Abstract
Background
Osteogenesis imperfecta (OI), the commonest inherited bone fragility disorder, affects 1 in 15,000 live births resulting in frequent fractures and reduced mobility, with significant impact on quality of life. Early diagnosis is important, as therapeutic advances can lead to improved clinical outcome and patient benefit.
Report
Whole exome sequencing in patients with OI identified, in two patients with a multi-system phenotype, compound heterozygous variants in NBAS (Neuroblastoma amplified sequence). Patient 1: NBAS c.5741G>A p.(Arg1914His); c.3010C>T p.(Arg1004*) in a 10-year old boy with significant short stature, bone fragility requiring treatment with bisphosphonates, developmental delay and immunodeficiency. Patient 2: NBAS c.5741G>A p.(Arg1914His); c.2032C>T p.(Glu678*) in a 5-year old boy with similar presenting features, bone fragility, mild developmental delay, abnormal liver function tests and immunodeficiency.
Discussion
Homozygous missense NBAS variants cause SOPH syndrome (Short stature; Optic atrophy; Pelger-Huet anomaly), the same missense variant was found in our patients on one allele and a nonsense variant in the other allele. Recent literature suggests a multi-system phenotype. In this study, patient fibroblasts have shown reduced collagen expression, compared to control cells and RNAseq studies, in bone cells show that NBAS is expressed in osteoblasts and osteocytes of rodents and primates. These findings provide proof-of-concept that NBAS mutations have mechanistic effects in bone, and that NBAS variants are a novel cause of bone fragility, which is distinguishable from ‘Classical’ OI.
Conclusions
Here we report on variants in NBAS, as a cause of bone fragility in humans, and expand the phenotypic spectrum associated with NBAS. We explore the mechanism underlying NBAS and the striking skeletal phenotype in our patients.
Keywords: bone fragility, Osteogenesis imperfecta, NBAS, nonsense mediated decay (NMD), secretory pathway, collagen expression
Introduction
NBAS (Neuroblastoma-amplified sequence), also previously referred to as NAG (Neuroblastoma-amplified gene) contains 52 exons, spans 420 kb and is mapped to chromosome 2p24.3 [1] (Scott et al., 2003). It was initially identified as a gene co-amplified with the N-myc (MYCN) gene in neuroblastoma cell lines [2] (Wimmer et al., 1999). NBAS was initially described as a novel factor involved in the nonsense mediated (NMD) decay pathway in human cells, in zebrafish and in nematodes [3,4] (Longman et al., 2007; Anastasaki et al., 2010). It was shown that NBAS acts in concert with core NMD factors to co-regulate a large number of endogenous RNA targets [5] (Longman et al., 2013). Subsequently, NBAS was also identified as a component of the syntaxin 18 complex, which is involved in Golgi-to-endoplasmic reticulum (ER) retrograde transport [6, 7] (Aoki et al., 2009; Spang A., 2013). NBAS is also said to be an important component of the ER tethering complex [8] (Hong WJ and Lev S., 2014). Using whole exome sequencing, we identified compound heterozygous variants in NBAS in two patients with bone fragility: Patient 1 presented with significant short stature, bone fragility requiring treatment with bisphosphonates, developmental delay and immunodeficiency and was independently investigated for a novel cause of bone fragility (having tested negative for all published variants in OI). Patient 2 was recruited to the Deciphering Developmental Disorders (DDD) study and underwent trio whole exome sequencing.
Homozygous missense variants in NBAS have been implicated in a hereditary short stature syndrome referred to as SOPH syndrome (Short stature; Optic atrophy; Pelger-Huet anomaly) observed in the Yakut Siberian population isolate [9] (Maksimova et al., 2010). Compound heterozygous mutations in NBAS have also been described in acute onset liver failure [10] (Haack et al., 2015). Further reports have suggested a multi-system phenotype [11,12,13] (Segarra NG et al., 2015; Capo-Chichi et al., 2015; Staufner et al., 2016). Bone fragility severe enough to need bisphosphonate therapy has not reported as a feature, so far, in association with NBAS. Therefore, we propose that compound heterozygous mutations in NBAS can account for a significant form of bone fragility and needs to be considered in the differential diagnosis of osteogenesis imperfecta (OI).
Clinical Report
Patient 1
This 10-year old boy is the second child of healthy, non-consanguineous parents, of North European origin, with no significant family history. He was born by spontaneous breech delivery at 33 weeks gestation following a normal pregnancy, with a birth weight of 1.75kg (9th centile). He required continuous positive airway pressure ventilation (CPAP) for 24 hours and phototherapy for jaundice. He had recurrent infections and significant problems with his feeding, requiring percutaneous gastrostomy insertion to maintain adequate nutrition. He had recurrent admissions to hospital with infections including severe recurrent shingles with worsening of liver function during infectious episodes and consequent progressive lymphopenia and hypogammaglobulinaemia requiring immunoglobulin replacement. He was diagnosed with a horizontal nystagmus, bilateral optic atrophy and myopia, needing corrective glasses, but his hearing was reported as normal. He has moderate intellectual disability and growth parameters remain well below the 0.4th centile. In early childhood, he went on to sustain fractures of his tibia and metatarsals, following minimal trauma and has discoloured teeth. He attends an integrated play school, as he has been diagnosed with an autism spectrum disorder.
On examination at 9-years of age, he was dysmorphic, with proptosis, progeric appearance to his skin, bilateral low-set ears, dentinogenesis imperfecta, and bilateral 5th finger clinodactyly with bulbous tips to his fingers and toes (Figure 1a-e). He has a high-pitched voice and growth parameters at 7 years of age were: weight ~ 13.1kg (<0.4th centile), height ~ 88cms (<<0.4th centile), head circumference ~ 49.5cms (0.4th-2nd centile), with no evidence of asymmetry. Repeat cranial ultrasounds and MRI-brain scans did not show any evidence of ventricular dilatation despite initial concerns regarding a large anterior fontanelle. A dysplasia skeletal survey showed multiple Wormian bones, slender tubular bones and osteopenia (Figure 2a-d). A transiliac bone biopsy at 7-years of age, following recurrent low-trauma fractures, demonstrated osteoporosis with high bone turnover with marked periosteal bone resorption, which was different to appearances in classical OI (Figure 3a-b). In terms of his skeletal phenotype, he had a tender back with loss of vertebral height on lateral spine radiograph and a low lumbar bone mineral areal density (BMAD) with a Z-score of -3.5 at 9-years of age. He was commenced on Pamidronate with remarkable improvement to his bone health. Collagen species analysis on SDS-Page gel was normal with no obvious shift, but electron microscopy showed multiple small collagen flowers, variable shaped collagen fibrils with a mean collagen fibril diameter (CFD) of 75nm (Figure 3c-d). Genetic testing so far included: normal 60K ArrayCGH, FRAX, UPD7 and 11p15 methylation testing, targeted exome panel including all published genes known to cause bone fragility, all of which were reported as negative.
