Abstract
Date palm (Phoenix dactylifera L.) is one of the oldest fruit crops in the arid regions of the Middle East. However, little information is available regarding its plastid genomes. In this study, we sequenced the chloroplast (cp) genomes of two economically important but genomically unexplored date palm cultivars of Phoenix dactylifera (var. Naghal and Khanezi). The data assembly and genome annotation revealed a typical quadripartite structure similar to Arecaceae, and the genome sizes of Naghal and Khanezi were 158,210 bp and 158,211 bp, respectively. Structurally, both cp genomes were comprised of four regions: a pair of inverted repeats (27,273 bp for Khanezi and for Naghal 27,272 bp), a large single-copy region (86,090 bp and 86,092 bp) and a small single-copy region (17,575 bp and 17,574 bp). Both genomes had 138 representative genes, whereas 227 and 229 randomly distributed microsatellites were also observed in Khanezi and Naghal, respectively. Phylogenetic analysis based on the whole cp genomes and 68 shared genes showed identical phylogenetic trees of Khanezi and Naghal forming clades with Khalas and Aseel cultivars, respectively. The current study showed detailed comparative cp genome analysis, which could be essential for broader population genetics and molecular studies of these four date palm cultivars.
Introduction
Date palm, Phoenix dacylifera L., belongs to Arecaeae and is an ecologically, culturally and economically important fruit crops in North Africa, the Middle East and certain areas of the African sub-continent [1]. Date palm is a perennial, monocotyledon (2n = 36), dioecious, cross-pollinated tree that has been widely cultivated and domestically grown in semi-arid environments since ancient times [2], [3], [4]. Depending upon the variety, a tree usually takes 4 to 6 years to become sufficiently mature to produce fruit [5]. It has been estimated that there are approximately 3,000 different cultivars of data palm, and approximately 60 are commercially cultivated and traded in the international market [6]. The fruit of these varieties vary in shape, color, size, weight and taste. Morphological variations, which are heavily dependent on environmental factors and data variety, do exist among cultivars. These variations are reflected in the diversity of the chloroplast genome, as well. For example, Zehdi et al. [7] recently showed that chloroplast diversity is 70% in eastern Algeria, while the proportion of haplotypes were lower (11 to 42%) in Egypt, Tunisia and Morocco. Additionally, the nuclear and chloroplast sequence diversity across Algeria, Morocco, Tunisia and Egypt remains unexplored [1], which can be attributed to the lack of complete chloroplast genomic information.
The chloroplast is a metabolic epicenter for maintaining plant growth and development through the photosynthesis process[8]. The genome of the chloroplast (cp) encodes numerous essential proteins for photosynthesis and metabolic processes [9, 10]. The cp genome is also used for plant systematics and taxonomy11. In addition, this genome acts as a source of molecular markers to perform phylogenetics due to the lower level of recombination compared to the nuclear genome [11–15]. Currently, more than 850 chloroplast genomes have been sequenced, including more than 320 chloroplast genomes from crops and trees [16]. The composition and sequence of cp genomes show significant variations within and among species [17]. Understanding the cp genome can help elucidate genomic interactions among related species for conservation and to improve valuable features of crop species[18]. Recent reports suggest that the cp genome can be used to resolve the phylogenetics of species and help in understanding genetic diversity and population dynamics [17–20].
The cp genome is comprised of a conserved quadripartite structure, which consists of a large single-copy region, a small single-copy region, and a set of inverted repeats [16, 17, 20]. Recent developments in genome sequencing technologies have allowed researchers to efficiently utilize the cp genomics data set for designing molecular barcodes and markers for detailed taxonomical systematics and phylogenetics [21]. Previously, a partial date palm genome was reported to be 380 million bp with more than 25,000 gene models [22–26], including the cp genome composition and architecture of date palm cultivars Aseel and Khalas from Pakistan and Saudi Arabia, respectively. In the case of Arecacea as a whole, the NCBI genome database shows a total of 3 draft genomes and 34 organelle genomes. However, the detailed gene structures and comparative taxonomic differentiation of date palm is poorly explored and reported. In the current study, we aimed to sequence the complete chloroplast genomes of two date palm cultivars ‘Khanezi’ and ‘Naghal’ to better understand the genome architecture and to compare them with available date palm cultivars (Aseel and Khalas) and related species from Arecacea.
Materials and methods
Genome sequencing and assembly
The chloroplast (cp) DNA was extracted according to the protocol of Shi et al. [27] with several modifications, as described by Al-Dous et al.[22]. We carried out complete chloroplast genome sequencing of date palm (P. dactylifera L.) cv. ‘Khanezi’ and ‘Naghal’ using the Illumina HiSeq4000 sequencing platform at Duke University, USA. A total of 26,363,570,180 and 24,702,016,614 bp were generated for Khanezi and Naghal, respectively. The raw reads were later trimmed and filtered using CLC Genomics Workbench v7.0 (CLC Bio, Aarhus, Denmark), which was also used for preparing the de novo genome assembly. Reads were filtered using Trimmomatic 0.36. Leading and trailing nucleotides with a phred score lower than 20 or when the phred score dropped below 20 over a 4 bp sliding window were trimmed. Illumina adapters were clipped using TruSeq 4 adapter sequences. After quality filtering and adapter trimming, reads less than 50 bp were discarded. The first assembly was made with SPADESv3.9.0, with an additional switchover to SOAPdenovo v2.04, which built assemblies from every odd K-mer from 21 to 63 bp. Contiguity and the scaffold N50 of the assembly maximized at was K = 51.
Chloroplast genomes were assembled using NCBI references from pool of assemblies using a combination of MIRA v4.0 and mitobim v1.8. Reference assemblies were assembled via 8–10 iterations with mitobim. The resulting assembly was later compared with the previously reported date palm cp genomes. Primers were designed and prepared via Macrogen Inc., South Korea to perform PCR amplification and sanger sequencing to fill gaps as in a previous report[28]. After adding the results of Sanger sequencing, the completed cp genome was used as a reference to map the initial short reads to refine the assembly based on maximum sequence coverage.
Genome annotation and sequence architecture
A program (DOGMA) was used to annotate the Khanezi and Naghal cp genomes[29]. After the annotation, the results were compared and checked manually. Any errors in codon position were adjusted by comparing to homologs in the cp genome from NCBI. Transfer RNAs (tRNAs) were validated using tRNAscan-SE version 1.21[30] choosing the default setup. OGDRAW[8] was utilized to reveal structural features of the Khanezi and Naghal cp genomes. Relative synonymous codon usage (RSCU) was determined using MEGA7.0[31] to elucidate the divergence in synonymous codons while avoiding the influence of related amino acids. mVISTA software was utilized in Shuffle-LAGAN-mode to explore the variations in the whole cp genomes of Khanezi and Naghal cultivars compared to the two other cp genomes reported previously for Aseel and Khalas, using the Khanezi and Naghal annotation as a reference[32].
Characterization of repeat sequence and SSRs
The REPuter program was used to show repeat sequences, which included reverse, palindromic, and direct repeats [33]. In this case, the following settings were used: a) Hamming-distance of 3, b) 90% or greater sequence-identity and c) repeat size of 30 bp. Phobosv3.3.12[34] was used to assess SSR in the chloroplast genome. The search parameters were sat at ≥10 for mononucleotides, ≥8 for dinucleotides, ≥4 for trinucleotides and tetranucleotides, and ≥3 for hexanucleotides and pentanucleotides. Additionally, tandem repeats in the cp genomes of Naghal and Khanezi cultivars were identified using TandemRepeatsFinder v4.1b with default parameters [35].
