Abstract
Classical non-homologous end joining (C-NHEJ) and homologous recombination (HR) compete to repair mammalian chromosomal double strand breaks (DSBs). However, C-NHEJ has no impact on HR induced by DNA nicking enzymes. In this case, the replication fork is thought to convert the DNA nick into a one-ended DSB, which lacks a readily available partner for C-NHEJ. Whether C-NHEJ competes with HR at a non-enzymatic mammalian replication fork barrier (RFB) remains unknown. We previously showed that conservative “short tract” gene conversion (STGC) induced by a chromosomal Tus/Ter RFB is a product of bidirectional replication fork stalling. This finding raises the possibility that Tus/Ter-induced STGC proceeds via a two-ended DSB intermediate. If so, Tus/Ter-induced STGC might be subject to competition by C-NHEJ. However, in contrast to the DSB response, where genetic ablation of C-NHEJ stimulates HR, we report here that Tus/Ter-induced HR is unaffected by deletion of either of two C-NHEJ genes, Xrcc4 or Ku70. These results show that Tus/Ter-induced HR does not entail the formation of a two-ended DSB to which C-NHEJ has competitive access. We found no evidence that the alternative end-joining factor, DNA polymerase θ, competes with Tus/Ter-induced HR. We used chromatin-immunoprecipitation to compare Rad51 recruitment to a Tus/Ter RFB and to a neighboring site-specific DSB. Rad51 accumulation at Tus/Ter was more intense and more sustained than at a DSB. In contrast to the DSB response, Rad51 accumulation at Tus/Ter was restricted to within a few hundred base pairs of the RFB. Taken together, these findings suggest that the major DNA structures that bind Rad51 at a Tus/Ter RFB are not conventional DSBs. We propose that Rad51 acts as an “early responder” at stalled forks, binding single stranded daughter strand gaps on the arrested lagging strand, and that Rad51-mediated fork remodeling generates HR intermediates that are incapable of Ku binding and therefore invisible to the C-NHEJ machinery.
Author summary
Genomic instability is a significant contributor to human disease, ranging from hereditary developmental disorders to cancer predisposition. Two major triggers to genomic instability are chromosomal double strand breaks (DSBs) and the stalling of replication forks during the DNA synthesis (S phase) of the cell cycle. The “rules” that govern mammalian DSB repair are increasingly well understood, and it is recognized that the two major DSB repair pathways—classical non-homologous end joining (C-NHEJ) and homologous recombination (HR)—compete to repair a mammalian DSB. In contrast, we do not yet have equivalent insight into the regulation of repair at sites of mammalian replication fork stalling. Here, we explore the relationship between C-NHEJ and HR at a defined chromosomal replication fork barrier in mammalian cells. We show that, in contrast to DSB repair, repair at stalled forks does not entail competition between C-NHEJ and HR. We find that Rad51, a key mediator of HR, accumulates in an intense and highly localized fashion at the stalled fork. Based upon these findings, we propose a model of HR initiation at the stalled fork in which a Rad51-mediated fork remodeling step prevents access of C-NHEJ to the stalled fork.
Introduction
The stalling of replication forks at sites of abnormal DNA structure, following collisions with transcription complexes or due to nucleotide pool depletion—collectively termed “replication stress”—is a significant contributor to genomic instability. Inherited mutations in genes that regulate the replication stress response cause a number of human diseases, ranging from developmental disorders to highly penetrant cancer predisposition syndromes [1–5]. Replication stress is thought to be a near-universal phenomenon in tumorigenesis and some of the molecules that act upon the stalled fork are considered promising targets for cancer therapy [6]. Replication fork stalling provokes a diverse set of cellular responses, including: stabilization of the stalled replisome; regulated replisome disassembly (“fork collapse”); protection of the fork from deleterious nucleolytic processing; remodeling of DNA structure at the stalled fork; and engagement of repair or “replication restart” [5, 7–15]. The S phase checkpoint and the homologous recombination (HR) systems are intimately involved in coordinating these responses, collaborating to suppress deleterious genome rearrangements at the stalled fork [2, 16–20]. However, the mechanisms governing this coordination remain poorly understood in mammalian cells.
DNA structure at the stalled fork is remodeled by topological stresses on the chromosome at the site of stalling and by the direct action of remodeling enzymes [5, 12, 21]. The fork can be reversed to form a Holliday junction, generating a solitary DNA end which is extensively single stranded due to accompanying nascent lagging strand resection [20, 22, 23]. Other forms of template switching can also occur in the vicinity of the stall site [18, 24, 25]. Endonuclease-mediated fork breakage—either scheduled or unscheduled—can generate double strand breaks (DSBs), which might be either one-ended or two-ended [5, 20]. The DNA structures generated by fork remodeling presumably limit the repair pathways that can be engaged. Two-ended DSBs can potentially be repaired by end joining mechanisms as well as by recombination [26, 27]. In contrast, a one-ended DSB or a solitary DNA end lacks a readily available ligation partner for end joining, and may preferentially engage break-induced replication [28, 29]. Consistent with this, HR induced by a two-ended chromosomal DSB is subject to competition by classical non-homologous end joining (C-NHEJ), whereas HR induced by a nicking enzyme (“nickase”)—in which the replication fork converts the nick into a one-ended DSB—is unaffected by deletion of C-NHEJ genes [30–32]. Thus, in mammalian cells, the susceptibility of HR to competition by C-NHEJ in a particular cellular context is a useful “probe” with which to analyze the DNA structural intermediates of HR. Since the stalled fork response entails the formation of diverse DNA structures and is not restricted to two-ended DSBs, repair pathway “choice” at a stalled fork may differ from that at a defined two-ended DSB.
Study of replication-coupled repair of a covalent DNA inter-strand crosslink (ICL) in Xenopus laevis egg extracts has revealed some of the fundamental steps of stalled fork processing and repair [2, 20]. The Fanconi anemia (FA)/BRCA pathway plays a key role in detecting and processing forks bidirectionally arrested at the ICL [33–35]. The FANCD2/FANCI heterodimer orchestrates dual incisions of one of the sister chromatids on either side of the ICL. Importantly, efficient incision of the bidirectionally arrested forks is suppressed until the two opposing forks have each stalled at the ICL [36]. The resulting two-ended DSB is repaired by HR-mediated sister chromatid recombination, in which the BRCA gene products play canonical roles in promoting Rad51 loading and strand exchange functions [37–41]. HR repair of such a two-ended DSB intermediate could, in principle, be subject to competition by C-NHEJ or other end joining systems. However, recent evidence of fork reversal during ICL repair suggests that at least one of the two DSB ends is extensively single stranded [23].
Competition between HR and C-NHEJ is not a major feature of DSB repair in yeast, since C-NHEJ is a relatively low-flux pathway. Additionally, the Fanconi anemia pathway in yeast is limited to evolutionarily conserved homologs of FANCM [42, 43], suggesting that the innovation of FANCD2/FANCI-coordinated incision of bidirectionally arrested forks occurred relatively recently in evolution. Thus, although certain “core” elements of DSB repair and stalled fork metabolism are conserved between yeast and vertebrates, there are likely significant inter-species differences that remain to be fully defined. Studies in yeast, using non-enzymatic, locus-specific replication fork barriers (RFBs), show that stalled fork HR can mediate both conservative and deleterious repair, the latter including gross chromosomal rearrangements and more localized copy number changes at the site of stalling [14, 18, 19, 24, 44–48]. In contrast to the above-noted X. laevis ICL repair model, HR at an RTS1 RFB in Schizosaccharomyces pombe is not accompanied by evidence of DSB formation [19, 24]. Processing of the stalled fork in S. pombe may also trigger an aberrant form of “replication restart”, a rad22Rad52-dependent process in which the restarted fork is prone to collapse [45]. This aberrant fork restart mechanism is reminiscent of break-induced replication (BIR) in Saccharomyces cerevisiae, which is characteristically unstable and mutation-prone [49, 50]. Indeed, current models of aberrant replication restart in S. pombe invoke a migrating bubble mechanism equivalent to the mechanism of BIR in S. cerevisiae [49]. Rad52-dependent pathways have also been implicated in stalled fork repair in mammalian cells [51–53].
To facilitate analysis of mammalian stalled fork metabolism and repair, we adapted the Escherichia coli Tus/Ter RFB for use in mammalian cells [54–58]. A chromosomally integrated array of six 23 bp Ter sites mediates Tus-dependent, locus-specific replication fork stalling and HR on a mammalian chromosome, enabling direct quantitation of the repair products of mammalian replication fork stalling. We showed that conservative “short tract” gene conversion (STGC) at Tus/Ter is positively regulated by BRCA1, BRCA2, Rad51 and the Fanconi anemia pathway—consistent with the idea that STGC represents a physiological HR response to fork stalling [56, 58]. In contrast, “long tract” gene conversion (LTGC)—an error-prone HR outcome in which a replicative mechanism copies several kilobases from the partner sister chromatid—is suppressed by BRCA1 and appears to be Rad51-independent. We recently identified a novel product of stalled fork repair in primary mouse cells lacking the hereditary breast/ovarian cancer predisposition gene, Brca1: the formation of small (2–6 kb) non-homologous or microhomology-mediated tandem duplications (TDs) [58]. Tus/Ter-induced TDs in Brca1 mutant cells are mediated by a replication restart-bypass mechanism, which is completed by Xrcc4-dependent C-NHEJ. This finding, together with previous observations, suggests that C-NHEJ can access DNA ends positioned close to the site of fork stalling [59, 60]. Notably, Tus/Ter-induced STGC is a product of bidirectional replication fork stalling [56]. By analogy with the processing of forks bidirectionally arrested at an ICL in X. laevis, Tus/Ter-induced STGC might entail the formation of a two-ended DSB intermediate and might therefore be subject to competition by C-NHEJ. To test this hypothesis, we have analyzed the impact of deletion of the C-NHEJ genes Xrcc4 and Ku70 on Tus/Ter-induced HR.
Results
Impact of Xrcc4 deletion on Tus/Ter-induced HR
To determine whether C-NHEJ interacts with HR at Tus/Ter-stalled replication forks, we targeted a 6xTer-HR reporter as a single copy to the ROSA26 locus of mouse embryonic stem (mES) cells carrying biallelic conditional alleles of the C-NHEJ gene Xrcc4 (here termed “Xrcc4fl/fl”), as described in Materials and Methods [56, 61]. The 6xTer-HR reporter contains an I-SceI target site adjacent to the 6xTer array (Fig 1A). Thus, transfection of Tus enables analysis of HR in the stalled fork response, while transfection of I-SceI in parallel samples enables analysis of DSB-induced HR. The reporter also contains elements to distinguish short tract gene conversions (STGC) from long tract gene conversions (LTGC), the latter being rare HR products in wild type cells [62, 63]. Although HR by either STGC or LTGC converts the cell to GFP+, LTGC additionally converts the cell to RFP+, by replicative duplication of an RFP cassette within the reporter (Fig 1A) [56, 64].
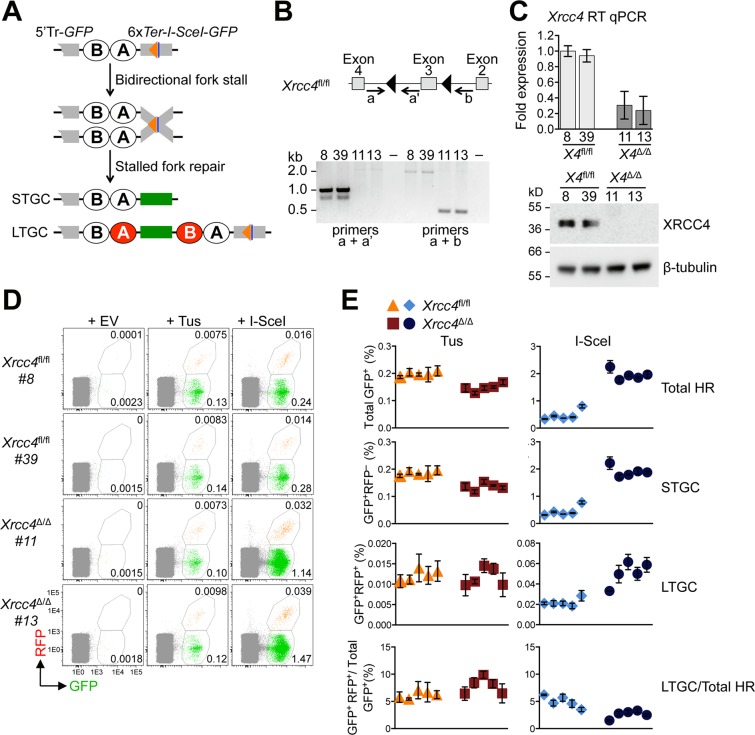
Fig 1. Impact of Xrcc4 deletion on Tus/Ter-induced and I-SceI-induced HR.
A, Schematic of 6xTer-HR reporter and HR repair products of Tus-Ter-induced fork stalling. Green box: wtGFP. Grey boxes: mutant GFP. Open ovals A and B: 5’ and 3’ artificial RFP exons. 5’Tr-GFP: 5’-truncated GFP. Orange triangle: 6xTer element array. Navy blue line: I-SceI endonuclease cut site. STGC, LTGC: short tract and long tract gene conversion HR repair outcomes. LTGC generates wtRFP through RNA splicing (red filled ovals). B, Xrcc4 gene structure in Xrcc4fl/fl ES cells. Xrcc4Δ/Δ allele lacks exon 3. Black triangles: loxP sites. Grey boxes: Xrcc4 Exons 2–4. Location and direction of Exon3 genotyping primers a, a’, and b as indicated by arrows. Gel: PCR products for Xrcc4fl/fl ES clones 8 and 39, and Xrcc4Δ/Δ clones 11 and 13. C, RT qPCR analysis of Xrcc4 expression in Xrcc4fl/fl or Xrcc4Δ/Δ clones. Xrcc4 expression normalized to GAPDH and displayed as fold difference from Xrcc4fl/fl clone 8 of the same experiment (x = -2ΔΔCt, with ΔΔCt = [CtXrcc4-CtGapdh]-[CtXrcc4-CtGAPDH]). Error-bars represent standard deviation of the ΔCt value (SDEV = √[SDEVXrcc42 + SDEVGAPDH2]). Xrcc4 abundance by Western blot in Xrcc4fl/fl clones 8 and 39, and Xrcc4Δ/Δ clones 11 and 13 cell protein extracts. D, Representative primary FACS data for two Xrcc4fl/fl and two Xrcc4Δ/Δ 6xTer-HR reporter clones, as indicated, transfected with empty, 3xMyc-NLS Tus or 3xMyc-NLS I-SceI expression vectors. FACS plots produced from pooled data of duplicate samples from three independent experiments. Numbers represent percentages. E, Frequencies of Tus/Ter-induced and I-SceI-induced repair in five independently derived Xrcc4fl/fl (orange triangles, red squares) or Xrcc4Δ/Δ (blue diamonds, navy blue circles) 6xTer-HR reporter clones transiently transfected with empty, Tus or I-SceI expression vectors. Each dot plot represents the mean of duplicate samples from three independent experiments (n = 3), values are corrected for transfection efficiency–see Materials and Methods. Error bars: standard error of the mean (s.e.m.). One-way ANOVA (Analysis of Variance) test comparing trend in HR between five Xrcc4fl/fl and five Xrcc4Δ/Δ clones: Tus-induced HR, total HR, p = 0.0017; STGC, p = 0.0015; LTGC, p = 0.7142; LTGC/(Total HR), p = 0.2636. I-SceI-induced HR, total HR, p<0.0001; STGC, p<0.0001; LTGC, p<0.0001; LTGC/(Total HR), p<0.0001. T-test comparing Xrcc4fl/fl vs. Xrcc4Δ/Δ clone pooled data, Tus-induced HR: total HR, p<0.0001; STGC, p<0.0001; LTGC, p = 0.6864; LTGC/(Total HR), p = 0.0332; I-SceI-induced HR, total HR, p<0.0001; STGC, p<0.0001; LTGC, p<0.0001; LTGC/(Total HR), p<0.0001.
We transduced a ROSA26-targeted Xrcc4fl/fl 6xTer-HR reporter clone with adenovirally-encoded Cre recombinase and screened for derivative clones that had either lost (Xrcc4Δ/Δ) or retained (Xrcc4fl/fl) Xrcc4. Xrcc4 loss or retention was detected by PCR on genomic (g)DNA and was confirmed in a subset of clones by western blotting (Fig 1B and 1C). We studied HR in five independent Cre-treated Xrcc4fl/fl 6xTer-HR reporter clones and five independent Cre-treated Xrcc4Δ/Δ 6xTer-HR reporter clones in response to either Tus or I-SceI—each transfected in parallel samples (see Materials and Methods). As expected, I-SceI-induced STGC and LTGC were elevated up to 4-fold in Xrcc4Δ/Δ cells in comparison to Xrcc4fl/fl cells (Fig 1D and 1E) [30]. Interestingly, deletion of Xrcc4 stimulated STGC more strongly than LTGC; as a result, the proportion of I-SceI-induced HR events that resolved as LTGC was reduced from ~5% in Xrcc4fl/fl cells to ~2–3% in Xrcc4Δ/Δ cells (Fig 1E). The impact of Xrcc4 deletion on Tus/Ter-induced HR was quite different. Tus/Ter-induced STGC was marginally reduced in Xrcc4Δ/Δ cells in comparison to Xrcc4fl/fl cells, while Tus/Ter-induced LTGC was unaffected by deletion of Xrcc4 (Fig 1D and 1E). These results suggest that the interaction between HR and C-NHEJ at a chromosomal DSB is not recapitulated in the regulation of HR at a stalled replication fork.
