Abstract
Aims: DsbA catalyzes disulfide bond formation in secreted and outer membrane proteins in bacteria. In pathogens, DsbA is a major facilitator of virulence constituting a target for antivirulence antimicrobial development. However, many pathogens encode multiple and diverse DsbA enzymes for virulence factor folding during infection. The aim of this study was to determine whether our recently identified inhibitors of Escherichia coli K-12 DsbA can inhibit the diverse DsbA enzymes found in two important human pathogens and attenuate their virulence.
Results: DsbA inhibitors from two chemical classes (phenylthiophene and phenoxyphenyl derivatives) inhibited the virulence of uropathogenic E. coli and Salmonella enterica serovar Typhimurium, encoding two and three diverse DsbA homologues, respectively. Inhibitors blocked the virulence of dsbA null mutants complemented with structurally diverse DsbL and SrgA, suggesting that they were not selective for prototypical DsbA. Structural characterization of DsbA-inhibitor complexes showed that compounds from each class bind in a similar region of the hydrophobic groove adjacent to the Cys30-Pro31-His32-Cys33 (CPHC) active site. Modeling of DsbL- and SrgA-inhibitor interactions showed that these accessory enzymes could accommodate the inhibitors in their different hydrophobic grooves, supporting our in vivo findings. Further, we identified highly conserved residues surrounding the active site for 20 diverse bacterial DsbA enzymes, which could be exploited in developing inhibitors with a broad spectrum of activity.
Innovation and Conclusion: We have developed tools to analyze the specificity of DsbA inhibitors in bacterial pathogens encoding multiple DsbA enzymes. This work demonstrates that DsbA inhibitors can be developed to target diverse homologues found in bacteria. Antioxid. Redox Signal. 29, 653–666.
Keywords: : infection, therapeutics, protein folding, thiol
Introduction
“Antibiotic resistance is happening right now, across the world” (1). Many common infections that until recently were readily treatable are becoming resistant to most, if not all available, antibiotics, heralding the beginning of a post-antibiotic era, in which common infections will become untreatable and once again lethal. In the era of bacterial multi-drug resistance, many promising new approaches are being investigated to combat these infections. One promising antibacterial strategy is to develop compounds that inhibit bacterial virulence rather than bacterial viability or growth (10, 11, 76). Enzymes and systems required for the biogenesis of virulence factors, the bacterial weaponry necessary for infection, represent attractive target candidates for inhibition as they have the capacity to inhibit a pathogen's ability to cause infection without necessarily affecting its growth or viability, thus reducing selection pressure in bacteria and subsequent development and spread of resistance (28, 67).
Innovation.
Disulfide bond (Dsb) enzymes are an essential part of the bacterial virulence factor assembly line. Their central role in bacterial pathogenesis makes Dsb oxidoreductases attractive targets for pharmacological intervention, and several drug discovery and development campaigns are currently underway for Dsb inhibitors as novel antibacterial agents. Many pathogens, however, encode multiple diverse DsbA homologues. Here, we studied DsbA inhibition in relevant pathogen-specific backgrounds and showed that low-affinity inhibitors can block diverse DsbA enzymes present in clinically important pathogens. This work could guide the future optimization of DsbA inhibitors, rationalization of pathogen-specific DsbA inhibitors, and elaboration of broad-spectrum DsbA antimicrobial leads.
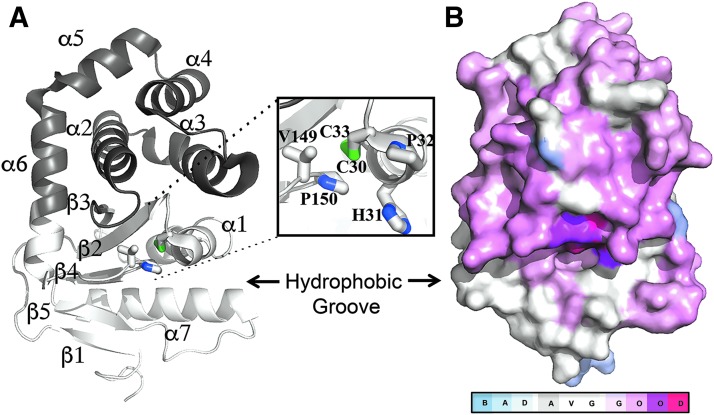
In Gram-negative pathogens, DsbA fulfills these criteria as it works as a folding catalyst for multiple classes of virulence factors but is not necessary for bacterial viability (29). In such pathogens, DsbA is a periplasmic enzyme that catalyzes disulfide bond formation in secreted and outer membrane proteins. As many of these proteins serve virulence functions during infection, DsbA proteins are major facilitators of bacterial pathogenesis (29, 42, 74) and, consequently, DsbA is being targeted for the development of antivirulence drugs (64). Currently, the DsbA oxidative system of Escherichia coli K-12 is the best-studied example of oxidative protein folding in bacteria. This pathway involves two enzymes: DsbA, which binds to unfolded polypeptides and oxidizes them via a disulfide exchange reaction, and DsbB, which restores the oxidizing activity of DsbA, allowing folding of subsequent substrates to occur. E. coli DsbA (EcDsbA) is a thioredoxin (TRX)-like thiol oxidase comprising the characteristic Cys30-Pro31-His32-Cys33 (CPHC) redox active site flanked by a hydrophobic groove required for binding the cognate oxidase EcDsbB (24, 46) (Fig. 1A). Not surprisingly, this groove is the binding site for most EcDsbA inhibitors described to date (4, 18).
FIG. 1.
Escherichia coli DsbA structure and sequence conservation. (A) Ribbon diagram of EcDsbA [PDB 1FVK (24)]. The TRX-fold and inserted helical domain are shown in light and dark gray, respectively. Secondary structural features are labeled, and the catalytic disulfide bond is shown in green. Inlet shows a close-up view of the active site of EcDsbA encompassing a Cys30-Pro31-His32-Cys33 redox-active motif and the adjacent cis-proline loop (V149-cisPro150). (B) Amino acid sequence conservation in diverse bacterial DsbA prototypes. The conservation scores obtained from an Expresso T-Coffee multiple sequence alignment of 20 DsbA protein sequences are depicted on the surface of EcDsbA by using a color gradient, from blue (poorly conserved), through to white (reliable conservation) and red (well conserved). EcDsbA, Escherichia coli disulfide bond; TRX, thioredoxin.
Although inhibitor development has focused on the prototypical E. coli K-12 DsbA, the increasing number of publicly available sequenced bacterial genomes has revealed that DsbA enzymes are diverse among bacteria (19, 29, 62). These proteins show variable conservation in amino acid sequence and variation in copy number among different bacterial genera, species, or even strains within the same species. For example, uropathogenic E. coli (UPEC) strains such as the pyelonephritis isolate CFT073 contain two oxidative systems: the DsbA/DsbB pair, as well as an additional DsbL/DsbI redox pair, which may be dedicated to a specific group of virulence substrates and is lacking from commensal or enteric E. coli pathotypes (23, 68). Similarly, certain Salmonella enterica serovars (e.g., S. Typhimurium) contain an extended number of Dsb proteins, including DsbA/DsbB, DsbL/DsbI, and another virulence plasmid-encoded DsbA homologue called SrgA (9, 30). Multiple copies of DsbA have also been described in other important pathogens such as Neisseria meningitidis (63), Neisseria gonorrhoeae (29), and Pseudomonas aeruginosa (7, 61), among others.
With such diversity, DsbA presents a unique opportunity as an antivirulence target to customize the specificity of inhibitors to individual pathogens (narrow spectrum) or groups of pathogens sharing a common target (broader spectrum); however, no study to date has investigated DsbA inhibitor specificity in pathogens with multiple diverse DsbA homologues.
The current work investigates the activity of EcDsbA inhibitors, representing two different chemical classes, against two important human pathogens containing multiple diverse DsbA enzymes. We examine the target and pathogen specificity, and we compare at atomic resolution the binding mode of these inhibitors with the diverse DsbAs. Our work provides evidence that low-affinity inhibitors can block diverse DsbA enzymes found in different bacteria or within the same pathogen, and that the extent of inhibition achieved is dependent on the full DsbA complement that each pathogen possesses. Further, this study also highlights conserved residues neighboring the active site of DsbA enzymes that may be exploited in broad-spectrum inhibitor development.
Results
Diversity among bacterial DsbA enzymes
DsbA enzymes are found in all classes of Proteobacteria and Chlamydiales as well as in many Fusobacteria and Actinobacteria species (29, 50). A previously reported comparative analysis of 13 structurally characterized DsbA proteins revealed that they form two main structural classes DsbA-I and DsbA-II, which primarily differ on their central β-sheet topology in the TRX-fold, which could be further divided into four subclasses (50). Currently, there are 22 DsbA homologues that have been characterized both structurally and functionally (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/ars). These are found in representatives of diverse bacterial classes (Actinobacteria, Bacilli, Alpha-, Beta-, and Gammaproteobacteria) and mostly share an overall low identity over their full-length amino acid sequence (between 10% and 56% identity by pair-wise sequence alignment analysis, Supplementary Fig. S1).
The variability at the amino acid level shown by these enzymes prompted us to combine sequence and phylogenetic analyses together with the available structural information to further define the diversity in this family of thiol oxidases. Twenty of the 22 characterized DsbA homologues constituted the group of structurally and functionally defined DsbA proteins used throughout the sequence comparison studies performed hereafter (defined as prototypes). We excluded (i) DsbA from S. enterica serovar Typhimurium (SeDsbA) as it is 85% identical to EcDsbA, and (ii) DsbL from UPEC that is 93% identical to its S. enterica positional orthologue SeDsbL (30).
