Abstract
In triploblastic animals, Par-proteins regulate cell-polarity and adherens junctions of both ectodermal and endodermal epithelia. But, in embryos of the diploblastic cnidarian Nematostella vectensis, Par-proteins are degraded in all cells in the bifunctional gastrodermal epithelium. Using immunohistochemistry, CRISPR/Cas9 mutagenesis, and mRNA overexpression, we describe the functional association between Par-proteins, ß-catenin, and snail transcription factor genes in N. vectensis embryos. We demonstrate that the aPKC/Par complex regulates the localization of ß-catenin in the ectoderm by stabilizing its role in cell-adhesion, and that endomesodermal epithelial cells are organized by a different cell-adhesion system than overlying ectoderm. We also show that ectopic expression of snail genes, which are expressed in mesodermal derivatives in bilaterians, is sufficient to downregulate Par-proteins and translocate ß-catenin from the junctions to the cytoplasm in ectodermal cells. These data provide molecular insight into the evolution of epithelial structure and distinct cell behaviors in metazoan embryos.
Research organism: Other
eLife digest
Most animals – including birds, fish and mammals – have symmetrical left and right sides, and are known as bilaterians. During early life, the embryos of animals in this group develop three distinct layers of cells: the ectoderm (outer layer), the endoderm (inner layer), and the mesoderm (middle layer). These layers then go on to form the animal’s tissues and organs. The ectoderm produces external tissues, such as the skin and the nervous system; the endoderm forms internal tissues, like the gut; and the mesoderm creates all tissues in between, like muscles and blood.
Another, smaller group of animals, called cnidarians, do not have left and right sides. Instead, they have a ‘radial symmetry’, meaning they have multiple identical parts arranged in a circle. These animals – which include corals, jellyfish and sea anemones – only develop two distinct layers of cells, equivalent to the outer and inner layers of bilaterians. Cnidarians evolved before bilaterians, but their genetic material is equally complex. So why did these two groups evolve to have different layers of cells? And how exactly do animal embryos develop these distinct layers?
To address these questions, Salinas-Saavedra et al. studied embryos of the sea anemone Nematostella vectensis. Molecules called Par-proteins play an important role in controlling how cells behave and attach to one another (and therefore how they form layers). So, using a technique called immunohistochemistry to look inside cells, Salinas-Saavedra et al. examined these proteins in the two layers of cells in sea anemone embryos.
The experiments found that in the sea anemones, Par-proteins are arranged differently in cells that form the ‘skin’ compared to cells that form the ‘gut’. In other words, cells in the outer layer attach to one another in a different way than cells in the inner layer, where the Par-proteins are degraded by ‘mesodermal’ genes. The findings also show that these sea anemones have all they need to form a third middle layer of cells. Like bilaterians, they could potentially move cells in and out of sheets that line surfaces inside the body – but they do not naturally do this.
Understanding how animals form different layers of cells is important for scientists studying evolution and the development of embryos. However, it also has wider applications. For instance, some cells involved in developing the mesoderm are also involved in forming tumors. Future research in this area could help scientists learn more about how cancer-like cells form in animals.
Introduction
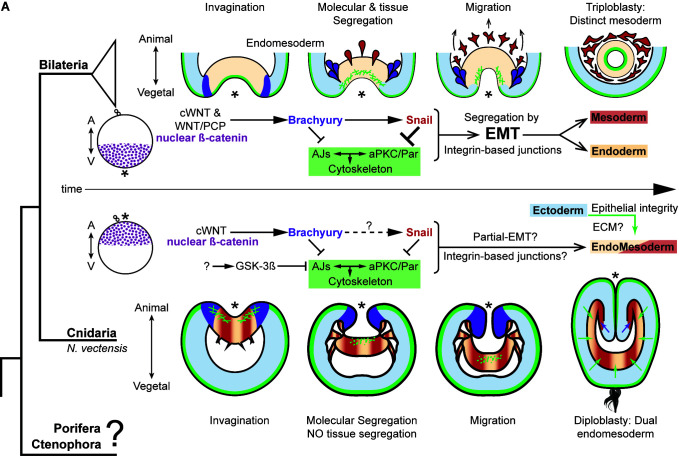
Bilaterian animals comprise more than the 95% of the extant animals on earth and exhibit enormous body plan diversity (Martindale and Lee, 2013). One of the most important morphological features in bilaterian evolution is the emergence of the mesoderm, an embryological tissue that gives rise important cell types such as muscle, blood, cartilage, bone, and kidneys in the space between ectoderm and endoderm. The emergence of mesoderm clearly contributed to the explosion of biological diversity throughout evolution (Martindale and Lee, 2013; Martindale, 2005). Cnidarians (e.g. sea anemones, corals, hydroids, and ‘jellyfish’) are the sister group to bilaterians, and despite their surprisingly complex genomes (Putnam et al., 2007), do not possess a distinct mesodermal tissue layer. Instead, the gastrodermal lining to their gut cavity consists of a bifunctional endomesodermal epithelium with molecular characteristics of both bilaterian endodermal and myoepithelial mesodermal cells (Martindale and Lee, 2013; Technau and Scholz, 2003; Martindale et al., 2004; Jahnel et al., 2014). For example, brachyury and snail, among other genes, contribute to the specification of the endomesodermal fates in both bilaterian and cnidarian embryos (Technau and Scholz, 2003; Martindale et al., 2004; Magie et al., 2007; Yasuoka et al., 2016; Servetnick et al., 2017). Yet in bilaterians, mesodermal cells segregate from an embryonic endomesodermal precursor to form both endoderm and a third tissue layer (mesoderm) not present in the embryos of diploblastic cnidarians (Martindale et al., 2004; Rodaway and Patient, 2001; Davidson et al., 2002; Maduro and Rothman, 2002; Solnica-Krezel and Sepich, 2012). How mesodermal cells originally segregated from an ancestral endomesodermal epithelium during animal evolution is still unclear (Martindale and Lee, 2013; Martindale, 2005; Technau and Scholz, 2003), particularly because virtually all of the genes required for mesoderm formation are present in cnidarian genomes (Putnam et al., 2007; Baumgarten et al., 2015; Chapman et al., 2010; Shinzato et al., 2011). During the last decade, several studies have described molecular and cellular characteristics related to the segregation of mesoderm during bilaterian development (Solnica-Krezel and Sepich, 2012; Keller et al., 2003; Darras et al., 2011; Schäfer et al., 2014). Here, we investigate the cellular basis of morphogenesis during embryogenesis of the ‘diploblastic’ sea anemone, Nematostella vectensis.
In most bilaterian embryos described to date, after a series of synchronous and stereotyped cleavage divisions, maternal determinants induce the localization of nuclear ß-catenin to blastomeres derived from the vegetal pole (Martindale and Lee, 2013). Hence, gastrulation and the specification of endomesodermal fates is restricted to the vegetal pole. In these species, brachyury is expressed at the border of the blastopore and snail is expressed in the prospective mesodermal tissues (Technau and Scholz, 2003). The formation of mesoderm involves a variety of cellular processes including the downregulation of E-cadherin, loss of apicobasal cell polarity, and in some cases, the induction of epithelial-to-mesenchymal transition (EMT) (Solnica-Krezel and Sepich, 2012; Schäfer et al., 2014; Acloque et al., 2009; Lim and Thiery, 2012).
Embryos of the cnidarian starlet sea anemone N. vectensis develop without a stereotyped cleavage pattern but cell fates become organized along the embryonic animal-vegetal axis (Fritzenwanker et al., 2007; Salinas-Saavedra et al., 2015). During blastula formation, embryonic cells of N. vectensis form a single hollow epithelial layer. Epithelial cells of the animal pole, characterized by the nuclear localization of Nvß-catenin prior to gastrulation (Wikramanayake et al., 2003; Lee et al., 2007), invaginate by apical constriction to form the endomesodermal epithelium (Magie et al., 2007; Tamulonis et al., 2011). The expression of Nvbrachyury around the presumptive border of the blastopore and Nvsnail genes in the presumptive endomesodermal gastrodermis of N. vectensis embryos occurs even before the morphological process of gastrulation begins (Scholz and Technau, 2003; Röttinger et al., 2012).
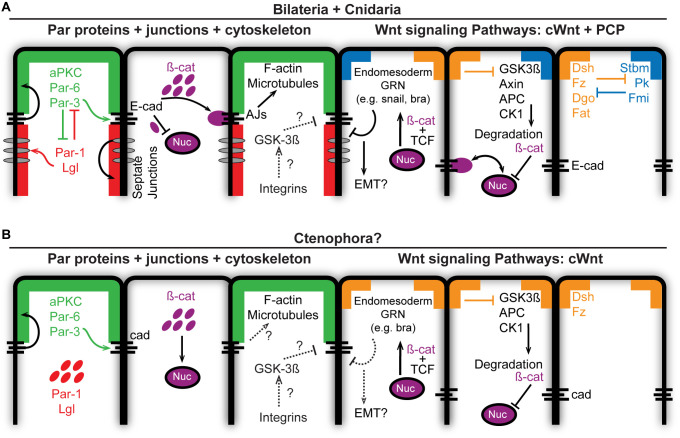
Interestingly, the components of the intracellular polarity Par system (NvaPKC, NvPar-6, NvPar-3, NvPar-1, and NvLgl), which show a highly polarized bilaterian-like subcellular distribution throughout all epithelial cells at the blastula stage in N. vectensis (Salinas-Saavedra et al., 2015), are specifically degraded and down-regulated from the endomesoderm during the gastrulation process (Figure 1A). We have previously suggested that the expression of bilaterian ‘mesodermal genes’ (e.g. Nvsnail) might induce the loss of apicobasal cell-polarity indicated by the absence of the components of the Par system in the endomesoderm of N. vectensis embryos (Salinas-Saavedra et al., 2015). Recent studies in N. vectensis and bilaterians have provided information that supports this hypothesis. For example, it has been shown that snail is necessary and sufficient to downregulate Par3 in Drosophila mesoderm, inducing the disassembly of junctional complexes in these tissues (Weng and Wieschaus, 2016, 2017). In addition, we have shown that Nvbrachyury regulates epithelial apicobasal polarity of N. vectensis embryos, suggesting some aspects of epithelial cell polarity are highly conserved (Servetnick et al., 2017). Together, this evidence suggests a plausible cellular and molecular mechanism for the segregation of a distinct cell layer in bilaterian evolution from an ancestral bifunctional endomesodermal tissue. Thus, in this study, we describe the functional association between the components of the Par system, apical junctions, epithelial integrity, and the nuclearization of Nvß-catenin in a cnidarian embryo. In addition, we demonstrate that the endomesoderm in N. vectensis is organized by different junctional complexes that confer different functional properties to this tissue than the overlying ectoderm. And finally, we investigate the putative interactions between the components of the Par system, the canonical Wnt signaling pathway, and snail gene expression, giving insights on the evolution of the mesoderm and EMT.
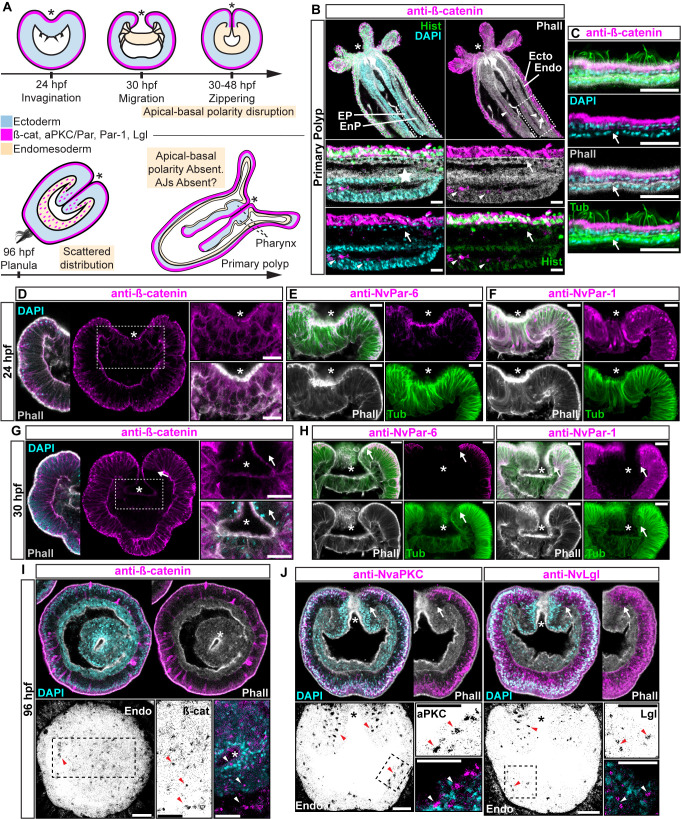
Figure 1. Components of the Par system and ß-catenin are downregulated from the N. vectensis endomesoderm during gastrulation.
(A–F) Confocal images of immunofluorescent staining (IFS) of lateral views of gastrulation embryos (animal pole up). The * marks the site of gastrulation in all cases. Samples are counterstained with Phalloidin (Phall) staining (white) to show cell boundaries, DAPI to visualize cell nuclei (blue), and Tubulin antibody (Tub) staining is shown as counterstain (green). All images are a single optical section from a z-stack confocal series. All scale bars, 20 µm. (A) Summary diagram depicting the localization of ß-catenin and Par proteins at the observed stages. Pale boxes denote changes observed in the endomesoderm. (B) IFS for ß-catenin (magenta) in primary polyps. High magnification images from boxed region (endomesoderm, Endo) are shown on the bottom. Arrows indicate the absence of ß-catenin expression in the endomesoderm. Arrowheads indicate the ß-catenin expression in the ectodermal pharynx (EP). Star indicates the endomesodermal pharynx (EnP). Histone antibody (Hist) staining is shown as counterstain to show the penetrability in the fixed tissue. See also Figure 1—figure supplement 1. (C) IFS for ß-catenin (magenta) in the ecto and endomesoderm (arrow) of primary polyps. (D) IFS for ß-catenin (magenta) at 24 hpf shows localization to the apical domain where adherens junctions reside in all cells of the blastula. High magnification images from boxed region (prospective endomesoderm) are shown on the right. (E) IFS for NvPar-6 (magenta) at 24 hpf showing the same sub-cellular localization as ß-catenin (A). High magnification images from boxed region in (A) (prospective endomesoderm) are shown on the right. Merged image shown on upper left. (F) IFS for NvPar-1 at 24 hpf shows a complementary basolateral expression. High magnification images from boxed region (prospective endomesoderm) are shown on the right. (G) IFS for ß-catenin at 30 hpf shows the loss of expression of ß-catenin (magenta) in invaginating endomesoderm (box). The arrow (D–F) marks the boundary between ectoderm and invaginating endomesoderm. High magnification images from boxed region (prospective endomesoderm) are shown on the right. (H) IFS for NvPar-6 and NvPar-1(magenta) at 30 hpf show that all Par proteins are down regulated at the site of gastrulation. IFS for NvPar-6 shows an even earlier down regulation than ß-catenin (D). High magnification images from boxed region (prospective endomesoderm) are shown on the right. Merged image shown on upper left. (I) Oral view of IFS for ß-catenin (magenta) at 96 hpf showing apical localization in overlying ectoderm, but absence in endomesodermal tissues. The two bottom panels show high magnifications of the endomesoderm region (image inverted). Arrowheads indicate the localization of ß-catenin expression (black) in some scattered endomesodermal cells. (J) Lateral view of IFS for NvaPKC and NvLgl (magenta) at 96 hpf showing loss of expression in invaginating epithelial cells. The four bottom panels show high magnifications of the endomesoderm region (image inverted). Arrowheads indicate the localization of NvaPKC and NvLgl proteins (black) in some scattered endomesodermal cells.
