Abstract
Purpose
BRCA1/2 mutations are frequent in patients with triple-negative breast cancer (TNBC). These patients are often treated with primary systemic chemotherapy. The aim of this study was to analyze the effects of BRCA1/2 mutations on pathologic complete response (pCR) and disease-free survival (DFS) in a cohort of patients with TNBC treated with anthracycline and taxane–containing chemotherapy, with or without bevacizumab.
Patients and Methods
Germline DNA was sequenced to identify mutations in BRCA1 and BRCA2 in 493 patients with TNBC from the GeparQuinto study. The pCR rates were compared in patients with and without mutation, as well as in patients treated with and without bevacizumab. In addition, the influence of BRCA1/2 mutation status and pCR status on DFS was evaluated relative to treatment.
Results
BRCA1/2 mutations were detected in 18.3% of patients with TNBC. Overall, patients with mutations had a pCR rate of 50%, compared with 31.5% in patients without a mutation (odds ratio [OR], 2.17; 95% CI, 1.37 to 3.46; P = .001). The pCR rate among patients treated with bevacizumab was 61.5% for BRCA1/2 mutation carriers and 35.6% for those without mutations (OR, 2.90; 95% CI, 1.43 to 5.89; P = .004). pCR was a strong predictor of DFS for patients without BRCA1/2 mutations (hazard ratio, 0.18; 95% CI, 0.11 to 0.31) but not for patients with BRCA1/2 mutations (hazard ratio, 0.74; 95% CI, 0.32 to 1.69).
Conclusion
The addition of bevacizumab may increase the pCR after standard neoadjuvant chemotherapy for patients with TNBC with BRCA1/2 mutations. In patients treated with anthracycline and taxane–based chemotherapy (with or without bevacizumab), pCR was a weaker predictor of DFS for BRCA1/2 mutation carriers than for patients without mutations.
INTRODUCTION
Treatment for patients with triple-negative breast cancer (TNBC) involves several challenges. The absence of hormone receptors and human epidermal growth factor receptor 2 (HER2) makes the tumors ineligible for antihormonal treatment and trastuzumab or pertuzumab—leaving chemotherapy, with the associated toxicity, as the most effective therapy.
Chemotherapy can achieve a good response and also leads to a favorable long-term prognosis in patients with TNBC. When given as neoadjuvant therapy, the standard chemotherapy regimen including taxane and anthracyclines results in a pathologic complete response (pCR) in 30% to 50% of patients.1,2 In addition, patients who achieve a pCR have an excellent long-term prognosis in comparison with women who do not achieve a pCR.1,2
Mutations in BRCA1 and BRCA2 are more common in TNBC than in other molecular subtypes, with 11% to 17% of patients with TNBC having germline mutations.3,4 Patients with TNBC with BRCA1/2 mutations are reported to have a higher pCR rate than noncarriers in response to standard chemotherapies, with or without platinum agents.4-9
A recent report has raised the question of whether patients with BRCA1/2 germline mutations may represent a subpopulation in which a pCR is not associated with any benefit in terms of the prognosis.10 As deliberations on whether and how to establish pCR as a surrogate marker for drug approval are ongoing, it will become important to identify subgroups with and without this association between pCR and prognosis.
BRCA1/2 mutations have been reported to result in higher clinical response rates in relation to chemotherapy in patients with breast cancer. However, the influence of antiangiogenic agents such as bevacizumab on clinical response among patients with BRCA1/2 mutations has not been assessed, despite evidence that tumors with BRCA1/2 mutations show overexpression of vascular endothelial growth factor, Ang-1, and Ang-2.11,12 Similarly, hypoxia-associated DNA damage,13 associated with inhibition of angiogenesis, may contribute to synthetic lethal responses to treatment in tumors with BRCA1/2 mutations.
The influence of bevacizumab on breast cancer outcome has been tested in two large neoadjuvant chemotherapy trials (NSABP-B40 and GeparQuinto), both of which showed an increased pCR.14,15 However, only the NSABP-B40 study showed improved disease-free survival (DFS) with the addition of bevacizumab.16,17 Also, although the benefit of bevacizumab was mainly seen in patients with TNBC in the GeparQuinto study, in the NSABP-B40 study this effect was most prominent in hormone receptor–positive patients.14,15 These results suggest that additional predictors interacting with the bevacizumab response and treatment-related prognosis are needed.
The aims of this study were to assess the extent to which the pCR rate in patients in the GeparQuinto clinical trial depended on germline BRCA1/2 mutation status, whether pCR rate interacts with bevacizumab treatment, and how pCR translates into DFS in patients with and without BRCA1/2 mutations.
PATIENTS AND METHODS
The GeparQuinto phase III trial (ClinicalTrials.gov identifier: NCT00567554) recruited patients into two different treatment settings. Patients with HER2-positive breast cancer were randomly assigned to an anti-HER2 therapy–related study arm,18 whereas HER2-negative patients were recruited with the aim of testing the efficacy of bevacizumab on neoadjuvant response.15,17 The current study reports on the patients with TNBC in the HER2-negative arm.
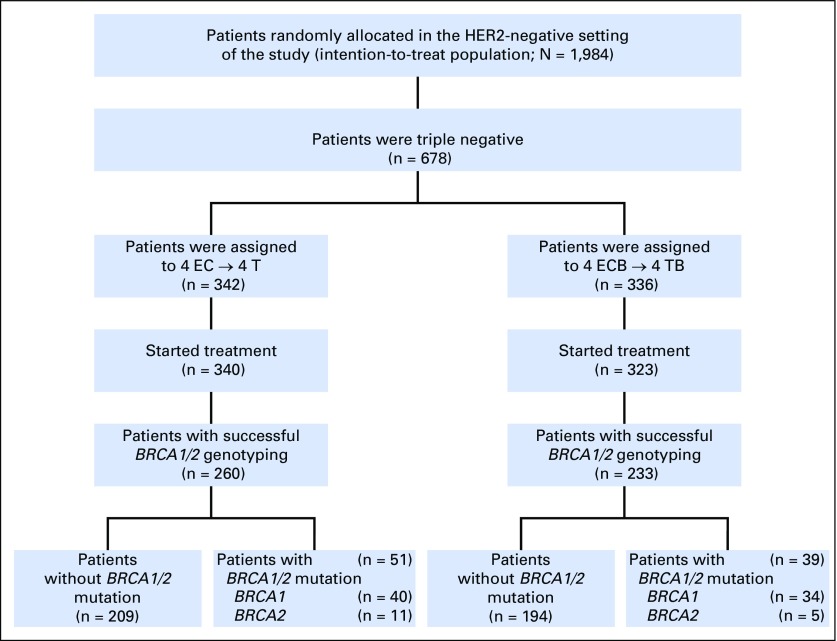
A total of 1,948 HER2-negative patients were randomly assigned in the main study.15 Among 663 patients with TNBC who received treatment,19 528 provided blood samples for prespecified pharmacogenetics studies. All patients provided written informed consent, and the relevant ethics committees at the participating study sites approved the study protocol (Data Supplement).