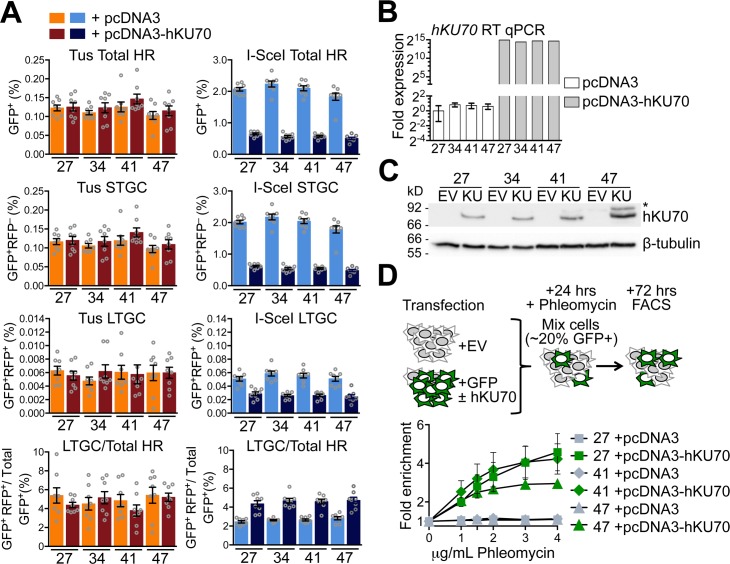
To determine whether the observed phenotypes are affected by re-expression of wtXrcc4, we used lentiviral transduction to express N-terminal influenza haemagglutinin (HA)-tagged wild type mouse (m)Xrcc4 in Xrcc4Δ/Δ 6xTer-HR reporter clones #11 and #13 and in Xrcc4fl/fl 6xTer-HR reporter clones #8 and #39. Briefly, we adapted the lentiviral vector pHIV-Zsgreen [65] by replacing the Zsgreen cDNA with a bicistronic cDNA encoding the enzyme nourseothricin (NTC) acetyl transferase (NAT) [66] fused via a self-cleaving T2A peptide to the human (h)CD52 antigen (S1A Fig) [67]. Transient expression of the empty pHIV-NAT-CD52 vector in mouse ES cells produced strong cell surface staining of hCD52, as revealed by immunostaining using an anti hCD52-specific monoclonal antibody [68] (S1B Fig). Transduction of mES cells with the empty pHIV-NAT-CD52 vector, followed by selection in NTC, generated pools of transduced cells that stained strongly and specifically with anti-hCD52, whereas transduction with pHIV-NAT (i.e., lacking hCD52 expression), followed by NTC selection, generated no CD52-specific cell surface signal (S1B Fig). CD52 expression levels in pHIV-NAT-CD52-mXrcc4-transduced, NTC-selected mES cells were lower than in control empty vector (pHIV-NAT-CD52)-transduced controls, possibly reflecting constraints imposed by Xrcc4 expression from the multicistronic lentiviral expression cassette. Nonetheless, exogenous wtXrcc4 was overexpressed in comparison to endogenous Xrcc4, as revealed by RT-qPCR and by western blotting in lentivirally transduced Xrcc4fl/fl cultures (Fig 2A and 2B). As expected, re-expression of wtXrcc4 complemented the sensitivity of Xrcc4Δ/Δ cells to the radiomimetic drug phleomycin (Fig 2C).
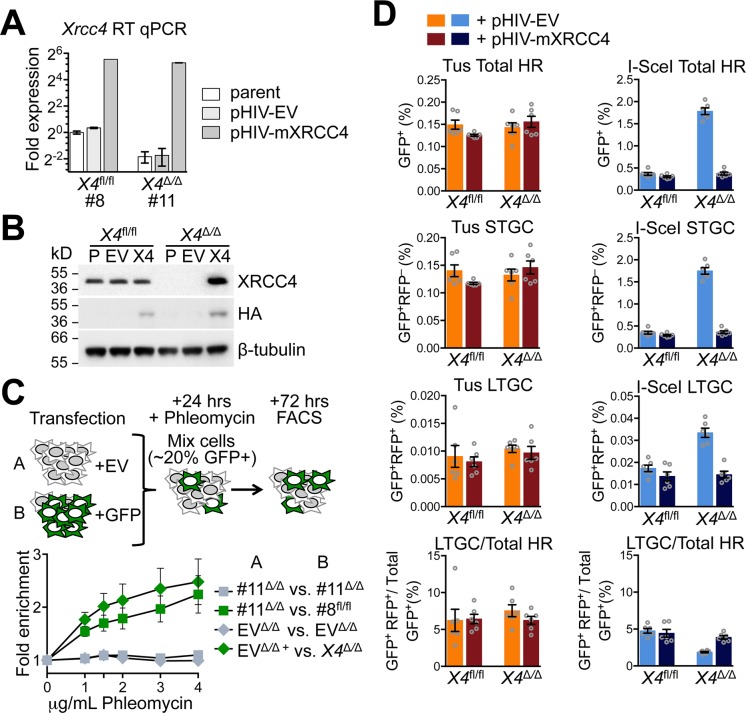
Fig 2. Stable re-expression of wtXrcc4 does not affect Tus/Ter-induced HR in Xrcc4Δ/Δ cells.
A, RT qPCR analysis of Xrcc4 expression in stably transduced Xrcc4fl/fl or Xrcc4Δ/Δ clones. Xrcc4 expression normalized to GAPDH and displayed as fold difference from Xrcc4fl/fl parental reporter clone 8 of the same experiment (x = -2ΔΔCt, with ΔΔCt = [CtXrcc4-CtGapdh]-[CtXrcc4-CtGAPDH]). Error-bars represent standard deviation of the ΔCt value (SDEV = √[SDEVXrcc42 + SDEVGAPDH2]). B, Xrcc4 protein abundance by Western blot in extracts of parental Xrcc4fl/fl clone #8 and Xrcc4Δ/Δ clone #11 and derivative cultures stably transduced with empty lentiviral vector (pHIV-NAT-hCD52, “EV”) or HA-tagged mouse Xrcc4 lentiviral expression vector (“X4”). C, Fold enrichment of cultures transiently expressing exogenous GFP. Results represent fold enrichment of cultures transiently co-transfected with pcDNA3beta and GFP-expression plasmid co-cultured cells transiently transfected with pcDNA3beta alone. Each plot represents the mean of triplicate samples from three independent experiments (n = 3), fold enrichment GFP+ cells normalized to 0 μg/mL phleomycin control. Error bars: s.e.m. D, Frequencies of Tus/Ter-induced and I-SceI-induced repair in Xrcc4fl/fl clone #8 or Xrcc4Δ/Δ clone #11 6xTer-HR reporter cells lentivirally transduced with pHIV-NAT-hCD52-EV (empty vector control) or pHIV-NAT-hCD52-mXrcc4 (expressing HA-tagged mouse Xrcc4 expression vector) with selection of transduced cells in 100 μg/ml NTC. Cells were transiently transfected with empty, 3xMyc-NLS Tus or 3xMyc-NLS I-SceI expression vectors. Each plot represents the mean of duplicate samples from six independent experiments (n = 6). Error bars: s.e.m. Tus-induced Total HR, t-test: flox8 +Xrcc4 vs. flox8 +EV p = 0.0662; del11 +Xrcc4 vs. del11 +EV p = 0.4509; del11 +EV vs. flox8 +EV p = 0.6719; del11 +Xrcc4 vs. flox8 +Xrcc4 p = 0.0588; del11 +Xrcc4 vs. flox8 +EV p = 0.5025. Tus-induced STGC, t-test: flox8 +Xrcc4 vs. flox8 +EV p = 0.0836; del11 +Xrcc4 vs. del11 +EV p = 0.4126; del11 +EV vs. flox8 +EV p = 0.6144; del11 +Xrcc4 vs. flox8 +Xrcc4 p = 0.0595; del11 +Xrcc4 vs. flox8 +EV p = 0.7215. Tus-induced LTGC, t-test: flox8 +Xrcc4 vs. flox8 +EV p = 0.6686; del11 +Xrcc4 vs. del11 +EV p = 0.5972; del11 +EV vs. flox8 +EV p = 0.5313; del11 +Xrcc4 vs. flox8 +Xrcc4 p = 0.3007; del11 +Xrcc4 vs. flox8 +EV p = 0.7870. Tus-induced LTGC/Total HR ratio, t-test: flox8 +Xrcc4 vs. flox8 +EV p = 0.9182; del11 +Xrcc4 vs. del11 +EV p = 0.2133; del11 +EV vs. flox8 +EV p = 0.4686; del11 +Xrcc4 vs. flox8 +Xrcc4 p = 0.8360; del11 +Xrcc4 vs. flox8 +EV p = 0.5771. I-SceI-induced Total HR, t-test: flox8 +Xrcc4 vs. flox8 +EV: p = 0.1292; del11 +Xrcc4 vs. del11 +EV p<0.0001; del11 +EV vs. flox8 +EV p<0.0001; del11 +Xrcc4 vs. flox8 +Xrcc4 p = 0.1030; del11 +Xrcc4 vs. flox8 +EV p = 0.8690. I-SceI-induced STGC, t-test: flox8 +Xrcc4 vs. flox8 +EV p = 0.1353; del11 +Xrcc4 vs. del11 +EV p<0.0001; del11 +EV vs. flox8 +EV p<0.0001; del11 +Xrcc4 vs. flox8 +Xrcc4 p = 0.0939; del11 +Xrcc4 vs. flox39 +EV p = 0.0081. I-SceI-induced LTGC, t-test: flox8 +Xrcc4 vs. flox8 +EV p = 0.1840; del11 +Xrcc4 vs. del11 +EV p<0.0001; del13 +EV vs. flox39 +EV p<0.0001; del11 +Xrcc4 vs. flox8 +Xrcc4 p = 0.7589; del11 +Xrcc4 vs. flox39 +EV p = 0.1347. I-SceI-induced LTGC/Total HR ratio, t-test: flox8 +Xrcc4 vs. flox8 +EV p = 0.5908; del11 +Xrcc4 vs. del11 +EV p = 0.0001; del11 +EV vs. flox8 +EV p = 0.0001; del11 +Xrcc4 vs. flox8 +Xrcc4 p = 0.3729; del11 +Xrcc4 vs. flox39 +EV p = 0.4615.
Xrcc4Δ/Δ 6xTer-HR reporter cells transduced with pHIV-NAT-CD52-Xrcc4 and selected in NTC revealed suppression of I-SceI-induced HR to levels equivalent to that observed in isogenic Xrcc4fl/fl 6xTer-HR reporter cells (Fig 2D). Indeed, I-SceI-induced STGC and LTGC were each restored to wild type levels and the ratio of LTGC:Total HR reverted from ~2% to ~4% in Xrcc4-transduced Xrcc4Δ/Δ cells. Parallel cultures transduced with pHIV-NAT-CD52 empty vector and selected in NTC retained the original Xrcc4Δ/Δ phenotype. These experiments confirm that Xrcc4 affects the balance between I-SceI-induced STGC and LTGC, suppressing STGC more strongly than LTGC. In contrast, all measures of Tus/Ter-induced HR were unaffected by re-expression of wtXrcc4 in Xrcc4Δ/Δ cells (Fig 2D). To confirm these findings, and to minimize opportunities for cellular adaptation during complementation with wtXrcc4, we used transient transfection to restore expression of wtXrcc4 in Xrcc4Δ/Δ cells. Consistent with the above-noted findings, transient Xrcc4 expression strongly suppressed I-SceI-induced HR in Xrcc4Δ/Δ 6xTer-HR reporter cells, but had no significant impact on Tus/Ter-induced STGC or LTGC in these cells (S2 Fig). Taken together, these experiments show that Xrcc4 status has no impact on Tus/Ter-induced HR in mouse ES cells.
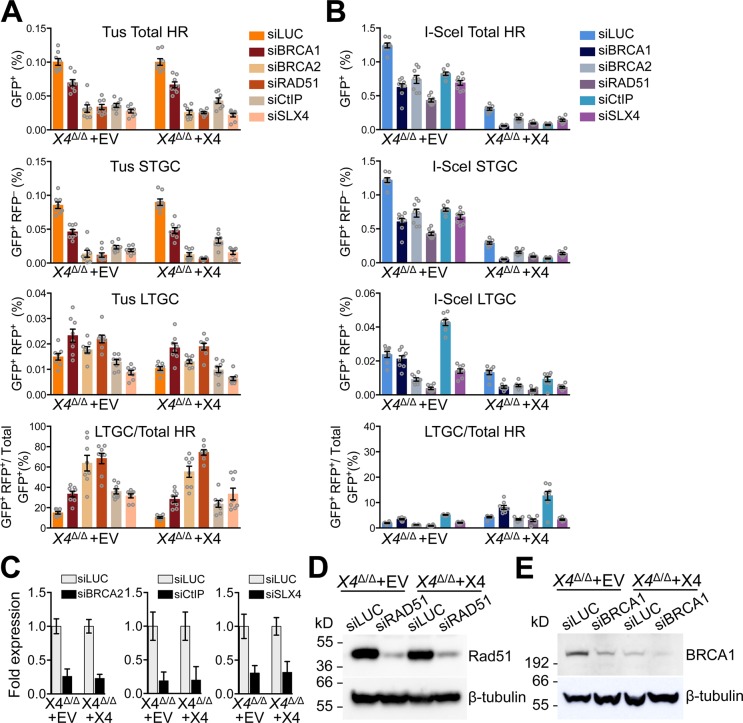
We showed previously that STGC at Tus/Ter-stalled forks is controlled by the HR proteins BRCA1, CtIP, BRCA2 and Rad51 and by the structure-specific nuclease scaffold SLX4 [56, 58]. In contrast, Tus/Ter-induced LTGC is suppressed by BRCA1 and is independent of BRCA2 or Rad51. We found that these relationships were unaffected by Xrcc4 status (Fig 3A). In the regulation of I-SceI-induced HR, we previously noted a specific role for BRCA1 and CtIP in suppressing an HR bias towards LTGC [64]. In contrast, loss of BRCA2 or Rad51 had little impact on the LTGC/Total HR ratio in response to an I-SceI-induced DSB. We observed similar effects on I-SceI-induced HR in Xrcc4Δ/Δ 6xTer-HR reporter cells (Fig 3B). Thus, although Xrcc4 deletion affects the ratio of LTGC:total HR in response to I-SceI, the interactions between HR mediators in execution of HR appear to be largely unaffected by loss of C-NHEJ.
Fig 3. Loss of Xrcc4 does not perturb HR regulation of Tus/Ter-induced STGC and LTGC.
