Antimicrobial resistance (AMR) is a big challenge threatening public health worldwide. Every year, AMR-associated bacterial infections result in over 70,000 people deaths in the United States [1,2]. Worryingly, it is estimated that AMR will claim for 10,000,000 lives globally per year by 2050 [3]. The major reasons for this macabre scenario are i) The rapid appearance of new AMR determinants; ii) The global transmission of epidemic pathogens with multi-drug resistance (MDR) or even extensive-drug resistance (XDR); and iii) The failure to invent new antimicrobial agents. In general, carbapenems are used as a last line of defense antibiotics against severe infections with MDR-possessing Enteriobactericeae. Colistin, a cationic antimicrobial polypeptide, is clinically regarded as a final line of defense to treat lethal challenges with carbapenem-resistant pathogens [4,5]. Unfortunately, it seems likely that the clinical use of carbapenems and colistin has been significantly challenged by the occurrence of mobilized colistin resistance determinant (MCR-1) in clinical carbapenem-resistant isolates producing New Delhi metallo-β-lactamase 1 (NDM-1) [6,7] or its variant NDM-5 [8,9]. In addition to the intrinsic determinants of colistin resistance (e.g., EptA [10,11] and ICR-Mo [12]), the transferable determinants (MCR-1 and MCR-2 [10,13,14]) consistently encode members of phosphoethanolamine (PEA)-lipid A transferases, which adopt a ‘ping-pong’ catalysis reaction in transferring of the PEA moiety to the 4ʹ-phosphate position of the lipopolysaccharide (LPS)-lipid A species anchored on bacterial surface [4,5].
In the two years since its first discovery in Southern China, in late of 2015 [15], mcr-1 has been detected to coexist with blaNDM-1-like genes in migratory birds (like the Muscovy duck [16] plus chickens [17]), in healthy people [18] and also in patients with bloodstream infections [6,7,9,19]. This indicates that the possible route of mcr-1/blaNDM-like genes is through the food-chain transmission. With the exception of Cronobacter sakazakii that carries NDM-9 [20], E. coli seems to be the predominant reservoir for the coexistence of MCR-1 and NDM-like enzymes [6–9,16–19,21,22]. Surprisingly, mcr-3, a new mcr-like gene, was found in an XDR-E. coli clinical isolate which harbored both mcr-1 and blaNDM-5 [23]. This might further highlight the importance of ‘one health’ approach in active surveillance of mcr-like genes in human, animal and environmental sectors. However, current knowledge on how MDR-bacterial pathogens emerge and evolve is relatively limited.
Here we report that antibiotic exposure elicits the emergence of a carbapenem- and colistin-resistant E. coli clinical isolate coharboring MCR-1 and NDM-5 in a hospitalized patient. On 3 June 2017, a 58-year-old man was admitted to the Lishui People’s Hospital of Zhejiang Province, China (Figure 1(A)), and was diagnosed to be suffering from Small Intestinal Perforation with Peritonitis (SIPWP). A routine examination of this patient suggested that neither immune-suppression nor specific comorbidity were present (Table S1). While in hospital, the inflammatory markers (temperature and Leukocyte count) of this patient were normal (Figure 1(B)). The patient was subjected to anti-infection therapy through 0.5 h of intravenous guttae containing 2.0g q6h of Piperacillin/Tazobactam (TZP) prior to surgical operation. To probe the suspected bacterial agent, the specimen of peritoneal fluid from the patient (Figure 1(A)) was inoculated on a petri plate with MacConkey agar. Antibiotic susceptibility assays revealed that all of the acquired E. coli isolates (designated as S-E. coli) are sensitive to most of the antibiotics including TZP (Table S2).
Figure 1.