Divergence among cp genome sequences and phylogenetic analysis
Whole cp genome and the 68 shared genes were analyzed to assess pairwise sequence divergence of the four date palm cultivars (Naghal, Khanezi, Aseel and Khalas). Missing, ambiguous and poorly annotated genes were re-confirmed by comparison and multiple sequence alignment using MAFFT(v7.222)[36] with the default settings. The Kimura-2-parameter method was used for calculating pairwise sequence divergence [37]. To resolve the Khanezi and Naghal phylogenetic positions within the family Areaceae, the 16 available cp genomes in NCBI database were used. Multiple alignments were done based on conserved structures and gene order in the cp genomes[37]. We used 4 different methods to make the trees: Bayesian-inference (MrBayes v3.1.2[38]), maximum parsimony (PAUP-4.0[39]), maximum-likelihood and neighbor joining (MEGA7.01[31]) according to the methods of Wu et al.[40, 41]. For Bayesian posterior probabilities (PP) in the BI analyses, the best substitution model GTR + G model was tested according to the Akaike information criterion (AIC) by jModelTest verion 2102. The Markov Chain Monto Carlo (MCMC) was run for 1,000,000 generations with 4 incrementally heated chains, starting from random trees and sampling 1 out of every 100 generations. The first 30% of trees were discarded as burn-in to estimate the value of posterior probabilities. Furthermore, parameters for the ML analysis were optimized with a BIONJ tree as the starting tree with 1000 bootstrap replicates using the Kimura 2-parameter model with gamma-distributed rate heterogeneity and invariant sites. MP was run using a heuristic search with 1000 random addition sequence replicates with the tree-bisection-reconnection (TBR) branch-swapping tree search criterion. In the second tier of phylogenies, a set of seventy shared genes from the cp genomes of the 16 Areaceae members were aligned in Clustal X with the default program settings and several manual adjustments to improve and preserve reading frames. The 4 previously mentioned phylogenetic inference models were utilized to build trees using 70 concatenated genes as described above and suggested in Asaf et al.[28]
Results and discussion
Sequencing and assembling the genomic data
The de novo assembly results showed the total sequences in data set of 1,493,007 sequences. In addition, the data set contained sequence data for 616,016,370 nucleotides. The sequencing coverage for Khanezi was 7826.8x and for Naghal was 7874.7x. The N50 values were 3,065 bp and 2,122 bp for Khanezi and Naghal, respectively.
Chloroplast genomes of P. dactylifera L. cv. ‘Khanezi’ and ‘Naghal’
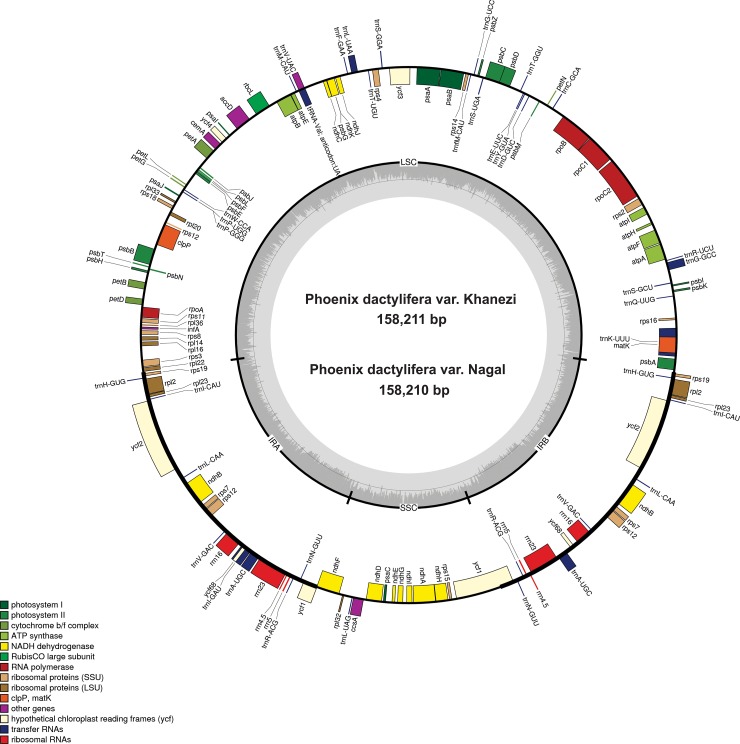
Date palm cp genomes are typical circular double-stranded DNA molecules, and they share a common quadripartite structure with the vast majority of other Arecaceae and angiosperms [26]. Sequence analysis and assembly revealed that Khanezi has a cp genome size of 158,211 bp, whereas the Naghal has 158,210 bp (Fig 1; Table 1). Previous cp genome analysis revealed that Khalas and Aseel have cp genome sizes of 158,462 bp and 158,458 bp, respectively [26]. This finding suggests close inter-linkage the two varieties [26]. Similar linkages and variations were also shown by Racchi et al.[42] and Khan et al.[43] for cp diversity of date palm varieties growing in Egypt, Tunisia, Morocco and Algeria. Structurally, both cp genomes in this study were comprised of four regions: a pair of inverted repeats (IR a and b), a large single copy (LSC) region and small single copy (SSC) region (Table 1; Fig 1) with varying sizes. For example, two IRs that mirrored each other showed a single bp difference in size (Khanezi—27,273 bp and Naghal—27,272 bp). In the case of LSC, the two cp genomes varied in size by two bp, i.e., 86,090 bp for Khanezi and 86,092 bp for Naghal. Similarly, the SSC was 17,575 bp and 17,574 bp for Khanezi and Naghal, respectively (Table 1). In contrast, the GC percentage was similar in the four regions. Similar patterns were also noted by Khan et al.[43] and Yang et al.[26] in Aseel and Khalas, respectively, suggesting similar GC content and differences of one to four bp across the four regions of the cp genomes (Table 1).
Fig 1. Gene map of the P. dactylifera var Khanezi and Naghal chloroplast genomes.
Genes drawn inside the circle are transcribed clockwise, and those outside the circle are transcribed counter-clockwise. Asterisks indicate intron-containing genes. Genes belonging to different functional groups are color-coded. Darker gray in the inner circle corresponds to GC content, and lighter gray corresponds to AT content.
Table 1. Summary of complete chloroplast genomes for Khanezi and Naghal.
| Region | P. dactylifera var Khanezi | P. dactylifera var Naghal | P. dactylifera var Khalas | P. dactylifera vr Aseel |
|---|---|---|---|---|
| LSC | ||||
| Length (bp) | 86090 | 86092 | 86197 | 86194 |
| GC(%) | 35.3 | 35.3 | 35.3 | 35.3 |
| Length (%) | 54.41 | 54.41 | 54.39 | 54.39 |
| SSC | ||||
| Length (bp) | 17575 | 17574 | 17712 | 17711 |
| GC(%) | 31 | 31 | 30.8 | 30.8 |
| Length (%) | 11.10 | 11.10 | 11.17 | 11.17 |
| IR | ||||
| Length (bp) | 27273 | 27272 | 27277 | 27276 |
| GC(%) | 42.4 | 42.4 | 42.4 | 42.4 |
| Length (%) | 17.23 | 17.23 | 17.21 | 17.21 |
| Total | ||||
| GC(%) | 37.3 | 37.3 | 37.2 | 37.2 |
| Length (%) | 158211 | 158210 | 158462 | 158458 |
The coding sequences in both Khanezi and Naghal possess similar GC content and length relative to the cp genome; however, the length in bp was longer for Naghal than for Khanezi (Table 2). The tRNA, rRNAs and intergenic spaces were similar across the two cp genomes. However, Khalas has higher levels of rRNAs, and Aseel has lower levels of rRNAs compared to the Khanezi and Naghal cp genomes. The protein coding sequences (CDS) were 82,144 and 82,153 bp in length in Khanezi and Naghal cp genomes, respectively, and were composed of protein-coding genes contain 27,381 and 27,384 bp of codons, respectively (Table 3). Similar to other cp genomes, such as Aseel, the date palm cp genome is also AT-rich (62.7%), and the values vary slightly among non-coding, protein-coding, tRNA, and rRNA sequences, which have A+T contents of 59.5%, 62.1%, 44.7%, and 47%, respectively [26, 43]. The AT content was higher (31.1–31.7%) than GC (18.3–19.0%) in both cp genomes, and the SSC region had the highest AT and lowest GC content (Table 3). The higher AT content at the 3rd position has often been used to differentiate cp DNA from nuclear and mitochondria sequences [44]. The codon utilization was estimated for tRNA and protein-coding gene sequences in both Khanezi (S1 Table) and Naghal (S2 Table) cp genomes. Most of the preferred synonymous codons (RSCU) ended with an A or U. In the cp genomes of Khanezi and Naghal, leucine (Leu; 10.2%) was the most common amino acid followed by Isoleucine and serine (8.6% and 8.1%), whereas cysteine (1.2%) was the lowest frequency amino acid (S2 and S3 Tables). Similar results were previously reported for P.dactylifera var aseel and Khalas cp genomes [26].These results also consistent with the cp genomes of other angiosperms, such as Lonicera japonica [45], Oryza minuta [46] and Glycine max [47].
