Peripheral T-cell lymphomas (PTCL) comprise usually aggressive neoplasms of mature T cells with a heterogeneous molecular background. Over the past years, several genomic studies stemming from different research groups have shown evidence that mutation-induced activation of the T-cell receptor (TCR) signaling pathway is involved in the pathogenesis of several PTCL entities.1–7 Notably, recurrent activating point mutations in CD28 which encodes a major TCR costimulatory receptor have been reported, mainly in angioimmunoblastic T-cell lymphoma (AITL) (10–15% of the cases).2,3 Recently, four different ribonucleic acid (RNA) fusions involving the car-boxy-terminal part of CD28 with the N-terminal part of CTLA4 or ICOS were discovered by RNA sequencing analysis of AITL,2,8 cutaneous T-cell lymphoma (CTCL)4,9,10 and adult T-cell lymphoma/leukemia (ATLL).1 While these data suggest that overall these fusions are rare (5–11%), a high frequency of CTLA4(ex3)_CD28(ex4) fusion (38% of the cases) was reported in a cohort of 120 PTCLs from Asia (including 50 AITL, 39 PTCL-not otherwise specified [-NOS] and 31 extranodal natural killer/T-cell lymphoma [ENKTCL]) via reverse transcription polymerase chain reaction (RT-PCR) analysis of formalin-fixed paraffin-embedded (FFPE) samples.8 However, by reanalyzing published whole exome sequencing and RNA sequencing (RNAseq) data (43 AITL, 16 PTCL-NOS and 43 ENKTCL), Gong et al. found only one AITL positive case for the CTLA4(ex3)_CD28(ex4) fusion.11 In order to clarify these discrepant findings and to assess the prevalence of all depicted CD28 RNA fusions, we designed a RT-PCR assay to detect these four CD28 fusions which we then applied to a large series (n=273) of diagnostic frozen biopsy samples representative of various PTCL entities (Figure 1A). We found that these rearrangements are generally rare in PTCL (prevalence 4.8%), relatively more frequent in follicular helper T (TFH)-derived entities (6.5%), and most commonly represented by the ICOS(ex1)_CD28(ex2) fusion (73% of the cases).
Figure 1.
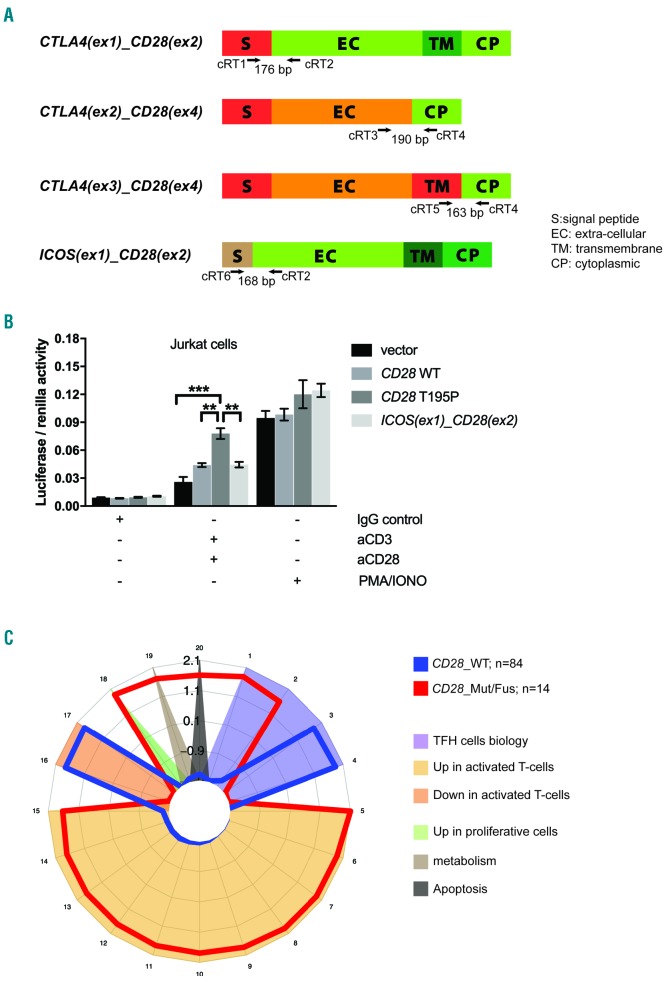
Biological impact of CD28 alterations. (A) Schematic representation of the different CD28 RNA fusions and PCR design for their screening. Arrows indicate the approximate positions of oligonucleotide primers on the indicated RNA fusions. (B) NF-κB luciferase reporter assay in Jurkat cells monitoring the activity of a CD28 T195P activating mutant and the ICOS(ex1)_CD28(ex2) fusion, compared with CD28 WT. Data are represented as mean+/−SEM from four independent experiments. (C) Spider plot representation of gene sets differentially enriched in patients with (red) (n=14) or without (blue) (n=84) alterations (mutations or fusions) in CD28. A total of 304 signatures, including 23 of 50 hallmark signatures and nine signatures from the curated and immunogenic signature collection of the Molecular Signatures Database (MSigDB), all signatures of interest in lymphoid biology (Signatures database, Staudt’s lab), and two manually annotated sequences linked to TCR signalling were tested. The list of signatures that reached statistical significance is provided in Online Supplementary Table S2. WT: wild-type; Mut/Fus: mutations or fusions.
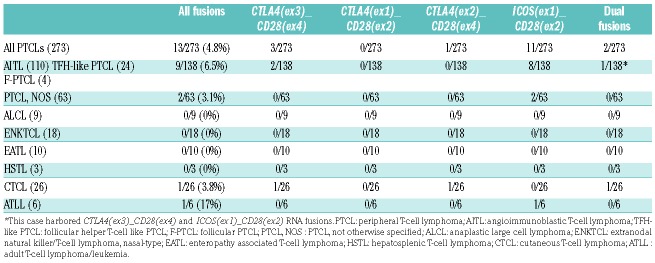
Our study group comprised 110 AITL and 28 other nodal lymphomas of TFH derivation (24 TFH-like PTCL12,13 and four follicular [F]-PTCLs) (Online Supplementary Figure S1) plus 135 samples of different PTCL entities (Table 1) collected in the frame of the genomic network from the Lymphoma Study Association (LYSA).14 Samples were divided equally between our two laboratories for molecular screening. RT-PCR amplifications of the CD28 fusions and ACTB (internal control) on total messenger (m)RNAs extracted from frozen tumor samples were analyzed by agarose gel electrophoresis (Online Supplementary Figure S2). All positive cases (n=13) and a subset of negative cases (n=58; 22%) were cross validated in both laboratories.
Table 1.
Distribution of CD28 RNA fusions detected by RT-PCR in different peripheral T-cell lymphoma entities.
Results are summarized in Table 1. Overall, CD28 fusions were detected in 13/273 (4.8%) cases. Among primarily nodal PTCLs, the highest prevalence was found in the group of AITL and other TFH-derived neoplasms with 9/138 positive samples (6/110 AITL, 3/24 TFH-like PTCL and none of the four F-PTCL; 6.8% in total), only 2/63 PTCL-NOS (3.2%) harbored a fusion, while no fusions were detected in any of the ALCLs (n=9). One of 26 CTCL and 1/6 ATLL cases were found to be positive. None of the non-cutaneous extranodal PTCL tested (18 ENKTCL, ten enteropathy associated T-cell lymphoma [EATL], three hepatosplenic T-cell lymphoma [HSTL]) were positive. A total of 15 fusions were amplified; the most common rearrangement being ICOS(ex1)_CD28(ex2) (11/15; 73% of the fusions), followed by CTLA4(ex3)_CD28(ex4) (3/15 fusions) while CTLA4(ex2)_CD28(ex4) was identified in a single CTCL sample. Interestingly, two cases (one TFH-derived PTCL and one CTCL) harbored two different CD28 RNA fusions, an observation previously made in one ATLL sample.1
All CD28 chimeric proteins are expected to comprise the cytoplasmic portion of CD28, accounting for signal transduction and a part or the totality of the transmembrane and extracellular domains of CTLA4, ICOS or CD28 (Figure 1A). Depending on the fusion partner, their expression is under the control of the regulatory elements of CTLA4 or ICOS.1,8 In normal T cells, following TCR activation, CD28 expression is downregulated while the expression of CTLA4 and ICOS is induced.15 In neoplastic cells harboring CD28 fusions, it can be anticipated that signals which normally induce the expression of CTLA4 or ICOS, would induce the expression of the fusion proteins whose triggering mimics CD28 signalling. Thus, the overall consequence of these fusions would be continuous or prolonged CD28 co-stimulatory signaling. It has been demonstrated that Jurkat cells transduced with the CTLA4(ex3)_CD28(ex4) fusion showed enhanced proliferation and interleukin (IL)-2 production upon CTLA4 binding.8,9 To further investigate the signaling induced by the ICOS(ex1)_CD28(ex2) fusion, we used a nuclear factor κ-light-chain-enhancer of activated B cells (NF-κB) luciferase-based reporter assay to assess its activity in comparison to mutant (T195P) and wild-type CD28 (Figure 1B). The chimeric protein is composed of the ICOS signal peptide fused to the extracellular, transmembrane and cytoplasmic domains of CD28. In the mature form, the signal peptide is cleaved, resulting in CD28 expression at the cell surface. Upon CD3 and CD28 co-stimulation, CD28_T195P induced higher levels of luciferase expression than CD28_WT (P=0.01) (as previously described),2 while the ICOS(ex1)_CD28(ex2) fusion delivered an activatory signal similar in amplitude to that of the CD28_WT form (Figure 1B).
Taking advantage of available transcript profiles for a large subset of AITL and other TFH lymphomas, we sought to assess the impact of CD28 genetic alterations (mutations and fusions) on gene expression (Figure 1C and Online Supplementary Figure S2). No significant difference in CD28, CTLA4 or ICOS mRNA levels was observed between six fusion-positive and 96 fusion-negative cases which had associated gene expression profiling data. Next, considering those samples harboring CD28 mutations or fusions (referred to as CD28_Mut/Fus) together versus those wild-type (WT) for CD28, the analysis of the top 100 genes differentially expressed showed the up-regulation of genes involved in PI3K, MAPK or NF-κB signalling pathways, in actin cytoskeleton remodeling, in metabolism or apoptosis in CD28_Mut/Fus cases (Online Supplementary Figure S3). Accordingly, gene set enrichment analyses (GSEA) showed that the molecular signatures of CD28_Mut/Fus were differentially enriched in 20 gene sets as compared to CD28_WT, reflecting higher T-cell activation, proliferation or metabolic activity (Figure 1C and Online Supplementary Table S2).
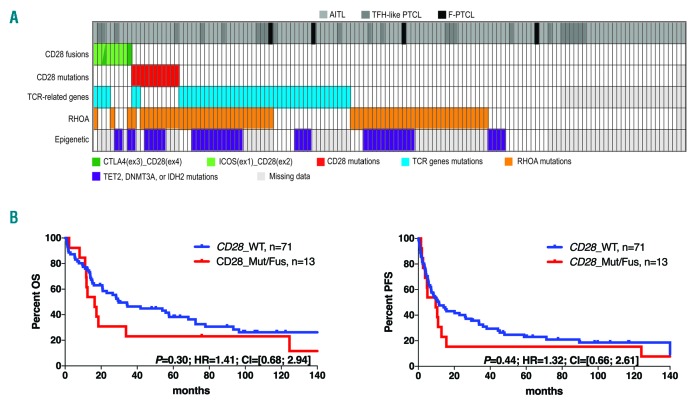
We then examined how CD28 fusions integrate into the mutational landscape and impact outcome in AITL and other PTCLs of TFH derivation. Figure 2A depicts the distribution of CD28 alterations and mutations in other TCR signaling-related genes (subsequently called TCR-related genes), RHOA and epigenetic modifiers (TET2 DNMT3A or IDH2) in the entire cohort of AITL, TFH-like PTCLs and F-PTCLs tested for CD28 fusions (138 cases in total). CD28 fusions (9/138) and point mutations (11/138) were mutually exclusive and altogether present in 15% of the samples. While the majority of CD28 point mutations (10/11 cases, 91%) were found in AITL, six of nine CD28 RNA fusions (67%) were present in other TFH PTCLs. CD28 mutations were mainly found in RHOA-mutated cases (10/11; 91%), but were virtually exclusive to mutations in other TCR-related genes (9/11; 82%). Conversely, CD28 fusions co-occurred with RHOA mutations in only three of nine cases (33%) whereas four of nine fusion-positive cases (44%) had mutations in other TCR-related genes.
