Abstract
DNA repair pathways are evolutionarily conserved molecular mechanisms that maintain the integrity of genomic DNA. In cancer therapies, the integrity and activity of DNA repair pathways predict therapy resistance and disease outcome. Members of the poly (ADP-ribose) polymerase (PARP) family initiate and organize the biologic process of DNA repair, which counteracts many types of chemotherapies. Since the first development in approximately 3 decades ago, PARP inhibitors have greatly changed the concept of cancer therapy, leading to encouraging improvements in tumor suppression and disease outcomes. Here we summaries both pre-clinical and clinical findings of PARP inhibitors applications, particularly for combination therapies.
Keywords: DNA repair, Chemoresistance, PARP inhibitor, Combination therapy, Cancer, Chemotherapy
1. Introduction
The integrity of the human genome is maintained by DNA repair pathways, which reverse the effects of DNA damage. Genomic DNA is continuously exposed to harmful factors, which cause sequence-independent damage to the nucleobases. Random DNA damage may result from endogenous causes, ranging from oxidative DNA damage to DNA replication errors and replication fork arrest. Exogenous damage is commonly introduced by chemical mutagens, ultraviolet light, radiation, or DNA-targeting chemotherapies. DNA lesions cause harmful changes in cells, such as genomic instability, the accumulation of mutations, cell transformation, or cell death. Throughout evolution, eukaryotic cells developed a set of molecular mechanisms that remove DNA lesions, in order to avoid the consequences of genomic deterioration. The activity and expression of the DNA repair machinery is dynamically regulated by a damage-sensing feedback loop, which enhances DNA repair when confronted with acutely introduced exogenous DNA lesions.
Cancer cells exploit endogenous DNA repair mechanisms to counteract DNA-targeting therapies. Radio- and chemotherapies result in DNA damage to the cancer genome in the form of DNA adducts and strand breaks. These changes in genomic DNA are effectively fixed by different intrinsic DNA repair pathways. Moreover, in drug-resistant cancer cells, enhancement of DNA repair pathways is frequently identified as the major molecular shift that alters their sensitivity to anti-cancer therapies, resulting in treatment resistance, cancer recurrence and poor disease outcome. Inhibiting DNA repair has long been proposed as a reasonable sensitization approach to improve genotoxic therapy. In this review, we discuss the recent advancements in poly (ADP-ribose) polymerase (PARP) inhibitors for cancer therapy, with a focus on the combination regimens that may potentiate traditional chemotherapy.
2. Genotoxic agents and DNA repair: yins and yangs in cancer therapy
The central strategy in cancer therapy is to eliminate fast-proliferating, transformed cells. Due to the frequent cellular division in cancer cells, DNA lesions in these cells are more likely to be recognized by cell cycle checkpoint, and translate to cell death, thereby making genotoxic treatments more harmful to cancer cells than non-replicating, terminally differentiated cells. However, in many cases, single agent treatment may not cause sufficient tumor suppression to impact a patient’s survival. The doses required for a monotherapy to kill cancer cells are often very high causing intolerable toxicity to various non-cancerous cells. Thus, combination treatments that base on synergistic effect may be a better strategy to control cancer progression and improve toxicity and ultimately disease outcome.
Chemotherapy causes DNA damage through a variety of mechanisms. Alkylating agents are the most common chemotherapies and act by introducing chemical modifications to DNA at the base pair level. For example, alkylating agents, such as temozolomide (TMZ) and dacarbazine, attach alkyl groups (CnH2n+1) to the N- and O- atoms in the nucleobases. Bifunctional alkylating agents, such as nitrogen mustards and chloroethylating agents (CCNU, BCNU), carry 2 reactive sites and introduce more complicated modifications to nucleobases, resulting in further DNA damage and blockage of replication forks (Kondo, Takahashi, Ono, & Ohnishi, 2010). Similarly, platinum-based compounds, such as cisplatin and carboplatin, transform into the aqueous complex cis- [PtCl(NH3)2(H2O)]+, which reacts with DNA base pairs. The major products include mono platinum adducts, intra-strand DNA crosslinks (>95% of all adducts), inter-strand DNA crosslinks, or protein–DNA crosslinks (Crul, van Waardenburg, Beijnen, & Schellens, 2002). These DNA nucleobase lesions may not cause immediate cytotoxic effects, but rather cause cell cycle arrest through disruption of replication fork progression, resulting in further cellular damage, such as replication-associated double-strand DNA breaks, mitotic catastrophe, and apoptosis.
Although chemotherapy-induced DNA lesions may be widespread, intrinsic DNA repair pathways are capable of detoxifying each type of DNA adduct, thereby enabling cancer cell survival, recurrence, and resistance. DNA lesions at the base pair level, such as alkylated nucleobases, platinum-associated intra-strand crosslinks, or single-strand DNA breaks, are effectively repaired by base-excision repair (BER), nucleotide-excision repair, and mismatch repair pathways. Some DNA adducts can be specifically reversed without base pair excision. For example, O-6-methylguanine-DNA methyltransferase (MGMT) removes the alkyl groups from the O6 position of guanine, thereby reducing the risk of DNA breaks and cell death. The DNA dioxygenases ABH2 and ABH3 remove 1-methyladenine and 3-methylcytosine from bases. Thus, base pair level DNA lesions are rapidly repaired, likely within 1 hour. Large-scale DNA damage, such as double-strand DNA breaks and clustered lesions, requires the involvement of either the non-homologous end joining (NHEJ) or homologous recombination (HR) pathway. Considering the key roles of DNA repair pathways in cancer therapy resistance, inhibitors targeting different DNA repair pathways have been developed as potential sensitizers for the traditional cancer therapies. In the following sections, we will discuss the latest advances in PARP DNA repair inhibitors, the most frequently used chemo-sensitizers, and their possible applications in cancer therapy. Detailed reviews about other DNA repair pathway inhibitors in cancer therapy can be found in other publications (Aziz, et al., 2012; Gavande, et al., 2016; Helleday, Petermann, Lundin, Hodgson, & Sharma, 2008).
3. PARP, a key mechanism in cancer resistance
In 1963, Chambon et al. first identified a catalytic reaction that involves DNA-dependent NAD+ consumption (Chambon, Weill, & Mandel, 1963). Subsequent studies identified poly(ADP-ribosyl)ation as an important post-translational modification of nuclear proteins, as well as a potential link between this process and DNA damage responses and chromatin modifications. The β-α-loop-β-α NAD+ fold was recognized as a signature domain, which is evolutionarily conserved through all PARP enzymes. To date, 18 proteins have been identified as members of the PARP superfamily, all of which contain this highly conserved catalytic site across species (Ame, Spenlehauer, & de Murcia, 2004; Kraus, 2015).
Among PARP family members, PARP-1 accounts for 75% of total PARP enzyme activity; it is the major player in sensing DNA damage and organizing the repair machinery. Similar to the rapid phosphorylation of histone H2A.X, PARP-mediated poly(ADP-ribosyl)ation of histones and other nuclear proteins is considered one of the earliest epigenetic events triggered by DNA strand breakage, highlighting DNA damage foci. PARP consumes NAD+ to catalyze the covalent attachment of ADP-ribose to the γ-carboxyl groups of the glutamate residues of acceptor proteins or PARP itself. The ADP-ribose polymer consists of a linear or branched polyanion, and initiates DNA repair by recruiting other repair enzymes, such as PARP-2, XRCC1, DNA polymerase β, and DNA ligase III, to sites of DNA damage (El-Khamisy, Masutani, Suzuki, & Caldecott, 2003; Masson, et al., 1998; Schreiber, et al., 2002). Although it is not directly involved in nucleobase modification, PARP-1 is a key spatial and temporal organizer for the entire repair process, as indicated by deficiency of the BER pathway when PARP-1 is genetically compromised (Dantzer, et al., 2000).
PARP-1 activity is critical for the establishment of resistance to genotoxic agents. Early studies showed that DNA lesions are more likely to accumulate in cells with genetically compromised PARP-1, which further translates into cell cycle arrest and loss of cell viability (de Murcia, et al., 1997; Trucco, Oliver, de Murcia, & Menissier-de Murcia, 1998). The role of PARP-mediated DNA repair in cancer resistance has been confirmed in cancer cell lines and xenografts. In 1996, Bernges and Zeller discovered that suppressing PARP activity with 3-aminobenzamide (3-AB), an NAD+ analog, reduced the resistance of ovarian cancer cell lines to genotoxic therapy (Bernges & Zeller, 1996). Further studies in pancreatic cancers similarly reached the conclusion that decreased PARP activity leads to better responses to genotoxic agents (Jacob, et al., 2007). Moreover, several clinical studies showed that hyperactivation of the PARP DNA repair pathway predicts therapeutic resistance in advanced tumors. For example, non-small cell lung carcinoma cells with high PARP-1 expression exhibit poor responses to cisplatin treatment (Michels, Vitale, Galluzzi, et al., 2013). In glioma, TMZ establishes N3-methyladnine and N7-methylguanine adducts to DNA, which are mainly repaired by the PARP/BER pathway. In glioblastoma-initiating cells, constitutive activity of PARP1 is essential for cancer resistance and progression (Sarkaria, et al., 2008; Venere, et al., 2014).
