Abstract
A kinesin family member 5b (KIF5B)-MET proto-oncogene, receptor tyrosine kinase (MET) rearrangement was reported in patients with lung adenocarcinoma but its oncogenic function was not fully evaluated. We used one-step reverse transcription-polymerase chain reaction for RNA samples to screen for the KIF5B-MET fusion in 206 lung adenocarcinoma and 28 pulmonary sarcomatoid carcinoma patients. Genomic breakpoints of KIF5B-MET were determined by targeted next-generation sequencing. Soft agar colony formation assays, proliferation assays, and a xenograft mouse model were used to investigate its oncogenic activity. In addition, specific MET inhibitors were administered to evaluate their anti-tumor activities. A KIF5B-MET fusion variant in a patient with a mixed-type adenocarcinoma and sarcomatoid tumor was identified, and another case was found in a pulmonary sarcomatoid carcinoma patient. Both cases carried the same chimeric gene, a fusion between exons 1–24 of KIF5B and exons 15–21 of MET. KIF5B-MET-overexpressing cells exhibited significantly increased proliferation and colony-forming ability. Xenograft tumors harboring the fusion gene demonstrated significantly elevated tumor growth. Ectopic expression of the fusion gene stimulated the phosphorylation of KIF5B-MET as well as downstream STAT3, AKT, and ERK1/2 signaling pathways. The MET inhibitors significantly repressed cell proliferation; phosphorylation of downstream STAT3, AKT, and ERK1/2; and xenograft tumorigenicity. In conclusion, the KIF5B-MET variant was demonstrated to have an oncogenic function in cancer cells. These findings have immediate clinical implications for the targeted therapy of subgroups of non-small cell lung cancer patients.
Abbreviations: ADC, adenocarcinoma; ALK, anaplastic lymphoma kinase; CT, computed tomography; HGF, hepatocyte growth factor; IHC, immunohistochemical; KIF5B, kinesin family member 5b; MET, MET proto-oncogene, receptor tyrosine kinase; NSCLC, non-small cell lung cancer; PTK, protein tyrosine kinase; RET, RET proto-oncogene; TKI, tyrosine kinase inhibitor; TTF-1, thyroid transcription factor-1
Introduction
In non-small cell lung cancer (NSCLC), a considerable proportion of patients present extensive genomic instability within their tumors. This instability occurs at different levels, ranging from simple nucleotide changes, gene amplifications, chromosomal and structural rearrangements to gains or losses of entire chromosomes [1]. Structural chromosome rearrangements have been shown to result in gene fusions and can be a major driving force for tumorigenesis [2]. Originating from chromosomal translocations, oncogenic fusion proteins are frequently observed in lung adenocarcinoma (ADC); these are typically composed of an N-terminal dimerization domain provided by the fusion partner protein fused to the kinase domain of a tyrosine kinase. These fusion proteins often lead to kinase domain activation and provide ideal targets for the development of anti-cancer therapies [3].
More than 80 chromosome rearrangements have been reported in NSCLC [4]. Anaplastic lymphoma kinase (ALK), ROS proto-oncogene tyrosine-protein kinase (ROS1) and RET proto-oncogene (RET) fusions are the most common chromosome translocations involving a kinase domain [5], [6]. Kinesin family member 5b (KIF5B) is the second frequent fusion partner of ALK [4], [7], and also serves as a fusion donor for RET [8]. The KIF5B gene is located on chromosome 10p11.22 and encodes the KIF5B protein. It has been suggested that KIF5B-protein tyrosine kinase (PTK) fusion proteins result in aberrant kinase activation of the PTK [6], [7], [9]. These fusion genes usually contain at least the N-terminal 1–15 exons of KIF5B, which encode a protein comprising the kinesin motor and coiled-coil domains that mediate homodimerization [7]. In lung tissue, KIF5B is ubiquitously and constitutively expressed, and its active promoter might consequently drive the activation of ALK/RET tyrosine kinases and subsequently enhance their downstream oncogenic effects [10], [11].
Recently, another KIF5B-PTK fusion transcript, KIF5B-MET, has been discovered in lung ADC patients; this consisted of a chimeric fusion of exons 1–24 of KIF5B to either exons 14–21 [12], [13] or exons 15–21 [14] of the MET proto-oncogene, receptor tyrosine kinase (MET), which was denoted as K24;M14 or K24:M15 based on the last KIF5B and first MET exons in the fusion, respectively. This is a remarkable finding given that MET is a known oncogenic driver of lung cancer. Given that MET gene alterations were not uncommon among lung ADC and pulmonary sarcomatoid carcinoma [15], [16], [17], we sought to screen KIF5B-MET fusions in such two histologic types of NSCLC in Taiwanese patients. We identified two patients with same KIF5B-MET fusion—K24:M15 variant. We further performed in vivo functional assays and applied a mouse xenograft model to corroborate its oncogenic activity, as well as its responsiveness to specific MET tyrosine kinase inhibitors.
Material and Methods
Sample Collection
A total of 206 diagnostic lung ADC and 28 pulmonary sarcomatoid carcinoma patient specimens, which had been identified as negative for EGFR, ALK, KRAS, HER2, or RET mutations in previous cohort studies [15], [18], [19], were included. Specimens were obtained from primary/metastatic surgical excisions, computed tomography-guided biopsies, and malignant pleural effusions at the National Taiwan University Hospital (Taipei, Taiwan). The histology of the lung cancer tissue was determined through pathological examination using immunohistochemical (IHC) staining for thyroid transcription factor-1 (TTF-1), CK7, CK5/6, p40, vimentin, and hematoxylin and eosin. Tumor specimen collection, preparation, and RNA extraction were performed as previously described [15]. Informed, written consent was obtained from all study participants prior to specimen collection. The institutional review board of the hospital approved the study.
Hematoxylin–Eosin and Immunohistochemical Staining
Tissue sections (4 μm thick) were dewaxed and rehydrated. For hematoxylin–eosin staining, sections were reacted with hemalum, which was followed by counterstaining with eosin. For IHC staining, slides were subjected to antigen retrieval and allowed to react with an anti-human c-MET C-terminus antibody (Spring Bioscience Corp., Pleasanton, CA; clone SP44, 1:50 dilution). The incubation procedure, counterstaining with hematoxylin, and negative controls were performed as described previously [15].
Reverse Transcription-Polymerase Chain Reaction Analysis of KIF5B-MET Transcript
The RT-PCR conditions were based on the manufacturer's protocol. Briefly, 50–100 ng of total RNA was used as template and the following components were added: (1) 10 ml 5× reaction buffer, (2) 2 ml dNTP mix (10 mM each), (3) 3 ml of 10 mM forward and reverse primer each, (4) 2 ml QIAGEN OneStep RT-PCR enzyme mix and (5) RNase-free water to reach a total volume of 50 ml. The RT-PCR reaction was initiated at 50 °C for 30 minutes, heated to 95 °C for 15 minutes, then followed by 40 cycles of denaturation at 94 °C for 50 seconds, annealing at 60 °C for 50 seconds, extension at 72 °C for 1 minutes, and a final extension at 72 °C for 10 minutes. The primers were forward primers for KIF5B exon 15 (KIF5B-E15F) (5′-TAAGGAAATGACCAACCACCAG-3′), KIF5B exon 20 (KIF5B-E20F) (5′-AGCCACAGATCAGGAAAAGA-3′), or KIF5B exon 24 (KIF5B-E24F) (5′-ATCGCAAACGCTATCAGCAAGA-3′), and a reverse primer for MET exon 15 (MET-E15R) (5′-TGCACAATCAGGCTACTGGGCC-3′). The resulting products were directly sequenced in both directions using the primers KIF5B-E15F/KIF5B-E20F/KIF5B-E24F and MET-E15R. The National Center for Biotechnology Information (NCBI) KIF5B (NM_004521.2; 963 amino acids) and MET (NM_000245.3; 1390 amino acids) were used as reference sequences.
