Erythroid nuclear maturation involves a complex process: massive transcription continues despite the nucleus globally condensing to one-tenth its original volume. We discovered that nuclear proteins such as core histones migrate into the cytoplasm during this process, and that histone migration and subsequent nuclear condensation and enucleation were abrogated by knockdown of the highly induced, erythroid specific export protein Exportin 7, or Xpo7.1 We previously showed that Xpo7 has an alternative, erythroid-specific promoter/start site,1 and ENCODE data2–6 show that this erythroid promoter is bound by the master erythroid regulators GATA1 and Tal1. However, these binding events only explain the induction of Xpo7 in a nucleus undergoing active transcription, not how Xpo7 exerts its function only after transcription has ended. In this study, we show that developmental downregulation of the microRNA miR-181a regulates enucleation by contributing to the timing of Xpo7 expression. We show that miR-181a is normally down-regulated during erythroid development and its expression inversely correlated with Xpo7 induction. MiR-181a directly binds the conserved mammalian microRNA binding site in the Xpo7 3′ UTR via reporter assay. Overexpression of miR-181a inhibits erythroid enucleation through repression of Xpo7 levels, both protein and mRNA expression. While the erythroid nucleus is undergoing active transcription, high miR-181a levels repress Xpo7 expression, but as miR-181a decreases during erythroid development, Xpo7 levels increase, allowing erythroid nuclear condensation to commence and ultimately enucleation to occur.
We previously reported that the nuclear export protein Xpo7 is highly induced during terminal erythroid differentiation and that the predominant Xpo7 transcript in terminal erythropoiesis contains an erythroid-specific start site located in the first intron.1 In order to determine the timing of Xpo7 expression, we looked to non-coding RNA regulation, and examined the 3′ and 5′ UTRs of the murine erythroid and non-erythroid Xpo7 transcripts. We found different length UTRs for the erythroid (from Ter119-positive cells) compared to the non-erythroid (from Ter119-negative cells) Xpo7 transcripts using 3′-and 5′-RACE (Figure 1A), as previously described.7 The erythroid-specific 3′ UTR of Xpo7 contains multiple predicted microRNA binding sites, among them a miR-181ab-c-d consensus binding site that is highly conserved only in mammals (Figure 1B). This specific microRNA is quite important for erythropoiesis because overexpression of the normally developmentally down-regulated miR-181a-1-miR-181b-1 cluster was previously shown to inhibit normal enucleation8 (though the specific mechanism was never uncovered). In addition, miR-181a-1 variants were revealed in a genome-wide association study (GWAS) in subjects with variation in RBC number.9