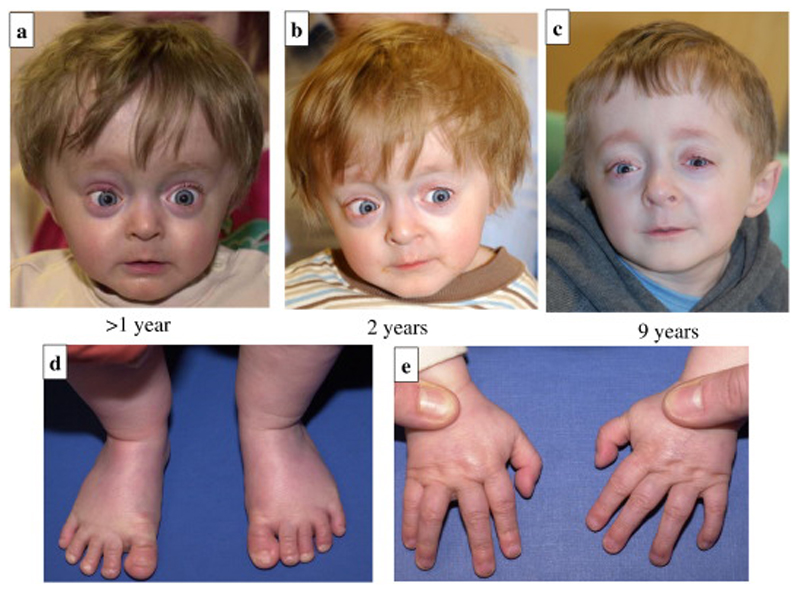
Figure 1.
a-e: 1a-c: Facial features as an infant and aged >1, 2 and 9 years showing grey sclerae, broad forehead, bilateral low-set ears, proptosis and progeric appearance; 1d-e: Hands and feet at 2-years of age.
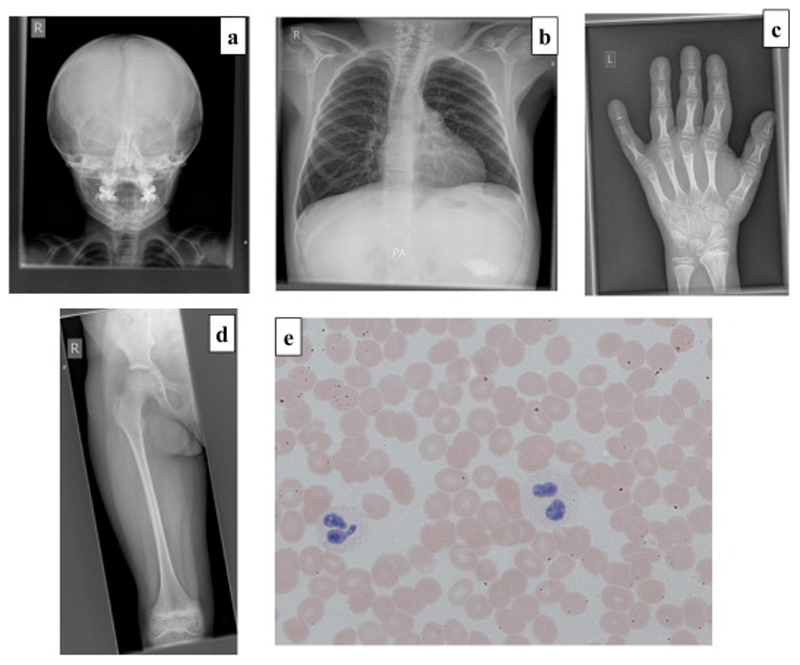
Figure 2.
a-d: Radiographs demonstrating slender ribs, tubular long bones with thin cortices and osteopenia consistent with a diagnosis of OI. 2a: AP skull radiograph (aged 6 years) There are multiple Wormian bones; the clavicles are slender and the erupted teeth are relatively dense. The anterior fontanelle remains open.
2b-d: Selected images from a full dysplasia skeletal survey (aged 9 years 8 months)
2b: Left hand. There is significant periarticular osteopenia (see Fig 2c). The metacarpals (and less marked) the proximal phalanges are overmodelled and there is an ivory epiphysis of the terminal phalanx of the fifth finger. The terminal tufts are prominent.
2c: AP Chest. The ribs and clavicles are slender; however, there are no fractures and vertebral body height is preserved. Note the presence of a gastrostomy.
2d. AP Right Femur: Overmodelled with slender diaphysis and relatively flared distal metaphysis. Periarticular osteopenia is again noted (see Fig 2a)
2e: Peripheral blood film in Patient 1 demonstrating a hypolobulated neutrophil (left) and a Pelger-Huet cell (right).
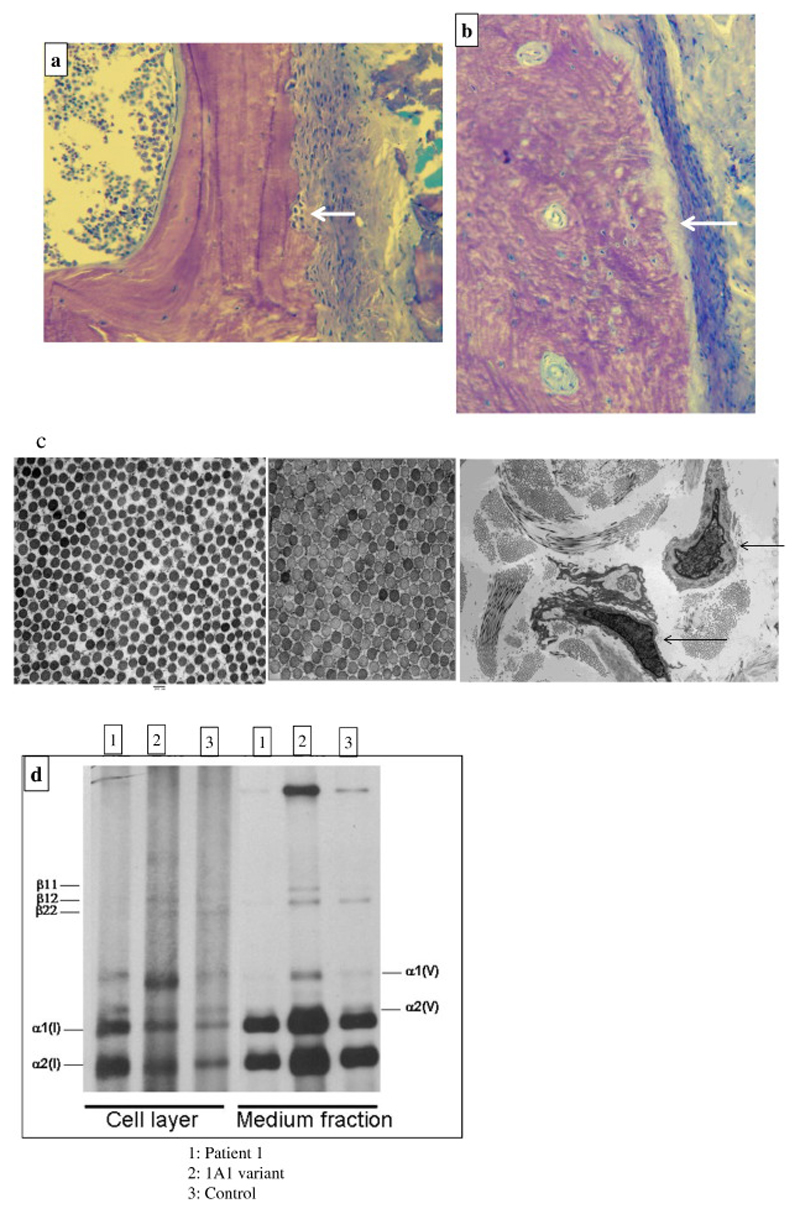
Figure 3.