A, Frequencies of Tus/Ter-induced repair Xrcc4Δ/Δ clone 11 6xTer-HR reporter clones stably transduced with pHIV-EV (lentiviral empty vector control) or with pHIV-mXrcc4 (HA-tagged mouse Xrcc4 lentiviral expression vector) with selection of transduced cells in 100 μg/mL NTC. Cells were transiently co-transfected with empty or 3xMyc-NLS Tus expression vectors and siRNAs as shown. Each plot represents the mean of duplicate samples from eight independent experiments (n = 8). Error bars: s.e.m. Xrcc4Δ/Δ clone #11 pHIV-EV: Tus-induced Total HR, t test: siLUC vs. siBRCA1 p = 0.0005; siLUC vs. siBRCA2 p<0.0001; siLUC vs. siRAD51 p<0.0001; siCtIP vs. siLUC p<0.0001; siSLX4 vs. siLUC p<0.0001. Tus-induced STGC, t test: siLUC vs. siBRCA1 p<0.0001; siLUC vs. siBRCA2 p<0.0001; siLUC vs. siRAD51 p<0.0001; siCtIP vs. siLUC p<0.0001; siSLX4 vs. siLUC p<0.0001. Tus-induced LTGC, t test: siLUC vs. siBRCA1 p = 0.0153; siLUC vs. siBRCA2 p = 0.1481; siLUC vs. siRAD51 p = 0.0034; siCtIP vs. siLUC p = 0.2292; siSLX4 vs. siLUC p = 0.0018. Tus-induced Ratio, t test: siLUC vs. siBRCA1 p = 0.0001; siLUC vs. siBRCA2 p = 0.0003; siLUC vs. siRAD51 p<0.0001; siCtIP vs. siLUC p<0.0001; siSLX4 vs. siLUC p<0.0001. Xrcc4Δ/Δ clone #11 pHIV-mXrcc4: Tus-induced Total HR, t test: siLUC vs. siBRCA1 p = 0.0002; siLUC vs. siBRCA2 p<0.0001; siLUC vs. siRAD51 p<0.0001; siCtIP vs. siLUC p<0.0001; siSLX4 vs. siLUC p<0.0001. Tus-induced STGC, t test: siLUC vs. siBRCA1 p<0.0001;siLUC vs. siBRCA2 p<0.0001; siLUC vs. siRAD51 p<0.0001; siCtIP vs. siLUC p<0.0001; siSLX4 vs. siLUC p<0.0001. Tus-induced LTGC, t test: siLUC vs. siBRCA1 p = 0.0023; siLUC vs. siBRCA2 p = 0.0240; siLUC vs. siRAD51 p = 0.0002; siCtIP vs. siLUC p = 0.7398; siSLX4 vs. siLUC p = 0.0022. Tus-induced Ratio, t test: siLUC vs. siBRCA1 p = 0.0004; siLUC vs. siBRCA2 p<0.0001; siLUC vs. siRAD51 p<0.0001; siCtIP vs. siLUC p = 0.0049; siSLX4 vs. siLUC p = 0.0051. B, Frequencies of I-SceI-induced repair Xrcc4Δ/Δ clone 11 6xTer-HR reporter clones stably transduced with pHIV-EV (lentiviral empty vector control, “EV”) or with pHIV-mXrcc4 (HA-tagged mouse Xrcc4 lentiviral expression vector, “X4”) with selection of transduced cells in 100 μg/ml NTC. Cells were co-transiently transfected with empty, or 3xMyc-NLS I-SceI expression vectors and siRNAs as shown. Each plot represents the mean of duplicate samples from eight independent experiments (n = 8). Error bars: s.e.m. Xrcc4Δ/Δ clone #11 pHIV-EV: I-SceI-induced total HR, t test: siLUC vs. siBRCA1 p<0.0001; siLUC vs. siBRCA2 p<0.0001; siLUC vs. siRAD51 p<0.0001; siCtIP vs. siLUC p<0.0001; siSLX4 vs. siLUC p<0.0001. I-SceI-induced STGC, t test: siLUC vs. siBRCA1 p<0.0001; siLUC vs. siBRCA2 p<0.0001; siLUC vs. siRAD51 p<0.0001 siCtIP vs. siLUC p<0.0001; siSLX4 vs. siLUC p<0.0001. I-SceI-induced LTGC, t test: siLUC vs. siBRCA1 p = 0.3335; siLUC vs. siBRCA2 p<0.0001; siLUC vs. siRAD51 p<0.0001; siCtIP vs. siLUC p<0.0001; siSLX4 vs. siLUC p = 0.0006. I-SceI-induced Ratio, t test: siLUC vs. siBRCA1 p = 0.0001; siLUC vs. siBRCA2 p = 0.0020; siLUC vs. siRAD51 p<0.0001; siCtIP vs. siLUC p<0.0001; siSLX4 vs. siLUC p = 0.6260. Xrcc4Δ/Δ clone 11 pHIV-mXrcc4: I-SceI-induced total HR, t test: siLUC vs. siBRCA1 p<0.0001; siLUC vs. siBRCA2 p<0.0001; siLUC vs. siRAD51 p<0.0001; siCtIP vs. siLUC p<0.0001; siSLX4 vs. siLUC p<0.0001. I-SceI-induced STGC, t test: siLUC vs. siBRCA1 p<0.0001; siLUC vs. siBRCA2 p<0.0001; siLUC vs. siRAD51 p<0.0001; siCtIP vs. siLUC p<0.0001; siSLX4 vs. siLUC p<0.0001. I-SceI-induced LTGC, t test: siLUC vs. siBRCA1 p = 0.0001; siLUC vs. siBRCA2 p = 0.0002; siLUC vs. siRAD51 p<0.0001; siCtIP vs. siLUC p = 0.0590; siSLX4 vs. siLUC p = 0.0001. I-SceI-induced Ratio, t test: siLUC vs. siBRCA1 p = 0.0011; siLUC vs. siBRCA2 p = 0.0330; siLUC vs. siRAD51 p = 0.0491; siCtIP vs. siLUC p = 0.0017; siSLX4 vs. siLUC p = 0.0136. C, RT qPCR analysis of BRCA1, BRCA2, CtIP and SLX4 mRNA in siRNA-treated Xrcc4Δ/Δ cells stably transduced with pHIV-EV (“EV”) or pHIV-mXrcc4 (“X4”) derived lentivirus. Data normalized to GAPDH and expressed as fold difference from siLUC sample from the same experiment (x = -2ΔΔCt, with ΔΔCt = [Ct target-CtGapdh]-[CtsiLUC-CtsiGAPDH]). Error-bars represent standard deviation of the ΔCt value (SDEV = √[SDEVTARGET2 + SDEVGAPDH2]). D, Western blot of RAD51 protein abundance in siRNA-treated stably transduced Xrcc4Δ/Δ cells; pHIV-empty vector control (“EV”) or pHIV-mXrcc4 (“X4”). E, Western blot of Brca1 protein abundance in siRNA-treated stably transduced Xrcc4Δ/Δ cells; pHIV-empty vector control (“EV”) or pHIV-mXrcc4 (“X4”).
Impact of DNA polymerase θ depletion on Tus/Ter- and I-SceI-induced HR
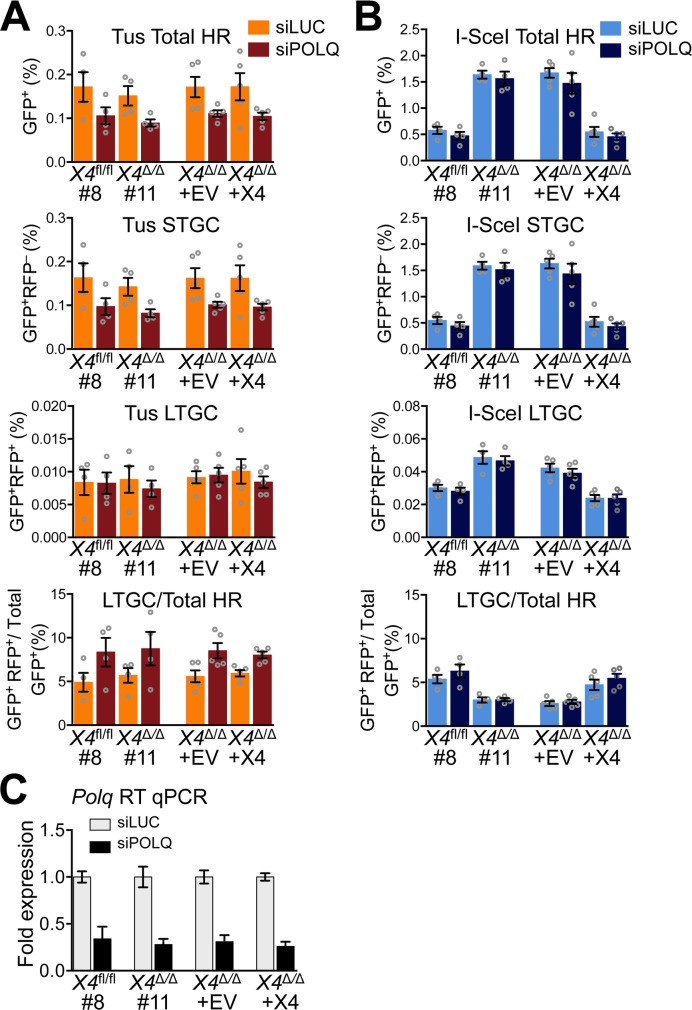
DNA polymerase θ, encoded by the POLQ gene, has been implicated in an alternative end joining (A-EJ) pathway and in the prevention of genomic instability at sites of replication fork stalling [69–72]. Polθ has also been found to suppress DSB-induced HR in some cell types [73, 74]. We therefore asked whether Polθ interacts with HR in mouse ES cells, either at a Tus/Ter RFB or in DSB repair. Interestingly, siRNA-mediated depletion of Polθ modestly suppressed Tus/Ter-induced STGC in multiple clones, but in each case the effect failed reach statistical significance (Fig 4). Depletion of Polθ had no impact on I-SceI-induced HR either in wild type or Xrcc4 null cells. These findings raise the possibility that Polθ supports conservative STGC at stalled forks. They also suggest that the previously reported competition between Polθ and HR in DSB repair is not a feature of mouse ES cells [73, 74].
Fig 4. Impact of DNA polymerase θ depletion on Tus/Ter- and I-SceI-induced HR.
A, Frequencies of Tus/Ter-induced repair in Xrcc4fl/fl clone 8, Xrcc4Δ/Δ clone 11, or Xrcc4Δ/Δ clone 11 stably transduced with pHIV-EV (lentiviral empty vector control) or with pHIV-mXrcc4 (HA-tagged mouse Xrcc4 lentiviral expression vector). Cells transiently co-transfected with empty or 3xMyc-NLS Tus expression vectors and siRNAs shown. Each plot represents the mean of duplicate samples from four (Xrcc4fl/fl clone 8, Xrcc4Δ/Δ clone 11) or five (Xrcc4Δ/Δ clone 11 stably transduced cultures) independent experiments (n = 4,5). Error bars: s.e.m. Tus-induced Total HR, siLUC vs. siPOLQ, t test: Xrcc4fl/fl #8, p = 0.1534; Xrcc4Δ/Δ #11, p = 0.0613; Xrcc4Δ/Δ pHIV-EV, p = 0.0578; Xrcc4Δ/Δ pHIV-mXrcc4, p = 0.0942. Total HR, siLUC, t test: Xrcc4fl/fl #8 vs. Xrcc4Δ/Δ #11, p = 0.6352; Xrcc4Δ/Δ pHIV-EV, vs. pHIV-mXrcc4, p = 0.9841. Total HR, siPOLQ, t test: Xrcc4fl/fl #8 vs. Xrcc4Δ/Δ #11, p = 0.4778; Xrcc4Δ/Δ pHIV-EV, vs. pHIV-mXrcc4, p = 0.7291. Tus-induced STGC, siLUC vs. siPOLQ, t test: Xrcc4fl/fl #8, p = 0.1437; Xrcc4Δ/Δ #11, p = 0.0510; Xrcc4Δ/Δ pHIV-EV, p = 0.0543; Xrcc4Δ/Δ pHIV-mXrcc4, p = 0.0864. STGC, siLUC, t test: Xrcc4fl/fl #8 vs. Xrcc4Δ/Δ #11, p = 0.6116; Xrcc4Δ/Δ pHIV-EV, vs. pHIV-mXrcc4, p = 0.9976. STGC, siPOLQ, t test: Xrcc4fl/fl #8 vs. Xrcc4Δ/Δ #11, p = 0.5041; Xrcc4Δ/Δ pHIV-EV, vs. pHIV-mXrcc4, p = 0.7757. Tus-induced LTGC, siLUC vs. siPOLQ, t test: Xrcc4fl/fl #8, p = 0.9647; Xrcc4Δ/Δ #11, p = 0.5780; Xrcc4Δ/Δ pHIV-EV, p = 0.8273; Xrcc4Δ/Δ pHIV-mXrcc4, p = 0.4575. LTGC, siLUC, t test: Xrcc4fl/fl #8 vs. Xrcc4Δ/Δ #11, p = 0.8797; Xrcc4Δ/Δ pHIV-EV, vs. pHIV-mXrcc4, p = 0.6780. LTGC, siPOLQ, t test: Xrcc4fl/fl #8 vs. Xrcc4Δ/Δ #11, p = 0.6918; Xrcc4Δ/Δ pHIV-EV, vs. pHIV-mXrcc4, p = 0.4764. Tus-induced LTGC/(Total HR), siLUC vs. siPOLQ, t test: Xrcc4fl/fl #8, p = 0.1359; Xrcc4Δ/Δ #11, p = 0.2154; Xrcc4Δ/Δ pHIV-EV, p = 0.0315; Xrcc4Δ/Δ pHIV-mXrcc4, p = 0.0043. LTGC/(Total HR), siLUC, t test: Xrcc4fl/fl #8 vs. Xrcc4Δ/Δ #11, p = 0.5835; Xrcc4Δ/Δ pHIV-EV, vs. pHIV-mXrcc4, p = 0.6703. LTGC/(Total HR), siPOLQ, t test: Xrcc4fl/fl #8 vs. Xrcc4Δ/Δ #11, p = 0.8795; Xrcc4Δ/Δ pHIV-EV, vs. pHIV-mXrcc4, p = 0.4704. B, Frequencies of I-SceI-induced repair in Xrcc4fl/fl clone 8, Xrcc4Δ/Δ clone 11, or Xrcc4Δ/Δ clone 11 stably transduced with pHIV-EV (lentiviral empty vector control) or with pHIV-mXrcc4 (HA-tagged mouse Xrcc4 lentiviral expression vector). Cells transiently co-transfected with empty or 3xMyc-NLS I-SceI expression vectors and siRNAs shown. Each plot represents the mean of duplicate samples from four (Xrcc4fl/fl clone 8, Xrcc4Δ/Δ clone 11) or five (Xrcc4Δ/Δ clone 11 stably transduced cultures) independent experiments (n = 4,5). Error bars: s.e.m. I-SceI-induced Total HR, siLUC vs. siPOLQ, t test: Xrcc4fl/fl #8, p = 0.3435; Xrcc4Δ/Δ #11, p = 0.6415; Xrcc4Δ/Δ pHIV-EV, p = 0.5332; Xrcc4Δ/Δ pHIV-mXrcc4, p = 0.6113. Total HR, siLUC, t test: Xrcc4fl/fl #8 vs. Xrcc4Δ/Δ #11, p<0.0001; Xrcc4Δ/Δ pHIV-EV, vs. pHIV-mXrcc4, p = 0.0004. Total HR, siPOLQ, t test: Xrcc4fl/fl #8 vs. Xrcc4Δ/Δ #11, p = 0.0010; Xrcc4Δ/Δ pHIV-EV, vs. pHIV-mXrcc4, p = 0.0178. I-SceI-induced STGC, siLUC vs. siPOLQ, t test: Xrcc4fl/fl #8, p = 0.3438; Xrcc4Δ/Δ #11, p = 0.6464; Xrcc4Δ/Δ pHIV-EV, p = 0.3965; Xrcc4Δ/Δ pHIV-mXrcc4, p = 0.4388. STGC, siLUC, t test: Xrcc4fl/fl #8 vs. Xrcc4Δ/Δ #11, p<0.0001; Xrcc4Δ/Δ pHIV-EV, vs. pHIV-mXrcc4, p<0.0001. STGC, siPOLQ, t test: Xrcc4fl/fl #8 vs. Xrcc4Δ/Δ #11, p = 0.0011; Xrcc4Δ/Δ pHIV-EV, vs. pHIV-mXrcc4, p = 0.0049. I-SceI-induced LTGC, siLUC vs. siPOLQ, t test: Xrcc4fl/fl #8, p = 0.5196; Xrcc4Δ/Δ #11, p = 0.6949; Xrcc4Δ/Δ pHIV-EV, p = 0.4229; Xrcc4Δ/Δ pHIV-mXrcc4, p = 0.9733. LTGC, siLUC, t test: Xrcc4fl/fl #8 vs. Xrcc4Δ/Δ #11, p = 0.0100; Xrcc4Δ/Δ pHIV-EV, vs. pHIV-mXrcc4, p = 0.0007. LTGC, siPOLQ, t test: Xrcc4fl/fl #8 vs. Xrcc4Δ/Δ #11, p = 0.0028; Xrcc4Δ/Δ pHIV-EV, vs. pHIV-mXrcc4, p = 0.0030. I-SceI-induced LTGC/(Total HR), siLUC vs. siPOLQ, t test: Xrcc4fl/fl #8, p = 0.3449; Xrcc4Δ/Δ #11, p = 0.9371; Xrcc4Δ/Δ pHIV-EV, p = 0.6062; Xrcc4Δ/Δ pHIV-mXrcc4, p = 0.3769. LTGC/(Total HR), siLUC, t test: Xrcc4fl/fl #8 vs. Xrcc4Δ/Δ #11, p = 0.0090; Xrcc4Δ/Δ pHIV-EV, vs. pHIV-mXrcc4, p = 0.0187. LTGC/(Total HR), siPOLQ, t test: Xrcc4fl/fl #8 vs. Xrcc4Δ/Δ #11, p = 0.0181; Xrcc4Δ/Δ pHIV-EV, vs. pHIV-mXrcc4, p = 0.0036. C, RT qPCR analysis of POLQ mRNA in siRNA-treated reporter cells. Data normalized to GAPDH and expressed as fold difference from siLUC sample from the same experiment (x = -2ΔΔCt, with ΔΔCt = [CtPolq-CtGapdh]-[CtsiLUC-CtsiGAPDH]). Error-bars represent standard deviation of the ΔCt value (SDEV = √[SDEVTARGET2 + SDEVGAPDH2]).