4. Combination treatment with PARP inhibitors and traditional chemotherapy
Upon understanding the critical role of PARP in cancer resistance approximately 3 decades ago, researchers began to develop specific PARP inhibitors. Nicotinamide, the first known PARP inhibitor, naturally exists in the cell (Rankin, Jacobson, Benjamin, Moss, & Jacobson, 1989). Nicotinamide establishes a core pharmacophore that anchors in the nicotinamide-binding pocket of PARP-1 (Ruf, de Murcia, & Schulz, 1998; Ruf, Mennissier de Murcia, de Murcia, & Schulz, 1996). The first synthetic PARP inhibitor, 3-AB, was designed to mimic the chemical structure of nicotinamide and showed promising synergistic effects with genotoxic agents (Chen & Zeller, 1992; Kehe, et al., 2008). However, the low potency (IC50=30 µM) and selectivity of 3-AB greatly limited its clinical application. Several generations of PARP inhibitors have been designed and tested, aiming to improve the chemical structure to facilitate better potency and selectivity.
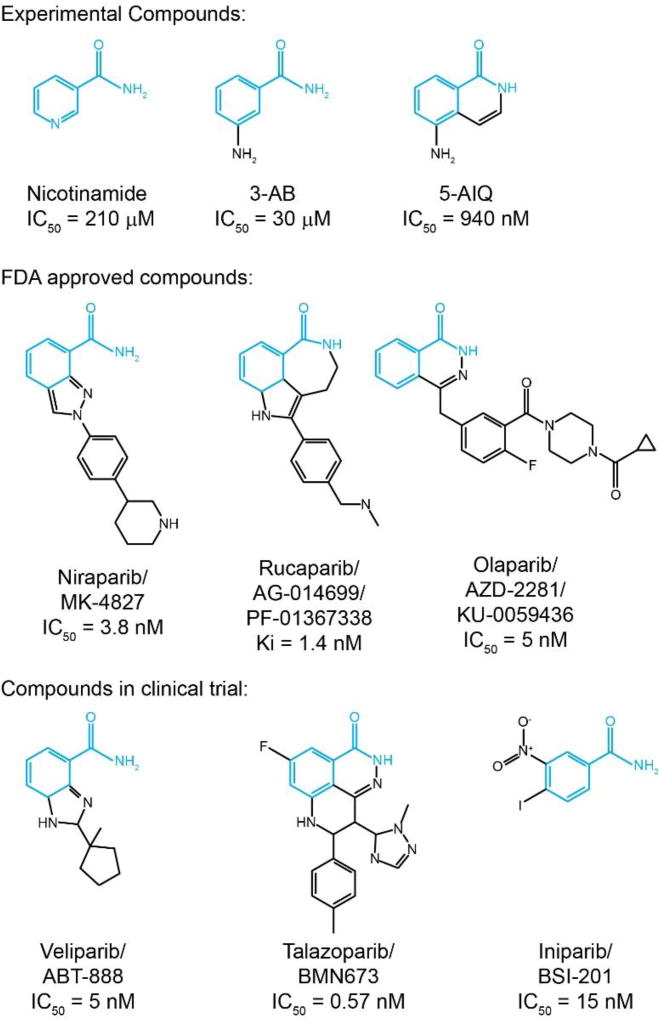
To date, the Food and Drug Administration has approved 3 PARP inhibitors for clinical application: niraparib, rucaparib, and olaparib. Additional candidates, such as veliparib, talazoparib, and iniparib, are currently being evaluated in preclinical studies and clinical trials (Figure 1).
Figure 1. The evolution of PARP inhibitors.
Schematic illustration for the chemical structures of PARP inhibitors. The nicotinamide pharmacophore is highlighted in blue.
Olaparib (AZD2281) is a potent PARP inhibitor, which contains a fluorine atom to improve its stability and potency. Olaparib was firstly tested in BRCA mutant cells as a single agent treatment, which successfully induce PARP inhibition and apoptotic changes (Farmer, et al., 2005). Several follow up studies examined the effect of a combination regimen including olaparib and traditional chemotherapeutic agents such as TMZ and platinum in BRCA-related and non-related cancer treatment. In Phase II study NCT00494442, olaparib was given as monotherapy at 400 mg twice daily does, which exhibitted antitumor activity in recurrent BRCA1 or BRCA2 mutated ovarian cancer. However, dose interruptions and reductions occurred due to adverse events caused by high doses of olaparib. (Audeh, et al., 2010; Yang, et al., 2011). In the metastatic prostate cancer that rarely respond to standard treatment, combination with olaparib synergized with intrinsic DNA-repair deficiency, resulting in high response rate to the treatment (Mateo, et al., 2015). With the outstanding efficacy and safety, olaparib has been included in more preclinical or clinical studies, including glioblastoma, melanoma, and Ewing's sarcoma (Brenner, et al., 2012; Krumm, et al., 2016; Ning, Wakimoto, Peters, Martuza, & Rabkin, 2017).
Rucaparib (AGO14699) is a potent, small molecule PARP inhibitor, which sensitizes cancer cells by suppressing phosphorylated signal transducer and transcription 3 activation (Ekblad, Camaioni, Schuler, & Macchiarulo, 2013). Compared with its lead compound AG14361, Rucaparib showed remarkably improved solubility and chemosensitization effect (Thomas, et al., 2007). Rucaparib was used as a stand-alone treatment for advanced ovarian and breast cancer patients with BRCA mutation. Strikingly, over 80% clinical cases responded completely/partially or stayed stably under rucaparib therapy (Drew, et al., 2016). Rucaparib shows stronger cytotoxicity than most of other PARP inhibitors, which may due to its effect to induce vasodilation and increase perfusion and drug aggregation in tumor tissue (Syed, 2017).
Niraparib (MK-4827) is a potent orally taken PARP1/2 inhibitor, which showed improved pharmacokinetic properties and selectivity on BRCA1/2 deficient cells (Jones, et al., 2009). Although niraparib exhibits modest suppression to PARP catalytic activity, it showed stronger PARP trapping effects, as compared to olaparib and veliparib (Murai, et al., 2012). Additional studies have demonstrated niraparib sensitized other solid tumors, such as ovarian cancer and colorectal cancer, to chemo- and radio-therapies (Genther Williams, et al., 2015; Kanjanapan, Lheureux, & Oza, 2017). However, in some patient-derived xenograft (PDX) models, niraparib failed to augment the effect of carboplatin/paclitaxel regimen in either homologous recombination (HR) deficient or proficient ovarian carcinoma (AlHilli, et al., 2016), indicating more assays are essential to guide the performance of PARP inhibitor, except HR status.
Besides the currently approved PARP inhibitor, many of the latest candidate compounds are currently tested in clinical trials. For examples, Veliparib (ABT-888) is a potent oral PARP1/2 inhibitor that has broad spectrum to sensitize tumor cells to radio-and chemotherapy (Donawho, et al., 2007). Despite comparable capability to suppress PARP catalytic activity, veliparib results in modest tumor suppressive effect as a single agent therapy, which was probably resulted from less effective in stabilizing PARP-DNA complex to impair DNA repair (Wagner, 2015). On the other hand, due to the superior blood-brain barrier permeability, veliparib was utilized in CNS tumors, such as glioblastoma as well (Su, et al., 2014).
Talazoparib (BMN 673) is another newly discovered PARP inhibitor that is effective to homologous recombination deficiency tumors (Murai, et al., 2014; Shen, et al., 2013). Murai et al., found that talazoparib had analogous effect on suppressing PARP catalytic activity, however, it showed 100-fold more cytotoxicity than olaparib and rucaparib, by trapping more PARP-DNA complexes (Murai, et al., 2014). Despite some cell lines showed resistant to talazoparib, more preclinical studies proved its tumor suppressive effects on Ewing’s sarcoma and chronic lymphocytic leukemia (Herriott, et al., 2015; Smith, et al., 2015).
Iniparib (BSI-201) is an irreversible PARP1 inhibitor with relatively smaller molecular weight (292.03 g/mol v.s. 435.08 g/mol for olaparib). As the first PARP inhibitor that reach the phase III clinical trial, iniparib was observed encouraging results in patients with solid tumors, including blioblastoma, non-small cell lung cancer and metastatic triple- negative breast cancer (Liang & Tan, 2010; Maxmen, 2010). Iniparib was originally considered as a non-competitive PARP inhibitor, however, it was later discovered as a modifier of cysteine-containing proteins, rather than a bona fide inhibitor of PARP (X. Liu, et al., 2012; Patel, De Lorenzo, Flatten, Poirier, & Kaufmann, 2012). However, a recent study showed that unlike other PARP inhibitors, BSI-201 showed minimal effect on enhancing DNA damage or pADPR formation, which raised the question the application of this compound (Chuang, Kapuriya, Kulp, Chen, & Shapiro, 2012). Moreover, a latest late stage clinical trial showed that BSI-201 did not meet predesignated criteria of primary endpoints of progression-free and overall survival, which highlight the requirement of a thorough understand in the mechanism of action in this PARP inhibitor (O'Shaughnessy, et al., 2014; O'Shaughnessy, et al., 2011).