a-b: 3a: Toluidine blue-stained section of an undecalcified trans-iliac bone biopsy, original magnification x400 demonstrating cortex, with periosteum on the right showing high turnover osteopenia with marked sub-periosteal bone resorption (arrow) and normal lamellar bone matrix structure in Patient-1, aged 9 years; 3b: appearance in ‘Classical OI’ with abnormal matrix pattern and increased periosteal bone formation surface (arrow). Toluidine blue; Original magnification of 400x.
c-d: 3c: Electron Microscopy of skin biopsy from Patient 1(left image) compared to normal control (middle image) showing normal collagen in mid-reticular dermis with mildly reduced mean collagen fibril diameter (CFD); Original magnification of 20,000x; Lower magnification (right image) with arrows indicating deep reticular dermal fibroblast with expanded protein filled rough endoplasmic reticulum; Original magnification x2,600.
3d: normal collagen species analysis with no deviation from control (1 and 3) in comparison to patient with a COL1A1 variant (2).
Patient 2
This 6-year old boy is the first child of healthy, non-consanguineous parents (mother is of Northern-Spanish origin whilst father is of Italian origin) with no significant family history. He was born at 38 weeks gestation, by spontaneous delivery following a normal pregnancy with a birth weight of 2.44kg (2nd centile). He did not need to go to Special Care Baby Unit but went on to develop a spiral fracture of his left femur whilst being positioned for a feed at 3-months of age. A skeletal survey performed at the time showed osteopenic slender bones, thin skull vault with a large anterior fontanelle, but no Wormian bones. These features were thought to be consistent with but not completely diagnostic of type IV OI. He had recurrent infections, problems with his feeding and poor weight gain. He also went on to develop recurrent episodes of ketotic hypoglycaemia, which resolved spontaneously and hypogammaglobulinaemia, needing 2-weekly immunoglobulin infusions. He was diagnosed with abnormal liver function when he had several episodes of temporarily elevated transaminases triggered by infections. He was diagnosed with a horizontal nystagmus and bilateral optic atrophy, but his hearing was reported to be normal. He has mild intellectual disability, managing in mainstream school with 1-to-1 help. His growth parameters remain below the 0.4th centile, with some preservation of head circumference (0.4th-2nd centile).
On examination at 5-years of age, he was dysmorphic, with brachycephaly, proptosis, progeric appearance to his skin, bilateral low-set ears, greyish sclerae, and bilateral 5th finger clinodactyly with bulbous tips to his fingers and toes (Figure 4a-c). He has a high-pitched, distinctive voice and growth parameters were: weight ~ 14.4kg (<0.4th centile), height ~ 96.9cms (<0.4th centile), head circumference ~ 49.6cms (0.4th-2nd centile), with no evidence of asymmetry. Repeat cranial ultrasounds, in view of persistent large anterior fontanelle, and a MRI-brain scan at one year of age showed non-specific white matter changes, but otherwise normal appearances. In terms of his skeletal phenotype, he has gone on to sustain stress fractures of his feet, diagnosed on bone scan and a low lumbar bone mineral areal density (BMAD) with a Z-score of -4.01 at 5-years of age. He has recently been started on Pamidronate with a good response to therapy. A dysplasia skeletal survey subsequently confirmed similar features as reported previously (Figure 4d-e). Genetic testing so far included: normal 60K ArrayCGH, targeted exome panel including genes known to cause OI, all of which were reported as negative. He was subsequently recruited to the DDD study (Decipher Patient ID: 264693).
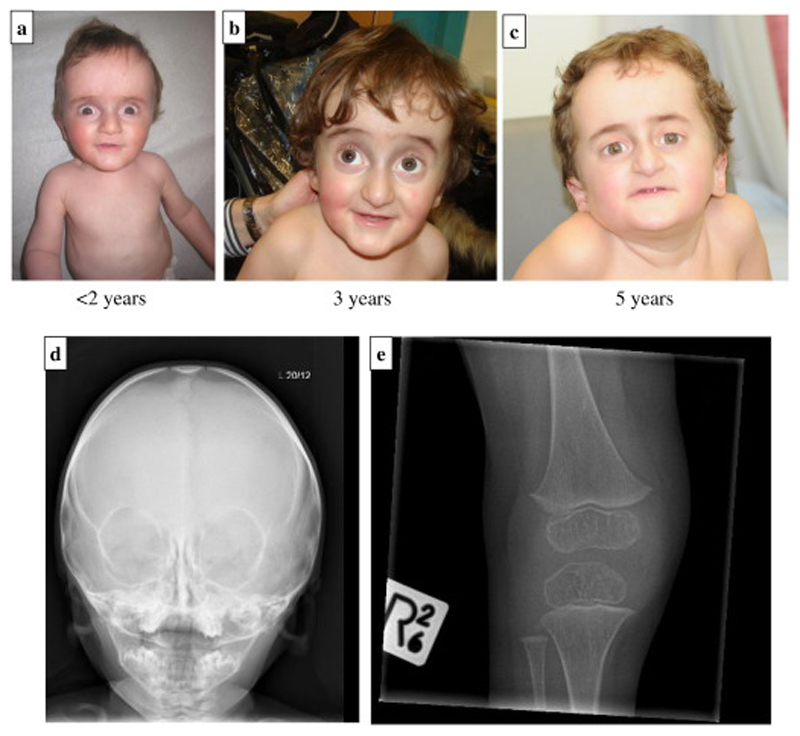
Figure 4.
a-e: 4a-c: Facial appearance of Patient 2 aged <2, 3 and 5 years demonstrating similar facial dysmorphism to Patient 1 with progeric appearance, grey sclerae, broad forehead, bilateral low-set ears, proptosis; 4d-e: Skull and right lower limb X-rays demonstrating thin skull vault, slender bones and osteopenia.
Materials and Methods
Patient 1 was recruited into a research project to study atypical forms of OI, to establish genotype: phenotype correlations therein. Ethical approval was obtained from the local regional ethics committee (LREC) to undertake phenotyping and genetic work-up in this group of patients. Patient 2 was recruited to the DDD study.
