Accumulation of immature megakaryoblasts with erythroid features defined as transient abnormal myelopoiesis (TAM) is diagnosed in approximately 10% of all newborns with Down syndrome (DS). TAM is characterized by somatic mutations in GATA1 leading to the exclusive expression of the short isoform of GATA1 (GATA1s).1,2 The transient nature and the occurrence already in utero imply a fetal origin of TAM. However, the developmental stage-specific consequences of GATA1s mutations in human megakaryopoiesis and erythropoiesis remain largely unknown. Here, we utilized lentiviral CRISPR-Cas9 genome editing3 to induce GATA1s mutations in fetal, neonatal and adult hematopoietic stem and progenitor cells (HSPCs). When introduced into fetal HSPCs, GATA1s caused accumulation of immature megakaryocytic progenitors, an effect that was unique to cells of fetal ontogeny. In neonatal HSPCs specifically, GATA1s triggered a transient accumulation of immature erythroid progenitor cells. Combined, these findings highlight the distinct effects of GATA1 and GATA1s on the different developmental stages of hematopoiesis and help to explain the transient nature of TAM.
Ten to twenty percent of TAM cases progress to myeloid leukemia associated with DS (ML-DS).1 Both TAM and ML-DS can carry the same mutations in the transcription factor GATA1,2 which is essential for megakaryopoiesis and erythropoiesis, as well as for the proper formation of eosinophils, basophils, and mast cells.4 Frameshift or nonsense mutations in exon 2 of GATA1 occur in approximately 90% of cases, leading to the introduction of a premature stop codon or loss of the adjoining splice site.5 Consequently, expression of full-length GATA1 is lost, and only GATA1s is retained by alternative splicing and initiation of translation from a second start codon in exon 3 (Figure 1A). GATA1s is a naturally occurring isoform of GATA1 that lacks the N-terminal transactivation domain,2 and is crucial for maintaining the proliferative status of megakaryocytic, erythroid and eosinophil precursors.6 Knock-in mice expressing GATA1s only experience hyperproliferation of a transient population of fetal megakaryocytic progenitors and exhibit normal hematopoiesis as adults.7 These data, along with findings that indicate the fetal origin of TAM, suggest that GATA1 mutations leading to the expression of GATA1s (hereafter referred to as GATA1s mutations) exert developmental stage-specific effects on megakaryocytic or erythroid progenitors.
Figure 1.
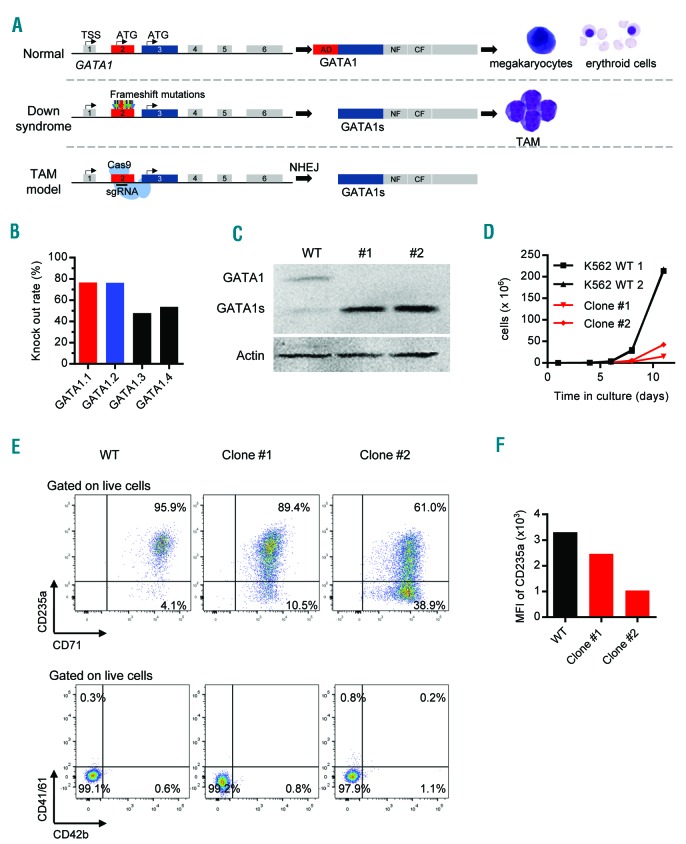
GATA1s mutation is essential for the development of transient abnormal myelopoiesis (TAM). (A) Schematic representation of the GATA1 locus, the translation of wild-type (WT) GATA1 product in normal fetal hematopoiesis (top), and the translation of GATA1s after the introduction of mutations as seen in TAM (middle) or as generated using CRISPR-Cas9 genome editing (bottom). (B) The efficacies of 4 different sgRNAs targeting exon 2 of GATA1 as assessed using a fluorescence-based reporter assay. The sgRNAs