Despite the original goal for use as a sensitizer, the first successful PARP inhibitor was used as a monotherapy. Certain cancers exhibit intrinsic deficiencies in DNA repair pathways that render them vulnerable to further DNA repair pathway suppression. Based on this concept, later known as “synthetic lethal therapy,” a PARP inhibitor was applied for the treatment of BRCA1/2-deficient ovarian and breast cancers. BRCA1/2-deficient cancers exhibit compromised HR DNA repair due to the loss of key repair enzymes. Application of a PARP inhibitor further diminishes DNA repair in those cancer cells, which effectively suppresses cancer cell proliferation, as the cells are likely to experience cell cycle arrest upon an overload of DNA damage (Ashworth, 2008).
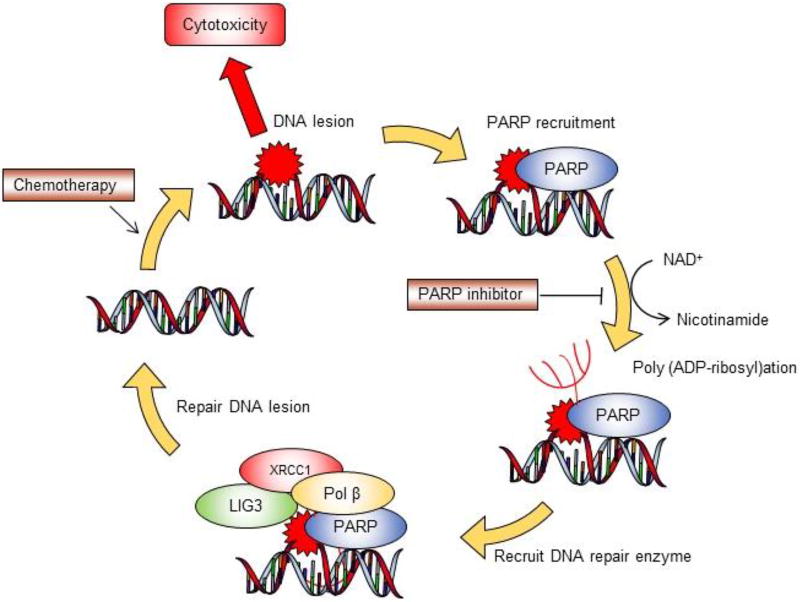
Combining PARP inhibitor with traditional chemotherapy has long been proposed in tissue culture studies as well as preclinical animal models. Mechanistically, PARP inhibition limits the capability of scavenging DNA lesions, and therefore leads to cytotoxicity and apoptotic changes (Figure 2). Fast proliferating cancer cells are more vulnerable to the combination regimen compared with normal cells, as frequent cell division ease translation from DNA lesions to cell death. However, the development of a PARP inhibitor combination therapy has been relatively slow, possibly due to the complicated nature of combination regimens, unexpected side effects, and difficulty establishing biologically appropriate dosages. In the following section, we will introduce the 2 most commonly used PARP inhibitor combination regimens used in recent clinical trials. We will also discuss the molecular mechanism of the synergy in these approaches.
Figure 2. PARP inhibitor sensitizes traditional chemotherapy.
Schematic illustration for the molecular mechanism of combination therapy involving PARP inhibitor and genotoxic agent. Traditional chemotherapy introduces DNA lesions and cytotoxicity to cancer cells. PARP plays an important role in forming poly (ADP-ribosyl)ation branches through consuming NAD+. DNA repair enzymes are recruited to the DNA lesion foci and remove adducts from nucleobases. Introducing PARP inhibitor compromises the scavenging of DNA adducts, resulting in more cell death with traditional chemotherapy.
TMZ/PARP inhibitor
TMZ is an oral DNA-alkylating agent that has been used in several types of cancer, most notably malignant gliomas. It was developed in the mid-1990s, through the modification on the chemical structure of mitozolomide and 5-(3-methyltriazen-1-yl)imidazole-4-carboxamide (MTIC) (Newlands, Stevens, Wedge, Wheelhouse, & Brock, 1997). TMZ is a monofunctional alkylating agent with good tissue distribution, including the ability to cross the blood-brain barrier. (O'Reilly, et al., 1993).
TMZ is extremely stable in solid form. In an aqueous environment, TMZ spontaneously undergoes hydrolysis into MTIC, which breaks down and releases reactive methyldiazonium ion. This eventually results in the formation of adducts to nucleobases in the form of N7-methylguanine (N7-MeG), O6-methylguanine (O6-MeG), and N3-methyladenine (N3-MeA). O6-MeG adducts can be reversed by MGMT, therefore MGMT-expressing tumors tend to be less responsive to genotoxic agents (Everhard, et al., 2006; Hegi, et al., 2005). Other types of DNA adducts are repaired through the PARP/BER DNA repair pathway. In fact, N7-MeG and N3-MeA comprise over 90% of DNA lesions that result from TMZ (Sarkaria, et al., 2008). However, these DNA adducts rarely translate into cytotoxicity and cancer suppression, presumably due to efficient detoxification through the PARP/BER DNA repair pathway. Pharmacologic inhibition of PARP has long been proposed as a rational strategy to sensitize cancer cells for TMZ treatment, considering the critical role of the PARP/BER pathway in counteracting the development of N7-MeG and N3-MeA lesions,
Several attempts have been made to evaluate the combined effects of TMZ and PARP inhibitors in cancer therapy. An early study showed that the PARP inhibitor 3-AB reverses the TMZ resistance of malignant glioma xenografts (C. L. Cheng, et al., 2005; Wedge, Porteous, & Newlands, 1996). In a preclinical model of primary central nervous system lymphoma, the combination of the PARP inhibitor NU1025 with TMZ improved overall survival and markedly reduced tumor growth in an animal model (Tentori, et al., 2002). The concept was later evaluated with more potent and specific PARP inhibitors. The combination of veliparib and TMZ has been evaluated in murine melanoma xenografts, and was found to delay tumor onset compared to monotherapy (Palma, et al., 2008). The combination of veliparib with radiation- or chemotherapy improved disease outcomes in glioblastoma-xenografted mice, whereas pre-exposure to TMZ ameliorated the sensitization effects of the PARP inhibitor (Clarke, et al., 2009). A recent finding showed that talazoparib synergizes with TMZ against Ewing sarcoma xenografts, implying a broader utility for a combination regimen in pediatric oncology (Smith, et al., 2015).
Given its success in preclinical models, the TMZ/PARP inhibitor combination regimen has been extensively evaluated in clinical trials. In the phase I/II trial NCT03212742, patients with unresectable, high-grade gliomas are receiving concomitant radiotherapy with a combination of olaparib and TMZ. In trial NCT01085422, veliparib has been used to sensitize patients with metastatic, castration-resistant prostate cancer to TMZ. The results indicated that combination regimen is well tolerated, with modest antitumor activity, decreasing circulating tumor cells, and tumor biomarker carcinoembryonic antigen (Hussain, et al., 2014). Lately, the therapeutic effect of veliparib and TMZ combination in the disease outcome of newly diagnosed glioblastoma was evaluated through a randomized Phase II/III study NCT02152982. The results indicated that the combination therapy significantly improved disease outcome in MGMT-hypermethylated GBM PDX models compared with TMZ alone. Interestingly, the combination was ineffective in MGMT-unmethylated lines, indicating MGMT promoter methylation status predicts the drug responding for TMZ/veliparib combination therapy (Gupta, et al., 2016). The veliparib/TMZ combination therapy is also being tested in metastatic breast cancer and BRCA1/2 breast cancer in the phase II study NCT01009788. In the phase II trial NCT01638546, the combination of veliparib with TMZ improved the response rate in relapsed/refractory small cell lung cancer patients, although an improvement in progression-free survival was not observed (Pietanza, et al., 2016).
Platinum/PARP inhibitor
Platinum compounds are established chemotherapies for a broad range of malignancies, such as testicular, ovarian, and breast cancers. Platinum compounds introduce prevalent intra-strand DNA adducts, leading to the collapse of replication forks and single- and double-strand DNA breaks. The major type of platinum-induced DNA lesions, intra-strand DNA crosslinks, are mainly removed via the PARP/BER DNA repair pathway (Burkle, Chen, Kupper, Grube, & Zeller, 1993; Zamble, Mu, Reardon, Sancar, & Lippard, 1996). Moreover, platinum resistance closely correlates with the activity of DNA repair pathways (Eastman, Schulte, Sheibani, & Sorenson, 1988). The combination of a PARP inhibitor with platinum therapy has been evaluated in preclinical studies for different types of cancers. An early study showed that the introduction of 3-AB overcame cisplatin resistance, resulting in enhanced cell cycle arrest and apoptosis (Nguewa, et al., 2006). Similarly, combination treatment with the PARP inhibitor PJ34 suppressed triple-negative breast cancer and hepatocellular carcinoma in vitro and in vivo (Hastak, Alli, & Ford, 2010; Huang, et al., 2008). Combination treatment with olaparib also improves the therapeutic effect of cisplatin in non-small cell lung cancer cells (H. Cheng, et al., 2013; Michels, Vitale, Senovilla, et al., 2013).