Mapping the Translocation Breakpoint by Targeted Next Generation Sequencing
DNA was extracted from fresh frozen cell pellets using QIAamp tissue DNA extraction kits (Qiagen, Valencia, CA). Based on human genome 19, NCBI build GRCh37, 94 probes for the targeted region (32,304,368–32,306,347 in chromosome 10 for KIF5B and 116,411,501–116,415,300 in chromosome 7 for MET) and four internal controls in chromosomes 6, 10, 16, and 17 were used for targeted capture reactions. In total, 2 μg of genomic DNA was sonicated into fragment sizes of approximately 800 base pairs, and used for library construction. We next performed target enrichment using the double capture protocol (Roche NimbleGen Inc., Madison, WI) following the manufacturer's instruction. Read sequences were analyzed using the Illumina Miseq system (Illumina, San Diego, CA) and with different tools (bwa.0.7.4, picard-tools-1.90, Genome Analysis-TK-2.5-2, and IGV2.1.16). The genomic KIF5B-MET translocation breakpoints were further confirmed by PCR using a forward primer specific for KIF5B intron 24, (5′-GGAACCTGGGAAGTGGAGAT-3′) and a reverse primer for MET intron 14 (5′-GAATGGAATCAGGGCAAAGA-3′), which was followed by Sanger sequencing.
Plasmids
KIF5B-MET (K24;M15) fusion cDNA was constructed by double PCR from available full-length human KIF5B and MET cDNA, which were purchased from GenScript Inc. (Piscataway, NJ) and Addgene (Cambridge, MA), respectively. Full-length flag-tagged KIF5B-MET cDNA was further cloned into the pLAS2w.Ppuro vector backbone for constitutive gene expression in cell lines and for use in functional assays. The pLAS2w.Ppuro vector was obtained from the RNA interference core laboratory, Academia Sinica, Taiwan.
Cell Lines
Ba/F3 and 293 T cell lines were obtained from the Center of Genomic Medicine, National Taiwan University. Ba/F3 cells were cultured in RPMI1640 containing 10% fetal bovine serum, 100 U/mL penicillin, 100 μg/mL streptomycin (Invitrogen Corporation, Carlsbad, CA) and 1 ng/mL IL-3 (R&D systems, Minneapolis, MN). 293 T cells were maintained in DMEM supplemented with 10% FBS. Both cell lines were cultured at 37 °C in a humidified atmosphere of 5% CO2.
Chemicals, Antibodies, and Western Blotting
The c-MET inhibitors crizotinib and capmatinib were purchased from Selleckchem (Houston, TX). Cell lysates were prepared and analyzed by western blotting as described previously [20]. The following primary antibodies were obtained from Cell Signaling Technology Inc. (Danvers, MA) and were used for these analyses: anti-MET (#3148), anti-phospho-Y1234/Y1235-MET (#3129), anti-STAT3 (#4904), anti-phospho-Y705-STAT3 (#9138), anti-AKT (#9272), anti-phospho-S473-AKT (#4051), anti-ERK1/2 (#4695), and anti-phospho-T202/Y204-ERK1/2 (#4376). The anti-flag antibody (#F3165) was purchased from Sigma (St. Louis, MO). The anti-α-tubulin and anti-β-actin antibodies were purchased from Millipore (Billerica, MA). Chemiluminescent signals on western blots were captured using the ImageQuant LAS 4000 system (GE Healthcare Life Sciences, Issaquah, WA).
Soft Agar Colony Formation Assay
293 T cells were transfected with plasmids encoding KIF5B-MET(K24;M15) or empty vector using Lipofectamine 2000 Reagent (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer's protocol. Transfected cells were grown for 48 hours, which was followed by subculturing cells with 1 μg/mL puromycin selection for 3 days. For soft agar colony formation, 3000 cells were mixed with 0.35% agarose and were plated onto 0.5% agar in six-well plates with triplicates. The cells were incubated for 3 weeks, stained with crystal violet and the number of transformed foci was counted. Three wells of cell colonies were scored.
Cell Proliferation and Viability Assays
For cell proliferation assay, 1 × 105 Ba/F3 cells harboring the empty vector or KIF5B-MET were re-suspended in 24-well plates. Cells were grown in the presence or absence of IL-3 and were collected at the indicated time, then cells were incubated MTT (3-(4,5-cimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide) to form formazan. The product was dissolved in 10% SDS (Sodium dodecyl sulfate) and the absorbance was recorded at 550 nm. For cell viability assay, 1 × 106 Ba/F3 cells were re-suspended into 24-well plates in the presence or absence of IL-3 and with different doses of crizotinib or capmatinib for 72 hours, and were analyzed as above. Cell viability was calculated as: (Test OD/ Control OD) x 100. The median inhibitory concentration (IC50) of the drugs (crizotinib and capmatinib) was calculated using polynomial regression analysis using Microsoft Excel. All the experiments were performed at least three independent experiments in triplicate.
Xenograft Tumor Model
Four-week-old athymic nude mice (BALB/c nude) were used in these studies under protocols approved by the Institutional Animal Care and Use Committee of the College of Medicine, National Taiwan University. The methods were performed in accordance with the approved guidelines. In total, 2 × 107 Ba/F3 cells harboring the empty vector or KIF5B-MET were resuspended in 200 μL PBS, and then separately subcutaneously injected into the left and right flanks of mice, respectively. Subcutaneous tumor volumes were calculated by the formula: tumor volume = 1/2(length × width2) [21]. The tumors were measured during the first 20 days or the indicated number of days after cell injection. The animals were sacrificed with carbon dioxide after the indicated number of days and the weights of the xenograft tumors were recorded and analyzed.
Statistical Analysis
Data are presented as the means ± SD from at least three independent experiments. Groups were compared using the Student's t test. Two-sided p-values less than 0.05 were considered significant. All analyses were performed using SPSS software version 15.0 for Windows (SPSS Inc., Chicago, IL).