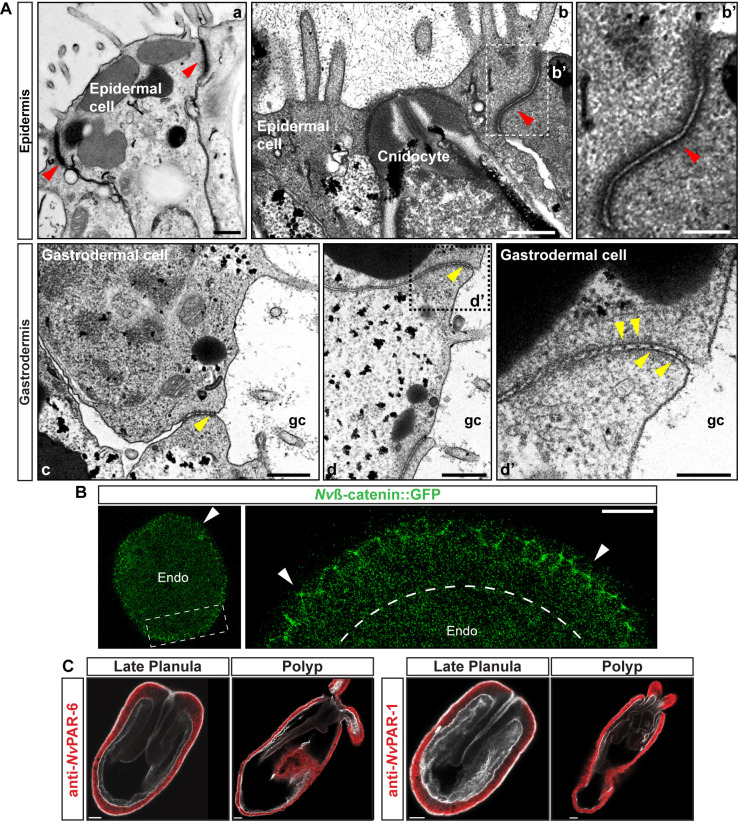
Figure 1—figure supplement 1. Epidermal and gastrodermal cells are joined by different set of junctional complexes.
Results
Ectodermal and endomesodermal epithelia are organized by different cell-cell adhesion complexes
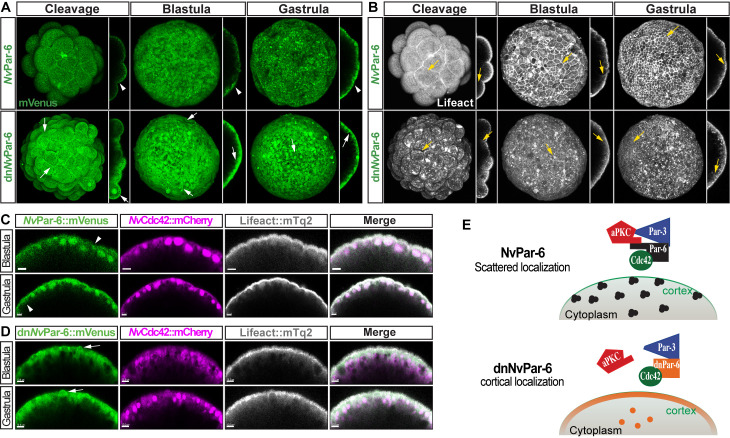
Components of the Par system are not present in the cells of endomesodermal epithelium of N. vectensis during gastrulation, even though the very same cells express these proteins during the blastula stage (Salinas-Saavedra et al., 2015) (Figure 1). This absence is consistent with the absence of apical Adherens Junctions (AJs) in the endomesoderm of N. vectensis (Figure 1—figure supplement 1) and other cnidarians (Magie et al., 2007; Chapman et al., 2010; Ganot et al., 2015). At polyp stages, neither ß-catenin (an AJ-associated protein) (Figure 1B and C) nor the Par proteins (Figure 1—figure supplement 1C) are detectable in endomesodermal cells of either the gastrodermis or the pharynx. When N. vectensis embryos are stained with antibodies to ß-catenin (Figure 1) or if Nvß-catenin::GFP mRNA is expressed in uncleaved zygotes (Figure 1—figure supplement 1B), clear localization of ß-catenin can be seen in the cortex of ectodermally derived epithelial cells (Figures 1B, C, D, G and I), but not in endomesodermal cells (Figure 1B and C). In pharyngeal cells that are located between the epidermis and gastrodermis, Nvß-catenin (Figure 1B and D), NvPar-6 (Figure 1E), and NvPar-1 (Figure 1F) expression begins to disappear, and is localized only in the most apical regions, indicating that AJs are being disassembled/degraded during the gastrulation process (Figure 1G and H). During later planula stages, ß-catenin and the components of the Par system display scattered patterns in the cytoplasm of a small subset of endomesodermal cells (Figure 1I and J). Even though we do not know the identity of these cells, this expression temporally coincides with the transient activation of Wnt signaling emanating from the oral pole (Kusserow et al., 2005; Marlow et al., 2013) at those developmental stages. In bilaterians (Acloque et al., 2009; Lim and Thiery, 2012) and N. vectensis (Kusserow et al., 2005; Marlow et al., 2013), the later activation of Wnt signaling is also associated with neurogenesis, and may cause the observed changes in protein localization.
Regardless of this scattered expression, it is clear that cells that undergo gastrulation in N. vectensis lose their polarized ectodermal cell-cell adhesion complex and components of the Par system, including ß-catenin, are downregulated from endomesodermal tissues (Figure 1). In bilaterians, the proper formation of an epithelial paracellular barrier (essential for tissue homeostasis) depends on the establishment of adhesive complexes between adjacent cells (Higashi et al., 2016; Jonusaite et al., 2016), which are regulated by the aPKC/Par complex (Ohno et al., 2015). To test if this absence of protein expression is correlated to differential cell-cell adhesion in the endomesodermal epithelium of N. vectensis, we assessed their role in regulating paracellular movements between ectodermal and endomesodermal epithelia by using a fluorescent tracer dye penetration assay (Figure 2A) (Higashi et al., 2016). For the purposes of these experiments, in order to avoid unwanted results related to tissue specification, cell proliferation, and cell movements, we used newly hatched juvenile polyps where the gastrodermis (endomesodermally derived) is fully differentiated.
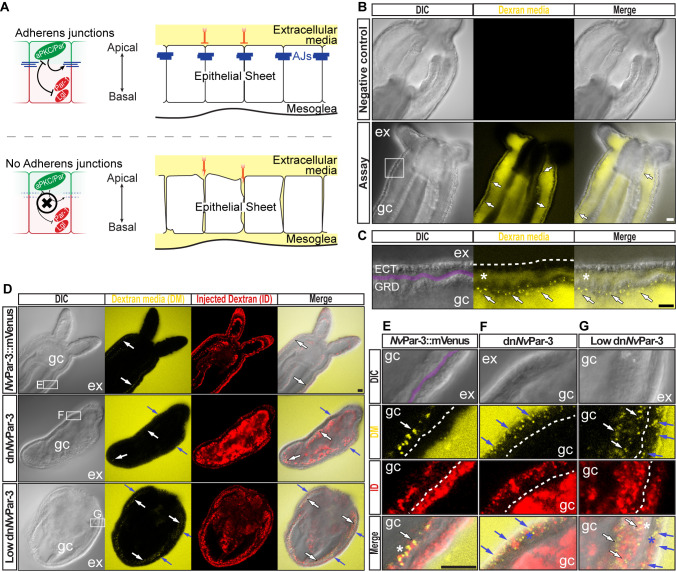
Figure 2. The aPKC/Par complex maintains Adherens Junctions (AJs) of ectodermal epithelial cells.
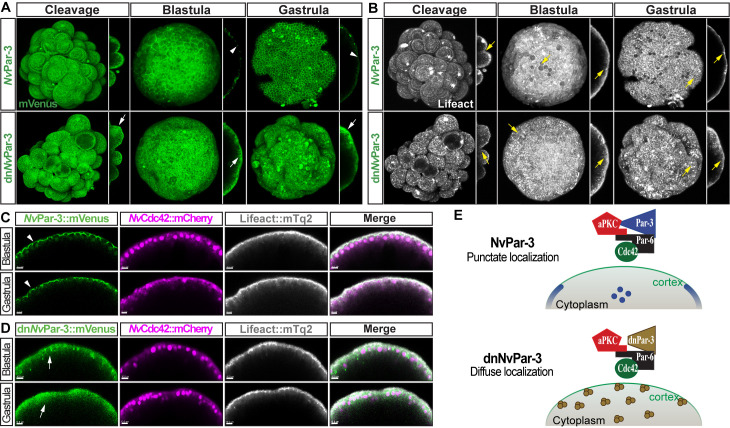
Arrows indicate the direction of the flow: from gastric cavity (gc) to mesoglea (white) and from external media (ex) to ectoderm (blue). Dashed lines indicate the base of the epidermis. All images are single optical section from the z-stack confocal series. Scale bars, 20 µm. (A) Diagram depicting the hypothesis that when the aPKC/Par complex is functional (top row), AJs are present (blue stripes) and a paracellular epithelial barrier is formed. When aPKC/Par complex is not functional (bottom row), AJs are disrupted, the epithelial barrier is perturbed, and the extracellular solution moves paracellularly into the mesoglea. (B) Penetration assay of wild type (uninjected) primary polyps at low magnification showing the movement of 10,000 MW fluorescent dextran. Top row, no dextran. Bottom row, dextran (yellow) in the gc moves in to the mesoglea through paracellular spaces between gastrodermal cells (arrows). (C) High magnification images from box shown in (B). *: mesoglea (purple band) that separates the ectoderm (ECT, dashed line) from gastrodermis (GDR). Note the dye moving between cells from the gc media (arrows). (D) Low magnification images comparing polyps expressing NvPar-3::mVenus and a dominant negative version of NvPar-3 (dnNvPar-3::mVenus) expressing-embryos. Dextran media (DM; extracellular) is pseudo-colored yellow. Dextran (red) was co-injected with mRNAs to label the cells and differentiate intracellular regions. mVenus channel was omitted for better visualization (shown in Figure 2—figure supplement 1). Lower concentrations of dnNvPar-3 were injected to preserve endomesodermal tissues. Note that the dextran media was found between the cells labeled in red. See also Figure 3—figure supplement 1 for dnNvPar-3 description. (E) High magnification images from (E) boxed region in (D). Purple band depicts Mesoglea. (F) High magnification images from (F) boxed region in (D). (G) High magnification images from (G) boxed region in (D). *: Paracellular spaces of both, the epidermis (blue) and gastrodermis (white).
Figure 2—figure supplement 1. mVenus protein expression related to Figure 2.
N. vectensis polyps were exposed to media containing 10,000 MW fluorescent dextran (Molecular Probes, Inc.). When juvenile polyps are incubated in dextran for 5–10 min (Figure 2B), fluorescent dextran solution moves into the gastric cavity and then spreads into the mesoglea through the gastrodermal epithelium (Figure 2C) but does not enter the mesoglea through the outer ectodermally-derived epidermis (Figure 2C and D). These results suggest that cell-cell adhesion is differentially regulated between the epidermis and gastrodermis and the absence/disruption of AJs may compromise Septate Junctions (SJs) in the gastrodermis. Similar results were obtained in N. vectensis polyps when we overexpressed NvPar-3::mVenus by injection of mRNA into uncleaved eggs which is normally expressed in ectodermal but not endodermal epithelial tissue (Figure 2D and E). However, in polyps expressing a dominant negative version of NvPar-3::mVenus (dnNvPar-3; microinjected into uncleaved eggs) dye penetrated between epithelial cells in both the gastrodermis and the outer epidermis (Figures 2D, F and G), demonstrating an ancestral role of the aPKC/Par complex in the maintenance of cell-cell adhesion and the paracellular boundary (SJs) of epithelial cells during animal development.
The NvaPKC/Par complex regulates the formation and maintenance of cell-cell junctions
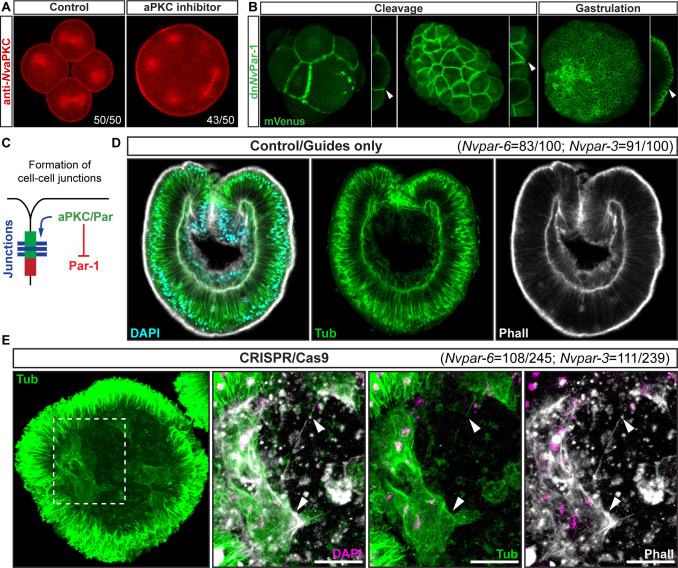
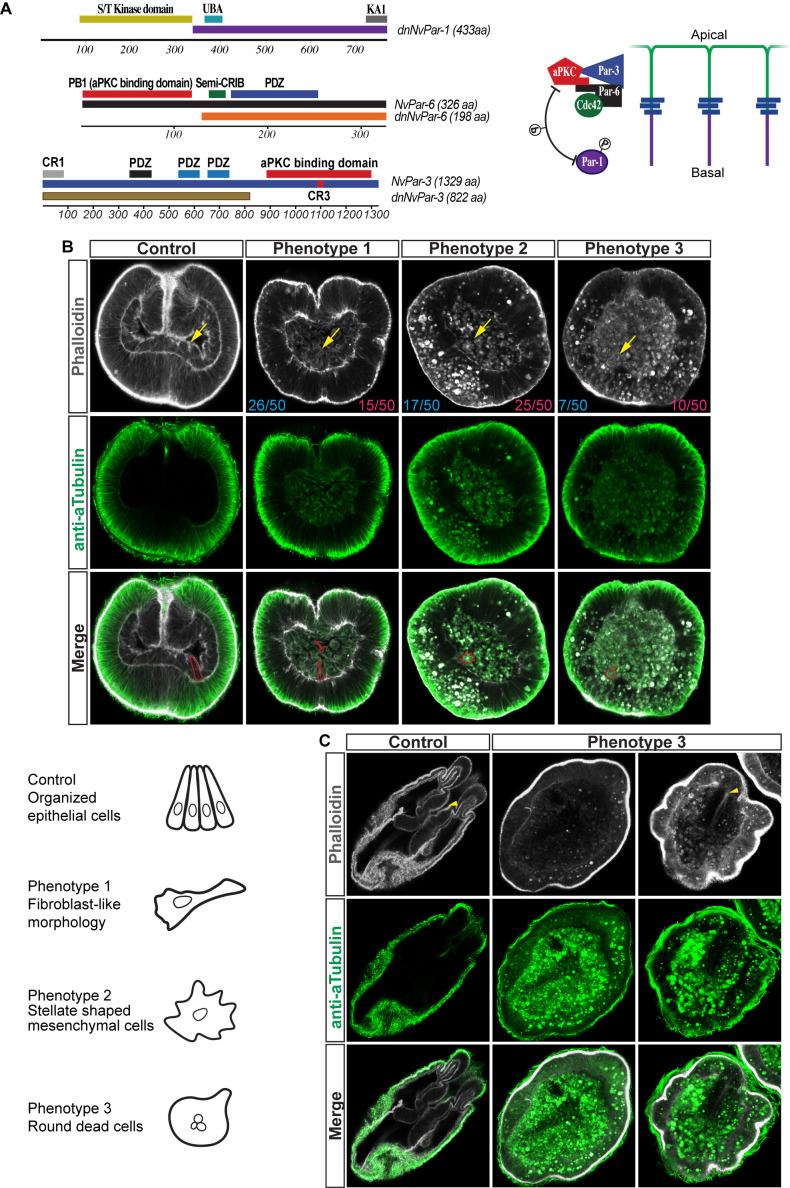
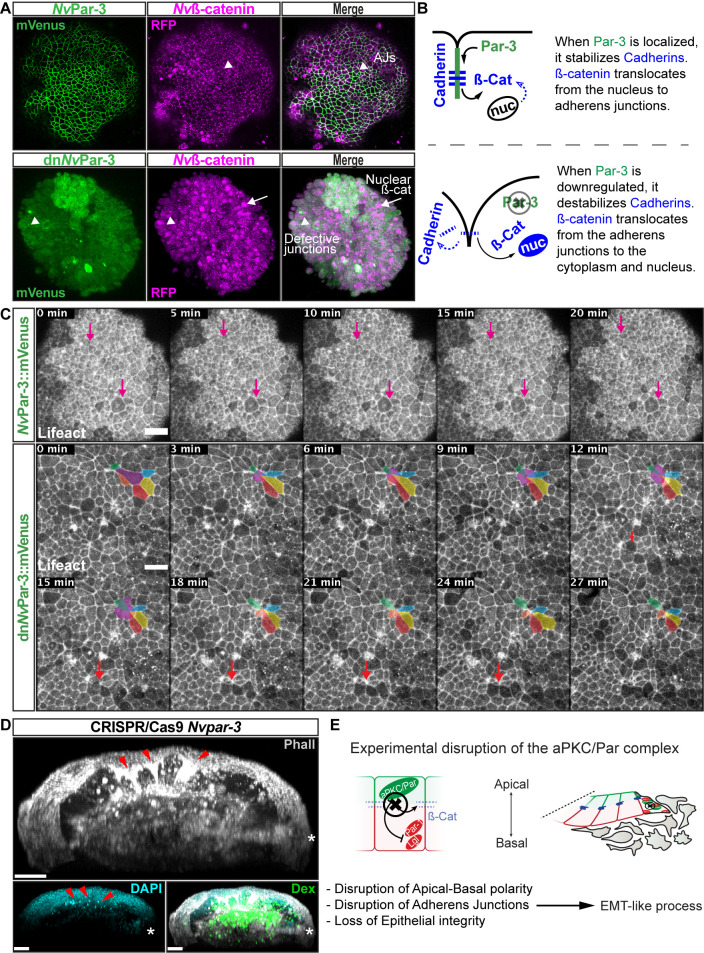
Our results suggest that the absence of Par proteins in the endomesoderm is associated with changes in cell-cell adhesion complexes. Pharmacological treatment of N. vectensis embryos with an aPKC activity inhibitor blocks cytokinesis but not mitosis in cleaving embryos (Figure 3A). In addition, a dominant negative version of NvPar-1 (dnNvPar-1), that lacks its kinase domain, localizes only to the cortex of cell–cell contacts (Figure 3B). Since NvPar-1 is phosphorylated by NvaPKC (Figure 3—figure supplement 2), we predict that, as in other systems, dnNvPar-1 could be phosphorylated by aPKC but would not phosphorylate the aPKC/Par complex (Vaccari et al., 2005; Böhm et al., 1997). Thus, dnNvPar-1 can localize to the cell cortex where aPKC may be inactive. These results together suggest that the formation of cell–cell contacts is regulated by the activity of the aPKC/Par complex in N. vectensis embryos (Figure 3C).