Table 2. Comparison of coding and non-codign region size among P. dactylifera four varieties.
| Region | P. dactylifera var Khanezi | P. dactylifera var Naghal | P. dactylifera var Khalas | P. dactylifera vr Aseel |
|---|---|---|---|---|
| Protein Coding | ||||
| Length (bp) | 82144 | 82153 | 83904 | 81408 |
| GC(%) | 37.9 | 37.9 | 37.9 | 37.8 |
| Length (%) | 51.92 | 51.92 | 52.94 | 51.37 |
| tRNA | ||||
| Length (bp) | 9050 | 9050 | 9050 | 9050 |
| GC(%) | 55.3 | 55.3 | 55.3 | 55.3 |
| Length (%) | 5.7 | 5.7 | 5.71 | 5.71 |
| rRNA | ||||
| Length (bp) | 2960 | 2960 | 3568 | 2735 |
| GC(%) | 53 | 53 | 48.2 | 53.2 |
| Length (%) | 1.87 | 1.87 | 2.25 | 1.72 |
| Intergenic | ||||
| GC(%) | 40.50 | 40.50 | 39.1 | 41.2 |
| Length (%) | 37.3 | 37.3 | 37.2 | 37.3 |
Table 3. Base compositions in the Khanezi and Naghal cp genome.
| T/U | C | A | G | Length (bp) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Khanezi | Naghal | Khanezi | Naghal | Khanezi | Naghal | Khanezi | Naghal | Khanezi | Naghal | |
| Genome | 31.7 | 31.7 | 19.0 | 19.0 | 31.1 | 31.1 | 18.3 | 18.3 | 158211 | 158210 |
| LSC | 32.9 | 32.9 | 18.1 | 18.1 | 31.8 | 31.8 | 17.2 | 17.2 | 86090 | 86092 |
| SSC | 34.5 | 34.5 | 16.2 | 16.2 | 34.5 | 34.5 | 14.7 | 14.7 | 17575 | 17574 |
| IR | 28.7 | 28.7 | 20.5 | 20.5 | 28.9 | 28.9 | 28.7 | 28.7 | 27273 | 27272 |
| tRNA | 24.9 | 24.7 | 23.7 | 24.0 | 22.1 | 22.3 | 29.3 | 29 | 2960 | 2960 |
| rRNA | 18.8 | 18.8 | 23.8 | 23.8 | 25.9 | 25.9 | 31.5 | 31.5 | 9050 | 9050 |
| Protein Coding genes | 31.3 | 31.3 | 17.8 | 17.8 | 30.7 | 30.7 | 20.1 | 20.1 | 82144 | 82153 |
| 1st position | 23.77 | 23.74 | 18.6 | 18.6 | 30.94 | 30.3 | 26.66 | 26.72 | 27381 | 27384 |
| 2nd position | 33.50 | 33.53 | 20.4 | 20.3 | 29.38 | 29.4 | 17.72 | 17.8 | 27381 | 27384 |
| 3rd position | 35.6 | 35.65 | 14.2 | 14.23 | 31.89 | 31.78 | 16.01 | 16.82 | 27381 | 27384 |
The Khanezi and Naghal cp genomes contain 111 unique genes and 19 duplicated genes in the IR. Among these unique genes, we identified 81 protein-coding, four ribosomal RNA and 29 transfer RNA genes (S4 and S5 Tables). The LSC region was comprised of sixty-two CDS and 23 t-RNA related genes. The SSC region was composed of twelve protein-coding genes and a tRNA gene. The protein-coding genes included 9 genes that encode large ribosomal proteins (rp14, rp20, rpl2, rp16, rp23, rp32, rp22, rp33, and rp36), twelve genes encoding small ribosomal proteins (rps8, rps2, rps11, rps3, rps7, rps12, rps14, rps16, rps4, rps15, rps18, and rps19), 5 genes encoding Photosystem-I (psaA, psaB, psaC, psaI, and psaJ), sixteen genes related to Photosystem-II (S6 Table), and 6 genes encoding ATP synthase and electron transport chain (atpB, atpA, atpF, atpE, atpI and atpH). Furthermore, approximately 51.92%, 5.7% and 1.87% of the cp genome sequences encoded proteins, tRNA, and rRNAs, respectively, whereas the remaining 37.4% was non-coding, including introns and intergenic spacers. The results showed that both Khanezi and Naghal cp genomes have 18 intron-containing genes, which is similar to previously reported results for the Khalas cp genome [48]. However, in the cp genome of the Aseel variety, there were 16 of these genes [43]. Among the genes that were similar to previous date palm cp genomes, almost all were single intron except for clpP, ycf3 and rps12, the exons of which are separated by two introns (S6 Table). Rps12 is a trans spliced gene, where an exon is in the LSC region and the other 2 reside in the IR regions separated by two introns. Similar results were reported for previously reported genomes, where the introns of all CDS shares similar splicing mechanisms as Group-II introns [49]. Among these genes, ndhA in SSC and trnK-UUU in LSC have the highest single intron size, whereas ycf68 (replicated as well) has the lowest single intron size (S4 and S5 Tables) in the Khanezi cp genome. The trnV-UAC (593 bp) has a longer intron than trnV-UAA (513 bp). It has been shown that these kinds of introns are essential for gene regulation, and they can also affect exo-gene expression patterns depending on their specific positions. Utilizing similar introns can also increase transformational efficiency[50]. It has been observed that ycf1, ycf2[51, 52], rpl23 [53] and accD [54, 55] are often absent from plants[53], but they were detected in the reported date palm cp genomes. A pair of genes (atpB-atpE) overlapped each other by ~4 bp. PsbC-psbD had a 53 bp overlap in the date palm cp genomes, whereas this overlap was 53 bp in A. thaliana, 17 bp in A. arenosa, 92 bp in A. halleri ssp gemmifere and A. lyrata ssp. petraea, 53 bp in Gossypium [56] and 52 bp in Camellia cp genomes [57]. As reported previously by Adachi et al. [58], there was a partial overlap of the psbD and psbC cistrons, where translation of the psbC cistron is dependent on the translation of the latter psbD cistron. This suggests independent translation of psbC. Likewise, the ndhC and ndhK cistrons of the tobacco chloroplast genome also overlap, and translation of ndhK is strictly dependent on the upstream termination codon[59].
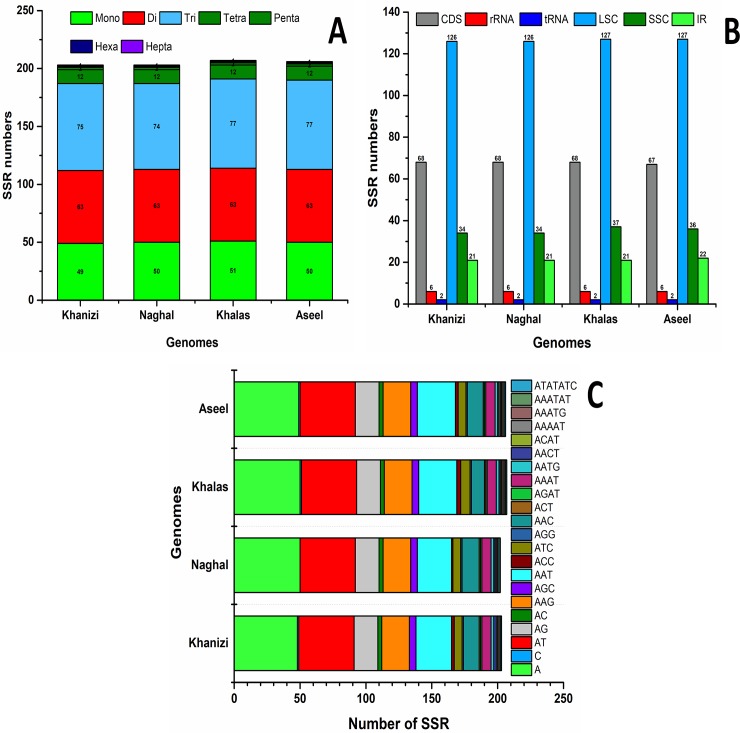
Simple sequence repeat (SSR) in Naghal and Khanezi
We analyzed the SSRs in the cp genomes of Khanezi and Naghal. During analysis, SSRs that were 10 bp or longer were defined as possible slipped strand mis-pairing due to mutational polymorphisms. From our SSR analysis, 227 and 229 microsatellites were found in the Khanezi and Naghal cp genomes, respectively (Fig 2). In Khanezi and Naghal, most mononucleotide SSRs were A motif (96.3% and 96.6%, respectively), with most SSR dinucleotides being T/A (69.54%, 71.06%) or G/A (26.31%, 27.77%) motifs (S7 Table). The chloroplast genome of Khanezi, similar to other species, contains different types of repeats that each have a specific function. The complete genome contains a different number of base pairs of the repeated sequence. Generally, as shown in S7 and S8 Tables, the whole genome more tri-base in the repeated sequence. However, there are 63 di-base pairs in the repeated sequence and 49 of mono-base pairs. The LSC region has the highest number of mono- and di-base pairs of the repeats, approximately 36 and 42, whereas Khalaas and Aseel are slightly higher at the mono level but lower at di level compared to Khanezi (S8 Table). For tri base pairs, the CDS region has a high frequency (37%) that is greater than Aseel and Khalas.