selected for later studies are marked red and blue. (C-F) K562 cells lentivirally transduced with GATA1-targeting (GATA1.1 and GATA1.2) or control (Luc.1) sgRNAs. (C) Western blot of GATA1 and GATA1s proteins in WT K562 cells and in two representative GATA1s-K562 clones (#1 and #2; used in subsequent experiments). (D) Number of WT and GATA1s-K562 cells (clone #1 and #2) grown in liquid culture. (E) Flow cytometric analysis of WT and GATA1s-K562 cells (clone #1 and #2) for expression of the indicated cell surface markers. (F) Expression of CD235a represented as mean fluorescence intensity (MFI), in WT K562 cells and GATA1s-K562 clone #1 and #2.
Although human induced pluripotent stem cell (iPSC) models of TAM/ML-DS have been established,8 technical hurdles precluded the investigation of endogenous GATA1s in primary HSPCs from different ontogenetic stages and their behavior during megakaryopoiesis and erythropoiesis.9 With the aim of overcoming these obstacles, we developed an all-in-one lentiviral CRISPR-Cas9 vector3 enabling the induction of GATA1s mutations in transduced cells. CRISPR-Cas9-mediated DNA cleavage in exon 2 of GATA1, followed by the cellular non-homologous end-joining repair induces frameshift mutations in the open reading frame (ORF) upstream of the GATA1s start codon in exon 3 (Figure 1A). Using this strategy, we can mimic in vitro the de novo acquisition of GATA1s mutations in exon 2 in the ignition of TAM (Figure 1A).
Two sgRNAs (sgGATA1.1 and sgGATA1.2) were deemed suitable for experiments based on a high knockout rate in a fluorescence-based reporter assay (Figure 1B) and no detectable off-target activity (Online Supplementary Figure S1). Accordingly, both sgRNAs efficiently induced GATA1s mutations in lentivirally transduced K562 cells (Figure 1C and Online Supplementary Figure S2). Sixty-eight K562 clones were established through single-cell sorting, of which twelve clones (17.6%) exhibited GATA1s expression and loss of full-length GATA1 via Western blotting (Figure 1C and data not shown). Mutational analysis of the GATA1 locus revealed that five out of twelve clones (42%) contained frameshift mutations in both alleles (hereafter referred to as GATA1s-K562 cells) (Online Supplementary Figure S2 and data not shown). To determine the proliferation potential and differentiation status of these cells, two GATA1s-K562 clones (#1 and #2) were analyzed. Both exhibited a 10-fold reduced growth rate compared to normal K562 cells (Figure 1D). Since the K562 cell line was derived from an erythroleukemia, we speculated that these cells may be dependent on full-length GATA1 to maintain their erythroid phenotype. Indeed, flow cytometry revealed reduced expression of the common erythroid markers CD235a and CD71 in GATA1s-K562 cells (Figure 1E and F). In contrast, expression of the megakaryocytic markers CD41/CD61 and CD42b was not affected (Figure 1E and Online Supplementary Figure S3), indicating that the differentiation status of the GATA1s-K562 clones did not shift towards this lineage.