We first examined the relationship of the 20 DsbA prototypes at the protein sequence level by constructing a structural multiple sequence alignment (MSA) using the Expresso T-Coffee algorithm. Despite the variable sequence identity (S.I.) in DsbA proteins, the two catalytic cysteines and the cis-proline in the active motif are 100% conserved among our set (Fig. 1B). Notably, the hydrophobic binding groove is present within all proteins that are adjacent to the redox active site. There were a number of conserved regions, including the amino acids spanning the catalytic cysteines (Cys30-Pro31-His32-Cys33 in EcDsbA) and the residues preceding the cis-proline (Gly149-Val150-cisPro151 in EcDsbA). Other highly conserved hydrophobic residues (Val22, Phe25, Phe26, Leu92, Phe93, and Val155 in EcDsbA) form the core of the DsbA fold and this conservation is probably required for structural stability. Above the groove, the C-terminus of helix α6 and residues neighboring the catalytic cysteines were moderately conserved. Surprisingly, residues at the C-terminus of helix α1 and the β5-α7 region in the TRX domain, which map to the center of the groove, were poorly conserved across DsbA homologues, except for an aromatic residue (Phe36 in EcDsbA) that was found in 16 out of the 20 DsbA prototype structures.
Phylogenetic analysis assigns bacterial DsbA proteins to three main clades
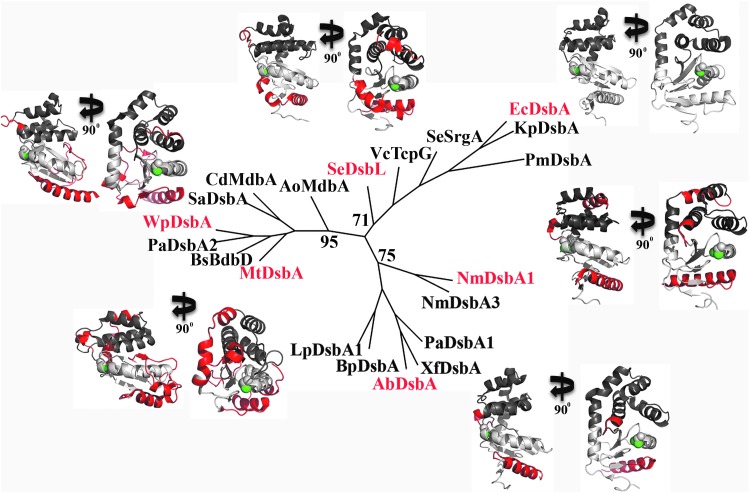
A consensus Neighbor-Joining tree of the 20 DsbA prototypes showed that they mostly grouped into clades that reflected the taxonomic group of their encoding organism, with some exceptions (Fig. 2 and Supplementary Fig. S1). The sequence and structural conservation is clearly apparent for proteins that cluster with EcDsbA [previously defined as DsbA-Ia, (50)], which include homologues from other Gammaproteobacteria such as Proteus mirabilis DsbA (PmDsbA, 85% SI and r.m.s.d of 1.1 Å), Klebsiella pneumoniae DsbA (KpDsbA, 56% S.I. and r.m.s.d of 0.8 Å), and SeSrgA (35% S.I. and r.m.s.d of 1.4 Å) (30, 33 38, 39). All of these proteins are characterized by long hydrophobic grooves flanking the active site, and they mostly share the same active site residues (Cys-Pro-His-Cys) (30).
FIG. 2.
Diversity in DsbA sequence and structure. Unrooted Neighbor-Joining consensus tree of 20 DsbA proteins encoded in 17 bacterial species. The tree was based on 1000 bootstrap replicates reconstructed by using protdist and neighbor, the protein distance-based method implemented by PHYLIP (21), using a multiple sequence alignment generated with the Expresso algorithm of the T-Coffee package (66). Proteins are grouped into three well-supported clades (% bootstrap values are shown) and structures from clade representatives (names in red font) are shown, highlighting the active site (shown in green) and the main structural differences from EcDsbA (shown in red). These differences primarily localize in the β5-loop-α7 region, which for more proteins is truncated and results in a smaller hydrophobic groove. In addition, all proteins clustering with WpDsbA have their β1 strand interacting with β3 (opposite site of the protein) rather than β5. Protein accession numbers and PDB codes are listed in Supplementary Figure S1.
A second clade (previously defined as DsbA-Ib) included DsbA proteins from Betaproteobacteria Burkholderia pseudomallei (BpDsbA, 27% S.I. and r.m.s.d of 2.1 Å) and Neisseria meningitidis (NmDsbA1, 23% S.I. and r.m.s.d of 2.2 Å; NmDsbA3, 22% S.I. and r.m.s.d of 2.2 Å) (35, 41, 71, 72). This clade also contains homologues from Gammaproteobacteria Xylella fastidiosa (XfDsbA, 19% S.I. and r.m.s.d of 2.1 Å), Legionella pneumophila (LpDsbA1, 25% S.I. and r.m.s.d of 2.3 Å), and one of the DsbA homologues of Pseudomonas aeruginosa (PaDsbA1, 30% S.I. and r.m.s.d of 2.2 Å) (37, 56, 60, 61).
The homologues most divergent to EcDsbA (in both sequence and structure) formed a third more taxonomically diverse clade (Fig. 2). These included: Actinomyces oris MdbA (AoMdbA, 18% S.I. and r.m.s.d of 3.3 Å), Mycobacterium tuberculosis DsbA (MtDsbA, 19% S.I. and r.m.s.d of 2.8 Å), and Corynebacterium diptheriae MdbA (CdMdbA, 16% S.I. and r.m.s.d of 2.4 Å) from Actinobacteria; Bacillus subtilis BdbD (BsBdbD, 15% S.I. and r.m.s.d of 2.7 Å) and Staphylococcus aureus DsbA (SaDsbA, 16% S.I. and r.m.s.d of 1.3 Å) from Gram-positive Firmicutes; Wolbachia pipientis DsbA from Alphaproteobacteria (WpDsbA, 10% S.I. and r.m.s.d of 2.9 Å); and the recently described second DsbA homologue of Pseudomonas aeruginosa from Gammaproteobacteria (PaDsbA2, 12% S.I. and r.m.s.d of 2.8 Å) (7, 15, 16, 27, 40, 55, 57, 58). These proteins display distinct topology to EcDsbA, where strand β1 is hydrogen bonded to the opposite edge of the core β-sheet (β2–β5) in the TRX domain (Fig. 2). Phylogenetic analysis using maximum likelihood produced an identical tree topology supporting that obtained with distance-based methods (data not shown).
Interestingly, most DsbA prototypes displayed structural features that differed from the archetypal E. coli enzyme, in particular, a substantially truncated hydrophobic groove that in E. coli DsbA interacts with the partner protein DsbB (34) (Fig. 2). This is more striking in DsbL, the accessory DsbA homologue found in UPEC and S. Typhimurium, which has a bent α6 connecting helix and a six-residue truncation in the β5–α7 motif, which together result in a severely reduced groove. DsbL enzymes additionally show a uniquely electropositive surface as compared with other DsbA homologues that are more hydrophobic (30, 62).
Taken together, phylogenetic and structural comparison of 20 DsbA homologues from diverse bacterial species revealed unique idiosyncrasies despite an overall conserved structure. Such sequence and structural features differentiate DsbA proteins into distinct groups, which are often congruent with bacterial taxonomy (Fig. 2 and Supplementary Fig. S1). This information can serve as a critical guide for predicting the spectrum of activity of DsbA inhibitors.
Activity of E. coli K-12 DsbA inhibitors against pathogens with multiple DsbA enzymes
The high degree of diversity among bacterial DsbA enzymes, in particular among homologues found within the same pathogen, poses the question as to whether our recently developed inhibitors of the archetypal E. coli K-12 DsbA (4) will also inhibit diverse DsbA enzymes found in pathogenic E. coli strains or other Gram-negative pathogens. To address this, we tested the effect of four small-molecule EcDsbA inhibitors (Fig. 3A) representing two different chemical classes (phenylthiophenes and phenoxyphenyls) on the motility of (i) the reference E. coli K-12 strain MG1655 (8) encoding only the prototypical EcDsbA; (ii) the reference UPEC strain CFT073 (51), which encodes DsbA (99.5% identical to K-12 EcDsbA) and EcDsbL (28% identity to EcDsbA); and (iii) the S. Typhimurium reference strain SL1344 (31) that encodes SeDsbA (85% identical to EcDsbA), SeDsbL (28% identity to EcDsbA and 93% identity to EcDsbL), and the plasmid-encoded SeSrgA (35% identity to EcDsbA). We have previously shown that, similar to K-12, motility in UPEC CFT073 and S. Typhimurium SL1344 can be mediated by the different DsbA homologues found in these pathogens (30, 68), and, therefore, this virulence phenotype can be used as a surrogate method to test the inhibition of DsbA proteins.
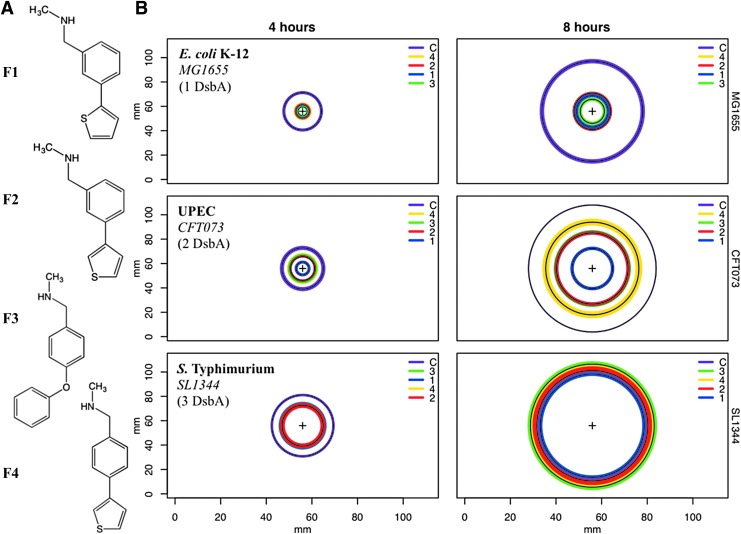
FIG. 3.