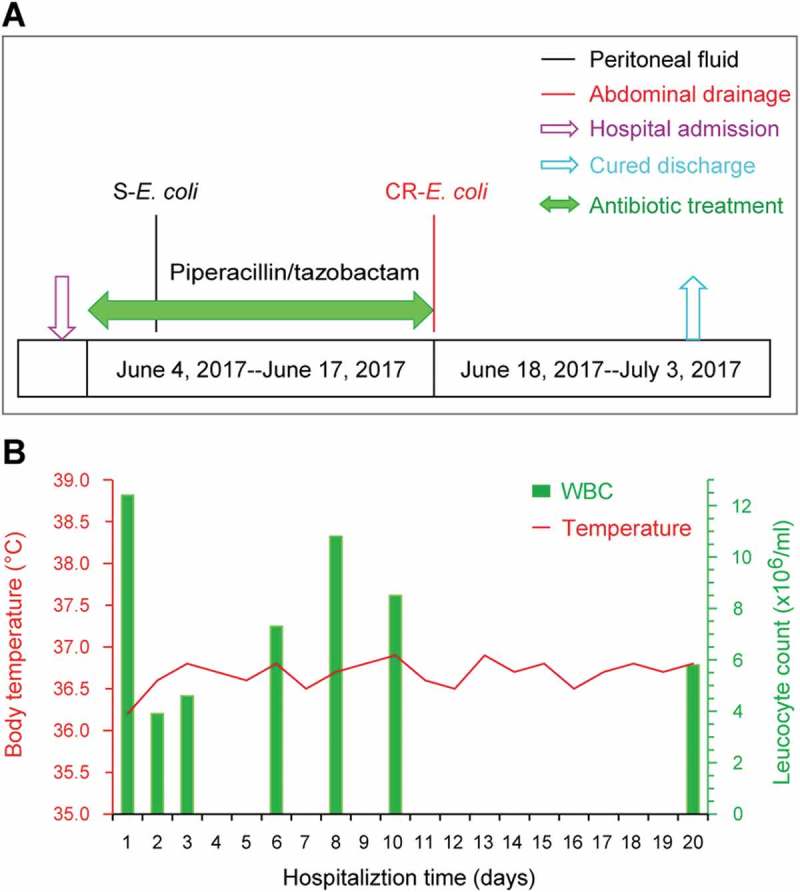
Clinical description of a patient undergoing antibiotic treatment.
A. The history of antibiotic exposure for a patient admitted into the hospitalThe colored text and bars represent the sources from which the bacterial species were isolated. S-E. coli, sensitive-E. coli; CR-E. coli, Carbapenem-resistant E. coli B. Dynamic alterations of body temperature and leukocyte counts of the patient recorded during the hospitalization period
Following 2-weeks of antibiotic therapy, which proved to be a clinically-successful anti-infection treatment (Figure 1(A)), the abdominal drainage and the blood of the patient were sampled for bacterial cultivation. Although there were no E. coli isolates recovered from the patient’s blood samples during the entire treatment period, a MDR-E. coli isolate was unexpectedly detected in the abdominal drainage sample (Table S2). This purulent culture was not only found to be highly-resistant to imipenem and meropenem, two widely-used carbapenems, but also it displayed an appreciable level of resistance to colistin (Table S2). Thus, it was appropriately designated as CR-E. coli LSB54 (Figure 1(A)). In addition, we favored to speculate that the LSB54 strain is a representative of a small sub-population of local microbiota on this hospitalized patient, and it was elicited by an over-exposure to antibiotics. After the evaluation of the patient’s inflammatory index and the infective wound, the antibiotic treatment was immediately discontinued, but surgical debridement and drainage were resumed. As a result, the patient received a suitable course of treatment and was discharged after being hospitalized for 30 days.
To further address the purulent culture (CR-E. coli LSB54), we integrated multiple approaches ranging from microbial genetics, molecular biology, to comparative genomics. The diversity of genetic backgrounds was observed among the antibiotic-susceptible clinical strains from patients with similar durations of hospitalization (Table S3). In contrast, sequence typing showed that CR-E. coli belongs to ST2179 (Table S3). This data seems to rule out the possibility that the MDR-E. coli was acquired in the hospital. Of note, a ST2179 E. coli isolate with multi-drug resistance was recently found to claim for extra-intestinal infections in horses in Brazil [24]. Therefore, it seems likely that a possible transmission route for the zoonotic agent ST2179 is present between animals and humans. As we recently described with mcr-1-bearing plasmids carried by commensal E. coli [25], whole genome sequencing was performed, giving evidence that two unique plasmids coexist in the MDR-E. coli strain LSB54 (Figure 2–4). Among the two plasmids, pLSB54-mcr-1 is the mcr-1-harboring plasmid (Figures 2 and 4(A)), and pLSB54-NDM-5 is the NDM-5-carrying plasmid (Figures 3 and 4(A)). Subsequent plasmid replicon typing (https://cge.cbs.dtu.dk/services/PlasmidFinder/) revealed that pLSB54-mcr-1 (Acc. no.: MG773376) is a IncHI2-type plasmid with a genome size of 251.657 kb (Figure 2), whereas pLSB54-NDM-5 (Acc. no.: MG773377) is a IncX3-like plasmid whose genome is 46.19 kb long (Figure 3).
