Abstract
This Commentary addresses the FDA approval of pembrolizumab and the changes necessary in clinical practice to identify patients who would benefit from such treatment.
As part of several large‐scale efforts [1], the U.S. Food and Drug Administration (FDA) approval of Keytruda (pembrolizumab; Merck, Kenilworth, NJ) for solid tumors demonstrating certain biomarker signatures represents a milestone in immuno‐oncology. The broad approval was partly due to the high overall objective response rate [2]. Numerous testing strategies for mismatch repair deficiency (dMMR) and programmed cell death ligand 1 (PD‐L1) expression are available (Fig. 1A); however, these require technical‐ and context‐specific analysis, which most practicing oncologists do not have. Thus, pembrolizumab therapy poses the challenge of how to effectively identify patients using locally available testing methodologies.
Figure 1.
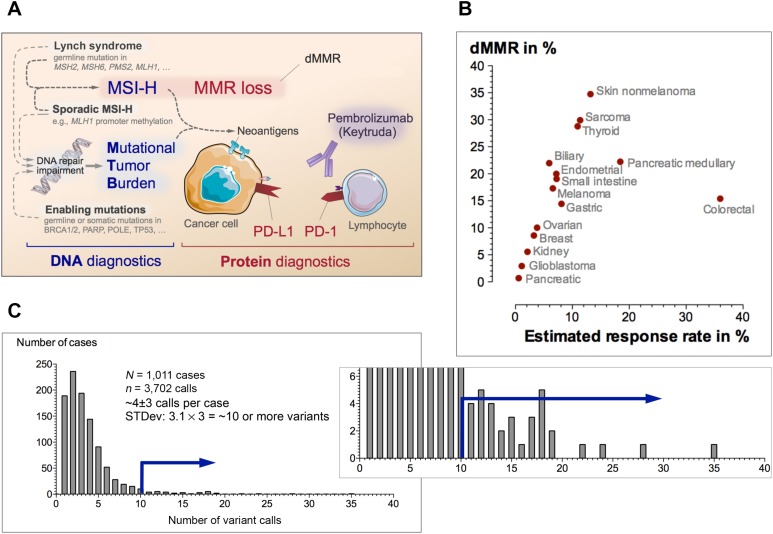
Keytruda testing in clinical practice. (A): Simplified concept of pembrolizumab testing strategy. Microsatellite instability or other causes may contribute to the overall mutational burden that is associated with increased levels of neoantigens. To prevent detection by the immune system, the tumor must mount immunologic camouflage (e.g., PD‐L1). Several assays at the DNA and protein level have been employed as biomarkers for pembrolizumab response. (B): Percentage of dMMR and response rates in 15 selected malignancies. When taking the objective response rate from the Keytruda trials into account, one can estimate the expected response as the fraction of responders within the subset of patients with dMMR (see supplemental references for details). The relationship between dMMR rate and response illustrate clinical expectations. (C): Example of a frequency distribution of number of variants per case in a next‐generation sequencing panel. “Hypermutant” was defined as number of variants per case >3 standard deviations above the mean. The blue arrow represents the cutoff for high mutational tumor burden.
Abbreviations: dMMR, deficient mismatch repair deficiency (defined by either IHC, or PCR; see text); MMR, mismatch repair; MSI‐H, high microsatellite instability; PD‐1, programmed cell death 1; PD‐L1, programmed cell death ligand 1; STDev, standard deviation.
Here, we attempt to answer 10 relevant questions about pembrolizumab testing.
1. What is the response rate across solid tumors?
The overall response rate is ∼39.7%, defined via objective radiographic responses [3].
2. What does dMMR mean?
dMMR stands for “mismatch repair deficiency” identified by any method. Microsatellite instability (MSI) is determined using polymerase chain reaction (PCR) and sizing analysis of mono‐ or dinucleotide markers (e.g., BAT25, BAT26, D2S123, D5S346, D17S250), whereas loss of mismatch repair (MMR) proteins MLH1, MSH2, MSH6, and/or PMS2 is assessed using immunohistochemistry (IHC) on tumor tissues.
3. Does it matter whether I test using PCR or IHC?
Each method is acceptable and has ∼90% sensitivity if performed alone; combining both methods increases the sensitivity [4].
4. What is the expected dMMR detection and response rate for my tumor?
The response rate depends on the dMMR rate by tumor and histotype (Fig. 1B). The estimates of median dMMR frequency for the selected malignancies do not account for possible population‐specific variations in dMMR rates [5] or various cancer subtypes (e.g., shown for medullary pancreatic cancer).
5. Is it sufficient if my tumor was screened for Lynch syndrome?
Yes. Lynch syndrome screening often entails immunohistochemistry for mismatch repair proteins followed by genotyping.
6. How can I decide between PD‐L1 and dMMR testing?
The package insert specifies which tumor should be tested and how; for example, the FDA approved PD‐L1 testing for gastric cancer and ALK‐negative and EGFR‐negative non‐small cell lung cancer.
7. What is the test cost, and will my patient's insurance cover testing?
Currently, the cost ranges between $150 and $550, and coverage depends on the tumor, stage, test method (Current Procedural Technology code), and insurance plan; many payers require prior authorization.