Based on the encouraging findings in preclinical models, the combination of a PARP inhibitor and platinum therapy has been evaluated in clinical trials. In the phase II study NCT01081951, patients with platinum-sensitive, recurrent, high-grade serous ovarian cancer underwent combination therapy with olaparib. This treatment improved progression-free survival from 9.6 months to 12.2 months compared with the paclitaxel and platinum agent alone. However, the benefit did not translate into improvements in overall survival, which might due to an imbalance in early censoring of the study (Oza, et al., 2012). A similar combination regimen was tested in the phase II/III trial NCT03150576, in which the combination of olaparib with paclitaxel and carboplatin was evaluated in 527 patients with triple-negative breast cancer and/or germline BRCA-positive breast cancers. Additionally, in the phase I/II trials NCT02855697 and NCT02489006, a combination regimen including olaparib and platinum-based chemotherapy is being evaluated in patients with progressive, high-grade serous/endometrioid ovarian cancer, or fallopian tumors. In a latest phase III study NCT02032277, six hundred and thirty-four triple-negative breast cancer patients was recruited to evaluate the sensitizing effect of PARP inhibitor veliparib in combination with carboplatin and paclitaxel, followed by doxorubicin and cyclophosphamide. The combination regimen improved the proportion of patients with triple-negative breast cancer who achieved a pathological complete response, although the progression free survival or overall survival was not conclusive in this study. The inclusion of veliparib in the treatment did not benefit for triple-negative breast cancer at the does used in this trial suggesting a higher does or using greater PARP trapping efficiency inhibitor such as olaparib and talazoparib might have been possible. (Loibl, et al., 2018).
5. Future directions for PARP inhibitor combination therapies
Explore the molecular basis of synergy
Combining PARP inhibitors with anti-cancer therapies is a promising strategy in many preclinical studies. Numerous clinical studies have been designed and executed to determine if adding PARP inhibitor leads to superior therapeutic effects over monotherapy. In addition to the well-accepted combination regimen involving a PARP inhibitor and genotoxic therapy, the application of PARP inhibitors has been expanded to combinations with non-genotoxic therapies. For example, in a study in patients with BRCA1-deficient cancer, the combination of ABT-888 with CTLA-4 blockade, an immune checkpoint therapy, resulted in immune-mediated tumor clearance and improved long-term survival (Higuchi, et al., 2015). This concept is currently being evaluated in the phase I/II clinical trial NCT02571725, in which the dosage and objective response rate will be analyzed upon treatment with a combination of olaparib and tremelimumab. Similarly, in the phase I/II study NCT02953457, the combination of olaparib, durvalumab, and tremelimumab will be evaluated for the treatment of patients with ovarian, fallopian tube, or peritoneal cancer. PARP inhibitors are also frequently combined with growth factor receptor inhibitors. For example, the phase II study NCT01116648 showed that combining olaparib with cediranib, a VEGFR inhibitor, improved progression-free survival in recurrent, platinum-sensitive, high-grade serous or endometrioid ovarian cancer (J. F. Liu, et al., 2014). The same combination regimen is currently being tested in the phase III study NCT03278717 and the clinical study NCT02681237 for ovarian cancer patients.
Overall, these novel combination regimens appear to improve disease outcome and suppress tumor expansion. However, rather than synergy, the improvements may result from the additive effects of combining different types of anti-tumor compounds. Each of the therapeutic agents may function according to its designated molecular mechanism, but minimally influence the other agents in the combination regimen. More research is required to explore the possible molecular mechanisms of synergy between the different types of anti-tumor compounds. Investigating drug synergy in preclinical models may provide important data to justify and guide future combinations, recognizing that additive effects are likely to provide limited benefit to patients. However, combination therapies increase toxicities such as myelosuppression, and fatigue, compounding the side effects associated with each of the therapeutic compounds.
Investigate baseline DNA repair mechanisms in different tumors
Understanding the intrinsic DNA repair activity in each type of tumor is another important consideration in PARP combination therapy. Expectedly, different types of tumors exhibit varied baseline DNA repair activity. However, even histologically identical tumors may have developed through completely different oncogenic routes, thereby imprinting them with a distinct spectrum of drug sensitivity and resistance mechanisms. For example, in BRCA1/2-deficient breast or ovarian cancers, the HR pathway deficiency creates a distinctive vulnerability to PARP inhibition (known as “BRCAness”) (Farmer, et al., 2005; McCabe, et al., 2006). In the case of PARP combination therapy, tumors with deficient DNA repair pathways may acquire “BRCAness”, and exhibit sensitivity to PARP combination therapy (Lord & Ashworth, 2016). Sporadic tumors may develop “BRCAness” through epigenetic silencing of key HR modules, such as members of the FANC-BRCA complex, and the establishment of sensitivity to PARP inhibition (Lyakhovich & Surralles, 2006; Marsit, et al., 2004; Turner, Tutt, & Ashworth, 2004). A recent finding showed that, in IDH1-mutated cancers, although no direct genetic abnormalities affect DNA repair pathways, the accumulated oncometabolite 2-hydroxyglutarate serves as an endogenous inhibitor of HR. The loss of functional HR enzymes establishes “BRCAness”, which predisposes tumors to sensitivity to PARP inhibition (Sulkowski, et al., 2017). Some anti-cancer therapies, such as bortezomib, may also induce “BRCAness” (Neri, et al., 2011). Exploring intrinsic deficiencies in DNA repair pathways may provide useful information to predict drug sensitivities and guide therapy strategies.
Several recent studies showed that alteration in intrinsic DNA repair pathway may shift the therapeutic effect of PARP inhibitors. For example, tumors with secondary mutation in BRCA2 acquire resistance to olaparib, which may due to the restoring homologous recombination through BRCA2 re-expression (Barber, et al., 2013). In mouse mammary tumor model, deficiency of 53BP1 was discovered to introduce resistance to PARP inhibitor (Jiao, et al., 2012), indicating more genetic and molecular changes involved in DNA damage/repair pathways should be considered when applies PARP inhibitor clinically.
Understanding the correlation between tumor-distinctive metabolism and PARP DNA repair
Metabolic reprogramming is a hallmark event during oncogenesis. The tumor-specific metabolic signature may influence DNA repair and therapy resistance. For example, hypoxic and acidic tumor microenvironments have lower DNA repair capacities (Bristow & Hill, 2008; Yuan, Narayanan, Rockwell, & Glazer, 2000). Therefore, the metabolic signature could be used to predict responses to PARP combination therapies. For PARP DNA repair, the catalytic activity of the enzyme relies on ATP and NAD+ to allow PAR formation and on DNA repair enzyme recruitment. Fluctuations in NAD+ metabolism is likely to influence the efficiency of PARP DNA repair by changing the availability of the key substrate NAD+ (Wang, et al., 2011). Several lines of evidence showed that, in the setting of compromised NAD+ metabolism, cancer cells exhibited extreme vulnerability to NAD+ depletion and PARP inhibitor treatment (Lu, et al., 2017; Tateishi, et al., 2015). The development of the NAMPT inhibitor FK866 confirmed the importance of the abundance of NAD+ in PARP-related chemoresistance. FK866 synergizes with intrinsic PARP deficiency or the presence of a PARP inhibitor, leading to a greater cytotoxic effect (Bajrami, et al., 2012; Tateishi, et al., 2015). Additional research into the relationship between cancer metabolism and DNA repair pathways, which will provide insight into possible combination regimens to improve upon traditional therapies, is warranted.
6. Conclusion and Future Directions
DNA repair pathways compromise the therapeutic effects of traditional anti-cancer therapies. The potential of DNA repair inhibitors in future of cancer therapy is becoming apparent. Since the first discovery of 3-AB around three decades ago, small molecular compound inhibitors targeting PARP/BER DNA repair pathway have become an effective sensitizing approach to genotoxic therapies, which lead to a large body of laboratory studies and clinical trials. Understanding the distinctive alterations in tumor biology, especially their intrinsic deficiency in DNA repair enzymes, as well as the signature metabolic pattern, would facilitate the development of PARP inhibitor combination regimen. With the expanding knowledge of drug synergy, we are poised for a rapid expansion of DNA repair inhibitors that move from based research to clinical application.
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, CCR, NCI. We appreciate the in-depth editing by Dr. Mark R. Gilbert.
Abbreviations
- PARP
poly (ADP-ribose) polymerase
- TMZ
temozolomide
- BER
base-excision repair
- MGMT
O-6-methylguanine-DNA methyltransferase
- NHEJ
non-homologous end joining
- HR
homologous recombination
- 3-AB
3-aminobenzamide
- MTIC
5-(3-methyltriazen-1-yl)imidazole-4-carboxamide
- N7-MeG
N7-methylguanine
- O6-MeG
O6-methylguanine
- N3-MeA
N3-methyladenine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement
The authors declare that there are no conflicts of interest.
