Summary
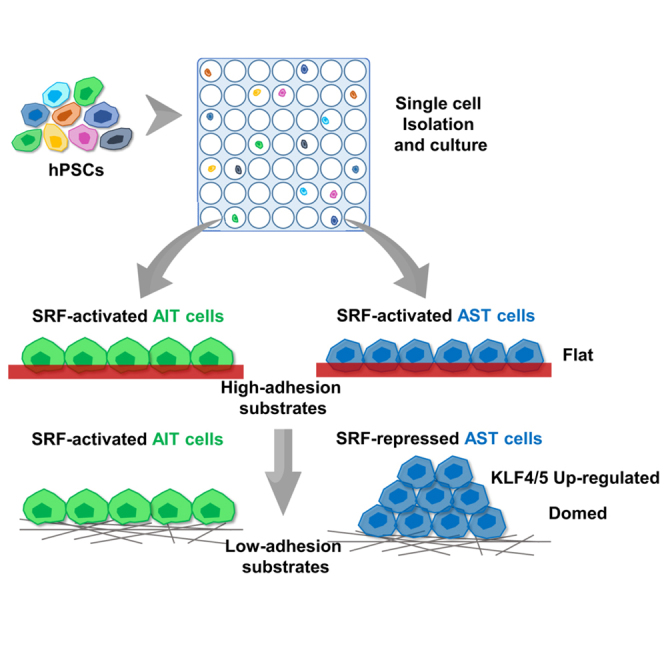
We show that a human pluripotent stem cell (hPSC) population cultured on a low-adhesion substrate developed two hPSC subtypes with different colony morphologies: flat and domed. Notably, the dome-like cells showed higher active proliferation capacity and increased several pluripotent genes’ expression compared with the flat monolayer cells. We further demonstrated that cell-matrix adhesion mediates the interaction between cell morphology and expression of KLF4 and KLF5 through a serum response factor (SRF)-based regulatory double loop. Our results provide a mechanistic view on the coupling among adhesion, stem cell morphology, and pluripotency, shedding light on the critical role of cell-matrix adhesion in the induction and maintenance of hPSC.
Keywords: human pluripotent stem cells, cell-matrix adhesion, stem cell morphology, single-cell culture, heterogeneity, serum response factor, nanofiber, mathematical model
Graphical Abstract
Highlights
-
•
Low-adhesion substrates reveal two different subtypes co-exist in the hPSC population
-
•
SRF-based regulatory loop-coupled adhesion, cell morphology, and KLF4/5 expression
-
•
The low-adhesion substrates are more suitable for high-pluripotency cell culture
When culturing hPSCs on low-adhesion substrate (gelatin nanofiber), Dr. Liu Li and her colleagues found two subtypes with different colony morphologies. The dome-like cells showed higher proliferation capacity and KLF4/5 and NANOG expression than the monolayer cells. A serum response factor-based regulatory double loop was proposed to explain how cell-matrix adhesion mediates the interaction between cell morphology and pluripotency genes.
Introduction
Somatic cells can be reprogrammed to induced pluripotent stem cells (iPSCs) by ectopic expression of four transcription factors (Oct4, Sox2, Klf4, and c-Myc [OSKM]) (Takahashi and Yamanaka, 2006). During the reprogramming process of iPSCs, the endogenous expression of OCT4, SOX2, and KLF4 increased sharply in the early phase and dropped moderately in the late phase, while the cell-matrix adhesion (represented by the focal adhesion (FA) proteins which link cells with their surrounding matrix) demonstrated exactly the opposite variation trend: downregulated in the early phase and upregulated in the late phase (Hansson et al., 2012). In the pluripotent state, iPSCs showed low adhesive strength to surrounding matrix compared with their parental somatic cells (Singh et al., 2013) and their lineage differentiated cells (Narve et al., 2017, Singh et al., 2013), indicating that pluripotency is associated with altered cell-matrix adhesion and motility. On the other hand, in contrast with cell-matrix adhesion, the cell-cell adhesion-related protein epithelial-cadherin (E-cadherin) was upregulated during the reprogramming process (Hansson et al., 2012). It has been reported that elimination of E-cadherin prevents somatic cells from reprogramming to pluripotency (Li et al., 2009) and that enhancement of E-cadherin can elevate the reprogramming efficiency of iPSCs (Chen et al., 2010) and can even replace the need for Oct4 (the most critical factor in OSKM) during iPSC reprogramming (Redmer et al., 2011, Sakurai et al., 2014). In addition, compact colonies and E-cadherin-mediated cell-cell adhesion are required for iPSC survival and stemness (Ohgushi et al., 2010). Furthermore, substantial remodeling of cell adhesive microenvironment is a prerequisite for reprogramming. For example, high cell-matrix adhesion represents a barrier toward iPSC reprogramming (Qin et al., 2014), while cell-cell adhesion promotes iPSC reprogramming (Caiazzo et al., 2016, Downing et al., 2013). Collectively, the above studies suggested that low cell-matrix adhesion and strong cell-cell adhesion are hallmarks of high pluripotency, and that both features are intimately coupled to the reprogramming process.
To further investigate how cell adhesion properties affect the hPSC culture and pluripotency, in this work, we employed a nanofibrous substrate (Liu et al., 2014) and platform for single-cell isolation and culture. We revealed that there exist two distinct subtypes of cells in the conventional PSC population, which differ in their morphology, gene expression pattern, cell-matrix and cell-cell adhesion, pluripotent level, and sensitivity to the adhesion. On the mechanistic level, we demonstrated that a serum response factor (SRF)-based regulation loop is the key to understand the observed interplay among the substrate adhesion, pluripotent stem cell morphology, and pluripotency.
Results
Two Morphologically Different Type Cells Co-exist in hPSC Populations
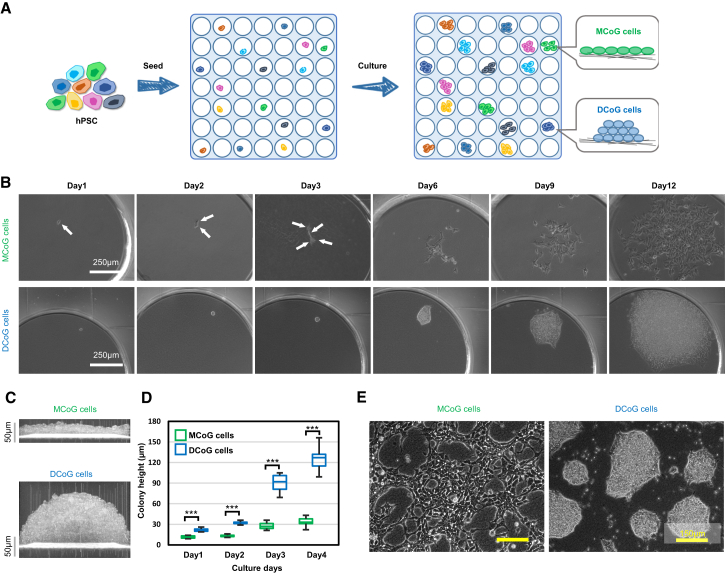
An increasing evidence suggests that stem cell cultures do not comprise a homogeneous cellular population (Drukker et al., 2012, Enver et al., 2009, Masaki et al., 2008). By using the single-cell isolation and analysis method, the heterogeneity of hPSCs has been scrutinized (Narsinh et al., 2011, Yan et al., 2013). However, single stem cell culture has not been reported previously. Here, we developed a culture platform to gain single hPSC-derived clones (Figure 1A). This platform comprised two layers: the upper layer is a polydimethylsiloxane (PDMS) multi-well membrane, which is used to separate the single cells, and the lower layer is a low-adhesion gelatin nanofibrous (GNF), which was developed for long-term culture of hPSCs without enzymatic dissociation (Liu et al., 2014). Based on this single-cell culture platform, several single-cell-derived clones have been successfully isolated. Interestingly, we found that these clones could be morphologically classified into two types (Figures 1B and S1A). The existence of these two subtypes was also confirmed in different hPSC lines (hiPSC lines, 253G1 and IMR; hESC lines, H1 and H9) (Figure S1B). Importantly, short tandem repeat analysis has obviated the possibility of the cross-contamination between cell lines (Figure S2).
Figure 1.
Two Subtypes of Clones Exist in an hPSC Population
(A) Schematic representation of single-cell dissociation and culture strategy. In the following figures, green represents MCoG cells and blue represents DCoG cells.
(B) Single hPSCs grew into multicellular clones during the culture course from day 1 to 12. These clones demonstrated two types of morphologies: flat monolayer cells (top panel) and domed-multilayer cells (bottom panel). To prevent single-cell apoptosis, ROCK inhibitor (10 μM) was added on the first 5 days and removed from day 6. White arrows indicate the individual cells.
(C) Cross-section images of colonies of two types of cells were observed by optical coherence tomography (OCT) microscopy system in real time.
