A4 expression and activity was minimal based on mRNA expression and the pattern of hydroxylated testosterone metabolites formed (Leeder et al., 2005) and 2) CYP3A7 has reduced activity toward MDZ relative to CYP3A5 and CYP3A4, and preferentially catalyzes the formation of 4-OH MDZ over 1ʹ-OH MDZ (Gorski et al., 1994; Williams et al., 2002).
Materials and Methods
Materials and Reagents.
MDZ, glucose-6-phospate, glucose-6-phosphate dehydrogenase, β-NADP, MgCl2, and EDTA were purchased from Sigma-Aldrich (St. Louis, MO). We purchased 1ʹ-OH MDZ and 4-OH MDZ from Ultrafine Chemicals (Manchester, England). We prepared 15N3-1ʹ-OH MDZ and 15N3-4-OH MDZ as previously described by Paine et al. (1997). All other reagents were of analytic grade. Microsomes prepared from baculovirus-infected insect cells (Supersomes) expressing human CYP enzymes (CYP3A4, CYP3A5, and CYP3A7) or control vector were purchased from Corning GENTEST (Woburn, MA). Recombinant enzymes were coexpressed with human NADPH-cytochrome P450 reductase and in the case of CYP3A4 and CYP3A7, cytochrome b5. Pooled adult human liver microsomes were purchased from XenoTech (Lenexa, KS). The vials of microsomes were stored at −80°C until use.
Liver Samples.
A total of 382 anonymous prenatal liver samples with estimated ages of 7 weeks to 32 weeks postconception were obtained through two tissue retrieval programs supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development: 342 from the Central Laboratory for Human Embryology at the University of Washington (Seattle, WA) and 40 from the Brain and Tissue Bank for Developmental Disorders at the University of Maryland (Baltimore, MD). The prenatal liver samples were from fetuses with the following genders: 164 females (ages 7.7–27 weeks after conception), 173 males (ages 7–32 weeks after conception), and 45 of unknown/unavailable gender (ages 7.6–15.4 weeks after conception). No difference in the estimated age postconception was observed between the females and males (Student t test, P = 0.62). All tissues were maintained at −80°C before extraction of nucleic acids or preparation of subcellular fractions. The use of these tissues was declared nonhuman subjects research by the University of Missouri–Kansas City Pediatric Health Sciences Review Board.
Genomic DNA Isolation and Genotyping for CYP3A5 Allelic Variants.
Genomic DNA was isolated from 5–25 mg tissue using a DNeasy Tissue Kit (Qiagen, Valencia, CA). Genotyping was performed with commercially available TaqMan-based allelic discrimination assays for CYP3A5*3, CYP3A5*6, and CYP3A5*1D (Life Technologies, Grand Island, NY) and KAPA Probe Fast qPCR master mix (Kapa Biosystems, Boston, MA).
RNA Extraction and Quantitative Real-Time Polymerase Chain Reaction Analysis.
Samples of frozen liver tissue (20–30 µg) were homogenized and total RNA extracted according to the Qiagen RNeasy protocol (Qiagen) with an on-column DNase I treatment. The quality of the isolated RNA was assessed using an Experion Automated Electrophoresis Station (Bio-Rad Laboratories, Hercules, CA); and RNA quantity was determined spectrophotometrically.
One-step quantitative real-time polymerase chain reaction (qRT-PCR) to detect total CYP3A5, normally spliced CYP3A5, and exon 3B-containing CYP3A5 transcripts was performed using qScript One-Step SYBR Green or qScript One-Step Fast qRT-PCR reagents (Quanta Biosciences, Inc., Gaithersburg, MD) according to manufacturer’s recommendations with the primers in Table 1. Reactions were performed in triplicate using 15 ng total RNA. Standard curves were generated by serial dilutions of plasmids containing CYP3A5 cDNA from 10 to 107 copies from which transcript numbers were calculated by linear regression. For all qRT-PCR assays, the minimal acceptable correlation coefficient for the standard curve from a single run was 0.985. The qRT-PCR for GAPDH and PPIA (peptidylprolyl isomerase A) was performed using TaqMan Endogenous Control Reagents (Life Technologies). CYP3A5 transcript levels are expressed as relative expression/ng total RNA after correction with the geometric mean of PPIA and GAPDH.
TABLE 1.
Primers and probes for quantitation of CYP3A5 transcripts
| Primers/Probes | Sequence |
|---|---|
| For total CYP3A5 | |
| CYP3A5 Ex 1 forward | 5′-GACCTCATCCCAAATTTGGCGG |
| CYP3A5 Ex 4 reverse | 5′-CAGGGAGTTGACCTTCATACGTT |
| For normally spliced CYP3A5 | |
| CYP3A5 Ex 2/3 forward | 5′-CCTATCGTCAGGGTCTCTGGAA |
| CYP3A5 Ex 4 reverse | 5′-TGATGGCCAGCACAGGGA |
| CYP3A5 Ex 3/4 probe | 5′-[6FAM]ATGTGGGGAACGTATGAA[BHQ1] |
| For CYP3A5 SV1 | |
| CYP3A5 Ex 3B forward | 5′-GTGAGACTCTTGCTGTGTGTCACA |
| CYP3A5 Ex 3B/4 reverse | 5′-GGAGTTGACCTTCATACGTTCCC |
| CYP3A5 Ex 3B probe | 5′-[6FAM]CTGTGTGTCGTACAACT[BHQ1] |
Preparation of Fetal Liver Microsomes.
Microsomes were prepared from frozen samples of fetal liver by differential centrifugation essentially as described by Lu and Levin (1972). Frozen liver samples were placed in homogenizing buffer (∼3 ml/g liver; 50 mM Tris-HCl, pH 7.4 at 4°C, containing 150 mM KCl and 2 mM EDTA) and allowed to thaw at 4°C. Liver samples were homogenized in Potter-Elvehjem-type glass mortars with Teflon pestles utilizing a motor-driven tissue homogenizer (Caframo Model BDC-3030, Wiarton, Ontario, Canada). Nuclei and lysosomes were removed from the homogenate by low-speed centrifugation (∼800gmax for 15 minutes at 4°C). The supernatant was subjected to further centrifugation (∼12,000gmax for 20 minutes at 4°C) to remove mitochondria and the resulting supernatant fraction was subjected to ultracentrifugation (104,000 to 109,000gmax for 70 minutes at 4°C). The resulting cytosolic fraction was stored at −80°C and the microsomal pellet was removed from the centrifuge tube with a Teflon-coated spatula, and manually resuspended in 0.25 M sucrose with a tapered, Teflon pestle and low-volume glass mortar and stored at −80°C until use. Protein concentrations were determined using the Micro BCA Protein Assay kit (Pierce Chemical, Rockford, IL) using bovine serum albumin (Sigma-Aldrich) as the standard.