Figure 3. Ectodermal NvaPKC/Par complex polarity regulates the epithelial integrity of both ecto- and endomesoderm.
(A) IFS for NvaPKC at 4 hpf showing that the aPKC inhibitor (Sigma P1614) blocks cytokinesis but not cell cycle. (B) in vivo expression of dnNvPar-1 shows precocious localization to zones of cell contact during cleavage stages, well before wild-type NvPar genes do. See also Figure 3—figure supplement 1A and Figure 3—figure supplement 2. (C) Diagram depicting the suggested the working hypothesis. (D) CRISPR/Cas9 knock-out for NvPar-6 and NvPar-3 at 48 hpf. Controls show no effect on gastrulation. Tubulin (Tub), Phalloidin (Phall), and DAPI are used as counterstains. (E) CRISPR/Cas9 mutants: tubulin stained low magnification of CRISPR phenotype. High magnification images from boxed region shows mesenchymal-like cells. Arrowheads indicate filopodia-like structures. Number of cases observed for each gene are shown. See also Figure 3—figure supplement 1, Figure 3—figure supplements 3–6, and Figure 3—video 1. Morphology is shown by DAPI, Tub, and Phall IFS. Except for 3B and 3D, all images are single optical sections from the z-stack confocal series. (B) and (D) are 3D reconstructions from a z-stack confocal series. All scale bars, 20 µm.
Figure 3—figure supplement 1. Disruption of the aPKC/Par complex in N. vectensis embryos.
Figure 3—figure supplement 2. Co-immunoprecipitation of NvPar proteins using a NvPar-1 specific antibody.
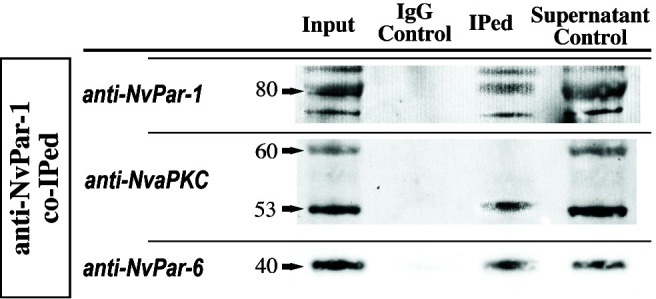
Figure 3—figure supplement 3. Different ectodermal thickness observed in CRISPR/Cas9 mutants.
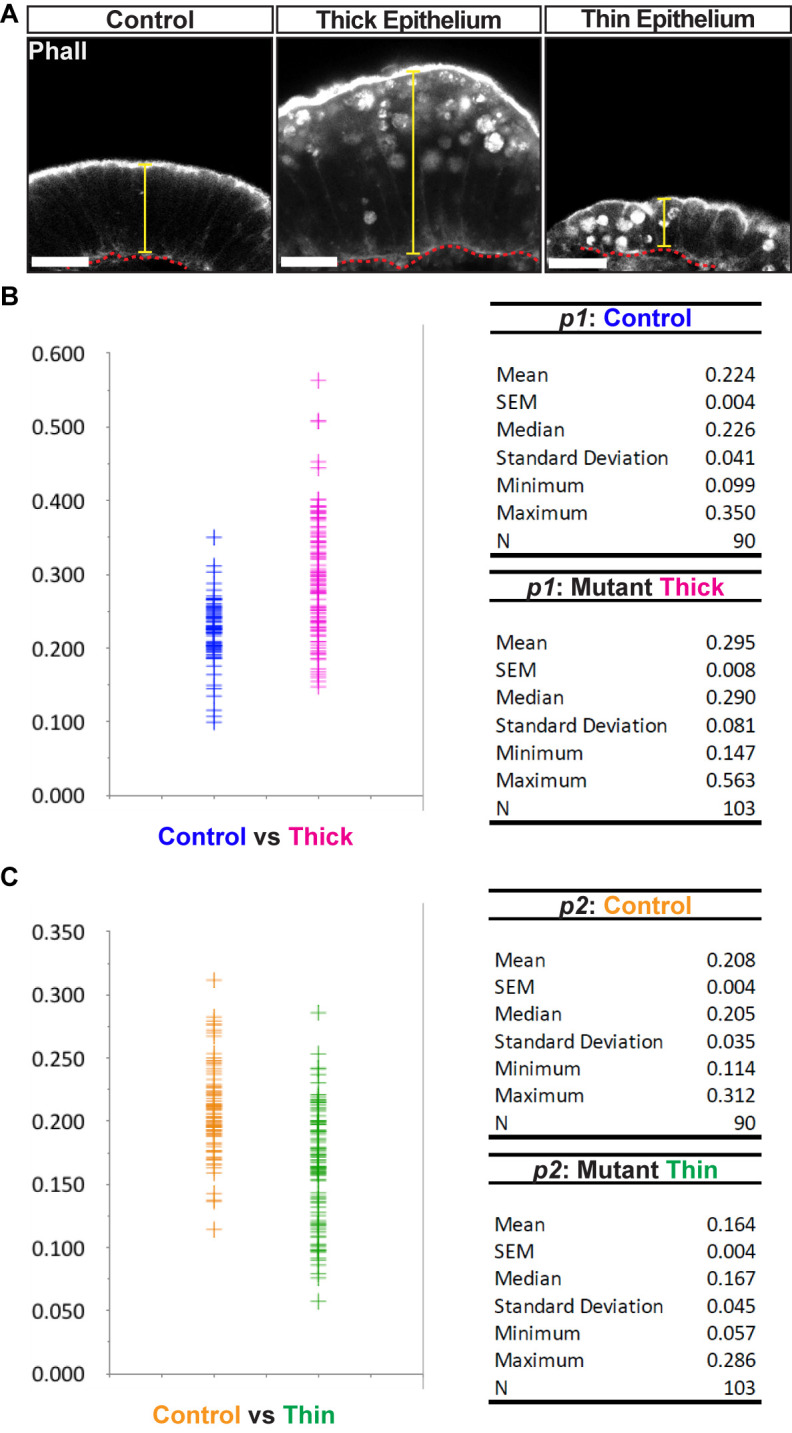
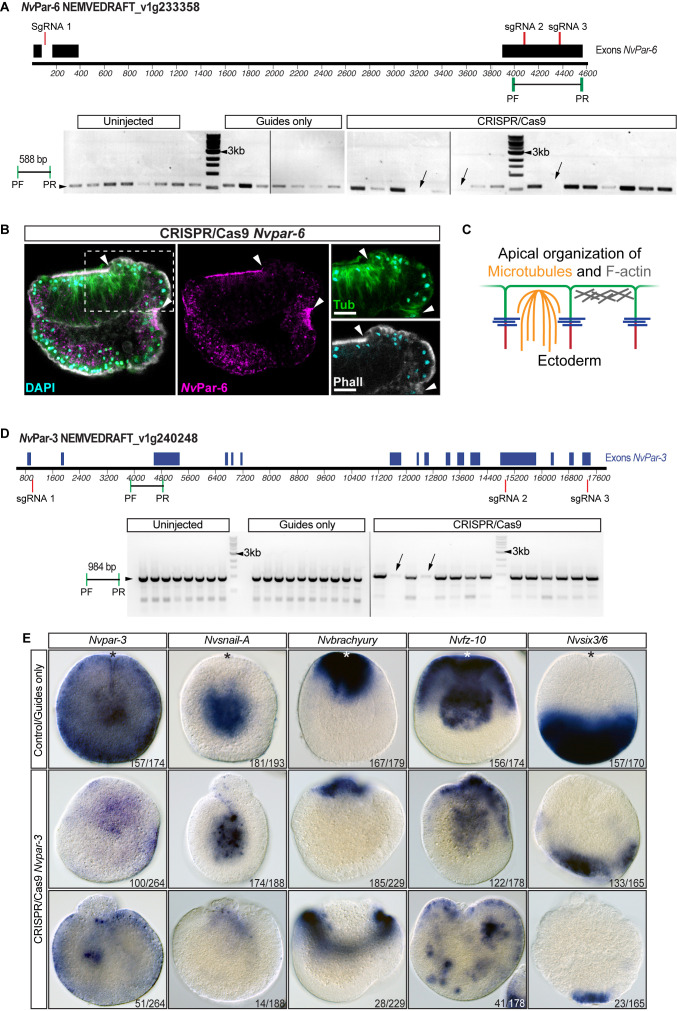
Figure 3—figure supplement 4. CRISPR/Cas9 mediated mutagenesis of Nvpar-6 and Nvpar-3.
Figure 3—figure supplement 5. In vivo localization of dnNvPar-6 protein at different embryonic stages.
Figure 3—figure supplement 6. In vivo localization of dnNvPar-3 protein at different embryonic stages.
Figure 3—figure supplement 7. Morphometric measurements methodology.
Figure 3—video 1. Related to Figure 3E.
We further tested this hypothesis by using genome editing by CRISPR/Cas9 targeting Nvpar-6 and Nvpar-3 genes (Figure 3D). We did not observe any effects on the embryo until 36 hpf at 16°C (late blastula stage), indicating the activity of maternally loaded proteins up until that stage. When NvPar-6 and NvPar-3 are mutated, the ectodermal epithelium loses its integrity, presenting changes in thickness (Figure 3—figure supplement 1B and Figure 3—figure supplement 3), and interestingly, the endomesoderm (which does not express these proteins) generates cells with mesenchymal-like morphotypes that are never normally seen in this species (Figure 1D and E). In Nvpar-6 and Nvpar-3 mutant embryos, we also observed the disruption of microtubules and actin cytoskeleton (Figure 3—figure supplement 4B), and AJs (visualized with the ß-catenin antibody in Figure 4) that confirms our previous observations of their role in regulating ectodermal cell polarity. Although it was difficult to dissect significant changes in the expression of germ layer markers (e.g. Nvbra, Nvsnail, NvSix3/6, and Nvfz10) from the morphological changes associated with epithelial integrity when these genes were disrupted (Figure 3—figure supplement 4E), it is clear that the primary defect in NvPar3 KO were aspects of cell adhesion and not cell type specification. Similar results were obtained when we overexpressed the mRNA encoding for a dominant negative version NvPar-6 (dnNvPar-6) and NvPar-3 (dnNvPar-3) into N. vectensis eggs (Figure 3—figure supplement 1 and Figure 3—figure supplements 5 and 6). However, dominant negative effects on the injected embryos were observed at earlier stages (10–12 hpf) than the CRISPR/Cas9 mutants (zygotic expression) because the mutant proteins compete with the wild type proteins (maternally loaded). Hence, in these experiments, embryonic lethality (90%) and cell death were higher.
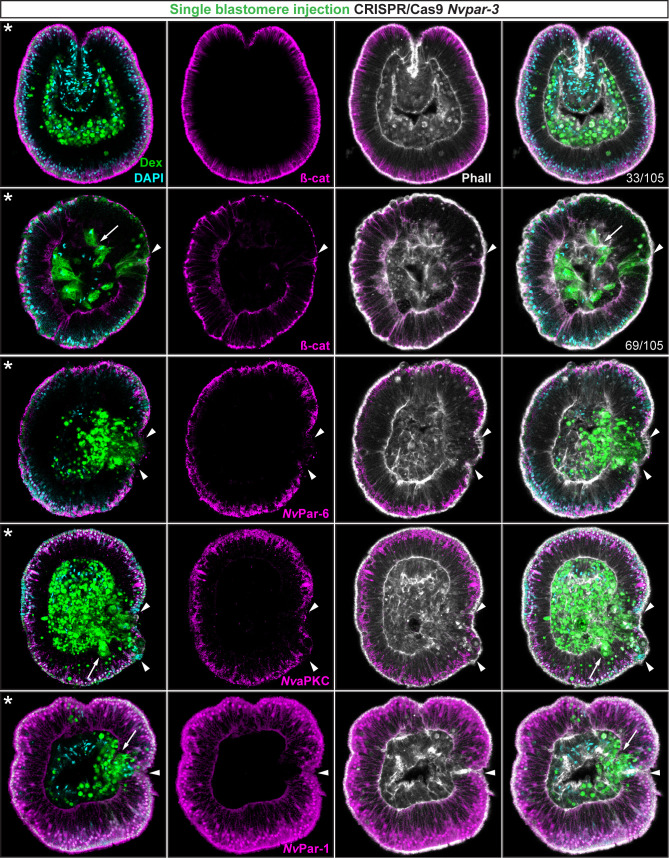
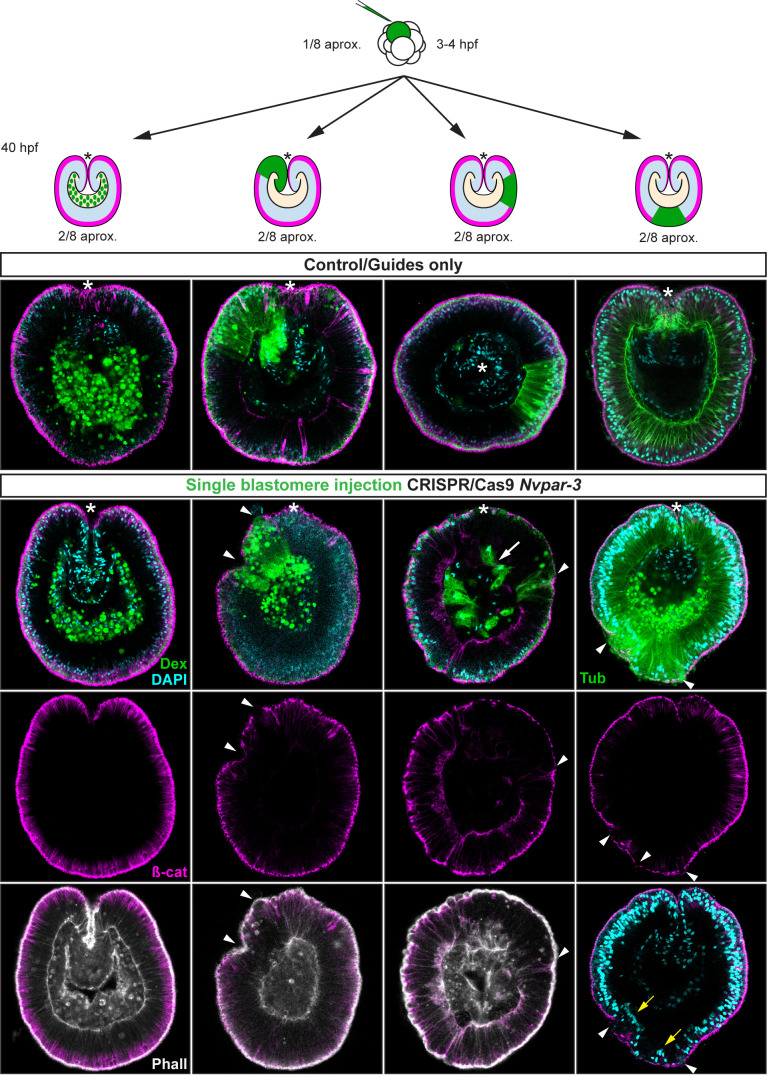
Figure 4. Ectodermal- but not endomesodermal-lineages are affected by single injected-blastomere CRISPR/Cas9 Nvpar-3 knock-outs.