References
- AlHilli MM, Becker MA, Weroha SJ, Flatten KS, Hurley RM, Harrell MI, Oberg AL, Maurer MJ, Hawthorne KM, Hou X, Harrington SC, McKinstry S, Meng XW, Wilcoxen KM, Kalli KR, Swisher EM, Kaufmann SH, Haluska P. In vivo anti-tumor activity of the PARP inhibitor niraparib in homologous recombination deficient and proficient ovarian carcinoma. Gynecol Oncol. 2016;143:379–388. doi: 10.1016/j.ygyno.2016.08.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ame JC, Spenlehauer C, de Murcia G. The PARP superfamily. Bioessays. 2004;26:882–893. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- Ashworth A. A synthetic lethal therapeutic approach: poly(ADP) ribose polymerase inhibitors for the treatment of cancers deficient in DNA double-strand break repair. J Clin Oncol. 2008;26:3785–3790. doi: 10.1200/JCO.2008.16.0812. [DOI] [PubMed] [Google Scholar]
- Audeh MW, Carmichael J, Penson RT, Friedlander M, Powell B, Bell-McGuinn KM, Scott C, Weitzel JN, Oaknin A, Loman N, Lu K, Schmutzler RK, Matulonis U, Wickens M, Tutt A. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. Lancet. 2010;376:245–251. doi: 10.1016/S0140-6736(10)60893-8. [DOI] [PubMed] [Google Scholar]
- Aziz K, Nowsheen S, Pantelias G, Iliakis G, Gorgoulis VG, Georgakilas AG. Targeting DNA damage and repair: embracing the pharmacological era for successful cancer therapy. Pharmacol Ther. 2012;133:334–350. doi: 10.1016/j.pharmthera.2011.11.010. [DOI] [PubMed] [Google Scholar]
- Bajrami I, Kigozi A, Van Weverwijk A, Brough R, Frankum J, Lord CJ, Ashworth A. Synthetic lethality of PARP and NAMPT inhibition in triple-negative breast cancer cells. EMBO Mol Med. 2012;4:1087–1096. doi: 10.1002/emmm.201201250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber LJ, Sandhu S, Chen L, Campbell J, Kozarewa I, Fenwick K, Assiotis I, Rodrigues DN, Reis Filho JS, Moreno V, Mateo J, Molife LR, De Bono J, Kaye S, Lord CJ, Ashworth A. Secondary mutations in BRCA2 associated with clinical resistance to a PARP inhibitor. J Pathol. 2013;229:422–429. doi: 10.1002/path.4140. [DOI] [PubMed] [Google Scholar]
- Bernges F, Zeller WJ. Combination effects of poly(ADP-ribose) polymerase inhibitors and DNA-damaging agents in ovarian tumor cell lines--with special reference to cisplatin. J Cancer Res Clin Oncol. 1996;122:665–670. doi: 10.1007/BF01209029. [DOI] [PubMed] [Google Scholar]
- Brenner JC, Feng FY, Han S, Patel S, Goyal SV, Bou-Maroun LM, Liu M, Lonigro R, Prensner JR, Tomlins SA, Chinnaiyan AM. PARP-1 inhibition as a targeted strategy to treat Ewing's sarcoma. Cancer Res. 2012;72:1608–1613. doi: 10.1158/0008-5472.CAN-11-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bristow RG, Hill RP. Hypoxia and metabolism. Hypoxia, DNA repair and genetic instability. Nat Rev Cancer. 2008;8:180–192. doi: 10.1038/nrc2344. [DOI] [PubMed] [Google Scholar]
- Burkle A, Chen G, Kupper JH, Grube K, Zeller WJ. Increased poly(ADP-ribosyl)ation in intact cells by cisplatin treatment. Carcinogenesis. 1993;14:559–561. doi: 10.1093/carcin/14.4.559. [DOI] [PubMed] [Google Scholar]
- Chambon P, Weill JD, Mandel P. Nicotinamide mononucleotide activation of new DNA-dependent polyadenylic acid synthesizing nuclear enzyme. Biochem Biophys Res Commun. 1963;11:39–43. doi: 10.1016/0006-291x(63)90024-x. [DOI] [PubMed] [Google Scholar]
- Chen G, Zeller WJ. Multiple effects of 3-aminobenzamide on DNA damage induced by cisplatin (DDP) in DDP-sensitive and -resistant rat ovarian tumor cell lines. Cancer Lett. 1992;67:27–33. doi: 10.1016/0304-3835(92)90005-g. [DOI] [PubMed] [Google Scholar]
- Cheng CL, Johnson SP, Keir ST, Quinn JA, Ali-Osman F, Szabo C, Li H, Salzman AL, Dolan ME, Modrich P, Bigner DD, Friedman HS. Poly(ADP-ribose) polymerase-1 inhibition reverses temozolomide resistance in a DNA mismatch repair-deficient malignant glioma xenograft. Mol Cancer Ther. 2005;4:1364–1368. doi: 10.1158/1535-7163.MCT-05-0128. [DOI] [PubMed] [Google Scholar]
- Cheng H, Zhang Z, Borczuk A, Powell CA, Balajee AS, Lieberman HB, Halmos B. PARP inhibition selectively increases sensitivity to cisplatin in ERCC1-low non-small cell lung cancer cells. Carcinogenesis. 2013;34:739–749. doi: 10.1093/carcin/bgs393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang HC, Kapuriya N, Kulp SK, Chen CS, Shapiro CL. Differential anti-proliferative activities of poly(ADP-ribose) polymerase (PARP) inhibitors in triple-negative breast cancer cells. Breast Cancer Res Treat. 2012;134:649–659. doi: 10.1007/s10549-012-2106-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke MJ, Mulligan EA, Grogan PT, Mladek AC, Carlson BL, Schroeder MA, Curtin NJ, Lou Z, Decker PA, Wu W, Plummer ER, Sarkaria JN. Effective sensitization of temozolomide by ABT-888 is lost with development of temozolomide resistance in glioblastoma xenograft lines. Mol Cancer Ther. 2009;8:407–414. doi: 10.1158/1535-7163.MCT-08-0854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crul M, van Waardenburg RC, Beijnen JH, Schellens JH. DNA-based drug interactions of cisplatin. Cancer Treat Rev. 2002;28:291–303. doi: 10.1016/s0305-7372(02)00093-2. [DOI] [PubMed] [Google Scholar]
- Dantzer F, de La Rubia G, Menissier-De Murcia J, Hostomsky Z, de Murcia G, Schreiber V. Base excision repair is impaired in mammalian cells lacking Poly(ADP-ribose) polymerase-1. Biochemistry. 2000;39:7559–7569. doi: 10.1021/bi0003442. [DOI] [PubMed] [Google Scholar]
- de Murcia JM, Niedergang C, Trucco C, Ricoul M, Dutrillaux B, Mark M, Oliver FJ, Masson M, Dierich A, LeMeur M, Walztinger C, Chambon P, de Murcia G. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc Natl Acad Sci U S A. 1997;94:7303–7307. doi: 10.1073/pnas.94.14.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donawho CK, Luo Y, Luo Y, Penning TD, Bauch JL, Bouska JJ, Bontcheva-Diaz VD, Cox BF, DeWeese TL, Dillehay LE, Ferguson DC, Ghoreishi-Haack NS, Grimm DR, Guan R, Han EK, Holley-Shanks RR, Hristov B, Idler KB, Jarvis K, Johnson EF, Kleinberg LR, Klinghofer V, Lasko LM, Liu X, Marsh KC, McGonigal TP, Meulbroek JA, Olson AM, Palma JP, Rodriguez LE, Shi Y, Stavropoulos JA, Tsurutani AC, Zhu GD, Rosenberg SH, Giranda VL, Frost DJ. ABT-888, an orally active poly(ADP-ribose) polymerase inhibitor that potentiates DNA-damaging agents in preclinical tumor models. Clin Cancer Res. 2007;13:2728–2737. doi: 10.1158/1078-0432.CCR-06-3039. [DOI] [PubMed] [Google Scholar]
- Drew Y, Ledermann J, Hall G, Rea D, Glasspool R, Highley M, Jayson G, Sludden J, Murray J, Jamieson D, Halford S, Acton G, Backholer Z, Mangano R, Boddy A, Curtin N, Plummer R. Phase 2 multicentre trial investigating intermittent and continuous dosing schedules of the poly(ADP-ribose) polymerase inhibitor rucaparib in germline BRCA mutation carriers with advanced ovarian and breast cancer. Br J Cancer. 2016;114:723–730. doi: 10.1038/bjc.2016.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastman A, Schulte N, Sheibani N, Sorenson C. Platinum and other metal coordination compounds in cancer chemotherapy. Springer; 1988. Mechanisms of resistance to platinum drugs; pp. 178–196. [Google Scholar]
- Ekblad T, Camaioni E, Schuler H, Macchiarulo A. PARP inhibitors: polypharmacology versus selective inhibition. FEBS J. 2013;280:3563–3575. doi: 10.1111/febs.12298. [DOI] [PubMed] [Google Scholar]