DNA Sequencing and mutation analysis in Patient 1
Total genomic DNA was isolated from 2 to 5 ml peripheral blood using standard extraction methods. Whole exome sequencing was performed by Personalis using their ACE Exome™ Assay. The data were analysed with the Personalis ACE pipeline; this uses BWA (version number 0.7.5a-r405) for alignment, GATK’s UnifiedGenotyper for variant calling and GATK’s VQSR to provide a site quality score using the single patient sample. The average depth in the target region was 115X. Variants were extensively annotated with AnnoL, a Personalis tool. The annotation includes frequencies by subpopulation from 1000 genomes and the Exome Sequencing Project; information from dbNSFP; predicted mutational impacts from tools including SIFT, Polyphen2 and MutationTaster; known associations from disease databases such as OMIM, ClinVar and the GWAS Catalog; and data from pathway and network tools including Reactome and MINT. Calls were quality filtered by Personalis and variants reported at >1% in any of the subpopulations were excluded. Personalis was supplied with clinical features of the patient and used these in ranking variants, taking into account the similarity in features with reported disease-causing mutations in the same gene. Evidence for the top ranking variants was manually reviewed by Personalis to reduce the list to potentially causative variants.
Additionally, the full set of variants reported by Personalis was investigated using their annotation. Quality control was carried out using the following hard filters, as recommended by the GATK best practice guidelines when there are exome sequences for less than 30 samples plus an extra filter on genotype quality: for SNVs, QD < 2, MQ < 40, FS > 60, HaplotypeScore > 13, MQRankSum < -12.5, ReadPosRankSum < -8; for Indels, QD < 2, ReadPosRankSum < -20, FS > 200. Population frequencies from the Exome Aggregation Consortium (ExAC, version 0.3) were used in filtering. Only passed ExAC variants were used and alleles at multiallelic sites were left aligned to ensure they could be matched with the patient data. The number of ExAC individuals in a sub-population that are genotyped at a site can be low, meaning that the frequency is estimated with a large error. Therefore, from the ExAC set only variants that had a frequency >5% in any subpopulation (excluding the ‘other’ population), or of >1% with a total allele count >1000 were retained. These variants were filtered out from the Personalis dataset. Variants reported with a frequency > 1% in any of the 1000 genomes, NHLBI ESP or UK10K cohort populations were also excluded. After filtering, those variants annotated as frameshift, splice site acceptor, splice site donor, or stop gained were extracted and are described as loss of function (LoF) variants. Finally, a larger group of variants annotated within a targeted gene list was compiled, which are described as targeted missense variants. The targeted gene list consisted of genes reported in connection with skeletal dysplasia, identified in GWAS studies to be associated with a change in bone density, implicated in bone metabolism in mouse models, and genes with human phenotype ontology terms relating to increased susceptibility to fracture (HPO:0002659), and totalled approximately 600 genes. The LoF and targeted missense variants were manually reviewed, including looking for compound heterozygotes, to assess whether they might contribute to the patient’s phenotype.
DNA Sequencing and mutation analysis in Patient 2
Trio-based exome sequencing was performed for Patient 2 and his parents as part of the DDD study, as previously described [14, 15] (Wright et al., 2015; DDD Study., Nature 2015). Putative de novo and inherited variants were identified from exome data using DeNovoGear software [15, 16] (Ramu et al., 2013; DDD Study., Nature 2015) and were validated using targeted Sanger sequencing.
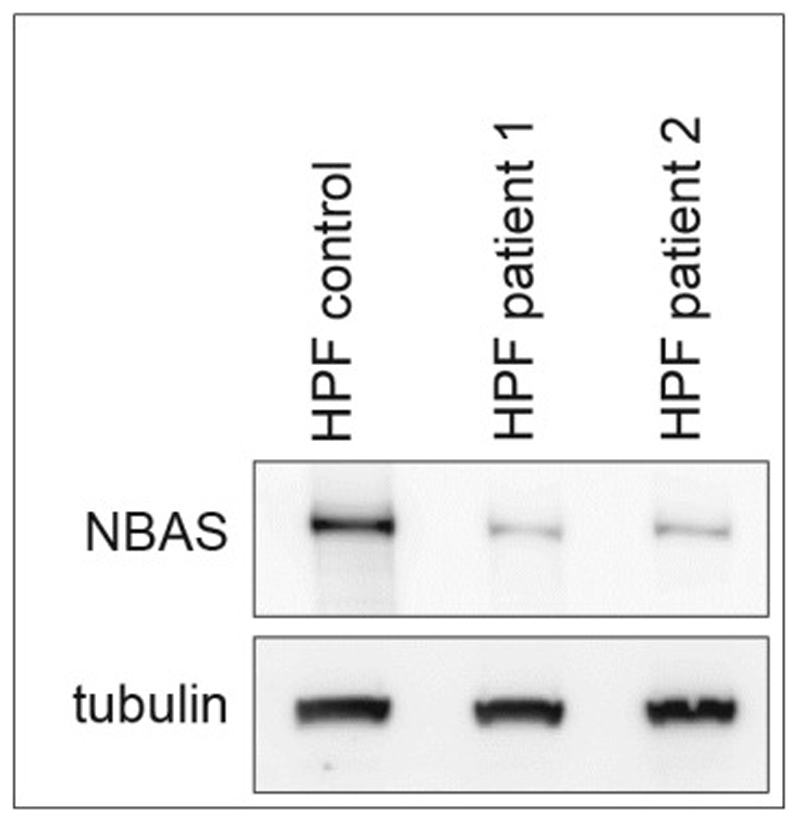
Western Blot Human primary fibroblasts (HPF) on Patient 1
HPF was grown in fully supplemented AmnioMAX C-100 media (Gibco) in hypoxic conditions. Prior to lysis, cells were washed twice with PBS and lysed for 20 min at 4 °C in IP buffer (10 mM Tris-HCl [pH 8], 150 mM NaCl, 1 mM EGTA, 1% NP-40, 0.2% Na-Deoxycholate, Complete Protease Inhibitor (Roche), 1 mM DTT. Proteins were resolved by SDS-PAGE using 3%–8% Tris-Acetate gel (Life Technologies) and analysed for the presence of NBAS by probing with NBAS antibody (Abcam). Uniform protein loading was confirmed by probing with tubulin antibody (TUB 2.1, Sigma-Aldrich).