Preparation of Anti-CYP3A5 Antibody.
The primary antibody was raised in rabbits against the C-terminal pentapeptide of CYP3A5, Thr-Leu-Ser-Gly-Glu that had been conjugated to keyhole limpet hemocyanin (KLH) using Sulfo-SMCC chemistry via a cysteine residue added to the N-terminus. Pathogen-free New Zealand White rabbits (HsdOkd:NZW) were immunized with 1 mg of peptide-KLH antigen emulsified in Complete Freund’s Adjuvant followed by secondary immunizations on days 28, 56 and 84. Production bleeds (∼25 ml) were obtained on day 98 and day 105 followed by complete exsanguination on day 112. Antigen preparation and antibody production was carried out by Harlan Bioproducts (Indianapolis, IN).
To minimize nonspecific binding, the antipeptide antibody was affinity purified using the PinPoint Xa Protein Purification System (Promega, Madison, WI). Oligonucleotides (sense, 5′-AGC TTT GCA CCT TAA GCG GCG AAT AAG-3′ and antisense, 5′-GAT CCT TAT TCG CCG CTT AAG GTG CAA-3′) encoding the CYP3A5 pentapeptide were synthesized (Sigma-Genosys, The Woodlands, TX) and ligated into the PinPoint Xa-3 vector to allow expression of the CYP3A5 pentapeptide in Escherichia coli as the C-terminus of a biotinylated fusion protein. An affinity column was prepared by immobilization of the biotinylated fusion protein on TetraLink Tetrameric Avidin Resin (Promega). Antipeptide antibodies were allowed to bind to the fusion protein solid phase, the column was washed with >5 bed volumes of buffer, and bound antibodies were eluted with 100 mM glycine, pH 2.8. The antibody was specific for CYP3A5 and did not react with heterologously expressed CYP1A1, CYP1A2, CYP2A6, CYP2B6, CYP2C8, CYP2C9, CYP2C19, CYP2D6, CYP2E1, CYP3A4, or CYP3A7.
Immunoquantitation of Microsomal CYP3A5 Protein.
Microsomal proteins (2 µg) were separated on 4%–15% Criterion 26-well gels in running buffer (25 mM Tris, 192 mM glycine, and 0.1% w/v SDS, pH 8.3) at 200 V for 45 minutes. Proteins were transferred to nitrocellulose membranes using a Bio-Rad semidry transfer unit with transfer buffer containing 39 mM glycine, 48 mM Tris, 0.0375% SDS, and 20% methanol. After blocking overnight at room temperature on a rocker in 10 mM Tris, 150 mM NaCl, 0.2% Tween-20, pH 8.0 (TNT), containing 4% skim milk powder (M-TNT), membranes were incubated for 1 hour with affinity-purified anti-CYP3A5 antibody diluted 1:20,000-fold in M-TNT and washed with TNT (5 minutes × 6). To detect bound antibody, blots were incubated with horseradish peroxidase-conjugated donkey anti-rabbit antibody (1:50,000) in M-TNT for 30 minutes, washed (5 minutes × 6), incubated with ECL Plus chemiluminescence reagents according to the manufacturer’s directions (Amersham Biosciences, Little Chalfont, United Kingdom), and exposed on Hyperfilm ECL for 3 minutes. Films were scanned using a flat-bed scanner, and densitometric analysis of immunoreactive protein was conducted using Kodak Digital Science 1D Image Analysis Software, version 3.6 (Eastman Kodak Company, Rochester, NY). Standard curves (0.01–0.2 pmol per lane heterologously expressed CYP3A5; BD Gentest, BD Biosciences, Woburn, MA) were present on each membrane. Standard curves were linear over the range of standards, and the coefficients of determination (r2 values) ranged from 0.972 to 0.999. Values presented are the mean of duplicate determinations.
Formation of 1ʹ-OH MDZ and 4-OH MDZ by Human Fetal Liver Microsomes and Recombinant Enzymes.
Human liver microsomes (0.1 mg protein/ml) or recombinant CYP3A enzymes (2.5 pmol) were incubated at 37 ± 1°C in 0.25 ml (final volume) incubation mixtures containing potassium phosphate buffer (100 mM, pH 7.4), MgCl2 (3 mM), EDTA (1 mM), and MDZ (8 µM) at the final concentrations indicated. In the case of recombinant CYP3A5, cytochrome b5 was added to the incubation mixture at a final concentration of 10 nM. Incubation mixtures were prewarmed to 37 ± 1°C for 5 minutes in a shaking water bath, and the reactions were initiated by the addition of a NADPH-generating system consisting of NADP (1 mM), glucose-6-phosphate (5 mM), and glucose-6-phosphate dehydrogenase (1 U/ml) and incubated for 0 or 5 minutes at 37 ± 1°C before termination with 0.25 ml of ice-cold Na2CO3 (100 mM, pH 11.0). The formation of 1ʹ-OH MDZ and 4-OH MDZ was linear over the incubation period under these conditions. Samples were placed on dry ice; after freezing, samples were stored at −80°C until analysis.
On the day of analysis, terminated microsomal incubations were thawed, 50 µl of a solution containing the internal standards (15N3-1ʹ-OH MDZ and 15N3-4-OH MDZ) was added to each sample and briefly vortexed. Following the addition of 3 ml of ethyl acetate, the samples were shaken for 20 minutes on a horizontal shaker and subjected to centrifugation (3700g for 20 minutes). The supernatant was transferred to a clean test tube and evaporated to dryness under a steady stream of nitrogen. After evaporation to dryness, the extracted samples were reconstituted in 200 µl of a 50:50 mixture of acetonitrile/water and transferred to a 96-well plate. Concentrations of 1ʹ-OH MDZ and 4-OH MDZ were quantified by high-pressure liquid chromatography with tandem mass spectrometry, as previously described elsewhere (Shuster et al., 2014).
Pooled adult human liver microsomes (0.1 mg/ml) were incubated for 0 and 5 minutes, with and without the NADPH-generating system that served as positive and negative controls, respectively. All incubations were conducted in duplicate except those incubated for 0 minutes, which were performed as single determinations.
