Abstract
Bacteriophages (phages), or bacterial viruses, are the most abundant and diverse biological entities that impact the global ecosystem. Recent advances in metagenomics have revealed their rampant abundance in the biosphere. A fundamental aspect of bacteriophages that remains unexplored in metagenomic data is the process of recombination as a driving force in evolution that occurs among different viruses within the same bacterial host. Here, we systematically examined signatures of recombination in every gene from 211 species-level viral groups in a recently obtained dataset of the Earth’s virome that contain corresponding information on the host bacterial species. Our study revealed that signatures of recombination are widespread (84%) among the diverse viral groups. We identified 25 recombination-intense viral groups, widely distributed across the viral taxonomy, and present in bacterial species living in the human oral cavity. We also revealed a significant inverse association between the recombination-intense viral groups and Type II restriction endonucleases, that could be effective in reducing recombination among phages in a cell. Furthermore, we identified recombination-intense genes that are significantly enriched for encoding phage morphogenesis proteins. Changes in the viral genomic sequence by recombination may be important to escape cleavage by the host bacterial immune systems.
Introduction
Bacteriophages (phages), or bacterial viruses, are the most abundant and diverse group of biological entities on the planet1,2. Their abundance and lytic lifecycle impact global ecosystems, including nutrient and energy cycles as well as structures of microbial communities3–5. Temperate phages, which not only can integrate their genomes into their host’s chromosome but also transfer bacterial DNA to a new host by transduction, alter the biology of their hosts by introducing novel functions, such as virulence factors and drug resistance6. As a result, phages impact bacterial genome evolution and ecosystem biogeochemistry. Recent metagenomic studies have shown that phages are dominant members in the human microbiome and are suggested to have potential roles in maintaining health7,8. Thus, phages are of great interest in a number of disciplines such as medicine, virology, ecology, medicine, and environmental sciences.
Metagenomics have also revealed that a large virome is present anywhere in the biosphere, which is primarily composed of phages (i.e., the phageome)9. Moreover, the current classification of bacterial and archaeal viruses, curated by the International Committee on Taxonomy of Viruses (ICTV), only reflects a fraction of viral diversity; in particular, those currently present in public databases10. A recent, large-scale study of the Earth’s virome analyzed a huge amount of metagenomic sequence data from 3,042 geographically diverse samples to assess the global distribution, phylogenetic diversity, and host specificity of viruses11. The study discovered over 125,000 partial DNA viral genomes, increased the number of viral genes registered in public databases by 16-fold, and linked species-level viral groups to bacterial hosts using CRISPR spacers and transfer RNA matches12,13. The data have been utilized to develop a bacteria-phage interaction database14 and a bioinformatics pipeline for metagenomics-virus sequence discovery and virus clustering15.
Recombination is a fundamental driving force in evolution16–19, and occurs among different viruses inside the same bacterial host (e.g., co-infecting invasive viruses20, temperate phages and defective prophages21, or an invasive virus and a resident prophage21). Although phage genomes are generally known to be mosaic, with active recombination or horizontal genetic exchange22, recombination does not necessarily increase the average fitness of offspring23,24. It remains unexplored, in the rapidly increasing metagenomic data, whether such signatures of recombination are observed across various phylogenetic groups of phages. In addition, it is also unclear whether specific phylogenetic groups, or genes, are recombination-intense and show signatures of increased recombination due to natural selection. Thus, the fundamental aspect of recombination among phage genomes based on metagenomic data should be explored.
Here, we systematically examined signatures of recombination in every gene from 211 species-level viral groups in the Earth’s virome dataset that contain corresponding information on host bacterial species; (i) we determined recombination-intense viral groups throughout the virome dataset, (ii) we examined the relationship between recombination-intense viral groups and the potential strength of host immunity, and (iii) we closely examined the recombination-intense genes of the viral groups.