Results
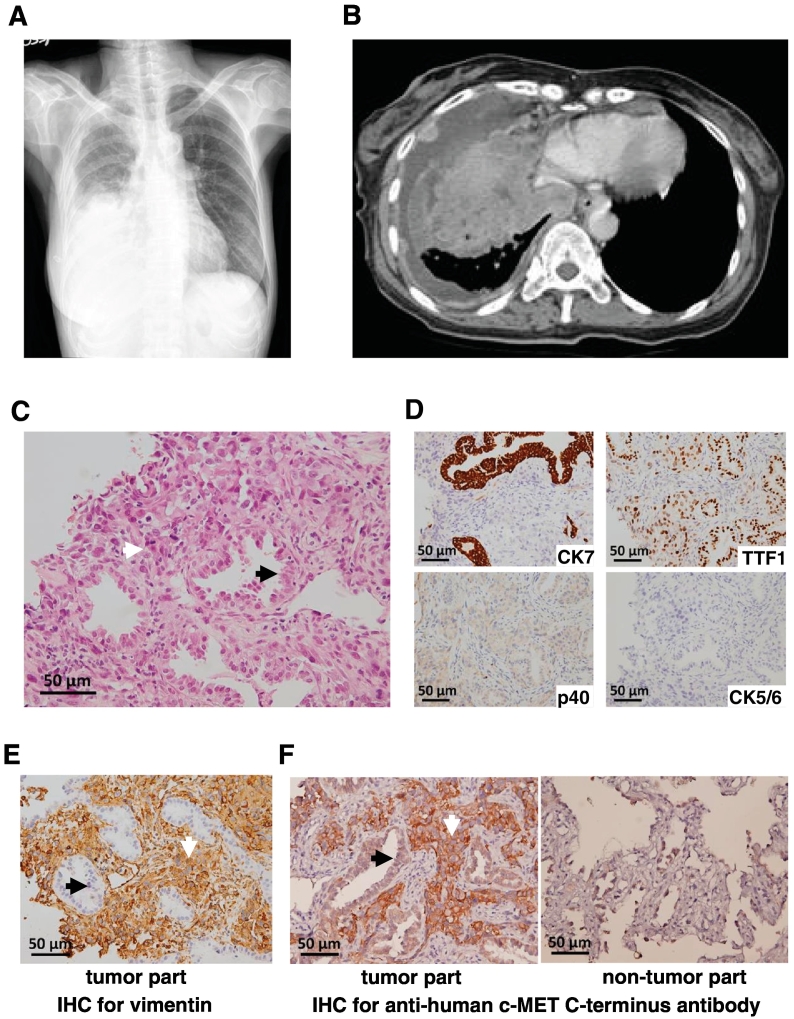
Among 206 lung ADC patients, the KIF5B-MET(K24;M15) fusion variant was only detected in one person (1/206, 0.5%). The 61-year-old woman, who had never smoked, presented with a huge mass and massive pleural effusion on her chest roentgenogram (Figure 1A) as well as multiple pleural-based tumors in the right thoracic cavity based on chest computed tomography (CT) (Figure 1B). We performed chest sonography-guided biopsy and her pathological examination revealed a mixed type of both adenocarcinoma and sarcomatoid components (Figure 1C). The adenocarcinoma component was positive for CK7 and TTF-1 but negative for p40, CK5/6, and vimentin; only the sarcomatoid component was positive for vimentin (Figure 1D and E). IHC staining using an anti-c-MET carboxyl-terminal antibody revealed that the high-grade tumor cells with sarcomatoid features were strongly positive, whereas the adenocarcinoma cells were equivocally positive for c-MET (Figure 1F, left panel), with a similar intensity to the background reactive pneumocytes (Figure 1F, right panel). The patient received six courses of pemetrexed plus cisplatin treatment. Follow-up chest CT showed that lung tumors rapidly progressed with extended pleural seeding. The patient then received hospice care and expired 5 months after initial diagnosis.
Figure 1.
Radiological images and pathology of the adenocarcinoma cancer patient analyzed in this study. (A) Chest roentgenogram showing a mass in the right lower lung with massive pleura effusion. (B) Chest computerized tomography revealed a huge heterogeneous mass and multiple pleural-based tumors in the right thoracic cavity. (C) Pathologic examination showing both adenocarcinoma (acinar pattern, black arrow) and sarcomatoid component (interstitial growth pattern, white arrow) with mitotic figures (hematoxylin–eosin stain, original magnification 400×). (D) Immunohistochemical analyses of the cancer. From left to right, upper, CK7 (positive in adenocarcinoma only), TTF-1 (positive in both adenocarcinoma and sarcomatoid component); lower, p40 (negative), CK5/6 (negative) (original magnification 400×). (E) Positive immunohistochemical staining of vimentin in sarcomatoid component only. (F) Immunohistochemical staining for c-MET showing strong positivity in sarcomatoid component but not in adenocarcinoma cells of the tumor (left panel); adenocarcinoma cells showing a similar intensity to the background reactive pneumocytes of the non-tumor part (right panel) (original magnification 400×). Black arrows indicate adenocarcinoma cells and white arrows mark sarcomatoid component in this figure.
We next examined 28 available pulmonary sarcomatoid carcinomas from the cohort in our institute. We discovered another case with KIF5B-MET(K24;M15) fusion variant (3.6%, 1 of 28). The second case was a 76-year-old man, who had a 20 pack-year history of smoking and presented with a protruding mass over his right anterior chest wall. His chest CT showed a soft tissue mass over the right upper lung with pleural effusion. Chest sonography-guided biopsy diagnosed a stage IV pulmonary sarcomatoid carcinoma. Due to poor performance status and rapidly progressed tumor invasion, he received supportive and hospice care only. He passed away approximately 1 month after initial diagnosis.
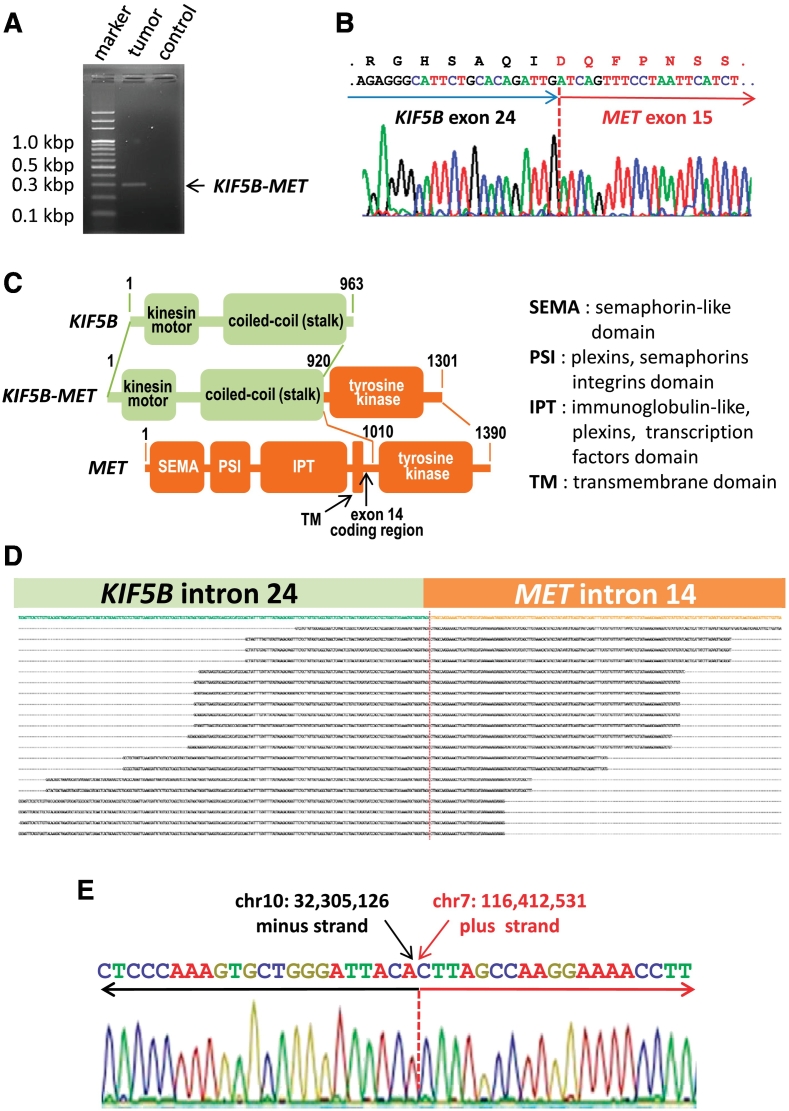
For KIF5B-MET fusion gene analysis, we directly amplified the fusion point of the KIF5B-MET transcript using one-step RT-PCR and a single product of 298 base pairs was obtained from the first adenocarcinoma-sarcomatoid mixed-type tumor (Figure 2A). Nucleotide sequencing of the product confirmed the fusion point of KIF5B-MET to be the end of exon 24 of KIF5B and the first nucleotide of MET exon 15 (Figure 2B). The second identified sarcomatoid tumor carried the same KIF5B-MET(K24;M15) fusion as the first case and its pathological feature were shown in Supplementary Figure S1. This KIF5B-MET fusion protein contained the complete N-terminal kinesin motor and coil-coiled domains of KIF5B and the entire C-terminal tyrosine kinase domain of MET (Figure 2C). We further mapped the genomic translocation breakpoint by targeted next generation sequencing (Figure 2D). Sanger sequencing confirmed the breakpoints of the translocation with nucleotide resolution (chr10:32,305,126–chr7:116,412,531), and the KIF5B-MET gene was created from fusion of the minus strand of chromosome 10 (p11.22) to the plus stand of chromosome 7 (q31.2) (Figure 2E).