Treatment and Assessment of Clinical Data
The assessment of the clinical data from the GeparQuinto study has been described elsewhere.15 Briefly, HER2-negative patients received epirubicin and cyclophosphamide on day 1 and every 3 weeks for a total of four cycles, followed by docetaxel on day 1 and every 3 weeks for a total of four cycles. Patients were randomly assigned to receive either bevacizumab from the beginning of chemotherapy until surgery or no additional treatment. If there was no sonographic response after four cycles, the patients were taken off treatment and provided with alternative treatment.20 For the current study, patients were included in analyses if at least one cycle of chemotherapy or bevacizumab was completed. All clinical and histopathological data were assessed by local pathologists, including tumor stage, HER2 status, estrogen receptor/progesterone receptor status, and grade. Pathology reports were reviewed centrally for pCR.
DNA Extraction and Sequencing
Whole blood samples were collected in citrate-phosphate-dextrose-adenine tubes (Sarstedt AG, Nümbrecht, Germany) from patients at the time of random assignment. Germline DNA was extracted using the automated magnetic bead–based chemagic MSM I method (PerkinElmer chemagen, Baesweiler, Germany) in accordance with the manufacturer’s instructions. Quality control identified 35 poor-quality DNA samples among 528 samples available for the sequencing study.
The samples were genotyped as part of a multigene panel–based mutation analysis. The target regions included coding sequences and intron/exon boundaries of coding exons from 643 DNA repair genes, including BRCA1 and BRCA2. Baits for solution-based hybrid selection capture were designed using the Agilent SureSelect design tool. Germline DNA was fragmented and subjected to library preparation using the NEBNext Ultra DNA Library Prep kit with NEB Dual indexed adaptors and enriched for target regions by custom capture (Agilent eArray) using the Agilent SureSelect protocol. Products from each capture reaction were sequenced on a HiSEquation 2000 to 100 times mean coverage of target regions. All likely pathogenic mutations were validated using Sanger sequencing.
Bioinformatics Analysis
Paired end sequencing reads (100 bp) were aligned to the hg19 reference human genome using NovoAlign (Novocraft Technologies, Petaling Jaya, Malaysia). Realignment and recalibration were performed using GATK (VN 3.3.0). Samples with a read depth of < 20 times for > 10% of the target regions were excluded. Germline variations were called using GATK HaplotypeCaller. Variants with fewer than alternate allele reads from a minimum read depth of 20 times were excluded. Variants with a heterozygosity ratio of > 70:30 were excluded as possible mosaics. Annotations were defined using the bioR framework21 for population allele frequencies, 1,000 Genomes22 and dbSNP (VN 137),23 known mutation status (Human Gene Mutation database,24 ClinVar25), and protein sequence alterations (Clinical Annotation of Variants, VN 1.0).26
Statistical Considerations
In the primary analysis, pathologic response was defined as pathologic stage ypT0 and ypN0 after therapy. Analyses were repeated for pCR defined as ypT0/is and ypN0.2 DFS was defined as the time from random assignment until any invasive locoregional, invasive contralateral, or distant recurrence of breast cancer or any second primary invasive nonbreast cancer, or death as a result of any cause.17
Pearson’s χ2 test was used to compare genotyped participants and nonparticipants, as well as for comparisons between patients treated with and without bevacizumab. Fisher’s exact test was used to compare pCR rates between groups. Multivariate logistic regression including BRCA1/2 mutation status and treatment arm, as well as an interaction term, was performed to test for a different effect of BRCA1/2 mutation status on pCR in patients treated with and without bevacizumab. The Kaplan-Meier product-limit method was used to estimate disease-free survival and distant disease–free survival (DDFS). Univariate Cox proportional hazards models were used to calculate hazard ratios (HRs) and 95% CIs. In addition, Cox proportional hazard models including pCR, BRCA1/2 mutation status, and their interaction term, as well as BRCA1/2 mutation status, treatment arm, and their interaction term, were performed to test for the influence of differences in pCR and treatment on patient prognosis with and without a BRCA1/2 mutation.
All statistical tests were two sided, and a P value of < .05 was considered significant. No adjustment for multiple testing was performed. The analyses in this report were conducted using the statistical programming language R, version 3.2.2 (R Foundation for Statistical Computing, Vienna, Austria).