Figure 2.
CD28 mutations and fusions in AITL and other TFH-derived PTCLs. (A) Landscape of genetic alterations of CD28, other TCR signaling-related (PLCG1, CTNNB1, GTF2I, PIK3R1, PDPK1, VAV1, FYN, CARD11, KRAS; STAT3, LCK, TRAF6, AKT1, PIK3R5, VAV2, MAPK3, PIK3CA), RHOA, and epigenetic modifiers (TET2, DNMT3A or IDH2) in 138 AITL and other PTCLs of TFH derivation. (B) Overall survival (OS; left panel) and progression-free survival (PFS; right panel) of patients with (red) or without (blue) CD28 mutations or fusions. Analyses are restricted to the 84 patients with AITL or other PTCLs of TFH derivation treated with anthracyclin-based chemotherapy. TFH-like PTCL: follicular helper T-cell like peripheral T-cell lymphoma; F-PTCL: follicular PTCL; AITL: angioimmunoblastic T-cell lymphoma.
The 84 patients who received intent-to-treat anthracyclin-based induction chemotherapy were considered for clinical outcome analysis. Of these, seven patients had CD28 mutations and six harbored a CD28 fusion. Of six CD28 fusion-positive patients, five relapsed and four died within the first year after diagnosis (two from disease, one from treatment toxicity and one from unknown causes). Considering 13 CD28_Mut/Fus versus 71 CD28 WT patients, no significant difference was observed in overall survival (OS) or progression free survival (PFS) (Figure 2B). However, focusing on the subset of 59 TFH-derived PTCL previously characterized for mutations in a large panel of TCR-related genes who received an anthracyclin based regimen, three patients carrying at least one alteration (mutation or fusion) in one or more gene(s) related to TCR signalling other than RHOA (TCR_Mut/Fus), (36 patients, 61%) showed a trend towards shorter PFS (P=0.07) compared to other patients who carried RHOA mutations only (RHOA_Mut) (14 patients, 24%) or were WT for all tested genes (TCR_WT) (nine patients, 15%) (Online Supplementary Figure S4). These data expand our previous findings of the negative connotation of these alterations on patient outcome.
In conclusion, our findings definitively confirm that the CD28 RNA fusions with CTLA4 or ICOS are overall rare events in PTCL (4.8%). This is unambiguously demonstrated by the screening of a large cohort of frozen cases in two different laboratories. Specifically, the prevalence of the CTLA4(ex3)_CD28(ex4) fusion (found in two TFH-PTCLs and one CTCL) was only 1% in our cohort. The most common fusion was ICOS(ex1)_CD28(ex2) (73% of fusions), which was previously reported in one out of 20 AITL cases subjected to RNAseq.2 In AITL and other TFH-derived PTCL, where these fusions are most prevalent (6.5% of cases), they are mutually exclusive to CD28 mutations. Thus, taking into account both point mutations and fusions, CD28 ranks second after RHOA among the altered TCR-related gene in the group of PTCLs of TFH derivation, found in 15% of the patients. Considering the patients with mutations in TCR-related genes other than RHOA, 29% of them harbor CD28 alterations. Finally, within the limit of this retrospective series, the cases harboring CD28 Mut/Fus did not differ from others in terms of OS or PFS, and had gene expression programming (GEP) enriched in signatures reflecting higher T-cell activation and higher proliferation.