Figure 2.
Discovery of a transforming KIF5B-MET fusion variant in non-small cell lung cancer. (A) KIF5B-MET-specific one step RT-PCR using RNA extracted from the primary tumor samples and an amplification product of 298 base pairs was identified. The control sample was from another individual with lung adenocarcinoma individual. (B) cDNA sequencing chromatograms showing the conjoined regions. The break point exons and the amino acids are indicated. (C) Functional domain details of KIF5B-MET fusion variant kinase. Protein domains, gene partners, as well as numbers of amino acids are indicated. (D) Alignment of sequence reads mapping to genomic KIF5B and MET based on targeted next-generation sequencing. (E) DNA sequence chromatograms showing the conjoined regions at the genomic DNA sequence level of the fusion gene.
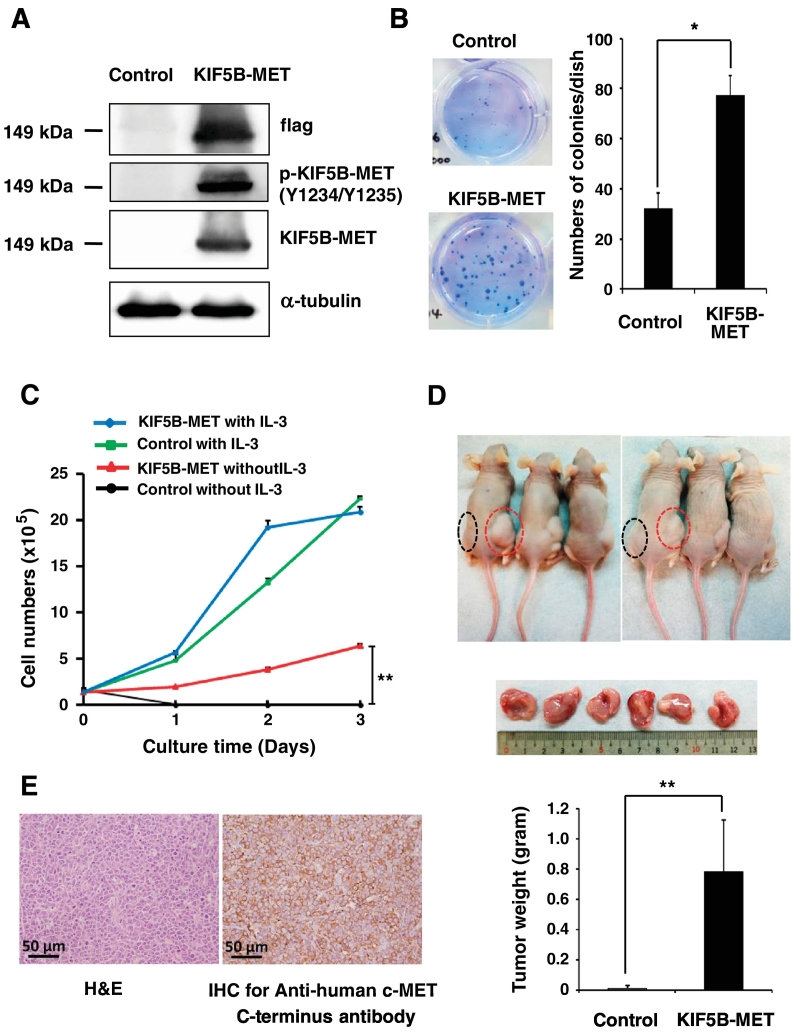
To examine the function of the KIF5B-MET fusion in cancer cells, we first cloned the flag tagged KIF5B-MET fusion cDNA into a plasmid, selected a single clone, and expressed it in 293 T cells. Based on the mRNA sequence, the size of predicted flag tagged KIF5B-MET protein is approximately 149 kDa, which was different from wild-type pro-c-Met of 175 kDa and mature c-Met β subunit of 145 kDa. Western blot analysis using anti-flag and anti-MET antibodies for KIF5B-MET-positive cells revealed a protein band, of which the size corresponded to the predicted value of KIF5B-MET and was different from that of wild-type MET (Supplementary Figure S2). We further observed the same size of protein bands for anti-phosphorylated MET and anti-flag in western blot analysis in KIF5B-MET-positive cells, indicating the constitutively expression of phosphorylated KIF5B-MET (Figure 3A). Soft agar colony formation assays showed that the enforced expression of KIF5B-MET in 293 T cells caused an increase in colony numbers compared to that in control cells carrying an empty vector (Figure 3B). For cell proliferation assays, KIF5B-MET-transfected Ba/F3 cells and empty vector-transfected Ba/F3 cells demonstrated similar proliferation ability in the presence of interleukin-3 (IL-3), whereas KIF5B-MET-positive cells maintained significant survival and proliferative ability in the absence of IL-3, indicating that KIF5B-MET promotes survival in Ba/F3 cells deprived of IL-3 (Figure 3C). We further determined that enforced expression of KIF5B-MET results in a significant increase in Ba/F3 xenograft tumor size and weight in nude mice; in contrast, the control group showed no detectable tumors (P < .001; Figure 3D). The xenograft tumors harboring the KIF5B-MET(K24;M15) fusion gene exhibited strong expression of human carboxy-terminal c-MET based on IHC examination (Figure 3E). An identical KIF5B-MET fusion transcript by RT-PCR in xenograft tumors with primary cancer tissue was detected (Supplementary Figure S3). These findings confirm that the KIF5B-MET fusion protein possesses oncogenic function and promotes tumor cell growth both in vitro and in vivo.
Figure 3.