Figure 1.
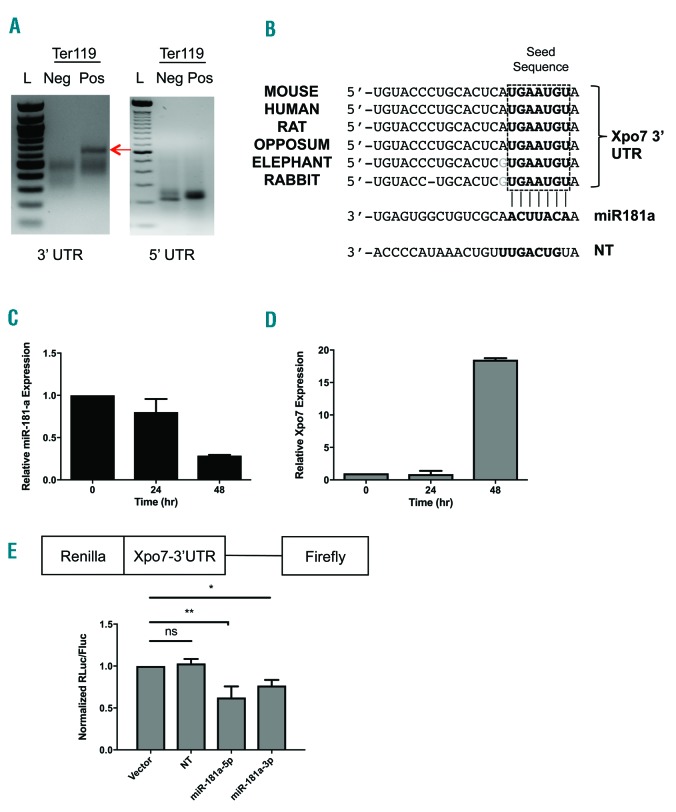
Xpo7 is regulated by erythroid-specific UTRs and is a direct target of miR-181a. (A) 3′- and 5′- RACE (Rapid Amplification of cDNA ends) of RNA isolated from Ter119-negative (non-erythroid) and Ter119-positive (erythroid) cells from murine fetal liver, using freshly isolated total RNA and a RACE kit (Roche). Prominent 3′- and 5′- UTRs change between Ter119-negative (Neg) and Ter119-positive (Pos) cells (representative gels). (B) Significant conservation of a predicted miR-181 site in Xpo7 3′UTR between numerous mammalian species (top). miR-181a complementary seed sequence is shown below that. Seed sequence of the non-targeting (NT) control miRNA (miR-223) is also shown (bottom). (C) qPCR against mature miR-181a transcript was performed using total RNA isolated using a miRNA kit (Qiagen) from erythroblasts during in vitro erythroid differentiation using erythropoietin (Epo) [from time 0–48 hours (hr)]. MiR-181a is developmentally down-regulated during erythroid differentiation. (D) qPCR against Xpo7 shows induction late in the in vitro erythroid differentiation culture. (E) Construct (above) for a luciferase assay using the erythroid (Ter119-positive) Xpo7-3′UTR fused to Renilla luciferase. Fluorescence after co-expression with either miR-181a-5p or -3p mimic was normalized to Firefly luciferase and shows direct binding of miR-181a to the Xpo7-3′UTR. Results are shown as the mean±Standard Error (n=3 per group; *P<0.05, **P<0.01).
Since miRNAs can control gene expression via repression of mRNA transcript stability, we hypothesized that mir-181a could regulate erythroid enucleation via repression of Xpo7 mRNA, thus explaining how Xpo7 could be transcribed in an active nucleus but not yet exert its effect. To examine the mechanism underlying this hypothesis, we first determined that miR-181a is developmentally down-regulated during erythropoiesis (Figure 1C) and its expression inversely correlated with Xpo7 levels (Figure 1D). Secondly, miR-181a-5p mimic binds directly to the predicted binding site in the Xpo7 3′UTR via reporter assay using a dual reporter with background firefly luciferase and Renilla luciferase fused to the Xpo7 erythroid 3′ UTR (Figure 1E). Similar effects were observed using the -3p mimic of miR-181a (Figure 1E).