(D) The colony height of MCoG and DCoG cells on culture days 1, 2, 3, and 4 (ROCK inhibitor was removed from day 2) (mean ± SD, n = 10 independent biological replicates, ∗∗∗p < 0.001). Colonies height were measured by OCT microscopy system.
(E) The bright-field images of two types of clones after 27 passages on GNF substrate, showing maintained morphologies, respectively.
See also Figures S1 and S2.
When grown on the low-adhesion GNF substrate, the majority of single-cell-derived clones showed a flat monolayer colony morphology, designated as a monolayer colony on GNF (MCoG), and the other subtype showed a compact dome-like colony morphology, designated as dome-like colonies on GNF (DCoG). Then we concretely investigated the morphologic differences between MCoG and DCoG cells. Using coherence tomography microscopy, we confirmed that the colony heights of the two types of cells are significantly different (Figures 1C and 1D). The three-dimensional scanning result indicated that the DCoG-type cells demonstrated a multi-layer structure without cavity (Video S1), in comparison with the monolayer structure of the MCoG-type cells (Video S2). Moreover, real-time observation of MCoG and DCoG cells revealed that the two types of cells demonstrated different morphologies after attachment, which later became more and more distinct during proliferation (Video S3). In addition, the morphology of these single-cell-derived clones could be stably maintained on the GNF substrate for more than 27 passages (Figure 1E). We also performed another isolation of single cells from these two subtypes of cells, and the resulting clones maintained their respective morphologies, which may indicate that these clones have been homogenized after single-cell culture. In summary, using a single-cell isolation and culture platform, we revealed two types of cells co-existing in the hPSC populations. The two types of cells demonstrated different morphologies, and each could be steadily maintained during proliferation.
Two Types of Cells Are Both Pluripotent but Possess Different Differentiation Potential
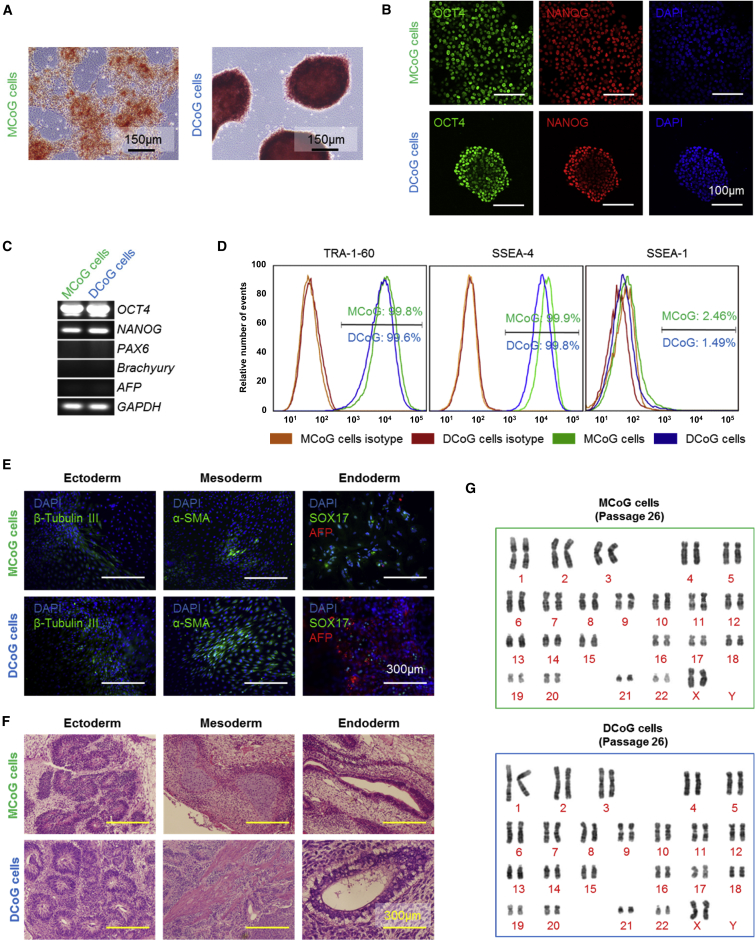
We next evaluated the pluripotency of these single-cell-derived clones. After more than 20 passages, the cells were positive for alkaline phosphatase and hPSC-specific markers (OCT4, NANOG, and surface markers SSEA4 and TRA-1-60), with minimal levels of lineage commitment markers (PAX6, Brachyury, AFP, and SSEA1) (Figures 2A–2D), indicating that both subtypes of cells could be maintained in undifferentiated states after a long-term culture. In addition, we also investigated whether the two subtypes could differentiate into all germ lineages both in vitro and in vivo. MCoG and DCoG cells formed embryonic bodies and teratomas, which could differentiate into cells of all three germ layers (Figures 2E and 2F). Furthermore, these clones still maintained a normal karyotype for over 27 passages (Figure 2G). We also found that, during spontaneous differentiation, the MCoG cells showed a tendency to differentiate more toward the ectoderm and mesoderm, and DCoG cells showed a tendency to differentiate more toward the endoderm (Figure S3). It has been reported that the differentiation efficiencies varies among different stem cell lines (Osafune et al., 2008, The International Stem Cell Initiative, 2007). Our finding indicated that the differentiation difference may also exist among stem cells within the same cell line. In summary, the above results indicate that both subtypes derived from a single hPSC could maintain their proper pluripotent status after long-term culture.
Figure 2.
The Two Types of Cells Are Both Pluripotent
(A) The images indicated that both MCoG and DCoG cells formed colonies and expressed AP.
(B) NANOG and OCT4 were expressed both in MCoG and DCoG cells after 26 passages.
(C) RT-PCR analysis of expression of pluripotency genes (OCT4 and NANOG) and differentiation genes (PAX6, ectoderm; BRACHYURY, mesoderm; and AFP, endoderm) in MCoG and DCoG cells.
(D) Flow cytometric analysis of pluripotency markers in MCoG and DCoG cells. The 50,000 cells analyzed express high levels of hPSC-specific cell surface markers (TRA-1-60, SSEA-4) and low levels of differentiation-specific cell surface markers (SSEA-1).
(E) Embryoid bodies formed by MCoG and DCoG cells differentiated into the three germ layers: ectoderm (β-tubulin), mesoderm (α-SMA), and endoderm (SOX17 and AFP).
(F) Teratomas formed by MCoG and DCoG cells in severe combined immunodeficiency mice, containing tissues that are representative of all three embryonic germ layers.
(G) Normal karyotype exhibited in both MCoG and DCoG cells.
See also Figure S3.
Two Types of Cells Showed Differences in Cell Adhesion and Proliferation
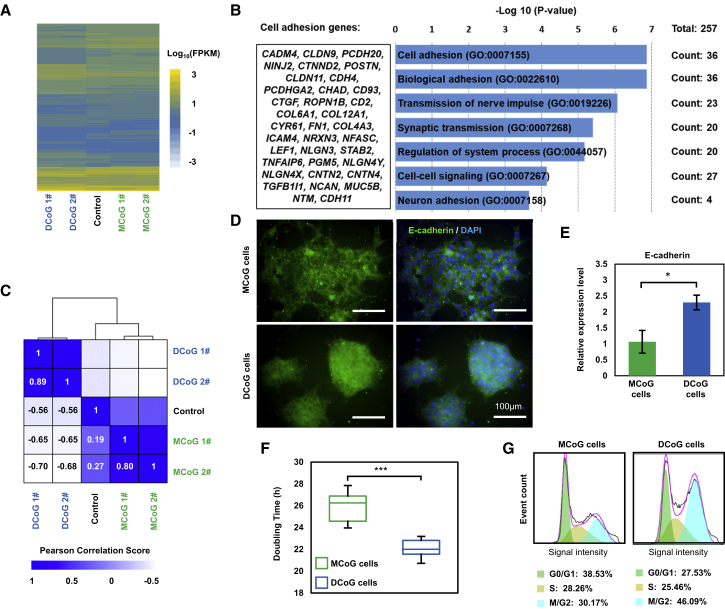
Besides pluripotency and differentiation potential, we also investigated the inherent difference of these two types of cells. The RNA sequencing (RNA-seq) result showed different (p ≤ 0.05) gene expression patterns (Figures 3A, S4A, and S4B and see Table S1), and functional annotation analysis with gene ontology (GO) clearly revealed that the downregulated genes (fold change > 2) in DCoG cells were mostly related to adhesion (Figure 3B), and indicated low cell adhesion of DCoG cells compared with McoG cells, which was also confirmed with a cell adhesion test (Figures S4C and S4D). Also, the expression pattern of standard iPSCs (without single-cell isolation) showed similarity with MCoG-type cells, and the pattern of DCoG-type cells is concealed in standard PSCs (Figures 3A and 3C). This is due to the fact that the majority of standard iPSCs are the MCoG type (in our experiments, >90% of the isolated single cells are MCoG type). On the other hand, the appearance of the two types of cells suggests that there may be substantial differences in the density of cell-cell contacts. The higher E-cadherin expression indicated a strong cell-cell interaction in DCoG cells compared with MCoG cells (Figures 3D and 3E). The increased E-cadherin further stabilizes the cortical cytoskeleton and maintains the domed colony morphology (Spencer et al., 2007). Moreover, the average doubling time of DCoG cells was 22 ± 0.7 hr, which was shorter than that of MCoG cells at 26.5 ± 1.5 hr (Figure 3F). Flow cytometric-mediated cell-cycle analysis showed a larger percentage (71.55%) of DCoG cells in the M/G2 and S phases compared with MCoG cells (58.43%) (Figure 3G). These results implied a higher active proliferation capacity of disseminated tumor cells than metastatic tumor cells. Also, we also observed a similar difference in teratoma formation: in immunodeficient mice, the DCoG cells generated teratomas of a larger volume in a shorter time than MCoG cells (the weight of MCoG-derived teratomas reached 1.96 ± 0.18 g after 57 ± 3 days, and the weight of DCoG-derived teratomas reached 2.48 ± 0.26 g after 43 ± 2 days). Taken together, the above findings indicated that the rare DCoG-type cells hold lower cell-matrix adhesion, higher cell-cell adhesion, and more active proliferation capacity in comparison with MCoG-type cells.