RESULTS
Patient Population
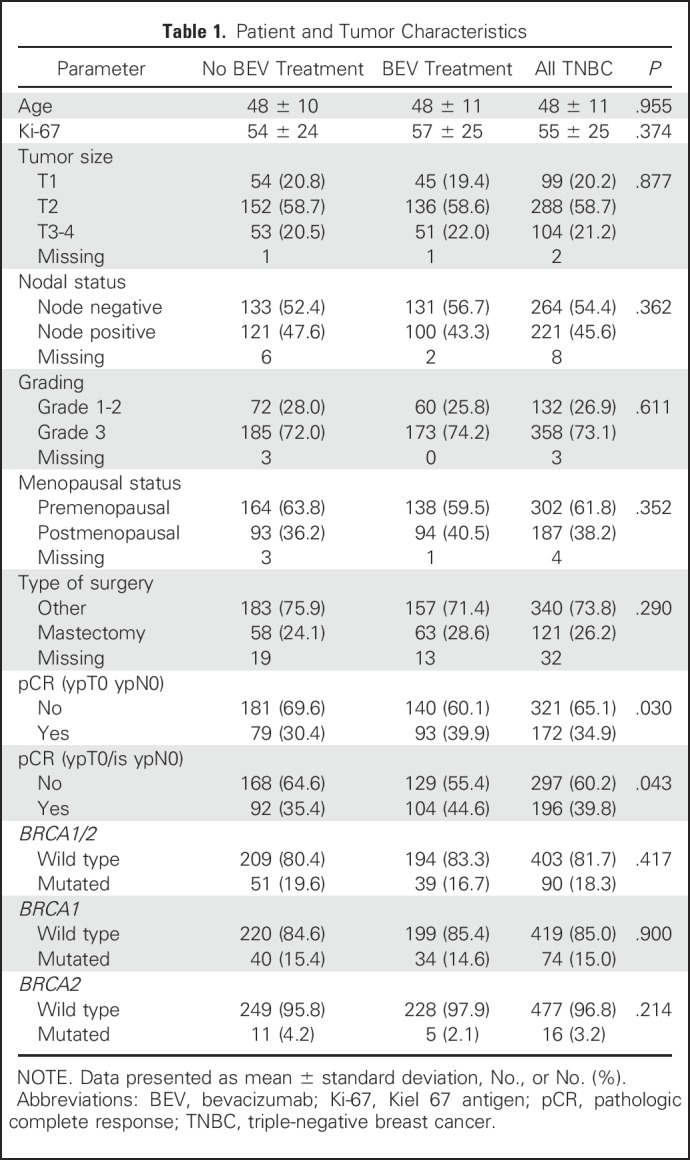
Genotype data were obtained for 493 patients from the 528 patients who provided germline DNA for this substudy (Fig 1). Genotyped patients had a more favorable tumor stage than nongenotyped participants (Appendix Table A1, online only). No significant differences in other parameters were observed (Appendix Table A1). In addition, there were no differences between patients treated with bevacizumab (n = 233) or not treated with bevacizumab (n = 260; Table 1). The pCR (ypT0/ypN0) rates were 30.4% (n = 79) and 39.9% (n = 93) for patients not treated and treated with bevacizumab, respectively. These pCR rates were similar to the pCR rates for the complete population (30.9% v 41.8%).19
Fig 1.
Patient selection. EC, epirubicin/cyclophosphamide; ECB, epirubicin/cyclophosphamide/bevacizumab; T, docetaxel; TB, docetaxel/bevacizumab.
Table 1.
Patient and Tumor Characteristics
Genotyping Results
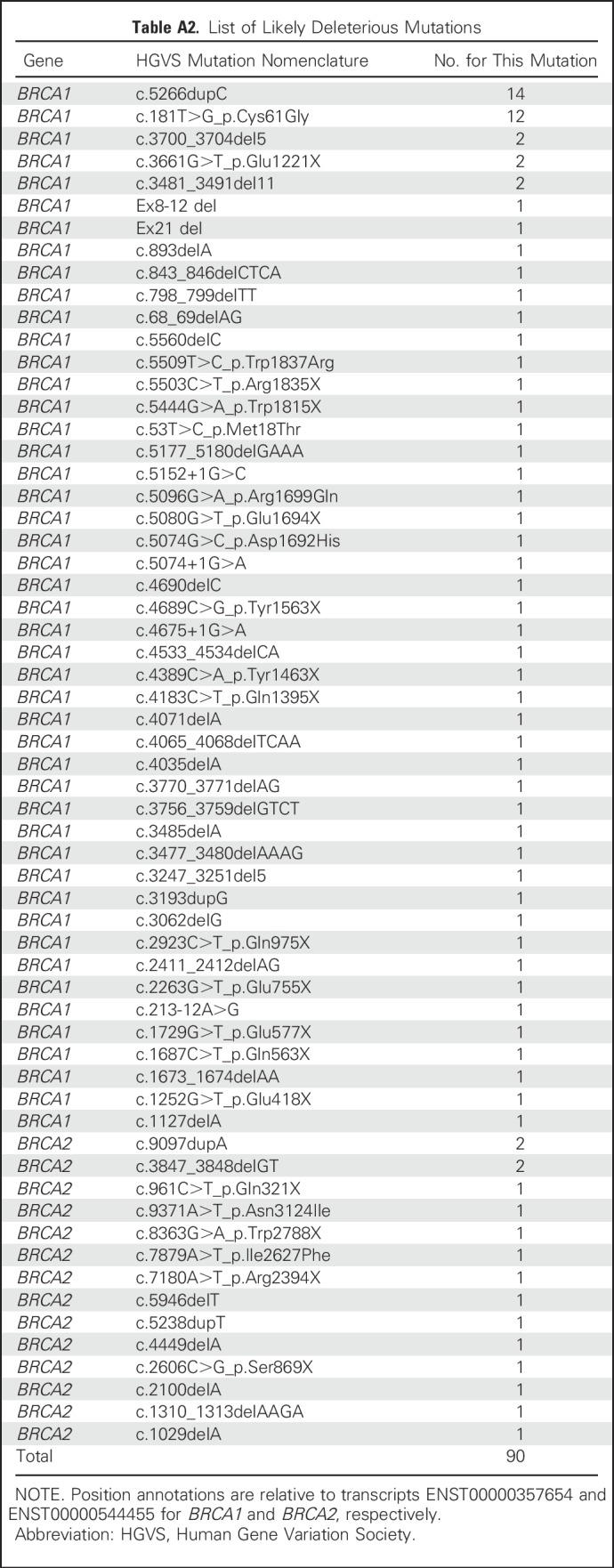
Deleterious BRCA1 and BRCA2 mutations were detected in 90 (18.3%) patients, including 74 (82.2%) with mutations in BRCA1 and 16 (17.8%) with mutations in BRCA2. No patients had mutations in both genes. Deleterious BRCA1 mutations included 49 frameshift mutations, 16 stop-gain mutations, five consensus splice site mutations, two deletions, one intronic variant, and 17 pathogenic missense mutations. Deleterious BRCA2 mutations included 10 frameshift, four stop-gain, and two pathogenic missense mutations. Mutations are listed in Appendix Table A2, online only. Among the patients with BRCA1/2 mutations, 51 (57%) were treated with standard chemotherapy alone and 39 (43%) with chemotherapy and bevacizumab (Fig 1). Similarly, among patients without mutations, 209 (52%) received chemotherapy alone and 194 (48%) were treated with bevacizumab.