- El-Khamisy SF, Masutani M, Suzuki H, Caldecott KW. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 2003;31:5526–5533. doi: 10.1093/nar/gkg761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everhard S, Kaloshi G, Criniere E, Benouaich-Amiel A, Lejeune J, Marie Y, Sanson M, Kujas M, Mokhtari K, Hoang-Xuan K, Delattre JY, Thillet J. MGMT methylation: a marker of response to temozolomide in low-grade gliomas. Ann Neurol. 2006;60:740–743. doi: 10.1002/ana.21044. [DOI] [PubMed] [Google Scholar]
- Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, Martin NM, Jackson SP, Smith GC, Ashworth A. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- Gavande NS, VanderVere-Carozza PS, Hinshaw HD, Jalal SI, Sears CR, Pawelczak KS, Turchi JJ. DNA repair targeted therapy: The past or future of cancer treatment? Pharmacol Ther. 2016;160:65–83. doi: 10.1016/j.pharmthera.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genther Williams SM, Kuznicki AM, Andrade P, Dolinski BM, Elbi C, O'Hagan RC, Toniatti C. Treatment with the PARP inhibitor, niraparib, sensitizes colorectal cancer cell lines to irinotecan regardless of MSI/MSS status. Cancer Cell Int. 2015;15:14. doi: 10.1186/s12935-015-0162-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta SK, Kizilbash SH, Carlson BL, Mladek AC, Boakye-Agyeman F, Bakken KK, Pokorny JL, Schroeder MA, Decker PA, Cen L, Eckel-Passow JE, Sarkar G, Ballman KV, Reid JM, Jenkins RB, Verhaak RG, Sulman EP, Kitange GJ, Sarkaria JN. Delineation of MGMT Hypermethylation as a Biomarker for Veliparib-Mediated Temozolomide-Sensitizing Therapy of Glioblastoma. J Natl Cancer Inst. 2016:108. doi: 10.1093/jnci/djv369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastak K, Alli E, Ford JM. Synergistic chemosensitivity of triple-negative breast cancer cell lines to poly(ADP-Ribose) polymerase inhibition, gemcitabine, and cisplatin. Cancer Res. 2010;70:7970–7980. doi: 10.1158/0008-5472.CAN-09-4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani L, Bromberg JE, Hau P, Mirimanoff RO, Cairncross JG, Janzer RC, Stupp R. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer. 2008;8:193–204. doi: 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
- Herriott A, Tudhope SJ, Junge G, Rodrigues N, Patterson MJ, Woodhouse L, Lunec J, Hunter JE, Mulligan EA, Cole M, Allinson LM, Wallis JP, Marshall S, Wang E, Curtin NJ, Willmore E. PARP1 expression, activity and ex vivo sensitivity to the PARP inhibitor, talazoparib (BMN 673), in chronic lymphocytic leukaemia. Oncotarget. 2015;6:43978–43991. doi: 10.18632/oncotarget.6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi T, Flies DB, Marjon NA, Mantia-Smaldone G, Ronner L, Gimotty PA, Adams SF. CTLA-4 Blockade Synergizes Therapeutically with PARP Inhibition in BRCA1-Deficient Ovarian Cancer. Cancer Immunol Res. 2015;3:1257–1268. doi: 10.1158/2326-6066.CIR-15-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SH, Xiong M, Chen XP, Xiao ZY, Zhao YF, Huang ZY. PJ34, an inhibitor of PARP-1, suppresses cell growth and enhances the suppressive effects of cisplatin in liver cancer cells. Oncol Rep. 2008;20:567–572. [PubMed] [Google Scholar]
- Hussain M, Carducci MA, Slovin S, Cetnar J, Qian J, McKeegan EM, Refici-Buhr M, Chyla B, Shepherd SP, Giranda VL, Alumkal JJ. Targeting DNA repair with combination veliparib (ABT-888) and temozolomide in patients with metastatic castration-resistant prostate cancer. Invest New Drugs. 2014;32:904–912. doi: 10.1007/s10637-014-0099-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob DA, Bahra M, Langrehr JM, Boas-Knoop S, Stefaniak R, Davis J, Schumacher G, Lippert S, Neumann UP. Combination therapy of poly (ADP-ribose) polymerase inhibitor 3-aminobenzamide and gemcitabine shows strong antitumor activity in pancreatic cancer cells. J Gastroenterol Hepatol. 2007;22:738–748. doi: 10.1111/j.1440-1746.2006.04496.x. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Killela PJ, Reitman ZJ, Rasheed AB, Heaphy CM, de Wilde RF, Rodriguez FJ, Rosemberg S, Oba-Shinjo SM, Nagahashi Marie SK, Bettegowda C, Agrawal N, Lipp E, Pirozzi C, Lopez G, He Y, Friedman H, Friedman AH, Riggins GJ, Holdhoff M, Burger P, McLendon R, Bigner DD, Vogelstein B, Meeker AK, Kinzler KW, Papadopoulos N, Diaz LA, Yan H. Frequent ATRX, CIC, FUBP1 and IDH1 mutations refine the classification of malignant gliomas. Oncotarget. 2012;3:709–722. doi: 10.18632/oncotarget.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P, Altamura S, Boueres J, Ferrigno F, Fonsi M, Giomini C, Lamartina S, Monteagudo E, Ontoria JM, Orsale MV, Palumbi MC, Pesci S, Roscilli G, Scarpelli R, Schultz-Fademrecht C, Toniatti C, Rowley M. Discovery of 2-{4-[(3S)-piperidin-3-yl]phenyl}-2H–indazole-7-carboxamide (MK-4827): a novel oral poly(ADP-ribose)polymerase (PARP) inhibitor efficacious in BRCA-1 and -2 mutant tumors. J Med Chem. 2009;52:7170–7185. doi: 10.1021/jm901188v. [DOI] [PubMed] [Google Scholar]
- Kanjanapan Y, Lheureux S, Oza AM. Niraparib for the treatment of ovarian cancer. Expert Opin Pharmacother. 2017;18:631–640. doi: 10.1080/14656566.2017.1297423. [DOI] [PubMed] [Google Scholar]
- Kehe K, Raithel K, Kreppel H, Jochum M, Worek F, Thiermann H. Inhibition of poly(ADP-ribose) polymerase (PARP) influences the mode of sulfur mustard (SM)-induced cell death in HaCaT cells. Arch Toxicol. 2008;82:461–470. doi: 10.1007/s00204-007-0265-7. [DOI] [PubMed] [Google Scholar]
- Kondo N, Takahashi A, Ono K, Ohnishi T. DNA damage induced by alkylating agents and repair pathways. J Nucleic Acids. 2010;2010:543531. doi: 10.4061/2010/543531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus WL. PARPs and ADP-Ribosylation: 50 Years … and Counting. Mol Cell. 2015;58:902–910. doi: 10.1016/j.molcel.2015.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumm A, Barckhausen C, Kucuk P, Tomaszowski KH, Loquai C, Fahrer J, Kramer OH, Kaina B, Roos WP. Enhanced Histone Deacetylase Activity in Malignant Melanoma Provokes RAD51 and FANCD2-Triggered Drug Resistance. Cancer Res. 2016;76:3067–3077. doi: 10.1158/0008-5472.CAN-15-2680. [DOI] [PubMed] [Google Scholar]
- Liang H, Tan AR. Iniparib, a PARP1 inhibitor for the potential treatment of cancer, including triple-negative breast cancer. IDrugs. 2010;13:646–656. [PubMed] [Google Scholar]
- Liu JF, Barry WT, Birrer M, Lee JM, Buckanovich RJ, Fleming GF, Rimel B, Buss MK, Nattam S, Hurteau J, Luo W, Quy P, Whalen C, Obermayer L, Lee H, Winer EP, Kohn EC, Ivy SP, Matulonis UA. Combination cediranib and olaparib versus olaparib alone for women with recurrent platinum-sensitive ovarian cancer: a randomised phase 2 study. Lancet Oncol. 2014;15:1207–1214. doi: 10.1016/S1470-2045(14)70391-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Shi Y, Maag DX, Palma JP, Patterson MJ, Ellis PA, Surber BW, Ready DB, Soni NB, Ladror US, Xu AJ, Iyer R, Harlan JE, Solomon LR, Donawho CK, Penning TD, Johnson EF, Shoemaker AR. Iniparib nonselectively modifies cysteine-containing proteins in tumor cells and is not a bona fide PARP inhibitor. Clin Cancer Res. 2012;18:510–523. doi: 10.1158/1078-0432.CCR-11-1973. [DOI] [PubMed] [Google Scholar]
- Loibl S, O'Shaughnessy J, Untch M, Sikov WM, Rugo HS, McKee MD, Huober J, Golshan M, von Minckwitz G, Maag D, Sullivan D, Wolmark N, McIntyre K, Ponce Lorenzo JJ, Metzger Filho O, Rastogi P, Symmans WF, Liu X, Geyer CE., Jr Addition of the PARP inhibitor veliparib plus carboplatin or carboplatin alone to standard neoadjuvant chemotherapy in triple-negative breast cancer (BrighTNess): a randomised, phase 3 trial. Lancet Oncol. 2018 doi: 10.1016/S1470-2045(18)30111-6. [DOI] [PubMed] [Google Scholar]
- Lord CJ, Ashworth A. BRCAness revisited. Nat Rev Cancer. 2016;16:110–120. doi: 10.1038/nrc.2015.21. [DOI] [PubMed] [Google Scholar]
- Lu Y, Kwintkiewicz J, Liu Y, Tech K, Frady LN, Su YT, Bautista W, Moon SI, MacDonald J, Ewend MG, Gilbert MR, Yang C, Wu J. Chemosensitivity of IDH1-Mutated Gliomas Due to an Impairment in PARP1-Mediated DNA Repair. Cancer Res. 2017;77:1709–1718. doi: 10.1158/0008-5472.CAN-16-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyakhovich A, Surralles J. Disruption of the Fanconi anemia/BRCA pathway in sporadic cancer. Cancer Lett. 2006;232:99–106. doi: 10.1016/j.canlet.2005.07.038. [DOI] [PubMed] [Google Scholar]
- Marsit CJ, Liu M, Nelson HH, Posner M, Suzuki M, Kelsey KT. Inactivation of the Fanconi anemia/BRCA pathway in lung and oral cancers: implications for treatment and survival. Oncogene. 2004;23:1000–1004. doi: 10.1038/sj.onc.1207256. [DOI] [PubMed] [Google Scholar]
- Masson M, Niedergang C, Schreiber V, Muller S, Menissier-de Murcia J, de Murcia G. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol Cell Biol. 1998;18:3563–3571. doi: 10.1128/mcb.18.6.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateo J, Carreira S, Sandhu S, Miranda S, Mossop H, Perez-Lopez R, Nava Rodrigues D, Robinson D, Omlin A, Tunariu N, Boysen G, Porta N, Flohr P, Gillman A, Figueiredo I, Paulding C, Seed G, Jain S, Ralph C, Protheroe A, Hussain S, Jones R, Elliott T, McGovern U, Bianchini D, Goodall J, Zafeiriou Z, Williamson CT, Ferraldeschi R, Riisnaes R, Ebbs B, Fowler G, Roda D, Yuan W, Wu YM, Cao X, Brough R, Pemberton H, A'Hern R, Swain A, Kunju LP, Eeles R, Attard G, Lord CJ, Ashworth A, Rubin MA, Knudsen KE, Feng FY, Chinnaiyan AM, Hall E, de Bono JS. DNA-Repair Defects and Olaparib in Metastatic Prostate Cancer. N Engl J Med. 2015;373:1697–1708. doi: 10.1056/NEJMoa1506859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxmen A. Beyond PARP inhibitors: agents in pipelines target DNA repair mechanisms. J Natl Cancer Inst. 2010;102:1110–1111. doi: 10.1093/jnci/djq294. [DOI] [PubMed] [Google Scholar]
- McCabe N, Turner NC, Lord CJ, Kluzek K, Bialkowska A, Swift S, Giavara S, O'Connor MJ, Tutt AN, Zdzienicka MZ, Smith GC, Ashworth A. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res. 2006;66:8109–8115. doi: 10.1158/0008-5472.CAN-06-0140. [DOI] [PubMed] [Google Scholar]
- Michels J, Vitale I, Galluzzi L, Adam J, Olaussen KA, Kepp O, Senovilla L, Talhaoui I, Guegan J, Enot DP, Talbot M, Robin A, Girard P, Orear C, Lissa D, Sukkurwala AQ, Garcia P, Behnam-Motlagh P, Kohno K, Wu GS, Brenner C, Dessen P, Saparbaev M, Soria JC, Castedo M, Kroemer G. Cisplatin resistance associated with PARP hyperactivation. Cancer Res. 2013;73:2271–2280. doi: 10.1158/0008-5472.CAN-12-3000. [DOI] [PubMed] [Google Scholar]
- Michels J, Vitale I, Senovilla L, Enot DP, Garcia P, Lissa D, Olaussen KA, Brenner C, Soria JC, Castedo M, Kroemer G. Synergistic interaction between cisplatin and PARP inhibitors in non-small cell lung cancer. Cell Cycle. 2013;12:877–883. doi: 10.4161/cc.24034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murai J, Huang SY, Das BB, Renaud A, Zhang Y, Doroshow JH, Ji J, Takeda S, Pommier Y. Trapping of PARP1 and PARP2 by Clinical PARP Inhibitors. Cancer Res. 2012;72:5588–5599. doi: 10.1158/0008-5472.CAN-12-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murai J, Huang SY, Renaud A, Zhang Y, Ji J, Takeda S, Morris J, Teicher B, Doroshow JH, Pommier Y. Stereospecific PARP trapping by BMN 673 and comparison with olaparib and rucaparib. Mol Cancer Ther. 2014;13:433–443. doi: 10.1158/1535-7163.MCT-13-0803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neri P, Ren L, Gratton K, Stebner E, Johnson J, Klimowicz A, Duggan P, Tassone P, Mansoor A, Stewart DA, Lonial S, Boise LH, Bahlis NJ. Bortezomib-induced "BRCAness" sensitizes multiple myeloma cells to PARP inhibitors. Blood. 2011;118:6368–6379. doi: 10.1182/blood-2011-06-363911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newlands ES, Stevens MF, Wedge SR, Wheelhouse RT, Brock C. Temozolomide: a review of its discovery, chemical properties, pre-clinical development and clinical trials. Cancer Treat Rev. 1997;23:35–61. doi: 10.1016/s0305-7372(97)90019-0. [DOI] [PubMed] [Google Scholar]
- Nguewa PA, Fuertes MA, Cepeda V, Alonso C, Quevedo C, Soto M, Perez JM. Poly(ADP-ribose) polymerase-1 inhibitor 3-aminobenzamide enhances apoptosis induction by platinum complexes in cisplatin-resistant tumor cells. Med Chem. 2006;2:47–53. doi: 10.2174/157340606775197697. [DOI] [PubMed] [Google Scholar]
- Ning J, Wakimoto H, Peters C, Martuza RL, Rabkin SD. Rad51 Degradation: Role in Oncolytic Virus-Poly(ADP-Ribose) Polymerase Inhibitor Combination Therapy in Glioblastoma. J Natl Cancer Inst. 2017;109:1–13. doi: 10.1093/jnci/djw229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Reilly SM, Newlands ES, Glaser MG, Brampton M, Rice-Edwards JM, Illingworth RD, Richards PG, Kennard C, Colquhoun IR, Lewis P, et al. Temozolomide: a new oral cytotoxic chemotherapeutic agent with promising activity against primary brain tumours. Eur J Cancer. 1993;29A:940–942. doi: 10.1016/s0959-8049(05)80198-4. [DOI] [PubMed] [Google Scholar]
- O'Shaughnessy J, Schwartzberg L, Danso MA, Miller KD, Rugo HS, Neubauer M, Robert N, Hellerstedt B, Saleh M, Richards P, Specht JM, Yardley DA, Carlson RW, Finn RS, Charpentier E, Garcia-Ribas I, Winer EP. Phase III study of iniparib plus gemcitabine and carboplatin versus gemcitabine and carboplatin in patients with metastatic triple-negative breast cancer. J Clin Oncol. 2014;32:3840–3847. doi: 10.1200/JCO.2014.55.2984. [DOI] [PubMed] [Google Scholar]
- O'Shaughnessy J, Schwartzberg LS, Danso MA, Rugo HS, Miller K, Yardley DA, Carlson RW, Finn RS, Charpentier E, Freese M, Gupta S, Blackwood-Chirchir A, Winer EP. A randomized phase III study of iniparib (BSI-201) in combination with gemcitabine/carboplatin (G/C) in metastatic triple-negative breast cancer (TNBC) Journal of Clinical Oncology. 2011;29:1007–1007. [Google Scholar]
- Oza AM, Cibula D, Oaknin A, Poole CJ, Mathijssen RHJ, Sonke GS, Colombo N, Špacek J, Vuylsteke P, Hirte HW, Mahner S, Plante M, Schmalfeldt B, Mackay H, Rowbottom J, Tchakov I, Friedlander M. Olaparib plus paclitaxel plus carboplatin (P/C) followed by olaparib maintenance treatment in patients (pts) with platinum-sensitive recurrent serous ovarian cancer (PSR SOC): A randomized, open-label phase II study. Journal of Clinical Oncology. 2012;30:5001–5001. [Google Scholar]
- Palma JP, Rodriguez LE, Bontcheva-Diaz VD, Bouska JJ, Bukofzer G, Colon-Lopez M, Guan R, Jarvis K, Johnson EF, Klinghofer V, Liu X, Olson A, Saltarelli MJ, Shi Y, Stavropoulos JA, Zhu GD, Penning TD, Luo Y, Giranda VL, Rosenberg SH, Frost DJ, Donawho CK. The PARP inhibitor, ABT-888 potentiates temozolomide: correlation with drug levels and reduction in PARP activity in vivo. Anticancer Res. 2008;28:2625–2635. [PubMed] [Google Scholar]