Impact of Ku70 deletion on Tus/Ter-induced HR
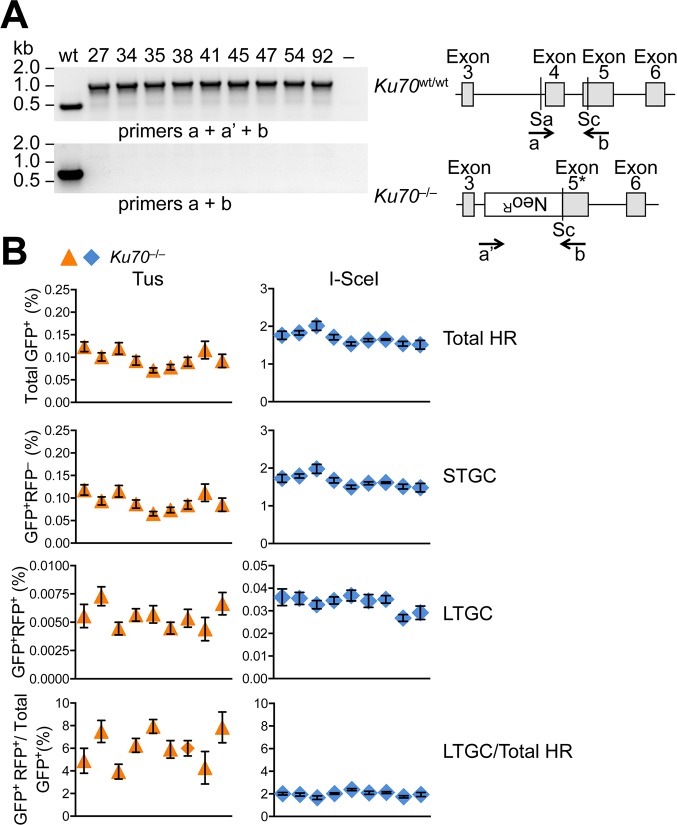
The binding of the Ku70/Ku80 heterodimer to DNA ends is required for engagement of C-NHEJ [75]. Ku has also been implicated in modulation of repair functions at forks stalled by the action of Topoisomerase I inhibitors, where one-ended breaks are thought to predominate [76, 77]. To determine whether Ku DNA end binding activity can influence Tus/Ter-induced HR independent of later steps of the C-NHEJ pathway, we targeted a single copy of the 6xTer-HR reporter to the ROSA26 locus of Ku70–/–mES cells [78]. Nine independent ROSA26-targeted Ku70–/– 6xTer-HR reporter clones revealed wild type levels of Tus/Ter-induced HR but greatly elevated levels of I-SceI-induced HR (Fig 5). To complement this phenotype, we co-transfected either Tus or I-SceI expression vectors with either empty vector or with a vector for expression of wt human KU70. Transient expression of wtKU70 suppressed I-SceI-induced HR and complemented phleomycin sensitivity of Ku70–/–cells, as expected (Fig 6). In contrast, wtKU70 expression had no impact on Tus/Ter-induced HR (Fig 6).
Fig 5. Impact of Ku70 deletion on Tus/Ter-induced HR.
A, Ku70 mutant cell genotyping and gene structure in Ku70–/–ES cells. Ku70 null allele lacks Exon 4 and partially Exon 5. Grey boxes: Ku70 Exons 3–6. Location and direction of Exons 4 and 5 and neoR genotyping primers a, a’, and b as indicated by arrows. Sa: SacI restriction site. Sc: ScaI restriction site. Gel: PCR products for single wild type control and nine Ku70–/–ES 6xTer-HR reporter clones (27, 34, 35, 38, 41, 45, 47, 54, and 92). B, Frequencies of Tus/Ter-induced and I-SceI-induced repair in nine independently derived Ku70–/– 6xTer-HR reporter clones (27, 34, 35, 38, 41, 45, 47, 54, and 92) transiently transfected with empty, 3xMyc-NLS Tus or 3xMyc-NLS I-SceI expression vectors. Each dot plot represents the mean of duplicate samples from six independent experiments (n = 6), values are corrected for transfection efficiency. Error bars: s.e.m. One-way ANOVA (Analysis of Variance) test comparing trend in HR between nine Ku70–/–clones: Tus-induced HR, total HR, p = 0.0205; STGC, p = 0.0173; LTGC, p = 0.1698; LTGC/(Total HR), p = 0.0261. I-SceI-induced HR, total HR, p = 0.0005; STGC, p = 0.0004; LTGC, p = 0.927; LTGC/(Total HR), p = 0.0081.
Fig 6. Transient KU70 expression does not affect Tus/Ter-induced HR in Ku70–/–cells.
A, Frequencies of Tus/Ter-induced and I-SceI-induced repair in four independently derived Ku70–/– 6xTer-HR reporter clones (clones 27, 34, 41, and 47) with and without transient expression of exogenous hKU70. Cells transiently co-transfected with empty pcDNA3beta or pcDNA3beta-hKU70 expression vector and either empty, 3xMyc-NLS Tus or 3xMyc-NLS I-SceI expression vectors. Each column represents the mean of duplicate samples from eight independent experiments (n = 8), values are corrected for transfection efficiency. Error bars: s.e.m. Tus-induced total HR, t test: #27 +EV vs. +hKU70, p = 0.8355; #34 +EV vs. +hKU70, p = 0.3799; #41 +EV vs. +hKU70, p = 0.2710; #47 +EV vs. +hKU70, p = 0.4703; Tus-induced STGC, t test: #27 +EV vs. +hKU70, p = 0.7892; #34 +EV vs. +hKU70, p = 0.4223; #41 +EV vs. +hKU70, p = 0.2345; #47 +EV vs. +hKU70, p = 0.4426; Tus-induced LTGC, t test: #27 +EV vs. +hKU70, p = 0.4375; #34 +EV vs. +hKU70, p = 0.2413; #41 +EV vs. +hKU70, p = 0.8608; #47 +EV vs. +hKU70, p = 0.9872; Tus-induced LTGC/(Total HR), t test: #27 +EV vs. +hKU70, p = 0.2701; #34 +EV vs. +hKU70, p = 0.4964; #41 +EV vs. +hKU70, p = 0.2507; #47 +EV vs. +hKU70, p = 0.8222. I-SceI-induced total HR, t test: #27 +EV vs. +hKU70, p<0.0001; #34 +EV vs. +hKU70, p<0.0001; #41 +EV vs. +hKU70, p<0.0001; #47 +EV vs. +hKU70, p<0.0001; I-SceI-induced STGC, t test: #27 +EV vs. +hKU70, p<0.0001; #34 +EV vs. +hKU70, p<0.0001; #41 +EV vs. +hKU70, p<0.0001; #47 +EV vs. +hKU70, p<0.0001; I-SceI-induced LTGC, t test: #27 +EV vs. +hKU70, p = 0.0002; #34 +EV vs. +hKU70, p<0.0001; #41 +EV vs. +hKU70, p<0.0001; #47 +EV vs. +hKU70, p<0.0001; I-SceI-induced LTGC/(Total HR), t test: #27 +EV vs. +hKU70, p = 0.0004; #34 +EV vs. +hKU70, p<0.0001; #41 +EV vs. +hKU70, p<0.0001; #47 +EV vs. +hKU70, p = 0.0003. One-way ANOVA (Analysis of Variance) test comparing trend in HR: Tus-induced HR, total HR, p = 0.2388; STGC, p = 0.1923; LTGC, p = 0.9660; LTGC/(Total HR), p = 0.5923. I-SceI-induced HR, total HR, p<0.0001; STGC, p<0.0001; LTGC, p<0.0001; LTGC/(Total HR), p<0.0001. B, RT qPCR analysis of hKU70 in transfected Ku70–/–clones. hKU70 expression normalized to GAPDH and displayed as fold difference from Ku70–/–reporter clone 27 of the same experiment (x = -2ΔΔCt, with ΔΔCt = [CtKU70-CtGAPDH]-[CtKU70-CtGAPDH]). Error-bars represent standard deviation of the ΔCt value (SDEV = √[SDEVKU702 + SDEVGAPDH2]). C, Western blot for abundance of hKU70 protein in Ku70–/–reporter clones transiently transfected with empty pcDNA3beta or pcDNA3beta-hKU70. D, Fold enrichment of cultures transiently expressing exogenous GFP. Results represent fold enrichment of cultures transiently co-transfected with GFP-expression plasmid and either pcDNA3beta or pcDNA3beta-hKU70 expression vector over co-cultured cells transiently transfected with pcDNA3beta alone. Each plot represents the mean of triplicate samples from three independent experiments (n = 3), fold enrichment GFP+ cells normalized to 0 μg/mL phleomycin control. Error bars: s.e.m.
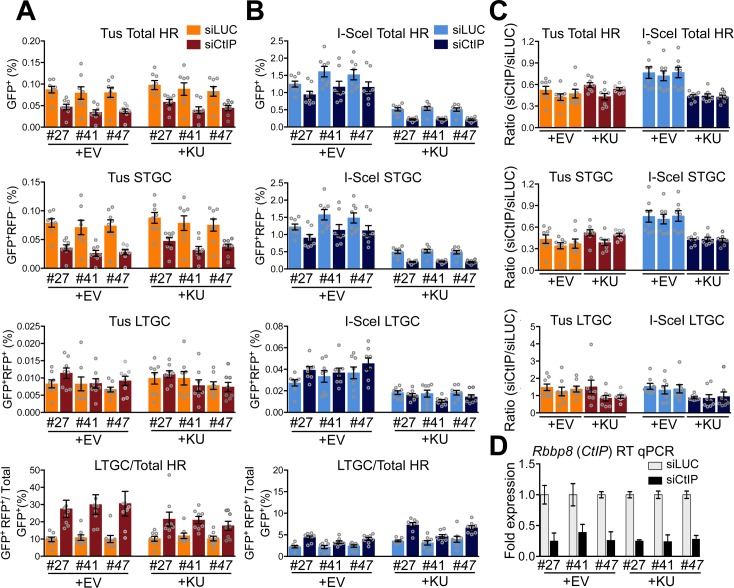
In the processing of a conventional DSB, Ku binding to the DNA end is a barrier to DNA end resection. DNA end resection activity, initiated by CtIP and the Mre11 nuclease, can displace Ku from the DNA end, providing a mechanism by which the HR machinery can overcome the barrier formed by Ku DNA end binding [79]. To further search for evidence of Ku interaction with stalled fork HR, we determined the impact of siRNA-mediated CtIP depletion on HR in Ku70–/–cells either uncomplemented or transiently complemented with wtKU70. As previously reported, CtIP depletion reduced HR in response to Tus/Ter or to an I-SceI-mediated DSB [58], and this effect was observed in both uncomplemented and Ku70-complemented Ku70–/–cells (Fig 7A and 7B). However, the proportional impact of CtIP depletion appeared less pronounced in uncomplemented I-SceI-transfected Ku70–/–cells than in the same cells complemented with wtKU70 (Fig 7B). We quantified this effect by calculating, for each test group, the induced HR in cells that received siCtIP as a proportion of induced HR in cells that received the control siRNA directed to luciferase. Notably, for I-SceI-induced HR, this ratio was increased in uncomplemented Ku70–/–cells in comparison to wtKU70-complemented cells (Fig 7C and 7D). In contrast, for Tus/Ter-induced HR, this ratio was unaffected by Ku70 status. We interpret these results as follows: at a DSB, Ku binding creates a barrier to end resection and CtIP plays a significant role in displacing Ku. This Ku-displacing role of CtIP is not required in Ku70–/–cells, and the relative importance of CtIP in HR at a DSB in Ku70–/–cells is correspondingly less. In contrast, at a Tus/Ter RFB, CtIP plays a significant role in HR that is fully independent of Ku70. Taken together with the above findings with regard to Xrcc4, the data indicate that C-NHEJ does not compete with HR at a mammalian Tus/Ter RFB.
Fig 7. Impact of CtIP depletion on repair frequencies in the presence or absence of hKU70.
A, Frequencies of Tus/Ter-induced repair in three independently derived Ku70–/– 6xTer-HR reporter clones (clones 27, 41, and 47) transiently expressing exogenous hKU70 and transfected with siRNAs shown. Cells transiently co-transfected with empty pcDNA3beta or pcDNA3beta-hKU70 expression vector and empty or 3xMyc-NLS Tus expression vectors treated with either siLUC or siCtIP. Each column represents the mean of duplicate samples from eight independent experiments (n = 8), values are corrected for transfection efficiency. Error bars: s.e.m. Tus-induced total HR, siLUC vs. siCtIP t test: #27 +EV, p = 0.0030; #41 +EV, p = 0.0207; #47 +EV, p = 0.0070; #27 +hKU70, p = 0.0047; #41 + hKU70, p = 0.0281; #47 + hKU70, p = 0.0148; Tus-induced STGC, t test: #27 +EV, p = 0.0011; #41 +EV, p = 0.0104; #47 +EV, p = 0.0070; #27 +hKU70, p = 0.0030; #41 + hKU70, p = 0.0207; #47 + hKU70, p = 0.0148; Tus-induced LTGC, t test: #27 +EV, p = 0.2786; #41 +EV, p = 0.5737; #47 +EV, p = 0.1304; #27 +hKU70, p = 0.8785; #41 + hKU70, p = 0.5737; #47 + hKU70, p = 0.5737; Tus-induced LTGC/(Total HR), t test: #27 +EV, p = 0.0002; #41 +EV, p = 0.0006; #47 +EV, p = 0.0006; #27 +hKU70, p = 0.0019; #41 + hKU70, p = 0.0030; #47 + hKU70, p = 0.0650; One-way ANOVA (Analysis of Variance) test comparing trend in Tus-induced Total HR: siLUC vs siCtIP all, p<0.0001; siLUC vs siCtIP +EV, p<0.0001; siLUC vs siCtIP +hKU70, p = 0.0002; siLUC all, p = 0.8904; siCtIP all, p = 0.1322. Tus-induced STGC, one-way ANOVA test: siLUC vs siCtIP all, p<0.0001; siLUC vs siCtIP +EV, p<0.0001; siLUC vs siCtIP +hKU70, p<0.0001; siLUC all, p = 0.9108; siCtIP all, p = 0.1155. Tus-induced LTGC, one-way ANOVA test: siLUC vs siCtIP all, p = 0.4334; siLUC vs siCtIP +EV, p = 0.3194; siLUC vs siCtIP +hKU70, p = 0.4144; siLUC all, p = 0.6254; siCtIP all, p = 0.2231. Tus-induced LTGC/(Total HR), one-way ANOVA test: siLUC vs siCtIP all, p<0.0001; siLUC vs siCtIP +EV, p = 0.0004; siLUC vs siCtIP +hKU70, p = 0.0012; siLUC all, p = 0.9449; siCtIP all, p = 0.2989. B, Frequencies of I-SceI-induced repair in three independently derived KU70Δ/Δ 6xTer-HR reporter clones. Cells transiently co-transfected with empty pcDNA3beta or pcDNA3beta-hKU70 expression vector and empty or 3xMyc-NLS I-SceI expression vectors treated with either siLUC or siCtIP. Each column represents the mean of duplicate samples from eight independent experiments (n = 8), values are corrected for transfection efficiency. Error bars: s.e.m. I-SceI-induced total HR, siLUC vs. siCtIP t test: #27 +EV, p = 0.0379; #41 +EV, p = 0.0281; #47 +EV, p = 0.0499; #27 +hKU70, p = 0.0003; #41 + hKU70, p = 0.0019; #47 + hKU70, p = 0.0011; I-SceI-induced STGC, siLUC vs. siCtIP t test: #27 +EV, p = 0.0379; #41 +EV, p = 0.0281; #47 +EV, p = 0.0379; #27 +hKU70, p = 0.0002; #41 + hKU70, p = 0.0019; #47 + hKU70, p = 0.0011; I-SceI-induced LTGC, siLUC vs. siCtIP t test: #27 +EV, p = 0.1104; #41 +EV, p = 0.7984; #47 +EV, p = 0.3282; #27 +hKU70, p = 0.3282; #41 + hKU70, p = 0.1949; #47 + hKU70, p = 0.1949; I-SceI-induced LTGC/(Total HR), siLUC vs. siCtIP t test: #27 +EV, p = 0.0011; #41 +EV, p = 0.0379; #47 +EV, p = 0.0070; #27 +hKU70, p = 0.0006; #41 + hKU70, p = 0.0650; #47 + hKU70, p = 0.0070; I-SceI-induced Total HR, one-way ANOVA test: siLUC vs siCtIP all, p<0.0001; siLUC vs siCtIP +EV, p = 0.0139; siLUC vs siCtIP +hKU70, p<0.0001; siLUC all, p<0.0001; siCtIP all, p<0.0001. I-SceI-induced STGC, one-way ANOVA test: siLUC vs siCtIP all, p<0.0001; siLUC vs siCtIP +EV, p = 0.0106; siLUC vs siCtIP +hKU70, p<0.0001; siLUC all, p<0.0001; siCtIP all, p<0.0001. I-SceI-induced LTGC, one-way ANOVA test: siLUC vs siCtIP all, p<0.0001; siLUC vs siCtIP +EV, p = 0.1503; siLUC vs siCtIP +hKU70, p = 0.1010; siLUC all, p = 0.0010; siCtIP all, p<0.0001. I-SceI-induced LTGC/(Total HR), one-way ANOVA test: siLUC vs siCtIP all, p<0.0001; siLUC vs siCtIP +EV, p<0.0001; siLUC vs siCtIP +hKU70, p<0.0001; siLUC all, p = 0.0147; siCtIP all, p<0.0001. C, Observed repair frequencies for Tus or I-SceI induced HR, STGC and LTGC expressed as the ratio of siCtIP frequency/siLUC frequency for data from clones 27, 41 and 47 shown in panels A and B. One-way ANOVA test: Tus-induced total HR, p = 0.0779; Tus-induced STGC, p = 0.0564; Tus-induced LTGC, p = 0.2067. One-way ANOVA test: I-SceI-induced total HR, p<0.0001; I-SceI-induced STGC, p<0.0001; I-SceI-induced LTGC, p = 0.0832. D, RT qPCR analysis of CtIP mRNA in siRNA-transfected Ku70–/–clones. Data normalized to GAPDH and expressed as fold difference from siLUC sample from the same experiment (x = -2ΔΔCt, with ΔΔCt = [CtsiCtIP-CtGapdh]-[CtsiLUC-CtsiGAPDH]). Error-bars represent standard deviation of the ΔCt value (SDEV = √[SDEVCtIP2 + SDEVGAPDH2]).