Figure 3.
The Differences between the Two Types of Cells
(A) Heatmap showing the expression levels (log10 transformed FPKM value) of standard PSC population (Control) compared with MCoG and DCoG cells.
(B) Gene ontology (GO) categories significantly enriched for genes downregulated in DCoG cells compared with MCoG cells. The genes with fold change > 2 and p < 0.05 were analyzed, and the genes of the top 1 enriched term are listed in the left column.
(C) Heatmap showing the hierarchically clustered correlation matrix resulting from comparing the expression values for each samples (MCoG and DCoG cells, and standard PSC population). Data are correlated using Pearson correlation.
(D) Immunofluorescence images of MCoG and DCoG cells cultured on GNF substrates on day 3. E-Cadherin is expressed and surrounds the cell body.
(E) Relative expression of E-cadherin in MCoG and DCoG cells cultured on GNF substrates on day 3 (mean ± SD, n = 3 independent experiments, ∗p < 0.05).
(F) Doubling time of MCoG and DCoG cells from 10 passages (n = 10 passages, ∗∗∗p < 0.001).
(G) Increased S and M/G2 phases population in DCoG cells. The cell cycles of MCoG and DCoG cells are analyzed by flow cytometric analysis (50,000 cells were analyzed) after propidium iodide staining. To avoid the interference of ROCK inhibitor on cell adhesion, data in this figure were obtained more than 48 hr after withdrawal of the ROCK inhibitor from cell cultures.
Cell-Matrix Adhesion Affects Cell Morphology and Pluripotency
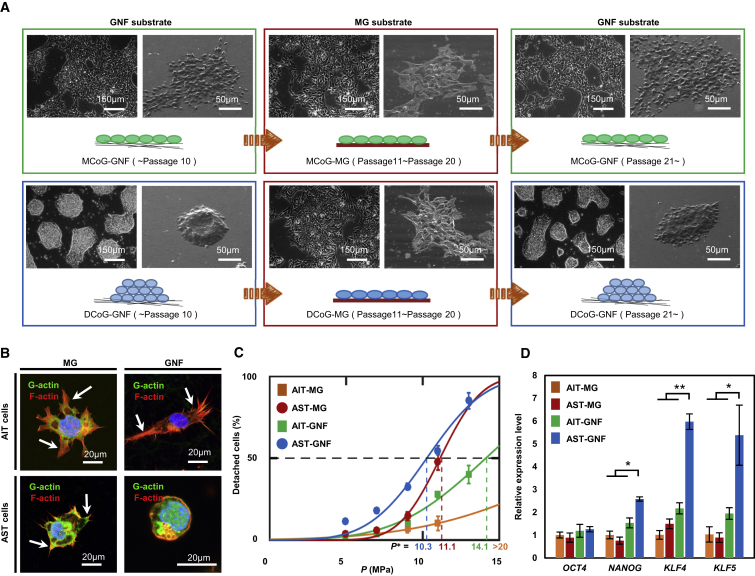
Cell-matrix adhesion (the link between cells and their surrounding matrix) has been reported to determine the morphology of cell colonies (dome-like or monolayer) (Chowdhury et al., 2010a, Chowdhury et al., 2010b). Here, we found that the morphologic difference is lost on high-adhesion Matrigel (MG) substrate (Figure 4A). Plating DCoG cells on MG resulted in a morphological change from domed to a flat monolayer. Interestingly, these cells formed domed colonies again when re-plated onto the GNF substrate. In contrast, the colony morphology of MCoG cells remained unchanged when plated on either the GNF or MG substrate. The result supports the notion that DCoG-type cells are sensitive to varying adhesion of substrates, but that MCoG-type cells are not, indicating some intrinsic differences between the two cell subtypes, which were concealed on the high-adhesion substrates. Thus, here we renamed DCoG-type cells as adhesion-sensitive-type (AST) cells, and MCoG-type cells as adhesion-insensitive-type (AIT) cells. We next observed the cell-matrix adhesion effect on AIT and AST cells at the single-cell level (Figures 4B and S5A). AST cells grown on the GNF substrate, without spreading, formed very few and short cell protrusions, and were hemispherical. By contrast, AIT cells were flat and spread, and formed long cell protrusions on both the GNF and MG substrates. However, AST cells were similar to AIT cells when plated on the MG substrate, where they spread well and formed long cell protrusions. Thus, the two types of cells have different cell-matrix adhesion properties on MG and GNF substrates (Chowdhury et al., 2010a, Chowdhury et al., 2010b).
Figure 4.
Substrate Regulates Cell Shape and Gene Expression
(A) Morphology change of MCoG cells and DCoG cells on different substrates during long-term passage. In each condition, the left panel is the phase contrast image and the right panel is the SEM image.
(B) Immunofluorescence images of single AIT and AST cells on the MG and GNF substrates, respectively. White arrows indicate cells engaged in spreading.
(C) Fraction of detached cells plotted as a function of hydrodynamic pressure P. Data points were fitted with the cumulative distribution function of normal distribution, and the critical pressure P∗ was determined as the required pressure at which 50% of cells were detached (mean ± SE, n ≥ 500 cells). Four conditions are investigated: AIT cells on MG (orange), AST cells on MG (red), AIT cells on GNF (green), and AST cells on GNF (blue).
(D) Relative expression of hPSC-specific genes in AIT and AST cells on MG and GNF substrates (mean ± SD, n ≥ 3 independent experiments, ∗p < 0.05, ∗∗p < 0.01). To avoid the interference of ROCK inhibitor on cell adhesion, data in this figure were obtained more than 48 hr after withdrawal of the ROCK inhibitor from cell cultures.
See also Figure S5.
So far, the cell-matrix organization and function of adhesion sites remain poorly defined in hPSCs. Recently some novel methodology has been proposed to investigate the traction between cells and matrix; here we quantified cell-matrix adhesion using a shock wave-based method (Yoshikawa et al., 2011) (Figure S5B). Among all four conditions tested (AST and AIT cells on GNF and MG substrates, respectively), the AST cells on the GNF substrate showed the lowest cell-matrix adhesion (Figure 4C), and it should be noted that only AST cells on GNF substrate formed domed colonies. Previous studies have indicated that the expression of reprogramming factors, in iPSCs, is strongly associated with altered cell-matrix adhesion (Hansson et al., 2012, Narve et al., 2017, Singh et al., 2013). Therefore, we next examined the hPSC-specific genes in these four conditions and found that the AST cells cultured on GNF showed higher expression levels of NANOG and KLF4/KLF5 compared with other conditions (Figure 4D). Furthermore, it has been reported that tight colony morphology is required for complete pluripotency (Narve et al., 2017, Ohgushi et al., 2010), and during the iPSC reprogramming process the expression level of pluripotent genes and cell adhesion genes showed completely opposite trends (Hansson et al., 2012, Singh et al., 2013). Therefore, congruent with our findings, it enables us to generate a view that there is some relevance among cell-matrix adhesion, cell morphology, and the expression of pluripotent genes, and the specific mechanisms contributing to these relationships have not been elucidated in detail.