The KIF5B-MET fusion kinase enhances cell proliferation. (A) 293 T cells were transfected with an empty vector (control) or KIF5B-MET. The expression levels of flag, KIF5B-MET, and KIF5B-MET phosphorylated at Tyr1234/Tyr1235 in cell lysates were analyzed by western blotting. (B) Expression of KIF5B-MET fusion protein enhances soft agar colony forming ability of positive cells in vitro. Colony numbers for 293 T cells transfected with KIF5B-MET or an empty vector (control) were quantitated; Student's t-test, average ± SD; n = 3, * P < .05. (C) IL-3-independent survival of Ba/F3 cells expressing KIF5B-MET. Ba/F3 cells stably expressing KIF5B-MET or empty vector (control) were grown and cells were collected at the time indicated and counted; Student's t-test, average ± SD; n = 6, ** P < .01. (D) Transforming activities of KIF5B-MET in mouse xenograft model. Nude mice were subcutaneously injected Ba/F3 cells carrying KIF5B-MET into the right flank (shown in red circles) and carrying empty vector into the left flank (control, shown in black circles). Tumor formation was examined after 20 days (upper panel). Xenograft tumors were not palpable or visible in the control group; tumors from the KIF5B-MET group were resected (middle panel), and weights were recorded (lower panel); Student's t-test, average ± SD; n = 12, ** P < .01. (E) Hematoxylin–eosin stain (left panel) and anti-human c-MET immunohistochemical stain (right panel) of the xenograft tumor Ba/F3 cells expressing KIF5B-MET.
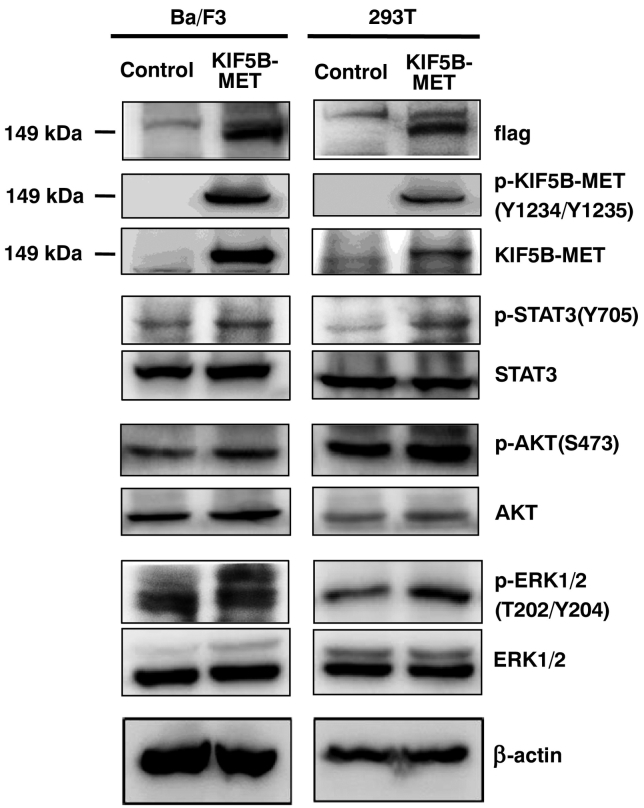
We hypothesized that the KIF5B-MET fusion protein could activate signaling pathways similar to those resulting from aberrant MET activation [22], [23], [24]. We therefore investigated the phosphorylation status of related signaling molecules in KIF5B-MET-overexpressing Ba/F3 and 293 T cells (Figure 4). We observed that this resulted in significantly higher levels of phosphorylation at Y1234/Y1235 of KIF5B-MET, in the absence of hepatocyte growth factor (HGF) stimulation. In addition, STAT3, AKT, and ERK1/2 signaling molecules were also activated (Figure 4).
Figure 4.
KIF5B-MET positively regulates Met downstream signaling pathways. Ba/F3 and 293 T cells expressing empty vector (control) or KIF5B-MET were analyzed for total KIF5B-MET, STAT3, AKT, and ERK1/2, as well as for phosphorylated Tyr1234/Try1235 KIF5B-MET, phosphorylated Tyr705 STAT3, phosphorylated Ser473 AKT, and phosphorylated Thr202/Try204 ERK1/2 by western blotting. Beta-actin was used as a loading control.
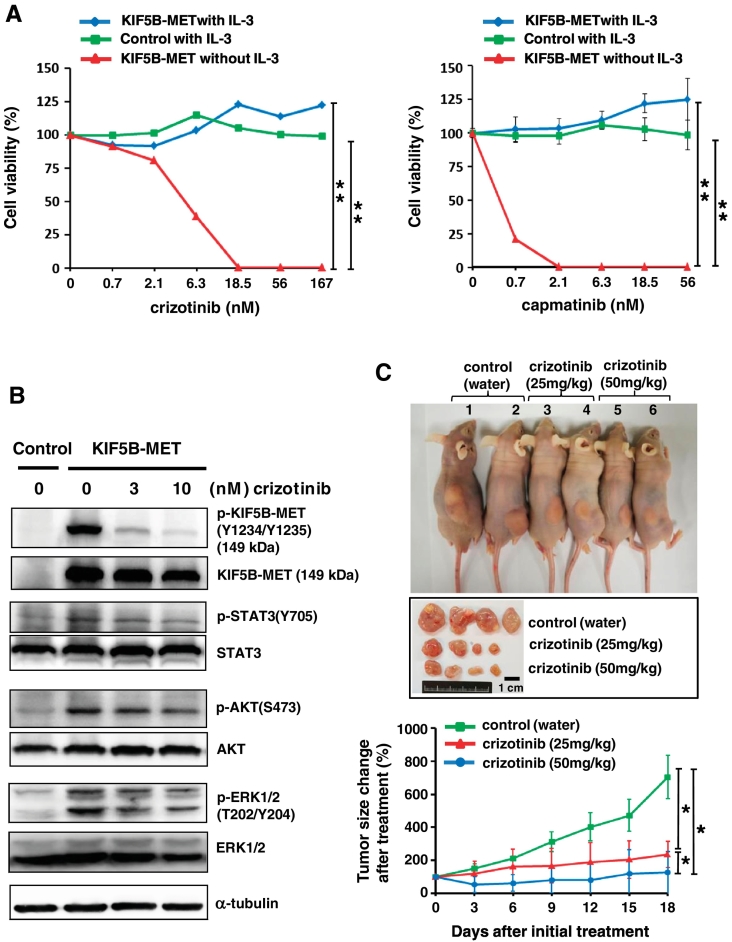
As KIF5B-MET induces KIF5B-MET tyrosine kinase phosphorylation, activates downstream signaling pathways, and promotes tumor growth in Ba/F3 cells, we further determined the efficacy of MET tyrosine kinase inhibitors in KIF5B-MET-positive cells. Two c-MET inhibitors—crizotinib and capmatinib—were selected. The IC50 value was 5.0 nM for crizotinib and 0.4 nM for capmatinib with KIF5B-MET-expressing Ba/F3 cells in IL-3-depleted culture media (Figure 5A). Western blotting showed that the phosphorylation of KIF5B-MET was significantly reduced by crizotinib in a concentration-dependent manner (Figure 5B). The suppression of KIF5B-MET kinase activity by 10 nM of crizotinib affected STAT3, AKT, and ERK signaling pathways. We next evaluated the efficacy of crizotinib in vivo using a Ba/F3 xenograft model (Figure 5C). Tumor-bearing mice were treated with either water, low-dose crizotinib (25 mg/kg), or high-dose crizotinib (50 mg/kg) once daily for 18 days. Both low-dose and high-dose crizotinib were well tolerated in this study. As expected, we observed marked tumor regression and growth inhibition in both crizotinib groups compared to those in the control group. In addition, the high dose crizotinib group showed better treatment response (Figure 5C, lower panel).
Figure 5.