Data Analysis.
Markers of CYP3A5 expression and activity were not normally distributed across the entire set of fetal liver samples (Shapiro-Wilks test, P < 0.05). After stratification by genotype, 1ʹ-OH MDZ and 4-OH MDZ formations were normally distributed for all CYP3A5 genotypes. Measures of CYP3A5 mRNA and 1ʹ-OH MDZ/4-OH MDZ ratios were not normally distributed for samples with genotypes of CYP3A5*1*3 and CYP3A5*3*3; therefore, nonparametric statistical tests were used where measures of CYP3A5 expression or activity were not normally distributed. The Wilcoxon test was employed to compare the mean between males and females and to test for differences in CYP3A5 expression between genotype groups. Student’s t test was employed to test for differences in MDZ metabolite formation between genotype groups. Multivariate analysis with Spearman’s rho was used to identify correlations between measures of CYP3A5 expression and activity. Linear regression was used to compare 1ʹ-OH MDZ and 4-OH MDZ formation within CYP3A5 genotype groups. P < 0.05 was considered statistically significant after Bonferroni correction for multiple hypothesis testing. All statistical analyses were performed using JMP 10.0.1 (SAS Institute, Cary, NC).
Results
Expression of CYP3A5 mRNA in Fetal Liver.
We used qRT-PCR of exons 1 thru 4 in combination with SYBR green detection to measure the total CYP3A5 mRNA levels in 382 prenatal (ages 49 to 224 days postconception) livers. Relative expression of total CYP3A5 mRNA (Table 2) varied approximately 235-fold among the samples that demonstrated mRNA levels above the limit of quantification (LOQ) for the assay (n = 380, 30 molecules input/reaction before correction with GAPDH and PPIA). There was no association between the expression of total CYP3A5 mRNA with respect to postconception age (Fig. 1) or gender (not shown).
TABLE 2.
Summary of CYP3A5 expression and activity in human fetal liver
Summary statistics for samples with levels greater than the limit of quantification (LOQ) for each assay.
| Samples |
Total mRNAa |
Normally spliced mRNAa |
SV1 mRNAa |
Protein |
1ʹ-OH MDZ |
4-OH MDZ |
1ʹ-OH MDZ/4-OH MDZ |
|---|---|---|---|---|---|---|---|
| pmol/mg | pmol/min/mg | ||||||
| All Samples | |||||||
| Mean ± SD | 464 ± 514 | 486 ± 606 | 1442 ± 921 | 16.0 ± 18.0 | 115.9 ± 89.2 | 70.8 ± 37.9 | 1.75 ± 0.93 |
| Range | 14–3285 | 28–3265 | 57–5387 | 1.0–88.2 | 6.5–513 | 5.6–202.3 | 0.74–5.66 |
| Fold range | 235 | 117 | 95 | 88 | 79 | 36 | 8 |
| N (N>LOQ) | 382 (381) | 382 (361) | 382 (351) | 46 (44) | 98 (93) | 98 (89) | 98 (89) |
| CYP3A5*1*1 | |||||||
| Mean ± SD | 1398 ± 589 | 1590 ± 785 | <LOQ | 42.1 ± 39.2 | 253.1 ± 130.5 | 93.2 ± 49.0 | 2.74 ± 0.58 |
| Range | 87–2596 | 72–3265 | 22 (0) | 14.3–69.8 | 98.0–513 | 44.5–202.3 | 2.05–3.68 |
| Fold range | 30 | 45 | 4.9 | 5 | 4.5 | 1.8 | |
| N (N>LOQ) | 22 (22) | 22 (22) | 2 (2) | 9 (9) | 9 (9) | 9 (9) | |
| CYP3A5*1*3 | |||||||
| Mean ± SD | 859 ± 511 | 923 ± 547 | 950 ± 732 | 29.2 ± 23.5 | 129.6 ± 71.5 | 69.0 ± 35.6 | 2.09 ± 0.88 |
| Range | 52–3285 | 49–2549 | 98–4960 | 5.5–88.2 | 15.5–317.8 | 17.5–144.7 | 0.74–5.66 |
| Fold range | 63 | 52 | 51 | 16 | 21 | 8 | 8 |
| N (N>LOQ) | 115 (115) | 114 (114) | 115 (115) | 13 (12) | 39 (38) | 39 (36) | 39 (36) |
| CYP3A5*1*6 | |||||||
| Mean ± SD | 861 ± 562 | 1081 ± 706 | <LOQ | 24.8 ± 3.4 | 166.2 ± 92.5 | 83.9 ± 35.3 | 2.55 ± 0.98 |
| Range | 249–1909 | 281–2141 | 8 (0) | 22.4–28.7 | 23.4–320 | 20.1–121.0 | 1.74–4.44 |
| Fold range | 8 | 8 | 1.3 | 14 | 6 | 2.5 | |
| N (N>LOQ) | 8 (8) | 8 (8) | 3 (3) | 7 (7) | 7 (6) | 7 (6) | |
| CYP3A5*3*3 | |||||||
| Mean ± SD | 158 ± 76 | 108 ± 58 | 1731 ± 894 | 6.7 ± 4.1 | 60.4 ± 30.9 | 65.3 ± 37.7 | 1.03 ± 0.45 |
| Range | 14–463 | 28–401 | 147–5388 | 1.0–15.6 | 12.2–151.5 | 5.6–175.2 | 0.77–3.5 |
| Fold range | 33 | 14 | 37 | 16 | 12 | 32 | 4.5 |
| N (N>LOQ) | 224 (222) | 224 (205) | 224 (224) | 25 (24) | 37 (34) | 37 (34) | 37 (34) |
| CYP3A5*3*6 | |||||||
| Mean ± SD | 387 ± 176 | 444 ± 316 | 765 ± 698 | 11.5 ± 5.2 | 69.7 ± 71.7 | 66.9 ± 36.5 | 1.23 ± 0.34 |
| Range | 116–681 | 110–1082 | 57–2313 | 7.8–15.1 | 6.5–170 | 35.7–107.1 | 0.91–1.59 |
| Fold range | 6 | 10 | 41 | 1.9 | 26 | 3 | 1.7 |
| N (N>LOQ) | 12 (12) | 12 (12) | 12 (12) | 2 (2) | 5(4) | 5 (3) | 5 (3) |
| CYP3A5*6*6 | |||||||
| Value (n = 1) | 325 | 113 | Not detected | 9.8 | 81.7 | 55.8 | 1.46 |
Relative expression corrected for PPIA and GAPDH.