Localized recruitment of Rad51 to a Tus/Ter replication fork barrier
Rad51 loading onto ssDNA is a key step in HR. In contrast to a DSB, where ssDNA is exposed following canonical DNA end resection, the stalled fork might present ssDNA for Rad51 loading through a number of different mechanisms. To determine whether Rad51 accumulates at Tus/Ter-stalled forks, we used chromatin-immunoprecipitation to study Rad51 accumulation at the ROSA26 locus, in cells transfected with a DSB-inducing nuclease, Tus, or appropriate negative controls. To induce a DSB at ROSA26, we used either I-SceI or Cas9 targeted to the I-SceI target site by a sgRNA specific to the I-SceI site. As a negative control for I-SceI and Tus, we transfected empty expression vector. As a negative control for Cas9/I-SceI sgRNA, we co-transfected wtCas9 with a non-targeting sgRNA. The chromatin-immunoprecipitation method is further described in Materials and Methods. We assessed Rad51 recruitment at 24 and 48 hours following transfection, and assayed its enrichment near the 6xTer array or neighboring I-SceI site by quantitative real-time PCR, using primers at different positions within the ROSA26 gene (Fig 8A).
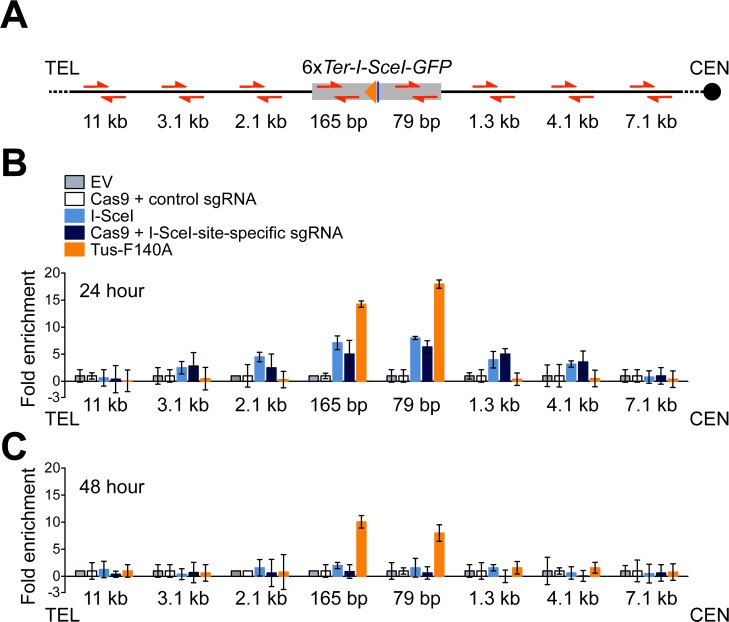
Fig 8. Distinct patterns of Rad51 recruitment to the Tus/Ter fork barrier and to a conventional DSB.
A, Schematic of the 1xGFP 6xTer reporter. Location of telomere (TEL) and centromere (CEN) shown. Red half-arrow heads: primer pairs. Grey box: mutant GFP allele (6xTer-I-SceI-GFP). Orange triangle: 6xTer element array. Navy blue line: I-SceI endonuclease target site and I-SceI site-specific guide RNA target site. Primer pair positions indicated as distance between the proximal end of the closer primer sequence to proximal edge of the 6xTer array. B and C, Rad51 protein abundance at sites near the 6xTer-I-SceI-GFP allele in response to Tus/Ter-induced replication fork stalling or DSB induction at 24 or 48 hours after transfection. Cells transiently transfected with empty vector (grey), pcDNA3β-myc NLS-Tus-F140A-3xHA (orange), pcDNA3β-myc NLS-I-SceI (royal blue), or co-transfected with spCas9 expression plasmid with control (white) or I-SceI site-specific (navy blue) guide RNA. Fold enrichment of Rad51 protein calculated as the mean 2-ΔΔCT from three independent experiments (n = 3) normalized against untreated controls (empty vector or guide RNA controls) and β-Actin control locus. Error bars indicate the standard deviation of the ΔCT measurement calculated as the change in Ct value obtained from the proximal-Ter locus and that obtained from β-Actin control locus.
24 hours after transfection with either I-SceI or Cas9/I-SceI sgRNA, Rad51 was detected maximally at sites in close proximity to the I-SceI site, and this signal spread up to ~4 kb either side of the DSB (Fig 8B). By the 48 hour time-point, a specific DSB-induced Rad51 signal was no longer detectable (Fig 8C). The Rad51 response to a Tus/Ter RFB differed markedly. Notably, Rad51 accumulation at Tus/Ter was more intense than in the response to a DSB, even though Tus/Ter consistently induces lower HR frequencies than I-SceI in our experiments. A second striking difference was the distribution of Rad51. At the Tus/Ter RFB, Rad51 was strictly localized to within a few hundred base pairs of the RFB, with no spreading of the Rad51 signal detectable even 1.3 kb from the RFB. Third, the Rad51 signal remained detectable at Tus/Ter up to 48 hours after transfection, at a time when the DSB-induced Rad51 signal had subsided. These findings reveal that Rad51 accumulation at the Tus/Ter RFB is more intense, more sustained and more specifically localized than in the DSB response. Taken together, these findings suggest that the major DNA structures that bind Rad51 at a Tus/Ter RFB are not conventional DSBs.
Discussion
In contrast to HR induced by a chromosomal DSB, where C-NHEJ competes to repair the two-ended break, we show here that HR induced by a Tus/Ter RFB in mammalian cells is unaffected by the status of the C-NHEJ genes Xrcc4 or Ku70. This shows that the fundamental mechanisms of repair pathway “choice” at a stalled replication fork and a chromosomal DSB differ markedly. The simplest explanation of these findings is that HR at Tus/Ter does not entail formation of a two-ended DSB intermediate. We recently used High Throughput Translocation Sequencing (HTGTS) to study translocation-competent DNA lesions at Tus/Ter [58]. In contrast to I-SceI-induced DSBs, where two-ended breaks predominate, the major lesions detected by HTGTS at Tus/Ter were solitary DNA ends. However, it is possible that two-ended DSB intermediates of STGC arise at Tus/Ter but are not readily detected by HTGTS. Indeed, in the X. laevis model of replication-coupled ICL repair, temporally coordinated dual incisions of one sister chromatid generate a two-ended DSB intermediate. Bidirectional replication fork stalling is a critical step in this repair process, the arrival of both forks being required for replisome disassembly, asymmetrical fork reversal, nascent lagging strand resection and FANCD2/FANCI-coordinated incisions flanking the ICL [20, 23, 36].
Significant parallels exist between Tus/Ter-induced STGC and the above-noted model of ICL repair, especially with regard to the role of bidirectional fork arrest. We previously used Southern blotting to show that Tus/Ter-induced STGC products are of a fixed size, identical to products of I-SceI-induced STGC [56]. In I-SceI-induced HR, where synthesis-dependent strand annealing (SDSA) is thought to be the dominant HR pathway, the fixed size of STGC products reflects the availability of a homologous second end of the two-ended break, which supports termination of gene conversion by annealing with the displaced nascent strand [26, 27]. Indeed, if I-SceI-induced STGC is denied a homologous second end, the STGC products retrieved are of variable size, reflecting termination of gene conversion at random sites within the reporter, without the assistance of homologous pairing/annealing [64]. These aberrant STGCs are likely completed by end joining with the non-homologous second end of the DSB [80]. In the case of Tus/Ter-induced HR, the stereotyped structure of the STGC products implies that a homologous second DNA end was available to enable termination of STGC by annealing. This second end, we believe, must originate from the second (opposing) fork that stalls at Tus/Ter [56]. In summary, the mechanism of STGC at Tus/Ter has paradoxical properties. The structure of Tus/Ter-induced STGC products and its dependency on the Fanconi/BRCA/HR pathway is suggestive of SDSA of a two-ended break. However, as shown here, C-NHEJ does not compete with Tus/Ter-induced HR. Several possible models could reconcile these paradoxical properties.
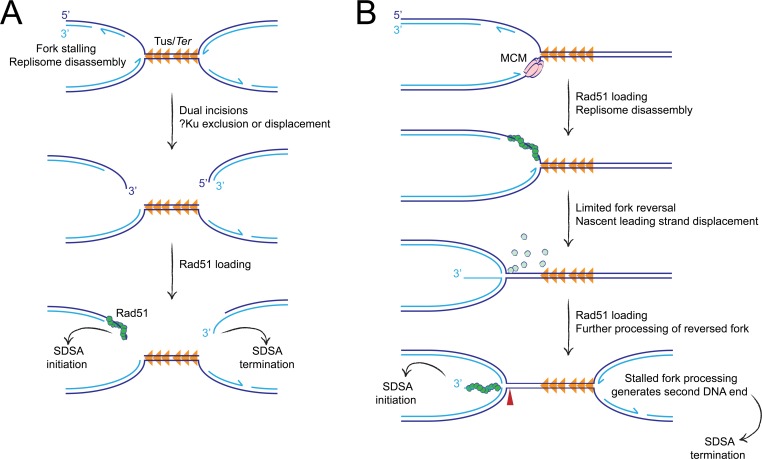
In one model, the processing of the stalled fork might entail production of a conventional DSB, but the ability of Ku to access the DNA ends productively might be impaired (Fig 9A). Indeed, unproductive binding of Ku to presumptive solitary DNA ends at Topoisomerase I inhibitor-induced DNA lesions has been reported [76, 77]. Notably, in these studies, DNA end binding by Ku was shown to modulate repair activity and to influence the requirement for early end resection activities regulated by CtIP and Mre11. In contrast, in our experiments, deletion of Ku70 had no impact on Tus/Ter-induced HR and we found no evidence of an interaction between CtIP and Ku70 in the regulation of Tus/Ter-induced HR. Thus, our findings do not fit readily with the idea that Ku binds unproductively to DSB intermediates during Tus/Ter-induced HR. In an alternative model, protein complexes at the stalled fork might deny Ku access to a conventional two-ended DSB intermediate by an as yet undefined steric exclusion mechanism. The process of V(D)J recombination in developing immune cells provides precedent for such a mechanism; the RAG protein recombination synapse both initiates incision of the recombining locus and helps to channel the DNA ends towards C-NHEJ, disfavoring engagement of alternative end joining pathways [81, 82]. However, none of our findings specifically support this model. Although inactivation of the Fanconi anemia pathway has been reported to promote C-NHEJ-mediated toxic chromosome rearrangements [59, 60], we have not yet found any genetic context in which an interaction between C-NHEJ and Tus/Ter-induced HR is “unmasked”.
Fig 9. Hypothetical models of Tus/Ter-induced HR.
A, Conventional DSB intermediate model. Dual incision of bidirectionally arrested forks generates DNA ends that are processed for HR. Unknown mechanisms prevent Ku access to the DNA ends at the stalled fork. Dark blue: parental strands. Light blue: nascent strands. Half arrows indicate direction of nascent strand synthesis. Orange triangles: Tus/Ter RFB. Green circles: Rad51 monomers. B, Template switch/fork reversal model. Rad51 is loaded onto exposed ssDNA lagging strand daughter strand gaps at the arrested fork. Following replisome disassembly, Rad51 mediates fork remodeling via a template switch mechanism. This process displaces the 3’ ssDNA end of the nascent leading strand, which is rapidly coated with RPA (not shown) followed by Rad51. The DNA end thus generated is incapable of binding Ku, excluding engagement of C-NHEJ. Further processing of the reversed fork may liberate the DNA end by more extensive fork reversal (not shown) and/or via incision of the 4-way reversed fork structure (red arrowhead). Although processing of the two opposing forks is depicted here as sequential, this model is also compatible with synchronous remodeling of both forks. Symbols as in panel A. Pale green circles, Rad51 monomers displaced from lagging strand.
A notable problem with the above-noted models, which invoke a conventional DSB intermediate, is their failure to account for the distinctive pattern of Rad51 accumulation we observe at Tus/Ter. We found that Rad51 accumulation at Tus/Ter is more intense, more sustained and more precisely localized than at a conventional DSB. These findings strongly suggest that the major DNA structures that recruit Rad51 to the Tus/Ter RFB are not conventional DSBs. We propose that Rad51 is recruited to non-DSB ssDNA structures at stalled forks and that the interaction of Rad51 with these structures accounts for the functional exclusion of C-NHEJ from stalled fork HR. A major trigger to Rad51 loading at Tus/Ter may be ssDNA gaps on the arrested lagging strand, present immediately adjacent to the Tus/Ter RFB (Fig 9B). Such ssDNA gaps would be present, albeit transiently, within a normally processive fork. However, fork stalling would render these same DNA structures abnormal, by virtue of their persistence. A static ssDNA signal at the site of stalling could provide a stable platform for the loading of Rad51. By this model, Rad51 might act as an “early responder” during stalled fork repair, as has been suggested previously [83]. If Rad51 deposition were a scheduled, early response to fork stalling, this might explain the intensity and localization of the Rad51 signal we observe at Tus/Ter.
Rad51 supports fork reversal in mammalian cells in response to a variety of DNA damaging agents [83]. Rad51-mediated template switching at the site of stalling could drive limited reversal of the collapsed fork. If initiated by Rad51-coated lagging strand gaps, this process would displace the unresected nascent leading strand as a 3’ ssDNA tail (Fig 9B). Rapid coating of the displaced ssDNA tail by RPA and Rad51 could render it inaccessible to binding by Ku and, hence, “invisible” to the C-NHEJ pathway. The hypothetical limited fork reversal intermediate envisioned by this model might be subject to further processing, leading to more extensive fork reversal and potentially enabling HR initiation without formation of a DSB. Alternatively, incision of the cruciate structure of the reversed fork could liberate a one-ended DSB with a long 3’ ssDNA tail formed by the displaced nascent leading strand. It is not yet clear whether Tus/Ter-induced HR entails the formation of such a DSB intermediate. In summary, a template switch/fork reversal model of HR initiation satisfies two of the key findings reported here: first, the intense, distinctively localized recruitment of Rad51 to the Tus/Ter RFB; second, the functional exclusion of C-NHEJ during Tus/Ter-induced HR. This hypothetical model makes a number of additional predictions, which it will be relevant to test in future studies.
An interesting feature of I-SceI-induced HR was revealed in this study. Specifically, although deletion of Xrcc4 elevated the frequencies of both STGC and LTGC, LTGC products as a proportion of all HR products were reduced from ~4% to ~2% in Xrcc4 null cells. Xrcc4 deletion did not perturb the fundamental relationships of I-SceI-induced HR control reported previously for BRCA1, CtIP, BRCA2 and Rad51 [64]. This suggests that Xrcc4 loss influences the balance between STGC and LTGC via an HR-independent mechanism. I-SceI-induced LTGCs, generated by the HR reporter used here, can be considered a type of gap repair [26]. Thus, I-SceI-induced LTGC might entail repair synthesis in one of two directions. The first would entail Rad51-mediated invasion of the misaligned GFP copy while the second would entail Rad51-mediated invasion of the correctly aligned, unbroken I-SceI site-containing GFP copy. (In the latter case, wtGFP would be generated by annealing at the point of SDSA termination.) In Xrcc4Δ/Δ cells, the loss of high flux error-free religation of I-SceI-induced DSBs might increase the proportion of cells in which I-SceI sites on both sister chromatids are broken simultaneously. In such a circumstance, the second mechanism of LTGC noted above would be suppressed. This, in turn, could lead to the observed reduction in the proportion of I-SceI-induced HR events that resolve as LTGCs in Xrcc4Δ/Δ cells.