Pathogen motility inhibition by small-molecule EcDsbA inhibitors. (A) Chemical structures of phenylthiophene (F1, F2, and F4) and phenoxyphenyl (F3) derivatives previously shown to bind to K-12 EcDsbA (4). (B) Circle plots of bacterial motility zones (mm) for E. coli K-12 MG1655 (top), UPEC CFT073 (middle), and S. Typhimurium SL1344 (bottom) cultured for 4 h (left) or 8 h (right) on soft agar containing inhibitors F1, F2, F3, or F4 at 1 mM or 0.1% DMSO (“C,” carrier control). Concentric circles (black lines) represent the mean motility diameter (in mm) of n = 4 independent replicates, with the standard deviation drawn as a colored zone around each circle (purple for DMSO control, blue for F1, red for F2, green for F3, and yellow for F4). The thickness of the colored zone represents ± one standard deviation value, and the color key is in descending order of the circle diameters graphed in each plot. Mean circle diameters for SL1344 (8 h) on DMSO (purple) and F3 (green) plates overlap and were the only two groups that were not statistically significant by ANOVA. UPEC, uropathogenic Escherichia coli; DMSO, dimethyl sulfoxide.
The diameter of each strain's motility zone was measured on soft agar containing 1 mM of each inhibitor over time (Fig. 3B). As expected, all compounds dramatically inhibited motility in E. coli K-12, with MG1655 motility zones being 60–76% smaller on soft agar containing inhibitors than 0.1% dimethyl sulfoxide (DMSO) control plates (Fig. 3B top). Inhibitor impact on UPEC motility was more variable, with relative motility inhibition of CFT073 ranging from ∼70% to 30% with activity following the order: F1>F2>F3>F4 (Fig. 3B middle). For S. Typhimurium strain SL1344, with three DsbA homologues, motility inhibition by the four inhibitors was less pronounced, resulting in relative motility zones that were on average 30% smaller than the DMSO control zone (Fig. 3B bottom). By 8 h, SL1344 motility on any inhibitor-supplemented plate only differed from the control plate by an average of <20%. Motility inhibition was not due to inhibition of bacterial growth, as shown for the most active inhibitor F1 at 1 mM (Supplementary Fig. S2), strongly suggesting that motility is reduced because of inhibition of DsbA-mediated FlgI folding; however, we cannot exclude that the tested inhibitors may have additional off-target effects that could contribute to the observed phenotype. Taken together, these findings suggest that the four EcDsbA inhibitors are also active against UPEC and S. Typhimurium but display a variable activity spectrum, likely due to differences in their specificity for the diverse DsbA enzymes encoded by each of these pathogens.
Specificity of E. coli K-12 DsbA inhibitors against diverse DsbA enzymes
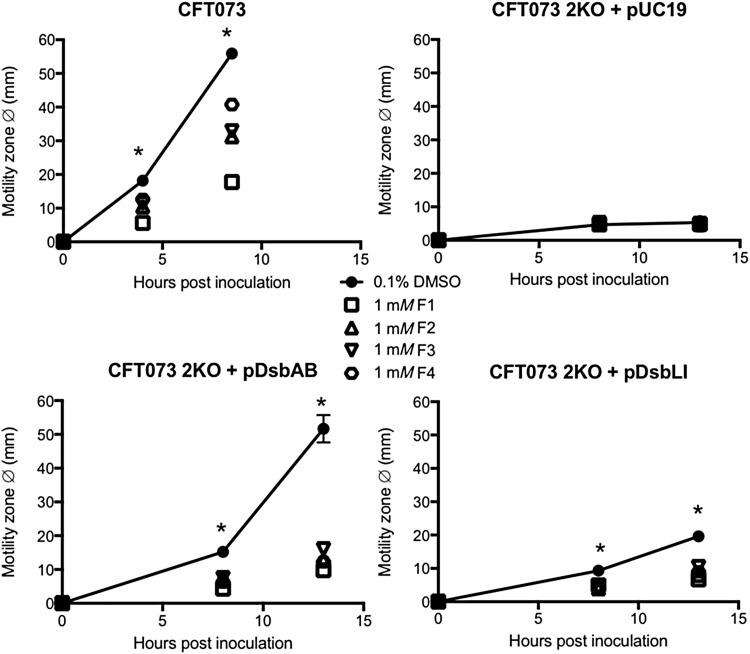
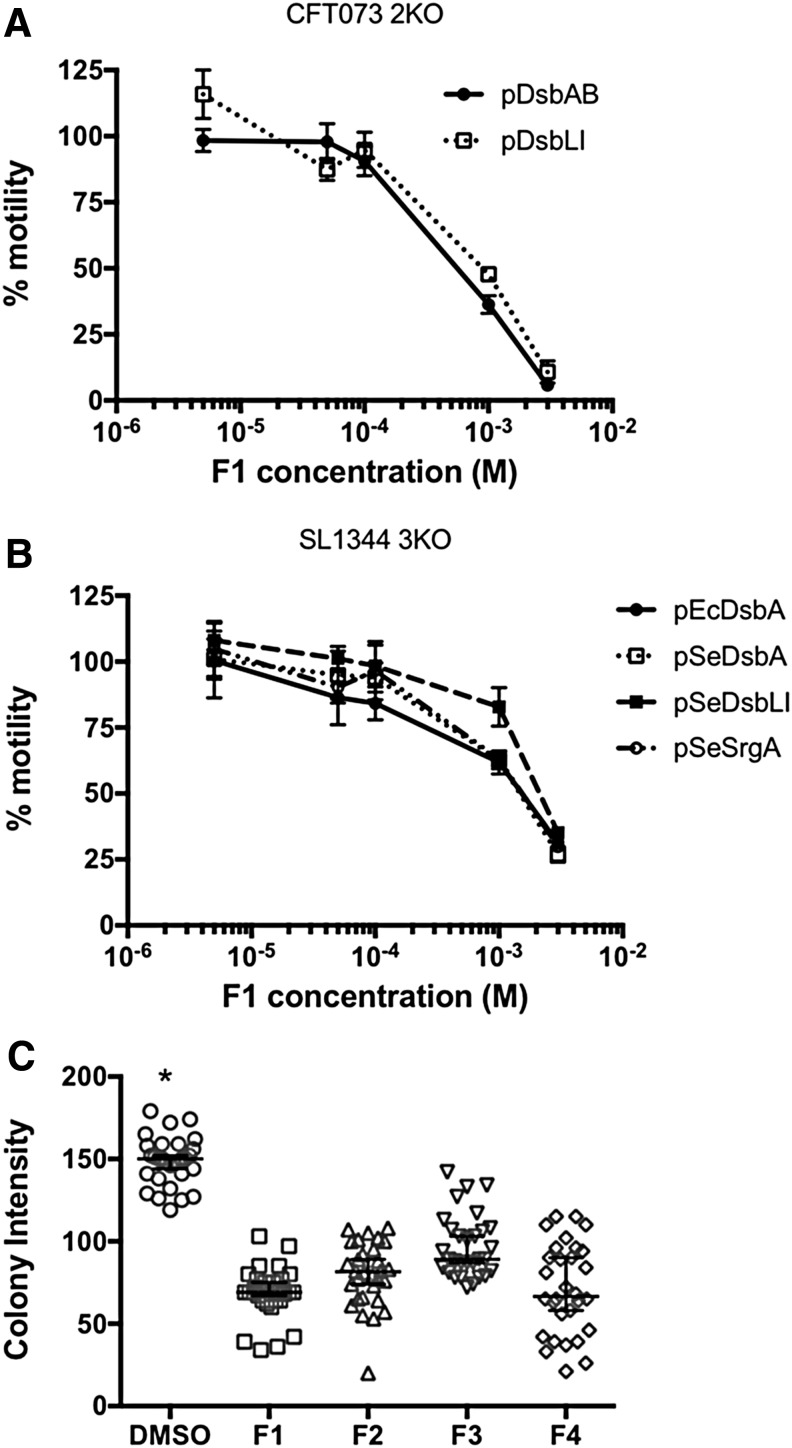
To investigate inhibitor specificity further, we utilized previously constructed and characterized sets of UPEC CFT073 and S. Typhimurium SL1344 mutants lacking the full complement of dsbA genes (two and three, respectively). The mutants were complemented with each missing homologue on plasmids that were under lac operon control (30, 68). Motility assays were conducted as described earlier, except ampicillin and isopropyl β-d-1-thiogalactopyranoside (IPTG) were also included in all soft agar plates to maintain complementation vectors and induce dsbA homologue expression. The lack of motility by UPEC mutant CFT073dsbAB,dsbLI (2KO) was complemented fully by pDsbAB and partially by pDsbLI, as previously reported (68), and this complementation was significantly inhibited by all four inhibitors at 1 mM (Fig. 4), suggesting that the inhibitors are active on both of these diverse enzymes.
FIG. 4.
Specificity of EcDsbA inhibitors for two diverse DsbA homologues in UPEC. UPEC strain CFT073 encodes DsbA and DsbL, both of which can complement the motility defect of the 2KO mutant when provided in trans (2KO + pUC19 empty vector vs. 2KO + pDsbAB/pDsbLI), fully or partially, respectively. Restoration of CFT073 2KO motility by either DsbA or DsbL was inhibited by all four inhibitors at a similar level. Dot plots represent mean motility zone diameter (mm) ± SEM of four independent replicates for each strain measured on motility plates containing 0.1% DMSO (carrier control; closed circles with line) or 1 mM inhibitor F1, F2, F3, or F4 (open symbols; squares, triangles, inverted triangles, and hexagons, respectively), *p < 0.05, ANOVA. SEM, standard error of the mean.