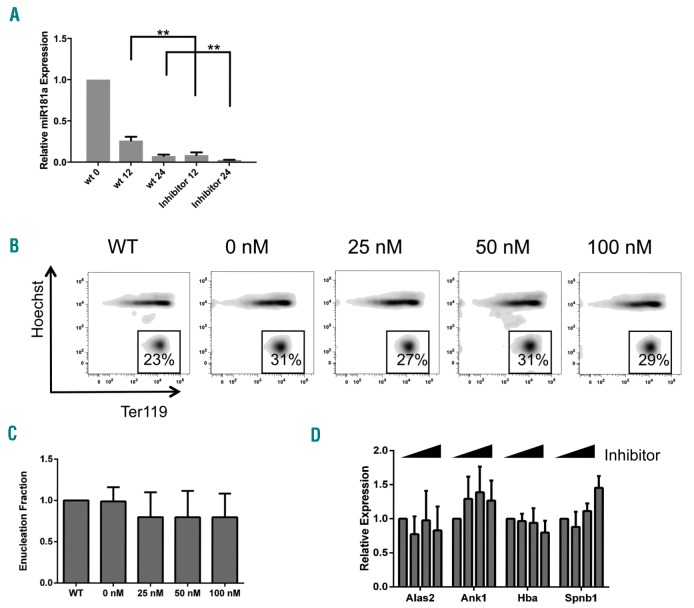
Given that miR-181a normally decreases during erythroid development, further decrease in miR-181a levels to one-tenth normal levels at 12 hours (h) and 24 h of differentiation using a synthetic inhibitor oligonucleotide (Figure 2A) does not significantly affect normal red blood cell (RBC) differentiation, including acquisition of cell surface marker Ter119 or enucleation (Figure 2B). There was no significant dose-response effect on enucleation (Figure 2C) or important erythroid transcripts (Figure 2D). These findings are as we expected, because miR-181a is normally down-regulated during erythropoiesis and plays a larger role in B-cell and myeloid development so, not surprisingly, decreasing it further does not inhibit erythropoiesis.
Figure 2.
Further downregulation of miR-181a, normally developmentally down-regulated during erythropoiesis, does not disrupt normal erythroid differentiation. Cultured murine fetal liver erythroid progenitors were transiently transfected with increasing concentrations of a synthetic miR-181a inhibitor oligonucleotide (RNAiMAX, Life Technologies), lowering endogenous miR-181a levels by up to one-tenth normal levels. (A) Expression of mature miR-181a in wild-type versus inhibitor-transfected cells at time 0 hours (h), 12 h, 24 h post transfection during in vitro erythropoiesis, showing up to one-tenth fold decreased miR-181a, using sample transfected with 50 nM inhibitor (optimal concentration as directed by the manufacturer). (B) Differentiation as measured by flow cytometry for Ter119 and Hoechst (DNA) at 48 h post transfection shows no observed change in Ter119 induction or loss of DNA staining (reticulocytes) even after increasing doses of inhibitor (representative plot). (C) Quantification of reticulocyte populations from (B) (Ter119-positive cells without nuclear DNA staining) also shows no dose response in enucleation. (D) Expression of key erythroid transcripts as measured by qRT-PCR shows no alteration. All results shown are the mean±Standard Error (n=3 per group; **P<0.01). Changes in enucleation (C) or relative expression (D) are not statistically significant.
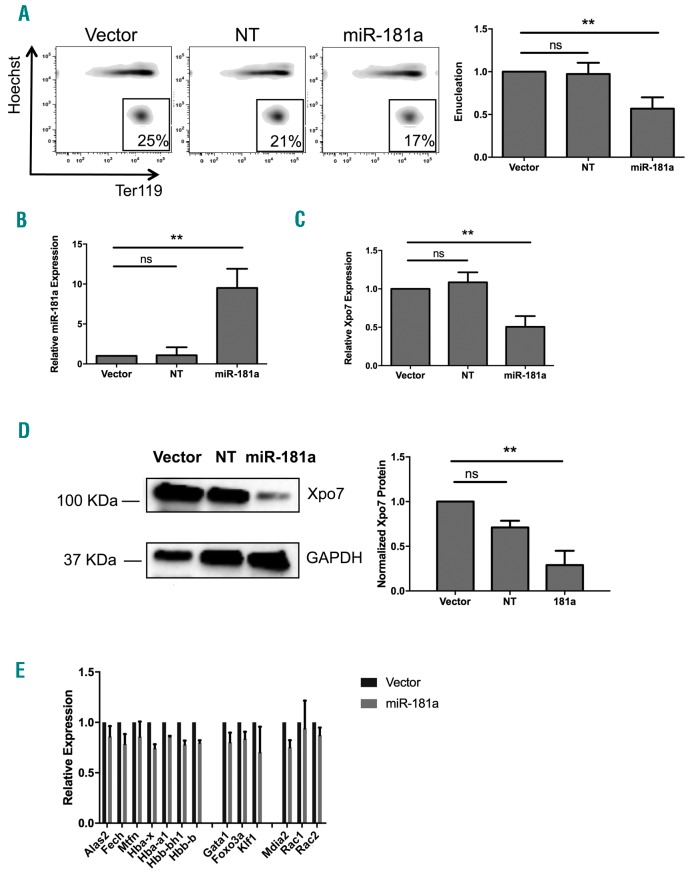
In contrast, overexpression of miR-181a at the time it is normally down-regulated does indeed significantly inhibit enucleation (Figure 3A). Increasing miR-181a to 10-fold higher than normal levels at 48 h of differentiation (Figure 3B) significantly represses Xpo7 both at the transcript (Figure 3C) and protein (Figure 3D) levels. Neither cell growth nor cell cycle exit during culture was significantly affected by overexpression of miR-181a (data not shown), showing this is not an artefact of cellular proliferation. Similar results were seen using transient transfection of an alternative construct, miR-181a-3p. Finally, hemoglobin production itself, and normal erythroid hemoglobin and enucleation transcript levels were not affected by miR-181a overexpression (Figure 3E).
Figure 3.