Fig 2. Analysis of simple sequence repeats (SSR) in the P. dactylifera var Khanezi and Naghal cp genomes.
A: Number of different SSR types detected in the four genomes; B: Frequency of identified SSRs in coding, non-coding, LSC, SSC and IR regions; C: Frequency of identified SSR motifs in different repeat class types.
We compared perfect SSRs in Khanezi and Naghal with cp genomes of two other Aseel and Khalas cultivars. SSR has been shown to have a higher rate of mutation compared to other neutral DNA regions because of slipped DNA strands [58]. SSRs with the highest genetic diversity occur in the cp genome and are known markers used for evolutionary, population genetics, and systematics studies [60]. In the current study, SSRs measuring 10 bp or longer were found and shown to be slipped strands or mis-paired. It has been shown that mutations can be a mechanism for SSR polymorphisms [61, 62]. In Khanezi and Naghal cp genomes, we found 227 and 229 microsatellites, respectively. The current results are consistent with previous reports, where the SSR are dominated by ‘A’ or ‘T’ mononucleotide repeats[63, 64]. These different kinds of SSR repeats (mononucleotide, pentanucleotide, and hexanucleotides) are comprised of A or T bases at higher frequencies, which corresponds to the biased-base composition and A/T richness of the cp genomes[65, 66]. These results are consistent with previous reports that show that the SSRs in cp genome contain polythymine (polyT) or polyadenine (polyA) repeats in addition to infrequent tandem cytosine and guanine repeats[66]. Therefore, the existence of such SSRs in the cp genome considerably contributes to the ‘AT’ ratio shown for the Khanezi and Naghal genomes. This phenomenon was also previously reported for different species [67, 68]. The current findings suggest that approximately 69% (Khanezi) and 77% (Naghal) SSRs were detected in non-coding regions. These results are consistent with previous studies determined that SSRs as unequally distributed in the chloroplast. In addition, these data might also provide information for designing targeted markers for detecting intra- and interspecific polymorphisms for date palm cultivars [69, 70].
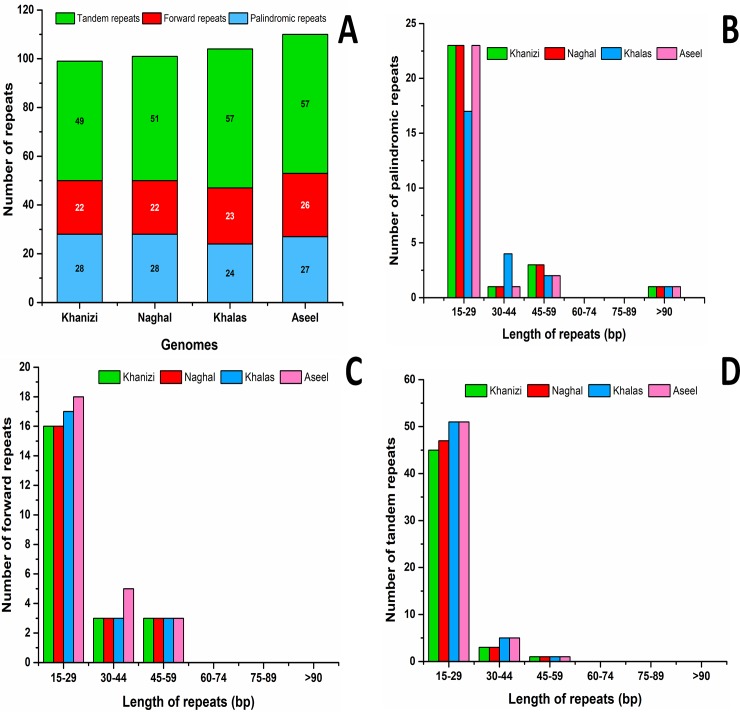
Repeat sequence and comparative distribution in date palm
The results showed that 99 and 101 repeats were found in the cp genomes of Khanezi and Naghal, respectively, which included 28 palindromic, 22 direct and 49 and 51 palindromic repeats (Fig 3). Among these repeats, 23 palindromic repeats were 15–29 bp in length, whereas there was one repeat each 30–44 bp and >90 bp in length and 3 palindromic repeats 45–59 bp in length. Another type of repeated sequence, forward repeats, occurred in these cp genomes at different frequencies. For example, there were 16 15–29 bp forward repeats, whereas there were 3 forward repeats, with each measuring 30–44 and 45–59 bp in length. In addition to these repeats, tandem repeats occurred in high numbers. Tandem repeats 15–29 bp in length were identified with frequency of 45 and 47 in the Naghal and Khanezi cp genomes, respectively. Similarly, 30–44 and 45–59 bp tandem repeats were found at a frequency of three and one, respectively (Fig 3). In comparison to both Naghal and Khanezi, the most tandem repeats were found in the cp genomes of the Aseel and Khalas varieties, with a frequency of 57. Repeat sequences are very helpful in phylogenetic studies and play a role in genome rearrangements[71, 72]. Analyses of various chloroplast genomes concluded that repeats are important in inducing indels and substitutions[73]. The length of direct and palindromic repeats in the Khanezi and Naghal cp genomes were considerably short ranging from 30–101 bp. In this case, similar results were previously shown for the cp genome of Camellia species, which have eighty-two repeats. In contrast, other reports have shown longer repeats of 132 bp and 287 bp in Poaceae and Fabaceae, respectively[74]. Recent studies have shown that variations in sequence and rearrangement of genomes can be due to slipped-strand un/mispairing and improper re-combination of repeats[75, 76]. Additionally, the occurrence of these repeats suggests that the regions are important hotspots for reconfiguration of the cp genome [76]. The data related to repeats could be utilized to develop molecular markers for understanding the population dynamics of Khanezi and Naghal [71].
Fig 3. Analysis of repeated sequences in P. dactylifera var Khanezi and Naghal cp genomes.
A: Totals of three repeat types; B: Frequency of palindromic repeats by length; C: Frequency of forward repeats by length; D: Frequency of tandem repeats by length.
Structural comparison of date palm cp genomes
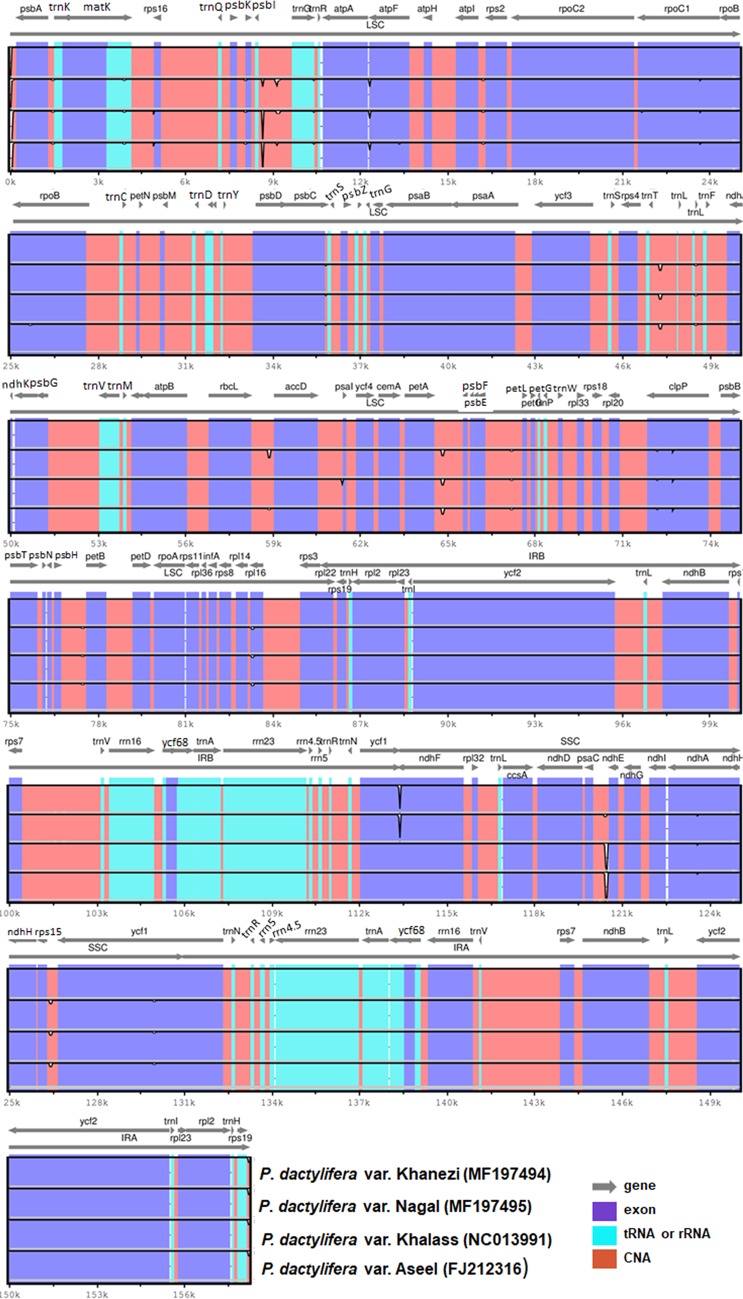
The date palm tree cp genomes evolve slowly, where the total rate of nucleotide substitution is approximately eightfold lower than observed in annual plants. Because previous cp genome studies were published almost five years ago with little focus on the comparative assessment among cp genomes of date palm cultivars, we analyzed two previously reported complete cp genomes from Aseel and Khalas cultivars together with the Naghal and Khanezi genomes from the current study. Among these cp genomes, Naghal was the smallest (158,210 bp), whereas Khalas had the largest cp genome size (158,462 bp). In addition, the difference in length between Naghal and Khanezi was a single base pair, whereas Khalas and Aseel had a 4 bp difference. Pairwise cp genomic alignment of these four cp genomes uncovered a high degree of synteny. Using the mVISTA algorithm, the sequences of the four available date palm cp genomes were compared (Fig 4).