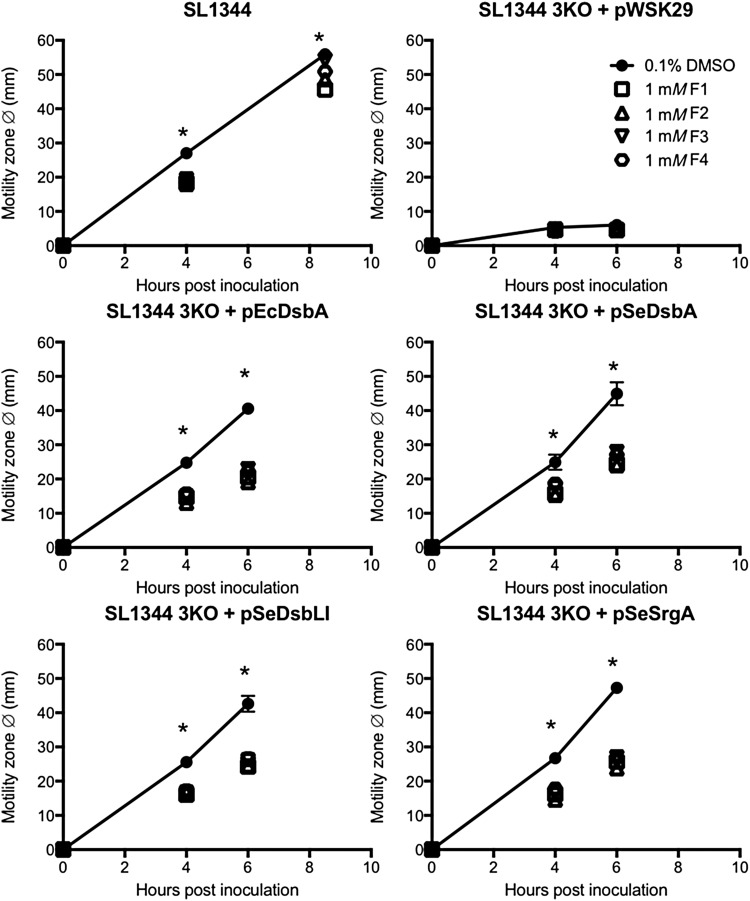
Similarly, the motility defect of the S. Typhimurium SL1344dsbA,dsbLI,srgA mutant (3KO) was fully complemented by either one of the three native Salmonella DsbA homologues (pSeDsbA, pSeDsbLI, pSeSrgA) or the E. coli DsbA (pEcDsbA), as previously reported (30), and in all cases, motility restoration was significantly impaired when inhibitors were present in the media at 1 mM concentration (Fig. 5). In fact, no differences were seen in the relative motility inhibition obtained with each compound across the different 3KO complemented strains.
FIG. 5.
Specificity of EcDsbA inhibitors for three diverse DsbA homologues in S. Typhimurium. S. Typhimurium strain SL1344 encodes DsbA, DsbL, and SrgA and each homologue can fully complement the motility defect of the 3KO mutant when provided in trans, as can the prototypical E. coli DsbA (3KO + pWSK29 empty vector vs. 3KO + pEcDsbA/pSeDsbA/pSeDsbLI/pSeSrgA). Restoration of SL1344 3KO motility by every homologue was inhibited by all four inhibitors at a similar level. Dot plots represent mean motility zone diameter (mm) ± SEM of four independent replicates for each strain measured on motility plates containing 0.1% DMSO (carrier control; closed circles with line) or 1 mM inhibitor F1, F2, F3, or F4 (open symbols; squares, triangles, inverted triangles, and hexagons, respectively), *p < 0.05, ANOVA.
To investigate whether motility inhibition was concentration dependent, inhibitors were further tested in dose-response motility assays by using both sets of UPEC and S. Typhimurium mutants and complemented strains. For UPEC, inhibitory effects were only observed at F1 concentrations of 1 mM or higher, with no motility inhibition seen at 100, 50, or 5 μM; this was true for both DsbA- and DsbL-mediated UPEC motility (Fig. 6A). For S. Typhimurium, motility inhibition by F1 followed a more gradual dose response in all complemented strains (Fig. 6B). Similar inhibitory effects were seen with inhibitors F2, F3, and F4 (Supplementary Fig. S3), suggesting that all compounds can act on the DsbA homologues in a dose-dependent fashion.
FIG. 6.
Dose-dependent inhibition of (A) UPEC and (B) S. Typhimurium motility by compound 1. Dose-response plots of relative motility for (A) UPEC CFT073 2KO transiently expressing DsbAB (closed circles) or DsbLI (open squares) and (B) S. Typhimurium SL1344 3KO complemented with EcDsbA (closed circles), SeDsbA (open squares), SeDsbLI (closed squares), or SeSrgA (open circles). Relative% motility was calculated by measuring the diameter of the motility zone for each strain in media containing 0.005, 0.05, 0.1, 1, or 3 mM of inhibitor F1 and dividing by the diameter of the same strain swimming in DMSO-containing media (carrier control). Dot plots represent mean relative motility ± SEM of four independent replicates. (C) Inhibition of S. Typhimurium DsbL by EcDsbA inhibitors. Intensity of fluorescent colonies of S. Typhimurium SL1344 3KO complemented with SeDsbLI. SeDsbLI catalyzes disulfide formation in arysulfate sulfotransferase, which can cleave MUS to release a fluorescent product. Bacteria were cultured in lysogeny broth-MUS agar plates with or without inhibitors (1 mM) and imaged in a BioRad GelDoc. The colony intensity of 30 colonies per plate was measured as adjusted volume (background-adjusted sum of all intensities within the colony boundaries) in Image Lab 5.0 and is shown as dot plots with group means and 95% confidence intervals. Addition of F1, F2, F3, or F4 in the agar resulted in a significant reduction in mean colony intensity compared with DMSO-containing plates, *p < 0.05, ANOVA. MUS, methylumbelliferyl sulfate; SeDsbA, DsbA from S. enterica serovar Typhimurium.
The ability of all compounds to inhibit the highly diverse DsbL homologue was further confirmed by monitoring the functional activity of arysulfate sulfotransferase (AssT) in SL1344, a native substrate of DsbL that requires disulfide bond formation for its functional folding and catalytic activity (30). Active AssT cleaves 4-methylumbelliferyl sulfate (MUS) to generate a fluorescent product. When plated on selective media containing IPTG and MUS, the intensity of 3KOpDsbLI fluorescent colonies was significantly decreased when inhibitors were present at 1 mM, as compared with the DMSO control (Fig. 6C). Taken together, these findings demonstrate that our EcDsbA inhibitors F1–F4 can interact and inhibit diverse DsbA homologues found in two species of pathogenic bacteria.
Atomic characterization of EcDsbA–inhibitor complexes
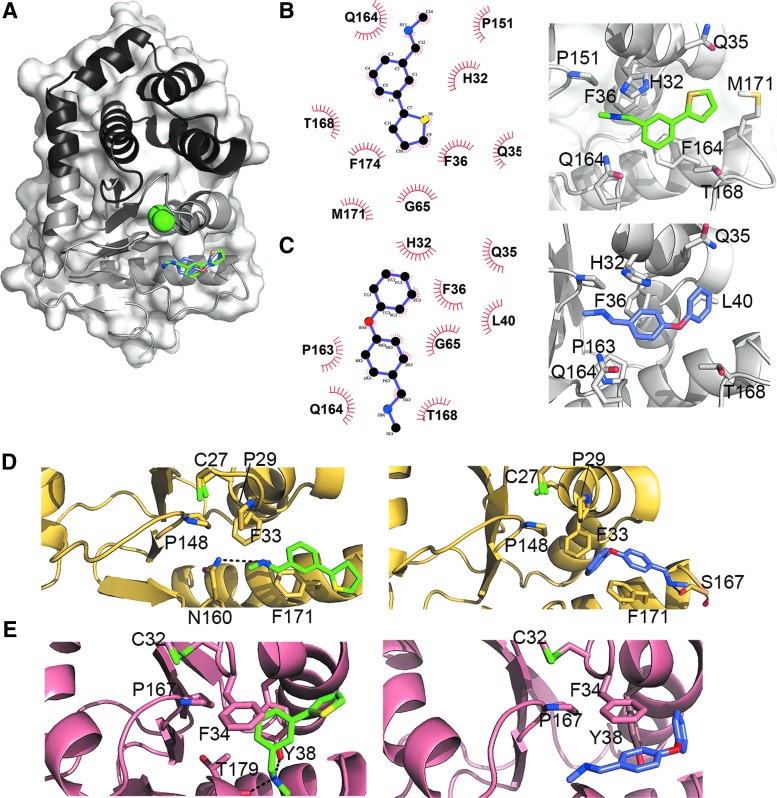
To better understand the mode of action of our EcDsbA inhibitors, we structurally characterized one phenylthiophene (F1) and one phenoxyphenyl (F3) derivative in complex with EcDsbA and defined their mode of binding (Fig. 7A). F1 and F3 were individually soaked at 10 mM concentration into preformed EcDsbA crystals, and the structures of these complexes were determined at 1.99 Å resolution (Supplementary Table S1). The co-crystal structures revealed that both molecules bind to EcDsbA in a similar region of the hydrophobic groove near the CPHC motif (Fig. 7A–C and Supplementary Fig. S4). To accommodate these compounds in the cleft, the loop linking β5 with α7 undergoes a substantial conformational change, which has been observed for other EcDsbA inhibitors on binding (4). F1 and F3 are stabilized primarily by hydrophobic contacts with His32, Gln35, Phe36, Gly65, Pro151, Gln164, and Thr168 (Fig. 7B and C). In addition, the phenyl ring of F1 forms a partial stacking with Phe174 and neighbors Met171 (Fig. 7B), whereas F3 flanks residues Leu40 and Pro163 (Fig. 7C). The binding of these small molecules to the hydrophobic groove of EcDsbA would obstruct the interaction with and reoxidation by EcDsbB, resulting in a pronounced reduction of EcDsbA function as shown by the dramatic inhibition of motility observed for E. coli K-12 (Fig 3B).
FIG. 7.