An SRF-Based Feedback Double Loop Regulating Cell Adhesion, Morphology, and Pluripotency
To investigate the mechanism underlying these effects, we focused on SRF, which has been well studied in other cell types such as epithelial cells, mouse embryonic fibroblasts, and mouse embryonic stem cells (ESCs). SRF is a transcription factor that regulates cell cytoskeleton structure, cell adhesion, motility, and fate decisions (Connelly et al., 2010, Medjkane et al., 2009, Schratt et al., 2002), and activated only when interacting with its co-factor megakaryocytic acute leukemia (MAL, also known as MRTF-A or MKL1), which is sensitive to variations of global-actin (G-actin) levels. In the nucleus, G-actin binds to MAL, preventing it from binding to SRF and activating G-actin-dependent nuclear export to reduce the amount of MAL in the nucleus (Vartiainen et al., 2007), resulting in the suppression of SRF transcription. G-actin exists as a monomeric form of the filamentous actin (F-actin) cytoskeleton, both of which can be interconverted. It has been reported that levels of F-actin may increase with the increase of adhesion and spreading of cells (Ihalainen et al., 2015, O’Connor and Gomez, 2013). On the other hand, depolymerization of F-actin can promote the reprogramming process, and thus enhances iPSC generation (Caiazzo et al., 2016, Sakurai et al., 2014).
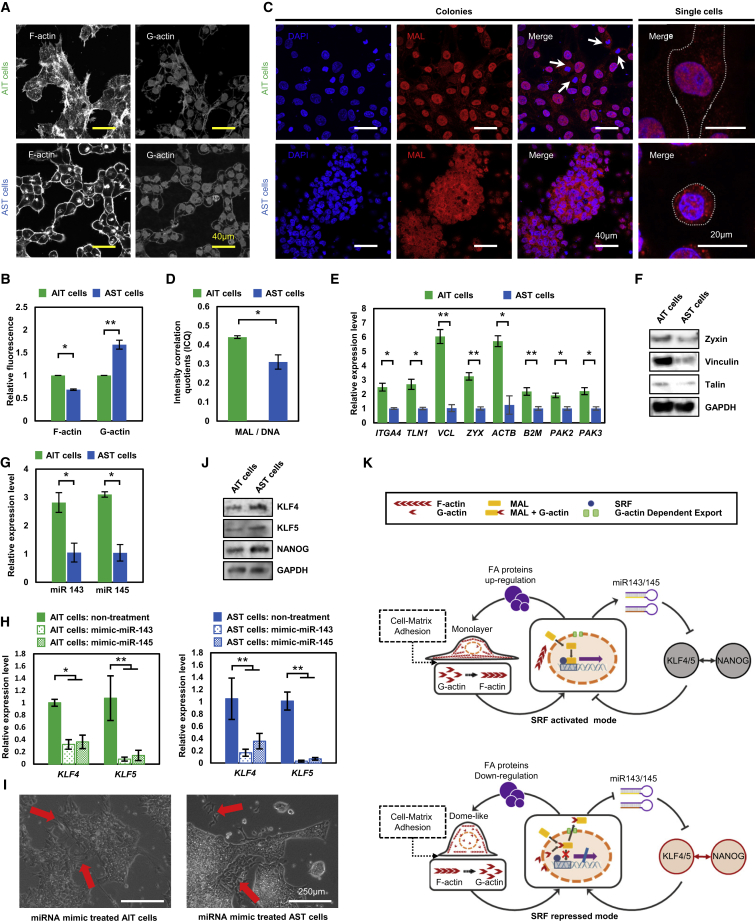
When cells were grown on the low-adhesion GNF substrate, the distinct F-actin stress fibers spread extensively in the cell protrusions and body of AIT cells. In contrast, F-actin was arranged in a cortical cell shell of AST cells (Figure 5A). Quantification of phalloidin and DNase I with fluorescence analysis indicated a significantly lower level of F-actin and higher level of G-actin in AST cells (Figure 5B). The increased level of G-actin resulted in greater export of MAL from the nucleus to the cytoplasm in AST cells (Figures 5C, 5D, S6A, and S6B). In comparison, MAL was concentrated in the nucleus of AIT cells. In addition, on the high-adhesion MG substrate there were no differences in the actin cytoskeleton and MAL distribution between AST and AIT cells (Figures S6C–S6F). Due to the decrease of binding with MAL in AST cells, the transcription of the SRF-related target genes was downregulated, including those encoding FA proteins and microRNA143/145 (miR143/145) (Cordes et al., 2009, Schratt et al., 2002, Xin et al., 2009). Next, an FA PCR array (84 genes included) was employed to investigate the expression level of FA proteins in two type cells. As the result shows, zyxin, vinculin, and talin (SRF target genes) (Schratt et al., 2002) were downregulated in AST cells (Figure 5E). The downregulation of these FA genes in AST cells was also confirmed at the protein level (Figure 5F). The different expression level of FA proteins would further affect the ratio of G/F-actin and consequently impact cell-matrix adhesion, spreading, and morphology (O’Connor and Gomez, 2013, Schratt et al., 2002).
Figure 5.
A Regulatory Circuitry in AST and AIT Cells
(A) Immunofluorescence images of F-actin and G-actin in AIT and AST cells on the GNF substrate. Confocal microscopy images show the basal surface.
(B) Quantification of F-actin and G-actin levels 2 hr after seeding. Total integrated fluorescence of phalloidin (anti-F-actin) and DNaseI (anti-G-actin) was normalized to the fluorescence of AIT cells (mean ± SD, n = 3 independent biological replicates, ∗p < 0.05, ∗∗p < 0.01).
(C) MAL localization in AIT and AST at the colony and single-cell levels. White arrows indicate dividing cells with a disassembled nuclear membrane and MAL distributed in the whole cell bodies.
(D) The intensity correlation quotients (ICQ) indicating the colocalization of MAL and the nucleus in AIT and AST cells (mean ± SD, n = 3 independent biological replicates, ∗p < 0.05).
(E) Relative expression of focal adhesion genes with 2-fold or larger changes in AIT and AST cells. These genes were selected from 84 genes involved in cellular adhesion (mean ± SD, n = 3 independent experiments, ∗p < 0.05, ∗∗p < 0.01).
(F) Western blot of SRF targeted focal adhesion proteins: zyxin, vinculin, and talin in AIT and AST cells.
(G) Relative expression of miRNA143 and miRNA145 in AIT and AST cells (mean ± SD, n = 3 independent experiments, ∗p < 0.05).
(H) The relative expression of KLF4 and KLF5 in AIT and AST cells under self-renewal conditions at 48 hr after transfection of miRNA-mimic-miR143 and 145 (mean ± SD, n = 3 independent biological replicates, ∗p < 0.05, ∗∗p < 0.01).
(I) Phase contrast images of AIT and AST cells at 72 hr after miR143/145-mimic transfection. The cells undergoing differentiation could be observed (red arrows), and the AST cells formed heterogeneous colonies with both flat and domed morphology. Scale bars, 250 μm.
(J) Western blot analysis of KLF4, KLF5, and NANOG expression in AIT and AST cells on GNF substrates.
(K) SRF-based double-loop-coupling cell morphology, adhesion property, and the expression of KLF4/5 and NANOG. To avoid the interference of ROCK inhibitor on cell adhesion, data in this figure were obtained more than 48 hr after withdrawal of the ROCK inhibitor from cell cultures.
See also Figure S6.
On the other hand, the expression levels of SRF-targeted miR143/145 were correspondingly decreased in AST cells (Figure 5G). The microRNAs (miRNAs) could partially bind to complementary target sites in mRNA 3′ UTRs, which results in degradation of the target mRNAs, or translational repression of the encoded proteins (Bartel et al., 2004). Previous studies have reported that miR143/145 could bind with the mRNA of KLF4/5 and suppress their expression (Cordes et al., 2009, Davis-Dusenbery et al., 2011, Xin et al., 2009, Yamaguchi et al., 2011). To verify the function of miR143/145 in hPSCs, we transfected the miRNA-mimic of miR143 and miR145 into the AIT and AST cells. As the result shows, 48 hr after the transfection the expression levels of KLF4/5 were dramatically decreased in two types of cells (Figure 5H). Seventy-two hours after the transfection, the AST cells could not maintain their homogeneous domed morphology and formed heterogeneous colonies with mixed flat and domed morphologies. Both AIT and AST cell start to differentiate even if under self-renewal culture conditions (Figure 5I), consistent with the previous report (Xu et al., 2009). Together with the previous findings, the above data indicated that the miR143/145 could inhibit the expression of KLF4/5 (Cordes et al., 2009, Xin et al., 2009, Xu et al., 2009), which can also repress SRF activity by forming a positive feedback loop (Cordes et al., 2009, Xin et al., 2009). Moreover, KLF4/5 have been shown to regulate numerous biological processes, including upregulation of NANOG (Chan et al., 2009, Jiang et al., 2008, Xu et al., 2009) and E-cadherin (Li et al., 2010, Lim and Thiery, 2012). Therefore, downregulated miR143/145 led to the high expression of KLF4/5 and NANOG at both gene and protein levels (Figures 4D and 5J).