Pathologic Complete Response and Prognosis Relative to BRCA Mutation Status
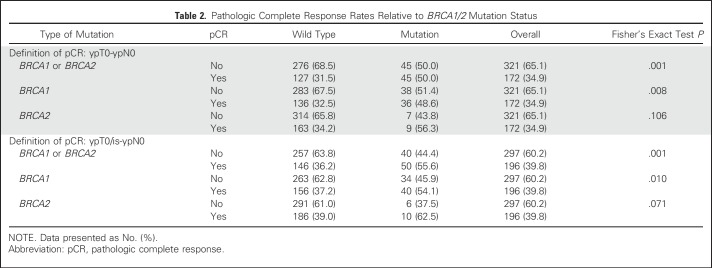
Irrespective of the treatment arm, the pCR (ypT0/ypN0) rates were 31.5% and 50.0% for patients without and with BRCA1/2 mutations, respectively (odds ratio [OR], 2.17; 95% CI, 1.37 to 3.46; P = .001). The pCR rates were similar for BRCA1 (48.6%) and BRCA2 (56.3%) mutation carriers (Table 2). Although the pCR rates in patients with BRCA1 mutations and patients with wild-type genotype were significantly different (OR, 1.97; 95% CI, 1.20 to 3.25; P = .008), the pCR rate in patients with BRCA2 mutations was similarly increased relative to the rate in patients with wild-type genotypes (OR, 2.48; 95% CI, 0.91 to 6.77; P = .106), although the difference was not statistically significant. The specific pCR rates for groups of BRCA1 and BRCA2 mutation carriers and different definitions of pCR are given in Table 2.
Table 2.
Pathologic Complete Response Rates Relative to BRCA1/2 Mutation Status
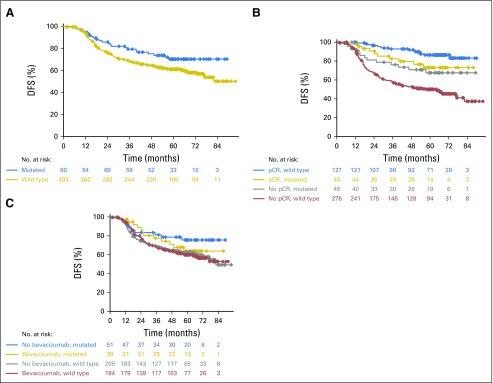
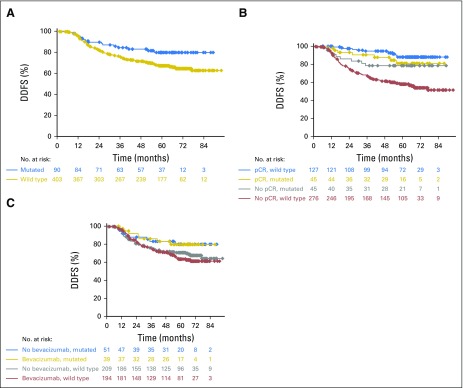
With regard to prognosis, patients with BRCA1/2 mutations had a significantly better DFS (HR, 0.644; 95% CI, 0.415 to 0.998; P = .047) than those with no mutations (Fig 2A). However, pCR in patients with BRCA1/2 mutations was not significantly associated with an improved DFS (HR, 0.74; 95% CI, 0.32 to 1.69; P = .472). In contrast, patients without mutations derived substantial benefit from pCR (HR, 0.18; 95% CI, 0.11 to 0.31; P < .001). Tests for interaction yielded a P value of .005, indicating that patients with BRCA1/2 mutations had a significantly different DFS after a pCR than patients without mutations. Kaplan-Meier curves are shown in Figure 2B. Analyses were repeated with DDFS as the outcome variable, and results were similar (Appendix Fig A1, online only).
Fig 2.
Kaplan-Meier curves for disease-free survival (DFS). (A) Comparison of DFS in all patients (hazard ratio [HR], 0.644; 95% CI, 0.415 to 0.998; P = .047). (B) Comparison of the effects of pathologic complete response (pCR) on the DFS. The HR for pCR in mutation carriers was 0.74 (95% CI, 0.32 to 1.69; P = .472); the HR for pCR in patients without mutations was 0.18 (95% CI, 0.11 to 0.31; P < .001). The interaction test showed a P value of .005. (C) Comparison of DFS relative to the treatment arm and mutation status. The HRs for bevacizumab treatment were 1.39 (95% CI, 0.61 to 3.15; P = .428) in patients with a BRCA1/2 mutation and 1.02 (95% CI, 0.74 to 1.40; P = .903) in patients without a BRCA1/2 mutation. P(interaction) = .451.
Pathologic Complete Response and Prognosis Relative to BRCA Mutations and Treatment Arm
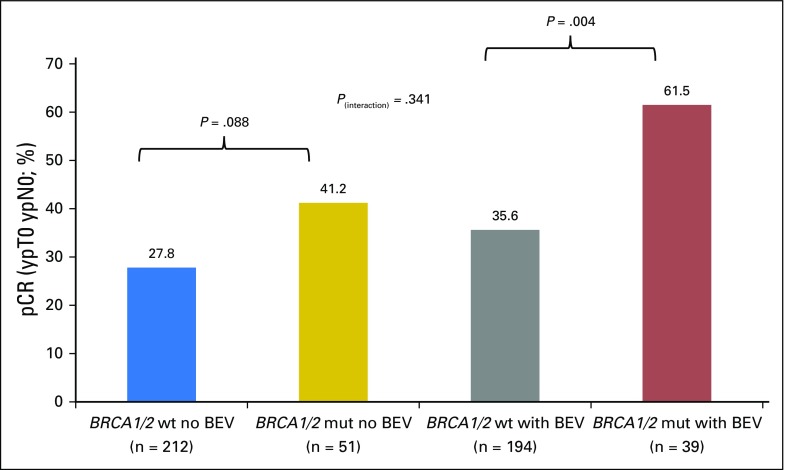
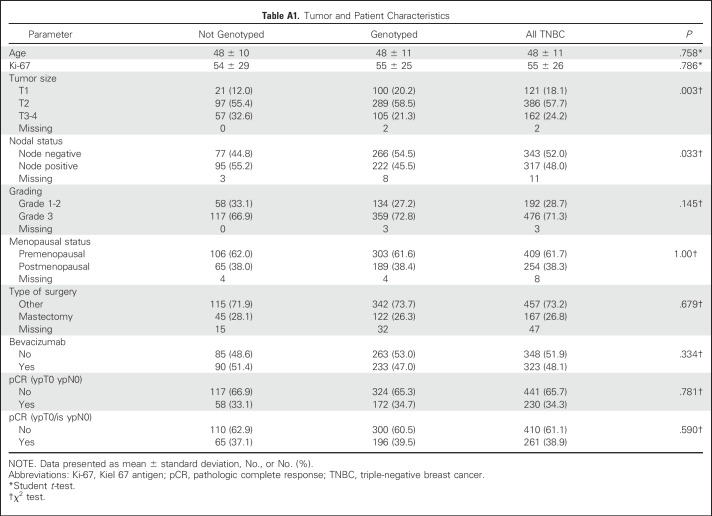
The relationship between pCR rates and both BRCA mutation status and treatment was also explored. In patients without bevacizumab treatment, the pCR rates were 27.8% and 41.2% in patients without and with BRCA1/2 mutations, respectively (OR, 1.82; 95% CI, 0.97 to 3.44; P = .088). Patients with bevacizumab treatment had corresponding pCR rates of 35.6% and 61.5%, respectively (OR, 2.90; 95% CI, 1.43 to 5.89; P = .004). The test for interaction was not significant (P = .341). pCR rates relative to mutation status and treatment arm are shown in Figure 3.