Structural characterization of EcDsbA in complex with inhibitors F1 and F3 and molecular docking of F1 and F3 with SrgA and DsbL. (A) Superposition of the co-crystal structures of EcDsbA-F1 and EcDsbA-F3 complexes showing that both inhibitors bind to a similar part of the EcDsbA hydrophobic groove. The position of the active site cysteine residues is shown in green. (B, C) PDBsum analysis (lig-plot, left panel) and close-up view (right panel) of the EcDsbA-F1 and EcDsbA-F3 complexes, respectively. (D, E) Binding mode of top ranked docked poses of F1 (green) and F3 (slate blue) into SrgA (D) and DsbL (E) hydrophobic clefts, obtained by using AutoDock Vina. For clarity, only interacting residues and the active site cysteines and cis-proline are displayed in sticks style. F1 and F3 are predicted to be stabilized by Π interactions with aromatic residues in both SrgA (F171) and DsbL (F34 and Y38). In addition, H-bonds can occur between F1 and SrgA N160, F3 and the carbonyl of SrgA S167. Similarly, F1 can form an H-bond with Y38 and the main chain carbonyl of T179 in DsbL. A likely shift in DsbL K37 would allow it to hydrogen bond to the sulfur and oxygen groups of F1 and F3, respectively. Similarly, a possible shift in the sidechain of DsbL Q164 would allow another interaction with F3 nitrogen. All hydrogen bonds are depicted by black-dashed lines.
Molecular docking of EcDsbA inhibitors F1 and F3 with DsbL and SrgA
F1 and F3 resulted in motility inhibition in UPEC and S. Typhimurium strains expressing the diverse DsbA homologues, DsbL and SrgA (Figs. 4 and 5). AutoDock Vina was used to explore the molecular docking of F1 and F3 with SeSrgA and SeDsbL. First, we assessed the ability of this molecular docking tool to reproduce the previously identified binding mode of these inhibitors to EcDsbA. In both cases, the top docked conformation out of a total of nine binding conformations, based on predicted affinity (kcal/mol), closely approximated the positioning of the inhibitors in the crystal structures (Supplementary Fig. S5A).
We then docked F1 and F3 into SeSrgA and SeDsbL by using the same parameters as for the control run with EcDsbA, except for the inclusion of some flexible amino acid side chains for the DsbA homologues. Specifically, DsbL Lys37 and Tyr38 were made flexible along with SrgA Asn160 as they partly occluded the hydrophobic cleft in their crystal structures. Docking conformations were ranked on the predicted binding affinities and resemblance to the experimental EcDsbA-F1/F3 binding modes seen in the crystal structures. In all cases, the final conformations selected (Fig. 7D, E and Supplementary Fig. S6) were in the top three out of nine binding modes based on predicted affinities, with the top three binding conformations being largely similar (Supplementary Fig. S5B, C). Resembling the EcDsbA-F1/F3 complexes, the hydrophobic nature of SeSrgA and SeDsbL clefts along with the aromatic nature of the F1 and F3 inhibitors means that Π–Π aromatic contacts are the predominate interactions (Π–Π interactions with Phe171 in SeSrgA and Phe34, Tyr38 in SeDsbL) that stabilize F1 and F3 in the hydrophobic groove of these enzymes. These docking models also show that backbone and side chain-mediated hydrogen bonds can also occur between the inhibitors and the diverse DsbA homologues (Fig. 7D, E, Supplementary Fig. S6).
Discussion
Inhibition of oxidative protein folding in bacteria is considered a promising antimicrobial approach that could provide urgently needed solutions to the global problem of rising drug resistance and the paucity in new antibiotic development. Most Gram-negative bacteria contain dedicated disulfide bond (Dsb) machinery to efficiently catalyze oxidative disulfide formation, which is an all-important step in the assembly of a wide array of bacterial virulence proteins, from adhesion factors and components of secretory machineries to toxins, flagella, and other critical virulence enzymes (29).
The thiol-oxidizing DsbA/DsbB system is currently the main target for pharmacological inhibition (64) as it constitutes the major player in virulence factor oxidative folding, and deletion of dsbA/dsbB genes in numerous pathogens results in significant or total reduction in virulence [reviewed in Refs. (29) and (62)]. Bacteria, however, display considerable diversity in their disulfide folding enzymes (17, 25, 59), particularly among DsbA thiol oxidases (29, 36, 50), raising questions about the spectrum of activity that can be achieved by some of the already developed inhibitors. Here, we utilized a combinatorial approach to explore the specificity of two classes of recently developed E. coli DsbA inhibitors against diverse DsbA targets found in two important human pathogens.
UPEC and S. Typhimurium are ideal organisms for exploring the therapeutic application of DsbA inhibitors. First, they are two of the top World Health Organization (WHO) listed pathogens of international concern due to their high antibiotic resistance rates and a combined global burden of more than 200 million infections annually (1). UPEC are the leading cause of urinary tract infections (UTIs) in hospitals and the community and a frequent cause of bloodstream infections (69). An estimated 150 million UTIs occur globally each year, costing more than 6 billion dollars in direct healthcare expenditure (22). Nontyphoidal S. enterica serotypes such as Typhimurium are the main cause of foodborne diarrhea, with an estimated burden of 94 million gastroenteritis cases and 1,55,000 global deaths each year (45).
Resistance to fluoroquinolones (one of the most widely used oral antibiotics) is globally high in E. coli and Salmonella, with rates exceeding 50% in 5/6 of the WHO regions and 25% in 3/6 WHO regions, respectively (1). Even higher rates of third-generation cephalosporin resistance in E. coli (>65% in 6/6 WHO regions), conferred by extended spectrum beta-lactamase enzymes, means that treatment of multidrug resistant (MDR) E. coli infections must now rely on last-line drugs, such as carbapenems or colistins. Worryingly, both carbapenem and colistin resistance has already been reported in MDR E. coli (44, 49, 54), highlighting the challenge that clinicians now face in treating pan-resistant E. coli infections worldwide (3).
UPEC and S. Typhimurium are also ideal pathogens for studying DsbA inhibitor specificity as they represent a large group of human, animal, and plant pathogens that encode not just one but also multiple and diverse DsbA homologues in their genome (29): DsbA and DsbL for UPEC, and DsbA, DsbL, and SrgA for S. Typhimurium. These homologues fulfil similar redox roles in vivo; however, they show differences with regard to their substrate repertoire in each pathogen (23, 68). SrgA is a close structural homologue of the prototypical E. coli K-12 DsbA but bears a different redox active site and redox properties (30). DsbL is a more distant homologue of EcDsbA in both sequence and 3D architecture, with one of the main differences being a truncated hydrophobic groove (23, 30). Given that the groove is the area of the enzyme where most described EcDsbA inhibitors were shown to bind to date [reviewed in Ref. (64)], this would suggest that such inhibitors might display variable activity against DsbL and SrgA. Our data for phenylthiophene and phenoxyphenyl class inhibitors, however, argue against this, as we have clearly evidenced that: (i) these inhibitors can be readily accommodated in the cleft of DsbA, SrgA, and DsbL and (ii) that they can block the function of all enzymes in UPEC and S. Typhimurium in vivo to an extent that attenuates virulence at similar levels.
Interestingly, when inhibitor impact on motility was assessed on wild-type strains of UPEC and S. Typhimurium (each encoding their full complement of DsbA homologues), the extent of motility inhibition was more variable (in the case of UPEC) or less pronounced (in the case of S. Typhimurium) than when assessed on complemented mutants encoding each homologue in trans. The explanation of a simply higher DsbA copy-number present in wild-type strains versus complemented mutants is probably unlikely to fully account for the observed differences, given that all mutants in our study were complemented with high copy-number plasmids, and hence expressed each DsbA homologue at higher levels than the wild-type strains (30, 68). This difference more likely suggests that the copy-number and biology of DsbA homologues found in each pathogen (i.e., enzyme sequence, structure, redox properties, gene expression, and full repertoire of substrates folded) determine the activity of each inhibitor. This highlights the significance of testing DsbA inhibitors directly on wild-type pathogenic isolates with a full complement of native DsbAs, such as those used in this study, instead of laboratory strains or non-pathogenic isolates, and this would also allow for testing whether resistance to such inhibitors could develop by homologue transfer.
Moreover, our work highlights that the abundance of structure-function information on bacterial DsbA proteins can allow insightful predictions to be made on their potential druggability based on their grouping into distinct clades. This would greatly assist the development of antimicrobials with a customized spectrum of activity. Although DsbA, DsbL, and SrgA are diverse homologues (in sequence and structure) found within the same pathogen, when examining their diversity in light of the wider diversity observed in 20 structurally and functionally characterized DsbA enzymes from distant bacterial species, we found that DsbA, SrgA, and DsbL all cluster together in clade 1 and are representative of the structural diversity observed within this clade. This would predict that EcDsbA inhibitors could be active against homologues within this clade, which was supported by our functional, structural, and modeling data. This notion is further supported by recent studies showing that PmDsbA and KpDsbA, which are also members of clade 1, interact and can be blocked by other EcDsbA inhibitors (38, 39), whereas homologues from clade 2 and 3 remain active (unpublished data). Similarly, inhibitors developed against any DsbA representative from clade 2 or 3 could be expected to display activity across organisms encoding other homologues for the same clade, and as clade 3 is highly taxonomically diverse, this would suggest that the development of broad-spectrum DsbA inhibitors may be a possibility.
We have demonstrated that phenylthiophene and phenoxyphenyl inhibitors of EcDsbA are active against DsbL and SrgA, which are distinct homologues encoded together with DsbA in the important human pathogens UPEC and S. Typhimurium. Our atomic resolution co-crystal structures revealed a binding mode for these small-molecule inhibitors to EcDsbA that would obstruct its interaction with the cognate oxidase EcDsbB, therefore altering DsbA redox homeostasis and markedly decreasing DsbA function. This mode of binding was closely mimicked by the modeled interactions seen for our inhibitors with the structurally diverse SrgA and DsbL, which would predict a similar inhibition of function to that of EcDsbA and that we confirmed functionally in different bacterial virulence assays.