Figure 2.
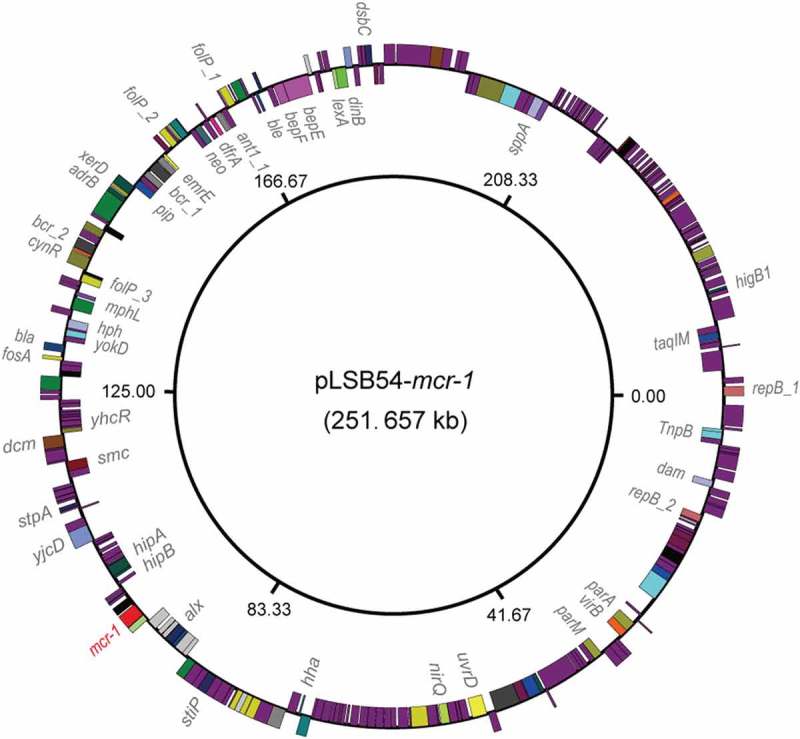
Genomic map of the mcr-1-carrying plasmid pLSB54-mcr-1.
The plasmid map was generated using GenomeVx [28] with automatic coloring.
Figure 4.
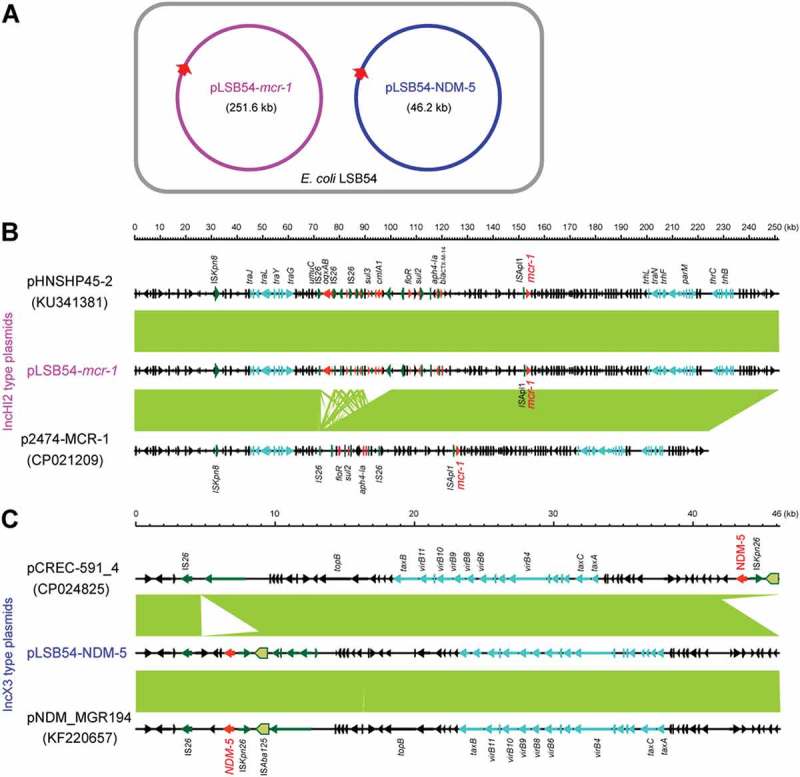
Comparative genomics of pLSB54-mcr-1 and pLSB54-NDM-5.