Results
Signatures of recombination are widespread among diverse viral groups
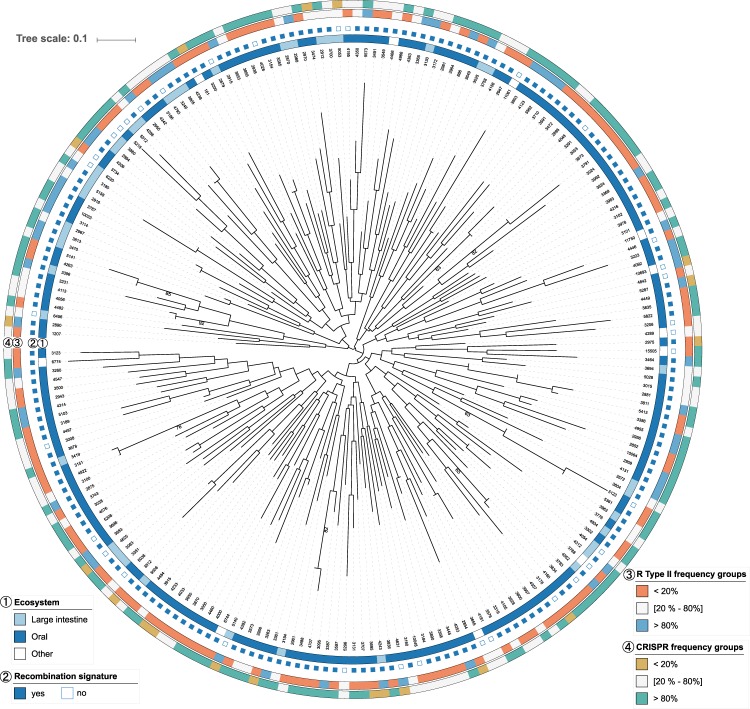
Among the more than 19,000 species-level viral groups defined in the Earth’s virome dataset composed of 3,042 metagenomic samples, we identified 211 viral groups that are usable for examining signatures of recombination and contain information on host bacterial species (Table S1). A proteomic tree of the 211 viral groups, constructed using the Virus Classification and Tree Building Online Resource (VICTOR) method10, is shown in Fig. 1, and a 16S rRNA gene maximum likelihood (ML) tree of the viral groups’ host bacterial species is shown in Fig. S1. The VICTOR tree (Fig. 1) revealed the presence of a very diverse dataset with long branches, almost no branch support (except for a few groups close to the tips of the tree), and an overall low phylogenetic signal. The host bacterial ML tree, on average, was well supported and reflected several major lineages (i.e., strains belonging to the phyla Firmicutes, Fusobacteria, Proteobacteria, and Bacteroidetes). About 75% of the viral groups originate from oral samples (Fig. 1) and account for 13.5% of all the metagenomic samples. For each viral group, we conducted a pan-genomic analysis and calculated the minimum number of recombination events (rmin) for every orthologous gene using the four-gamete test, a conservative method to locate pairs of closest segregating sites within 4 haplotypes that are likely to be generated by recombination between them. As a result, 88% of the viral groups originating from oral samples showed at least one recombination event per gene (Fig. 1), whereas the proportion was 69% of viral groups originating from other samples.
Figure 1.
Proteome-based VICTOR tree of the 211 viral groups. Scale bar indicates interproteomic distance inferred using the distance formula d4. Viral group IDs defined in the previous study11 are shown as leaf labels. The innermost colored ring (1) indicates oral and intestinal samples, respectively. The next ring (2) indicates the presence or absence of recombination signatures (i.e., minimum number of recombination events ≥1). The two outer rings (3,4) indicate frequency categories for Type II restriction endonucleases (3) and CRISPR arrays (4) in a host bacterial species.