From the positioning of our inhibitors in the protein structures and the residue conservation in neighboring areas, one can obtain information for the requirements of DsbA inhibitor design to increase inhibitor potency and/or customize the inhibitor activity spectrum. Both phenylthiophene and phenoxyphenyl inhibitors bound to the same region in the groove and were mostly surrounded by aromatic residues. Key amino acids included histidine 32 and phenylalanine 36 and 174 (E. coli DsbA numbering), which form hydrophobic interactions with the inhibitors, mainly Π–Π aromatic interactions. Some of these residues were found to be physicochemically conserved across diverse DsbA homologues; histidine 32 is present in 65% of the analyzed prototypes and in the remaining proteins this residue is most commonly aromatic (phenylalanine or tyrosine). Similarly, phenylalanine 36 is 65% conserved and the exceptions have mostly a tyrosine or leucine in this position. A conserved glutamine/asparagine at position 164 (50% conserved across prototypes) brings a polar group to interact with the inhibitors.
Conservation of these important residues across distinct DsbA homologues dictates that hydrophobic compounds that can maintain the conserved interactions would bind and partially block other diverse DsbA homologues. Future efforts for increasing the potency and spectrum of activity of the inhibitors should consider highly conserved residues surrounding the active site of DsbA. Expanding the structure of the inhibitors to interact with the conserved cysteines in the active site and/or the residues in the cis-proline loop ([Gly/Ala] [Val/Thr] cisPro) may be a useful approach to develop more potent inhibitors while maintaining a broad spectrum of activity.
Materials and Methods
Sequence and phylogenetic analysis of DsbA homologues
We carried out a Dali search (32) by using the structures of EcDsbA [PDB code 1FVK, (24)], SaDsbA [PDB code 3BCI, (27)], and DsbL [PDB code 3C7M (23)] as our query models to identify all DsbA homologues available in the RCSB Protein Data Bank. Given that DsbA-like proteins share low SI, we utilized different methods to curate our list of DsbA homologues and confirm that the identified proteins were, indeed, DsbA homologues. All identified DsbA structures were manually analyzed for the presence of the structural hallmarks of DsbA homologues (inserted alpha helical domain and the TRX-fold encompassing a CXXC motif and a cis-Proline loop). Further, structures and corresponding sequences were retained only where published experimental data confirmed that the identified enzymes functioned as thiol oxidases (Supplementary Fig. S1).
The corresponding full-length amino acid sequences of 22 DsbA proteins matching the criteria described earlier were retrieved, and a non-redundant set of 20 (prototypes) was used in subsequent analyses (DsbA from S. enterica and DsbL from UPEC were excluded due to high sequence and structural identity to their positional orthologues in E. coli and S. enterica, respectively; Supplementary Fig. S1). Multiple sequence alignments of the 20 DsbA prototypes were carried out by using three protein alignment methods of the T-Coffee MSA package: (1) Expresso structural alignment (6), (2) PSI-Coffee Homology extension (14), and (3) M-Coffee combining popular aligners (73). Alignment accuracy was evaluated by TCS (12, 13) and by confirming that the two completely conserved features of the DsbA active site (the characteristic CXXC catalytic motif and the cis-Proline loop), which are adjacent in three-dimensional space but distant in primary sequence, were aligned among all sequences. The Expresso structural alignment was selected and used to reconstruct an unrooted Neighbor-Joining and maximum likelihood consensus phylogenetic tree based on 1000 bootstrap replicates using the PHYLIP package (21). The accession number and PDB code for each DsbA homologue is shown in Supplementary Figure S1.
Bacterial strains, plasmids, and culture conditions
All bacterial strains used in this study (Supplementary Table S2) were routinely cultured at 37°C on solid or in liquid lysogeny broth (LB) medium supplemented, where necessary with kanamycin (km, 50 μg mL−1) or ampicillin (amp, 100 μg mL−1). Culture media were supplemented with 1 μM isopropyl IPTG to induce expression of DsbA, DsbL, and SrgA from plasmids pDsbA, pDsbLI, and pSrgA, respectively. Construction of UPEC and S. Typhimurium mutants and plasmids was previously described (30, 68).
Motility assays
Swimming motility of E. coli and S. Typhimurium strains was assessed as previously described (30, 68). Briefly, 2 μl of four independent liquid overnight cultures of each strain was inoculated onto the surface of LB semi-solid (0.3% w/v) agar containing DMSO or inhibitors (phenylthiophenes and phenoxyphenyl) at various concentrations (5–3 mM). Plates were incubated at 37°C, and the diameter of bacterial outward growth was measured in millimeters at various time-points. The mean motility zone diameter for each strain was calculated from four replicates tested under each condition, and group means were compared by one-way ANOVA (statistical significance set at p < 0.05). Circle plots of mean motility zone diameters for wild-type strains (Fig. 3B) were generated in R (2) by using the functions of the package “plotrix” (43). All other data graphs were generated in GraphPad Prism.
AssT activity assays
AssT enzyme activity was monitored at a colony level by using an agar plate assay, as previously described (30, 75). Briefly, strain SL1344 3KOpSeDsbLI was streaked on LB agar plates containing 0.1 mM 4-MUS (Sigma, Castle Hill, Australia) and 0.1% DMSO or 1 mM inhibitors F1–F4. The sulfate of the 4-MUS in the medium is cleaved by AssT forming 4-methylumbelliferone, a fluorescent product that can be detected under UV light (320 nm). Plates were incubated at 37°C overnight. Functional AssT production requires DsbL-mediated disulfide-bond formation and in conditions where functional AssT enzyme is produced S. Typhimurium colonies fluoresce brightly under UV light. Plates were UV exposed and imaged simultaneously in a BioRad GelDoc. The colony intensity of 30 colonies per plate was measured as adjusted volume (background-adjusted sum of all intensities within the colony boundaries) in Image Lab 5.0. Group means were compared by one-way ANOVA (statistical significance set at p < 0.05).
Expression and purification of EcDsbA
Recombinant EcDsbA was expressed and purified as previously described (4, 53). Briefly, E. coli BL21 (DE3) carrying native EcDsbA encoding plasmid were grown for 24 h at 30°C in ZYM-5052 autoinduction media (65) supplemented with 50 μg/mL kanamycin.
Cells were harvested by centrifugation, and the periplasmic fraction was obtained by cold osmotic shock (26). On addition of 0.8 M (NH4)2SO4, the periplasmic fraction was loaded onto a HiLoad 1610 Phenyl Sepharose HP column (GE Healthcare) equilibrated in 20 mM 2-amino-2-(hydroxymethyl)propane-1,3-diol (TRIS) (pH 8.0), 50 mM NaCl, and 1 M (NH4)2SO4. The bound proteins were eluted on a gradient from 1–0 M (NH4)2SO4. Fractions containing EcDsbA were buffer exchanged into 25 mM 4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES) (pH 6.8), and the protein was further purified by anion-exchange chromatography on a Mono Q 5/50 GL column (GE Healthcare). EcDsbA was then oxidized by the addition of 1.7 mM copper(II)[1,10-phenanthroline], and the protein was purified to homogeneity by using a HiLoad Superdex S-75 size-exclusion chromatography column (GE Healthcare) equilibrated in 25 mM HEPES (pH 6.8), 150 mM NaCl. Protein purity was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Crystallization and structure determination of EcDsbA-inhibitor co-crystals
Crystals of EcDsbA were grown by the hanging drop vapor-diffusion method using previously established conditions (47). Briefly, 1 μL of 30 mg/mL EcDsbA was mixed with 1 μL of crystallization buffer (11–13% polyethylene glycol [PEG] 8000, 5% glycerol, 1 mM CuCl2, 100 mM sodium cacodylate pH 6.1–6.4) and equilibrated against 0.5 mL of crystallization buffer at 18°C. Typically, large crystals (0.6 × 0.4 × 0.2 mm) were obtained after 2–3 days of incubation.
Crystal soaking was carried out by transferring EcDsbA crystals into 2 μL drops of 24% PEG 8000, 22% glycerol, 100 mM sodium cacodylate pH 6.1–6.4 containing compound 1 or compound 3 at a final concentration of 10 mM (2–5% of DMSO) and incubating them for 2 h. Crystals were mounted into loops and flash-cooled in liquid nitrogen before data collection. Diffraction data for complexes 1 and 3 were collected at the UQ ROCX facility (using a Rigaku FR-E Superbright X-ray generator and a Rigaku Saturn 944 CCD detector) and the Australian Synchrotron, respectively. Overall, 0.5° or 1° oscillation images were collected for a total of 180°. Diffraction data was indexed and integrated with CrystalClear 1.4. (Rigaku) or HKL2000. Phasing was carried out by molecular replacement with Phaser (48) by using the structure of EcDsbA (PDB code 1FVK) as a search model (24). The final models of EcDsbA in complex with compound 1 and 3 were completed by iterative cycles of model building and refinement by using Coot (20) and phenix.refine (5). Data collection and refinement statistics are summarized in Supplementary Table S1. Generation of molecular figures was carried out with PyMOL v1.7.0.5. The structures were submitted to the PDB under the codes 6BR4 and 6BQX.
Docking methods
The computational molecular docking tool AutoDock Vina (70) was used to predict the binding mode of F1 and F3 to DsbL and SrgA. The protocol was first validated by docking F1 and F3 into EcDsbA and comparing the docking results with the experimental crystal structures of EcDsbA in complex with those inhibitors. The docking protocol involved preparing the pdbqt files of proteins and ligands using AutoDockTools (52). The apo form of EcDsbA was set as a rigid structure, where a 24 × 16 × 24 Å search space was set up to cover the entire hydrophobic cleft. Standard chemical bond torsions were applied to F1 and F3, which were docked to EcDsbA by using AutoDock Vina to calculate binding affinity. For each inhibitor, the results were ranked on the basis of predicted free energy of binding and the conformations that most closely approximated the crystal structures. For docking F1 and F3 to SrgA and DsbL, the x-ray crystallographic structures of these proteins were recovered from the protein data bank (3TRK, and 3N41); any co-crystallized ligand and water molecules were removed; and F1 and F3 were docked to the macromolecules by using AutoDock Vina. The interaction between ligand and macromolecule was visualized by using the PyMOL molecular graphics system (PyMOL v1.7.0.5).