Effect of MET tyrosine kinase inhibitors on growth of KIF5B-MET-positive cancer cells in vivo and in vitro. (A) Specific effects of crizotinib and capmatinib on KIF5B-MET positive cells. Ba/F3 cells stably expressing KIF5B-MET or empty vector (control) were seeded and grown in the presence or absence of IL-3. Control group in the absence of IL-3 was not shown because of no cells growth under this condition. Cells were treated with different doses of crizotinib (left panel) or capmatinib (right panel). Crizotinib and capmatinib were serially diluted 1:3 from 500 nM, and the selected highest doses are indicated. Cell viability was analyzed by MTT assays. Student's t-test, average ± SD; n = 6, ** P < .01. (B) Suppression of KIF5B-MET kinase activity by crizotinib was found to affect KIF5B-MET and associated downstream signaling pathways by western blotting. (C) Therapeutic effect of crizotinib on KIF5B-MET positive tumor xenografts. Nude mice were injected subcutaneously with 1 x 107 Ba/F3 cells harboring an empty vector in the left flank and KIF5B-MET in the right flank. Different daily doses of crizotinib were administered orally after tumor growth reached 200 mm3. Tumor sizes were recorded and compared to that at initial treatment. The upper panel shows an individual group of mice harboring xenograft tumors on day 18. The middle panel shows representative resected tumors. The lower panel demonstrates serial changes in tumors size after initial treatment; Student's t-test, average ± SD; n = 7, * P < .05; ** P < .01.
Discussion
Some genomic alterations that drive lung carcinogenesis are gross chromosomal rearrangements, and most of these lead to constitutive activation of oncogenes encoding tyrosine kinases such as ALK, RET, and ROS1. In this study, we identified two patients with identical fusion variant involving the MET gene—KIF5B-MET(K24;M15)—in NSCLC. This variant was discovered in a case of mixed-type (ADC-sarcomatoid) NSCLC and another case of pulmonary sarcomatoid carcinoma. Our KIF5B-MET(K24;M15) variant was different from the previously identified KIF5B-MET(K24;M14) fusion with retained the entire exon 14 of MET, but was same as most recently reported KIF5B-MET(K24;M15) fusion variant. Clinical features of cases that were previously published and those described herein, consisting of NSCLC harboring KIF5B-MET fusions, are summarized in Table 1. The lung ADC patient harboring KIF5B-MET(K24;M14) fusion protein showed a dramatic tumor shrinkage under crizotinib treatment for 10 months [13]. Another case with KIF5B-MET(K24;M15) gene fusion also presented with clinical benefit more than 8 months after crizotinib initiation [14]. In this study, both of our cases did not receive MET inhibitors. The patient with mixed type of ADC-sarcomatoid tumor had poor conventional chemotherapy response and both patients had poor overall survival. However, our pre-clinical data corroborated the oncogenic functions and potential effect targeted tyrosine kinase inhibitors of MET on KIF5B-MET-expressing cancer cells. Therefore, our results provide evidence and suggest the clinical implications for the targeted therapy of subgroups of NSCLC patients with KIF5B-MET gene rearrangement.
Table 1.
Characteristics of Patients With Non-small Cell Lung Cancer Harboring KIF5B-MET Fusions
| No. Ref. | Age | Sex | Smoking status | Stage | KIF5B-MET variant | Histology | Treatment | OS Mon | Dead/Alive |
|---|---|---|---|---|---|---|---|---|---|
| 1. [12] | NA | NA | NA | NA | K24;M14 | ADC | NA | NA | NA |
| 2. [13] | 51 | F | never | IV | K24;M14 | ADC | Bvz/Pem-Cis and Pem (PD after 26 mon), SAIT301# (PD), crizotinib (10 mon), then other salvage therapies | >36 | Dead |
| 3. [14] | 33 | F | yes | IV | K24;M15 | ADC | Crizotinib (PR) | >8 | Alive |
| 4⁎ | 61 | F | never | IV | K24;M15 | ADC + PSC | Pem-Cis (PD) | 5 | Dead |
| 5⁎ | 76 | M | yes | IV | K24;M15 | PSC | supportive | 1 | Dead |
Abbreviations: ADC, adenocarcinoma; Bvz, bevacizuma, Mon, months; NA, not available; OS, overall survival; PD, progressive disease; PR, partial response; PSC, pulmonary sarcomatoid carcinoma; Pem-Cis, pemetrexed- cisplatin; Ref, reference.
Cases showing in this study.
SAIT301 is a c-MET inhibitor.
MET belongs to a family of receptor tyrosine kinases and was first identified as the chromosomal rearrangement product TPR (translocated promoter region)-MET in a carcinogen-induced human osteogenic sarcoma cell line [25]. The TPR-MET oncoprotein comprised a fusion of the N-terminal dimerization domains of TRP, which contain a leucine zipper motif, to the carboxy terminus of the MET. The oncogenic TRP-MET protein lacks the juxtamembrane tyrosine-binding site Y1003, recognized by the E3 ligase c-Cbl. As a result, the fusion protein fails to bind c-Cbl, and escapes from ubiquitin-dependent protein degradation [26]. Clinically, the TRP-MET rearrangement has been discovered in gastric carcinoma [20], [27], but not in lung cancer. In NSCLC, we demonstrated the existence of the KIF5B-MET(K24;M15) fusion, together with recently reported HLA-DRB1-MET gene fusion [28], were similar to TRP-MET in that it also lacks the c-Cbl binding site and might be one of the possible mechanism which drives its oncogenic function. Aside from this, other possible mechanism, such as auto-dimerization, could also drive tumor growth. Further study will need to be conducted in future. Nevertheless, MET fusions might be another class of actionable alteration in NSCLC.
KIF5B has unusual properties, in that it can be a donor gene for rearrangements in certain diseases. The KIF5B gene was originally identified as a fusion partner for platelet-derived growth factor receptor-α (PDGFRα) in a patient suffering from hypereosinophilia [29]. The donor, KIF5B, was also found to be fused to important driver genes of NSCLC encoding PTKs, including ALK, RET, and MET. Specific tyrosine kinase inhibitors have shown efficacy towards cells carrying KIF5B-associated fusion genes in the clinical setting or in animal studies. Idiopathic hypereosinophilic syndrome patients harboring KIF5B-PDGFα demonstrated a complete response to imatinib, a specific inhibitor targeting the tyrosine kinase domain of PDGFRα [29]. A lung ADC patient harboring a KIF5B-ALK fusion showed a partial response to the ALK-tyrosine kinase inhibitor (TKI) crizotinib, with 6 months of progression free survival [30]. In NIH3T3 cells expressing the KIF5B-RET protein, inhibition of anchorage-independent growth was observed after treatment with the RET kinase inhibitor vandetanib [31]. Transgenic mice specifically expressing human KIF5B-RET in the lung developed neoplasms in the pulmonary alveolar epithelial cells, and tumorigenesis was markedly suppressed by vandetanib treatment [31]. In this study, we demonstrated that KIF5B-MET-positive cells were sensitive to crizotinib and capmatinib, suggesting a potential target for existing TKIs.
In humans, MET acts as a membrane receptor for HGF [32]. Following HGF binding, MET subunits dimerize and transphosphorylate tyrosine residues, thereby activating signaling activity. Subsequently, phosphorylated tyrosine residues recruit many transducers and adaptors that mediate downstream Ras/Raf/MAPK, PI3K/AKT/mTOR, STAT3/5, and/or NF-κB complex signaling cascades [33], [34]. In addition, MET is rapidly internalized and is subsequently recruited to peripheral early endosomes, which triggers the sustained activation of downstream pathways [33]. These receptor activation cascades are observed in cancer cells with high-level MET amplification [35] or alterations in MET exon 14 splicing [36]. We demonstrated that our KIF5B-MET variant activates downstream signaling pathways, in a similar manner to that observed in the H1993 lung cancer cell harboring MET amplification [37]. These findings suggest patients with three different MET alterations might share concordant in vivo drug sensitivity to the MET-TKI crizotinib. Our study highlights the importance of conducting future, prospective clinical trials to clarify whether specific MET inhibitors can be used for the treatment of lung cancer patients harboring KIF5B-MET fusion mutations.