A. Cartoon description of the E. coli LSB54 strain carrying the two plasmids pLSB54-mcr-1 and pLSB54-NDM-5B. Colinear genome analyzes of three mcr-1-harboring plasmids pLSB54-mcr-1, pHNSHP45-2 (KU341381) and p2474-MCR1 (CP021209)C. Genomic comparison of the plasmid pLSB54-NDM-5 with two other NDM-5-expressing plasmids pCREC-591_4 (CP024825) and pNDM_MGR194 (KF220657)Boxed arrows represent the position and transcriptional direction of the ORFs. Regions of > 99% identity are given by green shading. Genes associated with the tra and pil loci are colored light blue, antibiotic resistance genes are colored red, insertion sequences are colored green, and other genes are colored black.
Figure 3.
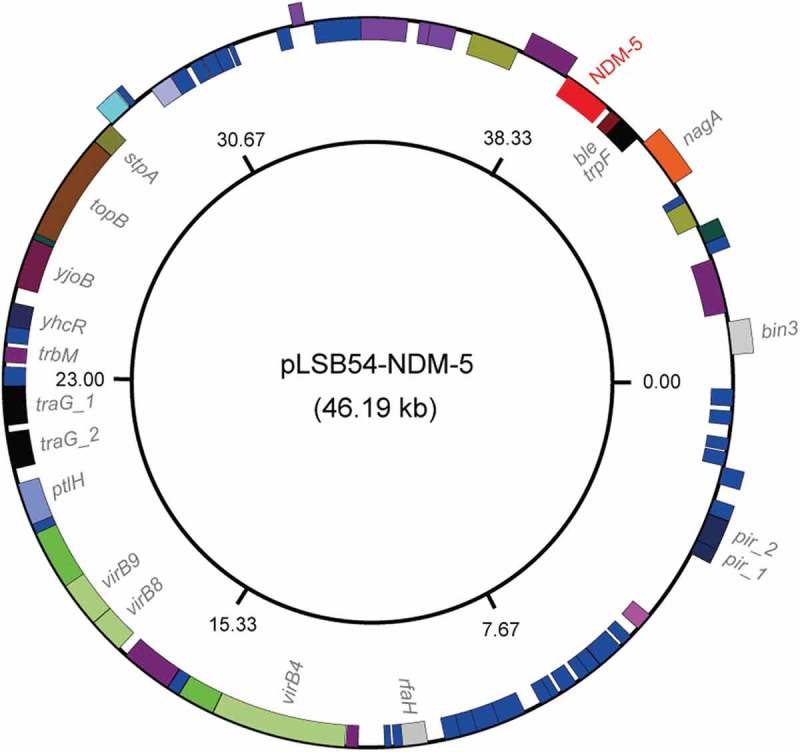
Genomic map of the NDM-5-producing plasmid pLSB54-NDM-5.
The plasmid map was generated using GenomeVx [28] with automatic coloring.
Colinear alignment analyzes of the plasmid genomes indicated that pLSB54-mcr-1 is almost identical with the prototypical mcr-1-harboring plasmid pHNSHP45-2 (Acc. no.: KU341381) (Figure 4(B)), and possesses 90% coverage and 99% identity with p2474-MCR1 (Acc. no.: CP021209) [26], another mcr-1-positive plasmid from a clinical E. coli isolate in China (Figure 4(B)) [6]. Given that pHNSHP45-2 is isolated from E. coli with origin of swine feces, it raised the possibility that origin of pLSB54-mcr-1 might lie in animal gut microbiota. Intriguingly, p2474-MCR1 was also detected to coexist with the blaNDM-1-carrying plasmid in a single isolate of E. coli [6]. Like most of the known IncHI2-type plasmids, pLSB54-mcr-1 is also positive for other antibiotic resistance genes (Table S4). Evidently, this is consistent with the phenotype of being resistant to multiple antibiotics which was observed with the strain LSB54 (Table S2).