Fig 3.
Pathologic complete response (pCR) rates relative to BRCA1/2 mutation status and treatment arm. BEV, bevacizumab; mut, mutation; wt, wild type.
Although substantially different pCR rates were observed, these did not translate into differential effects on DFS. The DFS associated with bevacizumab treatment in patients with BRCA1/2 mutations had a HR of 1.39 (95% CI, 0.61 to 3.15), whereas the DFS in patients without BRCA1/2 mutations had a HR of 1.02 (95% CI, 0.74 to 1.40). The test for interaction yielded a P value of 0.451 (Fig 2C). Again, analyses were repeated with DDFS as the outcome variable, and results were similar (Appendix Fig A1). In addition, all of the analyses were repeated based on the pCR definition ypT0/is yN0. Similar results were observed (data not shown).
DISCUSSION
This study shows that patients with TNBC and a BRCA1 or BRCA2 mutation had higher pCR rates than patients without a mutation (50% v 31.5%). In addition, pCR rates of > 60% were observed if patients with mutations were treated with neoadjuvant chemotherapy including anthracyclines and taxanes with the addition of bevacizumab. In contrast, pCR rates of only 40% were achieved in patients with germline BRCA1/2 mutations but no bevacizumab treatment.
A recent report evaluated BRCA1/2 mutations, pCR, and prognosis in the GeparSixto Study.4 The study compared the addition of carboplatin to treatment with weekly paclitaxel plus weekly liposome-encapsulated doxorubicin plus weekly bevacizumab. Although only 50% of the patients in GeparQuinto received bevacizumab, the drug was given to all patients with TNBC in the GeparSixto trial. pCR rates of 33.1% (n = 40) in patients without a BRCA1/2 mutation and 50% (n = 12) in patients with a BRCA1/2 mutation in the nonplatinum arm were observed in GeparSixto.4 These results were slightly attenuated compared with the findings in the GeparQuinto study. In addition, a smaller study of pCR in patients with TNBC treated mainly with dose-dense doxorubicin/cyclophosphamide and dose-dense paclitaxel or weekly paclitaxel without bevacizumab treatment recently suggested that BRCA1 mutations may represent a subgroup of patients with TNBC who do not benefit from a pCR.10 The GeparQuinto results also showed that pCR had a smaller influence on DFS in patients with mutations (HR, 0.74; P = .472). In contrast, a statistically significant influence of pCR on DFS (HR, 0.18; P < .001) was observed for patients with wild-type tumors. The test for interaction yielded a P value of .005, indicating that the effect of pCR on DFS was smaller in BRCA1/2 mutation carriers than in patients with wild-type tumors. However, these comparisons should be interpreted with caution because of small sample sizes.
One possible explanation for the absence of an influence of increased pCR on prognosis in patients with BRCA1/2 mutations is the inclusion of subsequent contralateral breast cancers in the DFS analyses. Patients with a BRCA1/2 mutation have a 5-year risk for contralateral breast cancer of 10% to 15%, compared with 3% in patients without mutations.27 To address this possibility, we repeated the analysis using DDFS as the outcome variable. Similar results were observed, suggesting that the finding is independent of contralateral breast cancer events. Interestingly, the GeparSixto study did not show a differential effect of pCR on prognosis in patients with and without BRCA1/2 mutations.4 Whether the effect of pCR on prognosis in GeparSixto was driven by the platinum treatment, possibly in combination with bevacizumab, remains to be determined.
At the molecular level, several preclinical studies and clinical data imply a connection between DNA repair, hypoxia, and vascular endothelial growth factor signaling pathways. Indeed, antiangiogenic therapy may contribute to synthetic lethality28,29 in patients with BRCA1/2 mutations. Treatment with antiangiogenic drugs has been shown to induce hypoxia in tumors,30 and hypoxia has been shown to downregulate different mechanisms of DNA repair.13 Treatment with chemotherapy and with bevacizumab as an agent that potentially catalyzes additional genetic instability and downregulation of DNA repair response pathways may synergistically result in high response rates. Thus, bevacizumab-induced hypoxia and the resulting impairment of several DNA repair pathways may be one component of synthetic lethality in the context of DNA repair and cell death.
The main limitation of this study is the low sample size and the small number of BRCA1/2 mutation carriers. Although almost 500 patients with TNBC breast cancer were genotyped, there were only 90 mutation carriers. Therefore, an analysis incorporating mutation status, treatment arm, pCR, and prognosis and addressing whether pCR relative to the treatment arm has an effect on prognosis could not be undertaken. However, combining data from GeparSixto, GeparQuinto, NSABP-B40 and CALGB 40603 could provide additional insight into this question. Another limitation is that not every patient included in GeparQuinto provided blood for this analysis. However, samples were collected at the time of random assignment, and there were no differences in patient or tumor characteristics with regard to treatment arm.
This study is the largest so far that has examined the effect of BRCA1/2 mutations in the neoadjuvant treatment of TNBC. pCR rates are clearly higher in patients with BRCA1/2 mutations. High pathologic complete response rates in patients treated with chemotherapy and bevacizumab imply a role of antiangiogenic-induced hypoxia in the synthetic lethality of tumor cells with BRCA1/2 mutations. In this study, with standard treatment with anthracyclines, cyclophosphamide, and taxanes (with or without bevacizumab), the data suggest that BRCA1/2 mutation carriers do not benefit as much from a pCR in comparison with patients with a BRCA1/2 wild-type genotype. Additional studies and analyses need to be conducted to clarify the population in which BRCA1/2 mutation status uncouples pCR from a favorable prognosis.