Conclusions
We identified two cases of NSCLC harboring the KIF5B-MET(K24;M15) fusion gene, one from lung ADC and another from pulmonary sarcomatoid carcinoma. In this pre-clinical data, cancer cells harboring the KIF5B-MET variant demonstrated oncogenic functions and activated MET-associated signaling pathways. Given the recent development of several MET inhibitors and their potential therapeutic efficacy for tumors expressing this KIF5B-MET fusion protein, we expect that further investigation of KIF5B-MET fusions will identify patients with NSCLC subtypes who are suitable for treatment using such targeted therapies.
Acknowledgments
Acknowledgments
We wish to thank the Department of Medical Research, the Center of Genomic Medicine and Department of Medical Genetics at the National Taiwan University (Taipei, Taiwan) for providing the laboratory facilities and technical support. We would also like to thank Editage (www.editage.com) for English language editing.
Funding
This work was supported by the Far Eastern Memorial Hospital National Taiwan University Hospital Joint Research Program grants (104-FTN09 and 106-FTN15); and the grant of Far Eastern Memorial Hospital (FEMH-2017-C-030), Taiwan.
Conflicts of Interest
Shih JY reports speaking honoraria from AstraZeneca, Roche, Boehringer Ingelheim, Eli Lilly, Pfizer, Novartis, Merck Sharp & Dohme, Ono Pharmaceutical and Bristol-Myers Squibb.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.neo.2018.06.007.
Contributor Information
Chien-Hung Gow, Email: gowchien@gmail.com.
Yi-Nan Liu, Email: benson1032@gmail.com.
Huei-Ying Li, Email: hueiying@ntu.edu.tw.
Min-Shu Hsieh, Email: mshsieh065@gmail.com.
Shih-Han Chang, Email: scottc07@gmail.com.
Sheng-Ching Luo, Email: scluo5052@gmail.com.
Tzu-Hsiu Tsai, Email: thtsai1971@ntu.edu.tw.
Pei-Lung Chen, Email: paylong@gmail.com.
Meng-Feng Tsai, Email: fj479340534@gmail.com.
Jin-Yuan Shih, Email: jyshih@ntu.edu.tw.
Appendix A. Supplementary data
Supplementary figures
References
- 1.Shen Z. Genomic instability and cancer: an introduction. J Mol Cell Biol. 2011;3:1–3. doi: 10.1093/jmcb/mjq057. [DOI] [PubMed] [Google Scholar]
- 2.Akbari Moqadam F, Lange-Turenhout EA, Aries IM, Pieters R, den Boer ML. MiR-125b, miR-100 and miR-99a co-regulate vincristine resistance in childhood acute lymphoblastic leukemia. Leuk Res. 2013;37:1315–1321. doi: 10.1016/j.leukres.2013.06.027. [DOI] [PubMed] [Google Scholar]
- 3.Gunby RH, Sala E, Tartari CJ, Puttini M, Gambacorti-Passerini C, Mologni L. Oncogenic fusion tyrosine kinases as molecular targets for anti-cancer therapy. Anti Cancer Agents Med Chem. 2007;7:594–611. doi: 10.2174/187152007784111340. [DOI] [PubMed] [Google Scholar]
- 4.Huret J. Lung: Translocations in Adenocarcinoma. Atlas Genet Cytogenet Oncol Haematol. 2015;19:311–315. [Google Scholar]
- 5.Shaw AT, Hsu PP, Awad MM, Engelman JA. Tyrosine kinase gene rearrangements in epithelial malignancies. Nat Rev Cancer. 2013;13:772–787. doi: 10.1038/nrc3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kohno T, Ichikawa H, Totoki Y, Yasuda K, Hiramoto M, Nammo T, Sakamoto H, Tsuta K, Furuta K, Shimada Y. KIF5B-RET fusions in lung adenocarcinoma. Nat Med. 2012;18:375–377. doi: 10.1038/nm.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takeuchi K, Choi YL, Togashi Y, Soda M, Hatano S, Inamura K, Takada S, Ueno T, Yamashita Y, Satoh Y. KIF5B-ALK, a novel fusion oncokinase identified by an immunohistochemistry-based diagnostic system for ALK-positive lung cancer. Clin Cancer Res. 2009;15:3143–3149. doi: 10.1158/1078-0432.CCR-08-3248. [DOI] [PubMed] [Google Scholar]
- 8.Takeuchi K, Soda M, Togashi Y, Suzuki R, Sakata S, Hatano S, Asaka R, Hamanaka W, Ninomiya H, Uehara H. RET, ROS1 and ALK fusions in lung cancer. Nat Med. 2012;18:378–381. doi: 10.1038/nm.2658. [DOI] [PubMed] [Google Scholar]
- 9.Daire V, Pous C. Kinesins and protein kinases: key players in the regulation of microtubule dynamics and organization. Arch Biochem Biophys. 2011;510:83–92. doi: 10.1016/j.abb.2011.02.012. [DOI] [PubMed] [Google Scholar]
- 10.Qian Y, Chai S, Liang Z, Wang Y, Zhou Y, Xu X, Zhang C, Zhang M, Si J, Huang F. KIF5B-RET fusion kinase promotes cell growth by multilevel activation of STAT3 in lung cancer. Mol Cancer. 2014;13:176. doi: 10.1186/1476-4598-13-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wong DW, Leung EL, Wong SK, Tin VP, Sihoe AD, Cheng LC, Au JS, Chung LP, Wong MP. A novel KIF5B-ALK variant in nonsmall cell lung cancer. Cancer. 2011;117:2709–2718. doi: 10.1002/cncr.25843. [DOI] [PubMed] [Google Scholar]
- 12.Stransky N, Cerami E, Schalm S, Kim JL, Lengauer C. The landscape of kinase fusions in cancer. Nat Commun. 2014;5:4846. doi: 10.1038/ncomms5846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho JH, Ku BM, Sun JM, Lee SH, Ahn JS, Park K, Ahn MJ. KIF5B-MET Gene rearrangement with robust antitumor activity in response to crizotinib in Lung adenocarcinoma. J Thorac Oncol. 2018;13:e29–e31. doi: 10.1016/j.jtho.2017.10.014. [DOI] [PubMed] [Google Scholar]
- 14.Plenker D, Bertrand M, de Langen AJ, Riedel R, Lorenz C, Scheel AH, Muller J, Bragelmann J, Dassler-Plenker J, Kobe C. Structural alterations of MET trigger response to MET kinase inhibition in lung adenocarcinoma patients. Clin Cancer Res. 2018;24:1337–1343. doi: 10.1158/1078-0432.CCR-17-3001. [DOI] [PubMed] [Google Scholar]