IFS for ß-catenin (ß-cat), NvPar-6, NvaPKC, and NvPar-1 in single injected-blastomere CRISPR/Cas9 NvPar-3 knock-outs at 40 hpf. Streptavidin-Biotin TxRed Dextran (Dex) is shown in green. Arrowheads indicate the absence of the protein and disrupted epithelium. Arrows indicate bottle-like shape cells. * indicate the orientation of the site of gastrulation. See also Figure 4—figure supplements 1 and 2, and Figure 4—video 1. Morphology is shown by DAPI and Phalloidin. All images are single optical sections from the z-stack confocal series.
Figure 4—figure supplement 1. Diagram depicting the hypothesis addressed in Figure 4.
Figure 4—figure supplement 2. Immunofluorescent staining for ß-catenin (ß-cat) in single injected-blastomere control and CRISPR/Cas9 Nvpar-3 knock-outs at 40 hpf.
Figure 4—video 1. z-stack series of CRISPR phenotype (ß-catenin panel).
The NvaPKC/Par complex regulates transepithelial signaling
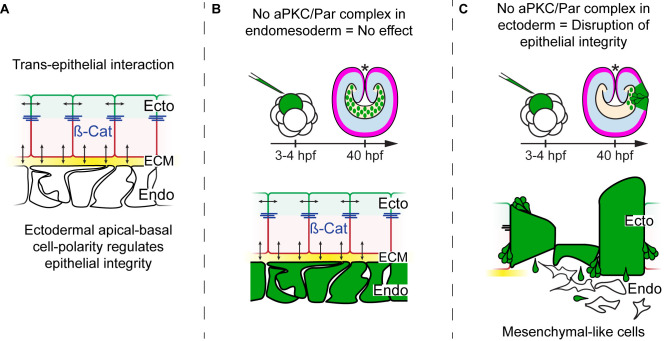
One surprising observation from the experiments described above show that the changes observed in the ectodermal and endodermal epithelium after disrupting NvPar-6 and NvPar-3 (Figure 3) suggests some sort of trans-epithelial regulation of cell–cell adhesion (most likely involving AJs) because these Par genes are not expressed in the endomesoderm. The polarizing activity of the aPKC/Par complex in the ectoderm is thus necessary to maintain the integrity of both ecto- and endodermal epithelia during cellular movements associated with gastrulation.
To assess whether the observed phenotypes on cell-cell adhesion are related to non-autonomous cell regulation (trans-epithelial interactions), we repeated the above experiments randomly injecting single blastomeres at 3–4 hpf (8–16 cell-stage) to make mutant clones in an otherwise wild type background (Figure 4). In these experiments, only the cell-lineage of the injected blastomere would be affected and would exhibit defective cell-cell adhesion in an otherwise undisturbed wild-type background. If endomesodermal cells derived from an injected blastomere display fibroblast/mesenchymal cell morphology, it would indicate that the organization of the endomesodermal epithelium is not dependent on the ectoderm but, rather, an intrinsic cell-autonomous activity of the aPKC/Par complex (Figure 4—figure supplement 1). Our results show that only ectodermal- but not endomesodermal-lineages are affected by these mutations (Figure 4 and Figure 4—figure supplement 2). Presumptive ectodermal cells derived from an injected blastomere fail to maintain AJs (and potentially SJs) and the resulting clone of epithelial cells loses its structural integrity inducing cell extrusion. In contrast, presumptive endomesodermal cells derived from an injected blastomere develop into a normal endomesodermal epithelium (Figure 3F). Our results complement the work of (Kirillova et al., 2018) and demonstrate that the proper cell-cell adhesion of the ectodermal layer somehow regulates trans-epithelially the integrity of the endomesodermal layer. This regulation may maintain the tension between cells during invagination at gastrula stages, or, in conjunction with the extracellular matrix (ECM) and basal cues, it may influence signaling patterns necessary to organize epithelial layers during N. vectensis embryogenesis.
Interaction between the NvaPKC/Par complex and the canonical Wnt signaling pathway
NvaPKC/Par complex regulates ß-catenin localization
In bilaterians, AJs recruit members of the aPKC/Par complex and the direct interaction between Par-3 and aPKC/Par-6 is required for the maintenance and maturation of AJs (Ohno et al., 2015; Ragkousi et al., 2017). AJs are characterized by the binding between cadherins and ß-catenin: cadherins sequester ß-catenin from the cytoplasm to the cortex, making it unavailable for nuclear signaling and endomesoderm specification (Wikramanayake et al., 2003; Kumburegama et al., 201124,41. Therefore, using ß-catenin as a marker for AJs, we separately co-injected NvPar-3::mVenus or a mutated dnNvPar-3::mVenus, with Nvß-catenin::RFP into uncleaved zygotes. We observed cortical co-localization of NvPar-3 and Nvß-catenin at the cell boundaries in the ectodermal epithelium of embryos co-injected with NvPar-3::mVenus and Nvß-catenin::RFP (Figure 5A). However, in embryos co-injected with Nvß-catenin::RFP and dnNvPar-3::mVenus, we observed an alteration of the sub-cellular expression of Nvß-catenin::RFP in all cells due to the translocation of ß-catenin from the cortical AJs into cell nuclei (Figure 5A).
Figure 5. NvaPKC/Par complex regulates ß-catenin localization and cell attachment.
(A) In vivo co-localization of NvPar-3venus co-injected with Nvß-cateninRFP, and dnNvPar-3venus co-injected with Nvß-cateninRFP. Arrowheads indicate junctions (AJs). Arrows indicate nuclear ß-catenin. (B) Diagram of the suggested interpretation for A. (C) In vivo time series of ectodermal epithelial layers of embryos injected with NvPar-3venus and dnNvPar-3venus mRNA demonstrating epithelial delamination in the absence of functional NvPar3. Lifeact::mTq2 mRNA was co-injected to visualize cell boundaries. Pink arrows indicate the absence cell detachments. A subset of cells was artificially colored. The purple cell detaches from the epithelium and the red arrow indicates a second celldetachment. See also Figure 5—video 1. (D) IFS of an embryo in which a single blastomere was injected with NvPar-3 guide RNAs and Cas9 and green dextran. Red arrowheads indicate the apical constriction and delamination of ectodermal cells in the mutated clone of cells. Note the different layers of nuclei stained with DAPI. Asterisks indicate the site of gastrulation. (E) Diagram of the suggested interpretation for D. All images are 3D reconstructions from a z-stack confocal series. All scale bars, 20 µm.
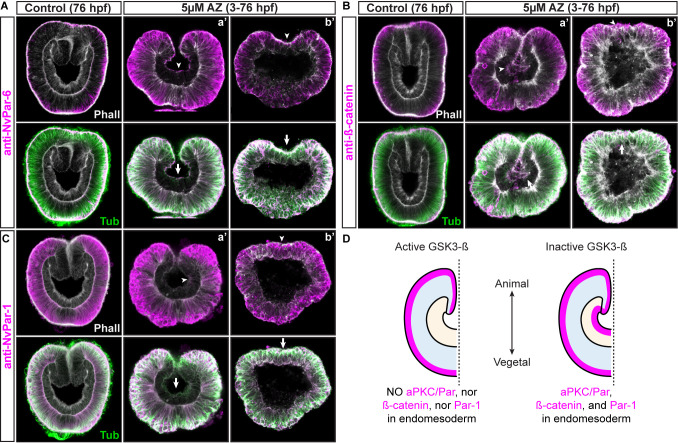
Figure 5—figure supplement 1. Apical junctions are regulated by GSK-3ß in epithelial cells during gastrulation in N. vectensis embryos.
Figure 5—video 1. Related to Figure 5C.
In addition, results from N. vectensis embryos treated with 5 µm 1-azakenpaullone (AZ; an inhibitor of GSK-3ß and a canonical Wnt agonist) suggest that GSK-3ß stabilizes AJs of epithelial cells in N. vectensis embryos (Figure 5—figure supplement 1). We observed an expansion of the expression domain of Par-6 (Figure 5—figure supplement 1) and a stabilization of AJs (labeled with ß-catenin in Figure 5—figure supplement 1) in endomesodermal cells of treated embryos, which was never observed in control embryos.
Interestingly, the association between the nuclearization of ß-catenin (canonical Wnt signaling pathway) and the Par system has been poorly studied. Two studies, one in Drosophila (Sun et al., 2001) and the another in Xenopus (Ossipova et al., 2005) embryos, have shown by immunoblotting that the kinase Par-1 is associated with Dishevelled protein and might act as a positive regulator of Wnt signaling. Here, we show in vivo embryonic evidence suggesting that NvPar-3 (whose cortical localization is normally inhibited by Par-1) recruits Nvß-catenin protein and stabilizes its localization at the apico-lateral cortex of ectodermal cells through the formation of AJs. Furthermore, the putative disassembly of the aPKC/Par complex induced by dnNvPar-3 overexpression, induces the nuclearization of Nvß-catenin protein (Figure 5A) due to its cytosolic availability caused by AJs disruption. Strikingly, we also observed the extrusion of individual cells from the ectodermal epithelium of dnNvPar-3 treated-embryos (Figure 5C) and single injected-blastomere CRISPR/Cas9 NvPar-3 knock-out (Figure 5D). This suggests that these treatments induce EMT-like processes, not observed under control conditions (Figure 5C).
Thus, our data suggest that preexisting mechanisms downstream to the induction of EMT may have been redeployed to segregate layers during the evolution to bilaterians. Bringing the question whether or not endomesodermal genes would induce similar effects when they are expressed in N. vectensis embryos.
We have recently showed that Nvbrachyury regulates apicobasal polarity of epithelial cells in N. vectensis embryos (Servetnick et al., 2017). We, therefore, examined the role of Nvsnail genes on the localization of ß-catenin, components of the Par system, and the stabilization of AJs. Our hypothesis is that expression of N. vectensis snail genes would destabilize AJs and induce the nuclearization of ß-catenin in ectodermal epithelial cells.
Nvsnail genes induce the translocation of Nvß-catenin from AJs to the cytoplasm
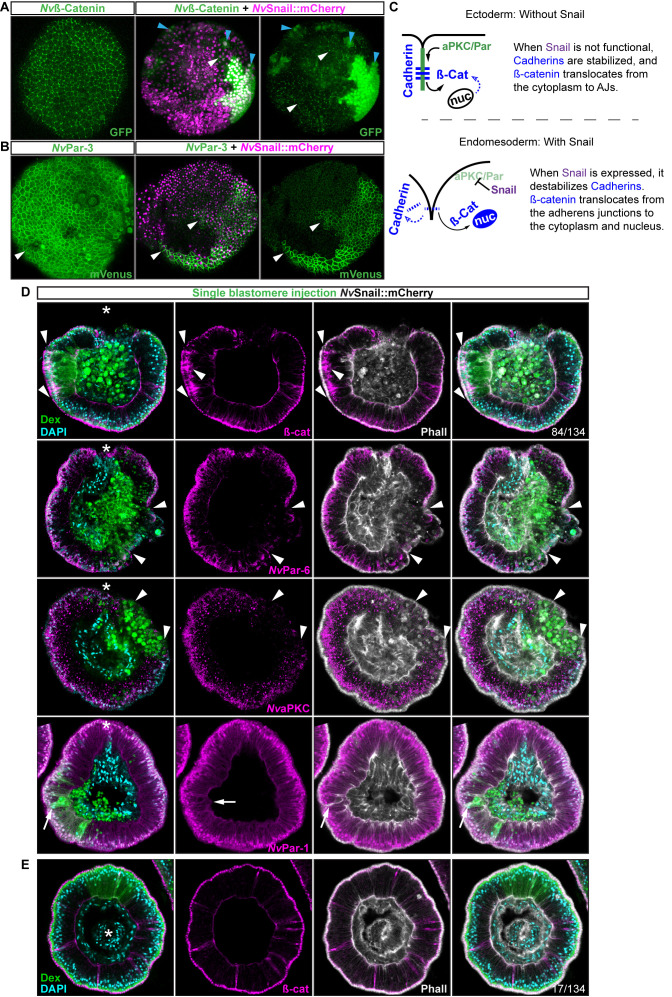
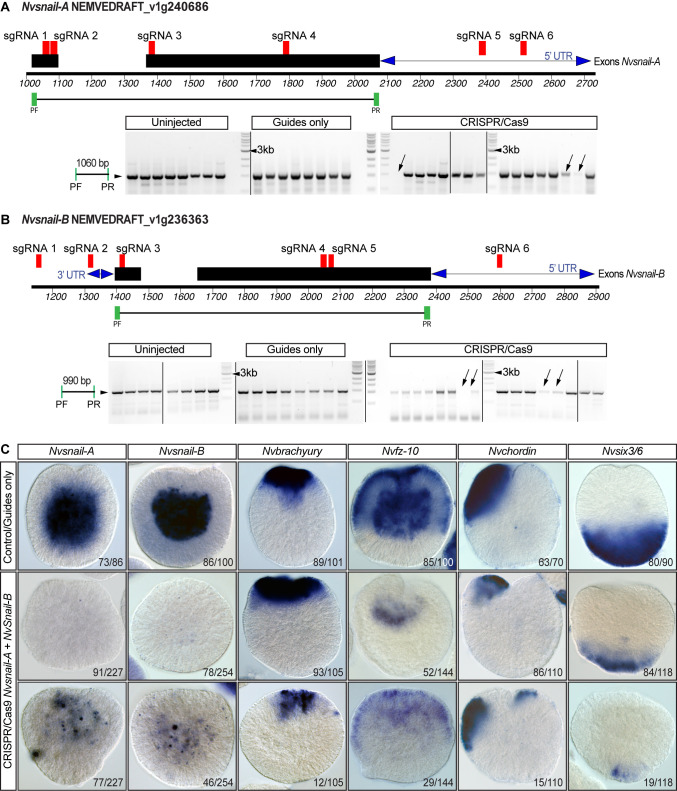
N. vectensis has two snail genes, Nvsnail-A and Nvsnail-B, which are both expressed in the endomesodermal plate prior to and throughout the gastrulation process, and which define the boundary between gastrodermis and ectodermal pharynx (Magie et al., 2007; Röttinger et al., 2012; Amiel et al., 2017). To determine the role of Nvsnail genes on ß-catenin nuclearization, we co-injected the mRNA of NvSnail-A::mCherry, NvSnail-B::mCherry, and Nvß-catenin::GFP into uncleaved eggs. The overexpression of both proteins NvSnail-A::mCherry and NvSnail-B::mCherry together induce the ectopic translocation of Nvß-catenin::GFP to the nuclei of ectodermal cells (Figure 6A). This treatment also delocalizes NvPar-3 from the cell cortex when both NvSnail::mCherry proteins are co-expressed with NvPar-3::mVenus (Figure 6B).
Figure 6. Nvsnail genes induce the translocation of Nvß-catenin and the disruption of epithelial integrity.
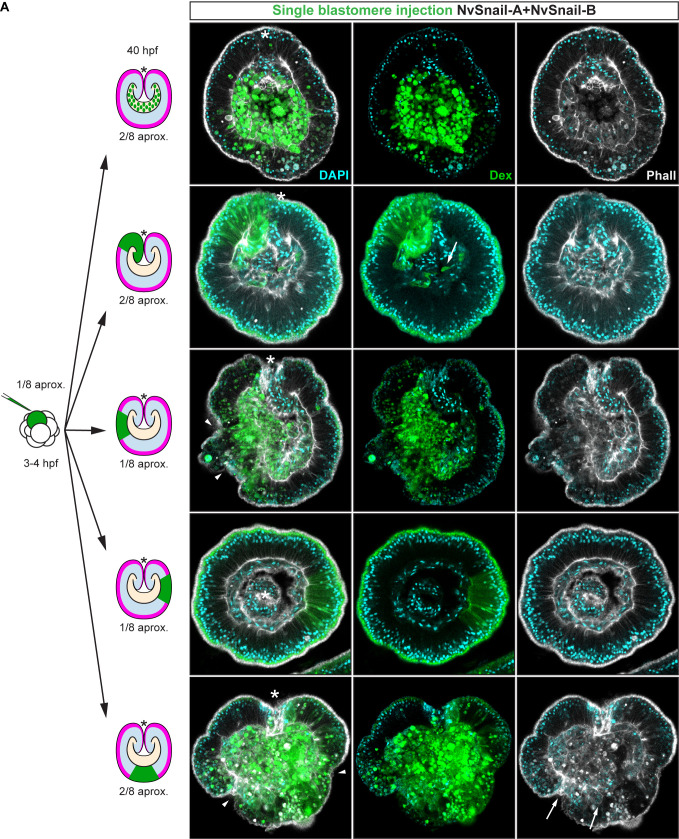
(A) in vivo localization of Nvß-cateninGFP co-injected with both NvSnail-A::mCherry and NvSnail-B::mCherry mRNA together in zygotes at 40 hpf. White arrowheads indicate AJs. Patched patterns of cytosolic and nuclear ß-catenin (Blue arrowheads) were observed. (B) in vivo localization of NvPar-3::mVenus co-injected with both NvSnail-A::mCherry and NvSnail-B::mCherry mRNA together at 40 hpf. Patched patterns of AJs (White arrowheads) were observed. (C) Diagram depicts the suggested interpretation for A and B. (D) IFS for ß-catenin (ß-cat), NvPar-6, NvaPKC, and NvPar-1 in embryos at 40 hpf where NvSnail-A::mCherry and NvSnail-B::mCherry mRNA were overexpressed together into a single ectodermal blastomere lineage (followed by green Streptavidin-TxRed Dextran (Dex). Arrowheads indicate the absence of the protein, cytosolic ß-cat, and disrupted epithelium. Arrows indicate bottle-like shape cells. (E) IFS for ß-cat in embryos at 40 hpf where NvSnail-A::mCherry and NvSnail-B::mCherry mRNA were overexpressed together into a single blastomere lineage and no affects were observed. See also Figure 6—figure supplement 1, Figure 6—video 1, and Figure 6—video 2. *site of gastrulation. Morphology is shown by DAPI and Phall IFS. Except from 6A and 6B, all images are single optical sections from the z-stack confocal series. (A) and (B) are 3D reconstructions from a z-stack confocal series.