8. What is the most effective strategy for Keytruda testing?
Although the overall effectiveness of testing is tumor‐ and context‐specific and can be estimated as a product of the dMMR rate and response rate (Fig. 1B), the testing strategy depends on local availability.
9. Which tests are available, and how can I get my patient's sample tested?
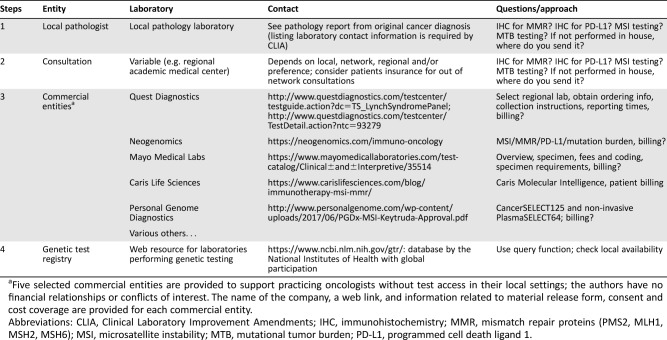
An outline to find out availability is provided in Table 1. Firstly, we recommend contacting your local pathologist because most pathology laboratories perform IHC, so PD‐L1 testing and MMR‐deficiency assessment or Lynch syndrome screening should be available. If your pathologist performs genotyping, MSI testing is also generally available. A second option is consulting regional academic medical centers. Thirdly, for many practicing oncologists without test access in their local hospitals, we provide a list of selected commercial testing laboratories. Finally, another excellent Web resource with global participation is the NIH's genetic testing registry at https://www.ncbi.nlm.nih.gov/gtr/.
Table 1. Practical approach to determine testing availability.
Five selected commercial entities are provided to support practicing oncologists without test access in their local settings; the authors have no financial relationships or conflicts of interest. The name of the company, a web link, and information related to material release form, consent and cost coverage are provided for each commercial entity.
Abbreviations: CLIA, Clinical Laboratory Improvement Amendments; IHC, immunohistochemistry; MMR, mismatch repair proteins (PMS2, MLH1, MSH2, MSH6); MSI, microsatellite instability; MTB, mutational tumor burden; PD‐L1, programmed cell death ligand 1.
10. Is mutational tumor burden a meaningful biomarker?
Many laboratories validate their strategy to identify tumors with a high mutational tumor burden (MTB; so‐called “hypermutant” tumors)—typically relative to their tested population (Fig. 1C). Several next‐generation‐sequencing‐based assays for determining MTB are currently actively reviewed by the FDA. In this context, we also want to draw attention to a concurrent article in this issue [6]. Briefly, Dizon et al. report a complete response to pembrolizumab in a woman with a mismatch repair deficient endometrial cancer carrying a germline BRCA1 mutation [6]. The case illustrates two important aspects: first, as shown in Figure 1A, several underlying genetic defects can contribute to the overall mutational burden—and the resulting increased levels of neoantigens require the tumor cells to equip themselves with a PD‐L1 camouflage, which we can now target; second, the case argues for assessment of the dMMR status (e.g., as part of Lynch syndrome screening strategies) when clinically indicated.
In summary, the high overall response rates of pembrolizumab received accelerated FDA approval. Accelerated adoption in clinical practice therefore means implementation of effective testing operations to appropriately screen and identify patients in order to deliver the pembrolizumab promise.
Footnotes
Editor's Note: See the related article, “Complete Remission Following Pembrolizumab in a Woman with Mismatch Repair‐Deficient Endometrial Cancer and a Germline BRCA1 Mutation,” by Don S. Dizon et al., on page 650 of this issue.
Disclosures
The authors reported no financial relationships.
References
- 1. Champiat S, Lambotte O, Barreau E et al. Management of immune checkpoint blockade dysimmune toxicities: A collaborative position paper. Ann Oncol 2016;27:559–574. [DOI] [PubMed] [Google Scholar]
- 2. Rizvi NA, Hellmann MD, Snyder A et al. Cancer immunology. Mutational landscape determines sensitivity to PD‐1 blockade in non‐small cell lung cancer. Science 2015;348:124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keytruda [package insert]. Kenilworth, NJ: Merck Sharp & Dohme Corp. Available at https://www.merck.com/product/usa/pi_circulars/k/keytruda/keytruda_pi.pdf. Accessed October 26, 2017.
- 4. Samowitz WS, Broaddus R, Iacopetta B et al. PCR versus immunohistochemistry for microsatellite instability. J Mol Diagn 2008;10:181–182; author reply 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nagahashi M, Ajioka Y, Lang I et al. Genetic changes of p53, K‐ras, and microsatellite instability in gallbladder carcinoma in high‐incidence areas of Japan and Hungary. World J Gastroenterol 2008;14:70–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dizon DS, Dias‐Santagata D, Bregar A et al. Complete remission following pembrolizumab in a woman with mismatch repair deficient endometrial cancer and a germline BRCA1 mutation. The Oncologist 2018. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]