Materials and methods
Molecular biology and siRNAs
The 6xTer-HR reporters used were assembled using standard cloning methods described previously for the 6xTer-HR reporter (REF). Stable Ter-containing plasmids were generated and manipulated in JJC33 (Tus–) mutant strains of E. coli. All primers for conventional and quantitative PCR were purchased from Life Technologies. All plasmids used for mouse embryonic stem (ES) cell transfection and 293T cell transfections were prepared by endotoxin-free maxiprep (QIAGEN Sciences, Maryland, MD). siRNA SMARTpools were purchased from GE Healthcare/Dharmacon.
Mouse cell lines and cell culture
Conditional Brca1 mutant mouse ES cell 1xGFP 6xTer reporters were previously described [58]. Conditional Xrcc4 mutant mouse ES cells (cells in which both Xrcc4 copies contained floxed Exon3 alleles) [61] or Ku70 mutant mouse ES cells (cells in which exon 4 and part of exon 5 is replaced with the neomycin resistance cassette [78] were thawed onto MEF feeders and subsequently maintained on gelatinized tissue culture plates in ES medium as described. 20 μg of Kpn I linearlized 6xTer/HR reporter ROSA26 targeting plasmid was introduced by electroporation of 2 x 107 cells. ES cells were plated onto 6-cm dishes containing Puromycin-resistant feeders and after 18 hours plates were supplemented with 4 μg/mL Puromycin for 24 hours. Individual colonies were picked for expansion between 9 and 14 days later. Multiple ROSA26 targeted lines were identified by PCR. HR cassette ROSA26 integration and overall structure was verified for targeted lines by Southern blotting. Multiple Xrcc4-deficient ES clones were generated by transient adenovirus-mediated Cre expression and excision of Xrcc4 Exon3. ROSA26 genotyping primers: ROSA26-sense-(CAT CAA GGA AAC CCT GGA CTA CTG); Ter-HR reporter antisense-(cct cgg cta ggt agg gga tc). KU70 status was verified by PCR: KU70 exon4 5’-sense-(CCA GTA AGA TCA TAA GCA GCG ATC G); KU70 exon5 3’-antisense-(CTC TTG TGA CTC ATC TTG AGC TGG); Exon 4/5-neo-deleted allele, KU70 3’- antisense-(GCC GAA TAG CCT CTC CAC CCA AGC G). Xrcc4 status was determined by PCR: Xrcc4 5’-sense-(ttc agc taa cca gca tca ata g); floxed allele, Xrcc4 3’-antisense-(gca cct ttg cct act aag cca tct cac); Exon 3-deleted allele, Xrcc4 3’- antisense-(taa gct att act cct gca tgg agc att atc acc). Exon3-deleted, Xrcc4-deficient mES cells were transduced with lentivirus expressing a single mRNA encoding nourseothricin acetyl transferase and human CD52 (the CAMPATH antigen), with or without wild type, hemagglutinin-epitope tagged mouse Xrcc4: pHIV-NAT-hCD52-EV (empty vector control) or pHIV-NAT-hCD52-mXrcc4. Stable cultures were selected and maintained in 100 μg/mL nourseothricin (Jenna Bioscience, AB-102L).
Lentivirus production and target mouse cell transduction
293T cells were propagated in standard DMEM media supplemented with 10% serum, glutamine and antibiotics. For lentivirus generation, 8 x 106 cells were seeded on 10 cm dishes and transfected 24 hours later with 5 μg pHIV, 4.45 μg psPAX2, and 0.55 μg pMD2G in antibiotic-free media using Lipofectamine 2000 (Invitrogen). Media was replaced 24 hours later, and supernatant harvested every 12 hours between 48 and 72 hours after transfection and stored at 4°C. Lentiviral particles were concentrated using Centricon Plus-70 filter devices (Millipore) per manufacturer’s instructions. 5 x 105 target mES cells were seeded per well in 6-well plate format, allowed to proliferate for 24 hours, transduced and placed under 100 μg/mL nourseothricin selection beginning 24 hours after transduction.
Recombination assays
1.6 x 105 cells were co-transfected in suspension with 0.35 μg empty vector, pcDNA3β-myc NLS-Tus, or pcDNA3β-myc NLS-I-SceI, and 20 pmol ONTargetPlus-smartpool using Lipofectamine 2000 (Invitrogen). GFP+RFP–, GFP+RFP+ and GFP–RFP+ frequencies were scored 72 hours after transfection by flow cytometry using a Becton Dickinson 5 Laser LSRII or or Beckman Coulter CytoFlex LX in duplicate. For each duplicate sample condition, 3–6 x 105 total events were scored. Repair frequencies presented are corrected for background events and for transfection efficiency (50–85%). Transfection efficiency was measured by parallel transfection with 0.05 μg wild type GFP expression vector, 0.30 μg control vector and 20 pmol siRNA. For transient mXrcc4 rescue experiments, 1.6 x 105 cells were co-transfected in suspension with 0.4 μg empty vector, pcDNA3β-myc NLS-Tus [56], or pcDNA3β-myc NLS-I-SceI [62], and either 0.1 μg empty vector, or pcDNA3β-HA-Xrcc4 using Lipofectamine 2000. For transient hKU70 rescue experiments, 1.6 x 105 cells were co-transfected in suspension with 0.35 μg empty vector, pcDNA3β-myc NLS-Tus, or pcDNA3β-myc NLS-I-SceI, and either 0.15 μg empty vector, or pcDNA3β-hKU70 using Lipofectamine 2000. For transient hKU70 rescue experiments including siRNA treatment, 1.6 x 105 cells were co-transfected in suspension with 0.35 μg empty vector, pcDNA3β-myc NLS-Tus, or pcDNA3β-myc NLS-I-SceI, and either 0.15 μg empty vector, or pcDNA3β-hKU70 using Lipofectamine 2000 and 20 pmol siRNA.
RT-qPCR analysis
RNA isolated from cells 48 hours after transfection was extracted using QIAGEN RNeasy Mini Kit (QIAGEN Sciences, Maryland, MD) 48 hours after transfection. All analyses of GAPDH and siRNA-targeted genes was performed using an Applied Biosystems 7300 Real time PCR System using Power SYBR Green RNA-to CTTM 1-Step Kit (Applied Biosystems, Foster City, CA). SYBR green RT-qPCR assays were performed using gene-specific primer sequences identified using the NIH NCBI Nucleotide utility for GAPDH, Slx4, Brca1, Brca2, CtIP, and Polq. Primers for RT-PCR: GAPDH-sense-(CGT CCC GTA GAC AAA ATG GT); GAPDH-antisense-(TCG TTG ATG GCA ACA ATC TC); Slx4-sense-(GTG GGA CGA CTG GAA TGA GG); Slx4-antisense-(GCA CCT TTT GGT GTC TCT GG); Brca1-sense-(ATG AGC TGG AGA GGA TGC TG); Brca1-antisense-(CTG GGC AGT TGC TGT CTT CT); Brca2-sense-(TCT GCC ACT GTG AAA AAT GC); Brca2-antisense-(TCA AGC TGG GCT GAA GAT T); CtIP-sense-(AGG AGA AGG AGG GGA CGC); CtIP-antisense-(TGA AAT ACC TCG GCG GGT G); Polq-sense-(TGC TTG GTC ACG TCT TGG AA); Polq-antisense-(CCT GAA ACA GAC TCT GGA GGT). mRNA was measured in triplicates. siRNA-target gene expression level was normalized to GAPDH and expressed as a fold difference from siLuciferase control treated samples analyzed in the same experiment (x = -2ΔΔCt, with ΔΔCt = [Ct target-CtGapdh]-[CtsiLUCIFERASE-CtsiGAPDH]). Error-bars represent the standard deviation of ΔCt (SDEV = √[SDEVTARGET2 + SDEVGAPDH2]). We used the Roche ProbeFinder utility based on Primer 3 software (Whitehead Institute, MIT) to generate gene-specific primer sequences for mouse Xrcc4 and human KU70: Xrcc4-sense-(AAA TGG CTC CAC AGG AGT TG); Xrcc4-antisense-(GGT GCT CTC CTC TTT CAA GG); KU70-sense-(ACA AGT ACA GGC GGT TTG CT); KU70-antisense-(TTC AGC AGT ACC AAC GGC TT). Xrcc4-specific primers mapped to exon 6 and the exon 6–7 boundary and hKU70-specific primers mapped to exon 7 and the exon 8, respectively. Xrcc4 gene expression level was normalized to GAPDH and expressed as a fold difference from a Xrcc4fl/fl reporter clone sample analyzed in the same experiment (x = -2ΔΔCt, with ΔΔCt = [CtXrcc4-CtGapdh]-[CtsiLUCIFERASE-CtsiGAPDH]). KU70 gene expression level was normalized to GAPDH and expressed as a fold difference from one Ku70Δ/Δ reporter clone sample analyzed in the same experiment (x = -2ΔΔCt, with ΔΔCt = [CtKu70-CtGapdh]-[CtsiLUCIFERASE-CtsiGAPDH]). Error-bars represent the standard deviation of the ΔCt value (SDEV = √[SDEVGene2 + SDEVGAPDH2]).
Western blotting
Cell lysates were prepared from cells 48 hours after transfection lysed in RIPA buffer (50mM Tris-HCl, pH 8.0, 250 mM NaCl, 0.1% sodium dodecyl sulfate, 1% NP-40 containing the protease inhibitors, PMSF, and Roche complete protease inhibitor tablet) and 10–30 μg resolved by 4–12% Bis-Tris SDS-PAGE (Invitrogen). Protein expression was analyzed by immunoblotting using the following antibodies; hRad51 (aliquot B32, 1:500), mXrcc4 (Abcam ab97351, 1:3,000), hKU70 (Thermofisher PA5-27538, 1:1000), mBrca1 (AB191042, 1:1000), HA (Abcam, ab18181, 1:500), beta-tubulin (Abcam ab6046, 1:4,000).
Cell staining
Live cells were prepared for measurement of cell surface expression of human CD52 as previously described. Cells were trypsinized and resuspended in FACS blocking buffer (PBS containing 1% BSA, 0.1% sodium-azide, and 5% heat-inactivated goat serum). Cells were stained for CD52 in blocking buffer: primary antibody, rat anti-hCD52 mAb YTH 34.5, 1:200 (Bio-Rad AbD Serotec Inc. MCA-1642); secondary antibody, Alexa-488 AffiniPure Goat anti-Rat IgG, 1:50 (Jackson Immunoresearch, 112-545-167). Stained cells were fixed in PBS containing 0.5% BSA, 0.05% sodium-azide, 1.5% paraformaldehyde, 1% sucrose prior to flow cytometric analysis. Cell staining was measured by flow cytometry using a Becton Dickinson 5 Laser LSRII or Beckman Coulter CytoFlex LX.
Competition assays
For Xrcc4 mutant cell competition experiments, 1.6 x 105 cells were co-transfected in suspension with 0.45 μg empty vector and either 50 ng empty vector or 50 ng GFP-expression plasmid using Lipofectamine 2000 (Invitrogen). For KU70 complementation cell competition experiments, 1.6 x 105 cells were co-transfected in suspension with 0.35 μg empty vector, 0.15 μg empty vector or hKU70-expression plasmid, and either 50 ng empty vector or 50 ng GFP-expression plasmid using Lipofectamine 2000 (Invitrogen). 18 hours after transfection, cells were counted, mixed 5:1, uncolored vs. GFP+ marked cells, and 5 x 104 cells plated in triplicate. 6 hours after cell plating growth medium was replaced with media containing phleomycin (Sigma-aldrich, P9564). After two days incubation, GFP+ frequencies were scored on a Beckman Coulter CytoFlex LX. Fold enrichment of cultures transiently co-transfected with GFP-expression plasmid normalized to 0 μg/mL phleomycin control. Plots represent the mean of triplicate samples from three independent experiments (n = 3).
Chromatin immunoprecipitation assays
24–48 parallel transfections of 1.6 x 105 cells were performed in suspension with 0.5 μg empty vector, pcDNA3β-myc NLS-Tus-F140A-3xHA, or pcDNA3β-myc NLS-I-SceI, or co-transfected with 0.45 μg spCas9 expression plasmid with either control (CAT CCT CGG CAC CGT CAC CC) or I-SceI-specific (GGA TAA CAG GGT AAT CAA GG) guide RNAs (in vitro transcribed, Engen sgRNA Synthesis kit, S. pyogenes, New England Biolabs E3322S, purified using RNA Clean and Concentrator Kit, Zymo Research, R1017, and quality assessed by denaturing 10% TBE-urea acrylamide gel run) using Lipofectamine 2000 (Invitrogen). 10 million 1xGFP 6xTer reporter cells [58] 24 or 48 hours after transfection were collected for chromatin immunoprecipitation (ChIP). Cells were fixed in serum free mES cell media containing 1% formaldehyde at room temperature, incubating for 15 min with gentle orbital shaking. Fixation was quenched by addition of glycine to 125 mM. Cells were lysed in lysis buffer (0.1% SDS, 20 mM EDTA, 50 mM Tris pH 8.1) containing protease inhibitor (PMSF supplemented with Roche protease inhibitor, Roche 13539320). All subsequent steps were performed in low DNA binding tubes (Fischer Scientific, 022431021). Chromatin shearing to 100–2000 bp was performed using Diagenode Bioruptor 300 with optional attached 4°C chiller. The predominant product size of ~500 bp as achieved by 20 sonication cycles, 15 seconds on and 30 seconds off. 100 μl lysate per ChIP reaction was precleared by addition of 10 μl Magna ChIP magnetic beads (Millipore Sigma, 16–663) in ChIP dilution buffer (1% Triton-X-100, 2mM EDTA, 150mM NaCl, 20mM Tris pH 8.1). Rad51 was immunoprecipitated by addition of 3 μg anti-Rad51 ChIP-grade antibody (Abcam, ab176458) and 12 hour incubation at 4°C on a Nutator mixer followed by addition of 10 μl Magna ChIP magnetic beads and additional 16 hour incubation at 4°C. Beads were washed six times using ice-cold ChIP RIPA buffer (50mM HEPES pH 7.6, 1mM EDTA, 7 mg/mL sodium deoxycholate, 1% NP-40). DNA was eluted in Elution buffer (1% SDS, 200mM sodium bicarbonate, 5.6 μg/mL RNAse A) and cross-links were reversed by 65°C overnight incubation. Protein was removed by proteinase K digest 30 min at 55°C. DNA purified by Qiagen PCR Purification column (Qiagen, 28106) was analyzed by qPCR using an ABI Prism 7300 sequence detection system and SYBR Green (Applied Biosystems, 4368702). Primers for qPCR: 79 bp CEN-sense-(CAA CAG CCA CAA CGT CTA TAT CAT G); 79 bp CEN-antisense-(ATG TTG TGG CGG ATC TTG AAG); 1.3 kp CEN-sense-(CAC CAC AAA TCG AGG CTG TA); 1.3 kp CEN-antisense-(GGA TCA AGG CAA AGG ATC AA); 4.1 kp CEN-sense-(TCC GGT GAA TAG GCA GAG TT); 4.1 kp CEN-antisense-(CAG GGA AAC CCA AAG AAG TG); 7.1 kp CEN-sense-(TGC AAA AAC CAT CCA AAC AA); 7.1 kp CEN-antisense-(GTG GAG GCT AGA AGC TGG TG); 165 bp TEL-sense-(TGG TGA GCA AGG GCG AGG AGC); 165 bp TEL-antisense-(TCG TGC TGC TTC ATG TGG TCG); 2.1 kp TEL-sense-(GGG AGG CTA ACT GAA ACA CG); 165 bp TEL-antisense-(GGT GGG GTA TCG ACA GAG TG); 3.1 kp TEL-sense-(GCA CGT TTC CGA CTT GAG TT); 165 bp TEL-antisense-(TCA GAG CGA CTT TGG GAG AG); 11 kp TEL-sense-(CAG GAA TTC TTT CCC CAC AA); 165 bp TEL-antisense-(TGC CAG GTC TCT AGG GCT TA). Data are presented as the mean calculated from three independent experiments (n = 3) normalized against untreated controls (empty vector or guide RNA controls) and control locus (beta-actin) using the 2-ΔΔCT method as described previously [84].