Overall, these results suggest that there is an SRF-based feedback double loop regulating cell morphology and the expression levels of KLF4 and KLF5, which work in either SRF-activated or -repressed modes (Figure 5K). When cultured on the high-adhesion substrate, both AIT and AST cells stayed in the SRF-activated status, whereas on the low-adhesion substrate, the AST cells shifted from SRF- activated to -repressed status, but AIT cells remained in SRF-activated status.
The SRF-Based Feedback Double Loop Made a Bi-stable Switch
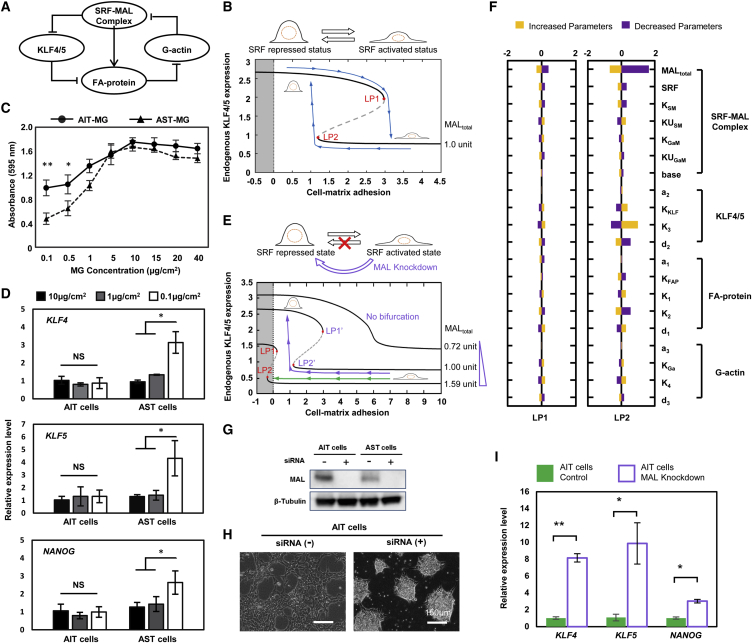
To better understand this phenomenon, a mathematical model was constructed to elucidate the behavior of the SRF-based regulatory double loop. The model focused on the core elements of the regulatory network consisting of four variables (nodes) (Figure 6A; see Supplemental Information for a detailed discussion). The cell's steady-state behavior (represented here by the expression level of KLF4/5) as a function of substrate adhesion is shown in the bifurcation analysis result (Figure 6B). In the case of low substrate adhesion, the cell is in the SRF-repressed state (upper branch), with high KLF4/5 expression and a dome-like morphology, whereas for high cell-matrix adhesion, the cell is in the SRF-activated state (lower branch) with low KLF4/5 expression and a flat monolayer morphology. Interestingly, by changing the adhesion of substrate matrix, one could, in principle, move the cell from one state to another. Suppose that one cell initially starts in the upper branch with high expression of KLF4/5. When the substrate adhesion property increases, the KLF4/5 expression level decreases and finally drops to the lower branch after it crosses the limit point (LP1). Namely, the cell transitions from an SRF-repressed state to an SRF-activated state. Conversely, for cells in the SRF-activated state, when the substrate adhesion property level decreases below another limit point (LP2), the KLF4/5 expression changes back to a high level corresponding to the SRF-repressed state.
Figure 6.
Positive Feedback Loop Makes a Bi-stable Switch
(A) The core regulatory network of the SRF-mediated double loop, comprising KLF4/5, MAL-SRF complex, focal adhesion proteins, and G-actin.
(B) Bifurcation diagram of the KLF4/5 expression level as a function of the cell-matrix adhesion, for a MAL expression level approximately corresponding to that of the AST cells. Solid black lines represent stable cell states, with the SRF-repressed state in the upper branch and the SRF-activated state in the lower branch. For cell-matrix adhesion between the two limiting points LP1 and LP2, the cell can be in either of the two stable states. With changes of cell-matrix adhesion, the cell can switch between the two states reversibly. The region of negative substrate adhesion (gray bar) is unreachable. Blue line indicates the transition of AST cells.
(C) AIT and AST cell adhesion curves on Matrigel-coated substrates with gradient of coating concentrations (0.1–40 μg/cm2) (mean ± SD, n = 4 independent biological replicates, ∗p < 0.05, ∗∗p < 0.01).
(D) Relative expression of KLF4, KLF5, and NANOG in AIT and AST cells on Matrigel-coated substrates with three different coating concentrations (0.1, 1, and 10 μg/cm2) (mean ± SD, n = 3 independent experiments, ∗p < 0.05, NS, not significant).
(E) Bifurcation diagrams of KLF4/5 expression level as a function of cell-matrix adhesion. The three diagrams correspond to the three values of MAL expression level indicated on the right. For a MAL expression level approximately that of an AIT cell (1.59 unit), the cell cannot switch to the upper branch by just changing the cell-matrix adhesion. With reduced total MAL expression, the upper branch can become reachable. Below a critical value (0.72 unit) of MAL expression, there is no bi-stability and the properties of the cell change smoothly. Green line indicates the irreversible transition of AIT cells. Violet line indicates the reversible transition of the small interfering RNA (siRNA)-treated AIT cells.
(F) The relative change of the position of the two limiting points LP1 and LP2, when each of the 20 parameters in the model is increased or decreased by 15%.
(G) Western blot result indicated the interference efficiency of MAL protein in AIT and AST cells.
(H) siRNAs were transfected with lipofection. Suspended AIT cells were treated with siRNA for 2 hr, and then seeded on the GNF substrate. These cells formed a dome-like morphology after MAL siRNA transfection. Samples with only addition of liposome were used as a control.
(I) Relative expression of KLF4, KLF5, and NANOG in AIT cells on low-adhesion GNF substrate with or without siRNA treatment (mean ± SD, n = 3 independent biological replicates, ∗p < 0.05, ∗∗p < 0.01). To avoid the interference of ROCK inhibitor on cell adhesion, data in this figure were obtained more than 48 hr after withdrawal of the ROCK inhibitor from cell cultures.
See also Figure S7.
To verify the mathematical model, we prepared different adhesion property substrates by varying coating concentration of hydrogels (Matrigel, fibronectin, vitronectin, and laminin) as previous studies described (Miyazaki et al., 2012). On substrates coated with the recommended concentration of hydrogels, the two types of cells showed similar cell-matrix adhesion (Figures 6C and S7A). However, the cell-matrix adhesion of AST cells dropped more rapidly than AIT cells with decreasing coating concentration. Interestingly, while AST cells cultured on sparsely MG-coated substrates (0.1 μg/cm2) demonstrated domed morphology and upregulation of KLF4/5 and NANOG expression (Figures 6D and S7B), on densely MG-coated substrates (1 and 10 μg/cm2) they showed monolayer morphology and an expression level of KLF4/5 and NANOG genes similar to AIT cells, indicating a transition suggested in Figure 6B (from the upper to the lower branch). In contrast, as for AIT cells we observed no obvious changes in the morphology and the expression pattern with varying MG coating concentration, indicating that the transition from the lower to the upper branch of Figure 6B is not possible for AIT cells. These results suggest that the AST cells can switch between the SRF-activated and -repressed states with varying substrate adhesion property, while AIT cells were kept in the SRF-activated state. In other words, the transition from the SRF-repressed state to the activated state is irreversible for AIT cells. This irreversibility for AIT cells would happen if the limit point LP2 is in the region that is unreachable by only changing substrate adhesion (Figure 6E, green arrows). To investigate which factors can influence the positions of LP2 and LP1, we carried out a parameter sensitivity analysis for the mathematical model, which showed that, among all the parameters in the model, the total amount of MAL (MALtotal) is the most sensitive, and the effect on LP2 is much bigger than that on LP1 (Figure 6F). Increasing MALtotal shifts LP1 and LP2 to the left, whereas decreasing it shifts LP1 and LP2 to the right (Figure S7C). Based on RNA-seq data, we plotted the cell's steady-state behavior given by the mathematical model for MALtotal values corresponding to AIT and AST cells (Figures 6B and 6F, blue arrows for AST cells and green arrows for AIT cells). Indeed, while AST cells can switch between the two states, for AIT cells the limit point LP2 is unreachable. This is consistent with the observed irreversibility of AIT cells, which indicated the AIT cells are always in an SRF-activated state regardless of the substrate adhesion. That is, AIT cells are not sensitive to the cell-matrix adhesion, while AST cells are sensitive to the cell-matrix adhesion, as their morphology and expression of pluripotent genes are changeable on substrates with varying matrix adhesion.
To further test the mathematical model, we noted that if the MAL protein in AIT cells is reduced then it is possible for the cells to move to the SRF-repressed state with high KLF4/5 expression (Figure 6F, violet arrows). We next performed small interfering RNA-mediated MAL knockdown on AIT cells. Indeed, we observed that in this case AIT cells showed domed morphology and upregulated KLF4/5 and NANOG expression on low-adhesion substrate (Figures 6G–6I), indicating a possible shift to the SRF-repressed state. Taken together, there seems to be some different cell-intrinsic characteristics in the two subtypes, which are possibly acquired during hPSC generation, and make the different bifurcation behavior for the two subtypes. More efforts are needed to understand how these differences occurred and were maintained.