Figure 6—figure supplement 1. Nvsnail-A and Nvsnail-B together regulate epithelial integrity.
Figure 6—video 1. Related to Figure 6E.
Figure 6—video 2. Related to Figure 6E.
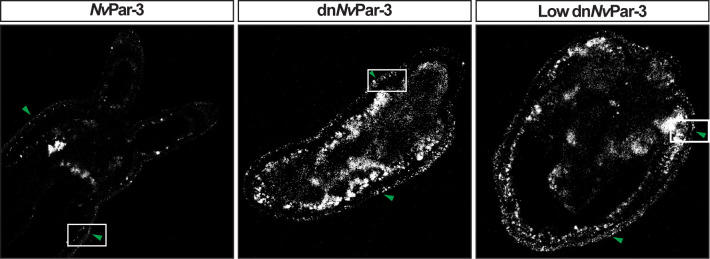
To determine the role of Nvsnail genes on cell adhesion/epithelial polarity, we randomly injected single blastomeres at the 8–32 cell-stage with mRNA from both NvSnail-A::mCherry and NvSnail-B::mCherry together. The fluorescent dextran that was co-injected with the mRNAs could be used to detect the clones where the over-expression of the co-injected mRNAs occurred in a ‘wild-type’ background (Figure 6D). Similar to the Nvpar-3 knock-out (Figure 4), the expression of Nvsnail genes is sufficient to induce the degradation of Par proteins and AJs (ß-catenin) from the ectoderm and disrupts its epithelial integrity; however, nuclear ß-catenin was not observed under these treatments (Figure 6D). Thus, nuclear Nvß-catenin::GFP observed in vivo when we overexpressed NvSnail proteins (Figure 6A) is a consequence of the high cytosolic availability generated by its ectopic overexpression and release from AJs.
Interestingly, not every ectodermal cell was affected by these treatments even though all of the cells expressed the injected mRNAs (Figure 6A and E, and Figure 6—figure supplement 1). This patched pattern suggests that the response to Nvsnail over-expression is spatially regulated. These results suggest that the role of Nvsnail genes on AJs and apicobasal cell polarity is constrained to the site of gastrulation in N. vectensis embryos under natural conditions, and that these genes may be required for gastrulation movements. Therefore, we predicted that ß-catenin (AJs) and Par proteins will be retained in the cells of the N. vectensis endomesodermal plate if both Nvsnail genes are disrupted.
Nvsnail genes downregulate apicobasal cell polarity and AJs in the endomesodermal epithelium of N. vectensis embryos
The snail genes temporally down-regulate E-cadherin during mesoderm segregation and EMT in bilaterian animals (Lim and Thiery, 2012). As we have shown here, as well as in previous studies (Magie et al., 2007; Magie and Martindale, 2008), the cells comprising the endomesodermal plate lose their cell-cell adhesion during gastrulation in N. vectensis embryos. It may be possible that temporal regulation of endomesodermal patterning might act upon the AJs. Our data suggest that once gastrulation is complete and the pharynx forms, components of the Par system and the ß-catenin components of the AJs are degraded from both the cortex and cytoplasm of endomesodermal cells (Figure 1 and Figure 1—figure supplement 1). Hence, it could be possible that Nvbrachyury induces the disruption of apicobasal polarity (Servetnick et al., 2017), remnant AJs maintain the endomesodermal-plate cells together, and Nvsnail genes degrades and prevents the reassembly of AJs in the endomesoderm of N. vectensis.
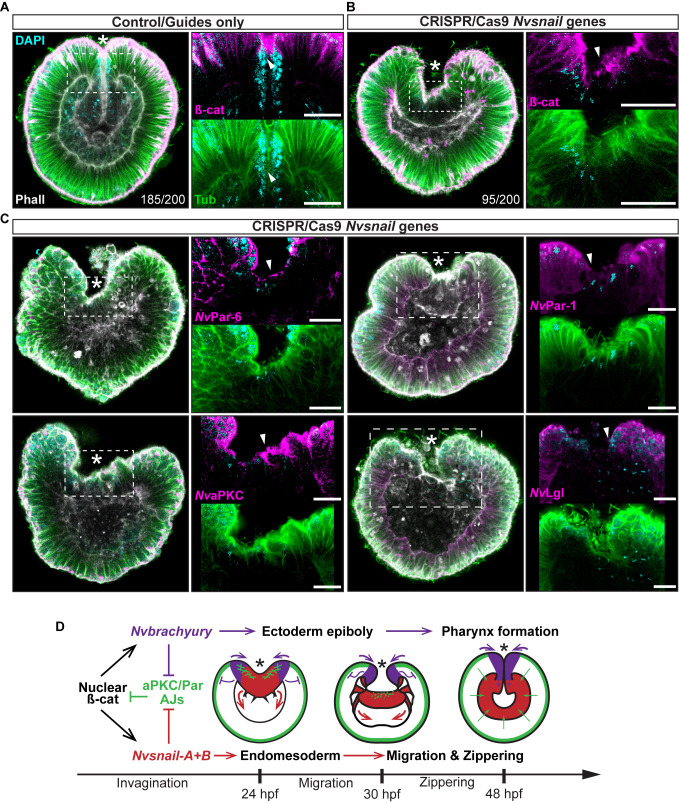
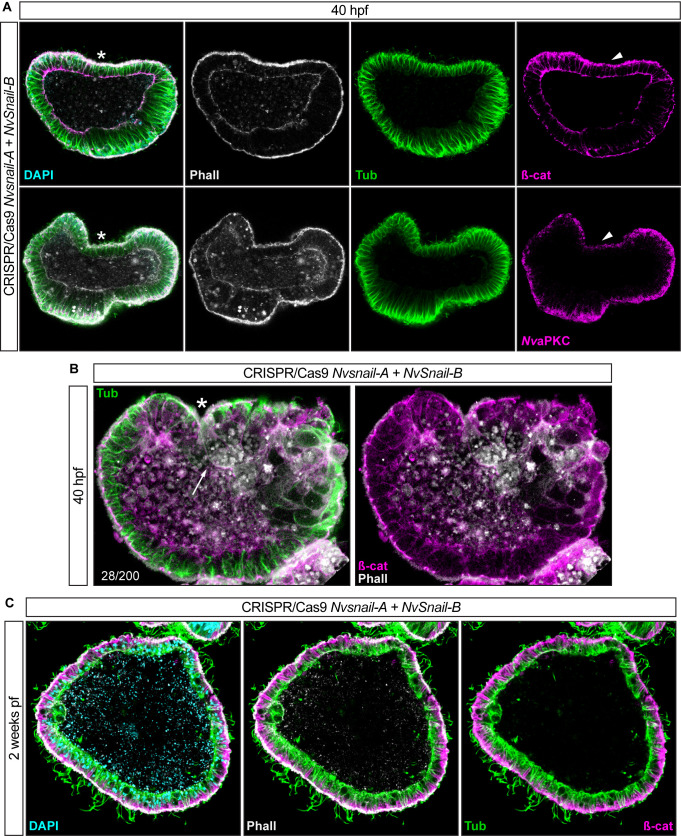
To address these issues, we used CRISPR/Cas9 knock-out of Nvsnail-A and Nvsnail-B genes together to inhibit zygotic function of these genes and investigate their role on the temporal regulation of AJs and cell polarity. In CRISPR/Cas9 mutants, the endomesodermal plate forms but it does not migrate further than its first invagination during gastrulation (Figure 7). Furthermore, AJs (labeled with ß-catenin) and apical Par proteins (labeled with antiNvPar-6 and antiNvaPKC) are retained at the apical cortex of the cells of the endomesodermal plate (Figure 7B and C and Figure 7—figure supplement 1). Surprisingly, NvPar-1 and NvLgl were not detected in those cells (Figure 7C), suggesting that the degradation of these basolateral proteins precede or do not depend on the activity of the Nvsnail genes. This suggests that Nvsnail regulates apical cell-polarity, AJs turnover, and the migration (‘zippering’) but not the invagination of the endomesodermal plate during gastrulation of N. vectensis embryos (Figure 7D). Interestingly, the invagination of the endomesodermal plate (controlled by the Wnt/PCP pathway) is uncoupled from its specification in N. vectensis embryos (Kumburegama et al., 2011; Wijesena et al., 2011), which is consistent with our observations.
Figure 7. Nvsnail genes prevents the reassembly AJs and NvaPKC/Par polarity allowing endomesodermal migration.
(A) Embryo wide CRISPR/Cas9 knock-out guides controls show no effect neither on gastrulation nor AJs (ß-catenin) localization. (B) Embryo wide CRISPR/Cas9 knock-out for both Nvsnail-A and Nvsnail-B at 40 hpf showing that AJs are retained in presumptive endomesodermal region similar to ectodermal cells. High magnification images from boxed region (endomesodermal plate) are shown on the right. (C) Embryo wide CRISPR/Cas9 knock-out for both Nvsnail-A and Nvsnail-B at 40 hpf showing that NvPar-6 and NvaPKC proteins are retained in in presumptive endomesodermal region similar to ectodermal cells. NvPar-1 and NvLgl were not detected in endomesodermal cells. High magnification images from boxed region (endomesodermal plate) are shown on the right. (D) Graphical summary of the observed results with previous published data (Servetnick et al., 2017). See also Figure 7—figure supplements 1 and 2. Morphology is shown by DAPI, Tubulin, and Phalloidin. All images are single optical sections from the z-stack confocal series. Arrowheads indicate protein localization. *site of gastrulation. All scale bars, 20 µm.
Figure 7—figure supplement 1. Nvsnail-A and Nvsnail-B together regulate AJs.
Figure 7—figure supplement 2. CRISPR/Cas9 mediated mutagenesis of Nvsnail-A and Nvsnail-B.
Discussion
AJs are down-regulated in mesoderm and neural crest of bilaterian animals
The segregation of different germ layers during embryogenesis of many bilaterian animals is carried out by similar cellular mechanisms. EMT is a shared mechanism utilized by mesoderm, neural crest cell (NCC), and tumorigenesis to delaminate cells in bilaterian animals (triploblastic animals). During EMT, the nuclearization of ß-catenin induces the expression of ‘endomesodermal’ genes like brachyury and snail (Acloque et al., 2009). The expression of these genes downregulates epithelial cadherins, disrupts apicobasal polarity (mediated by the aPKC/Par complex), disassembles AJs, and induces changes in cytoskeleton organization (Acloque et al., 2009; Lim and Thiery, 2012). A rearrangement of the actin-myosin cytoskeleton induces apical constriction of cells (generating a bottle-like shape), which detach from the epithelial sheet, break down the basal membrane, and invade a specific tissue as mesenchymal cells (Acloque et al., 2009; Lim and Thiery, 2012; Ohsawa et al., 2018).
Interestingly, mesoderm formation, tumorigenesis, and EMT have never been described as natural processes during N. vectensis (a diploblastic animal) embryogenesis. During N. vectensis gastrulation (Magie et al., 2007; Tamulonis et al., 2011), cells around the edge of the blastopore at the animal pole (which expresses Nvbrachyury) acquire a bottle-like shape by apical constriction, leading to epithelial buckling and the invagination of presumptive endomesoderm (which expresses Nvsnail). However, throughout this process the endomesoderm remains as a monolayer of epithelial cells and individual mesenchymal cells never detach and invade the blastocoel.
We have shown that by disrupting the aPKC/Par complex (apicobasal cell-polarity) in N. vectensis (Figures 3, 4 and 5), we are able to convert cells from the endomesodermal epithelium into mesenchymal-like cells, translocate Nvß-catenin (Figure 5A), and emulate EMT-like processes (apical constriction and individual cell-detachments) in the ectodermal epithelium of N. vectensis treated-embryos (Figure 5C and D). These results demonstrate that the cnidarian N. vectensis possesses mechanisms necessary to segregate individual germ layers (e.g. mesoderm and NCC) described in bilaterians; however, they do not do it.
Our working hypothesis is that the N. vectensis embryo is composed of two independent morphogenetic modules that are integrated and organized by the pharynx (Figure 7D). The first observation is that the ectoderm, whose apicobasal polarity (and thus, AJs and epithelial integrity) is regulated by Nvbrachyury that promotes ectodermal epithelial morphogenesis and pharynx formation (Servetnick et al., 2017), and the second module is generated by endomesodermal differentiation and cell-movements that are regulated by Nvsnail genes. This is supported by the expression Nvbrachyury in Nvsnail knock-out embryos (Figure 7—figure supplement 2), and Nvsnail knock-out phenotypes where ectodermal pharynx develops normally but no clear endomesoderm is formed (Figure 7—figure supplement 1). Additional work is required to elucidate any differences in function between Nvsnail-A and Nvsnail-B genes, however, both modules are specified by nuclear ß-catenin (Röttinger et al., 2012), suggesting that the nuclear ß-catenin (maternal) shift from the animal pole in cnidarians to the vegetal pole in bilaterians is mechanistically plausible and sufficient to re-specify the site of gastrulation and germ-layers along the animal-vegetal axis during Metazoan evolution (Martindale and Lee, 2013; Lee et al., 2007).
The dual identity and collective migration of the endomesodermal cells
Bilaterian-EMT has been a focus of study for decades as a mechanism to segregate different cell layers involved in a variety of different normal and pathological biological processes (Ohsawa et al., 2018; Nieto et al., 2016). This process appears to depend on the fine regulation of snail expression levels and their temporal activity. For example, during NCC migration, cells display ‘partial-EMT’ where cells remain attached to several neighboring cells but their apicobasal polarity and AJs are down-regulated, allowing collective-cell migration (Weng and Wieschaus, 2017; Nieto et al., 2016; Lee et al., 2006; Theveneau and Mayor, 2013; Ribeiro and Paredes, 2014). Our data suggest that ‘partial-EMT’ may be the mechanism by which the endomesodermal epithelium migrates into the blastocoel in N. vectensis embryos during normal gastrulation (Figure 8). In this scenario, upstream factors that regulate snail transcription may be critical for this process.
Figure 8. The differences between epithelial structure in ectoderm and endomesoderm in N. vectensis embryos are due to the lack of mechanisms to segregate a distinct mesoderm.
Diagram depicting key cellular and molecular mechanisms involved during gastrulation of bilaterian and N. vectensis (a cnidarian) embryos. See also Figure 8—figure supplement 1.
Figure 8—figure supplement 1. Suggested model for mesoderm specification in Metazoa.
In bilaterian animals, there are many other pathways in addition to the canonical Wnt pathway that activate snail transcription and induce the disruption of AJs and apicobasal cell polarity. For example, TGFß, BMP, NANOS, FGF, and MEK/ERK/ERG take on roles during the specification of mesoderm, NCC migration, tumorigenesis, and other EMT-related processes (Lim and Thiery, 2012; Nieto et al., 2016; Barrallo-Gimeno and Nieto, 2005). Concordantly in N. vectensis embryos, cells of the pharyngeal and endomesodermal tissues express components of all these pathways (Röttinger et al., 2012; Amiel et al., 2017; Extavour et al., 2005; Matus et al., 2006, 2007; Wijesena et al., 2017) that may modify their cellular characteristics. For example, one cadherin (NvCDH2 Clarke et al., 2016: 1g244010), and kinases that modify tubulin and histones are differentially regulated between ecto- and endomesodermal epithelium (Wijesena et al., 2017).