ACKNOWLEDGMENT
We thank the research groups that conducted this study (the German Breast Group and Arbeitsgemeinschaft für Gynäkologische Onkologie Breast Study Group), the participating study sites, and, first and foremost, the patients.
Appendix
Investigators in GeparQuinto
Aalen (K. Gnauert), Augsburg (B. Heinrich), Bad Mergentheim (T. Prätz), Bad Nauheim (U. Groh), Bad Reichenhall (H. Tanzer), Baden‐Baden (C. Villena), Bayreuth (A. Tulusan), Bergisch Gladbach (B. Liedtke), Berlin (J.‐U. Blohmer, K. Kittel, C. Mau, J. Potenberg, J. Schilling), Bielefeld (M. Just), Böblingen (E. Weiss), Bochum (U. Bückner), Bonn (M. Wolfgarten), Braunschweig (R. Lorenz), Bremen (G. Doering, S. Feidicker), Chemnitz (P. Krabisch), Cuxhaven (U. Deichert), Deggendorf (D. Augustin), Dortmund (G. Kunz), Dresden (K. Kast), Düsseldorf (von Minckwitz G, C. Nestle‐Krämling, M. Rezai), Ebersberg (C. Höss), Eggenfelden (J. Terhaag), Erlangen (P. Fasching), Eschweiler (P. Staib), Essen (B. Aktas), Esslingen (T. Kühn), Frankfurt am Main (F. Khandan, V. Möbus, C. Solbach, H. Tesch), Freiburg (E. Stickeler), Fürstenwalde (G. Heinrich), Fürth (H. Wagner), Gelsenkirchen (A. Abdallah), Gifhorn (T. Dewitz), Göttingen (G. Emons), Greifswald (A. Belau), Hagen (V. Rethwisch), Halle an der Saale (T. Lantzsch, C. Thomssen), Hamburg (U. Mattner, A. Nugent, V. Müller), Hameln (T. Noesselt), Hamm (F. Holms), Hanau (T. Müller), Hannover (J.‐U. Deuker, I. Schrader), Herne (D. Strumberg), Hildesheim (C. Uleer), Homburg/Saar (E. Solomayer), Jena (I. Runnebaum), Kaiserslautern (H. Link), Karlsruhe (O. Tomé, H.‐U. Ulmer), Kassel (B. Conrad, G. Feisel‐Schwickardi), Kiel (H. Eidtmann), Cologne (C. Schumacher, T. Steinmetz), Landshut (I. Bauerfeind), Lebach (S. Kremers), Leipzig (D. Langanke), Lich (U. Kullmer), Limburg (A. Ober), Lübeck (D. Fischer), Ludwigsfelde (A. Kohls), Ludwigshafen (W. Weikel), Magdeburg (J. Bischoff, K. Freese), Mainz (M. Schmidt, W. Wiest), Mannheim (M. Sütterlin), Marktredwitz (M. Dietrich) Minden (M. Griesshammer), Muniich (D.‐M. Burgmann, C. Hanusch, B. Rack, C. Salat, D. Sattler), Münster (J. Tio), Mutlangen (E. von Abel), Neuruppin (B. Christensen), Neuss (U. Burkamp), Oldenburg (C.‐H. Köhne), Paderborn (W. Meinerz), Quedlinburg (S.‐T. Grasshoff), Ravensburg (T. Decker), Recklinghausen (F. Overkamp), Reutlingen (I. Thalmann), Rheinfelden (A. Sallmann), Rosenheim (T. Beck), Rostock (T. Reimer), Rottweil (G. Bartzke), Saarbrücken (M. Deryal), Schweinfurt (M. Weigel), St. Gallen, Switzerland (J. Huober, P. Weder), Stade (C.‐C. Steffens), Stadthagen (S. Lemster), Stendal (A. Stefek), Stralsund (F. Ruhland), Stuttgart (M. Hofmann, J. Schuster, W. Simon), Traunstein (U. Kronawitter), Trier (M. Clemens), Tübingen (T. Fehm), Ulm (W. Janni), Unna (K. Latos), Villingen‐ Schwenningen (W. Bauer), Weiden (A. Rossmann), Weinheim (L. Bauer), Weissenfels (D. Lampe), Wiesbaden (V. Heyl, G. Hoffmann, F. Lorenz‐Salehi), Witten (J. Hackmann), Würzburg (R. Schlag).
Fig A1.
Kaplan-Meier curves for distant disease-free survival (DDFS). (A) Comparison of DDFS in all patients. Hazard ratio (HR), 0.554 (95% CI, 0.329 to 0.933; P = .024). (B) Comparison of the effects of pathologic complete response (pCR) on the DDFS. The HR for pCR in mutation carriers was 0.74 (95% CI, 0.27 to 1.98; P = .541); the HR for pCR in patients without mutations was 0.19 (95% CI, 0.10 to 0.34; P < .001). The interaction test showed a P value of 0.0195. (C) Comparison of DDFS relative to the treatment arm and mutation status. The HRs for bevacizumab treatment were 0.97 (95% CI, 0.36 to 2.60; P = .947) in patients with a BRCA1/2 mutation and 1.15 (95% CI, 0.81 to 1.64; P = .432) in patients without a BRCA1/2 mutation. P(interaction) = .756.
Table A1.
Tumor and Patient Characteristics
Table A2.
List of Likely Deleterious Mutations
Footnotes
Supported in part by a National Cancer Institute Specialized Program of Research Excellence grant in breast cancer to the Mayo Clinic (Grant No. P50 CA116201), National Institutes of Health Grant No. CA192393, and the Breast Cancer Research Foundation.
Clinical trial information: NCT00567554.