Statistical methods
Data presented represents the arithmetic mean and error bars represent the standard error of the mean (s.e.m.) of between three (n = 3) and nine (n = 9) independent experiments (n values given in figure legends). Figure legends specify the number of replicates within each independent experiment (performed in duplicate) and the number of independent experiments (n) that were performed to generate the data presented. The arithmetic mean of samples collected for groups of independent experiments for repair frequency statistical analysis, was calculated and data points for each independent experiment used to calculate the mean and standard error of the mean (s.e.m.), calculated as standard deviation/√n, (n indicates the number of independent experiments). Differences between sample pairs repair frequencies were analyzed by Student’s two-tailed unpaired t-test, assuming unequal variance. One-way ANOVA statistical analysis of greater than three samples was performed when indicated. P-values are indicated in the figure legends. All statistics were performed using GraphPad Prism v6.0d software.
Supporting information
A, Map of pHIV-NAT-hCD52 indicating key elements and location of mXrcc4 cDNA insert: EF1α: elongation factor alpha promoter; IRES: internal ribosomal entry signal; NAT: nourseothricin (NTC) acetyl transferase resistance gene: T2A, Thosea asigna viral self-cleaving peptide; CD52: human CAMPATH antigen cDNA. B, Primary FACS histogram data for mES reporter cells expressing human CD52. Left panel: Xrcc4Δ/Δ reporter cells transiently transfected with pHIV-NAT-hCD52 vector, stained with either no primary antibody (grey) or anti-hCD52 mAb YTH 34.5 (green). Right panel: hCD52 cell surface expression in Xrcc4Δ/Δ reporter cells following lentiviral transduction and selection in 100 μg/ml NTC. Gray histogram: pHIV-NAT-transduced cells. Green histogram: pHIV-NAT-hCD52-transduced cells. Inset numbers indicate median CD52 staining intensity.
(PDF)
A, Frequencies of Tus/Ter-induced and I-SceI-induced repair of Xrcc4fl/fl clone #8 or Xrcc4Δ/Δ clone #11 6xTer-HR reporter cells transiently expressing exogenous Xrcc4. Cells were transiently co-transfected with empty pcDNA3beta or pcDNA3beta-HA-mXrcc4 expression vector and empty, 3xMyc-NLS Tus or 3xMyc-NLS I-SceI expression vectors. Each plot represents the mean of duplicate samples from nine independent experiments (n = 9). Error bars: s.e.m. Tus-induced Total HR, t-test: flox8 +Xrcc4 vs. flox8 +EV p = 0.0048; del11 +Xrcc4 vs. del11 +EV p = 0.7251; del11 +EV vs. flox8 +EV p = 0.0002; del11 +Xrcc4 vs. flox8 +Xrcc4 p = 0.4659; del11 +Xrcc4 vs. flox8 +EV p = 0.0005. Tus-induced STGC, t-test: flox8 +Xrcc4 vs. flox8 +EV p = 0.0035; del11 +Xrcc4 vs. del11 +EV p = 0.7076; del11 +EV vs. flox8 +EV p = 0.0003; del11 +Xrcc4 vs. flox8 +Xrcc4 p = 0.4650; del11 +Xrcc4 vs. flox8 +EV p = 0.0005. Tus-induced LTGC, t-test: flox8 +Xrcc4 vs. flox8 +EV p = 0.5738; del11 +Xrcc4 vs. del11 +EV p = 0.8987; del11 +EV vs. flox8 +EV p = 0.3906; del11 +Xrcc4 vs. flox8 +Xrcc4 p = 0.8369; del11 +Xrcc4 vs. flox8 +EV p = 0.5332. Tus-induced Ratio, t-test: flox8 +Xrcc4 vs. flox8 +EV p = 0.0513; del11 +Xrcc4 vs. del11 +EV p = 0.7973; del11 +EV vs. flox8 +EV p = 0.2700; del11 +Xrcc4 vs. flox8 +Xrcc4 p = 0.7369; del11 +Xrcc4 vs. flox8 +EV p = 0.1734. I-SceI-induced Total HR, t-test: flox8 +Xrcc4 vs. flox8 +EV p = 0.0398; del11 +Xrcc4 vs. del11 +EV p<0.0001; del11 +EV vs. flox8 +EV p<0.0001; del11 +Xrcc4 vs. flox8 +Xrcc4 p = 0.5330; del11 +Xrcc4 vs. flox8 +EV p = 0.0034. I-SceI-induced STGC, t-test: flox8 +Xrcc4 vs. flox8 +EV p = 0.0391; del11 +Xrcc4 vs. del11 +EV p<0.0001; del11 +EV vs. flox8 +EV p<0.0001; del11 +Xrcc4 vs. flox8 +Xrcc4 p = 0.5491; del11 +Xrcc4 vs. flox8 +EV p = 0.0039. I-SceI-induced LTGC, t-test: flox8 +Xrcc4 vs. flox8 +EV p = 0.4390; del11 +Xrcc4 vs. del11 +EV p<0.0001; del11 +EV vs. flox8 +EV p = 0.0002; del11 +Xrcc4 vs. flox8 +Xrcc4 p = 0.6173; del11 +Xrcc4 vs. flox8 +EV p = 0.2036. I-SceI-induced Ratio, t-test: flox8 +Xrcc4 vs. flox8 +EV p = 0.9640; del11 +Xrcc4 vs. del11 +EV p<0.0001; del11 +EV vs. flox8 +EV p = 0.0012; del11 +Xrcc4 vs. flox8 +Xrcc4 p = 0.8666; del11 +Xrcc4 vs. flox8 +EV p = 0.9242. B, RT qPCR analysis of Xrcc4 in transfected Xrcc4fl/fl or Xrcc4Δ/Δ clones. Xrcc4 expression normalized to GAPDH and displayed as fold difference from Xrcc4fl/fl parental reporter clone 8 of the same experiment (x = -2ΔΔCt, with ΔΔCt = [CtXrcc4-CtGapdh]-[CtXrcc4-CtGAPDH]). Error-bars represent standard deviation of the ΔCt value (SDEV = √[SDEVXrcc42 + SDEVGAPDH2]). C, Western blot for abundance of HA-tagged mXrcc4 protein in Xrcc4fl/fl clone 8 or Xrcc4Δ/Δ clone 11 6xTer-HR reporter cells transiently transfected with empty pcDNA3beta or pcDNA3beta-mXrcc4.
(PDF)
Acknowledgments
We thank Dr. Johannes Walter and members of the Scully lab for helpful discussions, and Dr. Frederick Alt for providing Ku70–/–mouse ES cells. We thank Dr. Bryan Welm for helpful advice regarding lentiviral expression vectors.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by awards from the National Cancer Institute (https://www.cancer.gov/): P50CA168504 (NAW), R01CA095175 (RS), and R01CA217991 (RS). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Zeman MK, Cimprich KA. Causes and consequences of replication stress. Nat Cell Biol. 2014;16(1):2–9. 10.1038/ncb2897 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duxin JP, Walter JC. What is the DNA repair defect underlying Fanconi anemia? Current opinion in cell biology. 2015;37:49–60. 10.1016/j.ceb.2015.09.002 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.D'Andrea AD. Susceptibility pathways in Fanconi's anemia and breast cancer. The New England journal of medicine. 2010;362(20):1909–19. 10.1056/NEJMra0809889 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garcia-Muse T, Aguilera A. Transcription-replication conflicts: how they occur and how they are resolved. Nat Rev Mol Cell Biol. 2016. 10.1038/nrm.2016.88 . [DOI] [PubMed] [Google Scholar]
- 5.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40(2):179–204. Epub 2010/10/23. S1097-2765(10)00747-1 [pii] 10.1016/j.molcel.2010.09.019 ; PubMed Central PMCID: PMC2988877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319(5868):1352–5. 10.1126/science.1140735 . [DOI] [PubMed] [Google Scholar]
- 7.Lopes M, Cotta-Ramusino C, Pellicioli A, Liberi G, Plevani P, Muzi-Falconi M, et al. The DNA replication checkpoint response stabilizes stalled replication forks. Nature. 2001;412(6846):557–61. 10.1038/35087613 . [DOI] [PubMed] [Google Scholar]
- 8.Errico A, Costanzo V. Mechanisms of replication fork protection: a safeguard for genome stability. Crit Rev Biochem Mol Biol. 2012;47(3):222–35. 10.3109/10409238.2012.655374 . [DOI] [PubMed] [Google Scholar]
- 9.Long DT, Joukov V, Budzowska M, Walter JC. BRCA1 promotes unloading of the CMG helicase from a stalled DNA replication fork. Mol Cell. 2014;56(1):174–85. 10.1016/j.molcel.2014.08.012 ; PubMed Central PMCID: PMC4185004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011;145(4):529–42. Epub 2011/05/14. S0092-8674(11)00375-8 [pii] 10.1016/j.cell.2011.03.041 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schlacher K, Wu H, Jasin M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 2012;22(1):106–16. Epub 2012/07/14. 10.1016/j.ccr.2012.05.015 S1535-6108(12)00214-0 [pii]. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neelsen KJ, Lopes M. Replication fork reversal in eukaryotes: from dead end to dynamic response. Nat Rev Mol Cell Biol. 2015;16(4):207–20. 10.1038/nrm3935 . [DOI] [PubMed] [Google Scholar]
- 13.Petermann E, Helleday T. Pathways of mammalian replication fork restart. Nat Rev Mol Cell Biol. 2010;11(10):683–7. Epub 2010/09/16. nrm2974 [pii] 10.1038/nrm2974 . [DOI] [PubMed] [Google Scholar]
- 14.Carr AM, Lambert S. Replication Stress-Induced Genome Instability: The Dark Side of Replication Maintenance by Homologous Recombination. J Mol Biol. 2013. Epub 2013/05/07. S0022-2836(13)00271-4 [pii] 10.1016/j.jmb.2013.04.023 . [DOI] [PubMed] [Google Scholar]
- 15.Osman F, Whitby MC. Exploring the roles of Mus81-Eme1/Mms4 at perturbed replication forks. DNA Repair (Amst). 2007;6(7):1004–17. Epub 2007/04/06. S1568-7864(07)00070-5 [pii] 10.1016/j.dnarep.2007.02.019 . [DOI] [PubMed] [Google Scholar]
- 16.Chen C, Kolodner RD. Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nature genetics. 1999;23(1):81–5. 10.1038/12687 [DOI] [PubMed] [Google Scholar]
- 17.Myung K, Datta A, Kolodner RD. Suppression of spontaneous chromosomal rearrangements by S phase checkpoint functions in Saccharomyces cerevisiae. Cell. 2001;104(3):397–408. [DOI] [PubMed] [Google Scholar]
- 18.Lambert S, Mizuno K, Blaisonneau J, Martineau S, Chanet R, Freon K, et al. Homologous recombination restarts blocked replication forks at the expense of genome rearrangements by template exchange. Mol Cell. 2010;39(3):346–59. Epub 2010/08/14. 10.1016/j.molcel.2010.07.015 . [DOI] [PubMed] [Google Scholar]
- 19.Lambert S, Watson A, Sheedy DM, Martin B, Carr AM. Gross chromosomal rearrangements and elevated recombination at an inducible site-specific replication fork barrier. Cell. 2005;121(5):689–702. 10.1016/j.cell.2005.03.022 . [DOI] [PubMed] [Google Scholar]
- 20.Raschle M, Knipscheer P, Enoiu M, Angelov T, Sun J, Griffith JD, et al. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell. 2008;134(6):969–80. 10.1016/j.cell.2008.08.030 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Branzei D, Foiani M. Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol. 2010;11(3):208–19. Epub 2010/02/24. nrm2852 [pii] 10.1038/nrm2852 . [DOI] [PubMed] [Google Scholar]
- 22.Sogo JM, Lopes M, Foiani M. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science. 2002;297(5581):599–602. 10.1126/science.1074023 . [DOI] [PubMed] [Google Scholar]
- 23.Amunugama R, Willcox S, Wu RA, Abdullah UB, El-Sagheer AH, Brown T, et al. Replication Fork Reversal during DNA Interstrand Crosslink Repair Requires CMG Unloading. Cell reports. 2018;23(12):3419–28. 10.1016/j.celrep.2018.05.061 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mizuno K, Lambert S, Baldacci G, Murray JM, Carr AM. Nearby inverted repeats fuse to generate acentric and dicentric palindromic chromosomes by a replication template exchange mechanism. Genes Dev. 2009;23(24):2876–86. 10.1101/gad.1863009 ; PubMed Central PMCID: PMC2800087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hastings PJ, Ira G, Lupski JR. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS genetics. 2009;5(1):e1000327 Epub 2009/01/31. 10.1371/journal.pgen.1000327 ; PubMed Central PMCID: PMC2621351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiology and molecular biology reviews: MMBR. 1999;63(2):349–404. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hartlerode AJ, Scully R. Mechanisms of double-strand break repair in somatic mammalian cells. Biochem J. 2009;423(2):157–68. 10.1042/BJ20090942 ; PubMed Central PMCID: PMC2983087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mayle R, Campbell IM, Beck CR, Yu Y, Wilson M, Shaw CA, et al. Mus81 and converging forks limit the mutagenicity of replication fork breakage. Science. 2015;349(6249):742–7. 10.1126/science.aaa8391 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anand RP, Lovett ST, Haber JE. Break-induced DNA replication. Cold Spring Harb Perspect Biol. 2013;5(12). Epub 2013/07/25. 10.1101/cshperspect.a010397 a010397 [pii] cshperspect.a010397 [pii]. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pierce AJ, Hu P, Han M, Ellis N, Jasin M. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes & Development. 2001;15(24):3237–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vriend LE, Prakash R, Chen CC, Vanoli F, Cavallo F, Zhang Y, et al. Distinct genetic control of homologous recombination repair of Cas9-induced double-strand breaks, nicks and paired nicks. Nucleic Acids Res. 2016. 10.1093/nar/gkw179 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Delacote F, Han M, Stamato TD, Jasin M, Lopez BS. An xrcc4 defect or Wortmannin stimulates homologous recombination specifically induced by double-strand breaks in mammalian cells. Nucleic Acids Research. 2002;30(15):3454–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim H, D'Andrea AD. Regulation of DNA cross-link repair by the Fanconi anemia/BRCA pathway. Genes Dev. 2012;26(13):1393–408. 10.1101/gad.195248.112 ; PubMed Central PMCID: PMC3403008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knipscheer P, Raschle M, Smogorzewska A, Enoiu M, Ho TV, Scharer OD, et al. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science. 2009;326(5960):1698–701. Epub 2009/12/08. 10.1126/science.1182372 ; PubMed Central PMCID: PMC2909596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bunting SF, Callen E, Kozak ML, Kim JM, Wong N, Lopez-Contreras AJ, et al. BRCA1 functions independently of homologous recombination in DNA interstrand crosslink repair. Mol Cell. 2012;46(2):125–35. 10.1016/j.molcel.2012.02.015 ; PubMed Central PMCID: PMC3340543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang J, Dewar JM, Budzowska M, Motnenko A, Cohn MA, Walter JC. DNA interstrand cross-link repair requires replication-fork convergence. Nature structural & molecular biology. 2015;22(3):242–7. 10.1038/nsmb.2956 ; PubMed Central PMCID: PMC4351167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang H, Li Q, Fan J, Holloman WK, Pavletich NP. The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA-ssDNA junction. Nature. 2005;433(7026):653–7. 10.1038/nature03234 . [DOI] [PubMed] [Google Scholar]
- 38.San Filippo J, Chi P, Sehorn MG, Etchin J, Krejci L, Sung P. Recombination mediator and Rad51 targeting activities of a human BRCA2 polypeptide. J Biol Chem. 2006;281(17):11649–57. 10.1074/jbc.M601249200 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thorslund T, McIlwraith MJ, Compton SA, Lekomtsev S, Petronczki M, Griffith JD, et al. The breast cancer tumor suppressor BRCA2 promotes the specific targeting of RAD51 to single-stranded DNA. Nature structural & molecular biology. 2010;17(10):1263–5. Epub 2010/08/24. nsmb.1905 [pii] 10.1038/nsmb.1905 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jensen RB, Carreira A, Kowalczykowski SC. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 2010;467(7316):678–83. Epub 2010/08/24. nature09399 [pii] 10.1038/nature09399 ; PubMed Central PMCID: PMC2952063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao W, Steinfeld JB, Liang F, Chen X, Maranon DG, Jian Ma C, et al. BRCA1-BARD1 promotes RAD51-mediated homologous DNA pairing. Nature. 2017. 10.1038/nature24060 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whitby MC. The FANCM family of DNA helicases/translocases. DNA Repair (Amst). 2010;9(3):224–36. 10.1016/j.dnarep.2009.12.012 . [DOI] [PubMed] [Google Scholar]