Supplementary Material
Acknowledgments
The authors acknowledge Catherine Chapuis (Pathology, Lausanne) for her technical assistance.
Footnotes
Funding: this study was supported by grants received from the Association pour la Recherche contre le Cancer (ARC) (N.PJA 20151203507), the Fondation pour la Recherche Médicale (FRM) (N.DEQ20160334875), the MEDIC foundation and the Fond National Suisse (FNS) grant 310030_172954.
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Kataoka K, Nagata Y, Kitanaka A, et al. Integrated molecular analysis of adult T cell leukemia/lymphoma. Nat Genet. 2015;47(11):1304–1315. [DOI] [PubMed] [Google Scholar]
- 2.Rohr J, Guo S, Huo J, et al. Recurrent activating mutations of CD28 in peripheral T-cell lymphomas. Leukemia. 2016;30(5):1062–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vallois D, Dobay MPD, Morin RD, et al. Activating mutations in genes related to TCR signaling in angioimmunoblastic and other follicular helper T-cell–derived lymphomas. Blood. 2016;128(11):1490–1502. [DOI] [PubMed] [Google Scholar]
- 4.Wang L, Ni X, Covington KR, et al. Genomic profiling of Sezary syndrome identifies alterations of key T cell signaling and differentiation genes. Nat Genet. 2015;47(12):1426–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palomero T, Couronne L, Khiabanian H, et al. Recurrent mutations in epigenetic regulators, RHOA and FYN kinase in peripheral T cell lymphomas. Nat Genet. 2014;46(2):166–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sakata-Yanagimoto M, Enami T, Yoshida K, et al. Somatic RHOA mutation in angioimmunoblastic T cell lymphoma. Nat Genet. 2014;46(2):171–175. [DOI] [PubMed] [Google Scholar]
- 7.Yoo HY, Sung MK, Lee SH, et al. A recurrent inactivating mutation in RHOA GTPase in angioimmunoblastic T cell lymphoma. Nat Genet. 2014;46(4):371–375. [DOI] [PubMed] [Google Scholar]
- 8.Yoo HY, Kim P, Kim WS, et al. Frequent CTLA4-CD28 gene fusion in diverse types of T-cell lymphoma. Haematologica. 2016;101(6):757–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ungewickell A, Bhaduri A, Rios E, et al. Genomic analysis of mycosis fungoides and Sezary syndrome identifies recurrent alterations in TNFR2. Nat Genet. 2015;47(9):1056–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sekulic A, Liang WS, Tembe W, et al. Personalized treatment of Sézary syndrome by targeting a novel CTLA4:CD28 fusion. Mol Genet Genomic Med. 2015;3(2):130–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gong Q, Wang C, Rohr J, Feldman AL, Chan WC, McKeithan TW. Comment on: Frequent CTLA4-CD28 gene fusion in diverse types of T-cell lymphoma, by Yoo et al. Haematologica. 2016;101(6):e269–e270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dobay MP, Lemonnier F, Missiaglia E, et al. Integrative clinicopathological and molecular analyses of angioimmunoblastic T-cell lymphoma and other nodal lymphomas of follicular helper T-cell origin. Haematologica. 2017;102(4):e148–e151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lemonnier F, Couronne L, Parrens M, et al. Recurrent TET2 mutations in peripheral T-cell lymphomas correlate with TFH-like features and adverse clinical parameters. Blood. 2012;120(7):1466–1469. [DOI] [PubMed] [Google Scholar]
- 14.de Leval L, Parrens M, Le Bras F, et al. Angioimmunoblastic T-cell lymphoma is the most common T-cell lymphoma in two distinct French information data sets. Haematologica. 2015;100(9):e361–e364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen L, Flies DB. Molecular mechanisms of T cell co-stimulation and co-inhibition. Nat Rev Immunol. 2013;13(4):227–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.