Fig. 1.
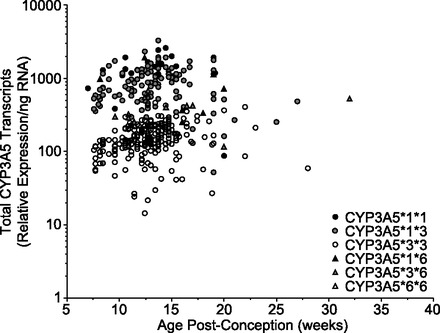
Expression of total CYP3A5 mRNA in human fetal liver relative to postconception age with stratification by genotype. Data are presented as relative corrected expression per nanogram total RNA.
The interindividual variability in the expression of total CYP3A5 mRNA was largely dependent upon CYP3A5 genotype (Fig. 2A; Table 2). CYP3A5 mRNA expression was highest in individuals with CYP3A5*1*1 genotypes. Prenatal livers that were homozygous for CYP3A5*3*3 exhibited the lowest relative expression of total CYP3A5 mRNA with 33-fold variability, and the expression levels were much lower than CYP3A5 expression levels in CYP3A5*1*1 samples (corrected P < 0.0001). Intermediate expression was observed in prenatal livers that were heterozygous for CYP3A5*1*3, with expression significantly higher than that in CYP3A5*3*3 livers (corrected P < 0.0001). No difference in relative expression of total CYP3A5 mRNA was observed in liver samples derived from individuals homozygous for CYP3A5*6*6 or heterozygous for CYP3A5*1*6 versus CYP3A5*1*1 (P > 0.05). Variability in relative total CYP3A5 mRNA expression ranged from 6-fold in livers from individuals with the CYP3A5*3*6 genotype (n = 12) to 63-fold in livers from individuals with the CYP3A5*1*3 genotype (n = 115).
Fig. 2.
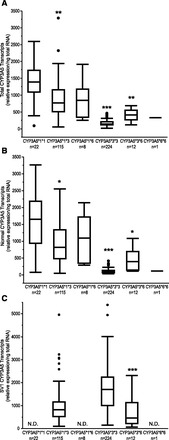
Expression of (A) CYP3A5 total transcripts, (B) normally spliced CYP3A5 transcripts, and (C) CYP3A5 SV1 transcripts with respect to CYP3A5 genotype in human fetal liver. CYP3A5 SV1 transcripts were below the limit of detection in all livers with genotypes of CYP3A5*1*1, CYP3A5*1*6, and CYP3A5*6*6. Statistically significant differences in average gene expression from the CYP3A5*1*1 genotype group for total (A) and normally spliced (B) transcripts are indicated with *P < 0.05, **P < 0.01, or ***P < 0.001. Statistically significant differences in average gene expression from the CYP3A5*1*3 genotype group for SV1 transcripts (C) are indicated with ***P < 0.001.
Although the CYP3A5 genotype accounts for a large proportion of the observed variability in CYP3A5 mRNA expression, extensive variability was observed among samples with the same genotype. To further characterize the interindividual variability in CYP3A5 mRNA expression, particularly in livers from individuals heterozygous for CYP3A5*1*3 and homozygous for CYP3A5*3*3, two assays were designed to specifically detect the relative levels of normally spliced CYP3A5 transcripts and the SV1 splice variant. Normally spliced CYP3A5 mRNA was present above the limit of detection (LOD) (80 molecules input/reaction) in all fetal liver samples, and was above the LOQ for the assay (220 molecules input/reaction) in 361 of 381 prenatal livers (Table 2). All samples in which normally spliced CYP3A5 mRNA was below the LOQ were genotyped as CYP3A5*3*3. The relative expression levels of normally spliced CYP3A5 were highly correlated with total CYP3A5 transcripts levels (Spearman’s ρ = 0.9, corrected P < 0.0001) across the entire cohort. Similar to the values for total CYP3A5 transcripts, the levels of the normally spliced CYP3A5 transcript varied highly at 117-fold across all samples, but were strongly associated with the presence of the CYP3A5*3 allele (Fig. 2B, corrected P < 0.0001).
In contrast to normally spliced transcripts, which were detected in all samples regardless of CYP3A5 genotype, the CYP3A5 SV1 transcript was only detectable above the LOD for the assay (50 molecules input/reaction) in samples carrying at least one copy of the CYP3A5*3 allele (Fig. 2C). As observed with total CYP3A5 and normally spliced CYP3A5 transcripts, expression of the CYP3A5 SV1 splice variant exhibited 95-fold variability among 350 prenatal livers containing at least one CYP3A5*3 allele, with expression above the LOQ for the assay (360 molecules input/reaction; Table 2). In prenatal liver samples from individuals heterozygous for CYP3A5*3, the CYP3A5 SV1 transcript expression varied 87-fold; in samples with two variant alleles, CYP3A5 SV1 transcript expression varied 37-fold. Across the entire set of prenatal livers, there was only a poor correlation between expression of the SV1 transcript and normally spliced or total CYP3A5 mRNA expression (Spearman’s ρ = −0.09 and 0.01, respectively).
Expression of CYP3A5 Immunoreactive Protein in Fetal Liver.
Immunoreactive CYP3A5 protein in microsomal fractions was determined for a subset of fetal livers (n = 46). Immunoreactive protein was detected in 44 of 46 samples, including 24 of 25 samples that were genotyped as CYP3A5*3/*3 (Fig. 3; Table 2). Two samples with CYP3A5*1*3 and CYP3A5*3*3 genotypes were below the LOD. Among samples with quantifiable protein, immunoreactive CYP3A5 protein levels varied nearly 90-fold (Table 2). Positive correlations between CYP3A5 protein and normally spliced and total CYP3A5 mRNA were observed (Spearman’s ρ = 0.77, corrected P < 0.001 and 0.66, corrected P < 0.001, respectively).
Fig. 3.
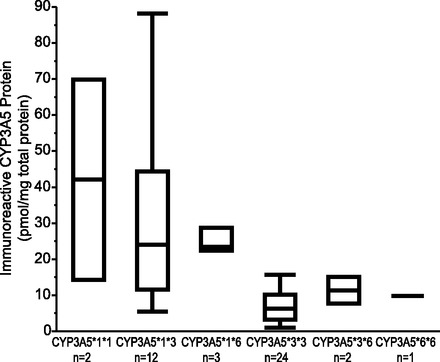
Expression of CYP3A5 immunoreactive protein/mg microsomal protein with respect to CYP3A5 genotype in human fetal livers.