The plasmid pLSB54-mcr-1 encodes the following genes conferring resistance to aminoglycosides, β-lactam (blaCTX-M-14), fluoroquinolones (oqxA and oqxB), fosfomycin (fosA3), phenicols (floR and cmlA1), sulphonamides (sul1, sul2 and sul3) and trimethoprim (dfr12) (Tables S2 and S4). As for pLSB54-NDM-5, it presents 100% coverage with the NDM-5-producing IncX3 plasmid pCREC-591_4 (Acc. no.: CP024825) of E. coli isolated from South Korea (Figure 4(C)) and exhibits a 99% identity to the plasmid pNDM_MGR194 (KF220657) of Klebsiella pneumoniae seen in India (Figure 4(C)). Whereas, the insertion loci of NDM-5 are quite different in the above three plasmids (Figure 4(C)). Unlike pLSB54-mcr-1, pLSB54-NDM-5 only contains a single resistance gene blaNDM-5 (Table S4). Because of the plasmid pNDM_MGR194 from human urine, we favored to anticipate that pLSB54-NDM-5 might originate from human microbiota.
Fine mapping of the genetic environment surrounding the mcr-1(blaNDM-5) gene further illustrated that i) In the IncHI2-type plasmid pLSB54-mcr-1, the insertion sequence ISApl1 is located upstream of the mcr-1 gene (Figure 5(A)) and the ISApl1-mcr-1-pap2 fragment is inserted downstream of the terF gene, just like the plasmids pHNSHP45-2 (Acc. no.: KU341381) and p2474-MCR (Acc. no.: CP021209) (Figure 5(A)); ii) The NDM-5-containing fragment is identical in all three IncX3-like plasmids, including pLSB54-NDM-5 (Figure 5(B)), and the blaNDM-5 is adjacent to the insertion sequence IS5, which is flanked by a truncated ISAba125 sequence (Figure 5(B)). Unlike the scenario where MCR-1 and NDM-5 is co-transferred by a single hybrid IncX3-X4 plasmid in E. coli [8], mcr-1 and blaNDM-5 are separately encoded by two distinct plasmids in the clinical E. coli isolate LSB54 (Figure 4(A)). In this clinical case, the MDR-E. coli which possesses resistance to colistin and carbapenem (two lines of last-resort antibiotics), emerges in a patient undergoing antibiotic treatment with TZP. This observation somewhat undermines rationality of anti-infection therapies (via inappropriate usage and/or overuse of antibiotics) to some extent. Very recently, we observed an increase in the antimicrobial activity of colistin against E. coli co-producing NDM-5 and MCR-1, when it was supplied with amikacin, which could be a promising therapeutic option for the treatment of lethal infections caused by MDR-bacterial pathogens [21].
Figure 5.
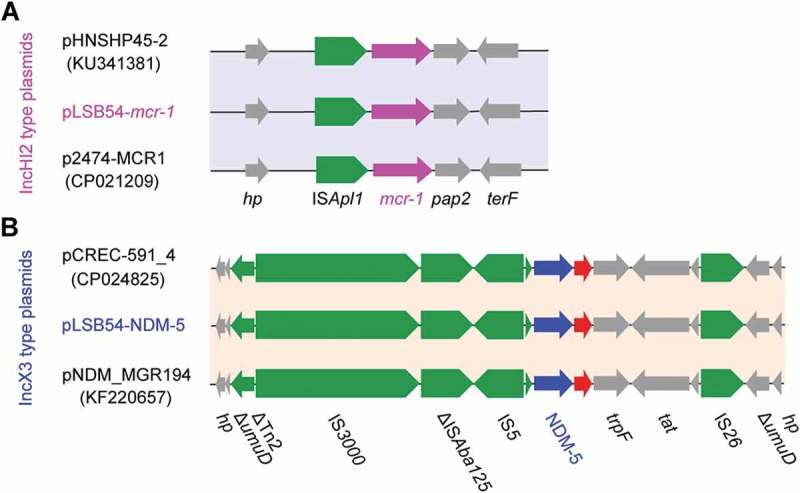
Genetic contents of mcr-1 (and/or blaNDM-5).