After validating the efficiency and functionality of our lentiviral GATA1s CRISPR-Cas9 system in a cell line, we subsequently generated GATA1s mutations in primary human fetal, neonatal and adult HSPCs from fetal livers, cord blood, and mobilized peripheral blood, respectively. Transduced cells were FACS sorted (Online Supplementary Figure S4) and subjected to megakaryocytic and erythroid differentiation assays. In sgGATA1.1- or sgGATA1.2-transduced cells, 78–92% of the GATA1 alleles possessed an insertion or deletion in exon 2 (Online Supplementary Figure S5).
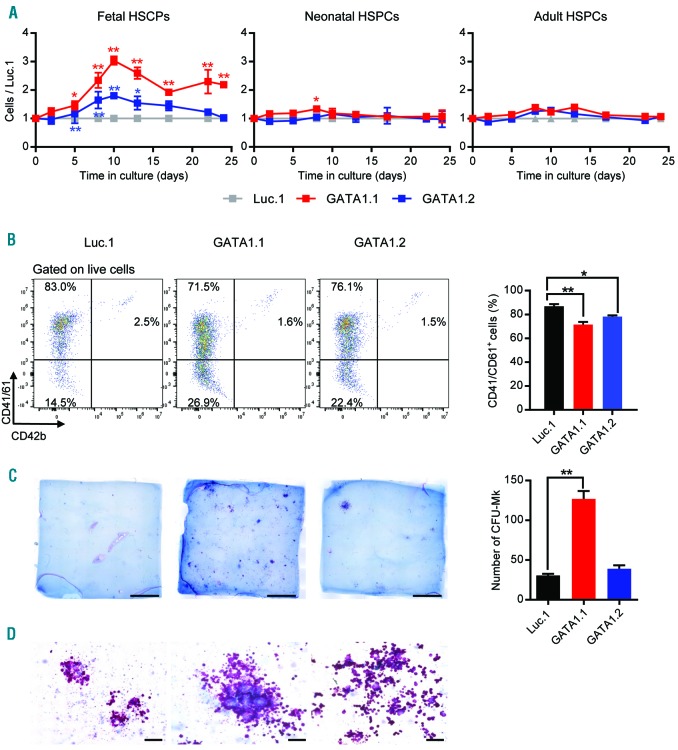
Under conditions that promote megakaryocytic differentiation, we observed a distinct growth advantage in GATA1s fetal HSPCs (i.e. sgGATA1.1- or sgGATA1.2-transduced HSPCs) compared to those with wild-type GATA1 (i.e. sgLuc.1-transduced HSPCs) (Figure 2A), suggesting that GATA1s causes hyperproliferation of fetal megakaryocytic progenitor cells. Interestingly, no such proliferative advantage was observed in neonatal or adult GATA1s HSPCs (Figure 2A). Although terminal megakaryocytic differentiation was not blocked by the GATA1s mutation, we observed a slight impairment (Figure 2B and Online Supplementary Figure S6A). To clarify whether the introduction of GATA1s into fetal HSPCs increases the frequency or proliferation of megakaryocytic progenitor cells, we performed collagen-based megakaryocyte colony-forming unit (CFU-MK) assays. We observed that more (Figure 2C) and larger (Figure 2C and D) CFU-MKs arose from GATA1s fetal HSPCs, implying that both parameters contribute to the hyperproliferative phenotype of these cells (Figure 2A). The higher frequency of megakaryocytic progenitors in GATA1s fetal HSPCs may also indicate GATA1s-mediated lineage skewing of fetal HSPCs towards megakaryopoiesis. Our data thus support a model where GATA1s specifically favors the accumulation of fetal megakaryocytic progenitors, but not those of neonatal or adult ontogeny.
Figure 2.
GATA1s mutations in fetal hematopoietic stem and progenitor cells (HSPCs) promote the hyperproliferation of megakaryocytic progenitors. Fetal, neonatal and adult HSPCs were transduced with GATA1-targeting (GATA1.1 and GATA1.2) or control (Luc.1) sgRNAs. (A) Relative number of transduced HSPCs (normalized to the number of Luc.1 cells) grown in liquid cultures supporting megakaryocytic differentiation. Data from 1 of 4 independent experiments performed in replicates are shown as mean±Standard Deviation (SD). *PANOVA<0.05, **PANOVA<0.01. (B) (Left) Representative flow cytometry plots (from 3 independent experiments), showing expression of the common megakaryocytic markers CD41/CD61 and CD42b in fetal HSPCs (pre-gated on live cells). Percentages of each population are indicated. (Right) Percentage of fetal CD41/CD61+ cells on day 10 of megakaryocytic differentiation. Data from 2 independent experiments performed in replicates are shown as mean±SD. *PANOVA<0.05, **PANOVA<0.01. (C) (Left) Photographs (scale bar 5 mm) of CFU-MK assays using lentivirally transduced fetal HSPCs. (Right) Number of CFU-MKs. **PANOVA<0.01. (D) Microscopic images of CFU-MK assays (400× original magnification; scale bar 100 μm).
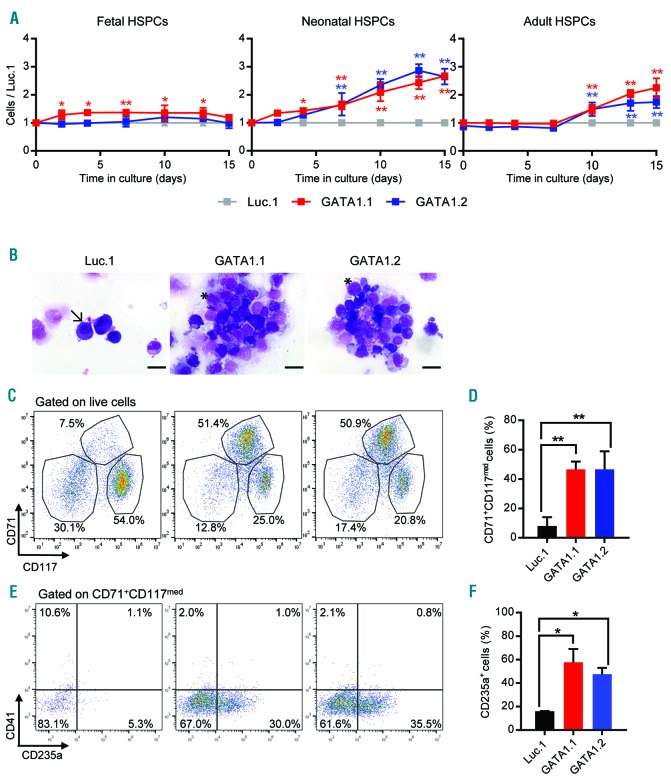
Next, we evaluated the CRISPR-Cas9-modified HSPCs under culture conditions that support both erythroid and megakaryocytic differentiation. GATA1s neonatal HSPCs showed enhanced proliferation (Figure 3A) and an accumulation of CD71+/CD117med/CD235amed erythroid progenitor cells (Figure 3C–F and Online Supplementary Figure S6B). This population was hardly present in wild-type GATA1 cells and increased 7-fold within ten days of culture. Morphological analysis further underlined their immature erythroblast-like phenotype, in contrast to the exclusively mature erythroid cells in the wild-type GATA1 control (Figure 3B). Interestingly, analysis at a later time point demonstrated the transient nature of this GATA1s-dependent accumulation of erythroid progenitors (Online Supplementary Figure S7A and B). In GATA1s adult HSPCs, a moderate growth advantage was also observed, but only after ten days of culture. This outgrowth coincided with the accumulation of eosinophilic granulocytes and mast cells (Online Supplementary Figure S7C), which was also described for the lentiviral overexpression of GATA1s.9
Figure 3.
GATA1s perturbs megakaryocytic/erythroid differentiation of neonatal hematopoietic stem and progenitor cells (HSPCs), resulting in accumulation of erythroid progenitors. Fetal, neonatal and adult HSPCs were transduced with GATA1-targeting (GATA1.1 and GATA1.2) or control (Luc.1) sgRNAs. (A) Relative number of transduced HSPCs (normalized to the number of Luc.1 cells) grown in liquid cultures supporting combined megakaryocytic/erythroid differentiation. Data from 1 of 4 independent experiments performed in replicates are shown as mean±Standard Deviation (SD). *PANOVA<0.05, **PANOVA<0.01. (B) Microscopic images (MGG staining; 1000x original magnification; scale bar 20 μm) of sorted neonatal CD34+ HSPCs on day 10 of combined megakaryocytic/erythroid differentiation. A representative mature erythroid cell is indicated by an arrow (left photograph), while characteristic immature erythroblast-like cells are indicated by an asterisk (middle and right photographs). (C) Representative flow cytometry plots (from 3 independent experiments), showing CD71+CD117−, CD71+CD117med and CD71+CD117+ neonatal HSPC-derived cells on day 10 of combined megakaryocytic/erythroid differentiation (pre-gated on live cells). Percentages of each population are indicated. (D) Percentage of neonatal CD71+CD117med cells on day 10 of combined megakaryocytic/erythroid differentiation. Data from 3 independent experiments are shown as mean±SD. **PANOVA<0.01. (E) Representative flow cytometry plots (from 2 independent experiments), depicting the CD41−CD235a− and CD41−CD235a+ cells within the neonatal CD71+CD117med population. Percentages of each population are indicated. (F) Percentage of the CD41−CD235a+ population. Data from replicates of 2 independent experiments are shown as mean±SD. *PANOVA<0.05.
In summary, by inducing endogenous GATA1s mutations in fetal, neonatal and adult HSPCs, our study demonstrates that GATA1s perturbs erythropoiesis and megakaryopoiesis in a developmental stage-specific manner. In fetal HSPCs specifically, GATA1s caused hyperproliferation of immature megakaryocytic progenitors. Furthermore, in neonatal HSPCs, GATA1s perturbed erythro-megakaryocytic differentiation and concomitantly caused a transient accumulation of immature erythroid progenitors. These findings highlight the distinct ontogenetic effects of GATA1 and GATA1s.
Our data were gathered from primary human cells of different developmental stages. As revealed by flow cytometric analysis, no major differences were observed in the distribution of progenitor populations at each developmental stage (Online Supplementary Figure S8). However, fetal HSPCs per se exhibited enhanced expression of both full length GATA1 and GATA1s compared to neonatal or adult HSPCs (Online Supplementary Figure S9), which may be attributed to the observed accumulation of megakaryocytic blasts upon CRISPR-Cas9 genome editing. Still, these data cannot explain the outgrowth of CD71+CD117med erythroid progenitors from GATA1s neonatal HSPCs. Thus, despite known developmental stage-specific functions of transcription factors, epigenetic regulators or cytokine receptors,10 the molecular basis for the differential response of fetal, neonatal and adult HSPCs to the GATA1s mutations remains elusive.
Our findings complement previous murine studies describing a transient population of megakaryocytic progenitors that are sensitive towards the oncogenic effects of GATA1s.7 Species-specific differences that hamper the modeling of TAM in mice10,11 mean that studies in human cells are required and these are essential in order to understand the interplay between fetal megakaryopoiesis, GATA1s-mutation and trisomy 21 during DS-associated leukemogenesis.12 While GATA1s-mutations and trisomy 21 are sufficient to induce TAM in humans, Ts65dn mice crossed with Gata1s-knock-in mice do not develop a neonatal hyperproliferative phenotype.10 Human iPSC models have partially closed this gap by showing impaired erythropoiesis and megakaryopoiesis due to GATA1s in synergy with trisomy 21.8,13–15 However, these models are restricted to primitive hematopoiesis or early stages of definitive hematopoiesis, precluding exploration of the developmental stage-specific effects of GATA1s. Our study complements these iPSC models by investigating the effects of GATA1s in CRISPR-modified HSPCs of different onto-genetic stages.
Notwithstanding these important discoveries related to GATA1s, TAM and ML-DS, the developmental stage-specific effects of trisomy 21 have yet to be defined. Our study paves the way for future efforts to clarify the specific interaction between trisomy 21 and GATA1 during DS fetal hematopoiesis, and the mechanisms by which GATA1s mutations can interfere with this interaction, leading to the development of TAM.
Supplementary Material
Acknowledgments:
We thank Michelle Ng for helpful discussions and D. Trono of EPFL, Lausanne, Switzerland, for kindly providing both pMD2.G (Addgene plasmid 12259) and psPAX2 (Addgene plasmid 12260).
Footnotes
Funding: JHK receives funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement n. 714226). DH is supported by the German Cancer Aid (111743). This work was supported by grants to JHK from the DFG (KL-2374/1-1 and KL-2374/1-3). SG, AKM, ML and LS were supported by the Hannover Biomedical Research School.
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Klusmann JH, Creutzig U, Zimmermann M, et al. Treatment and prognostic impact of transient leukemia in neonates with Down syndrome. Blood. 2008;111(6):2991–2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wechsler J, Greene M, McDevitt MA, et al. Acquired mutations in GATA1 in the megakaryoblastic leukemia of Down syndrome. Nat Genet. 2002;32(1):148–152. [DOI] [PubMed] [Google Scholar]
- 3.Reimer J, Knoss S, Labuhn M, et al. CRISPR-Cas9-induced t(11;19)/MLL-ENL translocations initiate leukemia in human hematopoietic progenitor cells in vivo. Haematologica. 2017; 102(9):1558–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132(4):631–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alford KA, Reinhardt K, Garnett C, et al. Analysis of GATA1 mutations in Down syndrome transient myeloproliferative disorder and myeloid leukemia. Blood. 2011;118(8):2222–2238. [DOI] [PubMed] [Google Scholar]
- 6.Halsey C, Tunstall O, Gibson B, Roberts I, Graham G. Role of GATA-1s in early hematopoiesis and differences between alternative splicing in human and murine GATA-1. Blood. 2010;115(16):3415–3416. [DOI] [PubMed] [Google Scholar]
- 7.Li Z, Godinho FJ, Klusmann JH, Garriga-Canut M, Yu C, Orkin SH. Developmental stage-selective effect of somatically mutated leukemogenic transcription factor GATA1. Nat Genet. 2005;37(6):613–619. [DOI] [PubMed] [Google Scholar]
- 8.Banno K, Omori S, Hirata K, et al. Systematic Cellular Disease Models Reveal Synergistic Interaction of Trisomy 21 and GATA1 Mutations in Hematopoietic Abnormalities. Cell Rep. 2016; 15(6):1228–1241. [DOI] [PubMed] [Google Scholar]
- 9.Maroz A, Stachorski L, Emmrich S, et al. GATA1s induces hyperproliferation of eosinophil precursors in Down syndrome transient leukemia. Leukemia. 2014;28(6):1259–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klusmann JH, Godinho FJ, Heitmann K, et al. Developmental stage-specific interplay of GATA1 and IGF signaling in fetal megakaryopoiesis and leukemogenesis. Genes Dev. 2010;24(15):1659–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alford KA, Slender A, Vanes L, et al. Perturbed hematopoiesis in the Tc1 mouse model of Down syndrome. Blood. 2010;115(14):2928–2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klusmann JH, Li Z, Bohmer K, et al. miR-125b-2 is a potential oncomiR on human chromosome 21 in megakaryoblastic leukemia. Genes Dev. 2010;24(5):478–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chou ST, Byrska-Bishop M, Tober JM, et al. Trisomy 21-associated defects in human primitive hematopoiesis revealed through induced pluripotent stem cells. Proc Natl Acad Sci USA. 2012;109(43):17573–17578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roy A, Cowan G, Mead AJ, et al. Perturbation of fetal liver hematopoietic stem and progenitor cell development by trisomy 21. Proc Natl Acad Sci USA. 2012;109(43):17579–17584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maclean GA, Menne TF, Guo G, et al. Altered hematopoiesis in trisomy 21 as revealed through in vitro differentiation of isogenic human pluripotent cells. Proc Natl Acad Sci USA. 2012; 109(43):17567–17572. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.