Midazolam Hydroxylation.
Previous studies have suggested that MDZ metabolite formation may be used to infer the identity of CYP3A isoforms present, particularly in adult human liver microsomes to determine the presence or absence of CYP3A5 (Gorski et al., 1994; Kuehl et al., 2001; Williams et al., 2002). We confirmed that ratios of MDZ metabolite formation can also be useful in discriminating the presence or absence of CYP3A5 in fetal human liver microsomes. Formation and metabolic ratios of 1ʹ-OH MDZ and 4-OH MDZ for recombinant CYP3A4, CYP3A5, and CYP3A7 enzymes using the experimental conditions employed for fetal human liver microsomes are shown in Table 3. The 1ʹ-OH MDZ/4-OH MDZ ratio observed for recombinant CYP3A7 of 1.03 is 4- and 8-fold lower than those observed for recombinant CYP3A4 and recombinant CYP3A5, respectively. To further understand the effects of CYP3A5/CYP3A7 mixtures on the MDZ metabolic ratio, incubations combining recombinant CYP3A5 and CYP3A7 were performed in which the relative amount of CYP3A5 in the reaction was increased from 10% to 90%. A shift in the 1ʹ-OH MDZ/4-OH MDZ is observed in reactions containing as little as 10% CYP3A5 making up the total CYP3A input (Table 3). These data indicate that the 1ʹ-OH MDZ/4-OH MDZ ratio is also useful as a phenotyping probe for samples predicted to express CYP3A7 and CYP3A5 in the absence of CYP3A4.
TABLE 3.
In vitro activity of recombinant CYP3A enzymes toward midazolam
| rCYP3A5 | rCYP3A7 | rCYP3A4 | Midazolam Metabolite Formation |
1ʹ-OH MDZ/4-OH MDZ Ratio | |
|---|---|---|---|---|---|
| 1ʹ-OH MDZ | 4-OH MDZ | ||||
| pmol | pmol/min/pmol P450 | ||||
| — | — | 2.5 | 4.20 | 1.04 | 4.05 |
| 2.5 | — | — | 10.16 | 1.15 | 8.85 |
| — | 2.5 | — | 0.72 | 0.70 | 1.03 |
| 2.25 | 0.25 | — | 6.86 | 0.91 | 7.52 |
| 1.875 | 0.625 | — | 6.62 | 1.00 | 6.65 |
| 1.25 | 1.25 | — | 4.20 | 0.85 | 4.96 |
| 0.625 | 1.875 | — | 2.38 | 0.74 | 3.22 |
| 0.25 | 2.25 | — | 1.20 | 0.66 | 1.82 |
Ninety-eight liver samples representing all CYP3A5 genotypes and mRNA expression levels were selected for microsome isolation and MDZ hydroxylation assays. Formation of 1ʹ-OH MDZ was detected in 93 of 98 samples of microsomes prepared from prenatal livers, and 4-OH MDZ formation was detected in 89 of 98 samples (Table 2). Five samples, where both metabolites were below the LOQ, were genotyped as CYP3A5*3*3 (n = 3), CYP3A5*1*3 (n = 1), or CYP3A5*3*6 (n = 1). In four samples genotyped as CYP3A5*1*3 (n = 2), CYP3A5*1*6 (n = 1), and CYP3A5*3*6 (n = 1), 1ʹ-OH MDZ formation was observed, but the absence of detectable 4-OH MDZ precluded calculation of the 1ʹ-OH MDZ/4-OH MDZ ratio.
On average, 1ʹ-OH MDZ formation was 115.9 ± 89.2 pmol/min/mg, and 4-OH MDZ formation was 70.8 ± 37.9 pmol/min/mg. Formation of 1ʹ-OH MDZ and 4-OH MDZ varied 79-fold and 36-fold, respectively. As observed with the results from the mRNA studies, 1ʹ-OH MDZ formation was significantly associated with CYP3A5 genotype (corrected P < 0.0001), whereas formation of 4-OH MDZ was not dependent upon CYP3A5 genotype (uncorrected P = 0.24, Table 2). A modest association was observed between the formation rates of 1ʹ-OH MDZ and 4-OH MDZ among all samples analyzed (r2 = 0.55, P < 0.0001), and stronger relationships were observed when the samples were stratified by CYP3A5 genotype (Fig. 4A, CYP3A5*1*1 samples, r2=0.91, P < 0.0001; and CYP3A5*3*3 samples, r2 = 0.98, P < 0.0001). There was no difference in metabolite formation between liver microsomes prepared from livers from males and females or with respect to age (P > 0.05; results not shown).
Fig. 4.
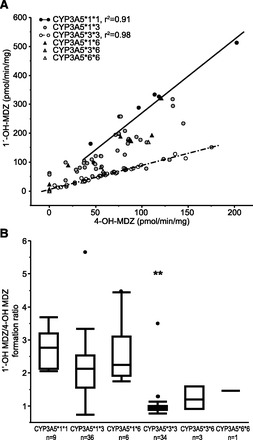
Correlation between 4-OH MDZ formation and 1ʹ-OH MDZ formation in human fetal liver microsomes (A) and 1ʹ-OH MDZ/4-OH MDZ formation ratios in human fetal liver microsomes based on CYP3A5 genotype (B). Data points in A represent the mean of duplicate determinations. The metabolic ratio (B) from the CYP3A5*3*3 genotype group was statistically significantly different from the CYP3A5*1*1 genotype group (corrected P < 0.01, indicated with **).
On average, the 1ʹ-OH MDZ/4-OH MDZ ratio was 1.75 ± 0.93 and varied 8-fold across the 89 fetal human liver microsome samples with quantifiable midazolam metabolite formation. The ratio of 1ʹ-OH MDZ/4-OH MDZ was dependent upon CYP3A5 genotype (Fig. 4B; Table 2) with livers from individuals with the CYP3A5*1*1 genotype exhibiting the highest ratios (2.75 ± 0.58) and CYP3A5*3*3 genotypes having the lowest ratios (1.03 ± 0.45). Three samples exhibited higher 1ʹ-OH MDZ/4-OH MDZ than might be predicted from their CYP3A5 genotype. In the cases with genotypes of CYP3A5*1*3 and CYP3A5*1*6, both samples had markedly reduced levels of 4-OH MDZ formation whereas the sample that was the CYP3A5*3*3 genotype had reduced levels of both metabolites. No difference in 1ʹ-OH MDZ/4-OH MDZ ratios was observed between males and females or with respect to postconception age (P > 0.05; results not shown).
Relationships among mRNA, protein, and catalytic activity for CYP3A5 in human fetal liver are shown in Table 4. We observed modest yet statistically significant relationships among 1ʹ-OH MDZ formation and normally spliced (Spearman’s ρ = 0.43, corrected P = 0.002) and total (Spearman’s ρ = 0.52, corrected P < 0.0001) CYP3A5 mRNA transcripts. The formation of 1ʹ-OH MDZ was also correlated with CYP3A5 immunoreactive protein (Spearman’s ρ = 0.6, corrected P = 0.001), but the formation of 4-OH MDZ was not statistically significantly associated with CYP3A5 mRNA or protein expression in fetal liver.
TABLE 4.
Spearman’s correlation coefficients with corrected P values
| CYP3A5 Expression/Activity | Normal splice CYP3A5 mRNA | CYP3A5 SV1 mRNA | CYP3A5 protein | 1ʹ-OH MDZ | 4-OH MDZ | 1ʹ-OH MDZ/ 4-OH MDZ | CYP3A7 mRNA |
|---|---|---|---|---|---|---|---|
| Total CYP3A5 mRNA | 0.9002a | 0.012 | 0.6577a | 0.5235a | 0.0761 | 0.656a | 0.377a |
| P < 0.0001a | P > 0.05 | P < 0.0001a | P < 0.0001a | P > 0.05 | P < 0.0001a | P > 0.0001a | |
| Normal spliced CYP3A5 mRNA | −0.0895 | 0.7678a | 0.4341a | −0.0513 | 0.6604a | 0.39a | |
| P > 0.05 | P < 0.0001a | P = 0.002a | P > 0.05 | P < 0.0001a | P = 0.0002a | ||
| CYP3A5 SV1 mRNA | −0.1431 | −0.1505 | −0.1169 | −0.1083 | 0.1628 | ||
| P > 0.05 | P > 0.05 | P > 0.05 | P > 0.05 | P > 0.05 | |||
| CYP3A5 protein | 0.5956a | 0.2585 | 0.6896a | 0.3131 | |||
| P = 0.0011a | P > 0.05 | P < 0.0001a | P > 0.05 | ||||
| 1ʹ-OH MDZ | 0.7576 | 0.5737a | 0.3465 | ||||
| P < 0.0001a | P < 0.0001a | P > 0.05 | |||||
| 4-OH MDZ | −0.0879 | 0.1309 | |||||
| P > 0.05 | P > 0.05 | ||||||
| 1ʹ-OH MDZ/ | 0.3092 | ||||||
| 4-OH MDZ | P > 0.05 |
Indicates statistically significant after Bonferroni correction for multiple hypothesis testing.
Discussion
Genetic variation in CYP3A5 and overlapping substrate specificity has hampered efforts to characterize the ontogeny and the relative contribution of CYP3A5 to the developmental changes in hepatic metabolism in humans. In adults, the CYP3A5*3 allele is presently considered to be the major determinant of polymorphic CYP3A5 expression in the liver and intestine. However, the overall contribution of CYP3A5 to total CYP3A activity in vivo in those individuals who express the enzyme remains a matter of debate and likely depends on the substrate under consideration. CYP3A5*3 alleles are defined by rs776746 in intron 3, which creates a cryptic splice site (Kuehl et al., 2001) and leads to formation of the SV1 splice variant that includes exon 3B from the intron 3 sequences. Transcripts that include exon 3B introduce a premature stop codon and are subject to nonsense-mediated decay, leading to reduced total CYP3A5 mRNA levels overall (Kuehl et al., 2001; Busi and Cresteil, 2005). However, splicing is leaky, and previous studies in adult (Kuehl et al., 2001) and fetal (Stevens et al., 2003) livers have observed expression of CYP3A5 protein and catalytic activity in livers with CYP3A5*3*3 genotype, albeit at very low levels.
Information theory analysis of the CYP3A5 gene splice donor and acceptor sites provides a partial explanation for the leaky alternative splicing observed in individuals who carry the CYP3A5*3 allele. The CYP3A5*3 mutation (rs776746, 6986 A>G) introduces a splice acceptor site with information content (Ri) of 8.2 bits that is 236 base pairs (bp) upstream of the canonical exon 4 acceptor site that remains unchanged with an Ri of 4.6 bits (Rogan et al., 2003). The difference in information content (3.6 bits) predicts that the CYP3A5*3 exon 3B acceptor is 12-fold stronger than the canonical exon 4 acceptor. Predictions based on information theory would suggest that in individuals homozygous for CYP3A5*3*3 approximately 8% (1 in 12) of all CYP3A5 transcripts produced would be normally spliced between exons 3 and 4. Likewise, CYP3A5*1*3 heterozygous individuals would produce approximately 54% normally spliced transcripts (6.5 in 12). In the cohort of fetal livers in this study, the average proportion of normally spliced CYP3A5 transcripts is 6% (range: 2%–20%) and 50% (range: 4%–85%) in CYP3A5*3 homozygous and heterozygous livers, respectively. These observations are in agreement with the approximations predicted computationally with information theory-based models of splice site donors and acceptors. In addition, Busi and Cresteil (2005) reported that normally spliced CYP3A5 transcripts have a half-life that is approximately 1.7-fold longer than SV1 transcripts from CYP3A5*3 alleles. The reduced stability of SV1 transcripts contributes to the overall reduction in total CYP3A5 transcripts observed in livers with CYP3A5*1*3 and CYP3A5*3*3 genotypes. However, considerable interindividual variability was observed in the relative levels of normal and alternatively spliced CYP3A5 transcripts in human fetal livers, suggesting that additional factors, such as tissue quality, environmental exposure, or epigenetic alterations, may contribute to alternative splicing and transcript stability, which ultimately lead to variable levels of functional CYP3A5 protein.
The CYP3A5*1 allele is in linkage disequilibrium with CYP3A7*2 (Rodriguez-Antona et al., 2005). The CYP3A7*2 (1226 C>G, rs2257401) variant produces the CYP3A7.2 enzyme with a T409R substitution which has higher 16α DHEA hydroxylase activity than the wild-type CYP3A7.1. Therefore, the CYP3A7*2/CYP3A5*1 haplotype is proposed to result in a higher metabolic turnover of exogenous and endogenous substances in fetuses carrying one or more copies of this haplotype as a result of the expression of CYP3A5 and higher catalytic activity of CYP3A7.2. The consequences of this higher metabolic capacity for the developing fetus are not yet clear and likely depend on the intrauterine environment, which can be influenced by a multitude of factors including maternal drug exposure/metabolism and maternal nutritional status. For example, antenatal corticosteroids are administered to women with preterm labor to improve respiratory outcomes in neonates. Corticosteroids are metabolized by the CYP3A family of enzymes, and it is conceivable that higher turnover as a result of the CYP3A7*2/CYP3A5*1 haplotype could reduce the efficacy of antenatal corticosteroids at preventing neonatal respiratory distress syndrome (RDS). In an investigation of the impact of polymorphisms in drug-metabolizing enzymes on the outcome of RDS after administration of maternal betamethasone, Haas et al. (2012) showed that maternal CYP3A5 activity score (calculated from genotype) and fetal CYP3A7*1E genotype were predictors of neonatal RDS. Although statistically significant associations with RDS were not observed for the fetal CYP3A5*1 and CYP3A7*2 alleles individually in this study, the investigators did not consider haplotypes across the CYP3A7/CYP3A5 genes, which may reveal an association. Additional studies are required to clarify the consequences of the CYP3A7*2/CYP3A5*1 haplotype for the developing child for this and other maternally administered compounds.
Stevens et al. (2003) investigated the ontogeny of the CYP3A enzymes in a bank of 212 human prenatal and pediatric liver samples (ages: 8 weeks of gestation through 18 years) and reported low but stable CYP3A5 protein expression throughout prenatal and postnatal development. CYP3A5 genotypes were not determined for their bank of human liver samples, so it is not clear whether the low levels of expression were a reflection of genetic variability or influenced by other factors (i.e., ontogeny, sample quality). Although overall immunoreactive CYP3A5 protein levels in a subset of fetal livers in this study were higher than that reported by Stevens et al., our results are consistent with the observation that CYP3A5 expression and catalytic activity do not vary with postconception age in fetal liver. In fetal livers with at least one CYP3A5*1 allele, CYP3A5 protein levels (28.31 ± 22.84 pmol/mg) were similar to average CYP3A5 protein levels reported for adult human liver microsomes in multiple studies (Kuehl et al., 2001; Lin et al., 2002; Dennison et al., 2007; Ohtsuki et al., 2012; Achour et al., 2014), suggesting that CYP3A5 expression may not change dramatically during development. However, given the high degree of variability observed between and within studies of CYP3A5 expression and that many studies did not account for CYP3A5 genotype, developmental changes in hepatic CYP3A5 expression may have been obscured. Therefore, a developmental change in CYP3A5 expression between prenatal and postnatal human livers cannot be dismissed entirely. Additional studies that account for genetic variation are required to comprehensively investigate the expression and catalytic activity of CYP3A5 between prenatal and postnatal human livers, particularly late in gestation and in the neonatal period when the most dynamic changes in CYP3A expression and activity occur.
In adults, the relative contribution of CYP3A5 to overall CYP3A catalytic activity has been a subject of debate. Here we report the expression and catalytic activity of CYP3A5 in human fetal liver. Relative to the CYP3A7 levels we previously reported (Leeder et al., 2005), the levels of immunoreactive CYP3A5 protein are low, comprising <1% to 20% of total immunoreactive CYP3A protein, and are related to CYP3A5 genotype. Although it is expressed at low levels relative to CYP3A7 in human fetal liver, we have reported the apparent contribution of CYP3A5 to the in vitro metabolism of testosterone (Leeder et al., 2005) and MDZ (present study) with human fetal liver microsomes. However, CYP3A4 expression has been shown to increase during the late second and third trimesters (Lacroix et al., 1997; Stevens et al., 2003). Given the paucity of liver samples from donors in the third trimester in this study, we may be overestimating the role of CYP3A5-mediated biotransformation. Nevertheless, as in human adult liver, the relative contribution of CYP3A5 expression to the overall metabolic capacity of human fetal liver is most likely substrate dependent.
In conclusion, we further explored the expression of CYP3A5 and the impact of alternative splicing on the variability of CYP3A5 functional activity in the context of fetal hepatic drug biotransformation. Here we report the mRNA expression, protein levels, and catalytic activity of CYP3A5 in a large cohort of prenatal human livers (7 to 32 weeks of age after conception). The data reported here demonstrate a high level of variability in CYP3A5 expression and catalytic activity, even after stratification by genotype. This interindividual variability is more pronounced in measures of CYP3A5 RNA versus catalytic activity. The substantial interindividual variability that remains even after stratification for CYP3A5 genotype suggests that factors such as environmental exposure and epigenetic alterations act in addition to genetic variation to contribute to variability of CYP3A5 expression in human prenatal livers.
Acknowledgments
The authors thank Kenda Worsfold for her technical expertise.
Abbreviations
- CYP3A
cytochrome P450 3A isoform
- DHEA
dehydroepiandrosterone
- LOD
limit of detection
- LOQ
limit of quantification
- MDZ
midazolam
- 1ʹ-OH MDZ
1ʹ-hydroxymidazolam
- PPIA
peptidylprolyl isomerase A
- qRT-PCR
quantitative real-time polymerase chain reaction
- RDS
respiratory distress syndrome
- TNT
Tris, NaCl, Tween-20
Authorship Contributions
Participated in research design: Vyhlidal, Pearce, Thummel, Leeder.
Conducted experiments: Vyhlidal, Pearce, Gaedigk, Calamia, Shuster.
Performed data analysis: Vyhlidal, Pearce.
Wrote or contributed to writing of the manuscript: Vyhlidal, Pearce, Gaedigk, Shuster, Thummel, Leeder.
Footnotes
References
- Achour B, Russell MR, Barber J, Rostami-Hodjegan A. (2014) Simultaneous quantification of the abundance of several cytochrome P450 and uridine 5′-diphospho-glucuronosyltransferase enzymes in human liver microsomes using multiplexed targeted proteomics. Drug Metab Dispos 42:500–510. [DOI] [PubMed] [Google Scholar]
- Birdwell KA, Decker B, Barbarino JM, Peterson JF, Stein CM, Sadee W, Wang D, Vinks AA, He Y, Swen JJ, et al. (2015) Clinical pharmacogenetics implementation consortium (CPIC) guidelines for CYP3A5 genotype and tacrolimus dosing. Clin Pharmacol Ther DOI: 10.1002/cpt.113 [published ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busi F, Cresteil T. (2005) CYP3A5 mRNA degradation by nonsense-mediated mRNA decay. Mol Pharmacol 68:808–815. [DOI] [PubMed] [Google Scholar]
- Dennison JB, Jones DR, Renbarger JL, Hall SD. (2007) Effect of CYP3A5 expression on vincristine metabolism with human liver microsomes. J Pharmacol Exp Ther 321:553–563. [DOI] [PubMed] [Google Scholar]
- Gorski JC, Hall SD, Jones DR, VandenBranden M, Wrighton SA. (1994) Regioselective biotransformation of midazolam by members of the human cytochrome P450 3A (CYP3A) subfamily. Biochem Pharmacol 47:1643–1653. [DOI] [PubMed] [Google Scholar]
- Guengerich FP. (1999) Cytochrome P-450 3A4: regulation and role in drug metabolism. Annu Rev Pharmacol Toxicol 39:1–17. [DOI] [PubMed] [Google Scholar]
- Haas DM, Lehmann AS, Skaar T, Philips S, McCormick CL, Beagle K, Hebbring SJ, Dantzer J, Li L, Jung J. (2012) The impact of drug metabolizing enzyme polymorphisms on outcomes after antenatal corticosteroid use. Am J Obstet Gynecol 206:447.e17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakkola J, Raunio H, Purkunen R, Saarikoski S, Vähäkangas K, Pelkonen O, Edwards RJ, Boobis AR, Pasanen M. (2001) Cytochrome P450 3A expression in the human fetal liver: evidence that CYP3A5 is expressed in only a limited number of fetal livers. Biol Neonate 80:193–201. [DOI] [PubMed] [Google Scholar]
- Hustert E, Haberl M, Burk O, Wolbold R, He YQ, Klein K, Nuessler AC, Neuhaus P, Klattig J, Eiselt R, et al. (2001) The genetic determinants of the CYP3A5 polymorphism. Pharmacogenetics 11:773–779. [DOI] [PubMed] [Google Scholar]
- Kuehl P, Zhang J, Lin Y, Lamba J, Assem M, Schuetz J, Watkins PB, Daly A, Wrighton SA, Hall SD, et al. (2001) Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat Genet 27:383–391. [DOI] [PubMed] [Google Scholar]
- Lacroix D, Sonnier M, Moncion A, Cheron G, Cresteil T. (1997) Expression of CYP3A in the human liver—evidence that the shift between CYP3A7 and CYP3A4 occurs immediately after birth. Eur J Biochem 247:625–634. [DOI] [PubMed] [Google Scholar]
- Leeder JS, Gaedigk R, Marcucci KA, Gaedigk A, Vyhlidal CA, Schindel BP, Pearce RE. (2005) Variability of CYP3A7 expression in human fetal liver. J Pharmacol Exp Ther 314:626–635. [DOI] [PubMed] [Google Scholar]
- Lin YS, Dowling AL, Quigley SD, Farin FM, Zhang J, Lamba J, Schuetz EG, Thummel KE. (2002) Co-regulation of CYP3A4 and CYP3A5 and contribution to hepatic and intestinal midazolam metabolism. Mol Pharmacol 62:162–172. [DOI] [PubMed] [Google Scholar]
- Lu AY, Levin W. (1972) Partial purification of cytochromes P-450 and P-448 from rat liver microsomes. Biochem Biophys Res Commun 46:1334–1339. [DOI] [PubMed] [Google Scholar]
- Ohtsuki S, Schaefer O, Kawakami H, Inoue T, Liehner S, Saito A, Ishiguro N, Kishimoto W, Ludwig-Schwellinger E, Ebner T, et al. (2012) Simultaneous absolute protein quantification of transporters, cytochromes P450, and UDP-glucuronosyltransferases as a novel approach for the characterization of individual human liver: comparison with mRNA levels and activities. Drug Metab Dispos 40:83–92. [DOI] [PubMed] [Google Scholar]
- Paine MF, Khalighi M, Fisher JM, Shen DD, Kunze KL, Marsh CL, Perkins JD, Thummel KE. (1997) Characterization of interintestinal and intraintestinal variations in human CYP3A-dependent metabolism. J Pharmacol Exp Ther 283:1552–1562. [PubMed] [Google Scholar]
- Rodríguez-Antona C, Jande M, Rane A, Ingelman-Sundberg M. (2005) Identification and phenotype characterization of two CYP3A haplotypes causing different enzymatic capacity in fetal livers. Clin Pharmacol Ther 77:259–270. [DOI] [PubMed] [Google Scholar]
- Rogan PK, Svojanovsky S, Leeder JS. (2003) Information theory-based analysis of CYP2C19, CYP2D6 and CYP3A5 splicing mutations. Pharmacogenetics 13:207–218. [DOI] [PubMed] [Google Scholar]
- Shuster DL, Risler LJ, Prasad B, Calamia JC, Voellinger JL, Kelly EJ, Unadkat JD, Hebert MF, Shen DD, Thummel KE, et al. (2014) Identification of CYP3A7 for glyburide metabolism in human fetal livers. Biochem Pharmacol 92:690–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens JC, Hines RN, Gu C, Koukouritaki SB, Manro JR, Tandler PJ, Zaya MJ. (2003) Developmental expression of the major human hepatic CYP3A enzymes. J Pharmacol Exp Ther 307:573–582. [DOI] [PubMed] [Google Scholar]
- Usmani KA, Rose RL, Hodgson E. (2003) Inhibition and activation of the human liver microsomal and human cytochrome P450 3A4 metabolism of testosterone by deployment-related chemicals. Drug Metab Dispos 31:384–391. [DOI] [PubMed] [Google Scholar]
- Westlind-Johnsson A, Malmebo S, Johansson A, Otter C, Andersson TB, Johansson I, Edwards RJ, Boobis AR, Ingelman-Sundberg M. (2003) Comparative analysis of CYP3A expression in human liver suggests only a minor role for CYP3A5 in drug metabolism. Drug Metab Dispos 31:755–761. [DOI] [PubMed] [Google Scholar]
- Williams JA, Ring BJ, Cantrell VE, Jones DR, Eckstein J, Ruterbories K, Hamman MA, Hall SD, Wrighton SA. (2002) Comparative metabolic capabilities of CYP3A4, CYP3A5, and CYP3A7. Drug Metab Dispos 30:883–891. [DOI] [PubMed] [Google Scholar]