- 15.Gow CH, Hsieh MS, Wu SG, Shih JY. A comprehensive analysis of clinical outcomes in lung cancer patients harboring a MET exon 14 skipping mutation compared to other driver mutations in an East Asian population. Lung Cancer. 2017;103:82–89. doi: 10.1016/j.lungcan.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 16.Qiu T, Li W, Zhang T, Xing P, Huang W, Wang B, Chu L, Guo L, Liu X, Li Y. Distinct MET protein localization associated with MET exon 14 mutation types in patients with non-small-cell Lung Cancer. Clin Lung Cancer. 2018;19:e391–e398. doi: 10.1016/j.cllc.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 17.Saffroy R, Fallet V, Girard N, Mazieres J, Sibilot DM, Lantuejoul S, Rouquette I, Thivolet-Bejui F, Vieira T, Antoine M. MET exon 14 mutations as targets in routine molecular analysis of primary sarcomatoid carcinoma of the lung. Oncotarget. 2017;8:42428–42437. doi: 10.18632/oncotarget.16403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gow CH, Chang HT, Lim CK, Liu CY, Chen JS, Shih JY. Comparable clinical outcomes in patients with HER2-mutant and EGFR-mutant lung adenocarcinomas. Genes Chromosomes Cancer. 2017;56:373–381. doi: 10.1002/gcc.22442. [DOI] [PubMed] [Google Scholar]
- 19.Tsai TH, Wu SG, Hsieh MS, Yu CJ, Yang JC, Shih JY. Clinical and prognostic implications of RET rearrangements in metastatic lung adenocarcinoma patients with malignant pleural effusion. Lung Cancer. 2015;88:208–214. doi: 10.1016/j.lungcan.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 20.Chang TH, Tsai MF, Su KY, Wu SG, Huang CP, Yu SL, Yu YL, Lan CC, Yang CH, Lin SB. Slug confers resistance to the epidermal growth factor receptor tyrosine kinase inhibitor. Am J Respir Crit Care Med. 2011;183:1071–1079. doi: 10.1164/rccm.201009-1440OC. [DOI] [PubMed] [Google Scholar]
- 21.Tomayko MM, Reynolds CP. Determination of subcutaneous tumor size in athymic (nude) mice. Cancer Chemother Pharmacol. 1989;24:148–154. doi: 10.1007/BF00300234. [DOI] [PubMed] [Google Scholar]
- 22.Gentile A, Trusolino L, Comoglio PM. The Met tyrosine kinase receptor in development and cancer. Cancer Metastasis Rev. 2008;27:85–94. doi: 10.1007/s10555-007-9107-6. [DOI] [PubMed] [Google Scholar]
- 23.Organ SL, Tsao MS. An overview of the c-MET signaling pathway. Ther Adv Med Oncol. 2011;3:S7–S19. doi: 10.1177/1758834011422556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frampton GM, Ali SM, Rosenzweig M, Chmielecki J, Lu X, Bauer TM, Akimov M, Bufill JA, Lee C, Jentz D. Activation of MET via diverse exon 14 splicing alterations occurs in multiple tumor types and confers clinical sensitivity to MET inhibitors. Cancer Discov. 2015;5:850–859. doi: 10.1158/2159-8290.CD-15-0285. [DOI] [PubMed] [Google Scholar]
- 25.Cooper CS, Blair DG, Oskarsson MK, Tainsky MA, Eader LA, Vande Woude GF. Characterization of human transforming genes from chemically transformed, teratocarcinoma, and pancreatic carcinoma cell lines. Cancer Res. 1984;44:1–10. [PubMed] [Google Scholar]
- 26.Peschard P, Park M. Escape from Cbl-mediated downregulation: a recurrent theme for oncogenic deregulation of receptor tyrosine kinases. Cancer Cell. 2003;3:519–523. doi: 10.1016/s1535-6108(03)00136-3. [DOI] [PubMed] [Google Scholar]
- 27.Soman NR, Correa P, Ruiz BA, Wogan GN. The TPR-MET oncogenic rearrangement is present and expressed in human gastric carcinoma and precursor lesions. Proc Natl Acad Sci U S A. 1991;88:4892–4896. doi: 10.1073/pnas.88.11.4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davies KD, Ng TL, Estrada-Bernal A, Le AT, Ennever PR, Camidge DR, Doebele RC, Aisner DL. Dramatic response to crizotinib in a patient with lung cancer positive for an HLA-DRB1-MET gene fusion. JCO Precis Oncol. 2017;(1):1–6. doi: 10.1200/PO.17.00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Score J, Curtis C, Waghorn K, Stalder M, Jotterand M, Grand FH, Cross NC. Identification of a novel imatinib responsive KIF5B-PDGFRA fusion gene following screening for PDGFRA overexpression in patients with hypereosinophilia. Leukemia. 2006;20:827–832. doi: 10.1038/sj.leu.2404154. [DOI] [PubMed] [Google Scholar]
- 30.Kim S, Kim TM, Kim DW, Go H, Keam B, Lee SH, Ku JL, Chung DH, Heo DS. Heterogeneity of genetic changes associated with acquired crizotinib resistance in ALK-rearranged lung cancer. J Thorac Oncol. 2013;8:415–422. doi: 10.1097/JTO.0b013e318283dcc0. [DOI] [PubMed] [Google Scholar]
- 31.Saito M, Ishigame T, Tsuta K, Kumamoto K, Imai T, Kohno T. A mouse model of KIF5B-RET fusion-dependent lung tumorigenesis. Carcinogenesis. 2014;35:2452–2456. doi: 10.1093/carcin/bgu158. [DOI] [PubMed] [Google Scholar]
- 32.Shiota G, Rhoads DB, Wang TC, Nakamura T, Schmidt EV. Hepatocyte growth factor inhibits growth of hepatocellular carcinoma cells. Proc Natl Acad Sci U S A. 1992;89:373–377. doi: 10.1073/pnas.89.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trusolino L, Bertotti A, Comoglio PM. MET signalling: principles and functions in development, organ regeneration and cancer. Nat Rev Mol Cell Biol. 2010;11:834–848. doi: 10.1038/nrm3012. [DOI] [PubMed] [Google Scholar]
- 34.Ponzetto C, Bardelli A, Zhen Z, Maina F, dalla Zonca P, Giordano S, Graziani A, Panayotou G, Comoglio PM. A multifunctional docking site mediates signaling and transformation by the hepatocyte growth factor/scatter factor receptor family. Cell. 1994;77:261–271. doi: 10.1016/0092-8674(94)90318-2. [DOI] [PubMed] [Google Scholar]
- 35.Smolen GA, Sordella R, Muir B, Mohapatra G, Barmettler A, Archibald H, Kim WJ, Okimoto RA, Bell DW, Sgroi DC. Amplification of MET may identify a subset of cancers with extreme sensitivity to the selective tyrosine kinase inhibitor PHA-665752. Proc Natl Acad Sci U S A. 2006;103:2316–2321. doi: 10.1073/pnas.0508776103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kong-Beltran M, Seshagiri S, Zha J, Zhu W, Bhawe K, Mendoza N, Holcomb T, Pujara K, Stinson J, Fu L. Somatic mutations lead to an oncogenic deletion of met in lung cancer. Cancer Res. 2006;66:283–289. doi: 10.1158/0008-5472.CAN-05-2749. [DOI] [PubMed] [Google Scholar]
- 37.Lutterbach B, Zeng Q, Davis LJ, Hatch H, Hang G, Kohl NE, Gibbs JB, Pan BS. Lung cancer cell lines harboring MET gene amplification are dependent on Met for growth and survival. Cancer Res. 2007;67:2081–2088. doi: 10.1158/0008-5472.CAN-06-3495. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures