Abstract
Protein-protein interactions (PPIs) that are part of the costimulatory and coinhibitory (immune checkpoint) signaling are critical for adequate T cell response and are important therapeutic targets for immunomodulation. Biologics targeting them have already achieved considerable clinical success in the treatment of autoimmune diseases or transplant recipients (e.g., abatacept, belatacept, and belimumab) as well as cancer (e.g., ipilimumab, nivolumab, pembrolizumab, atezolizumab, durvalumab, and avelumab). In view of such progress, there have been only relatively limited efforts toward developing small-molecule PPI inhibitors (SMPPIIs) targeting these cosignaling interactions, possibly because they, as all other PPIs, are difficult to target by small molecules and were not considered druggable. Nevertheless, substantial progress has been achieved during the last decade. SMPPIIs proving the feasibility of such approaches have been identified through various strategies for a number of cosignaling interactions including CD40-CD40L, OX40-OX40L, BAFFR-BAFF, CD80-CD28, and PD-1-PD-L1s. Here, after an overview of the general aspects and challenges of SMPPII-focused drug discovery, we review them briefly together with relevant structural, immune-signaling, physicochemical, and medicinal chemistry aspects. While so far only a few of these SMPPIIs have shown activity in animal models (DRI-C21045 for CD40-D40L, KR33426 for BAFFR-BAFF) or reached clinical development (RhuDex for CD80-CD28, CA-170 for PD-1-PD-L1), there is proof-of-principle evidence for the feasibility of such approaches in immunomodulation. They can result in products that are easier to develop/manufacture and are less likely to be immunogenic or encounter postmarket safety events than corresponding biologics, and, contrary to them, can even become orally bioavailable.
Keywords: costimulation, druggability, immune checkpoint, immunomodulation, ligand efficiency, molecular size, protein-protein interaction
1. INTRODUCTION
1.1. Challenges Faced by the Current Drug Discovery and Development Paradigm
During the past century, drug discovery and development has underpinned almost all of the clinically significant progress that occurred within the medical field. Nevertheless, an essentially stagnant number of about 20 new yearly drug approvals in the United States, despite steeply increasing research and development (R&D) costs, demonstrates a pressing need for innovation [1–6]. One of the most striking observations along these lines is that the number of new FDA-approved drugs per each (inflation-adjusted) $1 billion of R&D spending in the drug industry has been consistently decreasing being halved approximately every nine years since 1950 following a sort of reverse Moore’s law [5]. The original “Moore’s law”, formulated in the 1960s by Intel cofounder Gordon Moore [7] and having held true for more than 50 years since, states that the number of transistors that can be integrated into a microchip, and thus processor speed, doubles about every 1.5 years. There are multiple possible reasons for this decrease in drug discovery and development efficiency, including an increasing regulatory burden, an unrealistic public expectation of no side effects, and many others [5, 8, 9]; here, we will highlight briefly the depletion of effective new targets for traditional drug design.
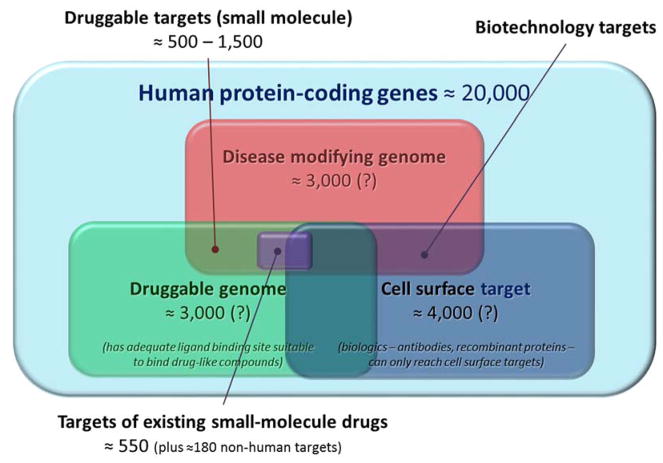
It is estimated that existing small-molecule drugs target only about 1% of the human proteome, which, following the completion of the human genome project, turned out to contain a rather low number of unique proteins (Figure 1). The number of protein-coding genes was originally estimated to be around 25,000 [10], but later lowered to only about 20,000 [11, 12]. Detailed distribution maps of the human proteome are now available [13, 14]. In the meantime, analyses of extensive collections of protein structures available in the Protein Data Bank (PDB) suggest that only about 10% are “druggable”, in the sense that they possess protein folds suitable for interactions with drug-like chemical compounds (i.e., possible ligand binding sites). This suggests only about 3,000 druggable human protein targets [15–17]. The human “pocketome” is a collection of curated co-crystallized binding sites, where each entry describes a druggable site on the protein known to interact with a peptide or a small molecule [18]. However, not all druggable proteins represent potential drug targets, since only ~10% of all genes seem to modify diseases. Only at the intersection of these two subsets (“druggable” and “disease modifying”) do true potential drug targets exist, with an estimated 500 to 1,500 members (Figure 1) [15, 16]. Along these lines, a 2012 review identified 364 successful drug targets, with an additional 286 clinical trial drug targets and 1331 research targets [19]. A more recent 2017 review compiled around 550 human protein targets for small-molecule drugs (up to a total of 670 if including biologics) plus another ~180 non-human proteins [20]. Biologics such as antibodies have an advantage in that they can interact potently with proteins along a broader surface spectrum and a variety of epitopes, not just druggable pockets. Thus, they have been increasingly pursued over the last decades. However, biologics usually cannot cross membranes to reach intracellular targets, leaving only the estimated <10% of all human proteins that are secreted or resident on the cell surface as feasible targets (Figure 1) [15, 17].
Figure 1.
According to current estimates, druggable disease-modifying targets represent only a relatively small subset of the total human protein targets and existing drugs target only a fraction of them. Data estimates are from references [9, 12, 15, 16, 20]; see text for details.
1.2. Protein-Protein Interactions as Drug Targets
Protein-protein interactions (PPIs) represent a large number of promising targets. This became quite evident as the advent of modern biotechnology methods allowed biologics to enter the scene. However, PPIs are difficult to modulate with small molecules as the corresponding interacting surfaces are relatively large and flat, with a tendency to lack deep and well-defined pockets that are suitable to bind a ligand with high-affinity. Nevertheless, the sheer number of such interactions (interactome), estimated to range in humans from ~300,000 [21] up to ~650,000 [22], implies that a significant number should still be druggable. There is already experimental evidence for about 80,000 PPIs in the human protein interactome with a distribution well characterized by a power law (i.e., a few proteins that have very high number of interactions and many that have only a few) [23].
Typically, PPIs involve relatively large protein surfaces lacking the well-defined binding pockets found in the traditional targets of most currently existing therapeutic drugs. Alongside fusion proteins, humanized monoclonal antibodies feature prominently as clinically approved PPI inhibitors (PPIIs), and while being serum-stable and target-specific, they also exhibit drawbacks inherent to all biologic-based therapies. Their inability to reach intracellular targets, propensity for immunogenicity, long elimination half-lives, lack of oral bioavailability, product heterogeneity, and possible manufacturing and storage stability issues present a set of problems to which development of small-molecule protein-protein interaction inhibitors (SMPPIIs) may present a viable solution [17, 24–26]. Notably, post-market safety events tend to be significantly more frequent among biologics than traditional small-molecule drugs [27]. For immunomodulatory biologics, a high incidence of additional unwanted adverse reactions, such as cytokine release syndrome, anaphylaxis, hypersensitivity, immunogenicity, infections, and malignancy, pose additional problems [28]. SMPPIIs can overcome many of these challenges, including that of oral bioavailability, while being superior in general cost and ease of manufacturing. These considerations are particularly important for prospective preventive therapies intended for those who are likely to develop autoimmune diseases. Since about 5% of the US population has an autoimmune disease, such as multiple sclerosis (MS), psoriasis, rheumatoid arthritis (RA), systemic lupus erythematous (SLE), and type 1 diabetes (T1D) among others [29], and these tend to be chronic, this is a significant therapeutic need and a considerable pharmaceutical market. Ultimately, prospective preventive therapies can only become successful here if they are sufficiently safe and patient-friendly (a) to be administrable in a wide enough population that is at elevated risk of developing the disease [30] and (b) to allow the long-term adherence and necessary compliance [31, 32]. This requires oral administration, and neither antibodies nor peptides are likely to be developable as such. Other alternatives to biologics including peptides and, more recently, nucleic acid-based aptamers, have been and are being explored as potential PPIIs; however, oral bioavailability is likely to remain a major challenge for them as well.
Some PPIs involve large interacting surfaces, such as those between pairs of globular proteins (e.g., IL-2R–IL-2) while others involve much smaller interacting surfaces, such as those between a globular protein and a single peptide chain (e.g., BCL-XL–BAD); the latter being much more susceptible to modulation by SMPPIIs [33, 34]. From a druggability perspective, it is encouraging that a computational analysis attempting to extract so-called small-molecule inhibitor starting points (SMISPs) from protein-ligand and protein-protein complexes in the Protein Data Bank (PDB) suggested that nearly half of all PPIs may be susceptible to small-molecule inhibition [35]. Historically, the success rate for different target types has been (in decreasing order): G-protein coupled receptors (GPCRs) (small ligands), enzymes (small substrates), ion channels, nuclear receptors, proteases, enzymes (large substrates), GPCRs (large ligands), cytotoxic/other, protein kinases, and protein–protein interactions [36]. From a financial standpoint, it also has to be noted that of the approximately 400 known diseases, only about 50 are considered as commercially attractive by current standards of viable return on investment (ROI) [36].
2. SMALL-MOLECULE PPI INHIBITORS
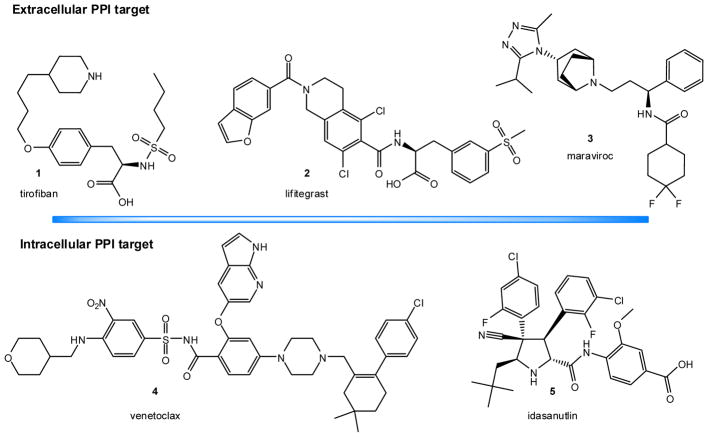
In the past couple decades, drug research has shown that small molecules can act as effective PPIIs. This is still a relatively novel field, although, progress holds promise. Effective small-molecule inhibitors have been discovered for a few important PPIs, and there are now >40 PPIs targeted by small molecules in preclinical development [33, 34, 37–46]. Tirofiban (1; Figure 2), a mimetic of the Arg-Gly-Asp tripeptide epitope of fibrinogen that binds to the αIIbβ3 integrin approved by the FDA in 2000, and maraviroc (3), an allosteric CCR5-receptor antagonist approved by the FDA in 2007, can be considered as the earliest examples of clinically approved SMPPIIs [47]. However, peptidomimetics targeting PPIs involving interactions between one protein and an isolated peptide loop or a strand of the other, which are not bona fide broad-surface PPIs and are more susceptible to small-molecule modulation [33, 34], are sometimes not considered SMPPIIs in a stricter sense [41]. Lifitegrast (SAR 1118; 2, Figure 2), a peptidomimetic small molecule LFA-1–ICAM-1 inhibitor developed first at Sunesis [48] from a series originating at Genentech [49] and later clinically by SARcode/Shire has also been approved by the FDA in 2016 for the treatment of dry eye [34]. Venetoclax (ABT-199; 4, Figure 2), part of a small-molecule series designed to target PPIs in the B cell lymphoma 2 (Bcl-2) family [50], has received FDA approval in 2015 [51]. Intriguingly, some of the data suggest that if the initial hurdles can be overcome, SMPPIIs tend to perform quite well in clinical development. Relatively few SMPPIIs have made it to clinical trials, but those that did have had a better than average chance of success [52]. For example, in phase 1, latest-generation PPIIs at the time of a recent review (those in development between 2005 and 2012) had an 82% probability of success, compared to 54% for all new molecular entities (NMEs), and in phase 2, their probability of success was 57% vs. 34% for all NMEs [52].
Figure 2.
Chemical structures of selected SMPPIIs that are FDA approved for clinical use or in advanced clinical development for extracellular and intracellular targets.
In most cases, small-molecule PPI interference is possible due to the presence of PPI “hot spots”, which are small areas of the protein-protein interface contributing most of the binding energy [53, 54]. Our focus will be solely on PPI inhibitors (antagonists) rather than activators (agonists) that stimulate activity or enhance binding, as so far there are only a very limited number of identified small-molecule PPI ‘agonists’ (i.e., enhancers or stabilizers) [44, 55, 56]. Examples include stabilizers of some of the PPIs in which protein 14-3-3 is involved [44, 57–59], a possible small-molecule activator of the tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL) receptor DR5 [60], and others. SMPPIIs can act either orthosterically (binding to hot spots on the PPI interface) or allosterically (binding to some other site not involved with direct PPI contact). SMPPII databases such as TIMBAL [61], 2P2I [62], or iPPI-DB [63] now contain 3D structures for more than 30 protein-protein complexes and several hundreds of protein-inhibitor complexes. With these developments, computationally enriched library selection is possible, and this has been shown to accelerate hit discovery [64]. Here, after briefly reviewing some representative examples and general physicochemical aspects relevant to the feasibility of PPIIs and their translatability to clinical applications in the interest of general context, we will focus on small-molecule inhibitors of cosignaling PPIs.
2.1. Illustrative Examples
Detailed reviews of SMPPIIs can be found in [33, 34, 37–46] and references therein. Here, we will only list briefly a few selected examples (Figure 2) grouped according to whether their target PPIs are extracellular or intracellular to highlight the major successes and the relevant concepts.
2.1.1. Extracellular PPIs
2.1.1.1. Integrins
Integrins were one of the very first successful SMPPIIs targets [40]. They are transmembrane proteins involved in cellular adhesion, migration, and signaling. Mimicry of the RGD (Arg-Gly-Asp) motif, a hot spot epitope involved in integrin binding, produced one of the first SMPPIIs and guided further development efforts for this protein family and other PPIs [40, 65]. Targeting of the αIIbβ3 receptor led to two molecules that achieved clinical approval for use in preventing platelet aggregation: epifibatide, which retains substantial peptide character, and tirofiban (Aggrastat, 2000) (1, Figure 2) [34, 40]. Later, lifitegrast (2, Figure 2), a peptidomimetic LFA-1–ICAM-1 inhibitor [48, 49] was approved for the treatment of dry eye (Xiidra, 2016).
2.1.1.2. CCR5–gp-120
This is a target to block HIV-1 entry into immune cells expressing the CD4 receptors (e.g., T cells, macrophages). HIV-1 infection of CD4-expressing cells is predicated on binding chemokine co-receptors, such as CCR5 or CXCR4, to induce membrane fusion of the virus and the host cell. The glycoprotein gp-120 acts as a chemokine mimic, and blockade of the CCR5–gp-120 interaction effectively prevents certain HIV strains from entering the cell [66]. Maraviroc (3, Figure 2), was identified as an allosteric CCR5-receptor antagonist that binds to the chemokine CCR5 receptor and blocks the binding of viral gp-120 [66]; hence, even if not a direct PPI inhibitor, it can be considered a SMPPII. It was approved by the FDA as an antiretroviral (Selzentry, 2007).
2.1.1.3. IL-2R–IL-2
Interleukin-2 is a soluble cytokine that potently activates T cells through its heterotrimeric receptor IL-2R; hence, this PPI is a therapeutic target for immunosuppression. Antibodies inhibiting this interaction (basiliximab, daclizumab) have shown clinical success. Shape and charge mimicry of the IL-2–binding epitope on IL-2R produced a high-affinity (~60 nM) orthosteric SMPPII, SP4206, and an alanine scan of IL-2 revealed a near identical hot-spot binding map for both [39, 67].
2.1.1.4. uPAR–uPA
The binding of urokinase-type plasminogen activator (uPA) to the cell-surface anchored urokinase receptor (uPAR) triggers a proteolytic cascade that promotes extracellular matrix degradation and integrin signaling. uPAR-targeted SMPPIIs (e.g., IPR-456) block this interaction (with relatively low, micromolar potency) reducing breast tumor cell invasiveness [68]. Another class of uPAR–uPA orthosteric SMPPIIs were shown to also diminish the cooperative vitronectin binding that mediates integrin contact [69].
2.1.2. Intracellular PPIs
The inhibition of intracellular PPIs is even more challenging than that of extracellular PPIs because the inhibitors have to be able to cross the cell membrane to reach their targets, which limits their physicochemical properties. For example, a potential SMPPI of the JNK-1–JIP-1 interaction turned out to be not cell-permeable, and its biological activity could not be evaluated [70]. On the other hand, small-molecule inhibitors are of particular interest for intracellular targets since antibodies generally cannot reach intracellular targets (Figure 1) [17]. Hence, the search for potential SMPPIIs is even more intense here, and several promising examples have been identified. Some that are in the most advanced development phase include:
2.1.2.1. Bcl-2/Bcl-XL–BAK/BAD
Members of the B cell lymphoma 2 (Bcl-2) family play important roles in mediating apoptosis, and several high affinity SMPPIIs have been identified for these PPIs. Venetoclax (ABT-199; 4, Figure 2), part of a small-molecule series designed at Abbott [50], which included ABT-737 and navitoclax (ABT-263), is now approved for the treatment of chronic lymphocytic leukemia (CLL) (Venclexta, 2015) [51].
2.1.2.2. MDM2–p53
In response to cellular stress and damage, transcription factor p53 arrests the cell cycle for DNA repair or promotes apoptosis. The p53 of undamaged cells is constitutively ubiquitinated and marked for proteasomal degradation via binding to the E3 ligase mouse double minute 2 homolog (MDM2, also called HDM2) [71]. Inhibition of this interaction could restore p53 function in cancerous cells leading to their growth arrest and apoptosis. Several SMPPIs have been developed for this PPI [72]. Nutlin-3 [73] and idasanutlin (5, Figure 2), one of its backup compound developed at Roche [74], have reached advanced clinical trials [34, 43]. JNJ-26854165 is another SMPPII that reached clinical development as an MDM2 ubiquitin ligase antagonist intended for oral administration [72]. A detailed review of small-molecule MDM2–p53 inhibitors in clinical development can be found in [75].
2.1.2.3. GPCR signaling components
Since GPCRs are a major target of many successful drugs, there has been considerable interest in modulating signaling that lies downstream of these receptors via the inhibition of either G-protein–effector or G-protein–regulator PPIs [76]. Several SMPPIIs have been identified and some promising results have been achieved, for example, for the inhibition of the binding of the Gβγ subunit to downstream effectors with M119 [76–79] and for that of the RGS (regulators of G-protein signaling) to the Gα subunit with CCG-4986 [80], CCG-63802 [81], CCG-50014 [82], CCG-203769 [83], and others [84]. Most of these turned out to be covalent binders, although with some selectivity [85–87].
2.2. SMPPII-Related General Physicochemical Considerations
2.2.1. Molecular size
The half-maximal (or median) effective dose (ED50), the half-maximal inhibitory concentration (IC50), or the related dissociation constant (Kd) are useful and typically used measures to quantitatively characterize the affinity and/or activity of therapeutic agents toward their targets on a molar scale. Existing small-molecule drugs average a value of around 20 nM, and it is generally desirable to have sub-micromolar (<1 μM) potency for lead compounds [88]. It is no coincidence that traditional drug targets, such as GPCRs, ion channels, or enzymes, have well-defined cavities or clefts for binding their natural ligand. Their relatively small volume allows ligands to focus multiple interactions (ionic, polar, hydrogen bond, and others) achieving a good enough ligand efficiency (binding energy per unit size, see below) [89]. The existence of such binding pockets also makes these targets more amenable to therapeutic modulation as well-fitting small-molecules can utilize the same binding strategy as the natural ligand(s) to effectively and selectively replace or displace them while still retaining favorable physicochemical properties. PPIs, on the other hand, tend to occur over a larger area and lack features that allow this kind of focused binding interactions. Since pockets suitable for small-molecule binding on protein-protein interfaces are generally smaller than those of traditional protein-ligand interactions, SMPPIIs must interfere with more than one such pocket to achieve adequate binding energy. According to one analysis, most existing clinically approved drugs target a single pocket occupying a volume that averages 271 Å3, whereas PPIIs target from 3 to 5 pockets with an average occupied volume of 100 Å3 [47]. As maximum binding energy is limited by pocket size, PPIIs must reach an appropriate number of binding pockets to achieve nanomolar potency [90]. Thus, SMPPIIs tend to be larger molecules, which can be undesirable in terms of ADME (absorption, distribution, metabolism, excretion) properties (e.g., decreased likelihood of oral bioavailability with MW > 500). In fact, SMPPIIs show a tendency toward larger molecular sizes when compared to existing drugs, such as receptor ligands, enzyme inhibitors, or ion channel modulators [91].
Screening out compounds with undesirable properties that break Lipinski’s “rule-of-five” [92] had been accepted practice for the past two decades, and the relatively large molecular sizes of SMPPIIs tend to result in molecular properties that fall outside of the acceptable criteria, i.e., high molecular weight (MW > 500), high number of hydrogen bond donors (HBD > 5) or acceptors (HBA > 10), and high lipophilicity (CLOGP > 5). Consequently, their size is also likely to be detrimental to ease of formulation, oral bioavailability, and membrane permeability. Nevertheless, an increasing number of new drugs have been launched recently that significantly violate these empirical rules proving that oral bioavailability is achievable even in the “beyond rule of five” chemical space [93].
2.2.2. Ligand efficiency
Ligand efficiency (LE) characterizes binding as a function of ligand size by normalizing binding energy to ligand size. LE is defined as binding energy per unit size, typically, the binding free energy (ΔG = –RTlnKd) per non-hydrogen atom, LE = ΔG0/N [89]. This metric is well-suited to characterize optimized ligand structures for any given target and can give an informative assessment of differences within ligand classes or binding sites. Typical protein–ligand interactions average LEs around 1.5 kJ/atom [39, 89, 90, 94, 95]; whereas, the most potent PPI-targeted small molecules or fragments average only around 1.0 kJ/atom [39, 42, 89, 90, 94–96]. Considering the definition of LE, a value of 1.5 kJ/atom means that the addition of one non-hydrogen atom has to be accompanied by an approximately two-fold increase in affinity to maintain the same LE [42, 90].
Given that SMPPIIs likely bind shallow pockets on the surface of their target protein, they can exhibit binding interactions only along part of their surface and are thus less efficient than drugs fully buried in traditional binding pockets. Therefore, larger structures are required to achieve the desired (i.e., nanomolar) affinities. To reach the average affinity of current small molecule drugs (Kd ≈ 20 nM [88]) with a LE of only ~1 kJ/atom, a SMPPII structure would need to have more than 45 non-hydrogen atoms, which puts it around the upper limit of drug-like compounds, as defined by rule-of-five type criteria. The sizes of SMPPIIs such as venetoclax (4; 868 Da), lifitegrast (2; 615 Da), and idasanutlin (5; 616 Da) (Figure 2), nicely illustrate this point as they are already above the rule-of-five limits discussed earlier, while still avoiding very large molecular sizes that would hinder their ADME characteristics.
2.2.3. Chemical space
There are not yet enough adequately potent and successful SMPPIIs to more definitively characterize their chemical space. As many examples illustrate already, PPI inhibition is achievable without blatant structural mimicry of proteins. However, it is also evident that the chemical space of these SMPPIIs lacks significant overlap with those of current drugs. From out of close to 5,000 compounds in the DrugBank small set, only seven showed structural similarity with one of 66 SMPPIIs analyzed in a 2010 study [97]. Aside from a larger molecular size, SMPPIIs tend to be more hydrophobic and contain more rigid aromatic scaffolds than existing drug structures. As a number of studies corroborated discrepant structural profiles between typical screening libraries and SMPPIIs [91, 97, 98], commonly utilized high-throughput screening (HTS) chemical libraries that give preference to “drug-like” criteria are not the best places to look for potential SMPPIIs. This may explain the failure of some early HTS efforts to find promising SMPPIIs. However, increasing SMPPII structural data has made it possible to perform computationally enriched library selection, which has been shown to accelerate hit discovery [64], and there are now PPII-focused screening libraries [99, 100]. Natural products and structures inspired by them could also provide additional diversity, since their chemical space is different from those of synthetic drugs and drug-like chemical libraries [101].
The need for alternative chemical spaces becomes even more apparent when considering that existing drugs exhibit relatively low structural diversity [102–105] and that bioactive molecules feature a relatively limited number of unique ring types [104]. An analysis of the basic ring-structure framework of existing drugs revealed surprisingly low diversity: half of the drugs had shapes described by only 32 of the 1179 possible frameworks [102]. Even the diversity that side chains provide to drug molecules is quite low with the average number of side chains per molecule being four, and the average number of heavy atoms per side chain being two [103]. In fact, this seems to be true for existing organic chemical compounds in general: an analysis of the molecular framework data from more than 24 million organic compounds in the CAS Registry found that half can be described by only 143 framework shapes [106]. The framework distribution conformed well to a power law with a rapidly decreasing probability of occurrence suggesting that the exploration of the chemical space is governed by a “rich get richer” type process, whereby the more often a framework has been used in the past, the more likely chemists are to use it to design and synthesize new compounds [106].
Regardless, some structural elements do exhibit good protein-binding, and privileged chemical spaces can be ascertained from frequent appearance among good protein binders, for example, biphenyl scaffolds or carboxyl moieties [107]. A number of studies suggested various scaffolds, many of them based on flat aromatic structures, as appropriate for design of effective SMPPIIs [108–110]. Notably, along these lines, we have identified certain xanthene-based organic dyes, such as erythrosine B and rose bengal, as the first promiscuous SMPPIIs [111, 112]. While their inhibitory mechanism is not entirely clear, it might be related to their rigid flat structures, since planar or partially planar aromatic and hydrophobic residues are over-represented at PPI interfaces [113], and they may make the binding of such structures here particularly likely.
3. SMALL-MOLECULE INHIBITORS FOR COSTIMULATORY AND COINHIBITORY PPIS
3.1. Costimulatory and Coinhibitory Signaling in Immune Responses
The status of immune responses is determined by costimulatory and coinhibitory signaling (cosignaling). T cell activation requires a primary signal in the form of an engagement of its T cell receptor (TCR) by the major histocompatibility complex (MHC) – peptide system of antigen-presenting cells (APCs). A secondary signal is provided by cosignaling protein molecules binding to their counterparts within this immunosynapse (Figure 3). In the absence of a secondary signal, T cells will not proliferate or differentiate into effector cells. While the underlying mechanisms are not yet fully elucidated, the two signal concept of T-cell activation has been around for a long time [114, 115], and it is also generally accepted that antigen recognition in the absence of costimulation alters the immune response and may ultimately lead to tolerance [116, 117]. Thus, the inhibition of costimulatory interactions can provide activation- and antigen-specific therapies for transplant recipients and autoimmune diseases that might avoid the broad and nonspecific immunosuppression caused by all existing immunosuppressive agents [118–125]. Conversely, coinhibitory (immune checkpoints) proteins contact their counterparts and promote attenuation of immune activation by T cell apoptosis, anergy, or functional exhaustion. As a therapeutic strategy, the inhibition of such coinhibitory interactions can be used to counteract the immunosuppressive properties of various cancer cells and boost intrinsic T cell surveillance or engineered anti-cancer activity (CAR-T).
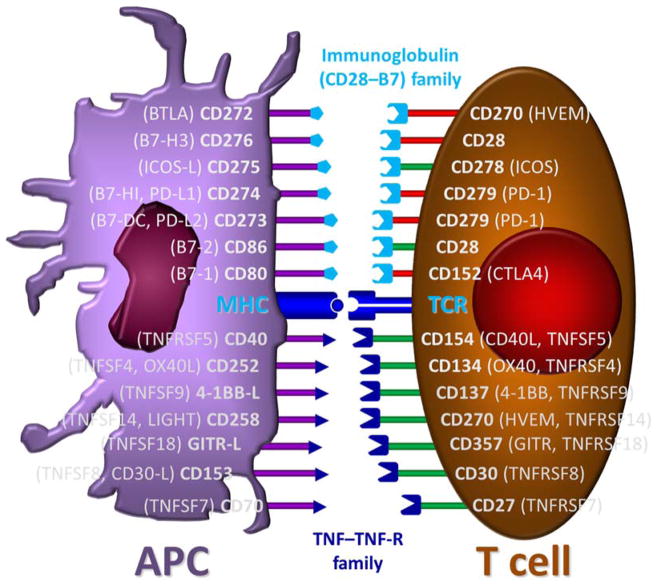
Figure 3.
Schematic illustration of some of the most important costimulatory and coinhibitory interacting pairs [121, 122]. They belong to two main families, the immunoglobulin superfamily (CD28–CD80/86 and ICOS–ICOS-L), shown in the top half of the figure, and the TNF superfamily, shown in the bottom half of the figure, each containing more than 25 already identified members [126]. The repertoire of cosignaling pathways contains both immune-stimulatory (green) and -inhibitory (red) specific pathways as well as some ambiguous ones.
Cosignaling proteins are distinguished by their general structural features and fall into two main classes: the tumor necrosis factor superfamily (TNFSF; e.g., CD40–CD40L, OX40–OX40L, and 4-1BB–4-1BB-L) and the immunoglobulin-like superfamily (IgSF; e.g., CD28–CD80/86, ICOS–ICOS-L, and PD-1–PD-L1) (Figure 3, Figure 4) [122]. There are now >25 IgSF as well as TNFRSF cosignaling molecules shown to be expressed on T cells; hence, a large number of possible therapeutic targets for immunomodulation [126]. TNFSF proteins are prominent members of the costimulatory repertoire, and feature >20 ligands and ≥30 receptors, involved in limited cross-talk, with each ligand engaging from one to five receptors [124, 127–132]. The importance of this superfamily is illustrated by the fact that there are modulatory biologics in clinical development targeting essentially all of its members. In fact, the development of biologics (both antibodies and fusion proteins) inhibiting the binding of TNF to one of its receptors resulted in five FDA-approved anti-TNF biologics (e.g., etanercept, infliximab, adalimumab), and this is one of the relatively few recent immunopharmacology and rational drug design success stories [132, 133].
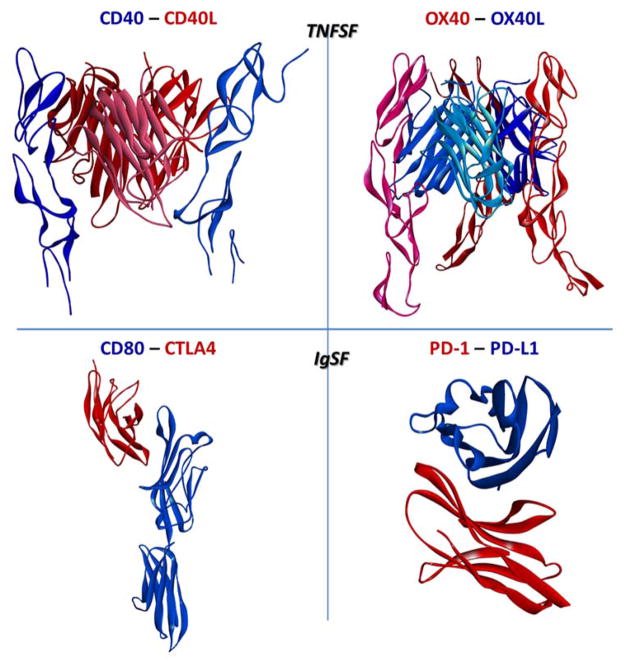
Figure 4.
Illustrative 3D structures of the protein-protein interacting surfaces for cosignaling PPIs. Structures shown are for CD40–CD40L, OX40–OX40L, CD80–CTLA4, and PD-1–PD-L1 based on PDB IDs 3QD6, 2HEV, 1I8L, and 4ZQK, respectively (the trimeric structure for CD40–CD40L is lacking one of the CD40 monomers). Proteins mainly expressed on T cells (Figure 3) are shown in reddish colors.
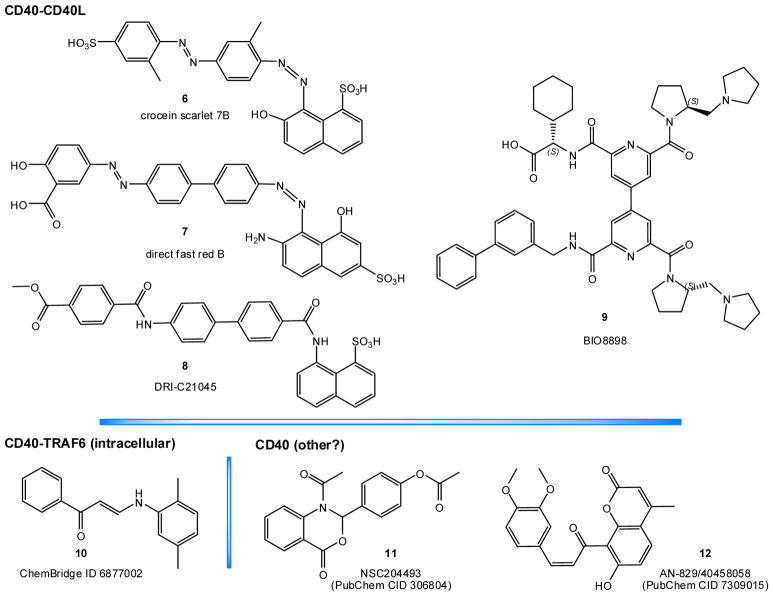
3.2. CD40–CD40L
The CD40 (TNFRSF5) – CD40L (CD154, TNFSF5) interaction is one of the most extensively studied among the TNFSF members, and they were, in fact, the first TNFSF costimulatory molecules identified [122]. CD40 (~48 kDa) is a type I transmembrane glycoprotein expressed constitutively on antigen-presenting cells (APCs; macrophages, dendritic cells, and B cells) and on certain non-immune cell types [134]. CD40L (~30 kDa) is a type II transmembrane protein that can exist in both membrane-bound and soluble (~18 kDa) form [135, 136]. Membrane-bound CD40L is found on mast cells, natural killer (NK) cells, macrophages, T cells, B cells, endothelial cells, vascular smooth muscle cells (VSMCs), and activated platelets, while soluble CD40L is derived mostly from platelet stores [137]. The broad expression of this PPI pair indicates its prominent role in various physiological processes. Its aberrant signaling is associated with pathologies, such as autoimmune thyroiditis, inflammatory bowel disease (IBD), MS, psoriasis, RA, SLE [123], cardiovascular disease [138], and possibly even obesity related insulin resistance [139–141]. Recent studies indicate that the CD40–CD40L interaction also plays an important role in glucolipotoxicity-induced β-cell death [142]. The therapeutic effects of inhibiting the CD40–CD40L interaction are largely due to suppression of B- and T cell-mediated inflammation and autoimmunity. An increasing number of studies confirmed a multitude of interactions between CD40 and CD40L on a wide variety of immune cell types as well as the critical role of this pathway in generating both humoral and cell-mediated alloreactive responses [143]. Furthermore, there is mounting evidence that CD40 could be a biomarker for auto-aggressive T cells and plays particularly important roles in autoimmune diseases such as MS (EAE) and T1D [125, 144, 145].
3.2.1. Structure and signaling
CD40L is fairly conserved across species; e.g., ~78% amino acid (AA) sequence similarity between human and mouse. However, like TNFSF ligands in general, it shares little sequence similarity with other TNFSF members (20–30%) [129]. Crystal structure studies show that the CD40L homotrimer has a truncated pyramid-like shape whose monomers are comprised of sandwiched anti-parallel β-sheets arranged in a Greek key topology (Figure 4). The interface between the trimer subunits is formed mainly by aromatic and hydrophobic residues. Mutations on CD40L that cause hyper-IgM syndrome, a condition marked by the inability of CD40L to properly engage CD40, have been mapped to its trimerization interface, the protein core, and the area contacting CD40 directly [146, 147]. The extracellular portion of CD40 is an elongated ladder-like structure comprised of three cysteine-rich domains (CRDs), all of which participate in binding CD40L along the crevice formed by two CD40L subunits. The CD40–CD40L binding interface itself contains a mixture of both hydrophilic and hydrophobic residues. Upon binding, two CD40L subunits interact with the extracellular domain of one CD40 subunit with most of the binding energy coming from hydrophilic and ionic interactions [148]. The CD40 contact area is dominated by interactions with its CRD2 and is twice the size on one CD40L subunit than the other [148]. A surface-distorting Ser132 point mutation on CD40L did not affect its binding affinity to CD40; however, it did significantly reduce ERK and p38 signaling without affecting JNK signaling. This showed that CD40L-induced CD40 trimerization was insufficient for full activation of its downstream effects, and a proper spatial orientation was also necessary.
Engagement by the CD40L trimer induces clustering of the CD40 protein and promotes the binding of adapter proteins called TNFR-associated factors (TRAFs) to its cytoplasmic domain. Ceramide enrichment of the cell-membrane further plays a critical role in the stabilization of these clusters on either end of the immunosynapse contact. The context-specific combination of CD40-bound TRAFs determines the utilized signaling pathway and its functional outcome. These include: mitogen-activated protein kinase (MAPK), extracellular signal-regulated kinase (ERK), NF-κB nuclear factor-κB (NF-κB), stress activated protein kinase (SAPK), and signal transducers and activators of transcription (STAT) [130, 149, 150]. Additionally, JAK3 can associate with CD40 directly and deliver anti-apoptotic signals via PI3K and STAT2 in B cells [151–153]. Besides CD40, CD40L can also bind to integrins, such as αIIbβ3 and Mac-1 (αMβ2), and mediate different inflammatory pathologies [154, 155].
3.2.2. Biologics
Identified as the first TNFSF costimulatory proteins, CD40L and CD40 are implicated in a myriad of immune-related pathological conditions, and have since become high profile targets [122, 143]. Blockade of this PPI has been confirmed as a potent immunomodulatory therapy, and a number of biologics have reached clinical development [124, 150, 156–158]: an anti-CD40 antagonist bleselumab (ASKP1240/4D11) for focal segmental glomerulosclerosis in kidney transplants and psoriasis [159, 160], a depleting anti-CD40 antagonist lucatumumab (HCD122) for Hodgkins and non-Hodgkins lymphoma [161] as well as a depleting anti-CD40 agonist dacetuzumab (SGN 40) for chronic lymphocytic leukemia and non-Hodgkin’s lymphoma [162, 163], a depleting anti-CD40 agonist Chi Lob 7/4 for non-Hodgkin’s lymphoma [164], and an anti-CD40 agonist CP-870893 for melanoma and pancreatic cancer [124, 165–168]. Efforts to target CD40L have been met with setbacks, as promising clinical trials of a humanized anti-CD40L antibody ruplizumab (hu5c8) had to be halted due to thromboembolic adverse effects [169–173]. The antibody was found not to activate CD40L-expressing platelets on its own, but rather as an immunocomplex whose platelet-aggregating effect was primarily driven by its Fc region via an FcyRIIa-dependent mechanism [174]. Identification of the mechanism causing the thromboembolic side effects has led to a resurgence of interest in CD40–CD40L blockade [143]. Along these lines, recently developed so-called Fc-silent domain antibodies (dAbs) that do not bind to FcgRIIa, including letolizumab and dapirolizumab pegol, were found to retain immunomodulatory activity, but do not activate platelets [175, 176]. Letolizumab (BMS-986004) is being developed by Bristol-Myers Squibb for the treatment of immune thrombocytopenic purpura. Dapirolizumab pegol (CDP7657) is a PEGylated anti-CD40 antagonist antibody fragment (Fab) developed by UCB Pharma currently in clinical trials for SLE [177].
Among possible non-antibody alternatives, a set of CD40 targeted RNA aptamers have been explored for the control of B lymphoma and bone-marrow aplasia in mouse models [178]. Nucleic acid-based aptamers are being explored as functionally comparable alternatives to antibodies that maintain the advantage of strong and specific binding to diverse targets, while also offering several further benefits, including smaller physical size, fast and inexpensive chemical synthesis, versatile chemical modification, and lack of immunogenicity [179–181]. Two sets of peptides have also been explored. A cyclic heptapetide (CLPTRHMAC) has been shown to functionally block the CD40–CD40L interaction without inducing platelet activation [182]. Another peptide mimicking the binding domain of CD40L (VLQWAKKGYYTMKSN, designated as KGYY15 mouse) showed some efficiency in preventing and even reversing early onset of T1D in NOD mice [183]. Activity was dependent on peptide length with the 15-mer being the most potent. A similar human KGYY15 peptide (VLQWAEKGYYTMSNN), which is 87% homologous, was also prepared as a first step toward translation to clinical relevance [183]. In our hands, this peptide showed only a relatively weak activity in inhibiting human CD40–CD40L interaction in a direct ELISA assay (IC50 of ~150 μM) [184].
3.2.3. Small-molecules
3.2.3.1. Organic dyes (naphthalenesulphonic acid derivatives)
Suramin was the first published small-molecule found to have CD40–CD40L inhibitory activity [185]. Using a cell-free direct ELISA assay, we found suramin to be a considerably more potent inhibitor of the CD40–CD40L PPI (IC50 = 15 μM) than of the TNFR1–TNF-α PPI (IC50 ≈ 500 μM). Its activity has also been confirmed in cell based assay with human B cells and pancreatic islets [185]. Using suramin as a structural guide, we have also identified a number of organic dye inhibitors with low micromolar activity and acceptable specificity over other TNFSF costimulatory PPIs [186–188]. Several of them, such as direct red 80, crocein scarlet 7B (6), direct fast red B (7) (Figure 5), and mordant brown 1, concentration-dependently reduced CD40L-induced activation in human B and THP-1 cells. Assays aimed at identifying the binding-partner of these compounds indicated CD40L and not CD40 as the likely binding partner [186, 187]. Molecular docking studies also indicated an allosteric site on the CD40L trimer as the likely binding site [189].
Figure 5.
SMPPIIs for the CD40–CD40L costimulatory PPI.
3.2.3.2. DRI-C21045 and other novel small-molecules
In general, such organic dye compounds, however, cannot be used for clinical applications not only because of their strong color, but also because most of them (e.g., azo dyes) are susceptible to quick metabolic degradation by intestinal microorganisms and hepatic enzymes [190, 191]. Therefore, we developed novel CD40–CD40L SMPPIIs starting from the chemical space of these organic dyes, but eliminating the aromatic azo chromophores responsible for their vivid color and metabolic susceptibility. We used an iterative design, synthesis, test, and redesign approach and were able to identify a set of new compounds such as DRI-C21045 (8, Figure 5) that showed inhibitory activity (IC50) in the high nanomolar / low micromolar range [184]. Activity was confirmed in cell assays, such as the inhibition of CD40L-induced activation in NF-κB sensor cells, THP-1 myeloid cells, and primary human B cells, and it was present at concentrations well below genotoxic or cytotoxic levels. More importantly, in vivo activity has also been confirmed. DRI-C21045 (30 mg/kg, s.c.) prolonged graft survival in a murine allogeneic skin transplant model with statistical significance. It dose-dependently suppressed alloantigen-induced T cell expansion in a draining lymph node experiment – with its highest dose approaching the efficacy of the MR-1 antibody used as the positive control [184]. While further optimization might still be required, these compounds provide clear proof-of-principle evidence that small molecule modulation of the CD40–CD40L costimulatory PPI is feasible with drug-like structures amenable to translation toward clinical applications.
These novel compounds such as DRI-C21045 (8) retained the aromatic ring scaffold of the original dye compounds, but the chromogenic azo linker groups were replaced with amide groups (compare 8 versus 6 and 7; Figure 5). A biphenyl linker markedly improved activity over a single ring one, and a naphthalene moiety and polar substitutions on both terminal aromatic rings were required to maintain activity [184]. Notably, because of the relatively small molecular weight (e.g., 580 Da for DRI-C21045), LE for these compounds is in line with most other known SMPPIIs. For the best DRI-C compounds, LE is in the 0.91–0.93 kJ/atom range, similar to that of typical PPI inhibitors (~1.0 kJ/atom), as discussed above [39, 42, 96].
3.2.3.3. BIO8898
BIO8898 (9, Figure 5) was identified to bind CD40L by an affinity selection-mass spectrometry method at Biogen Idec [192]. It is a large molecule, composed of a 4, 4′-bipyridine core and four arms: two 2-(N-pyrrolidinomethyl) pyrrolidine, one 2-cyclohexyl-2-aminoacetic acid, and one biphenyl arm (Figure 5). It was shown to inhibit the binding of mycCD40L to plated CD40-Ig fusion protein with an IC50 of 25 μM, as measured by a dissociation-enhanced lanthanide fluorescence immunoassay (DELFIA). Its ability to reduce CD40L-induced death in baby hamster kidney (BHK) cells was limited by its solubility, but the corresponding IC50 seemed to be within three-fold of the 25 μM value. Analysis of the crystal structure of the CD40L–BIO8898 complex revealed that BIO8898 binds in a deep pocket between two subunits of CD40L and disrupts the three-fold symmetry of the CD40L trimer [192], but it does not cause the dissociation of a CD40L monomer. Nevertheless, the interjection of BIO8898 is sufficient to negatively impact the CD40–CD40L binding because high affinity binding of CD40 to CD40L requires a CD40 protein to cooperatively contact two sites on adjacent CD40L monomers [192]. Hence, BIO8898 inhibits binding not by direct (orthosteric) interference at the CD40–CD40L interface, but by allosteric disruption of the overall CD40 binding sites on the CD40L trimer. Intriguingly, such allosteric binding can actually lead to improved LE for some SMPPIIs. In such cases, because the small molecule intercalates between the subunits in a deeper, permanent pocket, it can interact with the protein along its entire surface, while at orthosteric sites, it can only bind to shallower surface pockets making it more difficult to achieve adequately competitive binding affinities.
3.2.3.4. CD40–TRAF6 and CD40 downstream inhibitors
Some small-molecule inhibitors of the intracellular CD40–TRAF6 PPI, which is downstream from CD40–CD40L, have been identified via a screening approach of an existing chemical library that started with in silico drug docking and optimization [193]. The selected structures such as 10 (ChemBridge ID 6877002 and the similar 6860766; Figure 5) are relatively small, but very hydrophobic (CLOGP of 5.2 for 10) with activities in the micromolar range; e.g., IC50 of 16 and 50 μM for 10 in inhibiting the CD40-induced expression of IL1β and IL6 in bone marrow-derived macrophages [193]. In rodent models, they have been shown to improve obesity-associated insulin resistance [139, 141], sepsis [193], and neuroinflammation [194] (10 μmol/kg, s.i.d., i.p.).
In a separate study, some small-molecule inhibitors of the CD40-mediated NF-κB signaling pathway have been identified through a library screening approach using an NF-κB luciferase reporter assay in BL2 cells activated with trimerized CD40 ligand [195]. They include molecules such as 11 (NSC204493, PubChem ID 306804) and 12 (AN-829/40458058, PubChem ID 7309015) that previously were not implicated in inflammation, NF-κB signaling, or inflammatory arthritis. These molecules, however, seem more likely to be possible downstream regulators rather than direct CD40–CD40L inhibitors. Detailed reviews are not included here as they are beyond the scope of the present paper focusing on cosignaling PPIs.
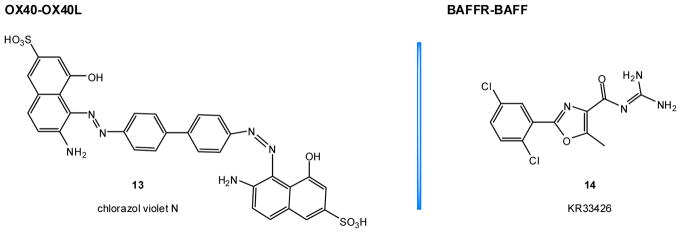
3.3. OX40–OX40L
OX40 (CD134, TNFRSF4) and OX40L (CD252, TNFSF4) are inducible costimulatory proteins [196–198]; detailed reviews of their critical role in immunity and autoimmunity have been published [199, 200]. OX40 (~48 kDa) is a type I transmembrane glycoprotein that is upregulated on CD4 and CD8 T cells following activation, while OX40L (~21kDa) is a type II transmembrane protein that is similarly upregulated on APCs (DCs, B cells, macrophages) and other cell types (NK cells, mast cells). Although OX40L lacks an extracellular proteolytic site, giving rise to thoughts that it exists only in membrane bound form, soluble OX40L has been detected in patients with RA and acute coronary syndrome [201, 202]. Ligation of OX40 by OX40L promotes the late-phase clonal expansion of effector T cell populations and their survival, thus also ensuring a greater pool of post-contraction memory T cells. Although OX40 is downregulated to basal levels on resting memory T cells, it is subsequently re-expressed upon re-exposure to antigen, and it was found necessary for a full in vivo response of CD4 memory T cells [203]. Concordant with other T cells, OX40 is also induced on immunosuppressive regulatory T (Treg) cells; however, signaling here antagonizes Treg activity to further promote T cell activation [199, 200]. Mouse studies have shown that inhibition of the OX40–OX40L PPI can control the development of inflammation in animal models of asthma, colitis, transplantation, graft-versus-host disease (GVHD), MS, RA, and T1D [204–209].
3.3.1. Structure and signaling
The brick-shaped OX40L monomer is very compact and exhibits the least amount of AA sequence similarity with other TNFSF ligands (~10–15%). OX40L has only about 132 residues in the entire extracellular region versus about 195 residues found in other TNFSF members [129]. It trimerizes into a non-pyramidal structure consisting of subunits that are sandwiched antiparallel β-sheets with a jelly roll motif [210, 211]. The peculiar shape and arrangement of OX40L (Figure 4) stands out also by its significantly smaller trimer interface versus other TNFSF members, as well as the replacement of traditionally alternating hydrophobic and hydrophilic residues in the trimer axis by generally hydrophobic ones [212]. The extracellular region of OX40 is comprised of three full CRDs and a partial vestigial one at the C-terminus. OX40 contacts OX40L extensively at the trimerization interface of subunits, with one OX40 protein bridging two OX40L monomers. The OX40 contact area is split evenly between each OX40L subunit, and mutational analysis of binding interface residues corroborated that hot-spots localize to these two different areas. The two critical OX40L residues located at diagonally opposite ends of the subunit trimerization interface, Phe180 and Asn166, interact with a hydrophobic region of CRD1 and via hydrogen bonds at the CRD2–CRD3 juncture, respectively [212].
Engagement by the OX40L trimer clusterizes OX40 into lipid-rich microdomains, promoting recruitment of TRAF2, TRAF3, and TRAF5 to its cytoplasmic tail by a TNFSF-conserved QEE motif [213–215]. OX40 acts either as an antigen-independent receptor or as a classic costimulator, synergizing with the TCR signal. Independently, OX40 activates the NF-κB1 pathway, leading to the expression of antiapoptotic Bcl-2 family proteins, such as Bcl-2, Bcl-xL, and Bfl-1 [214–217]. In the presence of antigen-dependent signaling, OX40 synergizes with TCR to promote and prolong PI3K/Akt activation as well as NFAT accumulation in the cell nucleus [218, 219].
3.3.2. Biologics
Although no biologics targeting the OX40–OX40L interaction have yet been approved for clinical use, several ongoing trials are evaluating agonistic anti-OX40 antibodies, such as MEDI-0562, MEDI-6469, PF-04518600, INCAGN0194, BMS-986178, and GSK3174998, in various cancers [220]. An anti-OX40L antagonist (oxelumab) failed Phase II trials on efficacy versus placebo in a mild allergic asthma trial. This may have been due to the short duration of the trial and the use of a non-stratified mild asthmatic patient population [124]. The transient expression of OX40 might have caused only a negligible effect on the acute phase and mildness of the disease, and this antibody might be better suited for treatment of a more chronic and severe asthmatic profile [221, 222].
3.3.3. Small-molecules
3.3.3.1. Organic dyes (naphthalenesulphonic acid derivatives)
We have identified four compounds within the chemical space of organic dyes by a direct ELISA assay as capable of blocking the OX40–OX40L interaction with chlorazol violet N (13, Figure 6) as one of the most promising [223]. They inhibited the binding of human OX40L to plated human OX40 with low micromolar potency (IC50 ≈ 2 μM) while also showing acceptable target specificity when tested across other TNFSF members (RANK–RANKL, 4-1BB–4-1BBL, CD40–CD40L, and TNF-R1–TNF-α). Functional studies with an OX40-expressing NF-κB SEAP reporter HEK cell line, showed 13 to act as a partial agonist of OX40. In other words, 13 can produce OX40 activation, but only to a lesser maximum degree than the natural OX40L ligand (~70%), while in the presence of sufficiently high OX40L, it actually slightly decreases activation by competing out the full agonist natural ligand [223]. Overall, data could be fitted well with a generalized minimal two-state theory (Del Castillo-Katz) model [224] for receptor activation. Cell-based assays have also shown that under polarizing conditions based on TGF-β, 13 (50 μM) was able to mimic the effects of an agonistic anti-OX40 mAb in suppressing Treg generation as well as in diverting central memory T cells (CD4+CD62L+Foxp3–) to a Th9 phenotype [223].
Figure 6.
SMPPIIs for the OX40–OX40L and BAFFR–BAFF costimulatory PPIs.
3.4. BAFFR–BAFF
The two ligands of this signaling network, BAFF (BLyS, CD257, TNFSF13B) and APRIL (CD256, TNFSF13), have different affinities toward three receptors, namely, BAFFR (CD268, TNFRSF13C), BCMA (CD269, TNFRSF17), and TACI (CD267, TNFRSF13B) [225–229]. Both BAFF (~32 kDa) and APRIL (~32 kDa) are type II transmembrane proteins expressed mainly by myeloid cells (macrophages, monocytes, neutrophils, and DCs), but also by malignant B cells, activated T cells, and epithelial cells. BAFFR (~19 kDa), BCMA (22 kDa), and TACI (32 kDa) are all type III transmembrane glycoproteins that are expressed mainly on B cells. BAFFR expression increases with the maturation stage of B cells, but is not found on bone marrow plasma cells. It is upregulated by activated T cells and constitutively expressed by Tregs. BCMA is restricted to antibody-producing cells, while TACI can be found on all peripheral B cells as well as monocytes and DCs [230–232].
Found as a membrane-bound or a proteolytically released soluble (~18 kDa) homotrimer, BAFF binds BAFFR and BCMA, while a multimerized “sixtymer” form is necessary to effectively activate TACI. Secreted soluble APRIL (~17 kDa) does not bind BAFFR, but possesses stronger affinity than BAFF for BCMA (Kd of 16 vs. 1600 nM) while both ligands bind TACI equally well [233–236]. For a biologically functional interaction with TACI, APRIL also requires a multimerized form, which it achieves by utilizing heparan sulfate proteoglycans as a scaffold, and their disruption can diminish general APRIL signaling [237, 238]. Ligation of BAFFR and BCMA provides survival signals to B cells, while ligation of TACI serves a more complex role, promoting survival and differentiation while countering proliferation [239]. The BAFFR–BAFF interaction is implicated in B cell-mediated autoimmunity and related lymphomas [240–242]. In addition of being a therapeutic target for autoimmune type 1 diabetes (T1D) [243–245], BAFF was also identified more recently as a possible therapeutic target for prevention of type 2 diabetes (T2D) and obesity-related insulin resistance [246, 247].
3.4.1. Structure and signaling
Protein crystal studies show that, following the general theme of other TNFSF ligands, monomeric BAFF is made up of sandwiched antiparallel β-sheets arranged in a Greek-key topology [248–250]. It exhibits low AA sequence similarity with other TNFSF members (22% TNFα, 17% CD40L, and 20% TRAIL), but preserves the well conserved residues found in hydrophobic TNFSF cores [129]. The BAFF trimerization interface is characteristically aromatic, and trimer assembly is driven by hydrophobic interactions with a few buried polar sites forming hydrogen bonds. BAFFR contains a single partial CRD that utilizes a β-hairpin DxL motif to contact two conserved arginines in a hydrophobic pocket on BAFF in a 1:1 molar ratio. The DxL motif was confirmed as a hot spot by mutations that abrogated binding to all three receptors. Other nearby residues were more ligand-specific and important for structural determination of affinity [251–253]. The BAFFR–BAFF contact surface is characterized by positive and negative charge complementarity, and the buried surface is rather small compared to other TNFSF members [250]. While this more focused interaction also implies a higher binding efficiency, it may actually confer an advantage to SMPPII design and discovery efforts targeting BAFFR–BAFF by the same principles that guide inhibition of focused interactions of traditional drug targets [254].
Engagement by BAFF trimerizes BAFFR at the cell surface, recruiting TRAF3 to its cytoplasmic domain. This releases NF-κB inducing kinase (NIK) from a TRAF3–TRAF2 complex and constitutive ubiquitinylation-driven proteosomal degradation. NIK subsequently activates the downstream alternative NF-κB pathway [255–257]. Furthermore, BAFFR can also independently activate the PI3K/Akt pathway as well as synergize with the BCR signal to promote the phosphorylation of Syk and activate the STAT3 pathway [258]. All three pathways seem to converge on upregulation of Bcl2-family survival genes, specifically, Mcl1 [259, 260].
3.4.2. Biologics
Several biologics targeting BAFF and APRIL have reached clinical development [124]. The anti-BAFF antagonist belimumab has been approved by the FDA in 2011 for the treatment of SLE, even though in the phase III clinical trials, only ~50% of the patients categorized as displaying B cell dysfunction in the form of circulating antinuclear antibodies responded to treatment [261]. Belimumab is also under evaluation for RA and Waldenstrom’s globulinemia (WG) [262, 263]. Several other biologics have reached different clinical evaluation phases for various indications. These include [124, 222]: an anti-BAFF antagonist tabalumab (LY2127399) for SLE, RA, MS and multiple myeloma (MM); a dual anti-BAFF/APRIL TACI-Ig fusion protein atacicept for SLE, RA, non-Hodgkins lymphoma, refractory MM, and WG [264–266]; and an anti-BAFF antagonist Ig-peptibody blisimimod (AMG 623) for SLE [267].
Progress has been relatively slow because, despite obvious signs of efficacy, BAFF and/or APRIL blockade in some cases showed only modest superiority compared to standard therapies, specifically, a substantially greater effect on decreasing IgM rather than IgG titers. The general consensus seems to be that compensatory (i.e., T cell dependent) mechanisms contribute toward disease heterogeneity and plasma cell survival, and that adverse events (i.e., infections) caused by excessive immunosuppression at higher dosages limit the ability to properly address disease severity [241]. Nevertheless, it is clear that there are specific patient subsets that benefit from BAFF and/or APRIL targeted therapies, which can also be useful as part of combinatorial therapy approaches. A recent antibody-design approach using phage-display based on nurse shark single variable domain new antigen receptor (VNAR) sequences yielded a DxL motif mimic and potently reduced B cell numbers in mice [268]. Furthermore, although designed as a delivery vehicle for STAT3 siRNA, a BAFFR-binding aptamer also showed nanomolar affinity and competitively inhibited BAFF-induced B cell proliferation [269].
3.4.3. Small-molecules
3.4.3.1. Guanidine derivatives (KR33426)
So far there seems to have been only a single attempt at identifying SMPPIIs for BAFFR–BAFF, which has been done by a group at Sejong University, Korea [270, 271]. They found a number of guanidine derivatives to inhibit BAFFR–BAFF binding by flow cytometry screening of their ability to diminish fluorescence induced by binding of a biotinylated human BAFF-murine CD8 construct (BAFF-muCD8) to BAFFR in WIL2-NS human B lymphoblasts [270]. The compounds showed activity in the low micromolar range at concentrations levels determined by an MTT assay to be non-cytotoxic. Their binding target was determined to be BAFFR, as only preincubation with WIL2-NS cell, but not BAFF, managed to diminish the fluorescent signal. The lone exception was KR33426 (14, Figure 6), which retained inhibitory activity in both settings.
KR33426 (14), the most promising compound of this series, was further evaluated for its potential to inhibit BAFFR–BAFF [271]. For example, it has been shown to inhibit the BAFF-induced increase of WIL2-NS cell density and to attenuate BAFF-induced anti-apoptotic activity on splenocytes. It has also been shown to have some beneficial effects in a SLE mouse model (MRLlpr/lpr mice) when injected for 4 weeks (5–10 mg/kg, i.p.) [271]. The biological targets of these effects seem to be a variety of B cell populations. It should be noted, however, that these compounds were selected from a set of compounds prepared originally as inhibitors of the sodium/hydrogen exchanger (NHE-1) [245], and these functional studies lack a proper control to differentiate the possible contribution of NHE inhibition of these compounds from BAFFR–BAFF inhibition with respect to their cell-based activity.
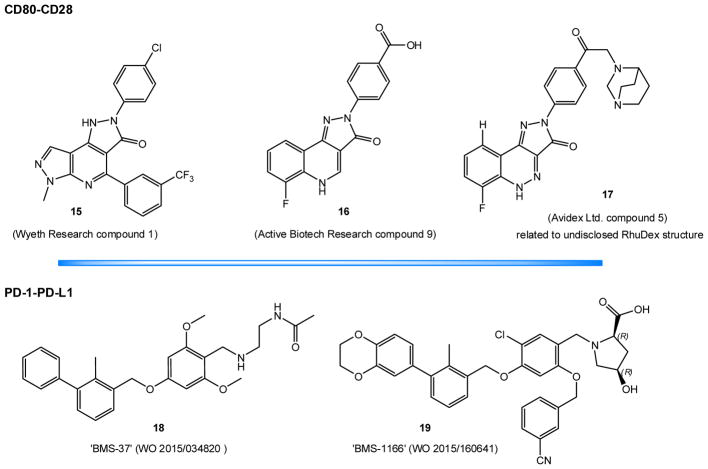
3.5. CD80/CD86–CD28/CTLA4
The CD28 (~44 kDa) and CTLA4 (CD152) (~33 kDa) protein ligands and their respective receptors CD80 (B7-1) (~60 kDa) and CD86 (B7-2) (~70 kDa) are all type I transmembrane glycoproteins from the immunoglobulin superfamily (IgSF) and are one of the most studied costimulatory interactions [29, 122]. Their main function is to prime the immune response and temper its activity upon antigenic stimulus, and their corresponding expression profiles are related to this. CD28 is a monovalent homodimer constitutively expressed on naïve and resting T cells, whereas CTLA4 is a bivalent homodimer that appears significantly only after T cell activation, with the exception of being constitutively expressed on Tregs [272, 273]. CD86 is constitutively expressed on APCs like dendritic cells, B cells, and monocytes, while CD80 appears more prominently on these cells post-activation. CD80 and CD86 can bind either of the ligands. However, CTLA4 interactions are stronger (0.4 and 4 μM with CD80 and CD86, respectively) than those of CD28 (4 and 40 μM). Engagement of CD28 in conjunction with the TCR signal co-stimulates T cell activation by upregulating antiapoptotic genes and stabilizing cytokine mRNA [274]. Conversely, engagement of CTLA4 attenuates this activation by repressing kinase activity and diminishing downstream signaling [275]. Thus, their collective expression profile is also functionally important, as CD86/CD80 first drive T cell activation, only to attenuate it when a higher-affinity inhibitory ligand appears. On Tregs, selective antagonism of CD28 prolonged contact with APCs and increased Treg immunosuppressive activity, as concurrent CTLA4 binding promoted immunosynapse formation and Treg activity [276].
3.5.1. Structure and signaling
CD28 features a single IgV-like domain in its extracellular region. This domain has a hydrophobic proline-rich MYPPPY loop motif in its complementarity determining region 3 - like (CDR3-like) region that is crucial for binding its cognate receptor [277]. CTLA4 shares ~31% AA sequence similarity with CD28. It also features a single IgV-like domain and an MYPPPY loop; thus, it is able to bind the same cognate receptors as CD28 [278, 279]. It has been proposed, however, that additional factors also mediate binding affinity, and so, specificity. The extended C-terminus region of the MYPPPY loop, also essential for binding, features a distinctly hydrophilic character in CD28 compared to the dominantly hydrophobic residues in CTLA4. This may explain the difference in CD80/CD86 binding affinities [280]. CD80 and CD86 share ~25% AA sequence similarity with each other and feature a single IgV-like and a single IgC-like domains in their extracellular regions [281]. For both proteins, it was determined that the IgV-like domains participate dominantly in binding of the cognate ligands; specifically, conserved residues on the GFFC′C″ face of the β-sheet fold. However, mutagenesis of certain residues on the ABED face of the β-sheet fold of the IgC-like domain completely prevented binding [282]. The IgC-like domain was then found to also contribute to binding by affecting quaternary structure and to be necessary for full co-signaling function [283].
Ligation of CD28 by CD80/CD86 localizes it to the immunosynapse and promotes the phosphorylation of tyrosine residues within recognition motifs on its cytoplasmic tail. PI3K and Grb2/Gads can localize to CD28 via their SH2 and SH3 domains to boost signaling through the Akt/JNK and ERK/MAPK pathways, thereby potentiating the TCR signal [284, 285]. The downstream effects of these are ultimately antiapoptotic and proliferative, most notably, the increased expression of Bcl-XL and the production of IL-2 and Glut-1 through the upregulation of NF-κB, AP-1, and NFAT transcription factors [286–288]. Ligation of CTLA4 by CD80/CD86 (Figure 4) counters the activity of CD28 by competitive inhibition within the immunosynapse and the attenuation of APC–T cell contact [289]. It can also interact with the serine/threonine phosphatase PP2A to reduce kinase signaling and possibly remove CD80/CD86 from the immunosynapse by transendocytosis for subsequent degradation using established intracellular trafficking of CTLA4 [290–292].
3.5.2. Biologics
An anti-CTLA4 antibody (ipilimumab) was approved by the FDA for the treatment of melanoma in 2011, and this was a turning point that initiated the recent expansion in the field of immuno-oncology, which has become the fastest-growing area not just within oncology, but within the entire pharmaceutical industry [293]. Since then, anticancer therapies targeting immune checkpoint (co-inhibitory) receptors such as CD80–CTLA4 and, to an even greater extent, PD-1–PD-L1 have witnessed a remarkable success [293, 294]. Another anti-CTLA4 antibody, tremelimumab, failed in a Phase III clinical trial, but has been reintroduced into clinical trials by AstraZeneca [293]. Together with surgery, radiation, and traditional chemotherapies, immune checkpoint therapies have now become a major approach used in the treatment of cancer, and they represent a novel approach (i) by targeting not the tumor cells, but molecules involved in the regulation of T cells and (ii) by focusing not on the activation of the immune system to attack particular targets on tumor cells, but on removing inhibitory pathways that block effective antitumor T cell responses [295].
On the other hand, looking at biologics targeting this pathway for immunosuppressive purposes, abatacept (CTLA4-Ig, the Fc region of the immunoglobulin IgG1 fused to the extracellular domain of CTLA4) was approved by the FDA for use in patients with RA in 2005, and it was the first selective costimulation modulator approved for clinical use [29]. It is in clinical trials for SLE arthritis and primary biliary cirrhosis [296, 297]. Abatacept has a considerably higher (~100-fold) affinity for CD80 than for CD86, and a search for a biologics that have a higher affinity to CD86 led to the discovery of belatacept, which differs by two AAs and was approved in 2011 for use in kidney transplant recipients. Detailed reviews can be found in [29, 298]. A more potent version of the CTLA4-Ig fusion protein, XPro9523, shows increased affinity for CD80/CD86/FcN and has demonstrated improved efficacy in a murine RA model and in cynomolgous monkey immunosuppression [299]. Bivalent anti-CD28 antibodies display cross-linking agonism; a monovalent Fab≈ CD28 antagonist, FR104, seeks to overcome this, and it has shown efficacy in nonhuman primate renal transplants [300]. This strategy also leaves CTLA4 signaling unaffected.
3.5.3. Small-molecules
One of the earliest efforts to block the CD28 costimulatory PPI were done at Schering-Plough by a scintillation proximity assay (SPA) based HTS testing for the ability to inhibit the binding of CD28-Ig to CD80-Ig using a natural product library [301]. A selected microbially-sourced cyclic polypeptide, NP1835-2, was shown to concentration-dependently inhibit T cell proliferation, surface activation marker expression, and the production of several T cell cytokines [302]. Here, we will focus on nonpeptidic SMPPIIs identified afterwards.
3.5.3.1. Dipyrazolopyridinone (Wyeth Research) compounds
A screen for CD80–CD28 binding antagonists from the Wyeth Research proprietary library identified several nanomolar potency inhibitors, two of which, C1 (15, Figure 7) (IC50 ≈ 60 nM) and C2 (IC50 ≈ 30 nM), were selected for further characterization [303]. They were shown to be specific for CD80 by selective enrichment of the CD80-Fc protein in equilibrium dialysis, while no binding to CD28-Fc, CTLA-4-Fc, or CD86-Fc was detected. They inhibited the binding of CD80 to both CD28 and CTLA4; however, in the direct ELISA assay they inhibited the CTLA4–CD80 binding at much higher concentrations than the CD28–CD80 binding (likely due to the slower off-rate of CTLA4). Fortuitously, they failed to bind mouse CD80; thus, the AA differences between species and known antibody epitopes could be used to guide mapping efforts. Both compounds were determined to bind to residues near or within two loops of T41-I49 and L85-E95 on the GFCC′C″ face of the N-terminus IgV-like domain, a site shared with a blocking antibody (EW3.1F1) and the binding interface of both CD28 and CTLA-4. In a cell-based assay of CD80–CD28 binding, these compounds however failed to show inhibition for concentrations of up to 100 μM [303].
Figure 7.
SMPPIIs for the CD80–C28 and PD-1–PD-L1 cosignaling PPIs.
3.5.3.2. Pyrazoloquinolinone (Active Biotech Research) compounds
Another set of compounds was identified as CD80–CD28 interaction inhibitors by first screening a commercial library of nearly 4,000 drug-like compounds in SPA [304]. Only one compound was shown to inhibit the interaction of CD28-transfected Chinese hamster ovary (CHO) cells and CD80-Ig by more than 50% with less than 20% inhibition in an LFA-3 transfected CHO cell counter-screen. The identified compound was then used to generate a sub-library of around 250 compounds which, along with 29 structurally related in-house compounds, were screened again by SPA. Selected hits were then screened by a cell-free homogeneous time-resolved fluorescence (HTRF) assay using CTLA4 as a positive control. It was determined that the substitution of a p-OH phenyl moiety by a p-COOH phenyl moiety on compound ‘1’ (IC50 = 1 μM) generated the most potent compounds ‘6’ (IC50 = 0.2 μM) and ‘9’ (16, Figure 7) (IC50 = 0.3 μM). Further analysis by surface plasmon resonance (SPR) showed that compounds ‘6’ (Kd = 0.59 μM) and ‘9’ (Kd = 0.28 μM) had dissociation constants 7–10 fold lower than ‘1’ (Kd = 3 μM). For both compounds, it was determined that the increased affinity was mainly related to a slower off-rate, and less so to a faster on-rate.
3.5.3.3. Pyrrazolocinnoline (Avidex/Medigene) compounds
Starting from the results of the above research, the chemical space of the most potent compounds identified there was used to generate another set of structural derivatives, which were then screened in a TR-FRET assay for inhibition of the sCD80-fAb–sCD28-Fc interaction [305]. SPR was also used to evaluate the dynamics of the inhibition of the binding of CD28-Fc to immobilized biotinylated CD80 on the sensor surface. Both experiments confirmed compounds with affinities in the low nanomolar range. The substitution of the hydroxyl group in the p-COOH phenyl moiety by a bulkier pyridine ring structure such as 17 (Figure 7) generated a dramatic increase in activity. These compounds were also specific for CD80, did not bind CD86 or CD28 in SPR, and had significantly slower off-rates from CD80 than CD28. Cell-based functional assays were used to investigate the ability of these compounds to inhibit CD80-induced IL-2 secretion by Jurkat and primary CD4+ T cells. The most potent ones were found to do so in the sub-micromolar range; hence, they were considered promising leads for the development of novel therapeutics for immune-mediated inflammatory diseases [305]. RhuDex (AV-1142742), a compound related to those investigated here whose structure has not been disclosed (but likely similar to 17), was investigated in clinical trials. It completed a Phase IIa study with 29 patients with RA (Medigene) and is currently being evaluated in primary biliary cirrhosis. Its activity has also been confirmed in a number of nonhuman primate and human assays [306–308]. For example, in a model of intestinal inflammation, RhuDex showed greater efficacy than abatacept in inhibiting the proliferation of lamina propria leukocytes and peripheral blood monocytes. Rhudex was also able to inhibit LPS-induced carotid artery plaque formation via reduced T cell activation and suppress protein-induced delayed type hypersensitivity (DTH) response in the skin of rhesus monkeys.
3.6. PD-1–PD-L1
PD-1 (CD279) (~50 kDa) is a monomeric type I transmembrane glycoprotein whose expression on T cells, B cells, and monocytes is induced by their activation. PD-L1 (CD274) (~40 kDa) is a type I transmembrane glycoprotein that is constitutively and inducibly expressed on T cells, B cells, monocytes, mast cells, and some non-immune cell types [309]. Ligation of PD-1 receptor by its ligand PD-L1 suppresses immune activity, as it functions to temper pre- and post-activation inflammatory signaling and limit T cell proliferation. Under normal physiological circumstances, the presence of these immune checkpoint proteins serves to maintain self-tolerance and minimize collateral tissue damage in an immune response. The PD-1–PD-L1 interaction is also implicated in autoimmunity, as a decreased ability to induce PD-L1 expression was seen in immune cells of SLE patients, and increased soluble PD-1 in RA patients is thought to potentially exacerbate disease severity [310, 311]. Sjögren’s syndrome (SSD) and RA patients have been found to have upregulated PD-1 in target tissues [312, 313]. Treatment of NOD mice by antagonistic biologics accelerated the onset of T1D and SSD [314, 315]. More importantly, this interaction is a major focus in cancer therapy since it was demonstrated that expression of PD-L1 by cancer cells allows evasion of T cell surveillance, and expression levels correlate with aggressiveness [316]. Inhibition of the PD-1–PD-L1 coinhibitory PPI can revert the exhausted phenotype of T cells to allow efficient elimination of cancer cells, and this approach of immune checkpoint (co-inhibitory) inhibition has led to remarkable clinical success recently [293, 295, 317–319]. Thus, there is great interest to develop inhibitors for this PPI, and several anti-PD-1–PD-L1 biologics are in clinical use as they have been shown to shrink solid tumors, repress metastasis, increase patient survival, and reduce treatment toxicity [320, 321].
3.6.1. Structure and signaling
PD-1 and PD-L1 belong to the B7 homology family. In its extracellular region, PD1 features a single IgV-like domain, while PD-L1 features both an IgV-like and an IgC-like domain. PD-L1 is relatively conserved between human and mouse with 70% AA sequence similarity, while human and mouse PD-1 share 64% AA sequence similarity [322, 323]. PD-L1 can also bind to CD80 with weaker affinity than PD-1 (Kds of 1.4 vs. 0.77 μM), but it does not bind CD86. The CD80–PD-L1 interaction is stronger than the CD80–CD28 one, but weaker than CD80–CTLA-4 (Kds of 4.0 vs. 0.4 μM) [324]. PD-L1 binds to PD-1 in a 1:1 molar ratio and their interaction (Figure 4) differs from those of other B7 family members in the sense that the IgV-like domains interact from the front of the GFCC′ β-sheets, specifically residues in the C′CFG strands, with no contribution from CDR-like loops, as opposed to the CTLA-4 and CD80 interaction. Structurally, this arrangement resembles the antigen-binding domain of antibodies and T cell receptors. The PD-1–PD-L1 PPI is mediated by both hydrophobic and polar contacts constructed around a partially solvent-accessible hydrophobic core [325]. The polar residues found at the periphery of the PPI site contribute to stability by forming hydrogen bonds as well. The hot spots are largely hydrophobic, with a distinctive cleft on PD-1 that relies on surface complementarity with PD-L1 featuring a Tyr123 residue inserting deep into it; a great spot for a phenyl anchor. A nearby hot spot groove offers an opportunity for a branched aliphatic moiety to anchor with a terminal hydrogen bond donor [326].
Ligation of PD-1 clusters it around the immunosynapse, essentially blocking the downstream signaling generated by the TCR-antigen complex and CD28 costimulation. The abrogation of related kinase activity is mediated by the recruitment of SHP-2 phosphatases to its intracellular ITHM motif, downregulating the expression of IL-2 and the antiapoptotic Bcl-xL protein [327, 328]. PD-1 can also activate Smad3 and arrest the cell cycle, as well as upregulate an E3 ligase Cbl-b to internalize the TCR and decrease its cell-surface presence [329, 330].
3.6.2. Biologics
Within the last three years (since 2014), two anti PD-1 antibodies (nivolumab and pembrolizumab) and three anti-PD-L1 antibodies (atezolizumab, durvalumab, and avelumab) have received FDA approval for clinical use in various forms of cancer, and several others are currently undergoing clinical trials [293, 317, 318]. This wave of PD1–PD-L1-blocking antibodies approved in 2014–2015 are the second generation therapies responsible for the current rapid growth of immuno-oncology; recent detailed reviews can be found in [293, 319]. Immunomodulation, however, is always a double-edged sword, and while this type of cancer immunotherapy has revolutionized cancer treatment, it is becoming increasingly clear that it is unavoidably associated with serious immune-related adverse effects and autoimmune diseases, most notably T1D, surface after such treatments [331, 332]. Among possible alternatives to PD-1–PD-L1 targeting biologics, peptides developed at Aurigene (Bangalore, India) such as ‘compound 8’, explored in a mouse model of sepsis [333], and NP-12/AUR-012, tested in models of melanoma, breast, kidney and colon cancers [334], have shown some efficacy. A set of macrocyclic-peptide inhibitors have also been explored recently [335].
3.6.3. Small molecules
3.6.3.1. Phenoxymethyl-biphenyl BMS series
The first published nanomolar potency SMPPIs of the PD-1–PD-L1 interaction were identified from an in-house library of synthesized derivatives based on a phenoxymethyl-biphenyl hydrophobic core at Bristol-Myers-Squibb [336, 337]. Several of these compounds were then resynthesized and evaluated for their binding and inhibitory properties at Jagiellonian University, Poland [317, 338–340]. Two representative structures, ‘BMS-37’ (18) and ‘BMS-1166’ (19) are shown in Figure 7. A number of these compounds were crystallized in their bound form with PD-L1, and they were found to bind in a hydrophobic pocket on PD-L1 shown to be at the core of the PD-1–PD-L1 PPI [338–340]. To determine the PD-1–PD-L1 inhibitory activity in a cell-based assay, Jurkat T cells constitutively expressing PD-1 and carrying a TCR-inducible NFAT-luciferase reporter construct were incubated with CHO-K1 cells constitutively expressing a TCR agonist and PD-L1. The validity of the assay was confirmed by PD-1 and PD-L1 antagonistic antibodies, which showed activity in the 0.33–1.15 nM range. Some compounds showed good activity in this assay; for example, 19 had EC50 around 0.3 μM, considerably below its cytotoxic concentration (LC50 ≈ 40 μM). Other compounds were more cytotoxic, e.g., 18 with an LC50 ≈ 3 μM [340].
3.6.3.2. CA-170
CA-170 (AUPM-170) is a small molecule antagonist that targets PD-L1 and VISTA (V-domain Ig-containing suppressor of T-cell activation). It was developed at Aurigene (Bangalore, India) and has been licensed for clinical development in patients with advanced tumors and lymphoma by Curis (Lexington, MA, USA) [334]. Structure has not been disclosed, but it is likely to be a peptidomimetic based on earlier peptide inhibitors from Aurigene. Preclinical (ex vivo) studies showed that CA-170 can induce effective proliferation and IFN-γ production by T cells that are specifically suppressed by PD-L1 or VISTA. CA-170 was claimed to have anti-tumor effects similar to anti-PD-1 or anti-VISTA antibodies in in vivo tumor models and to be orally bioavailable and safe in preclinical studies [334]. It is being investigating in a Phase 1 trial in patients with advanced solid tumors and lymphomas (NCT02812875, www.clinicaltrials.gov). Recent emerging clinical data seem to suggest that CA-170 has an acceptable safety profile and approximately dose-proportional PK profile with some preliminary evidence of immune modulation in tumor (http://www.curis.com/pipeline/ca-170) [334].
4. CONCLUSIONS
Cosignaling PPIs represent particularly valuable immunomodulatory therapeutic targets, and biologics targeting several of these interactions have already achieved considerable clinical success. Small-molecule modulators could represent possible alternatives that are orally bioavailable, easier to develop and manufacture, and less likely to be immunogenic or to encounter post-market safety problems. Several compounds proving the feasibility of SMPPII approaches have been identified through various drug discovery approaches for a number of important costimulatory and coinhibitory PPIs, including CD40–CD40L, OX40–OX40L, BAFFR–BAFF, CD80–CD28, and PD1–PD-L1. While so far only a few have advanced to preclinical (DRI-C21045 and KR33426) and clinical development (RhuDex and CA-170), there is proof-of-principle evidence that several cosignaling PPIs are susceptible to small-molecule modulation, and such SMPPIIs can lead to alternative immunomodulatory therapies.
Acknowledgments
Support by a grant from the National Institutes of Health National Institute of Allergy and Infectious Diseases (1R01AI101041, PI: P. Buchwald) is gratefully acknowledged.
5. LIST OF ABBREVIATIONS
- AA
amino acid
- ADME
absorption, distribution, metabolism, and excretion
- APC
antigen-presenting cells
- CRD
cysteine-rich domain
- GPCR
G-protein coupled receptor
- HTS
high-throughput screening
- IgSF
immunoglobulin-like superfamily
- JNK
c-Jun N-terminal kinase
- LE
ligand efficiency
- MHC
major histocompatibility complex
- MM
multiple myeloma
- MS
multiple sclerosis
- NME
new molecular entity
- PPI
protein-protein interaction
- PPII
protein-protein interaction inhibitor
- RA
rheumatoid arthritis
- RGS
regulators of G-protein signaling
- SLE
systemic lupus erythematous
- SSD
Sjögren’s syndrome
- SMPPII
small-molecule protein-protein interaction inhibitor
- SPA
scintillation proximity assay
- SPR
surface plasmon resonance
- TCR
T cell receptor
- T1D
type 1 diabetes
- T2D
type 2 diabetes
- TNF
tumor necrosis factor
- TNFSF
TNF superfamily
- TRAF
TNF receptor associated factor
- WG
Waldenstrom’s globulinemia
References
- 1.Proudfoot JR. Drugs, leads, and drug-likeness: an analysis of some recently launched drugs. Bioorg Med Chem Lett. 2002;12:1647–1650. doi: 10.1016/s0960-894x(02)00244-5. [DOI] [PubMed] [Google Scholar]
- 2.Reuben BG. The consumption and production of pharmaceuticals. In: Wermuth CG, editor. The Practice of Medicinal Chemistry. Academic Press; London: 2008. pp. 894–921. [Google Scholar]
- 3.Tufts Center for the Study of Drug Development Outlook 2007. Tufts Center for the Study of Drug Development; Boston, MA: 2007. pp. 1–6. [Google Scholar]
- 4.Munos B. Lessons from 60 years of pharmaceutical innovation. Nat Rev Drug Discov. 2009;8:959–968. doi: 10.1038/nrd2961. [DOI] [PubMed] [Google Scholar]
- 5.Scannell JW, Blanckley A, Boldon H, Warrington B. Diagnosing the decline in pharmaceutical R&D efficiency. Nat Rev Drug Discov. 2012;11:191–200. doi: 10.1038/nrd3681. [DOI] [PubMed] [Google Scholar]
- 6.Paul SM, Mytelka DS, Dunwiddie CT, Persinger CC, Munos BH, Lindborg SR, Schacht AL. How to improve R&D productivity: the pharmaceutical industry’s grand challenge. Nat Rev Drug Discov. 2010;9:203–214. doi: 10.1038/nrd3078. [DOI] [PubMed] [Google Scholar]
- 7.Moore G. Cramming more components onto integrated circuits. Electronics. 1965:38. [Google Scholar]
- 8.Walters WP, Green J, Weiss JR, Murcko MA. What do medicinal chemists actually make? A 50-year retrospective. J Med Chem. 2011;54:6405–6416. doi: 10.1021/jm200504p. [DOI] [PubMed] [Google Scholar]
- 9.Bodor N, Buchwald P. Retrometabolic Drug Design and Targeting. 1. Wiley; Hoboken, NJ: 2012. [Google Scholar]
- 10.International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 11.Clamp M, Fry B, Kamal M, Xie X, Cuff J, Lin MF, Kellis M, Lindblad-Toh K, Lander ES. Distinguishing protein-coding and noncoding genes in the human genome. Proc Natl Acad Sci USA. 2007;104:19428–19433. doi: 10.1073/pnas.0709013104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Encode Project Consortium. Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim MS, Pinto SM, Getnet D, Nirujogi RS, Manda SS, Chaerkady R, Madugundu AK, Kelkar DS, Isserlin R, Jain S, Thomas JK, Muthusamy B, Leal-Rojas P, Kumar P, Sahasrabuddhe NA, Balakrishnan L, Advani J, George B, Renuse S, Selvan LD, Patil AH, Nanjappa V, Radhakrishnan A, Prasad S, Subbannayya T, Raju R, Kumar M, Sreenivasamurthy SK, Marimuthu A, Sathe GJ, Chavan S, Datta KK, Subbannayya Y, Sahu A, Yelamanchi SD, Jayaram S, Rajagopalan P, Sharma J, Murthy KR, Syed N, Goel R, Khan AA, Ahmad S, Dey G, Mudgal K, Chatterjee A, Huang TC, Zhong J, Wu X, Shaw PG, Freed D, Zahari MS, Mukherjee KK, Shankar S, Mahadevan A, Lam H, Mitchell CJ, Shankar SK, Satishchandra P, Schroeder JT, Sirdeshmukh R, Maitra A, Leach SD, Drake CG, Halushka MK, Prasad TS, Hruban RH, Kerr CL, Bader GD, Iacobuzio-Donahue CA, Gowda H, Pandey A. A draft map of the human proteome. Nature. 2014;509:575–581. doi: 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thul PJ, Akesson L, Wiking M, Mahdessian D, Geladaki A, Ait Blal H, Alm T, Asplund A, Bjork L, Breckels LM, Backstrom A, Danielsson F, Fagerberg L, Fall J, Gatto L, Gnann C, Hober S, Hjelmare M, Johansson F, Lee S, Lindskog C, Mulder J, Mulvey CM, Nilsson P, Oksvold P, Rockberg J, Schutten R, Schwenk JM, Sivertsson A, Sjostedt E, Skogs M, Stadler C, Sullivan DP, Tegel H, Winsnes C, Zhang C, Zwahlen M, Mardinoglu A, Ponten F, von Feilitzen K, Lilley KS, Uhlen M, Lundberg E. A subcellular map of the human proteome. Science. 2017;356:eaal3321. doi: 10.1126/science.aal3321. [DOI] [PubMed] [Google Scholar]
- 15.Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discov. 2002;1:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 16.Russ AP, Lampel S. The druggable genome: an update. Drug Discov Today. 2005;10:1607–10. doi: 10.1016/S1359-6446(05)03666-4. [DOI] [PubMed] [Google Scholar]
- 17.Verdine GL, Walensky LD. The challenge of drugging undruggable targets in cancer: lessons learned from targeting BCL-2 family members. Clin Cancer Res. 2007;13:7264–7270. doi: 10.1158/1078-0432.CCR-07-2184. [DOI] [PubMed] [Google Scholar]
- 18.Kufareva I, Ilatovskiy AV, Abagyan R. Pocketome: an encyclopedia of small-molecule binding sites in 4D. Nucleic Acids Res. 2012;40:D535–D540. doi: 10.1093/nar/gkr825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu F, Shi Z, Qin C, Tao L, Liu X, Xu F, Zhang L, Song Y, Liu X, Zhang J, Han B, Zhang P, Chen Y. Therapeutic target database update 2012: a resource for facilitating target-oriented drug discovery. Nucleic Acids Res. 2012;40:D1128–D1136. doi: 10.1093/nar/gkr797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Santos R, Ursu O, Gaulton A, Bento AP, Donadi RS, Bologa CG, Karlsson A, Al-Lazikani B, Hersey A, Oprea TI, Overington JP. A comprehensive map of molecular drug targets. Nat Rev Drug Discov. 2017;16:19–34. doi: 10.1038/nrd.2016.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang QC, Petrey D, Deng L, Qiang L, Shi Y, Thu CA, Bisikirska B, Lefebvre C, Accili D, Hunter T, Maniatis T, Califano A, Honig B. Structure-based prediction of protein-protein interactions on a genome-wide scale. Nature. 2012;490:556–560. doi: 10.1038/nature11503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stumpf MP, Thorne T, de Silva E, Stewart R, An HJ, Lappe M, Wiuf C. Estimating the size of the human interactome. Proc Natl Acad Sci US A. 2008;105:6959–6964. doi: 10.1073/pnas.0708078105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klapa MI, Tsafou K, Theodoridis E, Tsakalidis A, Moschonas NK. Reconstruction of the experimentally supported human protein interactome: what can we learn? BMC Syst Biol. 2013;7:96. doi: 10.1186/1752-0509-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leader B, Baca QJ, Golan DE. Protein therapeutics: a summary and pharmacological classification. Nat Rev Drug Discov. 2008;7:21–39. doi: 10.1038/nrd2399. [DOI] [PubMed] [Google Scholar]
- 25.Firer MA, Gellerman G. Targeted drug delivery for cancer therapy: the other side of antibodies. J Hematol Oncol. 2012;5:70. doi: 10.1186/1756-8722-5-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huck BR, Kötzner L, Urbahns K. Small molecules drive big improvements in immuno-oncology therapies. Angew Chem Int Ed Engl. 2018;57:4412–4428. doi: 10.1002/anie.201707816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Downing NS, Shah ND, Aminawung JA, Pease AM, Zeitoun JD, Krumholz HM, Ross JS. Postmarket safety events among novel therapeutics approved by the US Food and Drug Administration between 2001 and 2010. J Am Med Assoc (JAMA) 2017;317:1854–1863. doi: 10.1001/jama.2017.5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sathish JG, Sethu S, Bielsky MC, de Haan L, French NS, Govindappa K, Green J, Griffiths CE, Holgate S, Jones D, Kimber I, Moggs J, Naisbitt DJ, Pirmohamed M, Reichmann G, Sims J, Subramanyam M, Todd MD, Van Der Laan JW, Weaver RJ, Park BK. Challenges and approaches for the development of safer immunomodulatory biologics. Nat Rev Drug Discov. 2013;12:306–324. doi: 10.1038/nrd3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adams AB, Ford ML, Larsen CP. Costimulation blockade in autoimmunity and transplantation: the CD28 pathway. J Immunol. 2016;197:2045–2050. doi: 10.4049/jimmunol.1601135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giannoukakis N, Phillips B, Trucco M. Toward a cure for type 1 diabetes mellitus: diabetes-suppressive dendritic cells and beyond. Pediatr Diabetes. 2008;9:4–13. doi: 10.1111/j.1399-5448.2008.00401.x. [DOI] [PubMed] [Google Scholar]
- 31.Cochrane GM, Horne R, Chanez P. Compliance in asthma. Respir Med. 1999;93:763–769. doi: 10.1016/s0954-6111(99)90260-3. [DOI] [PubMed] [Google Scholar]
- 32.Moia M, Mantovani LG, Carpenedo M, Scalone L, Monzini MS, Cesana G, Mannucci PM. Patient preferences and willingness to pay for different options of anticoagulant therapy. Intern Emerg Med. 2013;8:237–243. doi: 10.1007/s11739-012-0844-3. [DOI] [PubMed] [Google Scholar]
- 33.Arkin MR, Tang Y, Wells JA. Small-molecule inhibitors of protein-protein interactions: Progressing toward the reality. Chem Biol. 2014;21:1102–1114. doi: 10.1016/j.chembiol.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scott DE, Bayly AR, Abell C, Skidmore J. Small molecules, big targets: drug discovery faces the protein-protein interaction challenge. Nat Rev Drug Discov. 2016;15:533–550. doi: 10.1038/nrd.2016.29. [DOI] [PubMed] [Google Scholar]
- 35.Koes DR, Camacho CJ. Small-molecule inhibitor starting points learned from protein-protein interaction inhibitor structure. Bioinformatics. 2012;28:784–791. doi: 10.1093/bioinformatics/btr717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bartfai T, Lees GV. Drug Discovery: From Bedside to Wall Street. Academic Press; San Diego, CA: 2005. [Google Scholar]
- 37.Berg T. Modulation of protein-protein interactions with small organic molecules. Angew Chem Int Ed Engl. 2003;42:2462–2481. doi: 10.1002/anie.200200558. [DOI] [PubMed] [Google Scholar]
- 38.Arkin MR, Wells JA. Small-molecule inhibitors of protein-protein interactions: progressing towards the dream. Nat Rev Drug Discov. 2004;3:301–317. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- 39.Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007;450:1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- 40.Fry DC. Protein-protein interactions as targets for small molecule drug discovery. Biopolymers. 2006;84:535–552. doi: 10.1002/bip.20608. [DOI] [PubMed] [Google Scholar]
- 41.Whitty A, Kumaravel G. Between a rock and a hard place? Nat Chem Biol. 2006;2:112–118. doi: 10.1038/nchembio0306-112. [DOI] [PubMed] [Google Scholar]
- 42.Buchwald P. Small-molecule protein-protein interaction inhibitors: therapeutic potential in light of molecular size, chemical space, and ligand binding efficiency considerations. IUBMB Life. 2010;62:724–731. doi: 10.1002/iub.383. [DOI] [PubMed] [Google Scholar]
- 43.Mullard A. Protein-protein interaction inhibitors get into the groove Nat. Rev Drug Discov. 2012;11:173–175. doi: 10.1038/nrd3680. [DOI] [PubMed] [Google Scholar]
- 44.Milroy LG, Grossmann TN, Hennig S, Brunsveld L, Ottmann C. Modulators of protein-protein interactions. Chem Rev. 2014;114:4695–4748. doi: 10.1021/cr400698c. [DOI] [PubMed] [Google Scholar]
- 45.Villoutreix BO, Kuenemann MA, Poyet JL, Bruzzoni-Giovanelli H, Labbe C, Lagorce D, Sperandio O, Miteva MA. Drug-like protein-protein interaction modulators: challenges and opportunities for drug discovery and chemical biology. Mol Inform. 2014;33:414–437. doi: 10.1002/minf.201400040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Song Y, Buchwald P. TNF superfamily protein-protein interactions: feasibility of small-molecule modulation. Curr Drug Targets. 2015;16:393–408. doi: 10.2174/1389450116666150223115628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fuller JC, Burgoyne NJ, Jackson RM. Predicting druggable binding sites at the protein-protein interface. Drug Discov Today. 2009;14:155–161. doi: 10.1016/j.drudis.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 48.Zhong M, Gadek TR, Bui M, Shen W, Burnier J, Barr KJ, Hanan EJ, Oslob JD, Yu CH, Zhu J, Arkin MR, Evanchik MJ, Flanagan WM, Hoch U, Hyde J, Prabhu S, Silverman JA, Wright J. Discovery and development of potent LFA-1/ICAM-1 antagonist SAR 1118 as an ophthalmic solution for treating dry eye. ACS Med Chem Lett. 2012;3:203–206. doi: 10.1021/ml2002482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gadek TR, Burdick DJ, McDowell RS, Stanley MS, Marsters JC, Jr, Paris KJ, Oare DA, Reynolds ME, Ladner C, Zioncheck KA, Lee WP, Gribling P, Dennis MS, Skelton NJ, Tumas DB, Clark KR, Keating SM, Beresini MH, Tilley JW, Presta LG, Bodary SC. Generation of an LFA-1 antagonist by the transfer of the ICAM-1 immunoregulatory epitope to a small molecule. Science. 2002;295:1086–1089. doi: 10.1126/science.295.5557.1086. [DOI] [PubMed] [Google Scholar]
- 50.Souers AJ, Leverson JD, Boghaert ER, Ackler SL, Catron ND, Chen J, Dayton BD, Ding H, Enschede SH, Fairbrother WJ, Huang DC, Hymowitz SG, Jin S, Khaw SL, Kovar PJ, Lam LT, Lee J, Maecker HL, Marsh KC, Mason KD, Mitten MJ, Nimmer PM, Oleksijew A, Park CH, Park CM, Phillips DC, Roberts AW, Sampath D, Seymour JF, Smith ML, Sullivan GM, Tahir SK, Tse C, Wendt MD, Xiao Y, Xue JC, Zhang H, Humerickhouse RA, Rosenberg SH, Elmore SW. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat Med. 2013;19:202–208. doi: 10.1038/nm.3048. [DOI] [PubMed] [Google Scholar]
- 51.Mullard A. Pioneering apoptosis-targeted cancer drug poised for FDA approval. Nat Rev Drug Discov. 2016;15:147–149. doi: 10.1038/nrd.2016.23. [DOI] [PubMed] [Google Scholar]
- 52.Meier C, Cairns-Smith S, Schulze U. Can emerging drug classes improve R&D productivity? Drug Discov Today. 2013;18:607–609. doi: 10.1016/j.drudis.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 53.Bogan AA, Thorn KS. Anatomy of hot spots in protein interfaces. J Mol Biol. 1998;280:1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- 54.Darnell SJ, LeGault L, Mitchell JC. KFC Server: interactive forecasting of protein interaction hot spots. Nucleic Acids Res. 2008;36:W265–W269. doi: 10.1093/nar/gkn346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thiel P, Kaiser M, Ottmann C. Small-molecule stabilization of protein-protein interactions: an underestimated concept in drug discovery? Angew Chem Int Ed Engl. 2012;51:2012–2018. doi: 10.1002/anie.201107616. [DOI] [PubMed] [Google Scholar]
- 56.Andrei SA, Sijbesma E, Hann M, Davis J, O’Mahony G, Perry MWD, Karawajczyk A, Eickhoff J, Brunsveld L, Doveston RG, Milroy LG, Ottmann C. Stabilization of protein-protein interactions in drug discovery. Expert Opin Drug Discov. 2017;12:925–940. doi: 10.1080/17460441.2017.1346608. [DOI] [PubMed] [Google Scholar]
- 57.Rose R, Erdmann S, Bovens S, Wolf A, Rose M, Hennig S, Waldmann H, Ottmann C. Identification and structure of small-molecule stabilizers of 14-3-3 protein-protein interactions. Angew Chem Int Ed Engl. 2010;49:4129–4132. doi: 10.1002/anie.200907203. [DOI] [PubMed] [Google Scholar]
- 58.Milroy LG, Brunsveld L, Ottmann C. Stabilization and inhibition of protein-protein interactions: the 14-3-3 case study. ACS Chem Biol. 2013;8:27–35. doi: 10.1021/cb300599t. [DOI] [PubMed] [Google Scholar]
- 59.Ottmann C. Small-molecule modulators of 14-3-3 protein-protein interactions. Bioorg Med Chem. 2013;21:4058–4062. doi: 10.1016/j.bmc.2012.11.028. [DOI] [PubMed] [Google Scholar]
- 60.Wang G, Wang X, Yu H, Wei S, Williams N, Holmes DL, Halfmann R, Naidoo J, Wang L, Li L, Chen S, Harran P, Lei X, Wang X. Small-molecule activation of the TRAIL receptor DR5 in human cancer cells. Nat Chem Biol. 2013;9:84–89. doi: 10.1038/nchembio.1153. [DOI] [PubMed] [Google Scholar]
- 61.Higueruelo AP, Schreyer A, Bickerton GR, Pitt WR, Groom CR, Blundell TL. Atomic interactions and profile of small molecules disrupting protein-protein interfaces: the TIMBAL database. Chem Biol Drug Des. 2009;74:457–467. doi: 10.1111/j.1747-0285.2009.00889.x. [DOI] [PubMed] [Google Scholar]
- 62.Bourgeas R, Basse MJ, Morelli X, Roche P. Atomic analysis of protein-protein interfaces with known inhibitors: the 2P2I database. PLoS One. 2010;5:e9598. doi: 10.1371/journal.pone.0009598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Labbe CM, Kuenemann MA, Zarzycka B, Vriend G, Nicolaes GA, Lagorce D, Miteva MA, Villoutreix BO, Sperandio O. iPPI-DB: an online database of modulators of protein-protein interactions. Nucleic Acids Res. 2016;44:D542–D547. doi: 10.1093/nar/gkv982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Milhas S, Raux B, Betzi S, Derviaux C, Roche P, Restouin A, Basse MJ, Rebuffet E, Lugari A, Badol M, Kashyap R, Lissitzky JC, Eydoux C, Hamon V, Gourdel ME, Combes S, Zimmermann P, Aurrand-Lions M, Roux T, Rogers C, Muller S, Knapp S, Trinquet E, Collette Y, Guillemot JC, Morelli X. Protein-protein interaction inhibition (2P2I)-oriented chemical library accelerates hit discovery. ACS Chem Biol. 2016;11:2140–2148. doi: 10.1021/acschembio.6b00286. [DOI] [PubMed] [Google Scholar]
- 65.Fry DC. Drug-like inhibitors of protein-protein interactions: a structural examination of effective protein mimicry. Curr Protein Pept Sci. 2008;9:240–247. doi: 10.2174/138920308784533989. [DOI] [PubMed] [Google Scholar]
- 66.Dorr P, Westby M, Dobbs S, Griffin P, Irvine B, Macartney M, Mori J, Rickett G, Smith-Burchnell C, Napier C, Webster R, Armour D, Price D, Stammen B, Wood A, Perros M. Maraviroc (UK-427,857), a potent, orally bioavailable, and selective small-molecule inhibitor of chemokine receptor CCR5 with broad-spectrum anti-human immunodeficiency virus type 1 activity. Antimicrob Agents Chemother. 2005;49:4721–4732. doi: 10.1128/AAC.49.11.4721-4732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Raimundo BC, Oslob JD, Braisted AC, Hyde J, McDowell RS, Randal M, Waal ND, Wilkinson J, Yu CH, Arkin MR. Integrating fragment assembly and biophysical methods in the chemical advancement of small-molecule antagonists of IL-2: an approach for inhibiting protein-protein interactions. J Med Chem. 2004;47:3111–3130. doi: 10.1021/jm049967u. [DOI] [PubMed] [Google Scholar]
- 68.Khanna M, Wang F, Jo I, Knabe WE, Wilson SM, Li L, Bum-Erdene K, Li J, GWS, Khanna R, Meroueh SO. Targeting multiple conformations leads to small molecule inhibitors of the uPAR. PA protein-protein interaction that block cancer cell invasion. ACS Chem Biol. 2011;6:1232–1243. doi: 10.1021/cb200180m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu D, Zhou D, Wang B, Knabe WE, Meroueh SO. A new class of orthosteric uPAR. PA small-molecule antagonists are allosteric inhibitors of the uPAR.vitronectin interaction. ACS Chem Biol. 2015;10:1521–1534. doi: 10.1021/cb500832q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen T, Kablaoui N, Little J, Timofeevski S, Tschantz WR, Chen P, Feng J, Charlton M, Stanton R, Bauer P. Identification of small-molecule inhibitors of the JIP-JNK interaction. Biochem J. 2009;420:283–294. doi: 10.1042/BJ20081899. [DOI] [PubMed] [Google Scholar]
- 71.Fischer PM, Lane DP. Small-molecule inhibitors of the p53 suppressor HDM2: have protein-protein interactions come of age as drug targets? Trends Pharmacol Sci. 2004;25:343–346. doi: 10.1016/j.tips.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 72.Dömling A. Small molecular weight protein-protein interaction antagonists: an insurmountable challenge? Curr Opin Chem Biol. 2008;12:281–291. doi: 10.1016/j.cbpa.2008.04.603. [DOI] [PubMed] [Google Scholar]
- 73.Kojima K, Burks JK, Arts J, Andreeff M. The novel tryptamine derivative JNJ-26854165 induces wild-type p53- and E2F1-mediated apoptosis in acute myeloid and lymphoid leukemias. Mol Cancer Ther. 2010;9:2545–2557. doi: 10.1158/1535-7163.MCT-10-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ding Q, Zhang Z, Liu JJ, Jiang N, Zhang J, Ross TM, Chu XJ, Bartkovitz D, Podlaski F, Janson C, Tovar C, Filipovic ZM, Higgins B, Glenn K, Packman K, Vassilev LT, Graves B. Discovery of RG7388, a potent and selective p53-MDM2 inhibitor in clinical development. J Med Chem. 2013;56:5979–5983. doi: 10.1021/jm400487c. [DOI] [PubMed] [Google Scholar]
- 75.Zhao Y, Aguilar A, Bernard D, Wang S. Small-molecule inhibitors of the MDM2-p53 protein-protein interaction (MDM2 Inhibitors) in clinical trials for cancer treatment. J Med Chem. 2015;58:1038–1052. doi: 10.1021/jm501092z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Blazer LL, Neubig RR. Small molecule protein-protein interaction inhibitors as CNS therapeutic agents: current progress and future hurdles. Neuropsychopharmacology. 2009;34:126–141. doi: 10.1038/npp.2008.151. [DOI] [PubMed] [Google Scholar]
- 77.Bonacci TM, Mathews JL, Yuan C, Lehmann DM, Malik S, Wu D, Font JL, Bidlack JM, Smrcka AV. Differential targeting of Gbg-subunit signaling with small molecules. Science. 2006;312:443–446. doi: 10.1126/science.1120378. [DOI] [PubMed] [Google Scholar]
- 78.Lehmann DM, Seneviratne AM, Smrcka AV. Small molecule disruption of G protein beta gamma subunit signaling inhibits neutrophil chemotaxis and inflammation. Mol Pharmacol. 2008;73:410–418. doi: 10.1124/mol.107.041780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Casey LM, Pistner AR, Belmonte SL, Migdalovich D, Stolpnik O, Nwakanma FE, Vorobiof G, Dunaevsky O, Matavel A, Lopes CM, Smrcka AV, Blaxall BC. Small molecule disruption of G beta gamma signaling inhibits the progression of heart failure. Circ Res. 2010;107:532–539. doi: 10.1161/CIRCRESAHA.110.217075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Roman DL, Talbot JN, Roof RA, Sunahara RK, Traynor JR, Neubig RR. Identification of small-molecule inhibitors of RGS4 using a high-throughput flow cytometry protein interaction assay. Mol Pharmacol. 2007;71:169–175. doi: 10.1124/mol.106.028670. [DOI] [PubMed] [Google Scholar]
- 81.Blazer LL, Roman DL, Chung A, Larsen MJ, Greedy BM, Husbands SM, Neubig RR. Reversible, allosteric small-molecule inhibitors of regulator of G protein signaling proteins. Mol Pharmacol. 2010;78:524–533. doi: 10.1124/mol.110.065128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Turner EM, Blazer LL, Neubig RR, Husbands SM. Small molecule inhibitors of Regulator of G Protein Signalling (RGS) proteins. ACS Med Chem Lett. 2012;3:146–150. doi: 10.1021/ml200263y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Blazer LL, Storaska AJ, Jutkiewicz EM, Turner EM, Calcagno M, Wade SM, Wang Q, Huang XP, Traynor JR, Husbands SM, Morari M, Neubig RR. Selectivity and anti-Parkinson’s potential of thiadiazolidinone RGS4 inhibitors. ACS Chem Neurosci. 2015;6:911–919. doi: 10.1021/acschemneuro.5b00063. [DOI] [PubMed] [Google Scholar]
- 84.Roman DL, Traynor JR. Regulators of G protein signaling (RGS) proteins as drug targets: modulating G-protein-coupled receptor (GPCR) signal transduction. J Med Chem. 2011;54:7433–7440. doi: 10.1021/jm101572n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kimple AJ, Willard FS, Giguere PM, Johnston CA, Mocanu V, Siderovski DP. The RGS protein inhibitor CCG-4986 is a covalent modifier of the RGS4 Ga-interaction face. Biochim Biophys Acta. 2007;1774:1213–1220. doi: 10.1016/j.bbapap.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Roman DL, Blazer LL, Monroy CA, Neubig RR. Allosteric inhibition of the regulator of G protein signaling-Ga protein-protein interaction by CCG-4986. Mol Pharmacol. 2010;78:360–365. doi: 10.1124/mol.109.063388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Blazer LL, Zhang H, Casey EM, Husbands SM, Neubig RR. A nanomolar-potency small molecule inhibitor of regulator of G-protein signaling proteins. Biochemistry. 2011;50:3181–3192. doi: 10.1021/bi1019622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat Rev Drug Discov. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 89.Hopkins AL, Groom CR, Alex A. Ligand efficiency: a useful metric for lead selection. Drug Discov Today. 2004;9:430–431. doi: 10.1016/S1359-6446(04)03069-7. [DOI] [PubMed] [Google Scholar]
- 90.Buchwald P. Glucocorticoid receptor binding: a biphasic dependence on molecular size as revealed by the bilinear LinBiExp model. Steroids. 2008;73:193–208. doi: 10.1016/j.steroids.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 91.Neugebauer A, Hartmann RW, Klein CD. Prediction of protein-protein interaction inhibitors by chemoinformatics and machine learning methods. J Med Chem. 2007;50:4665–4668. doi: 10.1021/jm070533j. [DOI] [PubMed] [Google Scholar]
- 92.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development setting. Adv Drug Deliv Rev. 1997;23:3–25. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 93.DeGoey DA, Chen HJ, Cox PB, Wendt MD. Beyond the rule of 5: Lessons learned from AbbVie’s drugs and compound collection. J Med Chem. 2017;61:2636–2651. doi: 10.1021/acs.jmedchem.7b00717. [DOI] [PubMed] [Google Scholar]
- 94.Hajduk PJ. Fragment-based drug design: how big is too big? J Med Chem. 2006;49:6972–6976. doi: 10.1021/jm060511h. [DOI] [PubMed] [Google Scholar]
- 95.Reynolds CH, Bembenek SD, Tounge BA. The role of molecular size in ligand efficiency. Bioorg Med Chem Lett. 2007;17:4258–4261. doi: 10.1016/j.bmcl.2007.05.038. [DOI] [PubMed] [Google Scholar]
- 96.Laraia L, McKenzie G, Spring DR, Venkitaraman AR, Huggins DJ. Overcoming chemical, biological, and computational challenges in the development of inhibitors targeting protein-protein interactions. Chem Biol. 2015;22:689–703. doi: 10.1016/j.chembiol.2015.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Reynès C, Host H, Camproux AC, Laconde G, Leroux F, Mazars A, Deprez B, Fahraeus R, Villoutreix BO, Sperandio O. Designing focused chemical libraries enriched in protein-protein interaction inhibitors using machine-learning methods. PLoS Comput Biol. 2010;6:e1000695. doi: 10.1371/journal.pcbi.1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sperandio O, Reynès CH, Camproux AC, Villoutreix BO. Rationalizing the chemical space of protein-protein interaction inhibitors. Drug Discov Today. 2010;15:220–229. doi: 10.1016/j.drudis.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 99.Hamon V, Brunel JM, Combes S, Basse MJ, Roche P, Morelli X. 2P2Ichem: focused chemical libraries dedicated to orthosteric modulation of protein-protein interactions. Med Chem Comm. 2013;4:797–809. [Google Scholar]
- 100.Zhang X, Betzi S, Morelli X, Roche P. Focused chemical libraries--design and enrichment: an example of protein-protein interaction chemical space. Future Med Chem. 2014;6:1291–1307. doi: 10.4155/fmc.14.57. [DOI] [PubMed] [Google Scholar]
- 101.Bauer RA, Wurst JM, Tan DS. Expanding the range of ‘druggable’ targets with natural product-based libraries: an academic perspective. Curr Opin Chem Biol. 2010;14:308–314. doi: 10.1016/j.cbpa.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bemis GW, Murcko MA. The properties of known drugs. 1. Molecular frameworks. J Med Chem. 1996;39:2887–2893. doi: 10.1021/jm9602928. [DOI] [PubMed] [Google Scholar]
- 103.Bemis GW, Murcko MA. Properties of known drugs. 2. Side chains. J Med Chem. 1999;42:5095–5099. doi: 10.1021/jm9903996. [DOI] [PubMed] [Google Scholar]
- 104.Ertl P, Jelfs S, Muhlbacher J, Schuffenhauer A, Selzer P. Quest for the rings. In silico exploration of ring universe to identify novel bioactive heteroaromatic scaffolds. J Med Chem. 2006;49:4568–4573. doi: 10.1021/jm060217p. [DOI] [PubMed] [Google Scholar]
- 105.Triggle DJ. The chemist as astronaut: searching for biologically useful space in the chemical universe. Biochem Pharmacol. 2009;78:217–223. doi: 10.1016/j.bcp.2009.02.015. [DOI] [PubMed] [Google Scholar]
- 106.Lipkus AH, Yuan Q, Lucas KA, Funk SA, Bartelt WF, 3rd, Schenck RJ, Trippe AJ. Structural diversity of organic chemistry. A scaffold analysis of the CAS Registry. J Org Chem. 2008;73:4443–4451. doi: 10.1021/jo8001276. [DOI] [PubMed] [Google Scholar]
- 107.Hajduk PJ, Bures M, Praestgaard J, Fesik SW. Privileged molecules for protein binding identified from NMR-based screening. J Med Chem. 2000;43:3443–3447. doi: 10.1021/jm000164q. [DOI] [PubMed] [Google Scholar]
- 108.Che Y, Brooks BR, Marshall GR. Development of small molecules designed to modulate protein-protein interactions. J Comput Aided Mol Des. 2006;20:109–130. doi: 10.1007/s10822-006-9040-8. [DOI] [PubMed] [Google Scholar]
- 109.Fletcher S, Hamilton AD. Targeting protein-protein interactions by rational design: mimicry of protein surfaces. J R Soc Interface. 2006;3:215–233. doi: 10.1098/rsif.2006.0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hershberger SJ, Lee SG, Chmielewski J. Scaffolds for blocking protein-protein interactions. Curr Top Med Chem. 2007;7:928–942. doi: 10.2174/156802607780906726. [DOI] [PubMed] [Google Scholar]
- 111.Ganesan L, Margolles-Clark E, Song Y, Buchwald P. The food colorant erythrosine is a promiscuous protein-protein interaction inhibitor. Biochem Pharmacol. 2011;81:810–818. doi: 10.1016/j.bcp.2010.12.020. [DOI] [PubMed] [Google Scholar]
- 112.Ganesan L, Buchwald P. The promiscuous protein binding ability of erythrosine B studied by metachromasy (metachromasia) J Mol Recognit. 2013;26:181–189. doi: 10.1002/jmr.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Glaser F, Steinberg DM, Vakser IA, Ben-Tal N. Residue frequencies and pairing preferences at protein-protein interfaces. Proteins. 2001;43:89–102. [PubMed] [Google Scholar]
- 114.Lafferty KJ, Andrus L, Prowse SJ. Role of lymphokine and antigen in the control of specific T cell responses. Immunol Rev. 1980;51:279–314. doi: 10.1111/j.1600-065x.1980.tb00325.x. [DOI] [PubMed] [Google Scholar]
- 115.Bretscher P. The two-signal model of lymphocyte activation twenty-one years later. Immunol Today. 1992;13:74–76. doi: 10.1016/0167-5699(92)90138-W. [DOI] [PubMed] [Google Scholar]
- 116.Jenkins MK, Mueller D, Schwartz RH, Carding S, Bottomley K, Stadecker MJ, Urdahl KB, Norton SD. Induction and maintenance of anergy in mature T cells. Adv Exp Med Biol. 1991;292:167–176. doi: 10.1007/978-1-4684-5943-2_19. [DOI] [PubMed] [Google Scholar]
- 117.Sharpe AH. Mechanisms of costimulation. Immunol Rev. 2009;229:5–11. doi: 10.1111/j.1600-065X.2009.00784.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gao W, Demirci G, Li XC. Negative T cell costimulation and islet tolerance. Diabetes Metab Res Rev. 2003;19:179–185. doi: 10.1002/dmrr.345. [DOI] [PubMed] [Google Scholar]
- 119.Larsen CP, Knechtle SJ, Adams A, Pearson T, Kirk AD. A new look at blockade of T-cell costimulation: a therapeutic strategy for long-term maintenance immunosuppression. Am J Transplant. 2006;6:876–883. doi: 10.1111/j.1600-6143.2006.01259.x. [DOI] [PubMed] [Google Scholar]
- 120.Vincenti F, Luggen M. T cell costimulation: a rational target in the therapeutic armamentarium for autoimmune diseases and transplantation. Annu Rev Med. 2007;58:347–358. doi: 10.1146/annurev.med.58.080205.154004. [DOI] [PubMed] [Google Scholar]
- 121.Weaver TA, Charafeddine AH, Kirk AD. Costimulation blockade: towards clinical application. Front Biosci. 2008;13:2120–2139. doi: 10.2741/2829. [DOI] [PubMed] [Google Scholar]
- 122.Li XC, Rothstein DM, Sayegh MH. Costimulatory pathways in transplantation: challenges and new developments. Immunol Rev. 2009;229:271–293. doi: 10.1111/j.1600-065X.2009.00781.x. [DOI] [PubMed] [Google Scholar]
- 123.Peters AL, Stunz LL, Bishop GA. CD40 and autoimmunity: The dark side of a great activator. Semin Immunol. 2009;21:293–300. doi: 10.1016/j.smim.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Croft M, Benedict CA, Ware CF. Clinical targeting of the TNF and TNFR superfamilies. Nat Rev Drug Discov. 2013;12:147–168. doi: 10.1038/nrd3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wagner DH., Jr Overlooked mechanisms in type 1 diabetes etiology: How unique costimulatory molecules contribute to diabetogenesis. Front Endocrinol. 2017;8:208. doi: 10.3389/fendo.2017.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Giuroiu I, Weber J. Novel checkpoints and cosignaling molecules in cancer immunotherapy. Cancer J. 2017;23:23–31. doi: 10.1097/PPO.0000000000000241. [DOI] [PubMed] [Google Scholar]
- 127.Croft M. The role of TNF superfamily members in T-cell function and diseases. Nat Rev Immunol. 2009;9:271–285. doi: 10.1038/nri2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Locksley RM, Killeen N, Lenardo MJ. The TNF and TNF receptor superfamilies: integrating mammalian biology. Cell. 2001;104:487–501. doi: 10.1016/s0092-8674(01)00237-9. [DOI] [PubMed] [Google Scholar]
- 129.Bodmer JL, Schneider P, Tschopp J. The molecular architecture of the TNF superfamily. Trends Biochem Sci. 2002;27:19–26. doi: 10.1016/s0968-0004(01)01995-8. [DOI] [PubMed] [Google Scholar]
- 130.Aggarwal BB. Signalling pathways of the TNF superfamily: a double-edged sword. Nat Rev Immunol. 2003;3:745–756. doi: 10.1038/nri1184. [DOI] [PubMed] [Google Scholar]
- 131.Bossen C, Ingold K, Tardivel A, Bodmer JL, Gaide O, Hertig S, Ambrose C, Tschopp J, Schneider P. Interactions of tumor necrosis factor (TNF) and TNF receptor family members in the mouse and human. J Biol Chem. 2006;281:13964–13971. doi: 10.1074/jbc.M601553200. [DOI] [PubMed] [Google Scholar]
- 132.Tansey MG, Szymkowski DE. The TNF superfamily in 2009: new pathways, new indications, and new drugs. Drug Discov Today. 2009;14:1082–1088. doi: 10.1016/j.drudis.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 133.Palladino MA, Bahjat FR, Theodorakis EA, Moldawer LL. Anti-TNF-alpha therapies: the next generation. Nat Rev Drug Discov. 2003;2:736–746. doi: 10.1038/nrd1175. [DOI] [PubMed] [Google Scholar]
- 134.Foy TM, Aruffo A, Bajorath J, Buhlmann JE, Noelle RJ. Immune regulation by CD40 and its ligand GP39. Annu Rev Immunol. 1996;14:591–617. doi: 10.1146/annurev.immunol.14.1.591. [DOI] [PubMed] [Google Scholar]
- 135.Graf D, Muller S, Korthauer U, van Kooten C, Weise C, Kroczek RA. A soluble form of TRAP (CD40 ligand) is rapidly released after T cell activation. Eur J Immunol. 1995;25:1749–1754. doi: 10.1002/eji.1830250639. [DOI] [PubMed] [Google Scholar]
- 136.Mazzei GJ, Edgerton MD, Losberger C, Lecoanet-Henchoz S, Graber P, Durandy A, Gauchat JF, Bernard A, Allet B, Bonnefoy JY. Recombinant soluble trimeric CD40 ligand is biologically active. J Biol Chem. 1995;270:7025–7028. doi: 10.1074/jbc.270.13.7025. [DOI] [PubMed] [Google Scholar]
- 137.Henn V, Slupsky JR, Grafe M, Anagnostopoulos I, Forster R, Muller-Berghaus G, Kroczek RA. CD40 ligand on activated platelets triggers an inflammatory reaction of endothelial cells. Nature. 1998;391:591–594. doi: 10.1038/35393. [DOI] [PubMed] [Google Scholar]
- 138.Pamukcu B, Lip GY, Snezhitskiy V, Shantsila E. The CD40-CD40L system in cardiovascular disease. Ann Med. 2011;43:331–340. doi: 10.3109/07853890.2010.546362. [DOI] [PubMed] [Google Scholar]
- 139.Chatzigeorgiou A, Seijkens T, Zarzycka B, Engel D, Poggi M, van den Berg S, van den Berg S, Soehnlein O, Winkels H, Beckers L, Lievens D, Driessen A, Kusters P, Biessen E, Garcia-Martin R, Klotzsche-von Ameln A, Gijbels M, Noelle R, Boon L, Hackeng T, Schulte KM, Xu A, Vriend G, Nabuurs S, Chung KJ, Willems van Dijk K, Rensen PC, Gerdes N, de Winther M, Block NL, Schally AV, Weber C, Bornstein SR, Nicolaes G, Chavakis T, Lutgens E. Blocking CD40-TRAF6 signaling is a therapeutic target in obesity-associated insulin resistance. Proc Natl Acad Sci US A. 2014;111:2686–2691. doi: 10.1073/pnas.1400419111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yi Z, Bishop GA. Regulatory role of CD40 in obesity-induced insulin resistance. Adipocyte. 2015;4:65–69. doi: 10.4161/adip.32214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.van den Berg SM, Seijkens TT, Kusters PJ, Zarzycka B, Beckers L, den Toom M, Gijbels MJ, Chatzigeorgiou A, Weber C, de Winther MP, Chavakis T, Nicolaes GA, Lutgens E. Blocking CD40-TRAF6 interactions by small-molecule inhibitor 6860766 ameliorates the complications of diet-induced obesity in mice. Int J Obes. 2015;39:782–790. doi: 10.1038/ijo.2014.198. [DOI] [PubMed] [Google Scholar]
- 142.Bagnati M, Ogunkolade BW, Marshall C, Tucci C, Hanna K, Jones TA, Bugliani M, Nedjai B, Caton PW, Kieswich J, Yaqoob MM, Ball GR, Marchetti P, Hitman GA, Turner MD. Glucolipotoxicity initiates pancreatic b-cell death through TNFR5/CD40-mediated STAT1 and NF-kB activation. Cell Death Dis. 2016;7:e2329. doi: 10.1038/cddis.2016.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Pinelli DF, Ford ML. Novel insights into anti-CD40/CD154 immunotherapy in transplant tolerance. Immunotherapy. 2015;7:399–410. doi: 10.2217/imt.15.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wagner DH, Jr, Vaitaitis G, Sanderson R, Poulin M, Dobbs C, Haskins K. Expression of CD40 identifies a unique pathogenic T cell population in type 1 diabetes. Proc Natl Acad Sci USA. 2002;99:3782–3787. doi: 10.1073/pnas.052247099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Vaitaitis GM, Waid DM, Yussman MG, Wagner DH., Jr CD40-mediated signalling influences trafficking, T-cell receptor expression and T-cell pathogenesis, in the NOD model of type 1 diabetes. Immunology. 2017;152:243–254. doi: 10.1111/imm.12761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.An HJ, Kim YJ, Song DH, Park BS, Kim HM, Lee JD, Paik SG, Lee JO, Lee H. Crystallographic and mutational analysis of the CD40-CD154 complex and its implications for receptor activation. J Biol Chem. 2011;286:11226–11235. doi: 10.1074/jbc.M110.208215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Karpusas M, Hsu YM, Wang JH, Thompson J, Lederman S, Chess L, Thomas D. 2 Å Crystal structure of an extracellular fragment of human CD40 ligand. Structure. 1995;3:1031–1039. doi: 10.1016/s0969-2126(01)00239-8. [DOI] [PubMed] [Google Scholar]
- 148.Singh J, Garber E, Van Vlijmen H, Karpusas M, Hsu YM, Zheng Z, Naismith JH, Thomas D. The role of polar interactions in the molecular recognition of CD40L with its receptor CD40. Protein Sci. 1998;7:1124–1135. doi: 10.1002/pro.5560070506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Bishop GA, Hostager BS. Signaling by CD40 and its mimics in B cell activation. Immunol Res. 2001;24:97–109. doi: 10.1385/IR:24:2:097. [DOI] [PubMed] [Google Scholar]
- 150.Elgueta R, Benson MJ, de Vries VC, Wasiuk A, Guo Y, Noelle RJ. Molecular mechanism and function of CD40/CD40L engagement in the immune system. Immunol Rev. 2009;229:152–172. doi: 10.1111/j.1600-065X.2009.00782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Bishop GA, Hostager BS. The CD40-CD154 interaction in B cell-T cell liaisons. Cytokine Growth Factor Rev. 2003;14:297–309. doi: 10.1016/s1359-6101(03)00024-8. [DOI] [PubMed] [Google Scholar]
- 152.Dempsey PW, Doyle SE, He JQ, Cheng G. The signaling adaptors and pathways activated by TNF superfamily. Cytokine Growth Factor Rev. 2003;14:193–209. doi: 10.1016/s1359-6101(03)00021-2. [DOI] [PubMed] [Google Scholar]
- 153.Chatzigeorgiou A, Lyberi M, Chatzilymperis G, Nezos A, Kamper E. CD40/CD40L signaling and its implication in health and disease. Biofactors. 2009;35:474–483. doi: 10.1002/biof.62. [DOI] [PubMed] [Google Scholar]
- 154.André P, Prasad KS, Denis CV, He M, Papalia JM, Hynes RO, Phillips DR, Wagner DD. CD40L stabilizes arterial thrombi by a b3 integrin-dependent mechanism. Nat Med. 2002;8:247–252. doi: 10.1038/nm0302-247. [DOI] [PubMed] [Google Scholar]
- 155.Wolf D, Hohmann JD, Wiedemann A, Bledzka K, Blankenbach H, Marchini T, Gutte K, Zeschky K, Bassler N, Hoppe N, Rodriguez AO, Herr N, Hilgendorf I, Stachon P, Willecke F, Duerschmied D, von zur Muhlen C, Soloviev DA, Zhang L, Bode C, Plow EF, Libby P, Peter K, Zirlik A. Binding of CD40L to Mac-1’s I-domain involves the EQLKKSKTL motif and mediates leukocyte recruitment and atherosclerosis--but does not affect immunity and thrombosis in mice. Circ Res. 2011;109:1269–1279. doi: 10.1161/CIRCRESAHA.111.247684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Burkly LC. CD40 pathway blockade as an approach to immunotherapy. Adv Exp Med Biol. 2001;489:135–152. doi: 10.1007/978-1-4615-1277-6_12. [DOI] [PubMed] [Google Scholar]
- 157.Quezada SA, Jarvinen LZ, Lind EF, Noelle RJ. CD40/CD154 interactions at the interface of tolerance and immunity. Annu Rev Immunol. 2004;22:307–328. doi: 10.1146/annurev.immunol.22.012703.104533. [DOI] [PubMed] [Google Scholar]
- 158.Daoussis D, Andonopoulos AP, Liossis SN. Targeting CD40L: a promising therapeutic approach. Clin Diagn Lab Immunol. 2004;11:635–641. doi: 10.1128/CDLI.11.4.635-641.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Goldwater R, Keirns J, Blahunka P, First R, Sawamoto T, Zhang W, Kowalski D, Kaibara A, Holman J. A phase 1, randomized ascending single-dose study of antagonist anti-human CD40 ASKP1240 in healthy subjects. Am J Transplant. 2013;13:1040–1046. doi: 10.1111/ajt.12082. [DOI] [PubMed] [Google Scholar]
- 160.Wada T, Nangaku M. A circulating permeability factor in focal segmental glomerulosclerosis: the hunt continues. Clin Kidney J. 2015;8:708–715. doi: 10.1093/ckj/sfv090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Fanale M, Assouline S, Kuruvilla J, Solal-Celigny P, Heo DS, Verhoef G, Corradini P, Abramson JS, Offner F, Engert A, Dyer MJ, Carreon D, Ewald B, Baeck J, Younes A, Freedman AS. Phase IA/II, multicentre, open-label study of the CD40 antagonistic monoclonal antibody lucatumumab in adult patients with advanced non-Hodgkin or Hodgkin lymphoma. Br J Haematol. 2014;164:258–265. doi: 10.1111/bjh.12630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.de Vos S, Forero-Torres A, Ansell SM, Kahl B, Cheson BD, Bartlett NL, Furman RR, Winter JN, Kaplan H, Timmerman J, Whiting NC, Drachman JG, Advani R. A phase II study of dacetuzumab (SGN-40) in patients with relapsed diffuse large B-cell lymphoma (DLBCL) and correlative analyses of patient-specific factors. J Hematol Oncol. 2014;7:44. doi: 10.1186/1756-8722-7-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Furman RR, Forero-Torres A, Shustov A, Drachman JG. A phase I study of dacetuzumab (SGN-40, a humanized anti-CD40 monoclonal antibody) in patients with chronic lymphocytic leukemia. Leuk Lymphoma. 2010;51:228–235. doi: 10.3109/10428190903440946. [DOI] [PubMed] [Google Scholar]
- 164.Chowdhury F, Johnson PW, Glennie MJ, Williams AP. Ex vivo assays of dendritic cell activation and cytokine profiles as predictors of in vivo effects in an anti-human CD40 monoclonal antibody ChiLob 7/4 phase I trial. Cancer Immunol Res. 2014;2:229–240. doi: 10.1158/2326-6066.CIR-13-0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Beatty GL, Torigian DA, Chiorean EG, Saboury B, Brothers A, Alavi A, Troxel AB, Sun W, Teitelbaum UR, Vonderheide RH, O’Dwyer PJ. A phase I study of an agonist CD40 monoclonal antibody (CP-870,893) in combination with gemcitabine in patients with advanced pancreatic ductal adenocarcinoma. Clin Cancer Res. 2013;19:6286–6295. doi: 10.1158/1078-0432.CCR-13-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Vonderheide RH, Flaherty KT, Khalil M, Stumacher MS, Bajor DL, Hutnick NA, Sullivan P, Mahany JJ, Gallagher M, Kramer A, Green SJ, O’Dwyer PJ, Running KL, Huhn RD, Antonia SJ. Clinical activity and immune modulation in cancer patients treated with CP-870,893, a novel CD40 agonist monoclonal antibody. J Clin Oncol. 2007;25:876–883. doi: 10.1200/JCO.2006.08.3311. [DOI] [PubMed] [Google Scholar]
- 167.Vonderheide RH, Glennie MJ. Agonistic CD40 antibodies and cancer therapy. Clin Cancer Res. 2013;19:1035–1043. doi: 10.1158/1078-0432.CCR-12-2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Hassan SB, Sorensen JF, Olsen BN, Pedersen AE. Anti-CD40-mediated cancer immunotherapy: an update of recent and ongoing clinical trials. Immunopharmacol Immunotoxicol. 2014;36:96–104. doi: 10.3109/08923973.2014.890626. [DOI] [PubMed] [Google Scholar]
- 169.Kawai T, Andrews D, Colvin RB, Sachs DH, Cosimi AB. Thromboembolic complications after treatment with monoclonal antibody against CD40 ligand. Nat Med. 2000;6:114. doi: 10.1038/72162. [DOI] [PubMed] [Google Scholar]
- 170.Boumpas DT, Furie R, Manzi S, Illei GG, Wallace DJ, Balow JE, Vaishnaw ABG Lupus Nephritis Trial Group. A short course of BG9588 (anti-CD40 ligand antibody) improves serologic activity and decreases hematuria in patients with proliferative lupus glomerulonephritis. Arthritis Rheum. 2003;48:719–727. doi: 10.1002/art.10856. [DOI] [PubMed] [Google Scholar]
- 171.Koyama I, Kawai T, Andrews D, Boskovic S, Nadazdin O, Wee SL, Sogawa H, Wu DL, Smith RN, Colvin RB, Sachs DH, Cosimi AB. Thrombophilia associated with anti-CD154 monoclonal antibody treatment and its prophylaxis in nonhuman primates. Transplantation. 2004;77:460–462. doi: 10.1097/01.TP.0000110291.29370.C0. [DOI] [PubMed] [Google Scholar]
- 172.Roth GA, Zuckermann A, Klepetko W, Wolner E, Ankersmit HJ, Moser B, Volf I. Thrombophilia associated with anti-CD154 monoclonal antibody treatment and its prophylaxis in nonhuman primates. Transplantation. 2004;78:1238–1239. doi: 10.1097/01.tp.0000135457.69220.5b. [DOI] [PubMed] [Google Scholar]
- 173.Couzin J. Drug discovery. Magnificent obsession Science. 2005;307:1712–1715. doi: 10.1126/science.307.5716.1712. [DOI] [PubMed] [Google Scholar]
- 174.Mirabet M, Barrabes JA, Quiroga A, Garcia-Dorado D. Platelet pro-aggregatory effects of CD40L monoclonal antibody. Mol Immunol. 2008;45:937–944. doi: 10.1016/j.molimm.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 175.Xie JH, Yamniuk AP, Borowski V, Kuhn R, Susulic V, Rex-Rabe S, Yang X, Zhou X, Zhang Y, Gillooly K, Brosius R, Ravishankar R, Waggie K, Mink K, Price L, Rehfuss R, Tamura J, An Y, Cheng L, Abramczyk B, Ignatovich O, Drew P, Grant S, Bryson JW, Suchard S, Salter-Cid L, Nadler S, Suri A. Engineering of a novel anti-CD40L domain antibody for treatment of autoimmune diseases. J Immunol. 2014;192:4083–4092. doi: 10.4049/jimmunol.1303239. [DOI] [PubMed] [Google Scholar]
- 176.Kim SC, Wakwe W, Higginbotham LB, Mathews DV, Breeden CP, Stephenson AC, Jenkins J, Strobert E, Price K, Price L, Kuhn R, Wang H, Yamniuk A, Suchard S, Farris AB, 3rd, Pearson TC, Larsen CP, Ford ML, Suri A, Nadler S, Adams AB. Fc-Silent anti-CD154 domain antibody effectively prevents nonhuman primate renal allograft rejection. Am J Transplant. 2017;17:1182–1192. doi: 10.1111/ajt.14197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Tocoian A, Buchan P, Kirby H, Soranson J, Zamacona M, Walley R, Mitchell N, Esfandiari E, Wagner F, Oliver R. First-in-human trial of the safety, pharmacokinetics and immunogenicity of a PEGylated anti-CD40L antibody fragment (CDP7657) in healthy individuals and patients with systemic lupus erythematosus. Lupus. 2015;24:1045–1056. doi: 10.1177/0961203315574558. [DOI] [PubMed] [Google Scholar]
- 178.Soldevilla MM, Villanueva H, Bendandi M, Inoges S, Lopez-Diaz de Cerio A, Pastor F. 2-fluoro-RNA oligonucleotide CD40 targeted aptamers for the control of B lymphoma and bone-marrow aplasia. Biomaterials. 2015;67:274–285. doi: 10.1016/j.biomaterials.2015.07.020. [DOI] [PubMed] [Google Scholar]
- 179.Nozari A, Berezovski MV. Aptamers for CD antigens: from cell profiling to activity modulation. Mol Ther Nucleic Acids. 2017;6:29–44. doi: 10.1016/j.omtn.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Zhou J, Rossi J. Aptamers as targeted therapeutics: current potential and challenges. Nat Rev Drug Discov. 2017;16:181–202. doi: 10.1038/nrd.2016.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Keefe AD, Pai S, Ellington A. Aptamers as therapeutics. Nat Rev Drug Discov. 2010;9:537–550. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Deambrosis I, Lamorte S, Giaretta F, Tei L, Biancone L, Bussolati B, Camussi G. Inhibition of CD40-CD154 costimulatory pathway by a cyclic peptide targeting CD154. J Mol Med. 2009;87:181–197. doi: 10.1007/s00109-008-0416-1. [DOI] [PubMed] [Google Scholar]
- 183.Vaitaitis GM, Olmstead MH, Waid DM, Carter JR, Wagner DH., Jr A CD40-targeted peptide controls and reverses type 1 diabetes in NOD mice. Diabetologia. 2014;57:2366–2373. doi: 10.1007/s00125-014-3342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Chen J, Song Y, Bojadzic D, Tamayo-Garcia A, Landin AM, Blomberg BB, Buchwald P. Small-molecule inhibitors of the CD40-CD40L costimulatory protein-protein interaction. J Med Chem. 2017;60:8906–8922. doi: 10.1021/acs.jmedchem.7b01154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Margolles-Clark E, Jacques-Silva MC, Ganesan L, Umland O, Kenyon NS, Ricordi C, Berggren PO, Buchwald P. Suramin inhibits the CD40–CD154 costimulatory interaction: a possible mechanism for immunosuppressive effects. Biochem Pharmacol. 2009;77:1236–1245. doi: 10.1016/j.bcp.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 186.Margolles-Clark E, Umland O, Kenyon NS, Ricordi C, Buchwald P. Small molecule costimulatory blockade: organic dye inhibitors of the CD40-CD154 interaction. J Mol Med. 2009;87:1133–1143. doi: 10.1007/s00109-009-0519-3. [DOI] [PubMed] [Google Scholar]
- 187.Buchwald P, Margolles-Clark E, Kenyon NS, Ricordi C. Organic dyes as small molecule protein–protein interaction inhibitors for the CD40–CD154 costimulatory interaction. J Mol Recognit. 2010;23:65–73. doi: 10.1002/jmr.969. [DOI] [PubMed] [Google Scholar]
- 188.Margolles-Clark E, Kenyon NS, Ricordi C, Buchwald P. Effective and specific inhibition of the CD40-CD154 costimulatory interaction by a naphthalenesulphonic acid derivative. Chem Biol Drug Des. 2010;76:305–313. doi: 10.1111/j.1747-0285.2010.01014.x. [DOI] [PubMed] [Google Scholar]
- 189.Ganesan L, Vidović D, Schürer SC, Buchwald P. Exploratory computational assessment of possible binding modes for small molecule inhibitors of the CD40-CD154 costimulatory interaction. Pharmazie. 2012;67:374–379. [PubMed] [Google Scholar]
- 190.Levine WG. Metabolism of azo dyes: implication for detoxication and activation. Drug Metab Rev. 1991;23:253–309. doi: 10.3109/03602539109029761. [DOI] [PubMed] [Google Scholar]
- 191.Feng J, Cerniglia CE, Chen H. Toxicological significance of azo dye metabolism by human intestinal microbiota. Front Biosci. 2012;4:568–586. doi: 10.2741/400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Silvian LF, Friedman JE, Strauch K, Cachero TG, Day ES, Qian F, Cunningham B, Fung A, Sun L, Shipps GW, Su L, Zheng Z, Kumaravel G, Whitty A. Small molecule inhibition of the TNF family cytokine CD40 ligand through a subunit fracture mechanism. ACS Chem Biol. 2011;6:636–647. doi: 10.1021/cb2000346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Zarzycka B, Seijkens T, Nabuurs SB, Ritschel T, Grommes J, Soehnlein O, Schrijver R, van Tiel CM, Hackeng TM, Weber C, Giehler F, Kieser A, Lutgens E, Vriend G, Nicolaes GA. Discovery of small molecule CD40-TRAF6 inhibitors. J Chem Inf Model. 2015;55:294–307. doi: 10.1021/ci500631e. [DOI] [PubMed] [Google Scholar]
- 194.Aarts SABM, Seijkens TTP, Kusters PJH, van der Pol SMA, Zarzycka B, Heijnen P, Beckers L, den Toom M, Gijbels MJJ, Boon L, Weber C, de Vries HE, Nicolaes GAF, Dijkstra CD, Kooij G, Lutgens E. Inhibition of CD40-TRAF6 interactions by the small molecule inhibitor 6877002 reduces neuroinflammation. J Neuroinflammation. 2017;14:105. doi: 10.1186/s12974-017-0875-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Li G, Diogo D, Wu D, Spoonamore J, Dancik V, Franke L, Kurreeman F, Rossin EJ, Duclos G, Hartland C, Zhou X, Li K, Liu J, De Jager PL, Siminovitch KA, Zhernakova A, Raychaudhuri S, Bowes J, Eyre S, Padyukov L, Gregersen PK, Worthington J, Gupta N, Clemons PA, Stahl E, Tolliday N, Plenge RM Rheumatoid Arthritis Consortium I. Human genetics in rheumatoid arthritis guides a high-throughput drug screen of the CD40 signaling pathway. PLoS Genet. 2013;9:e1003487. doi: 10.1371/journal.pgen.1003487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Paterson DJ, Jefferies WA, Green JR, Brandon MR, Corthesy P, Puklavec M, Williams AF. Antigens of activated rat T lymphocytes including a molecule of 50,000 Mr detected only on CD4 positive T blasts. Mol Immunol. 1987;24:1281–1290. doi: 10.1016/0161-5890(87)90122-2. [DOI] [PubMed] [Google Scholar]
- 197.Tanaka Y, Inoi T, Tozawa H, Yamamoto N, Hinuma Y. A glycoprotein antigen detected with new monoclonal antibodies on the surface of human lymphocytes infected with human T-cell leukemia virus type-I (HTLV-I) Int J Cancer. 1985;36:549–555. doi: 10.1002/ijc.2910360506. [DOI] [PubMed] [Google Scholar]
- 198.Miura S, Ohtani K, Numata N, Niki M, Ohbo K, Ina Y, Gojobori T, Tanaka Y, Tozawa H, Nakamura M, et al. Molecular cloning and characterization of a novel glycoprotein, gp34, that is specifically induced by the human T-cell leukemia virus type I transactivator p40tax. Mol Cell Biol. 1991;11:1313–1325. doi: 10.1128/mcb.11.3.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Croft M. Control of immunity by the TNFR-related molecule OX40 (CD134) Annu Rev Immunol. 2010;28:57–78. doi: 10.1146/annurev-immunol-030409-101243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Webb GJ, Hirschfield GM, Lane PJ. OX40, OX40L and autoimmunity: a comprehensive review. Clin Rev Allergy Immunol. 2016;50:312–332. doi: 10.1007/s12016-015-8498-3. [DOI] [PubMed] [Google Scholar]
- 201.Yan J, Gong J, Chen G, Liu P, Wang C, Yang P. Evaluation of serum soluble OX40 ligand as a prognostic indicator in acute coronary syndrome patients. Clin Chim Acta. 2010;411:1662–1665. doi: 10.1016/j.cca.2010.06.016. [DOI] [PubMed] [Google Scholar]
- 202.Laustsen JK, Rasmussen TK, Stengaard-Pedersen K, Horslev-Petersen K, Hetland ML, Ostergaard M, Junker P, Hvid M, Deleuran B. Soluble OX40L is associated with presence of autoantibodies in early rheumatoid arthritis. Arthritis Res Ther. 2014;16:474. doi: 10.1186/s13075-014-0474-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Salek-Ardakani S, Song J, Halteman BS, Jember AG, Akiba H, Yagita H, Croft M. OX40 (CD134) controls memory T helper 2 cells that drive lung inflammation. J Exp Med. 2003;198:315–324. doi: 10.1084/jem.20021937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Weinberg AD, Wegmann KW, Funatake C, Whitham RH. Blocking OX-40/OX-40 ligand interaction in vitro and in vivo leads to decreased T cell function and amelioration of experimental allergic encephalomyelitis. J Immunol. 1999;162:1818–1826. [PubMed] [Google Scholar]
- 205.Higgins LM, McDonald SA, Whittle N, Crockett N, Shields JG, MacDonald TT. Regulation of T cell activation in vitro and in vivo by targeting the OX40-OX40 ligand interaction: amelioration of ongoing inflammatory bowel disease with an OX40-IgG fusion protein, but not with an OX40 ligand-IgG fusion protein. J Immunol. 1999;162:486–493. [PubMed] [Google Scholar]
- 206.Akiba H, Miyahira Y, Atsuta M, Takeda K, Nohara C, Futagawa T, Matsuda H, Aoki T, Yagita H, Okumura K. Critical contribution of OX40 ligand to T helper cell type 2 differentiation in experimental leishmaniasis. J Exp Med. 2000;191:375–380. doi: 10.1084/jem.191.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Tsukada N, Akiba H, Kobata T, Aizawa Y, Yagita H, Okumura K. Blockade of CD134 (OX40)-CD134L interaction ameliorates lethal acute graft-versus-host disease in a murine model of allogeneic bone marrow transplantation. Blood. 2000;95:2434–2439. [PubMed] [Google Scholar]
- 208.Yoshioka T, Nakajima A, Akiba H, Ishiwata T, Asano G, Yoshino S, Yagita H, Okumura K. Contribution of OX40/OX40 ligand interaction to the pathogenesis of rheumatoid arthritis. Eur J Immunol. 2000;30:2815–2823. doi: 10.1002/1521-4141(200010)30:10<2815::AID-IMMU2815>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 209.Croft M, Duan W, Choi H, Eun SY, Madireddi S, Mehta A. TNF superfamily in inflammatory disease: translating basic insights. Trends Immunol. 2012;33:144–152. doi: 10.1016/j.it.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Baum PR, Gayle RB, 3rd, Ramsdell F, Srinivasan S, Sorensen RA, Watson ML, Seldin MF, Baker E, Sutherland GR, Clifford KN, et al. Molecular characterization of murine and human OX40/OX40 ligand systems: identification of a human OX40 ligand as the HTLV-1-regulated protein gp34. EMBO J. 1994;13:3992–4001. doi: 10.1002/j.1460-2075.1994.tb06715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Godfrey WR, Fagnoni FF, Harara MA, Buck D, Engleman EG. Identification of a human OX-40 ligand, a costimulator of CD4+ T cells with homology to tumor necrosis factor. J Exp Med. 1994;180:757–762. doi: 10.1084/jem.180.2.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Compaan DM, Hymowitz SG. The crystal structure of the costimulatory OX40-OX40L complex. Structure. 2006;14:1321–1330. doi: 10.1016/j.str.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 213.Ye H, Park YC, Kreishman M, Kieff E, Wu H. The structural basis for the recognition of diverse receptor sequences by TRAF2. Mol Cell. 1999;4:321–330. doi: 10.1016/s1097-2765(00)80334-2. [DOI] [PubMed] [Google Scholar]
- 214.Kawamata S, Hori T, Imura A, Takaori-Kondo A, Uchiyama T. Activation of OX40 signal transduction pathways leads to tumor necrosis factor receptor-associated factor (TRAF) 2- and TRAF5-mediated NF-kB activation. J Biol Chem. 1998;273:5808–5814. doi: 10.1074/jbc.273.10.5808. [DOI] [PubMed] [Google Scholar]
- 215.Arch RH, Thompson CB. 4-1BB and OX40 are members of a tumor necrosis factor (TNF)-nerve growth factor receptor subfamily that bind TNF receptor-associated factors and activate nuclear factor kB. Mol Cell Biol. 1998;18:558–565. doi: 10.1128/mcb.18.1.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Rogers PR, Song J, Gramaglia I, Killeen N, Croft M. OX40 promotes Bcl-xL and Bcl-2 expression and is essential for long-term survival of CD4 T cells. Immunity. 2001;15:445–455. doi: 10.1016/s1074-7613(01)00191-1. [DOI] [PubMed] [Google Scholar]
- 217.Song J, So T, Croft M. Activation of NF-kB1 by OX40 contributes to antigen-driven T cell expansion and survival. J Immunol. 2008;180:7240–7248. doi: 10.4049/jimmunol.180.11.7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.So T, Song J, Sugie K, Altman A, Croft M. Signals from OX40 regulate nuclear factor of activated T cells c1 and T cell helper 2 lineage commitment. Proc Natl Acad Sci US A. 2006;103:3740–3745. doi: 10.1073/pnas.0600205103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Croft M, So T, Duan W, Soroosh P. The significance of OX40 and OX40L to T-cell biology and immune disease. Immunol Rev. 2009;229:173–191. doi: 10.1111/j.1600-065X.2009.00766.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Buchan SL, Rogel A, Al-Shamkhani A. The immunobiology of CD27 and OX40 and their potential as targets for cancer immunotherapy. Blood. 2018;131:39–48. doi: 10.1182/blood-2017-07-741025. [DOI] [PubMed] [Google Scholar]
- 221.Gauvreau GM, Boulet LP, Cockcroft DW, FitzGerald JM, Mayers I, Carlsten C, Laviolette M, Killian KJ, Davis BE, Larche M, Kipling C, Dua B, Mosesova S, Putnam W, Zheng Y, Scheerens H, McClintock D, Matthews JG, O’Byrne PM. OX40L blockade and allergen-induced airway responses in subjects with mild asthma. Clin Exp Allergy. 2014;44:29–37. doi: 10.1111/cea.12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Croft M, Siegel RM. Beyond TNF: TNF superfamily cytokines as targets for the treatment of rheumatic diseases. Nat Rev Rheumatol. 2017;13:217–233. doi: 10.1038/nrrheum.2017.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Song Y, Margolles-Clark E, Bayer A, Buchwald P. Small-molecule modulators of the OX40–OX40L costimulatory protein–protein interaction. Br J Pharmacol. 2014;171:4955–4969. doi: 10.1111/bph.12819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Buchwald P. A three-parameter two-state model of receptor function that incorporates affinity, efficacy, and signal amplification. Pharmacol Res Perspect. 2017;5:e00311. doi: 10.1002/prp2.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Moore PA, Belvedere O, Orr A, Pieri K, LaFleur DW, Feng P, Soppet D, Charters M, Gentz R, Parmelee D, Li Y, Galperina O, Giri J, Roschke V, Nardelli B, Carrell J, Sosnovtseva S, Greenfield W, Ruben SM, Olsen HS, Fikes J, Hilbert DM. BLyS: member of the tumor necrosis factor family and B lymphocyte stimulator. Science. 1999;285:260–263. doi: 10.1126/science.285.5425.260. [DOI] [PubMed] [Google Scholar]
- 226.Schneider P, MacKay F, Steiner V, Hofmann K, Bodmer JL, Holler N, Ambrose C, Lawton P, Bixler S, Acha-Orbea H, Valmori D, Romero P, Werner-Favre C, Zubler RH, Browning JL, Tschopp J. BAFF, a novel ligand of the tumor necrosis factor family, stimulates B cell growth. J Exp Med. 1999;189:1747–1756. doi: 10.1084/jem.189.11.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Yu G, Boone T, Delaney J, Hawkins N, Kelley M, Ramakrishnan M, McCabe S, Qiu WR, Kornuc M, Xia XZ, Guo J, Stolina M, Boyle WJ, Sarosi I, Hsu H, Senaldi G, Theill LE. APRIL and TALL-I and receptors BCMA and TACI: system for regulating humoral immunity. Nat Immunol. 2000;1:252–256. doi: 10.1038/79802. [DOI] [PubMed] [Google Scholar]
- 228.Mackay F, Silveira PA, Brink R. B cells and the BAFF/APRIL axis: fast-forward on autoimmunity and signaling. Curr Opin Immunol. 2007;19:327–336. doi: 10.1016/j.coi.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 229.Mackay F, Schneider P. Cracking the BAFF code. Nat Rev Immunol. 2009;9:491–502. doi: 10.1038/nri2572. [DOI] [PubMed] [Google Scholar]
- 230.Mackay F, Leung H. The role of the BAFF/APRIL system on T cell function. Semin Immunol. 2006;18:284–289. doi: 10.1016/j.smim.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 231.Darce JR, Arendt BK, Wu X, Jelinek DF. Regulated expression of BAFF-binding receptors during human B cell differentiation. J Immunol. 2007;179:7276–7286. doi: 10.4049/jimmunol.179.11.7276. [DOI] [PubMed] [Google Scholar]
- 232.Ng LG, Sutherland AP, Newton R, Qian F, Cachero TG, Scott ML, Thompson JS, Wheway J, Chtanova T, Groom J, Sutton IJ, Xin C, Tangye SG, Kalled SL, Mackay F, Mackay CR. B cell-activating factor belonging to the TNF family (BAFF)-R is the principal BAFF receptor facilitating BAFF costimulation of circulating T and B cells. J Immunol. 2004;173:807–817. doi: 10.4049/jimmunol.173.2.807. [DOI] [PubMed] [Google Scholar]
- 233.Medema JP, Planelles-Carazo L, Hardenberg G, Hahne M. The uncertain glory of APRIL. Cell Death Differ. 2003;10:1121–1125. doi: 10.1038/sj.cdd.4401291. [DOI] [PubMed] [Google Scholar]
- 234.Dillon SR, Gross JA, Ansell SM, Novak AJ. An APRIL to remember: novel TNF ligands as therapeutic targets. Nat Rev Drug Discov. 2006;5:235–246. doi: 10.1038/nrd1982. [DOI] [PubMed] [Google Scholar]
- 235.Bossen C, Cachero TG, Tardivel A, Ingold K, Willen L, Dobles M, Scott ML, Maquelin A, Belnoue E, Siegrist CA, Chevrier S, Acha-Orbea H, Leung H, Mackay F, Tschopp J, Schneider P. TACI, unlike BAFF-R, is solely activated by oligomeric BAFF and APRIL to support survival of activated B cells and plasmablasts. Blood. 2008;111:1004–1012. doi: 10.1182/blood-2007-09-110874. [DOI] [PubMed] [Google Scholar]
- 236.Day ES, Cachero TG, Qian F, Sun Y, Wen D, Pelletier M, Hsu YM, Whitty A. Selectivity of BAFF/BLyS and APRIL for binding to the TNF family receptors BAFFR/BR3 and BCMA. Biochemistry. 2005;44:1919–1931. doi: 10.1021/bi048227k. [DOI] [PubMed] [Google Scholar]
- 237.Reijmers RM, Groen RW, Kuil A, Weijer K, Kimberley FC, Medema JP, van Kuppevelt TH, Li JP, Spaargaren M, Pals ST. Disruption of heparan sulfate proteoglycan conformation perturbs B-cell maturation and APRIL-mediated plasma cell survival. Blood. 2011;117:6162–6171. doi: 10.1182/blood-2010-12-325522. [DOI] [PubMed] [Google Scholar]
- 238.Ingold K, Zumsteg A, Tardivel A, Huard B, Steiner QG, Cachero TG, Qiang F, Gorelik L, Kalled SL, Acha-Orbea H, Rennert PD, Tschopp J, Schneider P. Identification of proteoglycans as the APRIL-specific binding partners. J Exp Med. 2005;201:1375–1383. doi: 10.1084/jem.20042309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Mackay F, Schneider P. TACI, an enigmatic BAFF/APRIL receptor, with new unappreciated biochemical and biological properties. Cytokine Growth Factor Rev. 2008;19:263–276. doi: 10.1016/j.cytogfr.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 240.Ryan MC, Grewal IS. Targeting of BAFF and APRIL for autoimmunity and oncology. Adv Exp Med Biol. 2009;647:52–63. doi: 10.1007/978-0-387-89520-8_4. [DOI] [PubMed] [Google Scholar]
- 241.Liu Z, Davidson A. BAFF inhibition: a new class of drugs for the treatment of autoimmunity. Exp Cell Res. 2011;317:1270–1277. doi: 10.1016/j.yexcr.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Vincent FB, Saulep-Easton D, Figgett WA, Fairfax KA, Mackay F. The BAFF/APRIL system: emerging functions beyond B cell biology and autoimmunity. Cytokine Growth Factor Rev. 2013;24:203–215. doi: 10.1016/j.cytogfr.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Zekavat G, Rostami SY, Badkerhanian A, Parsons RF, Koeberlein B, Yu M, Ward CD, Migone TS, Yu L, Eisenbarth GS, Cancro MP, Naji A, Noorchashm H. In vivo BLyS/BAFF neutralization ameliorates islet-directed autoimmunity in nonobese diabetic mice. J Immunol. 2008;181:8133–8144. doi: 10.4049/jimmunol.181.11.8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Marino E, Walters SN, Villanueva JE, Richards JL, Mackay CR, Grey ST. BAFF regulates activation of self-reactive T cells through B-cell dependent mechanisms and mediates protection in NOD mice. Eur J Immunol. 2014;44:983–993. doi: 10.1002/eji.201344186. [DOI] [PubMed] [Google Scholar]
- 245.Wang Q, Racine JJ, Ratiu JJ, Wang S, Ettinger R, Wasserfall C, Atkinson MA, Serreze DV. Transient BAFF blockade inhibits type 1 diabetes development in nonobese diabetic mice by enriching immunoregulatory B lymphocytes sensitive to deletion by Anti-CD20 cotherapy. J Immunol. 2017;199:3757–3770. doi: 10.4049/jimmunol.1700822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Shen L, Chng MH, Alonso MN, Yuan R, Winer DA, Engleman EG. B-1a lymphocytes attenuate insulin resistance. Diabetes. 2015;64:593–603. doi: 10.2337/db14-0554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Kim B, Do MS, Hyun CK. B-cell-activating factor deficiency attenuates high-fat diet-induced glucose intolerance by potentiating adipose tissue function. Biochem Biophys Res Commun. 2015 doi: 10.1016/j.bbrc.2015.07.099. [DOI] [PubMed] [Google Scholar]
- 248.Karpusas M, Cachero TG, Qian F, Boriack-Sjodin A, Mullen C, Strauch K, Hsu YM, Kalled SL. Crystal structure of extracellular human BAFF, a TNF family member that stimulates B lymphocytes. J Mol Biol. 2002;315:1145–1154. doi: 10.1006/jmbi.2001.5296. [DOI] [PubMed] [Google Scholar]
- 249.Liu Y, Xu L, Opalka N, Kappler J, Shu HB, Zhang G. Crystal structure of sTALL-1 reveals a virus-like assembly of TNF family ligands. Cell. 2002;108:383–394. doi: 10.1016/s0092-8674(02)00631-1. [DOI] [PubMed] [Google Scholar]
- 250.Kim HM, Yu KS, Lee ME, Shin DR, Kim YS, Paik SG, Yoo OJ, Lee H, Lee JO. Crystal structure of the BAFF-BAFF-R complex and its implications for receptor activation. Nat Struct Biol. 2003;10:342–348. doi: 10.1038/nsb925. [DOI] [PubMed] [Google Scholar]
- 251.Bossen C, Schneider P. BAFF, APRIL and their receptors: structure, function and signaling. Semin Immunol. 2006;18:263–275. doi: 10.1016/j.smim.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 252.Kayagaki N, Yan M, Seshasayee D, Wang H, Lee W, French DM, Grewal IS, Cochran AG, Gordon NC, Yin J, Starovasnik MA, Dixit VM. BAFF/BLyS receptor 3 binds the B cell survival factor BAFF ligand through a discrete surface loop and promotes processing of NF-kB2. Immunity. 2002;17:515–524. doi: 10.1016/s1074-7613(02)00425-9. [DOI] [PubMed] [Google Scholar]
- 253.Hymowitz SG, Patel DR, Wallweber HJ, Runyon S, Yan M, Yin J, Shriver SK, Gordon NC, Pan B, Skelton NJ, Kelley RF, Starovasnik MA. Structures of APRIL-receptor complexes: like BCMA, TACI employs only a single cysteine-rich domain for high affinity ligand binding. J Biol Chem. 2005;280:7218–7227. doi: 10.1074/jbc.M411714200. [DOI] [PubMed] [Google Scholar]
- 254.Day ES, Cote SM, Whitty A. Binding efficiency of protein-protein complexes. Biochemistry. 2012;51:9124–9136. doi: 10.1021/bi301039t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Gardam S, Brink R. Non-canonical NF-kB signaling initiated by BAFF influences B cell biology at multiple junctures. Front Immunol. 2014;4:509. doi: 10.3389/fimmu.2013.00509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Morrison MD, Reiley W, Zhang M, Sun SC. An atypical tumor necrosis factor (TNF) receptor-associated factor-binding motif of B cell-activating factor belonging to the TNF family (BAFF) receptor mediates induction of the noncanonical NF-kB signaling pathway. J Biol Chem. 2005;280:10018–10024. doi: 10.1074/jbc.M413634200. [DOI] [PubMed] [Google Scholar]
- 257.Liao G, Zhang M, Harhaj EW, Sun SC. Regulation of the NF-kB-inducing kinase by tumor necrosis factor receptor-associated factor 3-induced degradation. J Biol Chem. 2004;279:26243–26250. doi: 10.1074/jbc.M403286200. [DOI] [PubMed] [Google Scholar]
- 258.Schweighoffer E, Vanes L, Nys J, Cantrell D, McCleary S, Smithers N, Tybulewicz VL. The BAFF receptor transduces survival signals by co-opting the B cell receptor signaling pathway. Immunity. 2013;38:475–488. doi: 10.1016/j.immuni.2012.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Do RK, Hatada E, Lee H, Tourigny MR, Hilbert D, Chen-Kiang S. Attenuation of apoptosis underlies B lymphocyte stimulator enhancement of humoral immune response. J Exp Med. 2000;192:953–964. doi: 10.1084/jem.192.7.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Batten M, Groom J, Cachero TG, Qian F, Schneider P, Tschopp J, Browning JL, Mackay F. BAFF mediates survival of peripheral immature B lymphocytes. J Exp Med. 2000;192:1453–1466. doi: 10.1084/jem.192.10.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Stohl W, Hilbert DM. The discovery and development of belimumab: the anti-BLyS-lupus connection. Nat Biotechnol. 2012;30:69–77. doi: 10.1038/nbt.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Bishton M, Spencer A, Dickinson M, Ritchie D. A single-arm, phase II study of the anti-Blys monoclonal antibody belimumab in symptomatic Waldenstrom macroglobulinemia. Clin Lymphoma Myeloma Leuk. 2013;13:575–578. doi: 10.1016/j.clml.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 263.Stohl W, Merrill JT, McKay JD, Lisse JR, Zhong ZJ, Freimuth WW, Genovese MC. Efficacy and safety of belimumab in patients with rheumatoid arthritis: a phase II, randomized, double-blind, placebo-controlled, dose-ranging study. J Rheumatol. 2013;40:579–589. doi: 10.3899/jrheum.120886. [DOI] [PubMed] [Google Scholar]
- 264.Rossi JF. Phase I study of atacicept in relapsed/refractory multiple myeloma (MM) and Waldenstrom’s macroglobulinemia. Clin Lymphoma Myeloma Leuk. 2011;11:136–138. doi: 10.3816/CLML.2011.n.031. [DOI] [PubMed] [Google Scholar]
- 265.Cogollo E, Silva MA, Isenberg D. Profile of atacicept and its potential in the treatment of systemic lupus erythematosus. Drug Des Devel Ther. 2015;9:1331–1339. doi: 10.2147/DDDT.S71276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.van Vollenhoven RF, Wax S, Li Y, Tak PP. Safety and efficacy of atacicept in combination with rituximab for reducing the signs and symptoms of rheumatoid arthritis: a phase II, randomized, double-blind, placebo-controlled pilot trial. Arthritis Rheumatol. 2015;67:2828–2836. doi: 10.1002/art.39262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Stohl W, Merrill JT, Looney RJ, Buyon J, Wallace DJ, Weisman MH, Ginzler EM, Cooke B, Holloway D, Kaliyaperumal A, Kuchimanchi KR, Cheah TC, Rasmussen E, Ferbas J, Belouski SS, Tsuji W, Zack DJ. Treatment of systemic lupus erythematosus patients with the BAFF antagonist “peptibody” blisibimod (AMG 623/A-623): results from randomized, double-blind phase 1a and phase 1b trials. Arthritis Res Ther. 2015;17:215. doi: 10.1186/s13075-015-0741-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Hasler J, Flajnik MF, Williams G, Walsh FS, Rutkowski JL. VNAR single-domain antibodies specific for BAFF inhibit B cell development by molecular mimicry. Mol Immunol. 2016;75:28–37. doi: 10.1016/j.molimm.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Zhou J, Tiemann K, Chomchan P, Alluin J, Swiderski P, Burnett J, Zhang X, Forman S, Chen R, Rossi J. Dual functional BAFF receptor aptamers inhibit ligand-induced proliferation and deliver siRNAs to NHL cells. Nucleic Acids Res. 2013;41:4266–4283. doi: 10.1093/nar/gkt125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Moon EY, Yi KY, Lee S. An increase in B cell apoptosis by interfering BAFF-BAFF-R interaction with small synthetic molecules. Int Immunopharmacol. 2011;11:1523–1533. doi: 10.1016/j.intimp.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 271.Lee GH, Oh JM, Kim HS, Yoon WK, Yi KY, Yang Y, Han SH, Lee S, Moon EY. KR33426, [2-(2,5-dichlorophenyl)-5-methyloxazol-4yl]carbonylguanidine, is a novel compound to be effective on mouse systemic lupus erythematosus. Eur J Pharmacol. 2011;668:459–466. doi: 10.1016/j.ejphar.2011.07.026. [DOI] [PubMed] [Google Scholar]
- 272.Harris NL, Ronchese F. The role of B7 costimulation in T-cell immunity. Immunol Cell Biol. 1999;77:304–11. doi: 10.1046/j.1440-1711.1999.00835.x. [DOI] [PubMed] [Google Scholar]
- 273.Beyersdorf N, Kerkau T, Hunig T. CD28 co-stimulation in T-cell homeostasis: a recent perspective. Immunotargets Ther. 2015;4:111–22. doi: 10.2147/ITT.S61647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Umlauf SW, Beverly B, Lantz O, Schwartz RH. Regulation of interleukin 2 gene expression by CD28 costimulation in mouse T-cell clones: both nuclear and cytoplasmic RNAs are regulated with complex kinetics. Mol Cell Biol. 1995;15:3197–205. doi: 10.1128/mcb.15.6.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Krummel MF, Sullivan TJ, Allison JP. Superantigen responses and co-stimulation: CD28 and CTLA-4 have opposing effects on T cell expansion in vitro and in vivo. Int Immunol. 1996;8:519–23. doi: 10.1093/intimm/8.4.519. [DOI] [PubMed] [Google Scholar]
- 276.Dilek N, Poirier N, Hulin P, Coulon F, Mary C, Ville S, Vie H, Clemenceau B, Blancho G, Vanhove B. Targeting CD28, CTLA-4 and PD-L1 costimulation differentially controls immune synapses and function of human regulatory and conventional T-cells. PLoS One. 2013;8:e83139. doi: 10.1371/journal.pone.0083139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Truneh A, Reddy M, Ryan P, Lyn SD, Eichman C, Couez D, Hurle MR, Sekaly RP, Olive D, Sweet R. Differential recognition by CD28 of its cognate counter receptors CD80 (B7) and B70 (B7.2): analysis by site directed mutagenesis. Mol Immunol. 1996;33:321–334. doi: 10.1016/0161-5890(95)00077-1. [DOI] [PubMed] [Google Scholar]
- 278.Stamper CC, Zhang Y, Tobin JF, Erbe DV, Ikemizu S, Davis SJ, Stahl ML, Seehra J, Somers WS, Mosyak L. Crystal structure of the B7-1/CTLA-4 complex that inhibits human immune responses. Nature. 2001;410:608–611. doi: 10.1038/35069118. [DOI] [PubMed] [Google Scholar]
- 279.Morton PA, Fu XT, Stewart JA, Giacoletto KS, White SL, Leysath CE, Evans RJ, Shieh JJ, Karr RW. Differential effects of CTLA-4 substitutions on the binding of human CD80 (B7-1) and CD86 (B7-2) J Immunol. 1996;156:1047–1054. [PubMed] [Google Scholar]
- 280.Sorensen P, Kussmann M, Rosen A, Bennett KL, da Thrige DG, Uvebrant K, Walse B, Roepstorff P, Bjork P. Identification of protein-protein interfaces implicated in CD80-CD28 costimulatory signaling. J Immunol. 2004;172:6803–6809. doi: 10.4049/jimmunol.172.11.6803. [DOI] [PubMed] [Google Scholar]
- 281.Bajorath J, Peach RJ, Linsley PS. Immunoglobulin fold characteristics of B7-1 (CD80) and B7-2 (CD86) Protein Sci. 1994;3:2148–2150. doi: 10.1002/pro.5560031128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Peach RJ, Bajorath J, Naemura J, Leytze G, Greene J, Aruffo A, Linsley PS. Both extracellular immunoglobin-like domains of CD80 contain residues critical for binding T cell surface receptors CTLA-4 and CD28. J Biol Chem. 1995;270:21181–21187. doi: 10.1074/jbc.270.36.21181. [DOI] [PubMed] [Google Scholar]
- 283.Girard T, Gaucher D, El-Far M, Breton G, Sekaly RP. CD80 and CD86 IgC domains are important for quaternary structure, receptor binding and co-signaling function. Immunol Lett. 2014;161:65–75. doi: 10.1016/j.imlet.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 284.Prasad KV, Cai YC, Raab M, Duckworth B, Cantley L, Shoelson SE, Rudd CE. T-cell antigen CD28 interacts with the lipid kinase phosphatidylinositol 3-kinase by a cytoplasmic Tyr(P)-Met-Xaa-Met motif. Proc Natl Acad Sci US A. 1994;91:2834–2838. doi: 10.1073/pnas.91.7.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Schneider H, Cai YC, Prasad KV, Shoelson SE, Rudd CE. T cell antigen CD28 binds to the GRB-2/SOS complex, regulators of p21ras. Eur J Immunol. 1995;25:1044–1050. doi: 10.1002/eji.1830250428. [DOI] [PubMed] [Google Scholar]
- 286.Boise LH, Minn AJ, Noel PJ, June CH, Accavitti MA, Lindsten T, Thompson CB. CD28 costimulation can promote T cell survival by enhancing the expression of Bcl-xL. Immunity. 1995;3:87–98. doi: 10.1016/1074-7613(95)90161-2. [DOI] [PubMed] [Google Scholar]; J Immunol. 2010;185:3788–99. [PubMed] [Google Scholar]
- 287.Frauwirth KA, Riley JL, Harris MH, Parry RV, Rathmell JC, Plas DR, Elstrom RL, June CH, Thompson CB. The CD28 signaling pathway regulates glucose metabolism. Immunity. 2002;16:769–777. doi: 10.1016/s1074-7613(02)00323-0. [DOI] [PubMed] [Google Scholar]
- 288.Miller J, Baker C, Cook K, Graf B, Sanchez-Lockhart M, Sharp K, Wang X, Yang B, Yoshida T. Two pathways of costimulation through CD28. Immunol Res. 2009;45:159–172. doi: 10.1007/s12026-009-8097-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Pentcheva-Hoang T, Egen JG, Wojnoonski K, Allison JP. B7-1 and B7-2 selectively recruit CTLA-4 and CD28 to the immunological synapse. Immunity. 2004;21:401–413. doi: 10.1016/j.immuni.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 290.Teft WA, Chau TA, Madrenas J. Structure-function analysis of the CTLA-4 interaction with PP2A. BMC Immunol. 2009;10:23. doi: 10.1186/1471-2172-10-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Gardner D, Jeffery LE, Sansom DM. Understanding the CD28/CTLA-4 (CD152) pathway and its implications for costimulatory blockade. Am J Transplant. 2014;14:1985–1891. doi: 10.1111/ajt.12834. [DOI] [PubMed] [Google Scholar]
- 292.Qureshi OS, Zheng Y, Nakamura K, Attridge K, Manzotti C, Schmidt EM, Baker J, Jeffery LE, Kaur S, Briggs Z, Hou TZ, Futter CE, Anderson G, Walker LS, Sansom DM. Trans-endocytosis of CD80 and CD86: a molecular basis for the cell-extrinsic function of CTLA-4. Science. 2011;332:600–603. doi: 10.1126/science.1202947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Hoos A. Development of immuno-oncology drugs - from CTLA4 to PD1 to the next generations. Nat Rev Drug Discov. 2016;15:235–247. doi: 10.1038/nrd.2015.35. [DOI] [PubMed] [Google Scholar]
- 294.Khalil DN, Smith EL, Brentjens RJ, Wolchok JD. The future of cancer treatment: immunomodulation, CARs and combination immunotherapy. Nat Rev Clin Oncol. 2016;13:273–290. doi: 10.1038/nrclinonc.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Sharma P, Allison JP. The future of immune checkpoint therapy. Science. 2015;348:56–61. doi: 10.1126/science.aaa8172. [DOI] [PubMed] [Google Scholar]
- 296.Ikeda K, Sanayama Y, Makita S, Hosokawa J, Yamagata M, Nakagomi D, Takabayashi K, Nakajima H. Efficacy of abatacept for arthritis in patients with an overlap syndrome between rheumatoid arthritis and systemic lupus erythematosus. Clin Dev Immunol. 2013;2013:697525. doi: 10.1155/2013/697525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Dyson JK, Hirschfield GM, Adams DH, Beuers U, Mann DA, Lindor KD, Jones DE. Novel therapeutic targets in primary biliary cirrhosis. Nat Rev Gastroenterol Hepatol. 2015;12:147–158. doi: 10.1038/nrgastro.2015.12. [DOI] [PubMed] [Google Scholar]
- 298.Crepeau RL, Ford ML. Challenges and opportunities in targeting the CD28/CTLA-4 pathway in transplantation and autoimmunity. Expert Opin Biol Ther. 2017;17:1001–1012. doi: 10.1080/14712598.2017.1333595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Bernett MJ, Chu SY, Leung I, Moore GL, Lee SH, Pong E, Chen H, Phung S, Muchhal US, Horton HM, Lazar GA, Desjarlais JR, Szymkowski DE. Immune suppression in cynomolgus monkeys by XPro9523: an improved CTLA4-Ig fusion with enhanced binding to CD80, CD86 and neonatal Fc receptor FcRn. MAbs. 2013;5:384–396. doi: 10.4161/mabs.23976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Poirier N, Dilek N, Mary C, Ville S, Coulon F, Branchereau J, Tillou X, Charpy V, Pengam S, Nerriere-Daguin V, Hervouet J, Minault D, Le Bas-Bernardet S, Renaudin K, Vanhove B, Blancho G. FR104, an antagonist anti-CD28 monovalent fab’ antibody, prevents alloimmunization and allows calcineurin inhibitor minimization in nonhuman primate renal allograft. Am J Transplant. 2015;15:88–100. doi: 10.1111/ajt.12964. [DOI] [PubMed] [Google Scholar]
- 301.Jenh CH, Zhang M, Wiekowski M, Tan JC, Fan XD, Hegde V, Patel M, Bryant R, Narula SK, Zavodny PJ, Chou CC. Development of a CD28 receptor binding-based screen and identification of a biologically active inhibitor. Anal Biochem. 1998;256:47–55. doi: 10.1006/abio.1997.2495. [DOI] [PubMed] [Google Scholar]
- 302.Fine JS, Macosko HD, Justice L, Chou CC, Jenh CH, Narula SK, Zavodny PJ. An inhibitor of CD28-CD80 interactions impairs CD28-mediated costimulation of human CD4 T cells. Cell Immunol. 1999;191:49–59. doi: 10.1006/cimm.1998.1403. [DOI] [PubMed] [Google Scholar]
- 303.Erbe DV, Wang S, Xing Y, Tobin JF. Small molecule ligands define a binding site on the immune regulatory protein B7. J Biol Chem. 2002;277:7363–7368. doi: 10.1074/jbc.M110162200. [DOI] [PubMed] [Google Scholar]
- 304.Uvebrant K, da Graca Thrige D, Rosén A, Åkesson M, Berg H, Walse B, Björk P. Discovery of selective small-molecule CD80 inhibitors. J Biomol Screen. 2007;12:464–472. doi: 10.1177/1087057107300464. [DOI] [PubMed] [Google Scholar]
- 305.Huxley P, Sutton DH, Debnam P, Matthews IR, Brewer JE, Rose J, Trickett M, Williams DD, Andersen TB, Classon BJ. High-affinity small molecule inhibitors of T cell costimulation: compounds for immunotherapy. Chem Biol. 2004;11:1651–1658. doi: 10.1016/j.chembiol.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 306.Heninger AK, Wentrup S, Al-Saeedi M, Schiessling S, Giese T, Wartha F, Meuer S, Schroder-Braunstein J. Immunomodulation of human intestinal T cells by the synthetic CD80 antagonist RhuDex. Immun Inflamm Dis. 2014;2:166–180. doi: 10.1002/iid3.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Doesch AO, Zhao L, Gleissner CA, Akhavanpoor M, Rohde D, Okuyucu D, Hakimi M, Dengler TJ, Katus HA, Erbel C. Inhibition of B7-1 (CD80) by RhuDex(R) reduces lipopolysaccharide-mediated inflammation in human atherosclerotic lesions. Drug Des Devel Ther. 2014;8:447–457. doi: 10.2147/DDDT.S59594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Haanstra KG, Endell J, Estevao D, Kondova I, Jonker M. Blocking T cell co-stimulation using a CD80 blocking small molecule reduces delayed type hypersensitivity responses in rhesus monkeys. Clin Exp Immunol. 2009;158:91–98. doi: 10.1111/j.1365-2249.2009.03994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Keir ME, Butte MJ, Freeman GJ, Sharpe AH. PD-1 and its ligands in tolerance and immunity. Annu Rev Immunol. 2008;26:677–704. doi: 10.1146/annurev.immunol.26.021607.090331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Wan B, Nie H, Liu A, Feng G, He D, Xu R, Zhang Q, Dong C, Zhang JZ. Aberrant regulation of synovial T cell activation by soluble costimulatory molecules in rheumatoid arthritis. J Immunol. 2006;177:8844–8850. doi: 10.4049/jimmunol.177.12.8844. [DOI] [PubMed] [Google Scholar]
- 311.Mozaffarian N, Wiedeman AE, Stevens AM. Active systemic lupus erythematosus is associated with failure of antigen-presenting cells to express programmed death ligand-1. Rheumatology. 2008;47:1335–1341. doi: 10.1093/rheumatology/ken256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Kobayashi M, Kawano S, Hatachi S, Kurimoto C, Okazaki T, Iwai Y, Honjo T, Tanaka Y, Minato N, Komori T, Maeda S, Kumagai S. Enhanced expression of programmed death-1 (PD-1)/PD-L1 in salivary glands of patients with Sjogren’s syndrome. J Rheumatol. 2005;32:2156–2163. [PubMed] [Google Scholar]
- 313.Hatachi S, Iwai Y, Kawano S, Morinobu S, Kobayashi M, Koshiba M, Saura R, Kurosaka M, Honjo T, Kumagai S. CD4+ PD-1+ T cells accumulate as unique anergic cells in rheumatoid arthritis synovial fluid. J Rheumatol. 2003;30:1410–1419. [PubMed] [Google Scholar]
- 314.Ansari MJ, Salama AD, Chitnis T, Smith RN, Yagita H, Akiba H, Yamazaki T, Azuma M, Iwai H, Khoury SJ, Auchincloss H, Jr, Sayegh MH. The programmed death-1 (PD-1) pathway regulates autoimmune diabetes in nonobese diabetic (NOD) mice. J Exp Med. 2003;198:63–69. doi: 10.1084/jem.20022125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Zhou J, Jin JO, Kawai T, Yu Q. Endogenous programmed death ligand-1 restrains the development and onset of Sjogren’s syndrome in non-obese diabetic mice. Sci Rep. 2016;6:39105. doi: 10.1038/srep39105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Thompson RH, Gillett MD, Cheville JC, Lohse CM, Dong H, Webster WS, Krejci KG, Lobo JR, Sengupta S, Chen L, Zincke H, Blute ML, Strome SE, Leibovich BC, Kwon ED. Costimulatory B7-H1 in renal cell carcinoma patients: Indicator of tumor aggressiveness and potential therapeutic target. Proc Natl Acad Sci US A. 2004;101:17174–17179. doi: 10.1073/pnas.0406351101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Konstantinidou M, Zarganes-Tzitzikas T, Magiera K, Holak TA, Dömling A. Immune checkpoint PD-1/PD-L1: is there life beyond antibodies? Angew Chem Int Ed Engl. 2018;57:ePub. doi: 10.1002/anie.201710407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Zhan MM, Hu XQ, Liu XX, Ruan BF, Xu J, Liao C. From monoclonal antibodies to small molecules: the development of inhibitors targeting the PD-1/PD-L1 pathway. Drug Discov Today. 2016;21:1027–1036. doi: 10.1016/j.drudis.2016.04.011. [DOI] [PubMed] [Google Scholar]
- 319.Li Y, Li F, Jiang F, Lv X, Zhang R, Lu A, Zhang G. A mini-review for cancer immunotherapy: molecular understanding of PD-1/PD-L1 pathway & translational blockade of immune checkpoints. Int J Mol Sci. 2016;17:1151. doi: 10.3390/ijms17071151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Naidoo J, Page DB, Li BT, Connell LC, Schindler K, Lacouture ME, Postow MA, Wolchok JD. Toxicities of the anti-PD-1 and anti-PD-L1 immune checkpoint antibodies. Ann Oncol. 2016;27:1362. doi: 10.1093/annonc/mdw141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Ohaegbulam KC, Assal A, Lazar-Molnar E, Yao Y, Zang X. Human cancer immunotherapy with antibodies to the PD-1 and PD-L1 pathway. Trends Mol Med. 2015;21:24–33. doi: 10.1016/j.molmed.2014.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Okazaki T, Honjo T. PD-1 and PD-1 ligands: from discovery to clinical application. Int Immunol. 2007;19:813–824. doi: 10.1093/intimm/dxm057. [DOI] [PubMed] [Google Scholar]
- 323.Zhang X, Schwartz JC, Guo X, Bhatia S, Cao E, Lorenz M, Cammer M, Chen L, Zhang ZY, Edidin MA, Nathenson SG, Almo SC. Structural and functional analysis of the costimulatory receptor programmed death-1. Immunity. 2004;20:337–347. doi: 10.1016/s1074-7613(04)00051-2. [DOI] [PubMed] [Google Scholar]
- 324.Butte MJ, Pena-Cruz V, Kim MJ, Freeman GJ, Sharpe AH. Interaction of human PD-L1 and B7-1. Mol Immunol. 2008;45:3567–3572. doi: 10.1016/j.molimm.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Zak KM, Kitel R, Przetocka S, Golik P, Guzik K, Musielak B, Domling A, Dubin G, Holak TA. Structure of the complex of human programmed death 1, PD-1, and its ligand PD-L1. Structure. 2015;23:2341–2348. doi: 10.1016/j.str.2015.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Zak KM, Kitel R, Przetocka S, Golik P, Guzik K, Musielak B, Domling A, Dubin G, Holak TA. Structure of the Complex of Human Programmed Death 1, PD-1, and Its Ligand PD-L1. Structure. 2015;23:2341–8. doi: 10.1016/j.str.2015.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Sheppard KA, Fitz LJ, Lee JM, Benander C, George JA, Wooters J, Qiu Y, Jussif JM, Carter LL, Wood CR, Chaudhary D. PD-1 inhibits T-cell receptor induced phosphorylation of the ZAP70/CD3zeta signalosome and downstream signaling to PKCtheta. FEBS Lett. 2004;574:37–41. doi: 10.1016/j.febslet.2004.07.083. [DOI] [PubMed] [Google Scholar]
- 328.Chemnitz JM, Parry RV, Nichols KE, June CH, Riley JL. SHP-1 and SHP-2 associate with immunoreceptor tyrosine-based switch motif of programmed death 1 upon primary human T cell stimulation, but only receptor ligation prevents T cell activation. J Immunol. 2004;173:945–954. doi: 10.4049/jimmunol.173.2.945. [DOI] [PubMed] [Google Scholar]
- 329.Karwacz K, Bricogne C, MacDonald D, Arce F, Bennett CL, Collins M, Escors D. PD-L1 co-stimulation contributes to ligand-induced T cell receptor down-modulation on CD8+ T cells. EMBO Mol Med. 2011;3:581–592. doi: 10.1002/emmm.201100165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Patsoukis N, Brown J, Petkova V, Liu F, Li L, Boussiotis VA. Selective effects of PD-1 on Akt and Ras pathways regulate molecular components of the cell cycle and inhibit T cell proliferation. Sci Signal. 2012;5:ra46. doi: 10.1126/scisignal.2002796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Couzin-Frankel J. Autoimmune diseases surface after cancer treatment. Science. 2017;358:852. doi: 10.1126/science.358.6365.852. [DOI] [PubMed] [Google Scholar]
- 332.Byun DJ, Wolchok JD, Rosenberg LM, Girotra M. Cancer immunotherapy - immune checkpoint blockade and associated endocrinopathies. Nat Rev Endocrinol. 2017;13:195–207. doi: 10.1038/nrendo.2016.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Shindo Y, McDonough JS, Chang KC, Ramachandra M, Sasikumar PG, Hotchkiss RS. Anti-PD-L1 peptide improves survival in sepsis. J Surg Res. 2017;208:33–39. doi: 10.1016/j.jss.2016.08.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Sasikumar PG, Ramachandra M. Small-molecule antagonists of the immune checkpoint pathways: concept to clinic. Future Med Chem. 2017;9:1305–1308. doi: 10.4155/fmc-2017-0107. [DOI] [PubMed] [Google Scholar]
- 335.Magiera-Mularz K, Skalniak L, Zak KM, Musielak B, Rudzinska-Szostak E, Berlicki L, Kocik J, Grudnik P, Sala D, Zarganes-Tzitzikas T, Shaabani S, Domling A, Dubin G, Holak TA. Bioactive macrocyclic inhibitors of the PD-1/PD-L1 immune checkpoint. Angew Chem Int Ed Engl. 2017;56:13732–13735. doi: 10.1002/anie.201707707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Chupak LS, Ding M, Martin SW, Zheng X, Hewawasam P, Connoly TP, Xu N, Yeung KS, Zhu J, Langley DR, Tenney DJ, Scola PM. Compounds useful as immunomodulators. 2015/160641 A2. WO. 2015
- 337.Chupak LS, Zheng X. Compounds useful as immunomodulators. 2015/034820 A1. WO. 2015
- 338.Zak KM, Grudnik P, Guzik K, Zieba BJ, Musielak B, Dömling A, Dubin G, Holak TA. Structural basis for small molecule targeting of the programmed death ligand 1 (PD-L1) Oncotarget. 2016;7:30323–30335. doi: 10.18632/oncotarget.8730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Guzik K, Zak KM, Grudnik P, Magiera K, Musielak B, Torner R, Skalniak L, Domling A, Dubin G, Holak TA. Small-molecule inhibitors of the programmed cell death-1/programmed death-ligand 1 (PD-1/PD-L1) interaction via transiently induced protein states and dimerization of PD-L1. J Med Chem. 2017;60:5857–5867. doi: 10.1021/acs.jmedchem.7b00293. [DOI] [PubMed] [Google Scholar]
- 340.Skalniak L, Zak KM, Guzik K, Magiera K, Musielak B, Pachota M, Szelazek B, Kocik J, Grudnik P, Tomala M, Krzanik S, Pyrc K, Dömling A, Dubin G, Holak TA. Small-molecule inhibitors of PD-1/PD-L1 immune checkpoint alleviate the PD-L1-induced exhaustion of T-cells. Oncotarget. 2017;8:72167–72181. doi: 10.18632/oncotarget.20050. [DOI] [PMC free article] [PubMed] [Google Scholar]