Fig 4. Alignment visualization of the P. dactylifera var Khanezi and Naghal chloroplast genome sequences.
VISTA-based identity plot showing sequence identity among the four-species using P. dactylifera var Khalas as a reference. The vertical scale indicates percent identity, ranging from 50% to 100%. The horizontal axis indicates the coordinates within the chloroplast genome. Arrows indicate the annotated genes and their transcription direction. The thick black lines show the inverted repeats (IRs).
The results showed comparatively low sequence identity among the cp genomes of the four varieties, especially in atpF, rpoC1, clpP, rpl16, ndhA, ycf1 and ndhF. Similar to previous reports on various cp genomes [26, 43], Naghal and Khanezi cp genomes also showed more divergence in the LSC, SSC and non-coding regions and compared to the IR and coding regions, respectively. Among the non-coding sequences, highly divergent regions included psbK-trnG, trnT-trnL, rbcL-accD, petA-psbJ and psaC-ndhE spacers as reported previously. In addition, previous studies have shown that coding and non-coding areas with high variation, such as trnS(GGA)-trnG(UCC), rpl16-rps3, trnT-trnL and atpB-rbcL, have led to the development of potential genetic markers in angiosperms. Furthermore, comparison of Khanezi and Naghal cp genomes with related varieties revealed various useful results, including that Khanezi showed 35 indels and 23 SNPs with Naghal. In contrast, Aseel and Khalaas have more indels, 293 and 299, respectively with Khanizi. In contrast, the number of SNPs in Aseel and Khalaas are 18 and 16. Similarly, Naghal revealed 292 and 296 indels and 10 and 12 SNPs in Aseel and Khalas, respectively (S9 Table). We further compared the Khanezi and Naghal cp genomes and calculated the average pairwise sequence divergence among the four varieties (S10 Table). Khanezi and Naghal exhibited 0.000120 and 0.000192 average sequence divergence, respectively. Khanezi showed more divergence from Khalas and Aseel (0.00108 and 0.000101, respectively) compared to the divergence of Naghal from these two varieties (0.000071 and 0.000076, respectively).
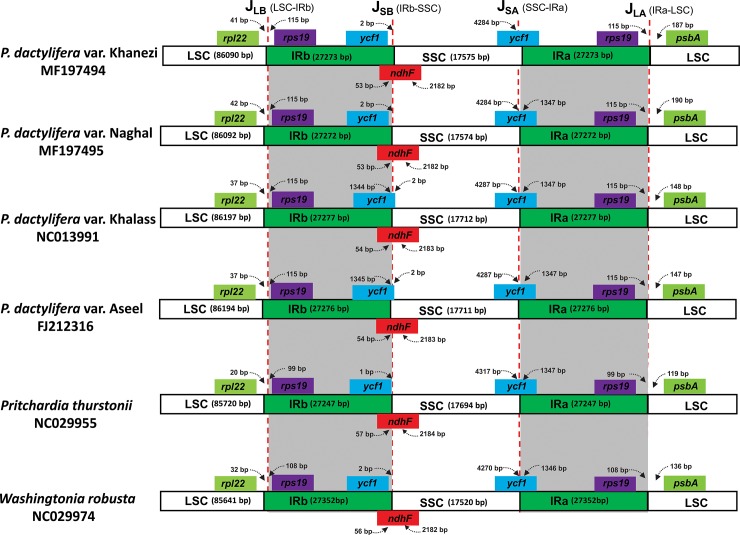
In the case of IRs, the contraction and expansion of the border regions have been posited as main features of cp genome size variation and have also been credited for evolution[77–79]. Considerable expansion and contraction of the IR region is mostly responsible for the size variation observed among chloroplast genomes [13, 80]. In this study, we compared the position of IR borders of four date palm varieties with two Arecaceae members Pritchardia thurstonii and Washingtonia robusta. Due to a characteristic expansion of IRB sequences into the LSC region, a specific rearrangement was acquired by monocot cp genomes early in their evolution. This expansion resulted in the inclusion of trnH and rps19 genes in the IR region. The distance between JLB and rps19 is 115 bp in all date palm verities, and is observed to be 99 bp and 108 bp in P. thurstonii and W. robusta, respectively (Fig 5). Similarly, JLA is located between rps19 and psbA, and the distance between psbA and JLA ranges from 147 to 187 bp among the four varieties. In Naghal and Khaneizi, this distance was 190 bp and 187 bp, respectively. However, in P. thurstonii and W. robusta, this distance was 119 bp and 136 bp, respectively. Similar results were obverted by Yang et al.[26] and Khan et al.[43] for the Khalas and Aseel cp genomes.
Fig 5. Comparison of border distance between adjacent genes and junctions of the LSC, SSC, and two IR regions among the chloroplast genomes of P. dactylifera var Khanezi and Naghal.
Boxes above or below the main line indicate the adjacent border genes. The figure is not to scale with respect to sequence length and only shows relative changes at or near the IR/SC borders.
Phylogenetic analysis of P. dactylifera var Khanezi and Naghal
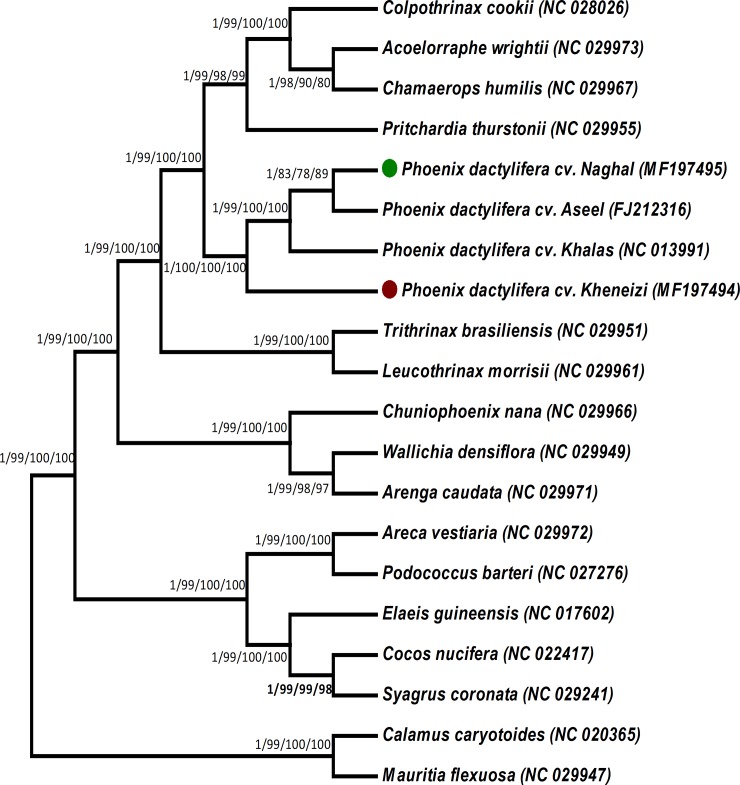
The phylogenetic position of P. dactylifera var Khanezi and Naghal within the family Arecaeae was established by analyzing multiple alignments of complete cp genomes and 68 shared genes of 16 Arecaeae members, representing seventeen genera (Fig 6 and S1 Fig). Phylogenetic analyses using maximum likelihood, Bayesian-inference, maximum-parsimony, and neighbor joining were performed. The results revealed that the complete cp genomes and 68 shared genes of P. dactylifera var Khanezi and Naghal contain the same phylogenetic signals; the complete genome sequence and the 68 shared genes (from all species) generated phylogenetic trees with identical topologies (Fig 6 and S1 Fig). In these phylogenetic trees based on the entire genome data set and the 68 shared genes, P. dactylifera var Khanezi and Naghal formed a single clade with Khalas with high Bayesian inference and bootstrap support using 4 phylogeny models (Fig 6 and S1 Fig). The results revealed that Naghal is closer to Aseel compared to Khalas and Khanezi. Most of the previous studies concerning the phylogenetic analysis of date palm cultivar used SSR, RAPD (random amplified polymorphic DNA) and DAMD (directed amplification of minisatellites DNA) markers to understand the genetic discrimination [81–83]. Akhtar et al. [84] showed rps14 for understanding the phylogenetic relationship among Pathri, Dhaddy, Makhi, Aseel, and Khudrawi date palm cultivars from Pakistan. Similarly, specific SSRs were used to differentiate among Khalas, Hillali, Khnaizi, and Jabri from Qatari date palm cultivars [85]. However, the current study for the first time used four different phylogenetic approaches and 68 shared genes to construct the phylogeny, suggesting a clear differentiation of the four cultivars.