Supplementary Material
Abbreviations Used
- ARC
Australian Research Council
- AssT
arysulfate sulfotransferase
- CPHC
Cys30-Pro31-His32-Cys33
- DMSO
dimethyl sulfoxide
- Dsb
disulfide bond
- EcDsbA
Escherichia coli DsbA
- HEPES
4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid
- IPTG
isopropyl β-D-1-thiogalactopyranoside
- LB
lysogeny broth
- MDR
multidrug resistant
- MSA
multiple sequence alignment
- MUS
methylumbelliferyl sulfate
- PEG
polyethylene glycol
- SeDsbA
DsbA from S. enterica serovar Typhimurium
- SEM
standard error of the mean
- S.I.
sequence identity
- TRX
thioredoxin
- UPEC
uropathogenic Escherichia coli
- UTIs
urinary tract infections
- WHO
World Health Organization
Acknowledgments
This work was supported by an Australian Research Council (ARC) Discovery Early Career Researcher Award (DE130101169) to MT and an ARC project grant (DP150102287) to B.H. B.H. is supported by an ARC Future Fellowship (FT130100580), and M.T. is supported by a Queensland University of Technology Vice-Chancellor's Senior Research Fellowship. The authors acknowledge the use of the Australian Synchrotron and the UQ ROCX Diffraction Facility.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.World Health Organization. Antimicrobial Resistance: Global Report on Surveillance. 2014. http://www.who.int/drugresistance/documents/surveillancereport/en/
- 2.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: URL www.R-project.org, 2016. (accessed April4, 2017) [Google Scholar]
- 3.World Health Organization. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. 2017. http://www.who.int/medicines/publications/global-priority-list-antibiotic-resistant-bacteria/en/
- 4.Adams LA, Sharma P, Mohanty B, Ilyichova OV, Mulcair MD, Williams ML, Gleeson EC, Totsika M, Doak BC, Caria S, Rimmer K, Horne J, Shouldice SR, Vazirani M, Headey SJ, Plumb BR, Martin JL, Heras B, Simpson JS, and Scanlon MJ. Application of fragment-based screening to the design of inhibitors of Escherichia coli DsbA. Angew Chem Int Ed Engl 54: 2179–2184, 2015 [DOI] [PubMed] [Google Scholar]
- 5.Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, and Adams PD. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr 68: 352–367, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Armougom F, Moretti S, Poirot O, Audic S, Dumas P, Schaeli B, Keduas V, and Notredame C. Expresso: automatic incorporation of structural information in multiple sequence alignments using 3D-Coffee. Nucleic Acids Res 34: W604–W608, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arts IS, Ball G, Leverrier P, Garvis S, Nicolaes V, Vertommen D, Ize B, Tamu Dufe V, Messens J, Voulhoux R, and Collet JF. Dissecting the machinery that introduces disulfide bonds in Pseudomonas aeruginosa. MBio 4: e00912–e00913, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blattner FR, Plunkett G, III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, and Shao Y. The complete genome sequence of Escherichia coli K-12. Science 277: 1453–1462, 1997 [DOI] [PubMed] [Google Scholar]
- 9.Bouwman CW, Kohli M, Killoran A, Touchie GA, Kadner RJ, and Martin NL. Characterization of SrgA, a Salmonella enterica serovar Typhimurium virulence plasmid-encoded paralogue of the disulfide oxidoreductase DsbA, essential for biogenesis of plasmid-encoded fimbriae. J Bacteriol 185: 991–1000, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brannon JR. and Hadjifrangiskou M. The arsenal of pathogens and antivirulence therapeutic strategies for disarming them. Drug Des Devel Ther 10: 1795–1806, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cegelski L, Marshall GR, Eldridge GR, and Hultgren SJ. The biology and future prospects of antivirulence therapies. Nat Rev Microbiol 6: 17–27, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang JM, Di Tommaso P, Lefort V, Gascuel O, and Notredame C. TCS: a web server for multiple sequence alignment evaluation and phylogenetic reconstruction. Nucleic Acids Res 43: W3–W6, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang JM, Di Tommaso P, and Notredame C. TCS: a new multiple sequence alignment reliability measure to estimate alignment accuracy and improve phylogenetic tree reconstruction. Mol Biol Evol 31: 1625–1637, 2014 [DOI] [PubMed] [Google Scholar]
- 14.Chang JM, Di Tommaso P, Taly JF, and Notredame C. Accurate multiple sequence alignment of transmembrane proteins with PSI-Coffee. BMC Bioinformatics 13 Suppl 4: S1, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chim N, Harmston CA, Guzman DJ, and Goulding CW. Structural and biochemical characterization of the essential DsbA-like disulfide bond forming protein from Mycobacterium tuberculosis. BMC Struct Biol 13: 23, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crow A, Lewin A, Hecht O, Carlsson Moller M, Moore GR, Hederstedt L, and Le Brun NE. Crystal structure and biophysical properties of Bacillus subtilis BdbD. An oxidizing thiol:disulfide oxidoreductase containing a novel metal site. J Biol Chem 284: 23719–23733, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Denoncin K. and Collet JF. Disulfide bond formation in the bacterial periplasm: major achievements and challenges ahead. Antioxid Redox Signal 19: 63–71, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duprez W, Premkumar L, Halili MA, Lindahl F, Reid RC, Fairlie DP, and Martin JL. Peptide inhibitors of the Escherichia coli DsbA oxidative machinery essential for bacterial virulence. J Med Chem 58: 577–587, 2015 [DOI] [PubMed] [Google Scholar]
- 19.Dutton RJ, Boyd D, Berkmen M, and Beckwith J. Bacterial species exhibit diversity in their mechanisms and capacity for protein disulfide bond formation. Proc Natl Acad Sci U S A 105: 11933–11938, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Emsley P. and Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132, 2004 [DOI] [PubMed] [Google Scholar]
- 21.Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.6. Distributed by the author. Department of Genome Sciences, University of Washington, Seattle: 2005 [Google Scholar]
- 22.Foxman B. The epidemiology of urinary tract infection. Nat Rev Urol 7: 653–660, 2010 [DOI] [PubMed] [Google Scholar]
- 23.Grimshaw JP, Stirnimann CU, Brozzo MS, Malojcic G, Grutter MG, Capitani G, and Glockshuber R. DsbL and DsbI form a specific dithiol oxidase system for periplasmic arylsulfate sulfotransferase in uropathogenic Escherichia coli. J Mol Biol 380: 667–680, 2008 [DOI] [PubMed] [Google Scholar]
- 24.Guddat LW, Bardwell JCA, Glockshuber R, HuberWunderlich M, Zander T, and Martin JL. Structural analysis of three His32 mutants of DsbA: support for an electrostatic role of His32 in DsbA stability. Protein Sci 6: 1893–1900, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hatahet F, Boyd D, and Beckwith J. Disulfide bond formation in prokaryotes: history, diversity and design. Biochim Biophys Acta 1844: 1402–1414, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heras B, Edeling MA, Byriel KA, Jones A, Raina S, and Martin JL. Dehydration converts DsbG crystal diffraction from low to high resolution. Structure 11: 139–145, 2003 [DOI] [PubMed] [Google Scholar]
- 27.Heras B, Kurz M, Jarrott R, Shouldice SR, Frei P, Robin G, Cemazar M, Thony-Meyer L, Glockshuber R, and Martin JL. Staphylococcus aureus DsbA does not have a destabilizing disulfide. A new paradigm for bacterial oxidative folding. J Biol Chem 283: 4261–4271, 2008 [DOI] [PubMed] [Google Scholar]
- 28.Heras B, Scanlon MJ, and Martin JL. Targeting virulence not viability in the search for future antibacterials. Br J Clin Pharmacol 79: 208–215, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heras B, Shouldice SR, Totsika M, Scanlon MJ, Schembri MA, and Martin JL. DSB proteins and bacterial pathogenicity. Nat Rev Microbiol 7: 215–225, 2009 [DOI] [PubMed] [Google Scholar]
- 30.Heras B, Totsika M, Jarrott R, Shouldice SR, Guncar G, Achard ME, Wells TJ, Argente MP, McEwan AG, and Schembri MA. Structural and functional characterization of three DsbA paralogues from Salmonella enterica serovar typhimurium. J Biol Chem 285: 18423–18432, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoiseth SK. and Stocker BA. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291: 238–239, 1981 [DOI] [PubMed] [Google Scholar]
- 32.Holm L, Kaariainen S, Rosenstrom P, and Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics 24: 2780–2781, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu SH, Peek JA, Rattigan E, Taylor RK, and Martin JL. Structure of TcpG, the DsbA protein folding catalyst from Vibrio cholerae. J Mol Biol 268: 137–146, 1997 [DOI] [PubMed] [Google Scholar]
- 34.Inaba K, Murakami S, Suzuki M, Nakagawa A, Yamashita E, Okada K, and Ito K. Crystal structure of the DsbB-DsbA complex reveals a mechanism of disulfide bond generation. Cell 127: 789–801, 2006 [DOI] [PubMed] [Google Scholar]
- 35.Ireland PM, McMahon RM, Marshall LE, Halili M, Furlong E, Tay S, Martin JL, and Sarkar-Tyson M. Disarming Burkholderia pseudomallei: structural and functional characterization of a disulfide oxidoreductase (DsbA) required for virulence in vivo. Antioxid Redox Signal 20: 606–617, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kimball RA, Martin L, and Saier MH., Jr Reversing transmembrane electron flow: the DsbD and DsbB protein families. J Mol Microbiol Biotechnol 5: 133–149, 2003 [DOI] [PubMed] [Google Scholar]