Overexpression of miR-181a inhibits erythroid enucleation and significantly decreases Xpo7. Cultured murine fetal liver erythroid progenitors were infected with retroviral constructs containing GFP with miR-181a-1, non-targeting (NT) miRNA (miR223), or empty vector. (A) Transduced cells over-expressing these constructs were sorted for GFP and analyzed by flow cytometry at 48 hours (h) post transduction using Hoechst 33342 and Ter119-APC. There is no observed change in Ter119 induction but there is a significantly decreased reticulocyte population after miR-181a overexpression: (left) representative plot; (right) quantification. (B and C) qRT-PCR performed on the mature transcript from RNA isolated at 48 h post induction shows that overexpression of miR-181a (B) was over ten-fold that of the vector control and Xpo7 transcript (C) was significantly decreased. Results are shown as the mean±Standard Error (n=6 per group; **P<0.01). (D) Representative immunoblot for Xpo7 in cultured murine fetal liver erythroid progenitors transduced with control, NT, or miR-181a retroviral overexpression constructs. (Upper panel) Blot is near 100KDa; (lower panel) blot is near 37KDa. Xpo7 protein is significantly decreased when miR-181a is over-expressed. Results are shown as the mean±Standard Error (n=3 per group; **P<0.01). (E) Expression of red cell hemoglobin or enucleation transcripts as measured by qRT-PCR was not significantly changed by miR-181a overexpression. Results are shown as the mean±Standard Error (n=2 per group).
Based on the previously published finding that overexpression of the miR-181a-1-miR-181b-1 cluster significantly inhibited murine erythroid nuclear condensation and enucleation, our results make clear the mechanism by which miR-181a regulates enucleation: 1) miR-181a directly binds the predicted microRNA binding site in the Xpo7 3′-UTR; and 2) overexpression of miR-181a inhibits enucleation and lowers Xpo7 transcript and protein levels. These findings support our conclusion that miR-181a partially regulates enucleation via the regulation of Xpo7 transcript and protein levels.
A complete understanding of the regulation of normal terminal RBC development is crucial in the development of novel therapies for erythropoietin (Epo)-refractory anemias, such as congenital dyserythropoietic anemia or myelodysplasia with refractory anemia. Our laboratory is focused on understanding the regulation of nuclear maturation of the developing erythroblast, including chromatin condensation and enucleation. We have previously shown that erythroid nuclear maturation involves nuclear histone migration to the cytoplasm and that the nuclear export protein Xpo7 facilitates this process. Regulation of the induction of Xpo7 transcript is straightforward. However, induction of mRNA transcript cannot explain how Xpo7 can be transcribed in an active nucleus but not yet exert its effect, so we looked to non-coding RNAs to better understand the timing of Xpo7 protein expression. We uncovered evidence of further regulation in different erythroid and non-erythroid 3′-and 5′-UTRs of Xpo7. The 3′ UTR of Xpo7 contains multiple predicted microRNA binding sites, among them an miR-181a-b-c-d consensus binding site that is highly conserved only in mammals. MiR-181a was previously implicated in the regulation of enucleation because its overexpression inhibited enucleation8 and it has since been discovered in a GWAS study in patients with variation in RBC number,9 suggesting that this microRNA involved in immune cell development may also play a role in terminal erythropoiesis via its downregulation.
In this study, we show that miR-181a is down-regulated during erythroid development and its expression inversely correlated with Xpo7 induction. We show that miR-181a directly binds the conserved mammalian microRNA binding site in the Xpo7 3′ UTR. Given the much higher levels and the more prominent role of mir-181a in B-cell and immune cell development,10 not surprisingly, a further decrease of miR-181a in terminal erythropoiesis does not disrupt erythropoiesis. In stark contrast, however, overexpression of miR-181a inhibits erythroid enucleation through repression of Xpo7 levels, both protein and mRNA expression. We recognize that there is only a modest decrease in Xpo7 levels and enucleation upon miR-181a overexpression, and this suggests that mir-181a downregulation alone does not solely control Xpo7 levels nor does it regulate nuclear maturation on its own. Given that miRNA regulation is often the fine-tuning of mRNA levels and not frank induction,11 this is consistent with the mechanism shown here. Taken together, our model shown here offers an intriguing insight into the role of non-coding RNAs in nuclear condensation and enucleation: that the developmental downregulation of a specific miRNA can affect a critical step in terminal erythroid development.
Supplementary Material
Footnotes
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Hattangadi SM, Martinez-Morilla S, Patterson HC, Shi J, et al. Histones to the cytosol: Exportin 7 is essential for normal terminal erythroid nuclear maturation. Blood. 2014;124(12):1931–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raney BJ, Dreszer TR, Barber GP, et al. Track data hubs enable visualization of user-defined genome-wide annotations on the UCSC Genome Browser. Bioinformatics. 2014;30(7):1003–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosenbloom KR, Sloan CA, Malladi VS, et al. ENCODE Data in the UCSC Genome Browser: Year 5 update. Nucleic Acids Res. 2013; 41(Database issue):D56–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng Y, Wu W, Kumar SA, et al. Erythroid GATA1 function revealed by genome-wide analysis of transcription factor occupancy, histone modifications, and mRNA expression. Genome Res. 2009; 19(12):2172–2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu M, Riva L, Xie H, et al. Insights into GATA-1-Mediated Gene Activation versus Repression via Genome-wide Chromatin Occupancy Analysis. Mol Cell. 2009;36(4):682–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu W, Cheng Y, Keller CA, et al. Dynamics of the epigenetic landscape during erythroid differentiation after GATA1 restoration. Genome Res. 2011;21(10):1659–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu W, Yuan B, Flygare J, Lodish HF. Long noncoding RNA-mediated anti-apoptotic activity in murine erythroid terminal differentiation. Genes Dev. 2011;25(24):2573–2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang L, Flygare J, Wong P, Lim B, Lodish HF. miR-191 regulates mouse erythroblast enucleation by down-regulating Riok3 and Mxi1. Genes Dev. 2011;25(2):119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van der Harst P, Zhang W, Mateo Leach I, et al. Seventy-five genetic loci influencing the human red blood cell. Nature. 2012;492(7429):369–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cuesta R, Martínez-Sánchez A, Gebauer F. miR-181a Regulates Cap-Dependent Translation of p27 kip1 mRNA in Myeloid Cells. Mol Cell Biol. 2009;29(10):2841–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartel DP. Review MicroRNAs: Target Recognition and Regulatory Functions. Cell. 2009;136(2):215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.