Discussion
In this study, using a single-cell isolation and culture platform we have successfully isolated and proliferated a single hPSC. We found two distinct subtypes of cells co-existing in the conventional hPSC population. Both types can be stably maintained without any application of small-molecule inhibitors and/or gene modification, and their properties inherited during proliferation. Our findings confirmed the inherent heterogeneity of the conventional hPSC population. It has been reported that the differentiation efficiencies vary among different stem cell lines (Osafune et al., 2008, The International Stem Cell Initiative, 2007). Here we revealed different differentiation potential between subtypes within the same cell line. Therefore, the suitably selected single-cell-derived clones of the two subtypes introduced herein may provide more appropriate candidates for lineage differentiation compared with the conventional hPSC population.
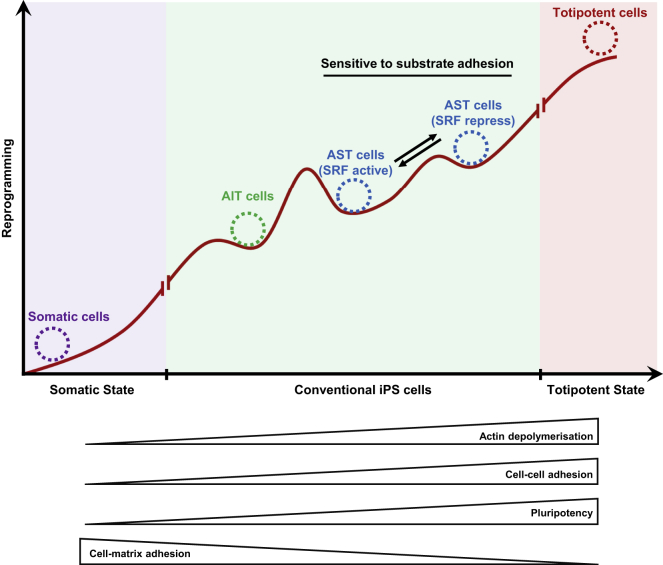
We found that the majority of the standard hPSC population are the AIT cells. Thus, when cultured on the conventional materials, the rare AST cells have always been overlooked and overwhelmed. In this study, using a low-adhesion substrate this adhesion-sensitive subpopulation was revealed. We further found that AST cells showed a reversible transition between the SRF-repressed and active modes when cultured on substrates with varying cell-matrix adhesion, while the other type AIT cells remained in the SRF-active mode. It is well known that overcoming the barrier in mesenchymal-epithelial transition (MET) process is necessary for reprograming fibroblasts into iPSCs (Li et al., 2010, Nieto et al., 2016, Spencer et al., 2007, Zhang et al., 2014). During the MET process, the cells demonstrated certain variations, such as actin depolymerization, decrease of cell-matrix adhesion, increase of cell-cell adhesion, and pluripotency (Lamouille et al., 2014, Li et al., 2010, Nieto et al., 2016, Sakurai et al., 2014, Singh et al., 2013). Interestingly, in our study, similar variations occurred for AST cells when shifting from SRF-active mode to SRF-repressed mode. Taking these together, we hypothesize that the AIT cells need to overcome a sizable barrier when transitioning from the SRF-active mode to the repressed mode, while this transition is much easier for the AST cells (Figure 7). This barrier may be the result of different inducing levels during PSC generation. However, the underlying mechanism for this barrier is so far unclear, and more work is needed to understand how the two types of cells originate.
Figure 7.
Barrier between AIT and AST Cells
A mathematical model was constructed and experimentally tested. It helped to provide a mechanistic view on the relationship among substrate adhesion, pluripotent stem cell morphology, and pluripotency. In particular, it explained the observed reversibility versus irreversibility of the morphological transition in AST and AIT cells, and correctly predicted the key parameter (MALtotal) that controls this behavior. Note that MAL concentration has different values in AIT and AST cells, respectively, suggesting that it itself is controlled by some other factors outside the current model. While the current model only explains the different behavior between AIT and AST cells in terms of their different MAL concentrations, it does not provide any clue why in the first place the two types of cells have distinct MAL expression levels. In future work, the model can be extended to include more players, especially those in the late stage of the reprogramming process, to elucidate how the two types of cells originate.
Furthermore, previous studies have reported that mouse naive ESCs can be maintained on a substrate with low cell-matrix adhesion, even without the application of leukemia inhibitory factor (Chowdhury et al., 2010a), that the adherent mouse ESCs are softer and much more sensitive to a local stress than their differentiated counterparts, and that the microenvironment may play a critical role in shaping embryogenesis during development (Chowdhury et al., 2010b). In light of our findings, it could be that the PSCs with higher pluripotency are more sensitive to the microenvironment.
In summary, our work gave insights on the important issue of the coupling between adhesion, stem cell morphology, and pluripotency, which are expected to facilitate further understanding of the PSC population and the interaction with their microenvironment. It may also contribute to creating a culture system for pluripotent stem cell maintenance and differentiation.
Experimental Procedures
Single-Cell Culture Device Fabrication
The lower layer, the GNF substrate, was generated by electrospinning (voltage, 11 kV; flow rate, 0.2 mL/hr) on culture cover glass slides. Gelatin (11 wt%, type B, from porcine skin; Sigma-Aldrich, USA) solutions were prepared by dissolving gelatin in a mixture of acetic acid, ethyl acetate, and distilled water (acetic acid:ethyl acetate, 3:2) for 16 hr prior to electrospinning. After electrospinning, the GNFs were crosslinked in 0.2 M N-ethyl-N'-(dimethylaminopropyl) carbodiimide and 0.2 M N-hydroxysuccinimide in ethanol for 4 hr. Before use, the GNFs were rinsed with 70% ethanol three times and dried. The upper layer, the multi-well membrane, was produced by spin-coating PDMS (Sylgard 184 from Dow Corning Toray, Japan) at a ratio of 1:10 on a silicon wafer containing an array of 100-μm-tall pillars. The mold was obtained by standard photolithography with SU8-100 (Microchem, Japan). After curing at 70°C for 10 min, the PDMS membranes were peeled from the mold. After rinsing with ethanol and drying, the micro-well was placed on the GNF samples.
Single hPSC Isolation and Culture
Cells were collected from the GNF substrate and then counted and diluted to establish cultures with a concentration of 1 × 103, 2 × 103, and 3 × 103 cells/mL, respectively. Cell suspensions (200 μL) were seeded on the single-cell culture platform. After 2 hr, 2 mL of conditioned medium was added and changed every 1–2 days. After formation of the single-cell-derived clones, they were dissociated by an enzyme-free solution and re-seeded on a new GNF substrate for proliferation.
Pluripotent Stem Cell Culture
The human iPSC lines 253G1 and IMR, and the human ESC lines H1 and H9, were used for this study. hESCs were used following the Kyoto University guidelines. Cells were seeded at 4 × 104 cells/cm2 in mTeSR-1 cell culture medium (STEMCELL Technologies, USA) supplemented with 10 μM of the ROCK inhibitor Y27632 (Wako, Japan) on GNF (eletrospinning time: 8 min) or MG-coated substrates (BD Biosciences, USA). Since day 2, the medium was changed to medium without Y27632. Then the culture medium was changed daily. Cells were dissociated to single cells and passaged every 3–4 days. Non-enzymatic cell dissociation ethylenediaminetetraacetic acid-based solution (Thermo Fisher Scientific, USA) was used to harvest cells cultured on the GNF substrate, and TrypLE Express (Gibco, USA) was used for dissociating cells cultured on the MG-coated substrate. For obtaining a close comparison, two types of cells were cultured in the same Petri dish with exactly the same culture condition. The images of different areas were captured by continuous scanning with a 20-min interval for 48 hr under a microscope (IX81, Olympus, Japan) at 37°C and 5% CO2. The separate images were combined and converted to video with a customized program (MATLAB, MathWorks, USA).