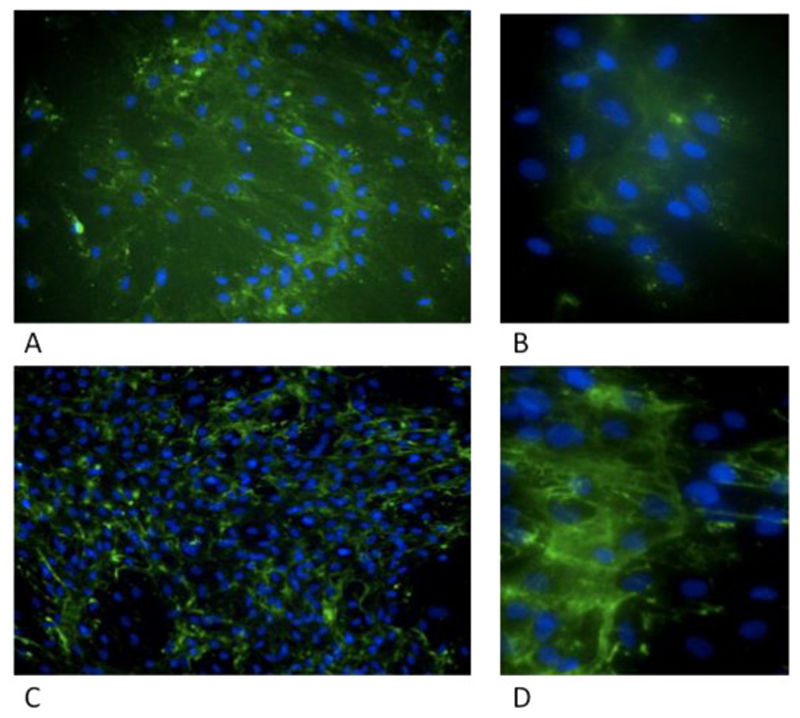
Microscopic analysis of collagen in cultured fibroblasts in Patient 1
NBAS patient 1 cultured fibroblasts and control sample were grown for 3 days in 96 well plates, fixed and stained with anti-Col1A1 antibody (green) and Hoechst (blue) and imaged using a high content microscope.
Results
Patient 1
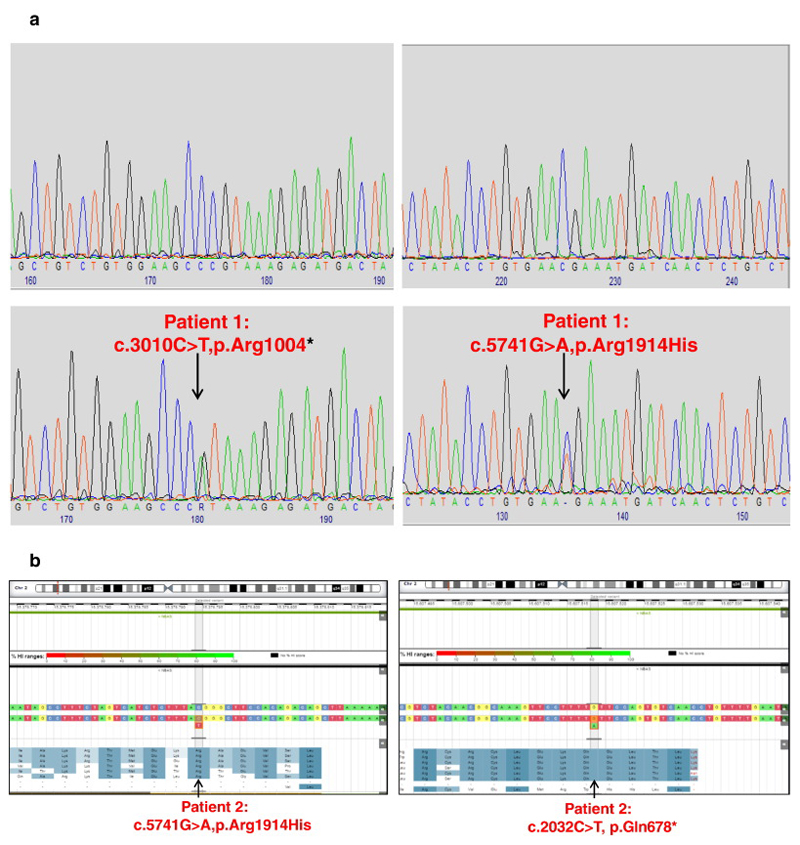
Personalis reported four top ranking variants. Two were heterozygous variants in the NBAS, c.5741G>A p.(Arg1914His) and c.3010C>T p.(Arg1004*) (Figure 5a). The impact of the variants was determined for transcript NM_015909.3. The potential pathogenic effect of the missense variant and nonsense variant were assessed using Alamut Visual version 2.6 (Interactive Biosoftware, Rouen, France) and the Association of Clinical Genetic Science best practice guidelines for the evaluation of pathogenicity and the reporting of sequence variants in clinical molecular genetics.
Figure 5.
a and b: Electropherograms (forward sequence) demonstrating NBAS variants in Patient 1 compared to normal control and Patient 2 (sequence variant plot).
The other two variants did not appear contributory to the phenotype and were of uncertain significance; a novel heterozygous variant of uncertain significance in ARID1B, c.1041_1043dupGGC (p.Ala350dup), and another heterozygous novel variant of uncertain significance in SKIV2L, c.3404T>C (p.Ile1135Thr). Previously reported pathogenic variants in ARID1B associated with autosomal dominant mental retardation type 12 include nonsense and frameshifting insertions/deletions which result in haploinsufficiency [17] (Hoyer et al., 2012). c.1041_1043dupGGC (p.Ala350dup), is an in-frame duplication variant in exon 1 of the ARID1B gene which results in the addition of one extra alanine to a stretch of 11 consecutive alanine residues. The product of the ARID1B gene plays a role in chromatin remodeling for transcriptional activation and repression of select genes. To our knowledge, variants in this polyalanine tract have not been reported in association with disease. Variants in SKIV2L have been previously reported in the literature in association with trichohepatoenteric syndrome-2 (THES2), an autosomal recessive disease [18] (Fabre et al., 2012). However, both the variants are unlikely to account for Patient 1’s phenotype.
The total number of detected variants was 189,298. After quality and frequency filtering, 19,772 remained. There were 89 LoF variants, none of which were in genes with an apparent function in bone. There were 23 targeted missense variants that were predicted to be likely benign following in-silico analysis.
The presence of the NBAS variants was confirmed by Sanger sequencing of the proband. Sequencing of the parents showed that c.3010C>T p.(Arg1004*) is present in the mother and c.5741G>A p.(Arg1914His) in the father. The c.5741G>A is the same missense variant which was described in homozygous form in patients with SOPH syndrome [5] (Maksimova et al., 2010). c.3010C>T was reported by Haack et al., 2015 in a compound heterozygous form to be associated with acute liver failure [10].
Pilot studies of collagen expression and transport in NBAS cells cultured from patients described in this study, show that collagen secretion appears reduced and collagen bundles appear more diffuse, as compared with control cells consistent with interference with trafficking and secretion (Figure 6). Furthermore, Western blot analysis shows reduced level of NBAS protein in patients, as compared to control cells (Figure 7). This implies that the compound heterozygous mutation reported in this patient, compromises the stability of the protein, resulting in a dramatic reduction in NBAS protein levels.
Figure 6.
NBAS patient 1 cultured fibroblasts (A,B) and control sample (C,D) were grown for 3 days in 96 well plates, fixed and stained with anti-Col1A1 antibody (green) and Hoechst (blue) and imaged using a high content microscope. (A, B) show increased diffuse cytoplasmic staining. Collagen bundles from control (C, D).
Figure 7.
Western blot on Patient 1 and 2 cultured fibroblasts showing reduced NBAS protein levels compared to controls; HPF: Human primary fibroblasts.