Recombination-intense viral groups
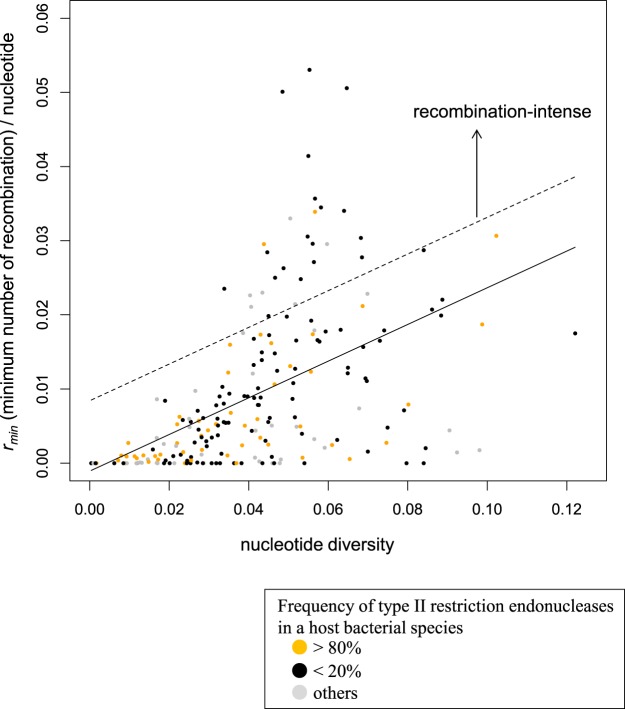
Next, we conducted a more quantitative analysis to explore the rates of recombination among viral groups. In order to account for the dependency of the rmin on gene length and nucleotide diversity, we took an approach similar to a previous study25. We plotted rmin per nucleotide versus nucleotide diversity of a viral group (Fig. 2), and calculated a linear regression line that captures the overall relationship between nucleotide diversity and the minimum number of recombination events per nucleotide. Using this relationship, we identified viral groups as recombination-intense that substantially deviate from the regression line. Based on the distribution of the deviation from the regression line among the 211 groups (Fig. S2), we identified the top 25 recombination-intense viral groups (above the dashed lines in Fig. 2 and Fig. S2, respectively). The host bacterial species of the 25 recombination-intense viral groups are shown in Table 1.
Figure 2.
Relationship between the minimum number of recombination events (rmin) per gene length and nucleotide diversity among the 211 viral groups. The x-axis and y-axis are nucleotide diversity and rmin per nucleotide, respectively. The solid line indicates the linear regression controlling for the number of sequences in each viral group. The dashed line indicates the threshold to focus on recombination-intense viral groups that considerably deviate from the regression line.
Table 1.
Recombination-intense viral groups.
| ID | Sample type | Host bacterial species | Nucleotide diversity | rmin (minimum number of recombinations)/nucleotide |
|---|---|---|---|---|
| 3050 | Oral (tongue dorsum) | Atopobium sp. ICM42b | 0.055 | 0.053 |
| 181 | Nasopharynx | Propionibacterium acnes | 0.048 | 0.050 |
| 2961 | Oral (tongue dorsum) | Streptococcus mitis | 0.065 | 0.051 |
| 4000 | Oral (tongue dorsum) | Atopobium sp. ICM42b | 0.055 | 0.041 |
| 3707 | Oral (tongue dorsum) | Atopobium sp. ICM42b | 0.057 | 0.036 |
| 3591 | Oral (supragingival plaque) | Actinomyces gerencseriae | 0.058 | 0.035 |
| 3319 | Oral (buccal mucosa) | Gemella haemolysans | 0.050 | 0.033 |
| 3549 | Oral (tongue dorsum) | Prevotella nanceiensis | 0.064 | 0.034 |
| 3189 | Oral (supragingival plaque) | Leptotrichia goodfellowii | 0.056 | 0.030 |
| 2890 | Oral (supragingival plaque) | Capnocytophaga granulosa | 0.057 | 0.034 |
| 2943 | Oral (tongue dorsum) | Campylobacter concisus | 0.068 | 0.030 |
| 3600 | Oral (attached/keratinized gingiva) | Gemella haemolysans | 0.060 | 0.030 |
| 3200 | Oral (buccal mucosa) | Streptococcus oligofermentans | 0.049 | 0.026 |
| 3358 | Oral (tongue dorsum) | Streptococcus gordonii | 0.047 | 0.025 |
| 4460 | Oral (tongue dorsum) | Atopobium sp. ICM42b | 0.056 | 0.027 |
| 3915 | Oral (buccal mucosa) | Gemella haemolysans | 0.040 | 0.023 |
| 3836 | Oral (tongue dorsum) | Oribacterium sinus | 0.069 | 0.028 |
| 3445 | Oral (buccal mucosa) | Streptococcus mitis | 0.053 | 0.025 |
| 2970 | Oral (supragingival plaque) | Capnocytophaga sp. CM59 | 0.055 | 0.031 |
| 2959 | Oral (buccal mucosa) | Actinomyces viscosus | 0.044 | 0.030 |
| 3184 | Oral (tongue dorsum) | Streptococcus mitis | 0.045 | 0.028 |
| 3776 | Oral (tongue dorsum) | Prevotella denticola | 0.052 | 0.021 |
| 2981 | Oral (tongue dorsum) | Lachnoanaerobaculum saburreum | 0.041 | 0.021 |
| 3576 | Oral (tongue dorsum) | Mogibacterium sp. CM50 | 0.045 | 0.020 |
| 4484 | Oral (tongue dorsum) | Lachnospiraceae bacterium oral taxon 082 | 0.084 | 0.029 |
Ordered by the extent of deviation from regression.