Immunofluorescence Staining and Quantification
Cells were fixed in 4% paraformaldehyde buffer for 15 min, and permeabilized with 0.5% Triton X-100/PBS for 5 min at room temperature. Samples were blocked with blocking buffer (5%, v/v, normal goat serum; 5%, v/v, normal donkey serum; 3%, v/v, BSA; and 0.1%, v/v, Tween 20 in PBS) for 1 hr and incubated with the following primary antibodies diluted in blocking buffer at room temperature for 2 hr or at 4°C overnight: anti-human OCT4 (2 μg/mL, sc-5279, Santa Cruz Biotechnology, USA), anti-human NANOG (9.4 μg/mL, 4903S, Cell Signaling Technology, UK), anti-human β-tubulin III (6 μg/mL, 5568, Cell Signaling Technology), anti-human α-SMA (2 μg/mL, ab5694, Abcam, UK), anti-human SOX17 (20 μg/mL, MAB1924, R&D Systems, USA), anti-E-cadherin (1/300 dilution, 3195S, Cell Signaling Technology), and anti-MKL1 (3 μg/mL, HPA030782, Sigma-Aldrich). Then, the sample was incubated with the following appropriate secondary antibodies diluted in blocking buffer at room temperature for 1 hr: Alexa Fluor 594 anti-rabbit or mouse immunoglobulin G (IgG) (0.35 μg/mL, 711-586-152 and 715-586-150, Jackson ImmnoResearch, USA) or Alexa Fluor 488 anti-mouse or rabbit IgG (0.35 μg/mL, 715-545-150 and 711-545-152, Jackson ImmnoResearch). Finally, samples were counterstained with 300 nM DAPI (4′-6-diamidino-2-phenylindole) (Wako) at room temperature for 20 min. After each step, samples were washed with D-PBS three times. For the quantification of G-actin and F-actin, phalloidin (1.5 μg/mL, P1951, Sigma-Aldrich) and DNaseI (3 μg/mL, D12371, Invitrogen, USA) double-stained z stack images for each sample (>120 cells) were taken by confocal microscopy, and the integrated density of cell areas were analyzed using ImageJ software (NIH, Bethesda, MD, USA).
qRT-PCR and RNA-Seq Analysis
Total RNA was isolated using the QIAshredder (QIAGEN, USA) or RNeasy Mini Kit (QIAGEN) following the manufacturer's instructions. Genomic DNA was degraded by RNase-Free DNase Set (QIAGEN). RNA concentrations were measured on a NanoDrop 1000 spectrometer (Thermo Fisher Scientific). cDNA of total RNA was synthesized with a first-strand synthesis kit (TaKaRa, Japan), and the cDNA of miRNA was synthesized with an miRNA cDNA Synthesis Kit, with poly(A) polymerase tailing (Applied Biological Materials, Canada), and amplified with SYBR Green PCR Master Mix (Thermo Fisher Scientific) on an Applied Biosystems 7300 Real-Time PCR system (Thermo Fisher Scientific). The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression level was used as an internal normalization control. Human FA PCR Array (PAHS-145Z, QIAGEN) in a 96-well format was used in real-time PCR analysis. The has-miR143 primer, has-miR145 primer, and universal 3′ miRNA reverse primer (MPH01164 and MPH01166, Applied Biological Materials) were used in real-time PCR analyses of miRNAs. The other primers used in this work are included in Table S2. RNA-seq libraries were prepared using 2 μg of total RNA according to the manufacturer’s instructions (TruSeq RNA Sample Preparation Kit v.2, Illumina, USA). Samples were multiplexed and sequenced (HiSeq 2000, Illumina). A total of 23,615 genes has been detected. A total of 2,964 significantly different genes (p <0.05) were selected to generate the heatmaps using the open-source R software packages. A total of 429 genes (p <0.05, fold change > 2) were selected for the GO analysis and 257 genes were found enriched. The GO analysis was carried out using DAVID 6.7 online analysis (https://david-d.ncifcrf.gov/home.jsp).
Author Contributions
L.Y., L.L., C.T., and H.K. conceived the project; L.Y., L.L., J.L., Y.T., K.O., C.L., F.T., Y.C., C.T., and H.K. designed the experiments; L.Y., J.L., L.L., J.H., Y.T., N.F., M.N., X.D., Y.D., Y.H., I.M., A.Y., M.T., and W.Y. carried out the experiments; L.Y., J.L., L.L., J.H., X.D., and Y.D. analyzed the data; L.Y., J.L., L.L., and C.T. wrote the paper.
Acknowledgments
We acknowledge Dr. Kouichi Hasegawa, Dr. Shinichiro Chuma, Mr. Kiyotaka Tusji, and Dr. Yulei Wei for their helpful suggestions and discussions. We also thank Panasonic for their support with the data analysis. Funding was provided by the Japan Society for the Promotion of Science (JSPS): Grants-in-Aid for Scientific Research (26289065, 23246045, and 17K14624) and a Ministry of Education, Culture, Sports, Science and Technology (MEXT) scholarship; the WPI-iCeMS is supported by the World Premier International Research Centre Initiative (WPI) MEXT, Japan; the Center of Innovation (COI) Program from MEXT and JST. Y.H. and C.T. acknowledge support from the Chinese Ministry of Science and Technology (2015CB910300) and the National Natural Science Foundation of China (91430217). M.T. thanks the German Science Foundation (SFB873B07) for support. I.M. is a shareholder of Myoridge.
Published: June 28, 2018
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, seven figures, two tables, and three videos and can be found with this article online at https://doi.org/10.1016/j.stemcr.2018.06.003.
Contributor Information
Chao Tang, Email: tangc@pku.edu.cn.
Hidetoshi Kotera, Email: kotera.hidetoshi.7e@kyoto-u.jp.
Li Liu, Email: li-liu@surg1.med.osaka-u.ac.jp.
Accession Numbers
The accession number for the RNA-seq data for the MCoG and DCoG cells reported in this paper is GEO: GSE115256.
Supplemental Information
References
- Bartel D.P., Lee R., Feinbaum R. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Caiazzo M., Okawa Y., Ranga A., Piersigilli A., Tabata Y., Lutolf M.P. Defined three-dimensional microenvironments boost induction of pluripotency. Nat. Mater. 2016;15:344–352. doi: 10.1038/nmat4536. [DOI] [PubMed] [Google Scholar]
- Chan K.K.-K., Zhang J., Chia N.-Y., Chan Y.-S., Sim H.S., Tan K.S., Oh S.K.-W., Ng H.-H., Choo A.B.-H. KLF4 and PBX1 directly regulate NANOG expression in human embryonic stem cells. Stem Cells. 2009;27:2114–2125. doi: 10.1002/stem.143. [DOI] [PubMed] [Google Scholar]
- Chen T., Yuan D., Wei B., Jiang J., Kang J., Ling K., Gu Y., Li J., Xiao L., Pei G. E-Cadherin-mediated cell–cell contact is critical for induced pluripotent stem cell generation. Stem Cells. 2010;28:1315–1325. doi: 10.1002/stem.456. [DOI] [PubMed] [Google Scholar]
- Chowdhury F., Li Y., Poh Y.-C., Yokohama-Tamaki T., Wang N., Tanaka T.S. Soft substrates promote homogeneous self-renewal of embryonic stem cells via downregulating cell-matrix tractions. PLoS One. 2010;5:e15655. doi: 10.1371/journal.pone.0015655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury F., Na S., Li D., Poh Y.-C., Tanaka T.S., Wang F., Wang N. Material properties of the cell dictate stress-induced spreading and differentiation in embryonic stem cells. Nat. Mater. 2010;9:82–88. doi: 10.1038/nmat2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly J.T., Gautrot J.E., Trappmann B., Tan D.W., Donati G., Huck W.T.S., Watt F.M. Actin and serum response factor transduce physical cues from the microenvironment to regulate epidermal stem cell fate decisions. Nat. Cell Biol. 2010;12:711–718. doi: 10.1038/ncb2074. [DOI] [PubMed] [Google Scholar]
- Cordes K.R., Sheehy N.T., White M.P., Berry E.C., Morton S.U., Muth A.N., Lee T.-H., Miano J.M., Ivey K.N., Srivastava D. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:1–7. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis-Dusenbery B.N., Chan M.C., Reno K.E., Weisman A.S., Layne M.D., Lagna G., Hata A. Down-regulation of Krüppel-like factor-4 (KLF4) by MicroRNA-143/145 is critical for modulation of vascular smooth muscle cell phenotype by transforming growth factor-β and bone morphogenetic protein 4. J. Biol. Chem. 2011;286:28097–28110. doi: 10.1074/jbc.M111.236950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downing T.L., Soto J., Morez C., Houssin T., Fritz A., Yuan F., Chu J., Patel S., Schaffer D.V., Li S. Biophysical regulation of epigenetic state and cell reprogramming. Nat. Mater. 2013;12:1154–1162. doi: 10.1038/nmat3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drukker M., Tang C., Ardehali R., Rinkevich Y., Seita J., Lee A.S., Mosley A.R., Weissman I.L., Soen Y. Isolation of primitive endoderm, mesoderm, vascular endothelial and trophoblast progenitors from human pluripotent stem cells. Nat. Biotechnol. 2012;30:531–542. doi: 10.1038/nbt.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enver T., Pera M., Peterson C., Andrews P.W. Stem cell states, fates, and the rules of attraction. Cell Stem Cell. 2009;4:387–397. doi: 10.1016/j.stem.2009.04.011. [DOI] [PubMed] [Google Scholar]