- 37.Kpadeh ZZ, Jameson-Lee M, Yeh AJ, Chertihin O, Shumilin IA, Dey R, Day SR, and Hoffman PS. Disulfide bond oxidoreductase DsbA2 of Legionella pneumophila exhibits protein disulfide isomerase activity. J Bacteriol 195: 1825–1833, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kurth F, Duprez W, Premkumar L, Schembri MA, Fairlie DP, and Martin JL. Crystal structure of the dithiol oxidase DsbA enzyme from proteus mirabilis bound non-covalently to an active site peptide ligand. J Biol Chem 289: 19810–19822, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kurth F, Rimmer K, Premkumar L, Mohanty B, Duprez W, Halili MA, Shouldice SR, Heras B, Fairlie DP, Scanlon MJ, and Martin JL. Comparative sequence, structure and redox analyses of Klebsiella pneumoniae DsbA show that anti-virulence target DsbA enzymes fall into distinct classes. PLoS One 8: e80210, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kurz M, Iturbe-Ormaetxe I, Jarrott R, Shouldice SR, Wouters MA, Frei P, Glockshuber R, O'Neill SL, Heras B, and Martin JL. Structural and functional characterization of the oxidoreductase alpha-DsbA1 from Wolbachia pipientis. Antioxid Redox Signal 11: 1485–1500, 2009 [DOI] [PubMed] [Google Scholar]
- 41.Lafaye C, Iwema T, Carpentier P, Jullian-Binard C, Kroll JS, Collet JF, and Serre L. Biochemical and structural study of the homologues of the thiol-disulfide oxidoreductase DsbA in Neisseria meningitidis. J Mol Biol 392: 952–966, 2009 [DOI] [PubMed] [Google Scholar]
- 42.Lasica AM. and Jagusztyn-Krynicka EK. The role of Dsb proteins of Gram-negative bacteria in the process of pathogenesis. FEMS Microbiol Rev 31: 626–636, 2007 [DOI] [PubMed] [Google Scholar]
- 43.Lemon J. Plotrix: a package in the red light district of R. R-News 6: 8–12, 2006 [Google Scholar]
- 44.Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu LF, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu JH, and Shen J. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16: 161–168, 2016 [DOI] [PubMed] [Google Scholar]
- 45.Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O'Brien SJ, Jones TF, Fazil A, Hoekstra RM; International Collaboration on Enteric Disease “Burden of Illness” Studies. The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis 50: 882–889, 2010 [DOI] [PubMed] [Google Scholar]
- 46.Martin JL, Bardwell JCA, and Kuriyan J. Crystal-structure of the Dsba protein required for disulfide bond formation in-vivo. Nature 365: 464–468, 1993 [DOI] [PubMed] [Google Scholar]
- 47.Martin JL, Waksman G, Bardwell JCA, Beckwith J, and Kuriyan J. Crystallization of Dsba, an Escherichia-coli protein required for disulfide bond formation in vivo. J Mol Biol 230: 1097–1100, 1993 [DOI] [PubMed] [Google Scholar]
- 48.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, and Read RJ. Phaser crystallographic software. J Appl Crystallogr 40: 658–674, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McGann P, Snesrud E, Maybank R, Corey B, Ong AC, Clifford R, Hinkle M, Whitman T, Lesho E, and Schaecher KE. Escherichia coli Harboring mcr-1 and blaCTX-M on a Novel IncF Plasmid: first Report of mcr-1 in the United States. Antimicrob Agents Chemother 60: 4420–4421, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McMahon RM, Premkumar L, and Martin JL. Four structural subclasses of the antivirulence drug target disulfide oxidoreductase DsbA provide a platform for design of subclass-specific inhibitors. Biochim Biophys Acta 1844: 1391–1401, 2014 [DOI] [PubMed] [Google Scholar]
- 51.Mobley HL, Green DM, Trifillis AL, Johnson DE, Chippendale GR, Lockatell CV, Jones BD, and Warren JW. Pyelonephritogenic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells: role of hemolysin in some strains. Infect Immun 58: 1281–1289, 1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, and Olson AJ. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30: 2785–2791, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paxman JJ, Borg NA, Horne J, Thompson PE, Chin Y, Sharma P, Simpson JS, Wielens J, Piek S, Kahler CM, Sakellaris H, Pearce M, Bottomley SP, Rossjohn J, and Scanlon MJ. The structure of the bacterial oxidoreductase enzyme DsbA in complex with a peptide reveals a basis for substrate specificity in the catalytic cycle of DsbA enzymes. J Biol Chem 284: 17835–17845, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Poirel L, Lagrutta E, Taylor P, Pham J, and Nordmann P. Emergence of metallo-beta-lactamase NDM-1-producing multidrug-resistant Escherichia coli in Australia. Antimicrob Agents Chemother 54: 4914–4916, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Premkumar L, Heras B, Duprez W, Walden P, Halili M, Kurth F, Fairlie DP, and Martin JL. Rv2969c, essential for optimal growth in Mycobacterium tuberculosis, is a DsbA-like enzyme that interacts with VKOR-derived peptides and has atypical features of DsbA-like disulfide oxidases. Acta Crystallogr D Biol Crystallogr 69: 1981–1994, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Premkumar L, Kurth F, Duprez W, Groftehauge MK, King GJ, Halili MA, Heras B, and Martin JL. Structure of the Acinetobacter baumannii dithiol oxidase DsbA bound to elongation factor EF-Tu reveals a novel protein interaction site. J Biol Chem 289: 19869–19880, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reardon-Robinson ME, Osipiuk J, Chang C, Wu C, Jooya N, Joachimiak A, Das A, and Ton-That H. A Disulfide bond-forming machine is linked to the sortase-mediated Pilus assembly pathway in the gram-positive bacterium actinomyces oris. J Biol Chem 290: 21393–21405, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reardon-Robinson ME, Osipiuk J, Jooya N, Chang C, Joachimiak A, Das A, and Ton-That H. A thiol-disulfide oxidoreductase of the gram-positive pathogen corynebacterium diphtheriae is essential for viability, pilus assembly, toxin production and virulence. Mol Microbiol 98: 1037–1050, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reardon-Robinson ME. and Ton-That H. Disulfide-bond-forming pathways in gram-positive bacteria. J Bacteriol 198: 746–754, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rinaldi FC, Meza AN, and Guimaraes BG. Structural and biochemical characterization of Xylella fastidiosa DsbA family members: new insights into the enzyme-substrate interaction. Biochemistry 48: 3508–3518, 2009 [DOI] [PubMed] [Google Scholar]
- 61.Shouldice SR, Heras B, Jarrott R, Sharma P, Scanlon MJ, and Martin JL. Characterization of the DsbA oxidative folding catalyst from Pseudomonas aeruginosa reveals a highly oxidizing protein that binds small molecules. Antioxid Redox Signal 12: 921–931, 2010 [DOI] [PubMed] [Google Scholar]
- 62.Shouldice SR, Heras B, Walden PM, Totsika M, Schembri MA, and Martin JL. Structure and function of DsbA, a key bacterial oxidative folding catalyst. Antioxid Redox Signal 14: 1729–1760, 2011 [DOI] [PubMed] [Google Scholar]
- 63.Sinha S, Langford PR, and Kroll JS. Functional diversity of three different DsbA proteins from Neisseria meningitidis. Microbiology 150: 2993–3000, 2004 [DOI] [PubMed] [Google Scholar]
- 64.Smith RP, Paxman JJ, Scanlon MJ, and Heras B. Targeting bacterial Dsb proteins for the development of anti-virulence agents. Molecules 21, 2016. DOI: 10.3390/molecules21070811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif 41: 207–234, 2005 [DOI] [PubMed] [Google Scholar]
- 66.Taly JF, Magis C, Bussotti G, Chang JM, Di Tommaso P, Erb I, Espinosa-Carrasco J, Kemena C, and Notredame C. Using the T-Coffee package to build multiple sequence alignments of protein, RNA, DNA sequences and 3D structures. Nat Protoc 6: 1669–1682, 2011 [DOI] [PubMed] [Google Scholar]
- 67.Totsika M. Benefits and challenges of antivirulence antimicrobials at the dawn of the post-antibiotic era. Drug Deliv Lett 6: 30–37, 2016 [Google Scholar]
- 68.Totsika M, Heras B, Wurpel DJ, and Schembri MA. Characterization of two homologous disulfide bond systems involved in virulence factor biogenesis in uropathogenic Escherichia coli CFT073. J Bacteriol 191: 3901–3908, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Totsika M, Moriel DG, Idris A, Rogers BA, Wurpel DJ, Phan MD, Paterson DL, and Schembri MA. Uropathogenic Escherichia coli mediated urinary tract infection. Curr Drug Targets 13: 1386–1399, 2012 [DOI] [PubMed] [Google Scholar]
- 70.Trott O. and Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31: 455–461, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vivian JP, Scoullar J, Rimmer K, Bushell SR, Beddoe T, Wilce MC, Byres E, Boyle TP, Doak B, Simpson JS, Graham B, Heras B, Kahler CM, Rossjohn J, and Scanlon MJ. Structure and function of the oxidoreductase DsbA1 from Neisseria meningitidis. J Mol Biol 394: 931–943, 2009 [DOI] [PubMed] [Google Scholar]
- 72.Vivian JP, Scoullar J, Robertson AL, Bottomley SP, Horne J, Chin Y, Wielens J, Thompson PE, Velkov T, Piek S, Byres E, Beddoe T, Wilce MC, Kahler CM, Rossjohn J, and Scanlon MJ. Structural and biochemical characterization of the oxidoreductase NmDsbA3 from Neisseria meningitidis. J Biol Chem 283: 32452–32461, 2008 [DOI] [PubMed] [Google Scholar]
- 73.Wallace IM, O'Sullivan O, Higgins DG, and Notredame C. M-Coffee: combining multiple sequence alignment methods with T-Coffee. Nucleic Acids Res 34: 1692–1699, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yu J. and Kroll JS. DsbA: a protein-folding catalyst contributing to bacterial virulence. Microbes Infect 1: 1221–1228, 1999 [DOI] [PubMed] [Google Scholar]
- 75.Yun HJ, Kwon AR, and Choi EC. Bacterial arylsulfate sulfotransferase as a reporter system. Microbiol Immunol 45: 673–678, 2001 [DOI] [PubMed] [Google Scholar]
- 76.Zucca M, Scutera S, Savoia D. New antimicrobial frontiers. Mini Rev Med Chem 11: 888–900, 2011 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.