AUTHOR CONTRIBUTIONS
Conception and design: Peter A. Fasching, Sibylle Loibl, Michael Untch, Bernd Gerber, Jens-Uwe Blohmer, Jens Huober, Volkmar Müller, Gunter von Minckwitz, Fergus J. Couch
Financial support: Fergus J. Couch
Administrative support: Sibylle Loibl
Provision of study materials or patients: Peter A. Fasching, Sibylle Loibl, Christian Schem, Hans Tesch, Michael Untch, Jörn Hilfrich, Mahdi Rezai, Bernd Gerber, Serban Dan Costa, Jens-Uwe Blohmer, Tanja Fehm, Jens Huober, Cornelia Liedtke, Richard M. Weinshilboum, Liewei Wang, Volkmar Müller, Brigitte Rack, Matthias Rübner, Gunter von Minckwitz
Collection and assembly of data: Peter A. Fasching, Sibylle Loibl, Chunling Hu, Steven N. Hart, Hermela Shimelis, Raymond Moore, Christian Schem, Hans Tesch, Michael Untch, Jörn Hilfrich, Mahdi Rezai, Bernd Gerber, Serban Dan Costa, Jens-Uwe Blohmer, Tanja Fehm, Jens Huober, Cornelia Liedtke, Valentina Nekljudova, Karsten E. Weber, Gunter von Minckwitz, Fergus J. Couch
Data analysis and interpretation: Peter A. Fasching, Sibylle Loibl, Chunling Hu, Steven N. Hart, Hermela Shimelis, Raymond Moore, Christian Schem, Hans Tesch, Michael Untch, Bernd Gerber, Richard M. Weinshilboum, Liewei Wang, James N. Ingle, Volkmar Müller, Valentina Nekljudova, Karsten E. Weber, Brigitte Rack, Matthias Rübner, Gunter von Minckwitz, Fergus J. Couch
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
BRCA1/2 Mutations and Bevacizumab in the Neoadjuvant Treatment of Breast Cancer: Response and Prognosis Results in Patients With Triple-Negative Breast Cancer From the GeparQuinto Study
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/site/ifc.
Peter A. Fasching
Honoraria: Roche, Amgen, Novartis, Pfizer, Celgene
Consulting or Advisory Role: Roche, Novartis
Research Funding: Novartis (Inst)
Sibylle Loibl
Research Funding: Pfizer, Roche, Celgene, Amgen, Novartis (Inst)
Chunling Hu
No relationship to disclose
Steven N. Hart
No relationship to disclose
Hermela Shimelis
No relationship to disclose
Raymond Moore
No relationship to disclose
Christian Schem
No relationship to disclose
Hans Tesch
Honoraria: Roche, GlaxoSmithKline
Michael Untch
No relationship to disclose
Jörn Hilfrich
No relationship to disclose
Mahdi Rezai
No relationship to disclose
Bernd Gerber
Consulting or Advisory Role: Roche
Travel, Accommodations, Expenses: AstraZeneca, Roche
Serban Dan Costa
No relationship to disclose
Jens-Uwe Blohmer
Consulting or Advisory Role: Roche
Travel, Accommodations, Expenses: Roche
Tanja Fehm
No relationship to disclose
Jens Huober
Honoraria: Novartis, Roche
Consulting or Advisory Role: Novartis, Roche
Travel, Accommodations, Expenses: Novartis, Roche
Cornelia Liedtke
Honoraria: Roche, Genomic Health, GlaxoSmithKline, AstraZeneca, Eisai, Celgene, Amgen, Gedeon Richter, Novartis, Pierre Fabre, Teva, Puma Biotechnology
Consulting or Advisory Role: Roche, Genomic Health, Teva, Pierre Fabre, Novartis
Speakers' Bureau: Genomic Health
Research Funding: Eisai, Roche, Novartis, Boehringer Ingelheim
Travel, Accommodations, Expenses: Roche, Teva, PharmaMar, Celgene, Daiichi Sankyo
Richard M. Weinshilboum
Stock or Other Ownership: OneOme
Liewei Wang
No relationship to disclose
James N. Ingle
No relationship to disclose
Volkmar Müller
Consulting or Advisory Role: AstraZeneca, Celgene, Daiichi Sankyo, Eisai, Nektar, Genentech, Novartis
Travel, Accommodations, Expenses: Pfizer, Roche Pharma AG
Valentina Nekljudova
No relationship to disclose
Karsten E. Weber
Stock or Other Ownership: Sividon Diagnostics
Patents, Royalties, Other Intellectual Property: EndoPredict
Brigitte Rack
Consulting or Advisory Role: Novartis, Genentech
Research Funding: AstraZeneca (Inst), Chugai Pharmaceutical (Inst), Eli Lilly (Inst), Novartis (Inst), BiPar/Sanofi-Aventis (Inst), Janssen (Inst)
Matthias Rübner
No relationship to disclose
Gunter von Minckwitz
Research Funding: Pfizer (Inst), GlaxoSmithKline (Inst), Roche (Inst), Sanofi (Inst), Amgen (Inst), Novartis (Inst), Celgene (Inst), Teva (Inst), Boehringer Ingelheim (Inst), AstraZeneca (Inst), Myriad Genetics (Inst)
Fergus J. Couch
Consulting or Advisory Role: AstraZeneca
Research Funding: GRAIL
Other Relationship: Ambry Genetics
REFERENCES
- 1.Cortazar P, Zhang L, Untch M, et al. : Pathological complete response and long-term clinical benefit in breast cancer: The CTNeoBC pooled analysis. Lancet 384:164-172, 2014 [DOI] [PubMed] [Google Scholar]
- 2.von Minckwitz G, Untch M, Blohmer JU, et al. : Definition and impact of pathologic complete response on prognosis after neoadjuvant chemotherapy in various intrinsic breast cancer subtypes. J Clin Oncol 30:1796-1804, 2012 [DOI] [PubMed] [Google Scholar]
- 3.Couch FJ, Hart SN, Sharma P, et al. : Inherited mutations in 17 breast cancer susceptibility genes among a large triple-negative breast cancer cohort unselected for family history of breast cancer. J Clin Oncol 33:304-311, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hahnen E, Lederer B, Hauke J, et al. : Germline mutation status, pathological complete response, and disease-free survival in triple-negative breast cancer: Secondary analysis of the GeparSixto randomized clinical trial. JAMA Oncol 3:1378-1385, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang C, Zhang J, Wang Y, et al. : Prevalence of BRCA1 mutations and responses to neoadjuvant chemotherapy among BRCA1 carriers and non-carriers with triple-negative breast cancer. Ann Oncol 26:523-528, 2015 [DOI] [PubMed] [Google Scholar]
- 6.Pfeifer W, Sokolenko AP, Potapova ON, et al. : Breast cancer sensitivity to neoadjuvant therapy in BRCA1 and CHEK2 mutation carriers and non-carriers. Breast Cancer Res Treat 148:675-683, 2014 [DOI] [PubMed] [Google Scholar]