Patient 2
Two heterozygous variants in the NBAS gene were identified, c.5741G>A p.(Arg1914His); c.2032C>T p.(Glu678*) (Figure 5b) and segregation analysis showed that c.5741G>A p.(Arg1914His) is present in the mother and c.2032C>T p.(Glu678*) is present in the father. The c.5741G>A is the same missense variant which was described in homozygous form in patients with SOPH syndrome [5] (Maksimova et al., 2010). The c.2032>T is a novel variant and has not been reported in HGMD or present in the Exome Aggregation Consortium (ExAC) dataset. Western blot analysis of human primary fibroblasts (HPF) cultured from patients showed reduced level of NBAS protein in patients, as compared to control cells (Figure 7).
Discussion
Osteogenesis imperfecta (OI) is a disease encompassing a group of disorders mainly characterised by bone fragility. There is a broad spectrum of clinical severity in OI, ranging from multiple fractures in-utero and perinatal death, to near-normal adult stature and low fracture incidence. Facial dysmorphism has been noted [19] (Gorlin Monographs). Sillence et al., in 1979 provided the clinical classification, which has been expanded [20,21] (Sillence et al., 1979; Forlino & Marini 2016). Defects in genes encoding type 1 collagen (COL1A1/A2) can be identified in 85% of patients with a clinical diagnosis of OI [21,22] (Forlino & Marini 2016; Marini et al., 2013). So far, several other genes have been implicated in rare forms of heritable bone fragility including autosomal dominant type V OI (IFITM5), X-linked osteoporosis (PLS3 and MBTPS2), autosomal recessive forms (BMP1/ CREB3L1/ CRTAP/ FKBP10/ P3H1/ P4HB/ PLOD2/ PPIB/ SEC24D/ SERPINF1/ SERPINH1/ SP7/ SPARC/ TMEM38B) and heterozygous mutations in WNT1/ LRP5.
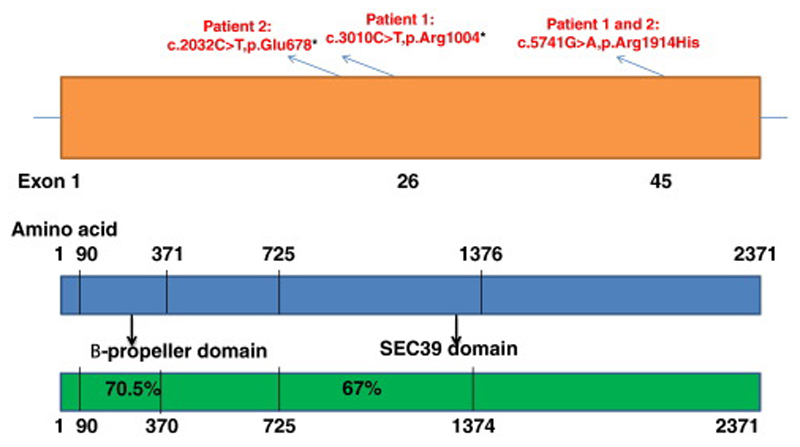
NBAS (the human neuroblastoma amplified sequence gene) was first isolated using genome scanning techniques in neuroblastoma cell lines [1] (Scott et al., 2003). The NBAS protein contains WD40 repeats (β-propeller domain; PFAM domain PF00400) in the N-terminal part of the protein and a SEC39 domain (PFAM domain PF08314), involved in the secretory pathway (Figure 8). Syntaxins play an important role in membrane fusion of transport vesicles with the acceptor compartment [23] (Jahn and Scheller. 2006). Although, the exact function of NBAS has not been clearly elucidated, it has been shown that a peripheral protein encoded by this gene is a sub-unit of the Syntaxin 18 complex [8] (Hong WJ and Lev S., 2014) involved in golgi-to-endoplasmic reticulum retrograde transport. [6] (Aoki et al., 2009).
Figure 8.
Schematic representation of NBAS structure with known protein domains in human (blue) and zebrafish (green) protein (shaded boxes represent the regions of sequence conservation within proteins; the level of conservation is indicated in percentage).
There are precedents for defects in the core secretory machinery resulting in human disease. Proteins exiting the ER are packaged into COPII transport vesicles which are made up of three protein complexes: SAR1, SEC23/SEC24 and SEC13/SEC31 [24] (Jensen and Schekman, 2011). Mammalian COPII proteins have been increasingly implicated in human disease, [25, 26, 27] (Jones et al., 2003; Boyadjiev et al., 2006; Singleton et al., 2015). In human phenotypes, the underlying pathogenesis appears to be due to failure to recruit the specific COPII protein, resulting in a large reduction in the packaging of specific cargo proteins in vitro, accompanied by swelling of the ER with untransported cargo in vivo in a specific tissue [28] (Fromme et al., 2007).
In the context of bone fragility, SEC24 is mainly responsible for sorting cargo molecules through interactions during COPII vesicle assembly. Mutations in SEC24D, a tissue specific isoform of the COPII coat is associated with a recessive form of OI [29] (Garbes et al., 2015). Evidence to-date suggests that similarly NBAS variants have a cargo-selective, tissue-specific phenotype. It has been shown to be localised to the ER; required for Golgi-ER retrograde transport and its loss is associated with defects in protein glycosylation. Collagen is a large extracellular matrix protein, synthesized in ER as a rigid rod precursor (procollagen which is approximately 300 nm in length) and packaged into COPII transport vesicles (which are typically 70-100 nm in size). This packaging of collagen requires the transmembrane (TM) protein, TANGO1 and the enzymatic activity of CUL3–KLHL12. Jin et al., 2012 showed that modification of a COPII protein allows the formation of transport vesicles large enough to hold a bulky cargo like procollagen [30,31] (Jin et al., 2012; Stephens et al., 2012).
Evidence also suggests that NBAS plays a role in nonsense mediated mRNA decay (NMD) [5] (Longman et al., 2013). The NMD pathway is a highly-conserved surveillance mechanism that selectively degrades mRNAs harbouring premature termination codons (PTC), acting to prevent the accumulation of truncated proteins that may interfere with cellular function [32,33] (Chang YF et al., 2007; Isken et al., 2008). NMD also has an important role in controlling the expression of many naturally occurring transcripts [34] (Hug et al., 2016). NBAS is a bona-fide NMD factor that acts together with core NMD factors to regulate expression of a large number of endogenous RNA targets [5] (Longman et al., 2013). Interestingly, NBAS-regulated genes harbour sequence features associated with protein trafficking and ER-coupled protein modifications. Multiple targets for NBAS appear to have a role in regulation of bone mineralisation, osteoblast differentiation and bone development. The target with strongest up-regulation upon NBAS-depletion was MGP gene (matrix Gla protein), which acts as an inhibitor of bone formation [5] (Longman et al., 2013). MGP variants are associated with Keutel syndrome which shows abnormal cartilage calcification [35] (Munroe PB et al., 1999). RNAseq studies in bone cells show that NBAS is expressed in osteoblasts and osteocytes of rodents and primates.