A. The genetic environment of mcr-1 and its neighboring lociB. The genetic environment of blaNDM-5 and its neighboring lociThe arrows indicate open reading frames, resistance genes, insertion sequences and accessory genes are separately shown with red, green and gray arrows.
In summary, our findings report the first clinical case that antibiotic exposure elicited the emergence of MDR-E. coli coharboring MCR-1 and NDM-5 in a patient. The fact that the co-existence of MCR-1 and NDM variants has been found in bacterial species isolated from humans, animals and environments [6,8,16,19,27], reminded us to employ the approach of ‘one health’ in combating potential challenges with MDR-pathogens. Given that inappropriate use of antibiotics is a potential threat for the rapid development of drug resistance and therapeutic failure, it is prerequisite to reconsider (and/or screen) the subpopulations of MDR-pathogens persisting in human microbiota prior to the formulation of appropriate antibiotic-based effective anti-infection therapies.
Funding Statement
This work was supported by National Key R&D Program of China (2017YFD0500202), National Key Basic Research Program of China (2016YFC1200100) and the National Natural Science Foundation of China (31570027 & 81772142). Dr. Feng is a recipient of the national “Young 1000 Talents” Award of China.
Acknowledgments
We would like to thank Mr. Vishnu Goutham Kota and Swaminath Sirinivas for critical reading this manuscript.
Author Contributions
YF designed and supervised this project; YF, JM, SL, WL, WW and JS performed experiments; YF, JM, SL and WL analyzed the data and prepared figures; YF drafted this manuscript.
Availability of data and materials
All the data are included in this manuscript. Complete sequences of the two plasmids were deposited into GenBank under the accession no. (MG773376 for pLSB54-mcr-1, MG773377 for pLSB54-NDM-5).
Ethics statement
Clinical data are recorded and approved by the local ethics committee from the Lishui People’s Hospital (2017-005-01).
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplementary data for this article can be accessed here.
References
- 1.Anonymous Centers for disease control and prevention, office of infectious disease. Antibiotic resistance threats in the United States, 2013. 2013 [updated 2013 Apr; cited 2016 September5]. Available from: http://www.cdc.gov/drugresistance/threat-report-2013
- 2.Laxminarayan R, Amabile-Cuevas CF, Cars O, et al. UN high-level meeting on antimicrobials–what do we need? Lancet. 2016;388:218–220. [DOI] [PubMed] [Google Scholar]
- 3.O’Neill LJ. Antimicrobial resistance: tackling a crisis for the health and wealth of nations. London: Review on Antimicrobial Resistance; 2014. [Google Scholar]
- 4.Sun J, Zhang H, Liu YH, et al. Towards understanding MCR-like colistin resistance. Trends Microbiol. 2018. Epub ahead of print DOI: 10.1016/j.tim.2018.02.006 [DOI] [PubMed] [Google Scholar]
- 5.Feng Y. Transferability of MCR-1/2 polymyxin resistance: complex dissemination and genetic mechanism. ACS Infect Dis. 2018;4:291–300. [DOI] [PubMed] [Google Scholar]
- 6.Zheng B, Dong H, Xu H, et al. Coexistence of MCR-1 and NDM-1 in clinical Escherichia coli isolates. Clin Infect Dis. 2016;63:1393–1395. [DOI] [PubMed] [Google Scholar]
- 7.Zheng B, Yu X, Xu H, et al. Complete genome sequencing and genomic characterization of two Escherichia coli strains co-producing MCR-1 and NDM-1 from bloodstream infection. Sci Rep. 2017;7:17885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun J, Yang RS, Zhang Q, et al. Co-transfer of blaNDM-5 and mcr-1 by an IncX3-X4 hybrid plasmid in Escherichia coli. Nat Microbiol. 2016;1:16176. [DOI] [PubMed] [Google Scholar]
- 9.Zheng B, Lv T, Xu H, et al. Discovery and characterisation of an Escherichia coli ST206 strain producing NDM-5 and MCR-1 from a patient with acute diarrhoea in China. Int J Antimicrob Agents. 2017. DOI: 10.1016/j.ijantimicag.2017.09.005 [DOI] [PubMed] [Google Scholar]