Almost all (24 out of 25) the recombination-intense viral groups were from oral samples, which is a statistically significant enrichment (P = 0.0066, Fisher’s exact test). Only viral group 181 originated from nasopharynx samples, but its host bacterial species, Propionibacterium acnes, forms part of the normal flora in the human oral cavity26. Namely, all the recombination-intense viral groups infect bacterial species that reside in the human oral cavity.
For the recombination-intense viral groups, we mapped reads back to the genomes to check their coverage (Fig. S3). Overall, the reads were mapped to the entire genome sequences, and the mean coverage per viral group was 24, eliminating possibilities that the elevated rates of recombination among the viral groups were primarily due to low coverage and metagenome mis-assembly in these sequences. There were a few viral groups showing some unusual patterns: sporadic low coverage typically in intergenic regions (viral groups 3050, 3776, 3576, 181, 3549, and 4000), and coverage difference along the genome (e.g. higher coverage at the 5′ end of viral group 4460 or 3184, and at the 3′ end of viral group 3200). The intergenic regions and the 3′ end of viral group 3200 clearly do not influence the results because they were not included in our pan-genomic analysis of orthologous genes. Regarding the higher coverage at the 5′ end of viral group 4460 or 3184, we confirmed that rates of recombination at the gene level (explained below in “Notable recombination-intense genes”) did not show significant increase in the regions (P = 1 and P = 0.33, one-sided Welch’s t-test).
Relationship between recombination-intense viral groups and host immunity
In order to deepen our understanding of species-level relationships between viral groups and their hosts, we examined the frequency of each host bacterial species’ immune systems (i.e., restriction-modification and CRISPR-Cas), self-protective mechanisms that cleave phage DNA when injected into a bacterial cell27. Type II restriction endonucleases, which recognize a specific nucleotide sequence and cleave at a fixed position28, and CRISPR arrays, which store the immunological memory of invading pathogens29, were included in the study. We classified three frequency categories (1: <20%, 2: between 20% and 80%, 3: >80%) of host immunity, which are shown in the outer rings of Fig. 1. As a result, we found a statistically significant inverse association between the recombination-intense viral groups and Type II restriction endonucleases; host bacteria species having Type II endonucleases at a frequency >80% (orange dots in Fig. 2) are rarely found (2/25) among the recombination-intense groups, compared to the other viral groups (P < 0.05 Fisher’s exact test). The result was the same when we used cutoff frequency 70% or 60%.There were only two such host bacterial species in the recombination-intense viral groups (Capnocytophaga granulosa and Actinomyces viscosus). This suggests that the high frequency of Type II endonucleases in host bacterial species could be effective in cleaving invading phages and reducing opportunities for recombination among phages in a cell. Furthermore, we did not find any other significant associations for either Type I or III restriction endonucleases, or CRISPR arrays in host bacterial species.