- 43.Xue X, Sung P, Zhao X. Functions and regulation of the multitasking FANCM family of DNA motor proteins. Genes Dev. 2015;29(17):1777–88. 10.1101/gad.266593.115 ; PubMed Central PMCID: PMC4573851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mizuno K, Miyabe I, Schalbetter SA, Carr AM, Murray JM. Recombination-restarted replication makes inverted chromosome fusions at inverted repeats. Nature. 2013;493(7431):246–9. 10.1038/nature11676 ; PubMed Central PMCID: PMC3605775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nguyen MO, Jalan M, Morrow CA, Osman F, Whitby MC. Recombination occurs within minutes of replication blockage by RTS1 producing restarted forks that are prone to collapse. eLife. 2015;4:e04539 10.7554/eLife.04539 ; PubMed Central PMCID: PMC4407270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Larsen NB, Hickson ID, Mankouri HW. Tus-Ter as a tool to study site-specific DNA replication perturbation in eukaryotes. Cell Cycle. 2014;13(19):2994–8. 10.4161/15384101.2014.958912 ; PubMed Central PMCID: PMC4614373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Larsen NB, Liberti SE, Vogel I, Jorgensen SW, Hickson ID, Mankouri HW. Stalled replication forks generate a distinct mutational signature in yeast. Proc Natl Acad Sci U S A. 2017;114(36):9665–70. 10.1073/pnas.1706640114 ; PubMed Central PMCID: PMC5594675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Larsen NB, Sass E, Suski C, Mankouri HW, Hickson ID. The Escherichia coli Tus-Ter replication fork barrier causes site-specific DNA replication perturbation in yeast. Nat Commun. 2014;5:3574 10.1038/ncomms4574 . [DOI] [PubMed] [Google Scholar]
- 49.Saini N, Ramakrishnan S, Elango R, Ayyar S, Zhang Y, Deem A, et al. Migrating bubble during break-induced replication drives conservative DNA synthesis. Nature. 2013;502(7471):389–92. Epub 2013/09/13. 10.1038/nature12584 nature12584 [pii]. ; PubMed Central PMCID: PMC3804423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sakofsky CJ, Malkova A. Break induced replication in eukaryotes: mechanisms, functions, and consequences. Crit Rev Biochem Mol Biol. 2017;52(4):395–413. 10.1080/10409238.2017.1314444 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Feng Z, Scott SP, Bussen W, Sharma GG, Guo G, Pandita TK, et al. Rad52 inactivation is synthetically lethal with BRCA2 deficiency. Proc Natl Acad Sci U S A. 2011;108(2):686–91. 10.1073/pnas.1010959107 ; PubMed Central PMCID: PMC3021033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bhowmick R, Minocherhomji S, Hickson ID. RAD52 Facilitates Mitotic DNA Synthesis Following Replication Stress. Mol Cell. 2016;64(6):1117–26. 10.1016/j.molcel.2016.10.037 . [DOI] [PubMed] [Google Scholar]
- 53.Sotiriou SK, Kamileri I, Lugli N, Evangelou K, Da-Re C, Huber F, et al. Mammalian RAD52 Functions in Break-Induced Replication Repair of Collapsed DNA Replication Forks. Mol Cell. 2016;64(6):1127–34. 10.1016/j.molcel.2016.10.038 ; PubMed Central PMCID: PMC5179496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mulcair MD, Schaeffer PM, Oakley AJ, Cross HF, Neylon C, Hill TM, et al. A molecular mousetrap determines polarity of termination of DNA replication in E. coli. Cell. 2006;125(7):1309–19. 10.1016/j.cell.2006.04.040 . [DOI] [PubMed] [Google Scholar]
- 55.Elshenawy MM, Jergic S, Xu ZQ, Sobhy MA, Takahashi M, Oakley AJ, et al. Replisome speed determines the efficiency of the Tus-Ter replication termination barrier. Nature. 2015;525(7569):394–8. 10.1038/nature14866 . [DOI] [PubMed] [Google Scholar]
- 56.Willis NA, Chandramouly G, Huang B, Kwok A, Follonier C, Deng C, et al. BRCA1 controls homologous recombination at Tus/Ter-stalled mammalian replication forks. Nature. 2014;510(7506):556–9. 10.1038/nature13295 ; PubMed Central PMCID: PMC4118467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Willis NA, Scully R. Spatial separation of replisome arrest sites influences homologous recombination quality at a Tus/Ter-mediated replication fork barrier. Cell Cycle. 2016;15(14):1812–20. 10.1080/15384101.2016.1172149 ; PubMed Central PMCID: PMC4968906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Willis NA, Frock RL, Menghi F, Duffey EE, Panday A, Camacho V, et al. Mechanism of tandem duplication formation in BRCA1-mutant cells. Nature. 2017;551(7682):590–5. 10.1038/nature24477 ; PubMed Central PMCID: PMC5728692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Adamo A, Collis SJ, Adelman CA, Silva N, Horejsi Z, Ward JD, et al. Preventing nonhomologous end joining suppresses DNA repair defects of Fanconi anemia. Mol Cell. 2010;39(1):25–35. 10.1016/j.molcel.2010.06.026 . [DOI] [PubMed] [Google Scholar]
- 60.Pace P, Mosedale G, Hodskinson MR, Rosado IV, Sivasubramaniam M, Patel KJ. Ku70 corrupts DNA repair in the absence of the Fanconi anemia pathway. Science. 2010;329(5988):219–23. 10.1126/science.1192277 . [DOI] [PubMed] [Google Scholar]
- 61.Yan CT, Kaushal D, Murphy M, Zhang Y, Datta A, Chen C, et al. XRCC4 suppresses medulloblastomas with recurrent translocations in p53-deficient mice. Proc Natl Acad Sci U S A. 2006;103(19):7378–83. 10.1073/pnas.0601938103 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Puget N, Knowlton M, Scully R. Molecular analysis of sister chromatid recombination in mammalian cells. DNA Repair (Amst). 2005;4(2):149–61. 10.1016/j.dnarep.2004.08.010 ; PubMed Central PMCID: PMC2967438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nagaraju G, Odate S, Xie A, Scully R. Differential regulation of short- and long-tract gene conversion between sister chromatids by Rad51C. Mol Cell Biol. 2006;26(21):8075–86. 10.1128/MCB.01235-06 ; PubMed Central PMCID: PMC1636746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chandramouly G, Kwok A, Huang B, Willis NA, Xie A, Scully R. BRCA1 and CtIP suppress long-tract gene conversion between sister chromatids. Nat Commun. 2013;4:2404 Epub 2013/09/03. 10.1038/ncomms3404 ncomms3404 [pii]. ; PubMed Central PMCID: PMC3838905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Welm BE, Dijkgraaf GJ, Bledau AS, Welm AL, Werb Z. Lentiviral transduction of mammary stem cells for analysis of gene function during development and cancer. Cell stem cell. 2008;2(1):90–102. 10.1016/j.stem.2007.10.002 ; PubMed Central PMCID: PMC2276651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kochupurakkal BS, Iglehart JD. Nourseothricin N-acetyl transferase: a positive selection marker for mammalian cells. PloS one. 2013;8(7):e68509 10.1371/journal.pone.0068509 ; PubMed Central PMCID: PMC3701686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xia MQ, Hale G, Lifely MR, Ferguson MA, Campbell D, Packman L, et al. Structure of the CAMPATH-1 antigen, a glycosylphosphatidylinositol-anchored glycoprotein which is an exceptionally good target for complement lysis. Biochem J. 1993;293 (Pt 3):633–40. ; PubMed Central PMCID: PMC1134413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hale G, Xia MQ, Tighe HP, Dyer MJ, Waldmann H. The CAMPATH-1 antigen (CDw52). Tissue Antigens. 1990;35(3):118–27. . [DOI] [PubMed] [Google Scholar]
- 69.Yousefzadeh MJ, Wood RD. DNA polymerase POLQ and cellular defense against DNA damage. DNA Repair (Amst). 2013;12(1):1–9. 10.1016/j.dnarep.2012.10.004 ; PubMed Central PMCID: PMC3534860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Roerink SF, van Schendel R, Tijsterman M. Polymerase theta-mediated end joining of replication-associated DNA breaks in C. elegans. Genome research. 2014;24(6):954–62. 10.1101/gr.170431.113 ; PubMed Central PMCID: PMC4032859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Koole W, van Schendel R, Karambelas AE, van Heteren JT, Okihara KL, Tijsterman M. A Polymerase Theta-dependent repair pathway suppresses extensive genomic instability at endogenous G4 DNA sites. Nat Commun. 2014;5:3216 10.1038/ncomms4216 . [DOI] [PubMed] [Google Scholar]
- 72.Lemmens B, van Schendel R, Tijsterman M. Mutagenic consequences of a single G-quadruplex demonstrate mitotic inheritance of DNA replication fork barriers. Nat Commun. 2015;6:8909 10.1038/ncomms9909 ; PubMed Central PMCID: PMC4654259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ceccaldi R, Liu JC, Amunugama R, Hajdu I, Primack B, Petalcorin MI, et al. Homologous-recombination-deficient tumours are dependent on Poltheta-mediated repair. Nature. 2015;518(7538):258–62. 10.1038/nature14184 ; PubMed Central PMCID: PMC4415602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mateos-Gomez PA, Gong F, Nair N, Miller KM, Lazzerini-Denchi E, Sfeir A. Mammalian polymerase theta promotes alternative NHEJ and suppresses recombination. Nature. 2015;518(7538):254–7. 10.1038/nature14157 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. 10.1146/annurev.biochem.052308.093131 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Balestrini A, Ristic D, Dionne I, Liu XZ, Wyman C, Wellinger RJ, et al. The Ku heterodimer and the metabolism of single-ended DNA double-strand breaks. Cell reports. 2013;3(6):2033–45. 10.1016/j.celrep.2013.05.026 ; PubMed Central PMCID: PMC3815622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chanut P, Britton S, Coates J, Jackson SP, Calsou P. Coordinated nuclease activities counteract Ku at single-ended DNA double-strand breaks. Nat Commun. 2016;7:12889 10.1038/ncomms12889 ; PubMed Central PMCID: PMC5031800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gu Y, Jin S, Gao Y, Weaver DT, Alt FW. Ku70-deficient embryonic stem cells have increased ionizing radiosensitivity, defective DNA end-binding activity, and inability to support V(D)J recombination. Proc Natl Acad Sci U S A. 1997;94(15):8076–81. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Symington LS. Mechanism and regulation of DNA end resection in eukaryotes. Crit Rev Biochem Mol Biol. 2016;51(3):195–212. 10.3109/10409238.2016.1172552 ; PubMed Central PMCID: PMC4957645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hartlerode AJ, Willis NA, Rajendran A, Manis JP, Scully R. Complex Breakpoints and Template Switching Associated with Non-canonical Termination of Homologous Recombination in Mammalian Cells. PLoS genetics. 2016;12(11):e1006410 10.1371/journal.pgen.1006410 ; PubMed Central PMCID: PMC5104497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lee GS, Neiditch MB, Salus SS, Roth DB. RAG proteins shepherd double-strand breaks to a specific pathway, suppressing error-prone repair, but RAG nicking initiates homologous recombination. Cell. 2004;117(2):171–84. . [DOI] [PubMed] [Google Scholar]
- 82.Boboila C, Alt FW, Schwer B. Classical and alternative end-joining pathways for repair of lymphocyte-specific and general DNA double-strand breaks. Advances in immunology. 2012;116:1–49. 10.1016/B978-0-12-394300-2.00001-6 . [DOI] [PubMed] [Google Scholar]
- 83.Zellweger R, Dalcher D, Mutreja K, Berti M, Schmid JA, Herrador R, et al. Rad51-mediated replication fork reversal is a global response to genotoxic treatments in human cells. The Journal of cell biology. 2015;208(5):563–79. 10.1083/jcb.201406099 ; PubMed Central PMCID: PMC4347635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3(6):1101–8. . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A, Map of pHIV-NAT-hCD52 indicating key elements and location of mXrcc4 cDNA insert: EF1α: elongation factor alpha promoter; IRES: internal ribosomal entry signal; NAT: nourseothricin (NTC) acetyl transferase resistance gene: T2A, Thosea asigna viral self-cleaving peptide; CD52: human CAMPATH antigen cDNA. B, Primary FACS histogram data for mES reporter cells expressing human CD52. Left panel: Xrcc4Δ/Δ reporter cells transiently transfected with pHIV-NAT-hCD52 vector, stained with either no primary antibody (grey) or anti-hCD52 mAb YTH 34.5 (green). Right panel: hCD52 cell surface expression in Xrcc4Δ/Δ reporter cells following lentiviral transduction and selection in 100 μg/ml NTC. Gray histogram: pHIV-NAT-transduced cells. Green histogram: pHIV-NAT-hCD52-transduced cells. Inset numbers indicate median CD52 staining intensity.
(PDF)
A, Frequencies of Tus/Ter-induced and I-SceI-induced repair of Xrcc4fl/fl clone #8 or Xrcc4Δ/Δ clone #11 6xTer-HR reporter cells transiently expressing exogenous Xrcc4. Cells were transiently co-transfected with empty pcDNA3beta or pcDNA3beta-HA-mXrcc4 expression vector and empty, 3xMyc-NLS Tus or 3xMyc-NLS I-SceI expression vectors. Each plot represents the mean of duplicate samples from nine independent experiments (n = 9). Error bars: s.e.m. Tus-induced Total HR, t-test: flox8 +Xrcc4 vs. flox8 +EV p = 0.0048; del11 +Xrcc4 vs. del11 +EV p = 0.7251; del11 +EV vs. flox8 +EV p = 0.0002; del11 +Xrcc4 vs. flox8 +Xrcc4 p = 0.4659; del11 +Xrcc4 vs. flox8 +EV p = 0.0005. Tus-induced STGC, t-test: flox8 +Xrcc4 vs. flox8 +EV p = 0.0035; del11 +Xrcc4 vs. del11 +EV p = 0.7076; del11 +EV vs. flox8 +EV p = 0.0003; del11 +Xrcc4 vs. flox8 +Xrcc4 p = 0.4650; del11 +Xrcc4 vs. flox8 +EV p = 0.0005. Tus-induced LTGC, t-test: flox8 +Xrcc4 vs. flox8 +EV p = 0.5738; del11 +Xrcc4 vs. del11 +EV p = 0.8987; del11 +EV vs. flox8 +EV p = 0.3906; del11 +Xrcc4 vs. flox8 +Xrcc4 p = 0.8369; del11 +Xrcc4 vs. flox8 +EV p = 0.5332. Tus-induced Ratio, t-test: flox8 +Xrcc4 vs. flox8 +EV p = 0.0513; del11 +Xrcc4 vs. del11 +EV p = 0.7973; del11 +EV vs. flox8 +EV p = 0.2700; del11 +Xrcc4 vs. flox8 +Xrcc4 p = 0.7369; del11 +Xrcc4 vs. flox8 +EV p = 0.1734. I-SceI-induced Total HR, t-test: flox8 +Xrcc4 vs. flox8 +EV p = 0.0398; del11 +Xrcc4 vs. del11 +EV p<0.0001; del11 +EV vs. flox8 +EV p<0.0001; del11 +Xrcc4 vs. flox8 +Xrcc4 p = 0.5330; del11 +Xrcc4 vs. flox8 +EV p = 0.0034. I-SceI-induced STGC, t-test: flox8 +Xrcc4 vs. flox8 +EV p = 0.0391; del11 +Xrcc4 vs. del11 +EV p<0.0001; del11 +EV vs. flox8 +EV p<0.0001; del11 +Xrcc4 vs. flox8 +Xrcc4 p = 0.5491; del11 +Xrcc4 vs. flox8 +EV p = 0.0039. I-SceI-induced LTGC, t-test: flox8 +Xrcc4 vs. flox8 +EV p = 0.4390; del11 +Xrcc4 vs. del11 +EV p<0.0001; del11 +EV vs. flox8 +EV p = 0.0002; del11 +Xrcc4 vs. flox8 +Xrcc4 p = 0.6173; del11 +Xrcc4 vs. flox8 +EV p = 0.2036. I-SceI-induced Ratio, t-test: flox8 +Xrcc4 vs. flox8 +EV p = 0.9640; del11 +Xrcc4 vs. del11 +EV p<0.0001; del11 +EV vs. flox8 +EV p = 0.0012; del11 +Xrcc4 vs. flox8 +Xrcc4 p = 0.8666; del11 +Xrcc4 vs. flox8 +EV p = 0.9242. B, RT qPCR analysis of Xrcc4 in transfected Xrcc4fl/fl or Xrcc4Δ/Δ clones. Xrcc4 expression normalized to GAPDH and displayed as fold difference from Xrcc4fl/fl parental reporter clone 8 of the same experiment (x = -2ΔΔCt, with ΔΔCt = [CtXrcc4-CtGapdh]-[CtXrcc4-CtGAPDH]). Error-bars represent standard deviation of the ΔCt value (SDEV = √[SDEVXrcc42 + SDEVGAPDH2]). C, Western blot for abundance of HA-tagged mXrcc4 protein in Xrcc4fl/fl clone 8 or Xrcc4Δ/Δ clone 11 6xTer-HR reporter cells transiently transfected with empty pcDNA3beta or pcDNA3beta-mXrcc4.
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.