- 7.Byrski T, Huzarski T, Dent R, et al. : Pathologic complete response to neoadjuvant cisplatin in BRCA1-positive breast cancer patients. Breast Cancer Res Treat 147:401-405, 2014 [DOI] [PubMed] [Google Scholar]
- 8.Byrski T, Gronwald J, Huzarski T, et al. : Pathologic complete response rates in young women with BRCA1-positive breast cancers after neoadjuvant chemotherapy. J Clin Oncol 28:375-379, 2010 [DOI] [PubMed] [Google Scholar]
- 9.Byrski T, Huzarski T, Dent R, et al. : Response to neoadjuvant therapy with cisplatin in BRCA1-positive breast cancer patients. Breast Cancer Res Treat 115:359-363, 2009 [DOI] [PubMed] [Google Scholar]
- 10.Paluch-Shimon S, Friedman E, Berger R, et al. : Neo-adjuvant doxorubicin and cyclophosphamide followed by paclitaxel in triple-negative breast cancer among BRCA1 mutation carriers and non-carriers. Breast Cancer Res Treat 157:157-165, 2016 [DOI] [PubMed] [Google Scholar]
- 11.Bindra RS, Schaffer PJ, Meng A, et al. : Down-regulation of Rad51 and decreased homologous recombination in hypoxic cancer cells. Mol Cell Biol 24:8504-8518, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bindra RS, Gibson SL, Meng A, et al. : Hypoxia-induced down-regulation of BRCA1 expression by E2Fs. Cancer Res 65:11597-11604, 2005 [DOI] [PubMed] [Google Scholar]
- 13.Huang LE, Bindra RS, Glazer PM, et al. : Hypoxia-induced genetic instability--a calculated mechanism underlying tumor progression. J Mol Med (Berl) 85:139-148, 2007 [DOI] [PubMed] [Google Scholar]
- 14.Bear HD, Tang G, Rastogi P, et al. : Bevacizumab added to neoadjuvant chemotherapy for breast cancer. N Engl J Med 366:310-320, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.von Minckwitz G, Eidtmann H, Rezai M, et al. : Neoadjuvant chemotherapy and bevacizumab for HER2-negative breast cancer. N Engl J Med 366:299-309, 2012 [DOI] [PubMed] [Google Scholar]
- 16.Bear HD, Tang G, Rastogi P, et al. : Neoadjuvant plus adjuvant bevacizumab in early breast cancer (NSABP B-40 [NRG Oncology]): Secondary outcomes of a phase 3, randomised controlled trial. Lancet Oncol 16:1037-1048, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.von Minckwitz G, Loibl S, Untch M, et al. : Survival after neoadjuvant chemotherapy with or without bevacizumab or everolimus for HER2-negative primary breast cancer (GBG 44-GeparQuinto). Ann Oncol 25:2363-2372, 2014 [DOI] [PubMed] [Google Scholar]
- 18.Untch M, Loibl S, Bischoff J, et al. : Lapatinib versus trastuzumab in combination with neoadjuvant anthracycline-taxane-based chemotherapy (GeparQuinto, GBG 44): A randomised phase 3 trial. Lancet Oncol 13:135-144, 2012 [DOI] [PubMed] [Google Scholar]
- 19.Gerber B, Loibl S, Eidtmann H, et al. : Neoadjuvant bevacizumab and anthracycline-taxane-based chemotherapy in 678 triple-negative primary breast cancers; results from the GeparQuinto study (GBG 44). Ann Oncol 24:2978-2984, 2013 [DOI] [PubMed] [Google Scholar]
- 20.Huober J, Fasching PA, Hanusch C, et al. : Neoadjuvant chemotherapy with paclitaxel and everolimus in breast cancer patients with non-responsive tumours to epirubicin/cyclophosphamide (EC) ± bevacizumab - results of the randomised GeparQuinto study (GBG 44). Eur J Cancer 49:2284-2293, 2013 [DOI] [PubMed] [Google Scholar]
- 21. doi: 10.1093/bioinformatics/btu137. Kocher JP, Quest DJ, Duffy P, et al: The Biological Reference Repository (BioR): A rapid and flexible system for genomics annotation. Bioinformatics 30:1920-1922, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. doi: 10.1038/nature15393. 1000 Genomes Project Consortium, Auton A, Brooks LD, et al: A global reference for human genetic variation. Nature 526:68-74, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. doi: 10.1093/nar/29.1.308. Sherry ST, Ward MH, Kholodov M, et al: dbSNP: The NCBI database of genetic variation. Nucleic Acids Res 29:308-311, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. doi: 10.1007/s00439-013-1358-4. Stenson PD, Mort M, Ball EV, et al: The Human Gene Mutation Database: Building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum Genet 133:1-9, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. doi: 10.1093/nar/gkv1222. Landrum MJ, Lee JM, Benson M, et al: ClinVar: Public archive of interpretations of clinically relevant variants. Nucleic Acids Res 44:D862-D868, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. doi: 10.1186/s13073-015-0195-6. Münz M, Ruark E, Renwick A, et al: CSN and CAVA: variant annotation tools for rapid, robust next-generation sequencing analysis in the clinical setting. Genome Med 7:76, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Molina-Montes E, Pérez-Nevot B, Pollán M, et al. : Cumulative risk of second primary contralateral breast cancer in BRCA1/BRCA2 mutation carriers with a first breast cancer: A systematic review and meta-analysis. Breast 23:721-742, 2014 [DOI] [PubMed] [Google Scholar]
- 28.Lord CJ, Tutt AN, Ashworth A: Synthetic lethality and cancer therapy: Lessons learned from the development of PARP inhibitors. Annu Rev Med 66:455-470, 2015 [DOI] [PubMed] [Google Scholar]
- 29.Dhillon KK, Bajrami I, Taniguchi T, et al. : Synthetic lethality: The road to novel therapies for breast cancer. Endocr Relat Cancer 23:T39-T55, 2016 [DOI] [PubMed] [Google Scholar]
- 30.Conley SJ, Gheordunescu E, Kakarala P, et al. : Antiangiogenic agents increase breast cancer stem cells via the generation of tumor hypoxia. Proc Natl Acad Sci USA 109:2784-2789, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]