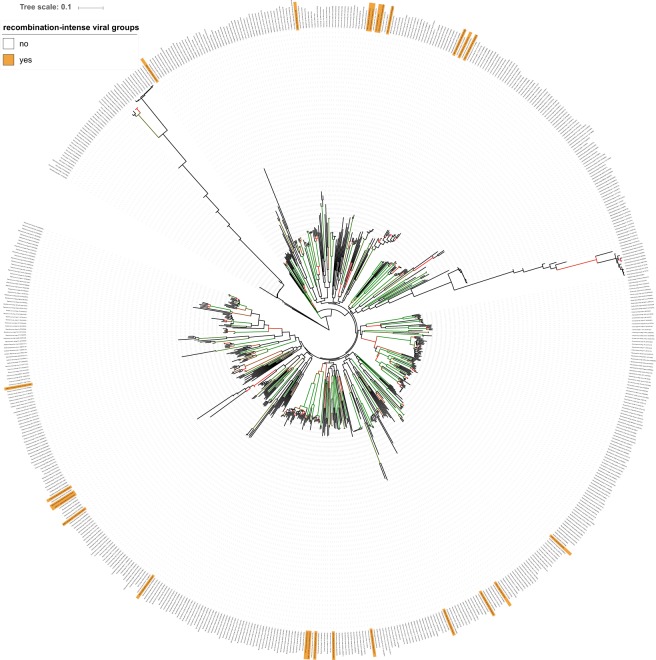
Additionally, we tried to reveal the phylogenetic nature of our metagenomic samples by conducting a joint phylogenetic analysis with an ICTV reference dataset using the VICTOR method10. The results of the analysis are shown in Fig. 3. Clearly, the recombination-intense viral groups are widely distributed across the ICTV taxonomy, even viral groups sharing the same (assumed) host bacterial species are not closely located in the phylogenic tree, indicating a diverse composition of the underlying metagenomic samples. However, at least some of the recombination-intense viral groups from this study were found in a well-supported subtree (e.g. groups 3319, 3707 and 4460), indicating a rather high phylogenetic relation to nearby ICTV reference phages. As observed earlier, the backbone of the tree is only weakly supported which is due to the nature of such diverse phage genomic datasets and is likely caused by multiple origins of prokaryotic viruses10,30.
Figure 3.
Phage proteomic tree based on the VICTOR method using a united dataset of comprehensive ICTV reference data and the recombination-intense viral groups. Leaf labels representing the recombination-intense viral groups are highlighted in orange. Branch support is indicated by color from red (50%) to green (100%). The vicinity of these metagenomic samples to actual ICTV phage species provides hints regarding their composition. Scale bar indicates interproteomic distances calculated via the distance formula d4. The tree was rooted at the midpoint69.
Notable recombination-intense genes
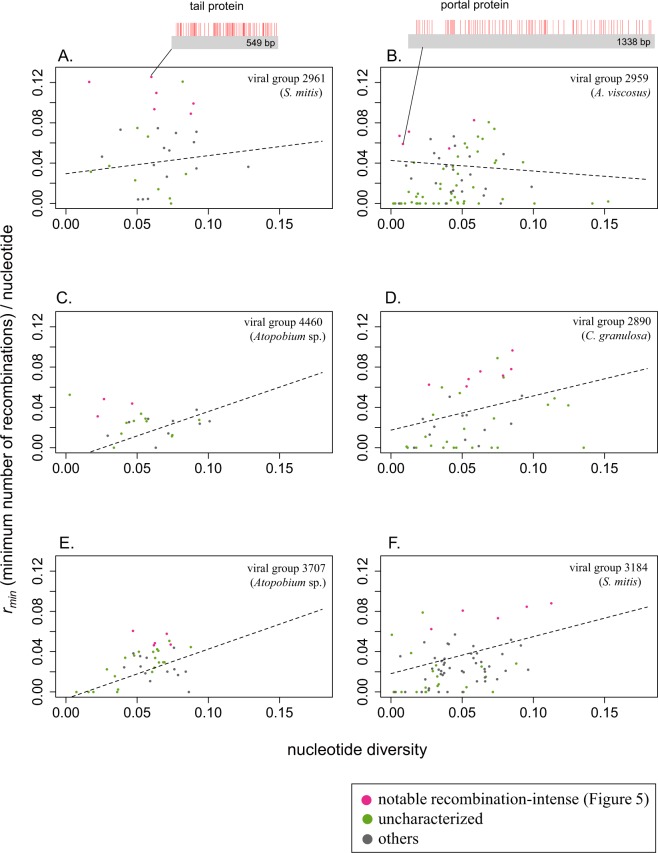
We further explored recombination-intense genes by similarly analyzing the relationship between rmin per nucleotide and nucleotide diversity at the gene level in each recombination-intense viral group (Figs 4 and S5). Substantial deviations from the regression were found; for example, in a gene encoding a phage tail protein (Fig. 4A) and a gene encoding a phage portal protein (Fig. 4B), with recombination breakpoints found throughout both genes. Because approximately 90% of such genes were initially annotated as hypothetical, we conducted iterative protein searches based on representing both query and database sequences by profile hidden Markov models31 using a UniProt database. We examined all hits with >99% probability of being true positives, and identified 89 notable genes in 24 viral groups (listed in Table S2).
Figure 4.