- Hansson J., Rafiee M.R., Reiland S., Polo J.M., Gehring J., Okawa S., Huber W., Hochedlinger K., Krijgsveld J. Highly coordinated proteome dynamics during reprogramming of somatic cells to pluripotency. Cell Rep. 2012;2:1579–1592. doi: 10.1016/j.celrep.2012.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihalainen T.O., Aires L., Herzog F.A., Schwartlander R., Moeller J., Vogel V. Differential basal-to-apical accessibility of lamin A/C epitopes in the nuclear lamina regulated by changes in cytoskeletal tension. Nat. Mater. 2015;14:1252–1261. doi: 10.1038/nmat4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J., Chan Y.-S., Loh Y.-H., Cai J., Tong G.-Q., Lim C.-A., Robson P., Zhong S., Ng H.-H. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat. Cell Biol. 2008;10:353–360. doi: 10.1038/ncb1698. [DOI] [PubMed] [Google Scholar]
- Lamouille S., Xu J., Derynck R. Molecular mechanisms of epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Zhou J., Wang L., Shin M.E., Su P., Lei X., Kuang H., Guo W., Yang H., Cheng L. Integrated biochemical and mechanical signals regulate multifaceted human embryonic stem cell functions. J. Cell Biol. 2009;191:631–644. doi: 10.1083/jcb.201006094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Liang J., Ni S., Zhou T., Qing X., Li H., He W., Chen J., Li F., Zhuang Q. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- Lim J., Thiery J.P. Epithelial-mesenchymal transitions: insights from development. Development. 2012;139:3471–3486. doi: 10.1242/dev.071209. [DOI] [PubMed] [Google Scholar]
- Liu L., Yoshioka M., Nakajima M., Ogasawara A., Liu J., Hasegawa K., Li S., Zou J., Nakatsuji N., Kamei K. Nanofibrous gelatin substrates for long-term expansion of human pluripotent stem cells. Biomaterials. 2014;35:6259–6267. doi: 10.1016/j.biomaterials.2014.04.024. [DOI] [PubMed] [Google Scholar]
- Masaki H., Ishikawa T., Takahashi S., Okumura M., Sakai N., Haga M., Kominami K., Migita H., McDonald F., Shimada F., Sakurada K. Heterogeneity of pluripotent marker gene expression in colonies generated in human iPS cell induction culture. Stem Cell Res. 2008;1:105–115. doi: 10.1016/j.scr.2008.01.001. [DOI] [PubMed] [Google Scholar]
- Medjkane S., Perez-Sanchez C., Gaggioli C., Sahai E., Treisman R. Myocardin-related transcription factors and SRF are required for cytoskeletal dynamics and experimental metastasis. Nat. Cell Biol. 2009;11:257–268. doi: 10.1038/ncb1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki T., Futaki S., Suemori H., Taniguchi Y., Yamada M., Kawasaki M., Hayashi M., Kumagai H., Nakatsuji N., Sekiguchi K. Laminin E8 fragments support efficient adhesion and expansion of dissociated human pluripotent stem cells. Nat. Commun. 2012;3:1210–1236. doi: 10.1038/ncomms2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narsinh K.H., Sun N., Sanchez-freire V., Lee A.S., Almeida P., Hu S., Jan T., Wilson K.D., Leong D., Rosenberg J. Single cell transcriptional profiling reveals heterogeneity of human induced pluripotent stem cells. J. Clin. Invest. 2011;121:1217–1221. doi: 10.1172/JCI44635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narve E., Stubb A., Guzman C., Blomqvist M., Balboa D., Lerche M., Saari M., Otonkoski T., Ivaska J. A strong contractile actin fence and large adhesions direct human pluripotent colony morphology and adhesion. Stem Cell Rep. 2017;9:67–76. doi: 10.1016/j.stemcr.2017.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto M.A., Huang R.Y., Jackson R.A., Thiery J.P. EMT: 2016. Cell. 2016;166:21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- O’Connor J.W., Gomez E.W. Cell adhesion and Shape regulate TGF-beta1-induced epithelial-myofibroblast transition via MRTF-a signaling. PLoS One. 2013;8:e83188. doi: 10.1371/journal.pone.0083188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohgushi M., Matsumura M., Eiraku M., Murakami K., Aramaki T., Nishiyama A., Muguruma K., Nakano T., Suga H., Ueno M. Molecular pathway and cell state responsible for dissociation-induced apoptosis in human pluripotent stem cells. Cell Stem Cell. 2010;7:225–239. doi: 10.1016/j.stem.2010.06.018. [DOI] [PubMed] [Google Scholar]
- Osafune K., Caron L., Borowiak M., Martinez R.J., Fitz-gerald C.S., Sato Y., Cowan C.A., Chien K.R., Melton D.A. Marked differences in differentiation propensity among human embryonic stem cell lines. Nat. Biotechnol. 2008;26:2007–2009. doi: 10.1038/nbt1383. [DOI] [PubMed] [Google Scholar]
- Qin H., Diaz A., Blouin L., Lebbink R.J., Patena W., Tanbun P. Systematic identification of barriers to human iPSC generation. Cell. 2014;158:449–461. doi: 10.1016/j.cell.2014.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redmer T., Diecke S., Grigoryan T., Quiroga-negreira A., Birchmeier W., Besser D. E-cadherin is crucial for embryonic stem cell pluripotency and can replace OCT4 during somatic cell reprogramming. EMBO Rep. 2011;12:720–726. doi: 10.1038/embor.2011.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai K., Talukdar I., Patil V.S., Dang J., Li Z., Chang K.Y., Lu C.C., Delorme-Walker V., Dermardirossian C., Anderson K. Kinome-wide functional analysis highlights the role of cytoskeletal remodeling in somatic cell reprogramming. Cell Stem Cell. 2014;14:523–534. doi: 10.1016/j.stem.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt G., Philippar U., Berger J., Schwarz H., Heidenreich O., Nordheim A. Serum response factor is crucial for actin cytoskeletal organization and focal adhesion assembly in embryonic stem cells. J. Cell Biol. 2002;156:737–750. doi: 10.1083/jcb.200106008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A., Suri S., Lee T., Chilton J.M., Cooke M.T., Chen W., Fu J., Stice S.L., Lu H., Mcdevitt T.C. Adhesion strength-based, label-free isolation of human pluripotent stem cells. Nat. Methods. 2013;10:438–444. doi: 10.1038/nmeth.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer H.L., Eastham A.M., Merry C.L.R., Southgate T.D., Perez-campo F., Soncin F., Ritson S., Kemler R., Stern P.L., Ward C.M. E-Cadherin inhibits cell surface localization of the pro-migratory 5T4 oncofetal antigen in mouse embryonic stem cells. Mol. Biol. Cell. 2007;18:2838–2851. doi: 10.1091/mbc.E06-09-0875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;2:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- The International Stem Cell Initiative Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat. Biotechnol. 2007;25:803–816. doi: 10.1038/nbt1318. [DOI] [PubMed] [Google Scholar]
- Vartiainen M.K., Guettler S., Larijani B., Treisman R. Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactor MAL. Science. 2007;316:1749–1752. doi: 10.1126/science.1141084. [DOI] [PubMed] [Google Scholar]
- Xin M., Small E.M., Sutherland L.B., Qi X., McAnally J., Plato C.F., Richardson J.A., Bassel-Duby R., Olson E.N. MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes Dev. 2009;23:2166–2178. doi: 10.1101/gad.1842409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N., Papagiannakopoulos T., Pan G., Thomson J.A., Kosik K.S. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137:647–658. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S., Yamahara K., Homma K., Suzuki S., Fujii S., Morizane R., Monkawa T., Matsuzaki Y., Kangawa K., Itoh H. The role of microRNA-145 in human embryonic stem cell differentiation into vascular cells. Atherosclerosis. 2011;219:468–474. doi: 10.1016/j.atherosclerosis.2011.09.004. [DOI] [PubMed] [Google Scholar]
- Yan L., Yang M., Guo H., Yang L., Wu J., Li R., Liu P., Lian Y., Zheng X., Yan J. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat. Struct. Mol. Biol. 2013;20:1131–1139. doi: 10.1038/nsmb.2660. [DOI] [PubMed] [Google Scholar]
- Yoshikawa H.Y., Rossetti F.F., Kaufmann S., Kaindl T., Madsen J., Engel U., Lewis A.L., Armes S.P., Tanaka M. Quantitative evaluation of mechanosensing of cells on dynamically tunable hydrogels. J. Am. Chem. Soc. 2011;133:1367–1374. doi: 10.1021/ja1060615. [DOI] [PubMed] [Google Scholar]
- Zhang J., Tian X., Zhang H., Teng Y., Li R., Bai F., Elankumaran S., Xing J. TGF-b-induced epithelial-to-mesenchymal transition proceeds through stepwise activation of multiple feedback loops. Sci. Signal. 2014;7:ra91. doi: 10.1126/scisignal.2005304. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.