In conclusion, N. vectensis has both up and downstream cellular and molecular mechanisms associated with EMT described in bilaterians. However, N. vectensis does not segregate a distinct mesodermal germ layer nor display EMT under natural conditions. In bilaterians, this mechanism must have evolved to segregate mesodermal cells from the endoderm to retain the tight cell-cell junctions required in endodermal epithelia. Interestingly, mesoderm segregation via EMT in Drosophila takes place after epithelial folding in response to snail expression. In these embryos, contractile myosin enhances the localization of AJs and Par-3 in the presumptive mesoderm and prevents their downregulation by Snail, thus delaying EMT (Weng and Wieschaus, 2016, 2017). Furthermore, the overexpression of Snail in Drosophila embryos is sufficient to disassemble ectodermal-AJs, but mesodermal-AJs are maintained by actomyosin contraction that antagonize Snail effects (Weng and Wieschaus, 2016, 2017). Our results suggest a similar mechanism since Nvsnail overexpression in endomesodermal lineages (Figure 6—figure supplement 1) is not sufficient to segregate cells and the endomesoderm remains as an epithelium. However, unlike Drosophila, Par proteins and AJs are not enhanced but degraded during the gastrulation of N. vectensis (Figure 1). As it is discussed in (Weng and Wieschaus, 2017), not only the degradation but also the turnover of AJs and Par proteins in adjacent epithelia is essential for EMT-mediated germ layer segregation in different animals. The dual identity of N. vectensis endomesoderm is characterized by the continuous expression of Nvsnail genes (Martindale et al., 2004) that repress the turnover of AJs and may play a role in inhibiting EMT from occurring (Figures 6 and 7).
Interestingly, components of the Wnt/PCP pathway are expressed only in the endomesoderm (Kumburegama et al., 2011; Wijesena et al., 2011), while components of the Par system are expressed only in the ectoderm (Salinas-Saavedra et al., 2015). It could be that NvSnail degrades AJs and inhibits their re-assembly in the endomesoderm, but the activation of contractile myosin by the Wnt/PCP pathway maintains the endomesodermal cells together in N. vectensis embryos. Hence in bilaterians, a mechanism (most likely downstream of Snail) that connects the cytoskeleton with cell-polarity may have evolved to tighten cell-cell adhesion in the endoderm and allow EMT.
To elucidate this, further comparative research and funding are needed to understand the cellular mechanisms that evolve to segregate mesoderm and control epithelial cell polarity at the base of the metazoan tree. For example, cnidarians, poriferans, and ctenophores present intriguing characteristics to study. In cnidarians, different modes of gastrulation have been described between species including unipolar and multipolar cell ingression and delamination (Kirillova et al., 2018; Byrum and Martindale, 2004; Marlow and Martindale, 2007). Poriferans display EMT-like processes and cell morphologies during regeneration and trans-differentiation (Nakanishi et al., 2014; Coutinho et al., 2017). However, whether or not these processes involve similar molecular and cellular mechanisms are still unclear. Interestingly, ctenophores segregate a mesodermal cell population during embryogenesis but do not have the genes that encode for all cell-cell adhesion complexes and specify for mesoderm in bilaterians (Figure 8—figure supplement 1) (Ganot et al., 2015; Ryan et al., 2013). Thus, there is much to be learned by the comparative study of cell biology to understand the evolutionary origins of EMT and germ layer formation.
Materials and methods
Key resources table.
| Reagent type (species) or resource |
Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Mouse Anti-alpha-Tubulin Monoclonal Antibody, Unconjugated, Clone DM1A |
Sigma-Aldrich | T9026; RRID:AB_477593 |
(1:500) |
| Antibody | Anti-beta-Catenin antibody produced in rabbit |
Sigma-Aldrich | C2206; RRID:AB_476831 |
(1:300) |
| Antibody | Anti-Histone, H1 + core proteins, clone F152.C25.WJJ antibody |
Millipore | MABE71; RRID:AB_10845941 |
(1:300) |
| Antibody | anti-NvaPKC custom peptide antibody produced in rabbit |
Bethyl labs; Salinas-Saavedra et al. (2015) |
Stored at MQ Martindale's lab; (1:100) |
|
| Antibody | anti-NvLgl custom peptide antibody produced in rabbit |
Bethyl labs; Salinas-Saavedra et al. (2015) |
Stored at MQ Martindale's lab; (1:100) |
|
| Antibody | anti-NvPar-1 custom peptide antibody produced in rabbit |
Bethyl labs; Salinas-Saavedra et al. (2015) |
Stored at MQ Martindale's lab; (1:100) |
|
| Antibody | anti-NvPar-6 custom peptide antibody produced in rabbit |
Bethyl labs; Salinas-Saavedra et al. (2015) |
Stored at MQ Martindale's lab; (1:100) |
|
| Antibody | Goat anti-Mouse IgG Secondary Antibody, Alexa Fluor 568 |
Thermo Fisher Scientific | A-11004; RRID:AB_2534072 |
(1:250) |
| Antibody | Goat anti-Rabbit IgG Secondary Antibody, Alexa Fluor 647 |
Thermo Fisher Scientific | A-21245; RRID:AB_2535813 |
(1:250) |
| Antibody | Sheep Anti-Digoxigenin Fab fragments Antibody, AP Conjugated, Roche |
Roche | 11093274910; RRID:AB_514497 |
|
| Other | Alexa Fluor 488 Phalloidin | Thermo Fisher Scientific | A12379; RRID:AB_2315147 |
(1:200) |
| Other | Texas Red Streptavidin | Vector Laboratories | SA-5006, RRID:AB_2336754 |
(1:200) |
| Other | DAPI (4',6-Diamidino- 2-Phenylindole, Dihydrochloride) |
Thermo Fisher Scientific | D1306; RRID:AB_2629482 |
(0.1 µg/µl) |
| Chemical compound, drug |
Protein kinase Cζ pseudosubstrate, myristoyl trifluoroacetate salt |
Sigma-Aldrich | P1614 | |
| Chemical compound, drug |
1-Azakenpaullone | Sigma-Aldrich | A3734 | |
| Chemical compound, drug |
Biotinylated Dextran Amine-Texas Red |
Vector Laboratories | SP-1140; RRID:AB_2336249 |
|
| Chemical compound, drug |
Dextran, Alexa Fluor 488; 10,000 MW, Anionic, Fixable |
Thermo Fisher Scientific | D22910 | |
| Chemical compound, drug |
Dextran, Alexa Fluor 555; 10,000 MW, Anionic, Fixable |
Thermo Fisher Scientific | D34679 | |
| Chemical compound, drug |
Dextran, Alexa Fluor 647; 10,000 MW, Anionic, Fixable |
Thermo Fisher Scientific | D22914 | |
| Chemical compound, drug |
Dextran, Cascade Blue, 10,000 MW, Anionic, Lysine Fixable |
Thermo Fisher Scientific | D1976 | |
| Deposited Data |
Nematostella vectensis
genome assembly 1.0 |
JGI | https://genome.jgi.doe.gov/Nemve1/Nemve1.home.html | |
| Biological sample | Nematostella vectensis | Whitney Laboratory for Marine Bioscience, FL, USA. |
RRID:SCR_005153 | |
| Sequence-based reagent |
Genomic PCR Nvpar-6 and
dnNvpar-6: F-AAAACCAC CATCAGCCGAGTCA; R-TATTGATAGAATACCAGTCTCA |
NEMVEDRAFT _v1g233358 |
||
| Sequence-based reagent |
sgRNAs Nvpar-6:
1-GGATGTTGCCGACTCGCAGT; 2-GGAGAAGGCGAACTCGTCTG; 3-GGATAACCCTGTGCCAGTCA |
CRISPRevolution sgRNA; Synthego |
NEMVEDRAFT _v1g233358 |
|
| Sequence-based reagent |
dnNvpar-3: F-ATGATGAAGGTTGTAGT;
R-TGCGCCCGATTCGAATCCATCT |
NEMVEDRAFT _v1g240248 |
||
| Sequence-based reagent |
sgRNAs Nvpar-3:
1-GGGTGTTCGAGGGACGCGAT; 2- GGGCAGGTTTATCCCGAAGG; 3- ACCAACGAUCUAGAUCCAGU |
CRISPRevolution sgRNA; Synthego |
NEMVEDRAFT _v1g240248 |
|
| Sequence-based reagent |
Genomic PCR Nvpar-3:
F-GTAGACGGGACTGGTTTGGA; R-AGGGACAGGTTGCTCCTTTT |
NEMVEDRAFT _v1g240248 |
||
| Sequence-based reagent |
dnNvpar-1:
F-AATATAAACTATGAACTTAACG; R-TTAAAGTTTTAATTCATTTGCA |
NEMVEDRAFT _v1g 139527 |
||
| Sequence-based reagent |
sgRNAs Nvsnail-A:
2-GGGGCCGGTAATGACGCGCG; 3-GGCGTAGAGTCACACCGCAA; 4-GGCGATGATATCGAGCTCGG; 5-GGGCATCTTGAGTGCACCCA |
CRISPRevolution sgRNA; Synthego |
NEMVEDRAFT _v1g240686 |
|
| Sequence-based reagent |
sgRNAs Nvsnail-A:
1-GGGCTCTCTTGCTCCGTAAC; 6-GGGTTTCCTGGCGCTGGGAT |
CRISPRevolution Modified sgRNA; Synthego |
NEMVEDRAFT _v1g240686 |
|
| Sequence-based reagent |
Genomic PCR Nvsnail-A (full length):
F-ATGCCCCGCTCGTTTCTAG; R-TCCTTGTGACGGGCAGCC |
NEMVEDRAFT _v1g240686 |
||
| Sequence-based reagent |
sgRNAs Nvsnail-B:
1-GGAAGAGGATGTGAGGTTTT; 2-GAGATGATATTAGGCTGGTG; 4-GAAAAGCTGTACGACTCCTT; 5-GGGCATCTTGAGAGCGCCCA |
CRISPRevolution sgRNA; Synthego |
NEMVEDRAFT _v1g236363 |
|
| Sequence-based reagent |
sgRNAs Nvsnail-B:
3-GGGTGAAGACTAAGACAGAG; 6-GGGCGATGAATCGTGTTTAA |
CRISPRevolution Modified sgRNA; Synthego |
NEMVEDRAFT _v1g236363 |
|
| Sequence-based reagent |
Genomic PCR Nvsnail-B (full length):
F-ATGCCGAGGTCCTTCCTGG; R-GCAGAGATTTTGCCGACACAT |
NEMVEDRAFT _v1g236363 |
||
| Recombinant DNA reagent |
pSPE3-mVenus | Roure et al. (2007) | Gateway vector | |
| Recombinant DNA reagent |
pSPE3-mCherry | Roure et al. (2007) | Gateway vector | |
| Recombinant DNA reagent |
pSPE3-Nvpar-6-mVenus | Salinas-Saavedra et al. (2015) | ||
| Recombinant DNA reagent |
pSPE3-Nvpar-3-mVenus | Salinas-Saavedra et al. (2015) | ||
| Recombinant DNA reagent |
Nvß-catenin expression
constructs |
Wikramanayake et al.
(2003); Röttinger et al., 2012 |
||
| Recombinant DNA reagent |
Nvsnail-A backbone to generate expression constructs |
Magie et al. (2007) | ||
| Software, algorithm |
Fiji (ImageJ) | NIH | http://fiji.sc | |
| Software, algorithm |
Imaris 7.6.4 | Bitplane Inc. | ||
| Software, algorithm |
CRISPRscan | Moreno-Mateos et al. (2015) | http://www.crisprscan.org/ | |
| Software, algorithm |
SPSS | IBM |
Culture and spawning of Nematostella vectensis
Spawning, gamete preparation, fertilization and embryo culturing of N. vectensis (RRID:SCR_005153) embryos was performed as previously described (Röttinger et al., 2012; Hand and Uhlinger, 1992; Layden et al., 2013; Wolenski et al., 2013). Adult N. vectensis were cultivated at the Whitney Laboratory for Marine Bioscience of the University of Florida (USA). Males and females were kept in separate glass bowls (250 ml) in 1/3x seawater (salinity: 12pp) reared in dark at 16°C. Animals were fed freshly hatched Artemia three times a week and macerated oyster the day before spawning. Spawning was induced by incubating the adults under an eight-hour light cycle at 25°C the night before the experiment. Distinct groups of animals were spawned once every 2 weeks. Oocytes and sperm were collected separately and fertilized in vitro by adding sperm to egg masses for 25 min. The jelly mass surrounding the fertilized eggs was removed by incubating the eggs in 4% L-Cysteine (in 1/3x seawater; pH 7.4) for 15–17 min and then washed 3 times with 1/3x seawater. De-jellied eggs were kept in glass dishes (to prevent sticking) in filtered 1/3 seawater at 16°C until the desired stage.
Immunohistochemistry
All immunohistochemistry experiments were carried out using the previous protocol for N. vectensis (Salinas-Saavedra et al., 2015) with a slight modification in the glutaraldehyde concentration to allow better antibody penetration. Embryos were fixed on a rocking platform at room temperature in two consecutive steps. Embryos of different stages were fixed for no longer than 3 min in fresh Fix-1 (100 mM HEPES pH 6.9; 0.05M EGTA; 5 mM MgSO4; 200 mM NaCl; 1x PBS; 3.7% Formaldehyde; 0.2% Glutaraldehyde; 0.5% Triton X-100; and pure water). Then, Fix-1 was removed and replace with fresh Fix-2 (100 mM HEPES pH 6.9; 0.05M EGTA; 5 mM MgSO4; 200 mM NaCl; 1x PBS; 3.7% Formaldehyde; 0.05% Glutaraldehyde; 0.5% Triton X-100; and pure water). Embryos were incubated in Fix-2 for 1 hr. Fixed embryos were rinsed at least five times in PBT (PBS buffer plus 0.1% BSA and 0.2% Triton X-100) for a total period of 3 hr. PBT was replaced with 5% normal goat serum (NGS; diluted in PBT) and fixed embryos were blocked for 1 to 2 hr at room temperature with gentle rocking. Primary antibodies were diluted in 5% NGS to desired concentration. Blocking solution was removed and replaced with primary antibodies diluted in NGS. All antibodies incubations were conducted over night on a rocker at 4°C. After incubation of the primary antibodies, samples were washed at least five times with PBT for a total period of 3 hr. Secondary antibodies were then applied (1:250 in 5% NGS) and samples were left on a rocker overnight at 4°C. Samples were then washed with PBT and left on a rocker at room temperature for an hour. To visualize F-actin, samples were incubated then for 1.5 hr in Phalloidin (Invitrogen, Inc. Cat. # A12379) diluted 1:200 in PBT. Samples were then washed once with PBT and incubated with DAPI (0.1 µg/µl in PBT; Invitrogen, Inc. Cat. # D1306) for 1 hr to allow nuclear visualization. Stained samples were rinsed again in PBS two times and dehydrated quickly into isopropanol using the gradient 50, 75, 90, and 100%, and then mounted in Murray’s mounting media (MMM; 1:2 benzyl benzoate:benzyl alcohol) for visualization. Note that MMM may wash DAPI out of your sample. For single blastomere microinjection experiments, after Phalloidin staining, samples were incubated with Texas Red Streptavidin (1:200 in PBT from 1 mg/ml stock solution; Vector labs, Inc. Cat.# SA-5006. RRID:AB_2336754) for 1 hr to visualize the injected dextran. We scored more than 1000 embryos per each antibody staining and confocal imaged more than 50 embryos at each stage.
The primary antibodies and concentrations used were: mouse anti-alpha tubulin (1:500; Sigma-Aldrich, Inc. Cat.# T9026. RRID:AB_477593), rabbit anti-ß-catenin (1:300; Sigma-Aldrich, Inc. Cat.# C2206. RRID:AB_476831), mouse anti-histone H1 (1:300; F152.C25.WJJ, Millipore, Inc. RRID:AB_10845941).
Rabbit anti-NvaPKC, rabbit anti-NvLgl, rabbit anti-NvPar-1, and rabbit anti-NvPar-6 antibodies are custom made high affinity-purified peptide antibodies that were previously raised by the same company (Bethyl Inc.). All these four antibodies are specific to N. vectensis proteins (Salinas-Saavedra et al., 2015) and were diluted 1:100.
Secondary antibodies are listed in Key resources table.