- 10.Xu Y, Wei W, Lei S, et al. An evolutionarily conserved mechanism for intrinsic and transferable polymyxin resistance. mBio. 2018;9:e02317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anandan A, Evans GL, Condic-Jurkic K, et al. Structure of a lipid A phosphoethanolamine transferase suggests how conformational changes govern substrate binding. Proc Natl Acad Sci USA. 2017;114:2218–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei W, Srinivas S, Lin J, et al. Defining ICR-Mo, an intrinsic colistin resistance determinant from Moraxella osloensis. PLoS Genet. 2018;14:e1007389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao R, Hu Y, Li Z, et al. Dissemination and mechanism for the MCR-1 colistin resistance. PLoS Pathog. 2016;12:e1005957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu Y, Lin J, Cui T, et al. Mechanistic insights into transferable polymyxin resistance among gut bacteria. J Biol Chem. 2018;293:4350–4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu YY, Wang Y, Walsh TR, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16:161–168. [DOI] [PubMed] [Google Scholar]
- 16.Yang RS, Feng Y, Lv XY, et al. Emergence of NDM-5 and MCR-1-producing Escherichia coli clone ST648 and ST156 from a single Muscovy duck (Cairina moschata). Antimicrob Agents Chemother. 2016;60(11):6899–6902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu BT, Song FJ, Zou M, et al. High incidence of Escherichia coli strains coharboring mcr-1 and blaNDM from chickens. Antimicrob Agents Chemother. 2017;61(3):pii: e02347-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhong LL, Zhang YF, Doi Y, et al. Coproduction of MCR-1 and NDM-1 by colistin-resistant Escherichia coli isolated from a healthy individual. Antimicrob Agents Chemother. 2017;61(1):pii: e01962-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mediavilla JR, Patrawalla A, Chen L, et al. Colistin- and carbapenem-resistant Escherichia coli harboring mcr-1 and blaNDM-5, causing a complicated urinary tract infection in a patient from the United States. mBio. 2016;7(4):pii: e01191-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu BT, Song FJ, Zou M, et al. Emergence of colistin resistance gene mcr-1 in Cronobacter sakazakii producing NDM-9 and in Escherichia coli from the same animal. Antimicrob Agents Chemother. 2017;61(2):pii: e01444-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou YF, Tao MT, Feng Y, et al. Increased activity of colistin in combination with amikacin against Escherichia coli co-producing NDM-5 and MCR-1. J Antimicrob Chemother. 2017;72(6):1723–1730. [DOI] [PubMed] [Google Scholar]
- 22.Yao X, Doi Y, Zeng L, et al. Carbapenem-resistant and colistin-resistant Escherichia coli co-producing NDM-9 and MCR-1. Lancet Infect Dis. 2016;16:288–289. [DOI] [PubMed] [Google Scholar]
- 23.Liu L, Feng Y, Zhang X, et al. New variant of mcr-3 in an extensively drug-resistant Escherichia coli clinical isolate carrying mcr-1 and blaNDM-5. Antimicrob Agents Chemother. 2017;61(12):pii: e01757-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leigue L, Warth JF, Melo LC, et al. MDR ST2179-CTX-M-15 Escherichia coli co-producing RmtD and AAC(6ʹ)-Ib-cr in a horse with extraintestinal infection, Brazil. J Antimicrob Chemother. 2015;70:1263–1265. [DOI] [PubMed] [Google Scholar]
- 25.Lv J, Mohsin M, Lei S, et al. Discovery of a mcr-1-bearing plasmid in commensal colistin-resistant Escherichia coli from healthy broilers in Faisalabad, Pakistan. Virulence. 2018;9:994–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhi C, Lv L, Yu LF, et al. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis. 2016;16:292–293. [DOI] [PubMed] [Google Scholar]
- 27.Delgado-Blas JF, Ovejero CM, Abadia Patino L, et al. Coexistence of mcr-1 and blaNDM-1 in Escherichia coli from Venezuela. Antimicrob Agents Chemother. 2016;60:6356–6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Conant GC, Wolfe KH. GenomeVx: simple web-based creation of editable circular chromosome maps. Bioinformatics. 2008;24:861–862. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data are included in this manuscript. Complete sequences of the two plasmids were deposited into GenBank under the accession no. (MG773376 for pLSB54-mcr-1, MG773377 for pLSB54-NDM-5).