Fig 6. Phylogenetic trees were constructed for twenty species from the family Areaceae using several different methods, and the tree shown is for the 68 shared protein-coding genes.
The following four methods were used for the 68 shared genes data set: Bayesian inference (BI), maximum parsimony (MP), maximum likelihood (ML) and neighbor-joining (NJ). Numbers above the branches are the posterior probabilities of BI and bootstrap values for NJ, MP and ML. Green and brown dots represent the positions of P. dactylifera var Khanezi and Nagha.
Conclusions
This study produced the first complete chloroplast genome of two important cultivars (Naghal and Khanezi) growing in the Sulatanate of Oman and the rest of the Arabian Peninsula. The genomic data were assembled and analyzed, and the genomes were compared with the only two other reported cultivars of date palm (Aseel and Khalas). Genome arrangements, gene content and order, and codon usage were consistent with the previously elucidated cp genomes from the genus Phoenix. The location and distribution of repeat sequences was determined, and sequence divergence of cp genomes and 68 shared genes were calculated for related species. The phylogenetic analysis based on whole cp genomes and 68 shared genes yielded identical phylogenetic trees, with Khanezi and Naghal forming single clades with Khalas and Aseel cultivars, respectively.
Supporting information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(XLS)
The following four methods were used for the entire genome data set: Bayesian inference (BI), maximum parsimony (MP), maximum likelihood (ML) and neighbor-joining (NJ). Numbers above the branches are the posterior probabilities of BI and bootstrap values for NJ, MP and ML. Green and brown dots represent the positions of P. dactylifera var Khanezi and Nagha.
(TIF)
Acknowledgments
The authors acknowledge the assistance of Ms. Asmahan in DNA extraction.
Data Availability
Data are available from the NCBI database with the ACCESSION NUMBERS (MF197494, MF197495).
Funding Statement
This work was supported by The Research Council Oman through their Open Research Grant (ORG/EBR/15/007). The funder had no role in study design and presentation.
References
- 1.Moussouni S, Pintaud J-C, Vigouroux Y, Bouguedoura N. Diversity of Algerian oases date palm (Phoenix dactylifera L., Arecaceae): Heterozygote excess and cryptic structure suggest farmer management had a major impact on diversity. PloS one. 2017;12(4):e0175232 10.1371/journal.pone.0175232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrow SC. A monograph of phoenix L.(palmae: Coryphoideae). Kew bulletin. 1998:513–75. [Google Scholar]
- 3.Terral JF, Newton C, Ivorra S, Gros‐Balthazard M, de Morais CT, Picq S, et al. Insights into the historical biogeography of the date palm (Phoenix dactylifera L.) using geometric morphometry of modern and ancient seeds. Journal of Biogeography. 2012;39(5):929–41. [Google Scholar]
- 4.Cherif E, Zehdi S, Castillo K, Chabrillange N, Abdoulkader S, Pintaud JC, et al. Male‐specific DNA markers provide genetic evidence of an XY chromosome system, a recombination arrest and allow the tracing of paternal lineages in date palm. New Phytologist. 2013;197(2):409–15. 10.1111/nph.12069 [DOI] [PubMed] [Google Scholar]
- 5.Pintaud J-C, Ludeña B, Aberlenc-Bertossi F, Zehdi S, Gros-Balthazard M, Ivorra S, et al. , editors. Biogeography of the date palm (Phoenix dactylifera L., Arecaceae): insights on the origin and on the structure of modern diversity. I International Symposium on Date Palm 994; 2011. [Google Scholar]
- 6.Bouguedoura N BM, Babahani S, Benziouche SE. Date Palm Status and Pespective in Algeria In Al-Khayri JM JS, Jhnson DV, editor. America Africa and the Americas; 2015 [Google Scholar]
- 7.Zehdi-Azouzi S, Cherif E, Moussouni S, Gros-Balthazard M, Abbas Naqvi S, Ludeña B, et al. Genetic structure of the date palm (Phoenix dactylifera) in the Old World reveals a strong differentiation between eastern and western populations. Annals of botany. 2015;116(1):101–12. 10.1093/aob/mcv068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lohse M, Drechsel O, Bock R. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Current Genetics. 2007;52(5–6):267–74. WOS:000250785100009. 10.1007/s00294-007-0161-y [DOI] [PubMed] [Google Scholar]
- 9.Corriveau JL, Coleman AW. Rapid screening method to detect potential biparental inheritance of plastid DNA and results for over 200 angiosperm species. American Journal of Botany. 1988:1443–58. [Google Scholar]
- 10.Zhang Q, Liu Y. Examination of the cytoplasmic DNA in male reproductive cells to determine the potential for cytoplasmic inheritance in 295 angiosperm species. Plant and Cell Physiology. 2003;44(9):941–51. [DOI] [PubMed] [Google Scholar]
- 11.Wolfe KH, Li W-H, Sharp PM. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proceedings of the National Academy of Sciences. 1987;84(24):9054–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Provan J, Powell W, Hollingsworth PM. Chloroplast microsatellites: new tools for studies in plant ecology and evolution. Trends in Ecology & Evolution. 2001;16(3):142–7. [DOI] [PubMed] [Google Scholar]
- 13.Ravi V, Khurana J, Tyagi A, Khurana P. An update on chloroplast genomes. Plant Systematics and Evolution. 2008;271(1–2):101–22. [Google Scholar]
- 14.Ohyama K, Fukuzawa H, Kohchi T, Shirai H, Sano T, Sano S, et al. Chloroplast gene organization deduced from complete sequence of liverwort Marchantia polymorpha chloroplast DNA. Nature. 1986;322(6079):572–4. [Google Scholar]
- 15.Shinozaki K, Ohme M, Tanaka M, Wakasugi T, Hayashida N, Matsubayashi T, et al. The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. The EMBO journal. 1986;5(9):2043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daniell H, Chan H-T, Pasoreck EK. Vaccination via chloroplast genetics: affordable protein drugs for the prevention and treatment of inherited or infectious human diseases. Annual review of genetics. 2016;50:595–618. 10.1146/annurev-genet-120215-035349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wambugu PW, Brozynska M, Furtado A, Waters DL, Henry RJ. Relationships of wild and domesticated rices (Oryza AA genome species) based upon whole chloroplast genome sequences. Scientific reports. 2015;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brozynska M, Furtado A, Henry RJ. Genomics of crop wild relatives: expanding the gene pool for crop improvement. Plant biotechnology journal. 2016;14(4):1070–85. 10.1111/pbi.12454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolodner R, Tewari K. Inverted repeats in chloroplast DNA from higher plants. Proceedings of the National Academy of Sciences. 1979;76(1):41–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuan L, Chen L, Qian K, Wang G, Lu M, Qian G, et al. A novel correlation between ATP5A1 gene expression and progression of human clear cell renal cell carcinoma identified by coexpression analysis. Oncology reports. 2017. 10.3892/or.2017.6132 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wambugu PW, Brozynska M, Furtado A, Waters DL, Henry RJ. Relationships of wild and domesticated rices (Oryza AA genome species) based upon whole chloroplast genome sequences. Scientific reports. 2015;5:13957 10.1038/srep13957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Al-Dous EK, George B, Al-Mahmoud ME, Al-Jaber MY, Wang H, Salameh YM, et al. De novo genome sequencing and comparative genomics of date palm (Phoenix dactylifera). Nature biotechnology. 2011;29(6):521 10.1038/nbt.1860 [DOI] [PubMed] [Google Scholar]