- Patel AG, De Lorenzo SB, Flatten KS, Poirier GG, Kaufmann SH. Failure of iniparib to inhibit poly(ADP-Ribose) polymerase in vitro. Clin Cancer Res. 2012;18:1655–1662. doi: 10.1158/1078-0432.CCR-11-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietanza MC, Krug LM, Waqar SN, Dowlati A, Hann CL, Chiappori A, Owonikoko TK, Woo K, Bensman Y, Hurtado B, Fujimoto J, Wistuba II, Travis WD, Chen AP, Heymach J, Kris MG, Fleisher M, Rudin CM, Byers LA. A multi-center, randomized, double-blind phase II study comparing temozolomide (TMZ) plus either veliparib (ABT-888), a PARP inhibitor, or placebo as 2nd or 3rd-line therapy for patients (Pts) with relapsed small cell lung cancers (SCLCs) Journal of Clinical Oncology. 2016;34:8512–8512. [Google Scholar]
- Rankin PW, Jacobson EL, Benjamin RC, Moss J, Jacobson MK. Quantitative studies of inhibitors of ADP-ribosylation in vitro and in vivo. J Biol Chem. 1989;264:4312–4317. [PubMed] [Google Scholar]
- Ruf A, de Murcia G, Schulz GE. Inhibitor and NAD+ binding to poly(ADP-ribose) polymerase as derived from crystal structures and homology modeling. Biochemistry. 1998;37:3893–3900. doi: 10.1021/bi972383s. [DOI] [PubMed] [Google Scholar]
- Ruf A, Mennissier de Murcia J, de Murcia G, Schulz GE. Structure of the catalytic fragment of poly(AD-ribose) polymerase from chicken. Proc Natl Acad Sci U S A. 1996;93:7481–7485. doi: 10.1073/pnas.93.15.7481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkaria JN, Kitange GJ, James CD, Plummer R, Calvert H, Weller M, Wick W. Mechanisms of chemoresistance to alkylating agents in malignant glioma. Clin Cancer Res. 2008;14:2900–2908. doi: 10.1158/1078-0432.CCR-07-1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber V, Ame JC, Dolle P, Schultz I, Rinaldi B, Fraulob V, Menissier-de Murcia J, de Murcia G. Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J Biol Chem. 2002;277:23028–23036. doi: 10.1074/jbc.M202390200. [DOI] [PubMed] [Google Scholar]
- Shen Y, Rehman FL, Feng Y, Boshuizen J, Bajrami I, Elliott R, Wang B, Lord CJ, Post LE, Ashworth A. BMN 673, a novel and highly potent PARP1/2 inhibitor for the treatment of human cancers with DNA repair deficiency. Clin Cancer Res. 2013;19:5003–5015. doi: 10.1158/1078-0432.CCR-13-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MA, Reynolds CP, Kang MH, Kolb EA, Gorlick R, Carol H, Lock RB, Keir ST, Maris JM, Billups CA, Lyalin D, Kurmasheva RT, Houghton PJ. Synergistic activity of PARP inhibition by talazoparib (BMN 673) with temozolomide in pediatric cancer models in the pediatric preclinical testing program. Clin Cancer Res. 2015;21:819–832. doi: 10.1158/1078-0432.CCR-14-2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su JM, Thompson P, Adesina A, Li XN, Kilburn L, Onar-Thomas A, Kocak M, Chyla B, McKeegan E, Warren KE, Goldman S, Pollack IF, Fouladi M, Chen A, Giranda V, Boyett J, Kun L, Blaney SM. A phase I trial of veliparib (ABT-888) and temozolomide in children with recurrent CNS tumors: a pediatric brain tumor consortium report. Neuro Oncol. 2014;16:1661–1668. doi: 10.1093/neuonc/nou103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulkowski PL, Corso CD, Robinson ND, Scanlon SE, Purshouse KR, Bai H, Liu Y, Sundaram RK, Hegan DC, Fons NR, Breuer GA, Song Y, Mishra-Gorur K, De Feyter HM, de Graaf RA, Surovtseva YV, Kachman M, Halene S, Gunel M, Glazer PM, Bindra RS. 2-Hydroxyglutarate produced by neomorphic IDH mutations suppresses homologous recombination and induces PARP inhibitor sensitivity. Sci Transl Med. 2017:9. doi: 10.1126/scitranslmed.aal2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed YY. Rucaparib: First Global Approval. Drugs. 2017;77:585–592. doi: 10.1007/s40265-017-0716-2. [DOI] [PubMed] [Google Scholar]
- Tateishi K, Wakimoto H, Iafrate AJ, Tanaka S, Loebel F, Lelic N, Wiederschain D, Bedel O, Deng G, Zhang B, He T, Shi X, Gerszten RE, Zhang Y, Yeh JR, Curry WT, Zhao D, Sundaram S, Nigim F, Koerner MV, Ho Q, Fisher DE, Roider EM, Kemeny LV, Samuels Y, Flaherty KT, Batchelor TT, Chi AS, Cahill DP. Extreme Vulnerability of IDH1 Mutant Cancers to NAD+ Depletion. Cancer Cell. 2015;28:773–784. doi: 10.1016/j.ccell.2015.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tentori L, Leonetti C, Scarsella M, d'Amati G, Portarena I, Zupi G, Bonmassar E, Graziani G. Combined treatment with temozolomide and poly(ADP-ribose) polymerase inhibitor enhances survival of mice bearing hematologic malignancy at the central nervous system site. Blood. 2002;99:2241–2244. doi: 10.1182/blood.v99.6.2241. [DOI] [PubMed] [Google Scholar]
- Thomas HD, Calabrese CR, Batey MA, Canan S, Hostomsky Z, Kyle S, Maegley KA, Newell DR, Skalitzky D, Wang LZ, Webber SE, Curtin NJ. Preclinical selection of a novel poly(ADP-ribose) polymerase inhibitor for clinical trial. Mol Cancer Ther. 2007;6:945–956. doi: 10.1158/1535-7163.MCT-06-0552. [DOI] [PubMed] [Google Scholar]
- Trucco C, Oliver FJ, de Murcia G, Menissier-de Murcia J. DNA repair defect in poly(ADP-ribose) polymerase-deficient cell lines. Nucleic Acids Res. 1998;26:2644–2649. doi: 10.1093/nar/26.11.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner N, Tutt A, Ashworth A. Hallmarks of 'BRCAness' in sporadic cancers. Nat Rev Cancer. 2004;4:814–819. doi: 10.1038/nrc1457. [DOI] [PubMed] [Google Scholar]
- Venere M, Hamerlik P, Wu Q, Rasmussen RD, Song LA, Vasanji A, Tenley N, Flavahan WA, Hjelmeland AB, Bartek J, Rich JN. Therapeutic targeting of constitutive PARP activation compromises stem cell phenotype and survival of glioblastoma-initiating cells. Cell Death Differ. 2014;21:258–269. doi: 10.1038/cdd.2013.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner LM. Profile of veliparib and its potential in the treatment of solid tumors. Onco Targets Ther. 2015;8:1931–1939. doi: 10.2147/OTT.S69935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Hasan MK, Alvarado E, Yuan H, Wu H, Chen WY. NAMPT overexpression in prostate cancer and its contribution to tumor cell survival and stress response. Oncogene. 2011;30:907–921. doi: 10.1038/onc.2010.468. [DOI] [PubMed] [Google Scholar]
- Wedge SR, Porteous JK, Newlands ES. 3-aminobenzamide and/or O6-benzylguanine evaluated as an adjuvant to temozolomide or BCNU treatment in cell lines of variable mismatch repair status and O6-alkylguanine-DNA alkyltransferase activity. Br J Cancer. 1996;74:1030–1036. doi: 10.1038/bjc.1996.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Khan S, Sun Y, Hess K, Shmulevich I, Sood AK, Zhang W. Association of BRCA1 and BRCA2 mutations with survival, chemotherapy sensitivity, and gene mutator phenotype in patients with ovarian cancer. JAMA. 2011;306:1557–1565. doi: 10.1001/jama.2011.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Narayanan L, Rockwell S, Glazer PM. Diminished DNA repair and elevated mutagenesis in mammalian cells exposed to hypoxia and low pH. Cancer Res. 2000;60:4372–4376. [PubMed] [Google Scholar]
- Zamble DB, Mu D, Reardon JT, Sancar A, Lippard SJ. Repair of cisplatin--DNA adducts by the mammalian excision nuclease. Biochemistry. 1996;35:10004–10013. doi: 10.1021/bi960453+. [DOI] [PubMed] [Google Scholar]