CRISPR-Cas9 technology has been used to generate stable knockout NBAS cell lines in human SAOS2 osteoblast cells. Further studies are underway to understand the precise mechanism of action of NBAS in NMD and secretion, resulting in a multi-system phenotype. Since NBAS has been proposed to function in the NMD pathway and Golgi-ER transport, the effect on bone fragility could be attributed to either pathway in isolation, or alternatively to a combination of both. We propose three possible models to explain how NBAS variants give rise to phenotypes and human disease based on the cell studies (WB and high content microscopy) and EM findings; 1) due to a compromised Golgi-ER retrograde transport, 2) due to an NMD phenotype, 3) a dual role i.e. a compromised Golgi-ER transport indirectly affecting NMD.
Homozygous missense variants c.5741G>A(p.Arg1914His) in NBAS were reported as being associated with SOPH syndrome in the Yakut Siberians (Short stature with optic atrophy and Pelger-Huet anomaly) [9] (Maksimova et al., 2010). Both patients reported here have features of SOPH i.e. short stature, granulocyte left shift with Pelger Huet cells but the facial dysmorphism is distinct as explained by the different ethnicity of our patients. Mutations in NBAS were reported in individuals with early-onset recurrent acute liver failure, but none of the individuals reported had a skeletal phenotype [10] (Haack et al., 2015). More recent literature is suggesting a multi-system phenotype with a skeletal phenotype and early-onset osteoporosis [11, 12, 13] (Garcia Segarra et al., 2015; Capo-Chichi JM et al., 2015; Staufner et al., 2015). Interestingly, our patients also have abnormal liver function tests of unexplained cause, since a young age, but have never had any episodes of acute onset liver failure (ALF). In addition, our patients also have a predominant skeletal phenotype with bone fragility, multiple vertebral (Patient 1) and long bone fractures (Patient 1 and 2) needing treatment with Pamidronate. Hence, mutations in NBAS are likely to be a novel cause of heritable bone fragility and should be included in the targeted gene panel testing for OI that is currently offered in diagnostic genetic testing, in order to clarify diagnosis, inform prognosis and discussions around recurrence risk (up to 25%).
Summary
Bone fragility, severe enough to need therapy in childhood, has not been previously reported as a feature associated with variants in NBAS. Therefore, we hypothesise that compound heterozygous variants in NBAS accounts for bone fragility and is a novel cause of Osteogenesis Imperfecta (OI). The increasing evidence pointing to a role for NBAS in liver, immune and connective tissue coupled with its extreme phenotypic variability make understanding NBAS function important.
Ackowledgments
We thank the families for their participation in this report.
Funding
Patient 1: This research was supported by The Sheffield Children’s Hospital Charity (TCHC) grant number CA15001.
Patient 2: The Deciphering Developmental Disorders (DDD) study presents independent research commissioned by the Health Innovation Challenge Fund [grant number HICF-1009-003], a parallel funding partnership between the Wellcome Trust and the Department of Health, and the Wellcome Trust Sanger Institute [grant number WT098051]. The views expressed in this publication are those of the author(s) and not necessarily those of the Wellcome Trust or the Department of Health. The study has UK Research Ethics Committee approval (10/H0305/83, granted by the Cambridge South REC, and GEN/284/12 granted by the Republic of Ireland REC). The research team acknowledges the support of the National Institute for Health Research, through the Comprehensive Clinical Research Network.
Footnotes
Competing Interests
No competing interest to declare.
References
- [1].Scott DK, Board JR, Lu X, Pearson AD, Kenyon RM, Lunec J. The neuroblastoma amplified gene, NAG: genomic structure and characterisation of the 7.3 kb transcript predominantly expressed in neuroblastoma. Gene. 2003;307:1–11. doi: 10.1016/s0378-1119(03)00459-1. [DOI] [PubMed] [Google Scholar]
- [2].Wimmer K, Zhu XX, Lamb BJ, Kuick R, Ambros PF, Kovar H, Thoraval D, Motyka S, Alberts JR, Hanash SM. Co-amplification of a novel gene, NAG, with the N-myc gene in neuroblastoma. Oncogene. 1999;18(1):233–8. doi: 10.1038/sj.onc.1202287. [DOI] [PubMed] [Google Scholar]
- [3].Longman D, Plasterk RH, Johnstone IL, Cáceres JF. Mechanistic insights and identification of two novel factors in the C. elegans NMD pathway. Genes Dev. 2007;21(9):1075–85. doi: 10.1101/gad.417707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Anastasaki C, Longman D, Capper A, Patton EE, Cáceres JF. Dhx34 and Nbas function in the NMD pathway and are required for embryonic development in zebrafish. Nucleic Acids Res. 2011 May;39(9):3686–94. doi: 10.1093/nar/gkq1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Longman D, Hug N, Keith M, Anastasaki C, Patton EE, Grimes G, Cáceres JF. DHX34 and NBAS form part of an autoregulatory NMD circuit that regulates endogenous RNA targets in human cells, zebrafish and Caenorhabditis elegans. Nucleic Acids Res. 2013;41(17):8319–31. doi: 10.1093/nar/gkt585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Aoki T, Ichimura S, Itoh A, Kuramoto M, Shinkawa T, Isobe T, Tagaya M. Identification of the neuroblastoma-amplified gene product as a component of the syntaxin 18 complex implicated in Golgi-to-endoplasmic reticulum retrograde transport. Mol Biol Cell. 2009;20(11):2639–49. doi: 10.1091/mbc.E08-11-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Spang A. Retrograde traffic from the Golgi to the endoplasmic reticulum. Cold Spring Harb Perspect Biol. 2013 Jun 1;5(6) doi: 10.1101/cshperspect.a013391. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Hong W, Lev S. Tethering the assembly of SNARE complexes. Trends Cell Biol. 2014 Jan;24(1):35–43. doi: 10.1016/j.tcb.2013.09.006. [DOI] [PubMed] [Google Scholar]
- [9].Maksimova N, Hara K, Nikolaeva I, Chun-Feng T, Usui T, Takagi M, Nishihira Y, Miyashita A, Fujiwara H, Oyama T, Nogovicina A, et al. Neuroblastoma amplified sequence gene is associated with a novel short stature syndrome characterised by optic nerve atrophy and Pelger-Huët anomaly. J Med Genet. 2010;47(8):538–48. doi: 10.1136/jmg.2009.074815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Haack TB, Staufner C, Köpke MG, Straub BK, Kölker S, Thiel C, Freisinger P, Baric I, McKiernan PJ, Dikow N, Harting I, et al. Biallelic Mutations in NBAS Cause Recurrent Acute Liver Failure with Onset in Infancy. Am J Hum Genet. 2015;97(1):163–9. doi: 10.1016/j.ajhg.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Garcia Segarra N, Ballhausen D, Crawford H, Perreau M, Campos-Xavier B, van Spaendonck-Zwarts K, Vermeer C, Russo M, Zambelli P-Y, Stevenson B, Royer-Bertrand B, et al. NBAS mutations cause a multisystem disorder involving bone, connective tissue, liver, immune system, and retina. Am J Med Genet Part A. 2015;167A(12):2902–12. doi: 10.1002/ajmg.a.37338. [DOI] [PubMed] [Google Scholar]