- 23.Al-Mssallem IS, Hu S, Zhang X, Lin Q, Liu W, Tan J, et al. Genome sequence of the date palm Phoenix dactylifera L. Nature communications. 2013;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khan I, Qayyum S, Ahmed S, Maqbool F, Tauseef I, Haleem KS, et al. Cloning and characterization of pyruvate carboxylase gene responsible for calcium malate overproduction in Penicillium viticola 152 and its expression analysis. Gene. 2017;605:81–91. 10.1016/j.gene.2016.12.036 . [DOI] [PubMed] [Google Scholar]
- 25.Yang C, Ma L, Ying Z, Jiang X, Lin Y. Sequence Analysis and Expression of a Blue-light Photoreceptor Gene, Slwc-1 from the Cauliflower Mushroom Sparassis latifolia. Curr Microbiol. 2017;74(4):469–75. 10.1007/s00284-017-1218-x . [DOI] [PubMed] [Google Scholar]
- 26.Yang M, Zhang X, Liu G, Yin Y, Chen K, Yun Q, et al. The complete chloroplast genome sequence of date palm (Phoenix dactylifera L.). PloS one. 2010;5(9):e12762 10.1371/journal.pone.0012762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi C, Hu N, Huang H, Gao J, Zhao Y-J, Gao L-Z. An improved chloroplast DNA extraction procedure for whole plastid genome sequencing. Plos one. 2012;7(2):e31468 10.1371/journal.pone.0031468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Asaf S, Khan AL, Khan MA, Waqas M, Kang S-M, Yun B-W, et al. Chloroplast genomes of Arabidopsis halleri ssp. gemmifera and Arabidopsis lyrata ssp. petraea: Structures and comparative analysis. Scientific Reports. 2017;7(1):7556 10.1038/s41598-017-07891-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wyman SK, Jansen RK, Boore JL. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004;20(17):3252–5. WOS:000225361400041. 10.1093/bioinformatics/bth352 [DOI] [PubMed] [Google Scholar]
- 30.Schattner P, Brooks AN, Lowe TM. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005;33:W686–W9. WOS:000230271400141. 10.1093/nar/gki366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar S, Nei M, Dudley J, Tamura K. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Briefings in bioinformatics. 2008;9(4):299–306. Epub 2008/04/18. 10.1093/bib/bbn017 ; PubMed Central PMCID: PMCPMC2562624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–W9. WOS:000222273100056. 10.1093/nar/gkh458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29(22):4633–42. 10.1093/nar/29.22.4633 WOS:000172378000014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kraemer L, Beszteri B, Gäbler-Schwarz S, Held C, Leese F, Mayer C, et al. STAMP: Extensions to the STADEN sequence analysis package for high throughput interactive microsatellite marker design. BMC Bioinformatics. 2009;10(1):41 10.1186/1471-2105-10-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27(2):573–80. Epub 1998/12/24. PubMed ; PubMed Central PMCID: PMCPMC148217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular biology and evolution. 2013;30(4):772–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. Journal of molecular evolution. 1980;16(2):111–20. Epub 1980/12/01. PubMed . [DOI] [PubMed] [Google Scholar]
- 38.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19(12):1572–4. 10.1093/bioinformatics/btg180 WOS:000184878700016. [DOI] [PubMed] [Google Scholar]
- 39.Swofford DL. PAUP*: Phylogenetic Analysis Using Parsimony (*and other methods), Version 4 Sunderland, MA: Sinauer Associates; 2002. [Google Scholar]
- 40.Wu ZQ, Tembrock LR, Ge S. Are Differences in Genomic Data Sets due to True Biological Variants or Errors in Genome Assembly: An Example from Two Chloroplast Genomes. Plos One. 2015;10(2). ARTN e0118019 10.1371/journal.pone.0118019. WOS:000349444900251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Meth. 2012;9(8):772–. http://www.nature.com/nmeth/journal/v9/n8/abs/nmeth.2109.html#supplementary-information. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Racchi M, Bove A, Turchi A, Bashir G, Battaglia M, Camussi A. Genetic characterization of Libyan date palm resources by microsatellite markers. 3 Biotech. 2014;4(1):21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khan A, Khan IA, Heinze B, Azim MK. The chloroplast genome sequence of date palm (Phoenix dactylifera L. cv.‘Aseel’). Plant molecular biology reporter. 2012;30(3):666–78. [Google Scholar]
- 44.Nie XL, Zhang Shuzuo, Du Yingxin, Wang Xianghong, Biradar Le, Tan Siddanagouda S, Wan Xiufang, Weining Fanghao, Song. Complete chloroplast genome sequence of a major invasive species, crofton weed (Ageratina adenophora). PloS one. 2012;7(5):e36869 10.1371/journal.pone.0036869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He L, Qian J, Li X, Sun Z, Xu X, Chen S. Complete chloroplast genome of medicinal plant lonicera japonica: genome rearrangement, intron gain and loss, and implications for phylogenetic studies. Molecules. 2017;22(2):249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Asaf S, Khan AL, Khan AR, Waqas M, Kang S-M, Khan MA, et al. Complete chloroplast genome of Nicotiana otophora and its comparison with related species. Frontiers in plant science. 2016;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Asaf S, Waqas M, Khan AL, Khan MA, Kang S-M, Imran QM, et al. The Complete Chloroplast Genome of Wild Rice (Oryza minuta) and Its Comparison to Related Species. Frontiers in Plant Science. 2017;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fang Y, Wu H, Zhang T, Yang M, Yin Y, Pan L, et al. A complete sequence and transcriptomic analyses of date palm (Phoenix dactylifera L.) mitochondrial genome. PloS one. 2012;7(5):e37164 10.1371/journal.pone.0037164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sugita M, Sugiura M. Regulation of gene expression in chloroplasts of higher plants. Plant molecular biology. 1996;32(1–2):315–26. [DOI] [PubMed] [Google Scholar]
- 50.Xu JW, Feng DJ, Song GS, Wei XL, Chen L, Wu XL, et al. The first intron of rice EPSP synthase enhances expression of foreign gene. Sci China Ser C. 2003;46(6):561–+. 10.1360/02yc0120 WOS:000186828500001. [DOI] [PubMed] [Google Scholar]
- 51.Wolf PG, Der JP, Duffy AM, Davidson JB, Grusz AL, Pryer KM. The evolution of chloroplast genes and genomes in ferns. Plant Mol Biol. 2011;76(3–5):251–61. WOS:000291172200005. 10.1007/s11103-010-9706-4 [DOI] [PubMed] [Google Scholar]
- 52.Oliver MJ, Murdock AG, Mishler BD, Kuehl JV, Boore JL, Mandoli DF, et al. Chloroplast genome sequence of the moss Tortula ruralis: gene content, polymorphism, and structural arrangement relative to other green plant chloroplast genomes. Bmc Genomics. 2010;11. Artn 143 10.1186/1471-2164-11-143. WOS:000275835900001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wicke S, Schneeweiss GM, dePamphilis CW, Müller KF, Quandt D. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol Biol. 2011;76(3):273–97. 10.1007/s11103-011-9762-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jansen RK, Cai Z, Raubeson LA, Daniell H, Leebens-Mack J, Müller KF, et al. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proceedings of the National Academy of Sciences. 2007;104(49):19369–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nakkaew A, Chotigeat W, Eksomtramage T, Phongdara A. Cloning and expression of a plastid-encoded subunit, beta-carboxyltransferase gene (accD) and a nuclear-encoded subunit, biotin carboxylase of acetyl-CoA carboxylase from oil palm (Elaeis guineensis Jacq.). Plant science. 2008;175(4):497–504. [Google Scholar]
- 56.Xu Q, Xiong G, Li P, He F, Huang Y, Wang K. Analysis of complete nucleotide sequences of 12 Gossypium chloroplast genomes: origin and evolution of allotetraploids. Plos One. 2012;7 10.1371/journal.pone.0037128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang H, Shi C, Liu Y, Mao SY, Gao LZ. Thirteen Camellia chloroplast genome sequences determined by high-throughput sequencing: genome structure and phylogenetic relationships. Bmc Evol Biol. 2014;14. Artn 151 10.1186/1471-2148-14-151. WOS:000339178500001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Adachi Y, Kuroda H, Yukawa Y, Sugiura M. Translation of partially overlapping psbD-psbC mRNAs in chloroplasts: the role of 5′-processing and translational coupling. Nucleic acids research. 2011;40(7):3152–8. 10.1093/nar/gkr1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Khakhlova O, Bock R. Elimination of deleterious mutations in plastid genomes by gene conversion. The Plant Journal. 2006;46(1):85–94. 10.1111/j.1365-313X.2006.02673.x [DOI] [PubMed] [Google Scholar]