Fluorescent tracer dye penetration assay
Primary polyps were incubated and mounted in 1/3 sea water with fluorescent dextran solution (0.5 mg/ml). For uninjected embryos we used Dextran, Alexa Fluor 555 (Molecular Probes, INC. Cat.# D34679). For injected embryos, expressing fluorescent proteins, we used Dextran, Alexa Fluor 647 (Molecular Probes, INC. Cat.# D22914). Animals were observed within 10 min of incubation. 15 animals were recorded per treatment. For better visualization of the dextran solution inside the gastric cavity as shown in Figure 2B, we delivered additional dextran solution by microinjecting dye through the polyp’s mouth. For the rest of the experiments, we mainly focused in the ectodermal permeability and we let the polyps to eat the solution by themselves as grown babies.
mRNA microinjections
The coding region for each gene of interest was PCR-amplified and cloned into pSPE3-mVenus or pSPE3-mCherry using the Gateway system (Roure et al., 2007). Eggs were injected directly after fertilization as previously described (Salinas-Saavedra et al., 2015; Layden et al., 2013; DuBuc et al., 2014) with the mRNA encoding one or more proteins fused in frame with reporter fluorescent protein (N-terminal tag) using final concentrations of 450 ng/µl for each gene. Fluorescent dextran was also co-injected to visualize the embryos. For single blastomere microinjections, we raised the embryos until 8–16 cell stages (3–4 hpf) and co-injected the mRNA solution with Biotinylated Dextran Amine-Texas Red (10 µg/µl; Vector labs, Inc. Cat.# SP-1140. RRID:AB_2336249). Live embryos were kept at 16°C and visualized after the mRNA of the FP was translated into protein (2–3 hr). To avoid lethality, lower mRNA concentrations of the mutant proteins (250 ng/µl) were used to image the specimens for Figures 2 and 4, and Figure 3—video 1. Live embryos were mounted in 1/3 sea water for visualization. Images were documented at different stages from 3 to 96 hr post fertilization. We injected and recorded more than 500 embryos for each injected protein and confocal imaged approximately 20 specimens for each stage for detailed analysis of phenotypes in vivo. We repeated each experiment at least five times obtaining similar results for each case. The fluorescent dextran and primers for the cloned genes are listed in Key resources table.
CRISPR/Cas9 knock-outs
To target our gene of interest, we used synthetic guide RNAs (sgRNA; Synthego, Inc.) and followed the instructions obtained from the manufacturer to form the RNP complex with Cas9 (Cas9 plus sgRNAs). Target sites, off-target sites, and CFD scores were identified and sgRNA were designed using CRISPRscan (Doench et al., 2014; Moreno-Mateos et al., 2015). We delivered the RNP complex by microinjection as previously described (Servetnick et al., 2017; Wijesena et al., 2017; Ikmi et al., 2014. Lyophilized Cas9 (PNA Bio., Inc. Cat.# CP01) was reconstituted in nuclease-free water with 20% glycerol to a final concentration of 2 µg/µl. Reconstituted Cas9 was aliquoted for single use and stored at −80°C. Embryos were injected, as described for mRNA microinjections, with a mixture (12.5 µl) containing sgRNAs (80 ng/μl of each sgRNA), Cas9 (3 μg), and Alexa Fluor 488-dextran (0.2 μg/μl; Molecular Probes, Inc. Cat.# D22910). Cas9 and sgRNA guides only controls were injected alongside each round of experiments. sgRNA guides controls are only shown in figures as Cas9 had no significative effects. 3 sgRNA were used to knock out Nvpar-3, 3 sgRNA were used to knock out Nvpar-6, 6 sgRNA were used to knock out Nvsnail-A, and 6 sgRNA were used to knock out Nvsnail-B. Single-embryo genomic DNA was analyzed as previously described (Servetnick et al., 2017). Gene expression was confirmed by in situ hybridization. We injected and recorded more than 1000 embryos for each treatment. We repeated each experiment at least six times obtaining similar results for each case. sgRNAs’ sequences and PCR primers flanking the targeted region are listed in Key resources table.
In situ hybridization
In situ hybridization was carried out following a previously published protocol for N. vectensis (Wolenski et al., 2013). Animals were fixed in ice-cold 4% paraformaldehyde with 0.2% glutaraldehyde in 1/3x seawater for 2 min, followed by 4% paraformaldehyde in PBTw for 1 hr at 4°C. Digoxigenin (DIG)-labeled probes, previously described (Salinas-Saavedra et al., 2015; Röttinger et al., 2012), were hybridized at 63°C for 2 days and developed with the enzymatic reaction of NBT/BCIP as substrate for the alkaline phosphatase conjugated anti-DIG antibody (Roche, Inc. Cat.#11093274910. RRID:AB_514497). Samples were developed until gene expression was visible as a purple precipitate.
Drug treatment
We incubated N. vectensis embryos in 20 µM of aPKC pseudosubstrate inhibitor (Protein kinase Cζ pseudosubstrate, myristoyl trifluoroacetate salt, Sigma, Cat.#P1614) from 0 to 4 hpf. Controls and 1-azakenpaullone (AZ; Sigma, Cat.#A3734) drug treatment of N. vectensis embryos was performed as previously described (Röttinger et al., 2012; Leclère et al., 2016). Embryos were developed in 5 µm AZ from 3 to 76 hpf. Controls were incubated in 0.08% DMSO.
Imaging of N. vectensis embryos
Images of live and fixed embryos were taken using a confocal Zeiss LSM 710 microscope using a Zeiss C-Apochromat 40x water immersion objective (N.A. 1.20). Pinhole settings varied between 1.2 and 1.4 A.U. according to the experiment. The same settings were used for each individual experiment to compare control and experimental conditions. Results from in situ hybridization studies were imaged using a Zeiss Imager.M2 with a Zeiss 425 HRc color digital camera run by Zeiss Zen 2012 software. Z-stack images were processed using Imaris 7.6.4 (Bitplane Inc.) software for three-dimensional reconstructions and FIJI for single slice and movies. Final figures were assembled using Adobe Illustrator and Adobe Photoshop.
Co-Immunoprecipitation
Tissue homogenization and protein extraction was performed as described in (Salinas-Saavedra et al., 2015; Suzuki et al., 2001; Wang et al., 2012). Briefly, embryos were homogenized in 200 µl of ice cold lysis buffer (30 mM HEPES, pH to 7.5, 1 mM EDTA, 150 mM NaCl, 50 mM NaF, 1 mM Na3VO4, 1 mM Na2MoO4, 1 mM MgCl2, 1% NP-40, 10% Glycerol, Protease Inhibitor cocktail (Sigma P8340) and PMSF. After 15 min’ incubation on ice, crude lysate was carefully laid on top of a 200 µl sucrose cushion (1M sucrose, 30 mM HEPES, pH to 7.5, 1 mM EDTA, 150 mM NaCl, 50 mM NaF, 1 mM Na3VO4, 1 mM Na2MoO4) and yolk pelleted by centrifugation at 1000 rpm for 10 min. The top layer was transferred to a clean microcentrifuge tube and 300 µl of lysis buffer was added. Approximately, 5 mg of protein was obtained from 60 µl of embryos (more than 15,000 embryos) homogenized in 500 µl of lysis buffer. 2 mg of total protein (in 500 µl of lysate buffer) was incubated with Par-specific antibodies cross-linked to Pierce Protein A/G Magnetic Beads (Pierce Biotechnology, Rockford, IL). (Pre-immune) IgG-IP pull downs were performed as a negative control for each experiment. Three antibodies, described previously (Salinas-Saavedra et al., 2015), against three different proteins (NvaPKC, NvPar-6, and NvPar-1) were utilized. We performed co-IP experiments using early cleavage (2–4 hpf) and for gastrula stages (24–30 hpf) lysates. Co-IP experiments were repeated four times for each stage using fresh lysates every time. For a detailed protocol, please, go to http://www.whitney.ufl.edu/research/faculty/mark-q-martindale/mark-q-martindale-lab-protocols/
Morphometric measurements
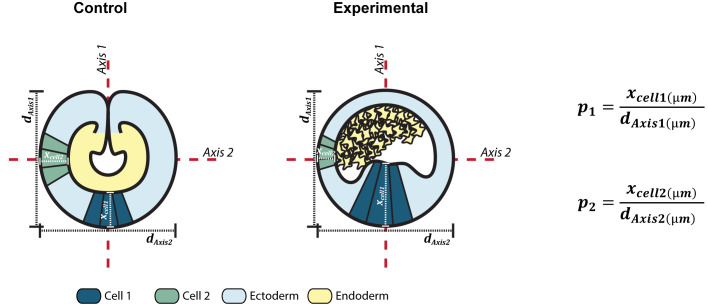
Epithelial thickness was measured using confocal images of embryos immunohistochemically labeled, processed with Imaris 7.6.4 (Bitplane Inc.). For detailed and graphical explanation, please see Figure 3—figure supplement 3 and Figure 3—figure supplement 7. For each treatment, epithelial thickness was determined by the average cell length (µm) along the apico-basal axis (A-B axis) of five individual cells. These values were made for two perpendicular axes Axis1 (A1) and Axis2 (A2) and normalized by the embryonic diameter (µm), in order to minimize technical artifacts (e.g. fixation and mounting) that could have affected the shape/size of the cell. Giving a proportion (p) that was calculated for A1 and A2, named p1 and p2, respectively. The values of p1 and p2 obtained for 90 control embryos, were statistically compared with the respective values obtained for 103 mutant-embryos using the Mann–Whitney U test (nonparametric; normality was tested using SPSS software) with a critical p-value of 0.05. The null hypothesis assumed no differences between the cell sizes of control and mutant-embryos. Values and statistics can be found in Figure 3—figure supplement 3—source data 1.
Acknowledgements
We thank Leslie S Babonis for TEM data in Figure 1—figure supplement 1, A Wikramanayake for Nvß-catenin expression constructs, and C Magie and E Röttinger for Nvsnail-A expression constructs. We thank E Seaver, CE Schnitzler, and the members of Martindale’s lab for helpful discussion. We also thank the NSF-IOS 1239422- Broadening participation of underrepresented groups in Developmental Biology and NSF REU (DBI-1156528) programs for AQR.
Funding Statement
The funders had no role in study design, data collection, and interpretation, or the decision to submit the work for publication.
Contributor Information
Miguel Salinas-Saavedra, Email: mssaavedra@whitney.ufl.edu.
Mark Q Martindale, Email: mqmartin@whitney.ufl.edu.
Alejandro Sánchez Alvarado, Stowers Institute for Medical Research, United States.
Patricia J Wittkopp, University of Michigan, United States.
Funding Information
This paper was supported by the following grants:
National Institutes of Health GM093116 to Mark Q Martindale.
National Science Foundation IOS-1755364 to Mark Q Martindale.
National Aeronautics and Space Administration 16-EXO16_2-0041 to Mark Q Martindale.
Synthego Grants to Mark Q Martindale.
Additional information
Competing interests
No competing interests declared.
Author contributions
Conceptualization, Supervision, Funding acquisition, Validation, Investigation, Visualization, Methodology, Writing—original draft, Project administration, Writing—review and editing.
Validation, Investigation, Writing—review and editing.
Conceptualization, Resources, Supervision, Funding acquisition, Validation, Investigation, Visualization, Methodology, Writing—original draft, Project administration, Writing—review and editing.
Additional files
Data availability
No major datasets were generated in this study. All data generated or analyzed during this study are included in the manuscript and supporting files. All raw images are available on request.
The following previously published dataset was used:
Putnam NH. Srivastava M. Hellsten U. Dirks B. Chapman J. Salamov A. Terry A. Shapiro H. Lindquist E. Kapitonov VV. Jurka J. Genikhovich G. Grigoriev IV. Lucas SM. Steele RE. Finnerty JR. Technau U. Martindale MQ. Rokhsar DS. 2007. Nematostella vectensis genome assembly JGI 1.0. http://genome.jgi.doe.gov/Nemve1/Nemve1.home.html Publicly available at the JGI Genome Portal.
References
- Acloque H, Adams MS, Fishwick K, Bronner-Fraser M, Nieto MA. Epithelial-mesenchymal transitions: the importance of changing cell state in development and disease. Journal of Clinical Investigation. 2009;119:1438–1449. doi: 10.1172/JCI38019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiel AR, Johnston H, Chock T, Dahlin P, Iglesias M, Layden M, Röttinger E, Martindale MQ. A bipolar role of the transcription factor ERG for cnidarian germ layer formation and apical domain patterning. Developmental Biology. 2017;430:346–361. doi: 10.1016/j.ydbio.2017.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrallo-Gimeno A, Nieto MA. The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development. 2005;132:3151–3161. doi: 10.1242/dev.01907. [DOI] [PubMed] [Google Scholar]
- Baumgarten S, Simakov O, Esherick LY, Liew YJ, Lehnert EM, Michell CT, Li Y, Hambleton EA, Guse A, Oates ME, Gough J, Weis VM, Aranda M, Pringle JR, Voolstra CR. The genome of Aiptasia, a sea anemone model for coral symbiosis. PNAS. 2015;112:11893–11898. doi: 10.1073/pnas.1513318112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhm H, Brinkmann V, Drab M, Henske A, Kurzchalia TV. Mammalian homologues of C. elegans PAR-1 are asymmetrically localized in epithelial cells and may influence their polarity. Current Biology. 1997;7:603–606. doi: 10.1016/S0960-9822(06)00260-0. [DOI] [PubMed] [Google Scholar]
- Byrum CA, Martindale MQ. Gastrulation From Cells to Embryo. Cold Spring Harbor Press; 2004. Gastrulation in the Cnidaria and Ctenophora; pp. 33–50. [Google Scholar]
- Chapman JA, Kirkness EF, Simakov O, Hampson SE, Mitros T, Weinmaier T, Rattei T, Balasubramanian PG, Borman J, Busam D, Disbennett K, Pfannkoch C, Sumin N, Sutton GG, Viswanathan LD, Walenz B, Goodstein DM, Hellsten U, Kawashima T, Prochnik SE, Putnam NH, Shu S, Blumberg B, Dana CE, Gee L, Kibler DF, Law L, Lindgens D, Martinez DE, Peng J, Wigge PA, Bertulat B, Guder C, Nakamura Y, Ozbek S, Watanabe H, Khalturin K, Hemmrich G, Franke A, Augustin R, Fraune S, Hayakawa E, Hayakawa S, Hirose M, Hwang JS, Ikeo K, Nishimiya-Fujisawa C, Ogura A, Takahashi T, Steinmetz PR, Zhang X, Aufschnaiter R, Eder MK, Gorny AK, Salvenmoser W, Heimberg AM, Wheeler BM, Peterson KJ, Böttger A, Tischler P, Wolf A, Gojobori T, Remington KA, Strausberg RL, Venter JC, Technau U, Hobmayer B, Bosch TC, Holstein TW, Fujisawa T, Bode HR, David CN, Rokhsar DS, Steele RE. The dynamic genome of Hydra. Nature. 2010;464:592–596. doi: 10.1038/nature08830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke DN, Miller PW, Lowe CJ, Weis WI, Nelson WJ. Characterization of the Cadherin-Catenin complex of the sea Anemone nematostella vectensis and implications for the evolution of metazoan Cell-Cell adhesion. Molecular Biology and Evolution. 2016;33:2016–2029. doi: 10.1093/molbev/msw084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutinho CC, Rosa IA, Teixeira JDO, Andrade LR, Costa ML, Mermelstein C. Cellular migration, transition and interaction during regeneration of the sponge Hymeniacidon heliophila. PLoS One. 2017;12:e0178350. doi: 10.1371/journal.pone.0178350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darras S, Gerhart J, Terasaki M, Kirschner M, Lowe CJ. β-catenin specifies the endomesoderm and defines the posterior organizer of the hemichordate Saccoglossus kowalevskii. Development. 2011;138:959–970. doi: 10.1242/dev.059493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH, Rast JP, Oliveri P, Ransick A, Calestani C, Yuh CH, Minokawa T, Amore G, Hinman V, Arenas-Mena C, Otim O, Brown CT, Livi CB, Lee PY, Revilla R, Schilstra MJ, Clarke PJ, Rust AG, Pan Z, Arnone MI, Rowen L, Cameron RA, McClay DR, Hood L, Bolouri H. A provisional regulatory gene network for specification of endomesoderm in the sea urchin embryo. Developmental Biology. 2002;246:162–190. doi: 10.1006/dbio.2002.0635. [DOI] [PubMed] [Google Scholar]