- [12].Capo-Chichi JM, Mehawej C, Delague V, Caillaud C, Khneisser I, Hamdan FF, Michaud JL, Kibar Z, Mégarbané A. Neuroblastoma Amplified Sequence (NBAS) mutation in recurrent acute liver failure: Confirmatory report in a sibship with very early onset, osteoporosis and developmental delay. Eur J Med Genet. 2015;58(12):637–641. doi: 10.1016/j.ejmg.2015.11.005. [DOI] [PubMed] [Google Scholar]
- [13].Staufner C, Haack TB, Köpke MG, Straub BK, Kölker S, Thiel C, Freisinger P, Baric I, McKiernan PJ, Dikow N, Harting I, et al. Recurrent acute liver failure due to NBAS deficiency: phenotypic spectrum, disease mechanisms, and therapeutic concepts. J Inherit Metab Dis. 2016;39(1):3–16. doi: 10.1007/s10545-015-9896-7. [DOI] [PubMed] [Google Scholar]
- [14].Wright CF, Fitzgerald TW, Jones WD, Clayton S, McRae JF, van Kogelenberg M, King DA, Ambridge K, Barrett DM, Bayzetinova T, Bevan AP, et al. Genetic diagnosis of developmental disorders in the DDD study: a scalable analysis of genome-wide research data. Lancet. 2015;385(9975):1305–14. doi: 10.1016/S0140-6736(14)61705-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Deciphering Developmental Disorders Study. Large-scale discovery of novel genetic causes of developmental disorders. Nature. 2015 Mar 12;519(7542):223–8. doi: 10.1038/nature14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ramu A, Noordam MJ, Schwartz RS, Wuster A, Hurles ME, Cartwright RA, Conrad DF. DeNovoGear: de novo indel and point mutation discovery and phasing. Nat Methods. 2013;10(10):985–7. doi: 10.1038/nmeth.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hoyer J, Ekici AB, Endele S, Popp B, Zweier C, Wiesener A, Wohlleber E, Dufke A, Rossier E, Petsch C, Zweier M, et al. Haploinsufficiency of ARID1B, a member of the SWI/SNF-a chromatin-remodeling complex, is a frequent cause of intellectual disability. Am J Hum Genet. 2012 Mar 9;90(3):565–72. doi: 10.1016/j.ajhg.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Fabre A, Charroux B, Martinez-Vinson C, Roquelaure B, Odul E, Sayar E, Smith H, Colomb V, Andre N, Hugot JP, Goulet O, et al. SKIV2L mutations cause syndromic diarrhea, or trichohepatoenteric syndrome. Am J Hum Genet. 2012 Apr 6;90(4):689–92. doi: 10.1016/j.ajhg.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Gorlin RJ, Cohen MM, Hennekam RCM. Syndromes of the head and neck. Oxford Monographs on Medical Genetics. 2001. Syndromes affecting bone: the osteogenesis imperfectas. [Google Scholar]
- [20].Sillence DO, Senn A, Danks DM. Genetic heterogeneity in osteogenesis imperfecta. J Med Genet. 1979;16(2):101–116. doi: 10.1136/jmg.16.2.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Forlino A, Marini JC. Osteogenesis imperfecta. Lancet. 2016;387(10028):1657–71. doi: 10.1016/S0140-6736(15)00728-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Marini JC, Blissett AR. New genes in bone development: what's new in osteogenesis imperfecta. J Clin Endocrinol Metab. 2013;98(8):3095–103. doi: 10.1210/jc.2013-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Jahn R, Scheller RH. SNAREs--engines for membrane fusion. Nat Rev Mol Cell Biol. 2006 Sep;7(9):631–43. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- [24].Jensen D, Schekman R. COPII-mediated vesicle formation at a glance. J Cell Sci. 2011;124(Pt 1):1–4. doi: 10.1242/jcs.069773. [DOI] [PubMed] [Google Scholar]
- [25].Jones B, Jones EL, Bonney SA, Patel HN, Mensenkamp AR, Eichenbaum-Voline S, Rudling M, Myrdal U, Annesi G, Naik S, Meadows N, et al. Mutations in a Sar1 GTPase of COPII vesicles are associated with lipid absorption disorders. Nat Genet. 2003;34(1):29–31. doi: 10.1038/ng1145. [DOI] [PubMed] [Google Scholar]
- [26].Boyadjiev SA, Fromme JC, Ben J, Chong SS, Nauta C, Hur DJ, Zhang G, Hamamoto S, Schekman R, Ravazzola M, Orci L, et al. Cranio-lenticulo-sutural dysplasia is caused by a SEC23A mutation leading to abnormal endoplasmic-reticulum-to-Golgi trafficking. Nat Genet. 2006;38(10):1192–7. doi: 10.1038/ng1876. [DOI] [PubMed] [Google Scholar]
- [27].Singleton B, Bansal D, Varma N, Das R, Naseem S, Saikia UN, Malhotra P, Varma S, Marwaha RK, King MJ, Ahmed M. Homozygosity mapping reveals founder SEC23B-Y462C mutations in Indian congenital dyserythropoietic anemia type II. Clin Genet. 2015;88(2):195–7. doi: 10.1111/cge.12527. [DOI] [PubMed] [Google Scholar]
- [28].Fromme JC, Ravazzola M, Hamamoto S, Al-Balwi M, Eyaid W, Boyadjiev SA, Cosson P, Schekman R, Orci L. The genetic basis of a craniofacial disease provides insight into COPII coat assembly. Dev Cell. 2007;13(5):623–34. doi: 10.1016/j.devcel.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Garbes L, Kim K, Rieß A, Hoyer-Kuhn H, Beleggia F, Bevot A, Kim MJ, Huh YH, Kweon HS, Savarirayan R, Amor D, et al. Mutations in SEC24D, encoding a component of the COPII machinery, cause a syndromic form of osteogenesis imperfecta. Am J Hum Genet. 2015;96(3):432–9. doi: 10.1016/j.ajhg.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Jin L, Pahuja KB, Wickliffe KE, Gorur A, Baumgärtel C, Schekman R, Rape M. Ubiquitin-dependent regulation of COPII coat size and function. Nature. 2012;482(7386):495–500. doi: 10.1038/nature10822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Stephens DJ. Cell biology: Collagen secretion explained. Nature. 2012;482(7386):474–5. doi: 10.1038/482474a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Hug N, Longman D, Cáceres JF. Mechanism and regulation of the nonsense-mediated decay pathway. Nucleic Acids Res. 2016;44(4):1483–95. doi: 10.1093/nar/gkw010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. Review. [DOI] [PubMed] [Google Scholar]
- [34].Isken O, Maquat LE. The multiple lives of NMD factors: balancing roles in gene and genome regulation. Nat Rev Genet. 2008 Sep;9(9):699–712. doi: 10.1038/nrg2402. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Munroe PB, Olgunturk RO, Fryns JP, Van Maldergem L, Ziereisen F, Yuksel B, Gardiner RM, Chung E. Mutations in the gene encoding the human matrix Gla protein cause Keutel syndrome. Nat Genet. 1999;21(1):142–4. doi: 10.1038/5102. [DOI] [PubMed] [Google Scholar]