- 60.Zhao YB, Yin JL, Guo HY, Zhang YY, Xiao W, Sun C, et al. The complete chloroplast genome provides insight into the evolution and polymorphism of Panax ginseng. Front Plant Sci. 2015;5. Artn 696 10.3389/Fpls.2014.00696. WOS:000348031400001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rose O, Falush D. A threshold size for microsatellite expansion. Mol Biol Evol. 1998;15(5):613–5. WOS:000073343800018. 10.1093/oxfordjournals.molbev.a025964 [DOI] [PubMed] [Google Scholar]
- 62.Huotari T, Korpelainen H. Complete chloroplast genome sequence of Elodea canadensis and comparative analyses with other monocot plastid genomes. Gene. 2012;508(1):96–105. WOS:000309249600014. 10.1016/j.gene.2012.07.020 [DOI] [PubMed] [Google Scholar]
- 63.Sato S, Nakamura Y, Kaneko T, Asamizu E, Tabata S. Complete structure of the chloroplast genome of Arabidopsis thaliana. DNA Res. 1999;6 10.1093/dnares/6.5.283 [DOI] [PubMed] [Google Scholar]
- 64.Qian J, Song J, Gao H, Zhu Y, Xu J, Pang X. The complete chloroplast genome sequence of the medicinal plant Salvia miltiorrhiza. Plos One. 2013;8 10.1371/journal.pone.0057607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang YJ, Ma PF, Li DZ. High-throughput sequencing of six bamboo chloroplast genomes: phylogenetic implications for temperate woody bamboos (Poaceae: Bambusoideae). Plos One. 2011;6 10.1371/journal.pone.0020596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yi X, Gao L, Wang B, Su YJ, Wang T. The Complete Chloroplast Genome Sequence of Cephalotaxus oliveri (Cephalotaxaceae): Evolutionary Comparison of Cephalotaxus Chloroplast DNAs and Insights into the Loss of Inverted Repeat Copies in Gymnosperms. Genome Biol Evol. 2013;5(4):688–98. WOS:000318557200006. 10.1093/gbe/evt042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kuang DY, Wu H, Wang YL, Gao LM, Zhang SZ, Lu L. Complete chloroplast genome sequence of Magnolia kwangsiensis (Magnoliaceae): implication for DNA barcoding and population genetics. Genome. 2011;54(8):663–73. WOS:000295395100006. 10.1139/G11-026 [DOI] [PubMed] [Google Scholar]
- 68.Chen JH, Hao ZD, Xu HB, Yang LM, Liu GX, Sheng Y, et al. The complete chloroplast genome sequence of the relict woody plant Metasequoia glyptostroboides Hu et Cheng. Front Plant Sci. 2015;6. Artn 447 10.3389/Fpls.2015.00447. WOS:000357288100001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Powell W, Morgante M, Mcdevitt R, Vendramin GG, Rafalski JA. Polymorphic Simple Sequence Repeat Regions in Chloroplast Genomes—Applications to the Population-Genetics of Pines. P Natl Acad Sci USA. 1995;92(17):7759–63. 10.1073/pnas.92.17.7759 WOS:A1995RP74800033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pauwels M, Vekemans X, Gode C, Frerot H, Castric V, Saumitou-Laprade P. Nuclear and chloroplast DNA phylogeography reveals vicariance among European populations of the model species for the study of metal tolerance, Arabidopsis halleri (Brassicaceae). New Phytol. 2012;193(4):916–28. WOS:000299778300013. 10.1111/j.1469-8137.2011.04003.x [DOI] [PubMed] [Google Scholar]
- 71.Nie XJ, Lv SZ, Zhang YX, Du XH, Wang L, Biradar SS, et al. Complete Chloroplast Genome Sequence of a Major Invasive Species, Crofton Weed (Ageratina adenophora). Plos One. 2012;7(5). ARTN e36869 10.1371/journal.pone.0036869. WOS:000305338200053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cavalier-Smith T. Chloroplast evolution: Secondary symbiogenesis and multiple losses. Curr Biol. 2002;12(2):R62–R4. 10.1016/S0960-9822(01)00675-3 WOS:000173485500011. [DOI] [PubMed] [Google Scholar]
- 73.Saski C, Lee SB, Fjellheim S, Guda C, Jansen RK, Luo H. Complete chloroplast genome sequences of Hordeum vulgare, Sorghum bicolor and Agrostis stolonifera, and comparative analyses with other grass genomes. Theor Appl Genet. 2007;115 10.1007/s00122-007-0567-4 [DOI] [PubMed] [Google Scholar]
- 74.Tangphatsornruang S, Sangsrakru D, Chanprasert J, Uthaipaisanwong P, Yoocha T, Jomchai N, et al. The chloroplast genome sequence of mungbean (Vigna radiata) determined by high-throughput pyrosequencing: structural organization and phylogenetic relationships. DNA research. 2009:dsp025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Timme RE, Kuehl JV, Boore JL, Jansen RK. A comparative analysis of the Lactuca and Helianthus (Asteraceae) plastid genomes: Identification of divergent regions and categorization of shared repeats. Am J Bot. 2007;94(3):302–12. WOS:000245097500003. 10.3732/ajb.94.3.302 [DOI] [PubMed] [Google Scholar]
- 76.Gao L, Yi X, Yang YX, Su YJ, Wang T. Complete chloroplast genome sequence of a tree fern Alsophila spinulosa: insights into evolutionary changes in fern chloroplast genomes. Bmc Evol Biol. 2009;9. Artn 130 10.1186/1471-2148-9-130. WOS:000268673300001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kode V, Mudd EA, Iamtham S, Day A. The tobacco plastid accD gene is essential and is required for leaf development. Plant J. 2005;44(2):237–44. WOS:000232885700005. 10.1111/j.1365-313X.2005.02533.x [DOI] [PubMed] [Google Scholar]
- 78.Yao X, Tang P, Li Z, Li D, Liu Y, Huang H. The first complete chloroplast genome sequences in Actinidiaceae: genome structure and comparative analysis. Plos One. 2015;10 10.1371/journal.pone.0129347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Raubeson LA, Peery R, Chumley TW, Dziubek C, Fourcade HM, Boore JL, et al. Comparative chloroplast genomics: analyses including new sequences from the angiosperms Nuphar advena and Ranunculus macranthus. Bmc Genomics. 2007;8(1):174 10.1186/1471-2164-8-174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chung S-M, Staub J, Chen J-F. Molecular phylogeny of Cucumis species as revealed by consensus chloroplast SSR marker length and sequence variation. Genome. 2006;49(3):219–29. 10.1139/g05-101 [DOI] [PubMed] [Google Scholar]
- 81.Elmeer K, Sarwath H, Malek J, Baum M, Hamwieh A. New microsatellite markers for assessment of genetic diversity in date palm (Phoenix dactylifera L.). 3 Biotech. 2011;1(2):91–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Khierallah HS, Bader SM, Hamwieh A, Baum M. Date Palm Genetic Diversity Analysis Using Microsatellite Polymorphism Date Palm Biotechnology Protocols Volume II: Springer; 2017. p. 113–24. [DOI] [PubMed] [Google Scholar]
- 83.Purayil FT, Robert GA, Gothandam KM, Kurup SS, Subramaniam S, Cheruth AJ. Genetic variability in selected date palm (Phoenix dactylifera L.) cultivars of United Arab Emirates using ISSR and DAMD markers. 3 Biotech. 2018;8(2):109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Akhtar W, Rasheed A, Shinwari ZK, Naqvi SMS, Mahmood T. Genetic characterization of different Pakistani date palm varieties. Pak J Bot. 2014;46(6):2095–100. [Google Scholar]
- 85.Elmeer K, Mattat I. Genetic diversity of Qatari date palm using SSR markers. Genet Mol Res. 2015;14:1624–35. 10.4238/2015.March.6.9 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(XLS)
The following four methods were used for the entire genome data set: Bayesian inference (BI), maximum parsimony (MP), maximum likelihood (ML) and neighbor-joining (NJ). Numbers above the branches are the posterior probabilities of BI and bootstrap values for NJ, MP and ML. Green and brown dots represent the positions of P. dactylifera var Khanezi and Nagha.
(TIF)
Data Availability Statement
Data are available from the NCBI database with the ACCESSION NUMBERS (MF197494, MF197495).