- Doench JG, Hartenian E, Graham DB, Tothova Z, Hegde M, Smith I, Sullender M, Ebert BL, Xavier RJ, Root DE. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nature Biotechnology. 2014;32:1–8. doi: 10.1038/nbt.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuBuc TQ, Dattoli AA, Babonis LS, Salinas-Saavedra M, Röttinger E, Martindale MQ, Postma M. In vivo imaging of Nematostella vectensis embryogenesis and late development using fluorescent probes. BMC Cell Biology. 2014;15:44. doi: 10.1186/s12860-014-0044-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Extavour CG, Pang K, Matus DQ, Martindale MQ. Vasa and Nanos expression patterns in a sea Anemone and the evolution of bilaterian germ cell specification mechanisms. Evolution & Development. 2005;7:201–215. doi: 10.1111/j.1525-142X.2005.05023.x. [DOI] [PubMed] [Google Scholar]
- Fritzenwanker JH, Genikhovich G, Kraus Y, Technau U. Early development and axis specification in the sea anemone Nematostella vectensis. Developmental Biology. 2007;310:264–279. doi: 10.1016/j.ydbio.2007.07.029. [DOI] [PubMed] [Google Scholar]
- Ganot P, Zoccola D, Tambutté E, Voolstra CR, Aranda M, Allemand D, Tambutté S. Structural molecular components of septate junctions in cnidarians point to the origin of epithelial junctions in eukaryotes. Molecular Biology and Evolution. 2015;32:44–62. doi: 10.1093/molbev/msu265. [DOI] [PubMed] [Google Scholar]
- Hand C, Uhlinger KR. The culture, sexual and asexual reproduction, and growth of the sea Anemone nematostella vectensis. The Biological Bulletin. 1992;182:169–176. doi: 10.2307/1542110. [DOI] [PubMed] [Google Scholar]
- Higashi T, Arnold TR, Stephenson RE, Dinshaw KM, Miller AL. Maintenance of the epithelial barrier and remodeling of Cell-Cell junctions during cytokinesis. Current Biology. 2016;26:1829–1842. doi: 10.1016/j.cub.2016.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikmi A, McKinney SA, Delventhal KM, Gibson MC. TALEN and CRISPR/Cas9-mediated genome editing in the early-branching metazoan Nematostella vectensis. Nature Communications. 2014;5:5486. doi: 10.1038/ncomms6486. [DOI] [PubMed] [Google Scholar]
- Jahnel SM, Walzl M, Technau U. Development and epithelial organisation of muscle cells in the sea Anemone nematostella vectensis. Frontiers in Zoology. 2014;11:15–44. doi: 10.1186/1742-9994-11-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonusaite S, Donini A, Kelly SP. Occluding junctions of invertebrate epithelia. Journal of Comparative Physiology B. 2016;186:17–43. doi: 10.1007/s00360-015-0937-1. [DOI] [PubMed] [Google Scholar]
- Keller R, Davidson LA, Shook DR. How we are shaped: the biomechanics of gastrulation. Differentiation. 2003;71:171–205. doi: 10.1046/j.1432-0436.2003.710301.x. [DOI] [PubMed] [Google Scholar]
- Kirillova A, Genikhovich G, Pukhlyakova E, Demilly A, Kraus Y, Technau U. Germ-layer commitment and axis formation in sea anemone embryonic cell aggregates. PNAS. 2018;115:1813–1818. doi: 10.1073/pnas.1711516115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumburegama S, Wijesena N, Xu R, Wikramanayake AH. Strabismus-mediated primary archenteron invagination is uncoupled from Wnt/β-catenin-dependent endoderm cell fate specification in Nematostella vectensis (Anthozoa, Cnidaria): Implications for the evolution of gastrulation. EvoDevo. 2011;2:2. doi: 10.1186/2041-9139-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusserow A, Pang K, Sturm C, Hrouda M, Lentfer J, Schmidt HA, Technau U, von Haeseler A, Hobmayer B, Martindale MQ, Holstein TW. Unexpected complexity of the Wnt gene family in a sea anemone. Nature. 2005;433:156–160. doi: 10.1038/nature03158. [DOI] [PubMed] [Google Scholar]
- Layden MJ, Röttinger E, Wolenski FS, Gilmore TD, Martindale MQ. Microinjection of mRNA or morpholinos for reverse genetic analysis in the Starlet Sea Anemone, nematostella vectensis. Nature Protocols. 2013;8:924–934. doi: 10.1038/nprot.2013.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclère L, Bause M, Sinigaglia C, Steger J, Rentzsch F. Development of the aboral domain in Nematostella requires β-catenin and the opposing activities of Six3/6 and Frizzled5/8. Development. 2016;143:1766–1777. doi: 10.1242/dev.120931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JM, Dedhar S, Kalluri R, Thompson EW. The epithelial-mesenchymal transition: new insights in signaling, development, and disease. The Journal of Cell Biology. 2006;172:973–981. doi: 10.1083/jcb.200601018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PN, Kumburegama S, Marlow HQ, Martindale MQ, Wikramanayake AH. Asymmetric developmental potential along the animal-vegetal Axis in the anthozoan cnidarian, Nematostella vectensis, is mediated by dishevelled. Developmental Biology. 2007;310:169–186. doi: 10.1016/j.ydbio.2007.05.040. [DOI] [PubMed] [Google Scholar]
- Lim J, Thiery JP. Epithelial-mesenchymal transitions: insights from development. Development. 2012;139:3471–3486. doi: 10.1242/dev.071209. [DOI] [PubMed] [Google Scholar]
- Maduro MF, Rothman JH. Making worm guts: the gene regulatory network of the Caenorhabditis elegans endoderm. Developmental Biology. 2002;246:68–85. doi: 10.1006/dbio.2002.0655. [DOI] [PubMed] [Google Scholar]
- Magie CR, Daly M, Martindale MQ. Gastrulation in the cnidarian Nematostella vectensis occurs via invagination not ingression. Developmental Biology. 2007;305:483–497. doi: 10.1016/j.ydbio.2007.02.044. [DOI] [PubMed] [Google Scholar]
- Magie CR, Martindale MQ. Cell-cell adhesion in the cnidaria: insights into the evolution of tissue morphogenesis. The Biological Bulletin. 2008;214:218–232. doi: 10.2307/25470665. [DOI] [PubMed] [Google Scholar]
- Marlow H, Matus DQ, Martindale MQ. Ectopic activation of the canonical wnt signaling pathway affects ectodermal patterning along the primary Axis during larval development in the anthozoan Nematostella vectensis. Developmental Biology. 2013;380:324–334. doi: 10.1016/j.ydbio.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlow HQ, Martindale MQ. Embryonic development in two species of scleractinian coral embryos: symbiodinium localization and mode of gastrulation. Evolution & Development. 2007;9:355–367. doi: 10.1111/j.1525-142X.2007.00173.x. [DOI] [PubMed] [Google Scholar]
- Martindale MQ, Lee PN. The development of form: causes and consequences of developmental reprogramming associated with rapid body plan evolution in the bilaterian radiation. Biological Theory. 2013;8:253–264. doi: 10.1007/s13752-013-0117-z. [DOI] [Google Scholar]
- Martindale MQ, Pang K, Finnerty JR. Investigating the origins of triploblasty: 'mesodermal' gene expression in a diploblastic animal, the sea anemone Nematostella vectensis (phylum, Cnidaria; class, Anthozoa) Development. 2004;131:2463–2474. doi: 10.1242/dev.01119. [DOI] [PubMed] [Google Scholar]
- Martindale MQ. The evolution of metazoan axial properties. Nature Reviews Genetics. 2005;6:917–927. doi: 10.1038/nrg1725. [DOI] [PubMed] [Google Scholar]
- Matus DQ, Thomsen GH, Martindale MQ. Dorso/ventral genes are asymmetrically expressed and involved in germ-layer demarcation during cnidarian gastrulation. Current Biology. 2006;16:499–505. doi: 10.1016/j.cub.2006.01.052. [DOI] [PubMed] [Google Scholar]
- Matus DQ, Thomsen GH, Martindale MQ. FGF signaling in Gastrulation and neural development in Nematostella vectensis, an anthozoan cnidarian. Development Genes and Evolution. 2007;217:137–148. doi: 10.1007/s00427-006-0122-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Mateos MA, Vejnar CE, Beaudoin JD, Fernandez JP, Mis EK, Khokha MK, Giraldez AJ. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nature Methods. 2015;12:982–988. doi: 10.1038/nmeth.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi N, Sogabe S, Degnan BM. Evolutionary origin of gastrulation: insights from sponge development. BMC Biology. 2014;12:26. doi: 10.1186/1741-7007-12-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto MA, Huang RY, Jackson RA, Thiery JP. EMT: 2016. Cell. 2016;166:21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- Ryan JF, Pang K, Schnitzler CE, Nguyen AD, Moreland RT, Simmons DK, Koch BJ, Francis WR, Havlak P, Smith SA, Putnam NH, Haddock SH, Dunn CW, Wolfsberg TG, Mullikin JC, Martindale MQ, Baxevanis AD, NISC Comparative Sequencing Program The genome of the ctenophore Mnemiopsis leidyi and its implications for cell type evolution. Science. 2013;342:1242592. doi: 10.1126/science.1242592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S, Goulas S, Hirose T. Cell Polarity 1 Biol Role Basic Mech. 2015. The PAR3-aPKC-PAR6 complex; pp. 3–23. [DOI] [Google Scholar]
- Ohsawa S, Vaughen J, Igaki T. Cell extrusion: a Stress-Responsive force for good or evil in epithelial homeostasis. Developmental Cell. 2018;44:284–296. doi: 10.1016/j.devcel.2018.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossipova O, Dhawan S, Sokol S, Green JB. Distinct PAR-1 proteins function in different branches of wnt signaling during vertebrate development. Developmental Cell. 2005;8:829–841. doi: 10.1016/j.devcel.2005.04.011. [DOI] [PubMed] [Google Scholar]
- Putnam NH, Srivastava M, Hellsten U, Dirks B, Chapman J, Salamov A, Terry A, Shapiro H, Lindquist E, Kapitonov VV, Jurka J, Genikhovich G, Grigoriev IV, Lucas SM, Steele RE, Finnerty JR, Technau U, Martindale MQ, Rokhsar DS. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- Ragkousi K, Marr K, McKinney S, Ellington L, Gibson MC. Cell-Cycle-Coupled oscillations in apical polarity and intercellular contact maintain order in embryonic epithelia. Current Biology. 2017;27:1381–1386. doi: 10.1016/j.cub.2017.03.064. [DOI] [PubMed] [Google Scholar]
- Ribeiro AS, Paredes J. P-Cadherin linking breast Cancer stem cells and invasion: a promising marker to identify an ‘Intermediate/Metastable’ EMT State. Frontiers in Oncology. 2014;4:371. doi: 10.3389/fonc.2014.00371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodaway A, Patient R. Mesendoderm. Cell. 2001;105:169–172. doi: 10.1016/S0092-8674(01)00307-5. [DOI] [PubMed] [Google Scholar]
- Roure A, Rothbächer U, Robin F, Kalmar E, Ferone G, Lamy C, Missero C, Mueller F, Lemaire P. A multicassette gateway vector set for high throughput and comparative analyses in Ciona and vertebrate embryos. PLoS ONE. 2007;2:e916. doi: 10.1371/journal.pone.0000916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röttinger E, Dahlin P, Martindale MQ. A framework for the establishment of a cnidarian gene regulatory network for ‘endomesoderm’ specification: the inputs of ß-catenin/TCF signaling. PLoS Genetics. 2012;8:e1003164. doi: 10.1371/journal.pgen.1003164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas-Saavedra M, Stephenson TQ, Dunn CW, Martindale MQ. Par system components are asymmetrically localized in ectodermal Epithelia, but not during early development in the sea Anemone nematostella vectensis. EvoDevo. 2015;6:20. doi: 10.1186/s13227-015-0014-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer G, Narasimha M, Vogelsang E, Leptin M. Cadherin switching during the formation and differentiation of the Drosophila mesoderm - implications for epithelial-to-mesenchymal transitions. Journal of Cell Science. 2014;127:1511–1522. doi: 10.1242/jcs.139485. [DOI] [PubMed] [Google Scholar]
- Scholz CB, Technau U. The ancestral role of brachyury: expression of NemBra1 in the basal cnidarian Nematostella vectensis (Anthozoa) Development Genes and Evolution. 2003;212:563–570. doi: 10.1007/s00427-002-0272-x. [DOI] [PubMed] [Google Scholar]
- Servetnick MD, Steinworth B, Babonis LS, Simmons D, Salinas-Saavedra M, Martindale MQ. Cas9-mediated excision of Nematostella brachyury disrupts endoderm development, pharynx formation and oral-aboral patterning. Development. 2017;144:2951–2960. doi: 10.1242/dev.145839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinzato C, Shoguchi E, Kawashima T, Hamada M, Hisata K, Tanaka M, Fujie M, Fujiwara M, Koyanagi R, Ikuta T, Fujiyama A, Miller DJ, Satoh N. Using the Acropora digitifera genome to understand coral responses to environmental change. Nature. 2011;476:320–323. doi: 10.1038/nature10249. [DOI] [PubMed] [Google Scholar]
- Solnica-Krezel L, Sepich DS. Gastrulation: making and shaping germ layers. Annual Review of Cell and Developmental Biology. 2012;28:687–717. doi: 10.1146/annurev-cellbio-092910-154043. [DOI] [PubMed] [Google Scholar]
- Sun TQ, Lu B, Feng JJ, Reinhard C, Jan YN, Fantl WJ, Williams LT. PAR-1 is a Dishevelled-associated kinase and a positive regulator of wnt signalling. Nature Cell Biology. 2001;3:628–636. doi: 10.1038/35083016. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Yamanaka T, Hirose T, Manabe N, Mizuno K, Shimizu M, Akimoto K, Izumi Y, Ohnishi T, Ohno S. Atypical protein kinase C is involved in the evolutionarily conserved par protein complex and plays a critical role in establishing epithelia-specific junctional structures. The Journal of Cell Biology. 2001;152:1183–1196. doi: 10.1083/jcb.152.6.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamulonis C, Postma M, Marlow HQ, Magie CR, de Jong J, Kaandorp J. A cell-based model of Nematostella vectensis gastrulation including bottle cell formation, invagination and zippering. Developmental Biology. 2011;351:217–228. doi: 10.1016/j.ydbio.2010.10.017. [DOI] [PubMed] [Google Scholar]
- Technau U, Scholz CB. Origin and evolution of endoderm and mesoderm. The International Journal of Developmental Biology. 2003;47:531–539. [PubMed] [Google Scholar]
- Theveneau E, Mayor R. Collective cell migration of epithelial and mesenchymal cells. Cellular and Molecular Life Sciences. 2013;70:3481–3492. doi: 10.1007/s00018-012-1251-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaccari T, Rabouille C, Ephrussi A. The Drosophila PAR-1 spacer domain is required for lateral membrane association and for polarization of follicular epithelial cells. Current Biology. 2005;15:255–261. doi: 10.1016/j.cub.2005.01.033. [DOI] [PubMed] [Google Scholar]
- Wang R, Liu X, Küster-Schöck E, Fagotto F. Proteomic analysis of differences in ectoderm and mesoderm membranes by DiGE. Journal of Proteome Research. 2012;11:4575–4593. doi: 10.1021/pr300379m. [DOI] [PubMed] [Google Scholar]
- Weng M, Wieschaus E. Myosin-dependent remodeling of adherens junctions protects junctions from Snail-dependent disassembly. The Journal of Cell Biology. 2016;212:219–229. doi: 10.1083/jcb.201508056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng M, Wieschaus E. Polarity protein Par3/Bazooka follows myosin-dependent junction repositioning. Developmental Biology. 2017;422:125–134. doi: 10.1016/j.ydbio.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijesena N, Simmons DK, Martindale MQ. Antagonistic BMP-cWNT signaling in the cnidarian Nematostella vectensis reveals insight into the evolution of mesoderm. PNAS. 2017;114:E5608–E5615. doi: 10.1073/pnas.1701607114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijesena NM, Kumburegama S, Xu R, Wikramanayake A. Wnt signaling in the cnidarian Nematostella vectensis: insights into the evolution of gastrulation. Developmental Biology. 2011;356:107. doi: 10.1016/j.ydbio.2011.05.027. [DOI] [Google Scholar]
- Wikramanayake AH, Hong M, Lee PN, Pang K, Byrum CA, Bince JM, Xu R, Martindale MQ. An ancient role for nuclear beta-catenin in the evolution of axial polarity and germ layer segregation. Nature. 2003;426:446–450. doi: 10.1038/nature02113. [DOI] [PubMed] [Google Scholar]
- Wolenski FS, Layden MJ, Martindale MQ, Gilmore TD, Finnerty JR. Characterizing the spatiotemporal expression of RNAs and proteins in the starlet sea Anemone, nematostella vectensis. Nature Protocols. 2013;8:900–915. doi: 10.1038/nprot.2013.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuoka Y, Shinzato C, Satoh N. The Mesoderm-Forming gene brachyury regulates Ectoderm-Endoderm demarcation in the coral Acropora digitifera. Current Biology. 2016;26:2885–2892. doi: 10.1016/j.cub.2016.08.011. [DOI] [PubMed] [Google Scholar]