Abstract
Esculetin (6,7-dihydroxycoumarin, ET) and 4-methylesculetin (6,7-dihydroxy-4-methylcoumarin, 4-ME) are typical coumarin derivatives that are attracting considerable attention because of their wide spectrum of biologic activities, but their metabolism remains unknown. This study aimed to elucidate the in vitro UDP-glucuronosyltransferase (UGT) metabolism characteristics of ET and 4-ME. 7-O-monoglucuronide esculetin (ET-G) and 7-O-monoglucuronide 4-methylesculetin (4-ME-G) were identified by liquid chromatography-mass spectrometry (LC-MS) and 1H-nuclear magnetic resonance (1HNMR) when ET or 4-ME was incubated with human liver (HLM) in the presence of UDP-glucuronic acid. Screening assays with 12 human expressed UGTs demonstrated that the formations of ET-G and 4-ME-G were almost exclusively catalyzed by UGT1A6 and UGT1A9. Phenylbutazone and carvacrol (UGT1A6 and UGT1A9 chemical inhibitors, respectively) at different concentrations (50, 100, and 200 μM) significantly inhibited the formation of glucuronidates of ET and 4-ME in HLM, UGT1A6, and UGT1A9 when the concentrations of ET and 4-ME ranged from 10 to 300 μM (P < 0.05). Clearance rates of ET in HLM, HIM, UGT1A6, and UGT1A9 were 0.54, 0.16, 0.69, and 0.14 ml/min/mg, respectively. Corresponding clearance rates values of 4-ME were 0.59, 0.03, 0.14, and 0.04 ml/min/mg, respectively. In conclusion, 7-O-monoglucuronidation by UGT1A6 and UGT1A9 was the predominant UGT metabolic pathway for both ET and 4-ME in vitro. The liver is probably the major contributor to the glucuronidation metabolism of ET and 4-ME. ET showed more rapid metabolism than 4-ME in glucuronidation.
Introduction
Coumarins (2H-1-benzopyran-2-one) and their derivatives are potentially useful leading compounds for the development of new chemical entities because of their wide spectrum of biologic activities, such as anti-inflammatory, anticoagulant, anticancer, anti-HIV, antiviral, antifungal, antioxidant, anti-Alzheimer, and neuroprotective properties (Venugopala et al., 2013). From 2008 to 2011, around 42 patent publications on biologically active coumarin derivatives emerged, suggesting that they are a promising source of new candidate drugs for multiple diseases (Kontogiorgis et al., 2012). However, the real mechanism of action of coumarins and their derivatives, as well as their pharmacokinetic properties, is unclear, which may explain why only a few new coumarins have been advanced to the clinical trial stage. Thus, studies on their pharmacokinetic profiles in humans may provide important information on their discovery and development.
Esculetin (6,7-dihydroxycoumarin, ET) is a derivative of coumarin that is present in various plants used as folk medicines, such as Fraxinus rhynchophylla, Rehmanniae glutinosa, and Artemisia capillaries (Prabakaran and Ashokkumar, 2013). ET is a proven antioxidant that protects hamster lung fibroblasts from lipid peroxidation, protein carbonylation, and DNA damage induced by hydrogen peroxide (Paya et al., 1992; Lin et al., 2000; Kim et al., 2008). ET has also been shown to promote analgesia (Tubaro et al., 1988), immunomodulatory function (Leung et al., 2005), and the apoptosis of various tumor cells by inhibiting signaling pathways or inducing apoptotic pathways (Park et al., 2008; Kok et al., 2009). In addition, ET protects DNA against oxidative stress (Kaneko et al., 2003); inhibits the synthesis of leukotriene B4, thromboxane B2 (Hoult et al., 1994), and platelet aggregation (Okada et al., 1995); hinders the growth of human leukemia cells; and prevents the production of IL-6 and IL-8 (Hu et al., 2009). The chemopreventive activity of ET has further been demonstrated in benzo[α]pyrene induced lung carcinogenesis in mice (Jay R et al., 2013). 4-Methylesculetin (6,7-dihydroxy-4-methylcoumarin, 4-ME) is a synthetic derivative of coumarin, also displays anti-inflammatory, potent reactive oxygen species scavenging, and neuroprotective properties. As a metal-chelating agent, 4-ME can be a promising candidate in the management of Alzheimer’s disease (Shirole et al., 2013). A recent study has shown the significant antigenotoxic effect of 4-ME in mouse cells, suggesting that it might be beneficial for cancer prevention (Edson and Rafael, 2012).
Most coumarins are poorly bioavailable (2 to 6%) because of extensive metabolism and rapid urine excretion, although they are completely absorbed after oral administration (Lake, 1999). The important pathway of coumarin metabolism is 7-hydroxylation mediated by hepatic cytochrome P450 (CYP) 2A6, and then further extensively conjugated with d-glucuronic acid and/or sulfate at 7-OH (Egan et al., 1990). ET and 4-ME are containing two hydroxyl groups at 6-C and 7-C, respectively, indicating that UDP-glucuronosyltransferase (UGT) conjugation is probably the main factor affecting their bioavailability, biologic activities, and toxicity in vivo. Glucuronidation is the primary phase II conjugation reaction that accounts for more than 35% of all phase II drug metabolites (Kiang et al., 2005). Many UGT isoforms (UGTs) have had broad and overlapping substrate specificities; and human UGTs are tissue specific because of their differences in expression (Liang et al., 2010). Identification of human UGTs responsible for the metabolism of ET and 4-ME can provide pivotal information to their pharmacokinetic properties.
Accordingly, this study aimed to elucidate the in vitro glucuronidation characteristics of ET and 4-ME using human microsomes and expressed UGTs. Glucuronidation metabolites of ET and 4-ME in human liver microsomes (HLM) have been identified by chromatography-mass spectrometry (LC-MS) and 1H-nuclear magnetic resonance (1HNMR). Glucuronidation pathways of ET and 4-ME were identified and determined by using incubation with a panel of 12 expressed recombinant UGTs (1A1, 1A3, 1A4, 1A6, 1A7, 1A8, 1A9, 1A10, 2B4, 2B7, 2B15, and 2B17) and HLM in the presence of specific inhibitors of UGTs. Enzyme kinetics of ET and 4-ME in HLM, human intestinal microsomes (HIM) and UGTs were also analyzed.
Materials and Methods
Chemicals and Reagents.
Esculetin (ET), 4-methylesculetin (4-ME), and propiophenone (internal standard, IS) (purity >98%, HPLC grade, confirmed by LC-MS) were purchased from Chengdu Mansite Pharmaceutical Co. Ltd. (Chengdu, China). Expressed human UGT isoforms (Supersomes Enzymes), pooled human liver/intestinal microsomes (HLM and HIM), and rat liver microsomes (RLM) were purchased from BD Biosciences (Woburn, MA). Uridine diphosphate glucuronic acid (UDPGA), alamethicin, magnesium chloride, d-saccharic-1,4-lactone monohydrate, and β-glucuronidase (Type HP-2 from helix pomatia) were purchased from Sigma-Aldrich (St. Louis, MO). All other chemicals and solvents were analytical grade or better.
Identification of Glucuronidation of ET and 4-ME.
The incubation mixture (200 μl) contained HLM (0.5 mg/ml), magnesium chloride (0.88 mM), alamethicin (0.022 mg/ml), saccharolactone (4.4 mM), substrates (80 μM), UDPGA (3.5 mM), and 50 mM potassium phosphate buffer (pH 7.4). After 20 minutes of incubation at 37°C, the reaction was terminated by the addition of 100 µl of 94% acetonitrile 6% glacial acetic acid containing 90 µM propiophenone as an internal standard, followed by centrifugation at 17,900 g for 30 minutes to obtain the supernatant for LC-MS analysis. Control incubation without UDPGA or without substrates or without microsomes was performed to ensure that the metabolites produced were microsome and UDPGA dependent. A quadrupole-time of flight tandem mass spectrometer (Bruker Daltonics, Billerica, MA) with an Agilent 1260 high-pressure liquid chromatography (HPLC) (Agilent Technologies, Santa Clara, CA) method was used to analyze the molecular weight of the glucuronides of ET and 4-ME. The glucuronides of ET and 4-ME were separated using Agilent ZORBAX SB-C18 column (4.6×150 mm, 5 µm). The mobile phase B was 100% acetonitrile, where the mobile phase A was 100% aqueous buffer (2.0 mM CH3COONH4, pH 6.8) with a flow rate of 1 ml/min. The gradient elution method was as follows: 0 to 4 minutes, 5 to 35% B, 4 to 8 minutes, 35 to 80% B, 8 to 10 minutes, 80% B, 10 to 12 minutes, 80 to 5% B. The detection wavelength was 350 nm. The injection volume was 200 µl. Ionization was achieved using electrospray ionization in the positive mode at a capillary voltage of +4500 V. The temperature of the dry heater was maintained at 200°C, and the nebulizer voltage was set at 1.5 bars. The dry gas was set at a flow rate of 8.0 l/min. The MS/MS spectra were produced by collision-induced dissociation of the selected precursor ions. Data were collected and analyzed using Bruker Daltonics software (version 4.0, Bruker).
To confirm that the metabolites are glucuronide compounds, 200 µl of glucuronidation incubation mixture containing glucuronide and aglycone was extracted three times with dichloromethane (sample/dichloromethane=2:5, v/v) to remove aglycone. The extracted aqueous sample was subsequently divided into two equal portions, one portion was analyzed after hydrolysis by β-glucuronidase (40 U/ml) at 37°C for 60 minutes, and the other portion was analyzed by ultra high-pressure liquid chromatography (UHPLC) directly.
Biosynthesis Metabolites and 1HNMR Spectrometry.
The glucuronidation metabolites of ET and 4-ME were biosynthesized and purified for structure elucidation and quantitative analysis. Enzymatic biosynthesis of ET-G and 4-ME-G was conducted using rat liver microsomes (RLM), because they resemble HLM in the metabolism of ET and 4-ME. In brief, 800 μM ET or 4-ME was, respectively, incubated with RLM (3 mg/ml), 5 mM UDPGA, 5 mM MgCl2, 25 μg/ml alamethicin, 10 mM d-saccharic acid 1,4-lactone and 50 mM potassium phosphate buffer solution (pH 7.4) in 10 ml final incubation for 6 hours at 37°C. The reaction was terminated by the addition of 2 ml of methanol in an ice bath. The supernatant was collected after being centrifuged at 17,900 g for 30 minutes. Then the supernatant was separated by Agilent 1200 HPLC (Agilent Technologies) equipped with an Agilent 1200 system controller, four G1310A LC pumps, a G1329B auto injector, a G1314B UV detector, and Agilent ZORBAXSB-C18 column (4.6×150 mm, 5 μm). The mobile phase consisted of methanol (A) and 0.1% (v/v) formic acid (B) with a linear gradient from initially 10 to 60% A over 11 minutes, then constantly 60% A, 11 to 15 minutes. The flow rate was 1 ml/min. The purity was greater than 99% for both metabolites by using HPLC-diode array detection. The glucuronic acid substitution positions of ET and 4-ME were determined by 1HNMR. All experiments were recorded on Bruker AVANCE III system (Bruker Daltonics). The purified metabolites of ET-G (1.6 mg) and 4-ME-G (1.3 mg) were stored at –20°C before dissolving in deuterated acetone (Sigma-Aldrich) for 1HNMR analysis. Chemical shifts are given on a δ scale and referenced to tetramethylsilane at 0 ppm for 1HNMR (500 MHz).
Assay with Expressed UGTs.
Human expressed UGTs (Supersomes Enzymes) are prepared from baculovirus-transfected insect cells with very high levels of catalytic activities (typically sixfold higher than an average HLM sample). This is ideal for identifying the metabolic pathways of drugs, screening high-throughput drug interactions, studying slowly metabolized chemicals, or manufacturing large-scale production of metabolites for structural identification (FDA, http://www.fda.gov/cder). The glucuronidation of ET and 4-ME were measured in reaction mixtures containing expressed human UGT1A1, 1A3, 1A4, 1A6, 1A7, 1A8, 1A9, 1A10, 2B4, 2B7, 2B15, and 2B17. The incubations were conducted as shown above for the HLM study. Three substrate concentrations (10, 30, and 100 μM) were performed in this study. All assays were conducted at 37°C for 30 minutes with the final protein concentration in range of ≈0.25∼1 mg/ml based on catalytic activities of individual expressed UGTs. All samples were pretreated as described above for HLM study and analysis by using UHPLC.
Chemical Inhibition Studies.
The glucuronidation activities of ET and 4-ME in microsomes (HLM and HIM) and expressed UGTs (UGT1A6 and UGT1A9) were determined in the absence or presence of phenylbutazone (UGT1A6 inhibitor) or carvacrol (UGT1A9 inhibitor). Glucuronidation of ET and 4-ME by HLM and UGT isoforms were measured at three concentrations of 10, 100, and 300 μM and were incubated in the absence or presence of phenylbutazone or carvacrol at low (50 μM), medium (100 μM), and high (200 μM) concentrations (final concentration in range of ≈0.013∼0.053 mg protein/ml based on optimization of the reaction). All protein concentrations were 0.5 mg/ml; incubation time for ET and 4-ME in microsomes and UGTs ranged from 15 to 60 minutes.
UHPLC Analysis of ET and 4-ME and Their Glucuronides.
The UHPLC condition used to quantify glucuronides of ET and 4-ME were as follows. Agilent 1290 UHPLC system coupled a photodiode array detector and the ChemStation software program (Agilent) was used with a SB C18 column (3.0×100 mm, 1.8 μm). The mobile phase B was 100% acetonitrile, where the mobile phase A was 100% aqueous buffer (2.0 mM CH3COONH4, pH 6.8) with a flow rate of 0.3 ml/min. The gradient program was as follows: 0 to 2 minutes, 5 to 35% B, 2.0 to 4.0 minutes, 35 to 80% B, 4.0 to 5 minutes, 80% B, 5.0 to 6.0 minutes, 80 to 5% B. The detection wavelength was 350 nm for ET, 4-ME, and their respective glucuronides and 250 nm for propiophenone (IS). The injection volume was 10 μl. The test linear response range for ET and 4-ME was 1.5625 to 800 μM.
Enzymes Kinetic Studies.
For estimating kinetic parameters, serial concentrations of ET or 4-ME were incubated with different enzymes (UGT1A6, UGT1A9, HLM, or HIM). The kinetic studies were conducted using microsomes or Supersomes at concentrations in the range of 0.013 to 0.053 mg/ml based on optimization of the reaction. Rates of the glucuronidation of ET and 4-ME by microsomes and expressed UGTs were expressed as amounts of glucuronides formed per milligram protein per minute (nmol/mg/min). Kinetic parameters were then obtained according to the profile of Eadie-Hofstee plots (Liu et al., 2007). If the Eadie-Hofstee plot was linear, Formation rates (V) of glucuronides at different substrate concentrations (C) were fit to the standard Michaelis-Menten equation:
| (1) |
where Km is the Michaelis-Menten constant and Vmax is the maximum rate of glucuronidation.
If Eadie-Hofstee plots showed characteristic profiles of atypical kinetic (autoactivation and biphasic kinetics) (Wang et al., 2006), the data from these atypical profiles were fit to eqs. 2 and 3, using the ADAPT II program (D’Argenio and Schumitzky, 1997). To confirm the best-fit model, the model candidates were discriminated using the minimum Akaike’s information criterion value (Yamaoka et al., 1978), and the rule of parsimony was employed.
The following eq. 2 describes enzyme reactions with auto activation:
| (2) |
where Vmax-0 is the intrinsic enzyme activity and Vmax-d is maximal induction of enzyme activity. R is the rate of enzyme activity induction, C is concentration of substrate, and Km is concentration of substrate needed to achieve 50% of (Vmax-0+Vmax-d).
The following eq. 3 describes enzyme reactions with biphasic kinetics:
| (3) |
where Vmax1 is the maximum enzyme velocity of the high-affinity phase, Vmax2 is the maximum velocity of the low-affinity phase, Km1 is concentration of substrate to achieve half of Vmax1 for high-affinity phase, and Km2 is concentration of substrate to achieve half of Vmax2 for low-affinity phase.
When Eadie-Hofstee plots revealed substrate inhibition kinetics, the reaction rate (V) were fit to eq. 4:
| (4) |
where C is the substrate concentration, V is the initial reaction rate, Vmax1 is the maximum enzyme velocity, Km1 is the substrate concentration required to achieve 50% of Vmax, and Ksi is the substrate inhibition constant.
Statistical Analysis.
One-way analyses of variance with or without Tukey-Kramer multiple comparison (post hoc) tests were used to evaluate statistical difference. Differences were considered significant when P < 0.05.
Results
Identification of Metabolites of ET and 4-ME.
LC-MS analysis showed that only mono-glucuronides were formed in HLM incubations with 80 μM ET or 4-ME in the presence of UDPGA. Retention times of ET, its metabolite (ET-G), and IS were 4.41, 4.13, and 5.11 minutes, respectively (Fig. 1A). The peak eluting at 4.59 and 4.39 minutes corresponded with 4-ME and its metabolite (4-ME-G) (Fig. 1B).
Fig. 1.
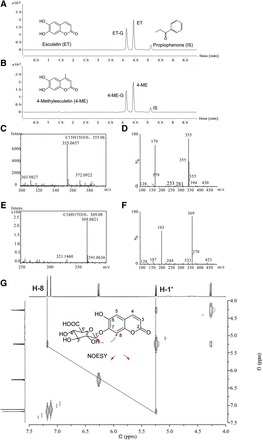
Identification of glucuronides of ET and 4-ME. UHPLC, HRMS, and MS2 scans were used to separate and identify ET, 4-ME, and their glucuronides in the experimental samples. 1HNMR was conducted to elucidate the glycosylation site of ET and 4-ME. (A and B) Chromatogram of ET, 4-ME and their glucuronides, as well as internal standard (IS), respectively. About 80 μM ET (or 4-ME) was incubated with HLM (final concentration = 0.5 mg/ml) in a phase II reaction system at 37°C for 30 minutes. (C and E) HRMS scans for glucuronides of ET and 4-ME, respectively. (D and F) MS2 scan for glucuronides of ET and 4-ME, respectively. Analysis of 1HNMR data also revealed that the added moiety belonged to glucuronic acid (Supplemental Figs. 1–4). The verified location of glucuronic acid at 7-OH position of ET was based on nuclear overhauser effect spectroscopy correlation between Glu-H-1′ (δH 5.24, 1H, d, J = 7.0 Hz) and H-8 (δH 7.18, 1H, s) (G).
The molecular formula of ET-G was established to be C15H14O10 through high-resolution mass spectrometry (HRMS) (m/z = 355.0657 [M+H]+, calculated for 355.0659) (Fig. 1C), which indicated that a C6H8O6 (m/z = 176) moiety was added by comparing the molecular formula of ET C9H7O4 (m/z = 179) (Fig. 1D). The MS2 spectrum of ET-G provided predominant characteristic fragment ions at m/z 179, which confirmed the pseudo-molecule ion [M+H]+ of ET. Inspection of 1HNMR spectroscopic data suggested that the moiety belongs to glucuronic acid, revealing that a proton of OH in ET was substituted by glucuronic acid (Supplemental Figs. 1 and 3). The substitution position of the proton of 7-OH can be confirmed by the nuclear overhauser effect spectroscopy [correlation between Glu-H-1′ (δH 5.24, 1H, d, J = 7.0 Hz) and H-8 (δH 7.18, 1H, s)] (Fig. 1G).
The molecular formula of 4-ME-G was established to be C16H16O10 through HRMS (m/z = 369.0821 [M+H]+, calculated for 369.0816), which also indicated that a C6H8O6 moiety was added by comparing the molecular formula of 4-ME C10H9O4 (m/z = 193) (Fig. 1E). In addition, the MS2 spectrum of 4-ME-G provided predominant characteristic fragment ions at m/z 193, which confirmed the pseudo-molecule ion [M+H]+ of 4-ME (Fig. 1F). Inspection of 1HNMR spectroscopic data also suggested that the moiety belongs to glucuronic acid, revealing that a proton of OH in 4-ME was substituted by glucuronic acid (Supplemental Figs. 2 and 4). The substitution position of the proton of 7-OH can be confirmed by the disappearance of broad singlet for 7-OH at δH 9.09 (1H, s) (Supplemental Figs. 2 and 4).
Main UGTs Responsible for the Glucuronidation of ET and 4-ME In Vitro.
Twelve expressed human UGTs including UGT1A1, 1A3, 1A4, 1A6, 1A7, 1A8, 1A9, 1A10, 2B4, 2B7, 2B15, and 2B17, were used to catalyze the glucuronidation of ET and 4-ME and to identify UGTs involved in their glucuronidation metabolism. Substrates of ET and 4-ME ranged from 10 to 100 μM in this experiment. We found that UGT1A6 produced the most rapid glucuronidation rates of ET (1.49 ± 0.12 to 2.98 ± 0.15 nmol/mg/min), followed by UGT1A9 (0.97 ± 0.06 to 1.81 ± 0.19 nmol/mg/min) (Fig. 2A). UGT1A7 and UGT2B15 also generated a detectable metabolite of ET (ET-G). Similarly, UGT1A6 produced the most rapid glucuronidation rates of 4-ME (4.10 ± 0.14 to 12.06 ± 0.51 nmol/mg/min) followed by UGT1A9 (2.46 ± 0.20 to 2.62 ± 0.05 nmol/mg/min) (Fig. 2B). UGT1A7 and UGT1A1 also generated a detectable metabolite of 4-ME (4-ME-G).
Fig. 2.
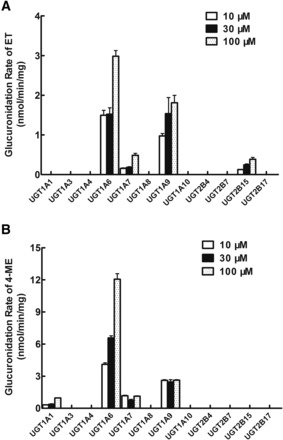
Glucuronidation of ET (A) and 4-ME (B) by 12 recombinant expressed human UGTs. Three concentrations (10, 30, and 100 μM) of ET (or 4-ME) were incubated with 12 commercially available recombinant expressed human UGTs (0.25∼1 mg/ml) at 37°C for 30 minutes. Glucuronides formed in the incubation samples were quantified by UHPLC-MS2, and glucuronidation rates were calculated and expressed as nmol/min/mg of protein. Each bar is the average of three determinations, and error bars are standard deviations of the mean (n = 3).
Effects of Chemical Inhibitors on the Glucuronidation of ET and 4-ME in HLM and Expressed UGTs.
To further confirm that UGT1A6 and UGT1A9 were the main UGT isoforms involved in ET and 4-ME glucuronidation in vitro, inhibitory effects of phenylbutazone (UGT1A6 inhibitor) and carvacrol (UGT1A9 inhibitor) on the glucuronidation of ET (Fig. 3, A–D) and 4-ME (Fig. 3, E–H) in pooled HLM, UGT1A6, and UGT1A9 were investigated (Aprile et al., 2010; Dong et al., 2012). About 100 and 200 μM of phenylbutazone and carvacrol significantly inhibited >50% ET-G formation in HLM using ET at a low concentration (10 μM) (Fig. 3, A and B) (P < 0.05). Results for UGT1A6 and UGT1A9 (Fig. 3, C and D) were highly consistent with those in HLM (P < 0.05). Similar results were also observed in the formation of 4-ME-G in microsomes and UGTs (Fig. 3, E–H). In addition, both phenylbutazone and carvacrol also significantly inhibited the glucuronidation of ET and 4-ME in almost all experiments, although the inhibition effects did not surpass 50% in some samples (P < 0.05).
Fig. 3.
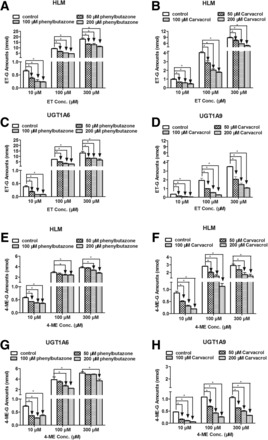
Effect of chemical inhibitors of UGT1A6 (phenylbutazone) and UGT1A9 (carvacrol) on the glucuronidation of ET (A, B, C, and D) and 4-ME (E, F, G, and H) in HLM, UGT1A6, and UGT1A9. Glucuronidation of ET and 4-ME by HLM and UGT isoforms were measured at three concentrations (10, 100, and 300 μM) and incubated in the presence of phenylbutazone or carvacrol at low three concentrations (50, 100, and 200 μM). All protein concentrations were 0.5 mg/ml, incubation time for ET and 4-ME in microsomes and UGTs ranged from 15 to 60 minutes. Control experiments were incubated without chemical inhibitors. Samples were processed and analyzed by UHPLC-MS2. All incubations were performed in triplicate. Each column represents mean percentage control of formation rates of these three metabolites, and error bars are standard deviations of the mean. An asterisk (*) indicates a statistically significant difference (P < 0.05) compared with the control according to Student’s t test.
Kinetics of ET and 4-ME Glucuronidation by Microsomes and Expressed UGT1A6 and UGT1A9.
Glucuronidation rates of ET and 4-ME by UGTs and microsomes were determined at different substrate concentrations. Within the tested concentration ranges, both UGTs and microsome-mediated glucuronidation of ET (Fig. 4, A–D) and 4-ME (Fig. 4, E–H) exhibited classic Michaelis-Menten kinetic characteristics, as evidenced by a linear Eadie-Hofstee plot (Fig. 4, a–h). All kinetic parameters of UGTs and microsome-catalyzed glucuronidation of ET and 4-ME are shown in Table 1.
Fig. 4.
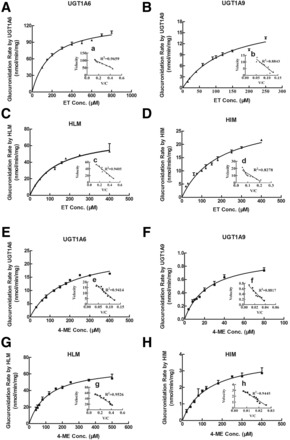
Kinetics of ET (A–D) and 4-ME (E–H) by UGT1A6, UGT1A9, HLM, and HIM. Kinetic assays were conducted using a microsomes or Supersomes in the concentrations range from 0.013 to 0.053 mg/ml based on optimization of the reaction. Glucuronidation rates of ET by UGT1A6 and UGT1A9 were determined at concentration ranges of 100 to 800 µM and 15 to 250 µM, respectively, within 60 minutes of reaction time. Glucuronidation rates of ET by HLM and HIM were determined at concentration ranges of 30 to 400 µM and 20 to 400 µM, respectively, within 30 minutes of reaction time. Glucuronidation rates of 4-ME by UGT1A6 and UGT1A9 were determined at concentration ranges of 30 to 400 µM and 4 to 80 µM, as well as reaction times of 30 and 60 minutes, respectively. Glucuronidation rates of 4-ME by HLM and HIM were determined at concentration ranges of 40 to 500 µM and 30 to 400 µM, respectively, within 30 minutes of reaction time. Curves were estimated based on fitted parameters generated using the Michaelis-Menten equation. An Eadie-Hofstee plot is shown in the inset to illustrate kinetics. Each data point was the average of three determinations, with error bars representing standard derivations. The apparent kinetic parameters are listed in Table 1.
TABLE 1.
Kinetic parameters of esculetin (ET) and 4-methylesculetin (4-ME) glucuronidation obtained from recombinant UGT1A6 and UGT1A9 and human liver and intestinal microsomes (HLM and HIM)
Kinetic parameters were obtained using simple Michaelis-Menten models as described in Materials and Methods.
| Compounds | Kinetic Parameters | UGT1A6 | UGT1A9 | HLM | HIM |
|---|---|---|---|---|---|
| ET | Km (µM) | 185.59 | 134.75 | 134.19 | 189.33 |
| Vmax (nmol/min/mg) | 127.40 | 19.23 | 72.61 | 30.45 | |
| CL(Vmax/Km, ml/min/mg) | 0.69 | 0.14 | 0.54 | 0.16 | |
| R2 | 0.9823 | 0.9775 | 0.9833 | 0.9738 | |
| AIC | 40.98 | 14.05 | 29.53 | 26.69 | |
| 4-ME | Km (µM) | 172.51 | 25.20 | 117.81 | 129.12 |
| Vmax (nmol/min/mg) | 24.70 | 0.99 | 69.91 | 3.88 | |
| CL(Vmax/Km, ml/min/mg) | 0.14 | 0.04 | 0.59 | 0.03 | |
| R2 | 0.9824 | 0.9793 | 0.9879 | 0.9918 | |
| AIC | 15.21 | −41.49 | 34.75 | −26.16 |
AIC, Akaike’s information criterion.
The intrinsic clearance (CL; Vmax/Km) of ET-G in UGT1A6, UGT1A9, HLM, and HIM were 0.69, 0.14, 0.54, and 0.16 ml/min/mg, respectively, accompanying the large variation in Vmax values (127.40 μM for UGT1A6, 19.23 μM for UGT1A9, 72.61 μM for HLM, and 30.45 μM for HIM). No large difference was observed among the Km values of ET glucuronidation in UGTs and microsomes (185.59 μM for UGT1A6, 134.75 μM for UGT1A9, 134.19 μM for HLM, and 189.33 μM for HIM).
CL of 4-ME-G in UGT1A6, UGT1A9, HLM, and HIM were 0.14, 0.04, 0.59, and 0.03 ml/min/mg, respectively, also accompanying the large variation in Vmax values (24.70 μM for UGT1A6, 0.99 μM for UGT1A9, 69.91 μM for HLM, and 3.88 μM for HIM). However, Km values of 4-ME in different enzymes also showed larger fluctuation: 172.51 μM for UGT1A6, 25.20 μM for UGT1A9, 117.81 μM for HLM, and 129.12 μM for HIM.
Discussion
The glucuronidation pathway is responsible for metabolism clearance of drugs that contain phenolic hydroxyl groups, such as coumarins and flavonoids. Most coumarin and flavonoid derivatives exert pleiotropic biologic activities and are also good substrates of human UGTs. ET and 4-ME are coumarin derivatives that contain two hydroxyl groups at 6-C and 7-C, respectively. Accordingly, the in vitro glucuronidation metabolism of ET and 4-ME in HLM, HIM, and human-expressed UGTs were investigated first in this study.
Mono-glucuronidation metabolites (ET-G and 4-ME-G) of ET and 4-ME were detected and identified in HLM. The glucuronidation position of ET and 4-ME was 7-OH. These results were consistent with a previous report demonstrating that 7-hydroxyl coumarins are easily metabolized by substitution with glucuronic acid at 7-OH (Egan et al., 1990). No diglucuronide was detected in any microsomal and UGT samples. Human-expressed UGTs and chemical inhibition assays were used to determine the main UGTs involved in the glucuronidation metabolism of ET and 4-ME. For both ET and 4-ME, UGT1A6 and UGT1A9 were the most capable of catalyzing their glucuronidation at tested substrate concentrations. In addition, a small amount of ET-G could be produced by UGT1A7 and UGT2B15, whereas a small quantity of 4-ME-G could be formed by UGT1A1 and UGT1A7. Different concentrations (50 to 200 μM) of UGT1A6 (phenylbutazone) and UGT1A9 (carvacrol) inhibitors were further used to confirm these results in inhibition assays with HLM or UGTs. Both phenylbutazone and carvacrol at different concentrations significantly inhibited the formation of glucuronidates of ET and 4-ME in HLM, UGT1A6, and UGT1A9 at ET and 4-ME concentrations of 10 to 300 μM (Fig. 3). These results demonstrated that the UGTs primarily responsible for the glucuronidation of ET and 4-ME in vitro were UGT1A6 and UGT1A9. It was demonstrated that UGT1A6, UGT1A9, and UGT2B7 can be involved in the glucuronidation of hydroxycoumarins (Antonio et al., 2003; Loureiro et al., 2006). However, UGT2B7 was not found to be involved in glucuronidating ET and 4-ME in our study.
To further elucidate the characteristics of glucuronidation metabolism of ET and 4-ME, rates of glucuronidation metabolism of ET and 4-ME by UGT1A6, UGT1A9, HLM, and HIM were determined at different substrate concentrations. Both UGTs and microsomes mediated ET and 4-ME glucuronidation following classic Michaelis-Menten kinetics. The comparable kinetic profiles of UGT1A6 and UGT1A9 were confirmed by evidence from Eadie-Hofstee plots observed in HLM and HIM, suggesting that UGT1A6 and UGT1A9 played primary roles in the formation of glucuronides of ET and 4-ME in these two organs, although 4-ME showed markedly higher affinity to UGT1A9 than to those of any other substrate and enzyme (Table 1). In addition, CL values of ET-G and 4-ME-G in HLM were 3.4-fold and 19.7-fold more than those in HIM, respectively (Table 1). UGT1A6 and UGT1A9, which showed the highest activity toward ET and 4-ME, were primarily expressed in the liver (Rowland et al., 2013). Therefore, the liver is probably the major contributor to the glucuronidation of ET and 4-ME. Moreover, CL and Vmax values of ET in UGTs and HIM were greater than those of 4-ME, suggesting that ET may possess more rapid excretion rate and less residence time in vivo than 4-ME.
It was demonstrated that carvacrol was a specific inhibitor of UGT1A9. Chemical inhibition experiments revealed that carvacrol dose dependently inhibited the formation of ET-G and 4-ME-G in both HLM and UGT1A9 at three tested substrate concentrations (Fig. 3, B, D, F, and H). Different from the inhibition effect of carvacrol, phenylbutazone showed attenuated inhibition on the formation of ET-G and 4-ME-G in both HLM and UGT1A9 with increased substrate concentration (Fig. 3, A, C, E, and G). Phenylbutazone is not a selective inhibitor for UGT1A6 because it exhibited an inhibition effect on UGT1As. In addition, the inhibition kinetics of phenylbutazone and carvacrol have been previously described to be substrate dependent (competitive and noncompetitive), but the mechanism of inhibition of drug-metabolizing enzymes is still unclear (Uchaipichat et al., 2006; Dong et al., 2012). Thus, the mechanism of inhibition of phenylbutazone and carvacrol require additional studies, and in vitro data on the inhibition of phenylbutazone and carvacrol should be interpreted with caution.
Glucuronidation is the primary phase II conjugation reaction for most coumarin derivatives. In addition to the liver, UGT1A6 and UGT1A9 are also expressed in the intestine, kidney, and stomach (Rowland et al., 2013). Their activity and expression regulation are vulnerable to genetic polymorphism, hepatocyte nuclear factor 1, aryl hydrocarbon receptor, and chemical inhibitors and inducers (Kiang et al., 2005). Thus, in vivo metabolism characteristics of ET and 4-ME in human subjects, as well as drug-drug interactions related to glucuronidation metabolism, require further study.
In conclusion, this study demonstrated that 7-O-glucuronidation, the main metabolic pathway of ET and 4-ME in vitro, was mainly catalyzed by UGT1A6 and UGT1A9 isoforms. The liver is probably the major contributor to the glucuronidation of ET and 4-ME. ET showed more rapid metabolism than 4-ME in glucuronidation.
Abbreviations
- CL
intrinsic clearance
- ET
esculetin
- ET-G
7-O-monoglucuronide esculetin
- HLM
human liver microsomes
- HIM
human intestinal microsomes
- HPLC
high-pressure liquid chromatography
- HRMS
high-resolution mass spectrometry
- 1HNMR
1H-nuclear magnetic resonance
- LC-MS
liquid chromatography-mass spectrometry
- 4-ME
4-methylesculetin
- 4-ME-G
7-O-monoglucuronide 4-methylesculetin
- RLM
rat liver microsomes
- UDPGA
uridine diphosphate glucuronic acid
- UGT
UDP-glucuronosyltransferase
- UHPLC
ultra high-pressure liquid chromatography
Authorship Contributions
Participated in research design: Zhu, Lu, Z. Liu.
Conducted experiments: Zeng, Luo, Dai.
Contributed new reagents or analytic tools: L. Liu, Hu.
Performed data analysis: Zhu, Zeng, Luo, Wu, Wang.
Wrote or contributed to the writing of the manuscript: Zhu, Lu, L. Liu, Hu, Z. Liu.
Footnotes
This work was supported by the Projects of National Natural Science Foundation of China [81120108025, 81473410] and the Platform Project of Department of Education of Guangdong Province.
There is no financial conflict of interests with the authors of this paper. Publication of this paper will not benefit or adversely affect the financial situations of the authors.
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
References
- Antonio L, Xu J, Little JM, Burchell B, Magdalou J, Radominska-Pandya A. (2003) Glucuronidation of catechols by human hepatic, gastric, and intestinal microsomal UDP-glucuronosyltransferases (UGT) and recombinant UGT1A6, UGT1A9, and UGT2B7. Arch Biochem Biophys 411:251–261. [DOI] [PubMed] [Google Scholar]
- Aprile S, Del Grosso E, Grosa G. (2010) Identification of the human UDP-glucuronosyltransferases involved in the glucuronidation of combretastatin A-4. Drug Metab Dispos 38:1141–1146. [DOI] [PubMed] [Google Scholar]
- D’Argenio DZ, Schumitzky A. (1997) ADAPT II User’s Guide: Pharmacokinetic/Pharmacodynamic System Analysis Software, Biomedical Simulations Resource, Los Angeles. [Google Scholar]
- Dong RH, Fang ZZ, Zhu LL, Liang SC, Ge GB, Yang L, Liu ZY. (2012) Investigation of UDP-glucuronosyltransferases (UGTs) inhibitory properties of carvacrol. Phytother Res 26:86–90. [DOI] [PubMed] [Google Scholar]
- Egan D, O’Kennedy R, Moran E, Cox D, Prosser E, Thornes RD. (1990) The pharmacology, metabolism, analysis, and applications of coumarin and coumarin-related compounds. Drug Metab Rev 22:503–529. [DOI] [PubMed] [Google Scholar]
- Hoult JR, Forder RA, de las Heras B, Lobo IB, Payá M. (1994) Inhibitory activity of a series of coumarins on leukocyte eicosanoid generation. Agents Actions 42:44–49. [DOI] [PubMed] [Google Scholar]
- Hu Y, Chen X, Duan H, Hu Y, Mu X. (2009) Chinese herbal medicinal ingredients inhibit secretion of IL-6, IL-8, E-selectin and TXB2 in LPS-induced rat intestinal microvascular endothelial cells. Immunopharmacol Immunotoxicol 31:550–555. [DOI] [PubMed] [Google Scholar]
- Jay RA, Harish R, Kanakadurga M, Parveen K, Kandarpa S, Mangala L. (2013) Anticancer activity of esculetin via-modulation of Bcl-2 and NF-κB expression in benzo[a]pyrene induced lung carcinogenesis in mice. Biomedicine & Preventive Nutrition 3:107–112. [Google Scholar]
- Kaneko T, Tahara S, Takabayashi F. (2003) Suppression of lipid hydroperoxide-induced oxidative damage to cellular DNA by esculetin. Biol Pharm Bull 26:840–844. [DOI] [PubMed] [Google Scholar]
- Kiang TK, Ensom MH, Chang TK. (2005) UDP-glucuronosyltransferases and clinical drug-drug interactions. Pharmacol Ther 106:97–132. [DOI] [PubMed] [Google Scholar]
- Kim SH, Kang KA, Zhang R, Piao MJ, Ko DO, Wang ZH, Chae SW, Kang SS, Lee KH, Kang HK, et al. (2008) Protective effect of esculetin against oxidative stress-induced cell damage via scavenging reactive oxygen species. Acta Pharmacol Sin 29:1319–1326. [DOI] [PubMed] [Google Scholar]
- Kok SH, Yeh CC, Chen ML, Kuo MY. (2009) Esculetin enhances TRAIL-induced apoptosis through DR5 upregulation in human oral cancer SAS cells. Oral Oncol 45:1067–1072. [DOI] [PubMed] [Google Scholar]
- Kontogiorgis C, Detsi A, Hadjipavlou-Litina D. (2012) Coumarin-based drugs: a patent review (2008 — present). Expert Opin Ther Pat 22:437–454. [DOI] [PubMed] [Google Scholar]
- Lake BG. (1999) Coumarin metabolism, toxicity and carcinogenicity: relevance for human risk assessment. Food Chem Toxicol 37:423–453. [DOI] [PubMed] [Google Scholar]
- Leung KN, Leung PY, Kong LP, Leung PK. (2005) Immunomodulatory effects of esculetin (6,7-dihydroxycoumarin) on murine lymphocytes and peritoneal macrophages. Cell Mol Immunol 2:181–188. [PubMed] [Google Scholar]
- Liang SC, Ge GB, Liu HX, Zhang YY, Wang LM, Zhang JW, Yin L, Li W, Fang ZZ, Wu JJ, et al. (2010) Identification and characterization of human UDP-glucuronosyltransferases responsible for the in vitro glucuronidation of daphnetin. Drug Metab Dispos 38:973–980. [DOI] [PubMed] [Google Scholar]
- Lin WL, Wang CJ, Tsai YY, Liu CL, Hwang JM, Tseng TH. (2000) Inhibitory effect of esculetin on oxidative damage induced by t-butyl hydroperoxide in rat liver. Arch Toxicol 74:467–472. [DOI] [PubMed] [Google Scholar]
- Liu X, Tam VH, Hu M. (2007) Disposition of flavonoids via enteric recycling: determination of the UDP-glucuronosyltransferase isoforms responsible for the metabolism of flavonoids in intact Caco-2 TC7 cells using siRNA. Mol Pharm 4:873–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loureiro AI, Bonifácio MJ, Fernandes-Lopes C, Almeida L, Wright LC, Soares-Da-Silva P. (2006) Human metabolism of nebicapone (BIA 3-202), a novel catechol-o-methyltransferase inhibitor: characterization of in vitro glucuronidation. Drug Metab Dispos 34:1856–1862. [DOI] [PubMed] [Google Scholar]
- Edson M, Rafael F. (2012) Antigenotoxic effect of 4-methylesculetin on mice cells exposed to doxorubicin. Toxicol Lett 211S:S64–S65. [Google Scholar]
- Okada Y, Miyauchi N, Suzuki K, Kobayashi T, Tsutsui C, Mayuzumi K, Nishibe S, Okuyama T. (1995) Search for naturally occurring substances to prevent the complications of diabetes. II. Inhibitory effect of coumarin and flavonoid derivatives on bovine lens aldose reductase and rabbit platelet aggregation. Chem Pharm Bull (Tokyo) 43:1385–1387. [DOI] [PubMed] [Google Scholar]
- Park C, Jin CY, Kim GY, Choi IW, Kwon TK, Choi BT, Lee SJ, Lee WH, Choi YH. (2008) Induction of apoptosis by esculetin in human leukemia U937 cells through activation of JNK and ERK. Toxicol Appl Pharmacol 227:219–228. [DOI] [PubMed] [Google Scholar]
- Payá M, Halliwell B, Hoult JR. (1992) Interactions of a series of coumarins with reactive oxygen species. Scavenging of superoxide, hypochlorous acid and hydroxyl radicals. Biochem Pharmacol 44:205–214. [DOI] [PubMed] [Google Scholar]
- Prabakaran D, Ashokkumar N. (2013) Protective effect of esculetin on hyperglycemia-mediated oxidative damage in the hepatic and renal tissues of experimental diabetic rats. Biochimie 95:366–373. [DOI] [PubMed] [Google Scholar]
- Rowland A, Miners JO, Mackenzie PI. (2013) The UDP-glucuronosyltransferases: their role in drug metabolism and detoxification. Int J Biochem Cell Biol 45:1121–1132. [DOI] [PubMed] [Google Scholar]
- Shirole T, Makhija D, Jagtap A. (2013) PP265—Attenuation of aluminium induced neurodegeneration by 4-methylesculetin. Clin Ther 35:e100–e101. [Google Scholar]
- Tubaro A, Del Negro P, Ragazzi E, Zampiron S, Della Loggia R. (1988) Anti-inflammatory and peripheral analgesic activity of esculetin in vivo. Pharmacol Res Commun 20 (Suppl 5):83–85. [DOI] [PubMed] [Google Scholar]
- Uchaipichat V, Mackenzie PI, Elliot DJ, Miners JO. (2006) Selectivity of substrate (trifluoperazine) and inhibitor (amitriptyline, androsterone, canrenoic acid, hecogenin, phenylbutazone, quinidine, quinine, and sulfinpyrazone) “probes” for human udp-glucuronosyltransferases. Drug Metab Dispos 34:449–456. [DOI] [PubMed] [Google Scholar]
- Venugopala KN, Rashmi V, Odhav B. (2013) Review on natural coumarin lead compounds for their pharmacological activity. Biomed Res Int 2013:963248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SW, Chen J, Jia X, Tam VH, Hu M. (2006) Disposition of flavonoids via enteric recycling: structural effects and lack of correlations between in vitro and in situ metabolic properties. Drug Metab Dispos 34:1837–1848. [DOI] [PubMed] [Google Scholar]
- Yamaoka K, Nakagawa T, Uno T. (1978) Application of Akaike’s information criterion (AIC) in the evaluation of linear pharmacokinetic equations. J Pharmacokinet Biopharm 6:165–175. [DOI] [PubMed] [Google Scholar]