Examples of notable recombination-intense genes deviating from the regression of rmin per gene length on nucleotide diversity in each viral group. The x-axis and y-axis are the same as in Fig. 2. Pink: notable recombination-intense genes (Fig. 5 and Table S2). Green: uncharacterized genes. Gray: others. Recombination breakpoints in a phage tail gene and a portal gene are shown as red vertical bars at the top. The dashed line indicates the linear regression controlling for the number of sequences in each gene.
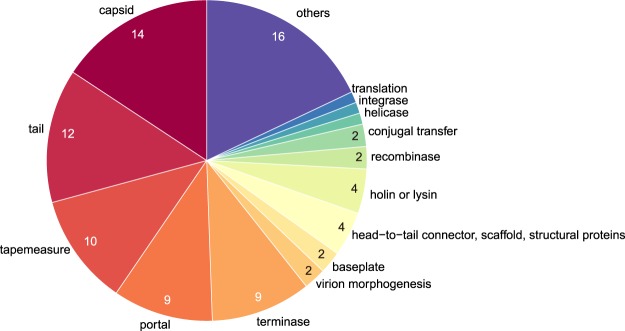
A breakdown of the notable recombination-intense genes is shown in Fig. 5. Approximately 75% of this set are genes that encode phage morphogenesis proteins. Among these, approximately 19% are associated with head morphogenesis, including genes for capsid, virion morphogenesis, and scaffold proteins32. Approximately 21% are associated with phage neck and DNA packaging, including portal protein, head-to-tail connector, and terminase33,34. Approximately 27% are associated with phage tail, including the genes for tail protein, tape measure protein, and baseplate33,35,36. Moreover, approximately 4% are DNA-associated genes, including the genes for integrase, helicase, and recombinase. Approximately 4% are lysis-associated genes, including holin and endolysin. Holin and endolysin are essential for host cell lysis in the lytic lifecycle37. Finally, the frequency of such genes in the pan-genome of other viral groups is approximately 27%, indicating significant enrichment among the notable recombination-intense genes (P < 10−15, chi-squared test).
Figure 5.
Breakdown of notable recombination-intense genes. The number of genes in each category is shown in the pie chart. More detailed information on each gene is shown in Table S2.
Discussion
The recombination events we detected and examined are those that have survived in the viral groups, and which are inevitably affected by natural selection17. These genes are likely to be under selective pressure from host bacterial immunity attacking foreign DNAs (i.e., restriction-modification and CRISPR-Cas systems), as this cleavage would effectively prevent survival and propagation of phages. These genes were found in the recombination-intense viral groups, all of which infect bacterial species living in human oral cavities, where hundreds of thousands of CRISPR spacer groups are transcribed38. Hence it is likely that there is a genetic conflict invoked by the coevolution of phages and host oral bacteria by means of classic Red Queen dynamics39. The signature of highly elevated recombination would reflect the evolutionary conflict in which phages continuously change their genomic sequences by recombination to escape cleavage by the host bacterial immune system in the human oral cavity. A recent study showed that acquisition of random mutations is not sufficient for phages to completely escape CRISPR-Cas targeting in a continuous co-culture27. Another study of long-term bacterium-phage coevolution experiments showed the presence of multiple phages increased phage persistence by enabling recombination-based formation of chimeric phage genomes in which sequences heavily targeted by CRISPR were replaced40. Therefore, recombination would play an important role, at least for recombination-intense oral phages, in escaping bacterial immunity.
Similarly, our analysis of Type II restriction endonucleases suggest that its high frequency in host bacterial species could be effective in cleaving invading phages and reducing opportunities for recombination among phages in a cell. This raises the interesting possibility that restriction-modification could be more effective than CRISPR-Cas in oral bacteria for dealing with phages. Recently, it was suggested that host bacteria are generally incapable of utilizing CRISPR-Cas to eradicate phages from the human oral cavity as well as the gut38. Further studies are warranted to elucidate the relative roles of restriction-modification and CRISPR-Cas in contributing to bacterial immunity. A recent study, in a different context, showed that CRISPR-Cas and restriction-modification function additively, at least against conjugative antibiotic resistance plasmid transfer in Enterococcus faecalis41.
Alternatively, the enrichment of recombination-intense viral groups in human oral cavities could be explained in terms of frequency of recombinases among the viral groups. However, there was no statistically significant difference in frequency of viral groups carrying a recombinase (P = 0.6, chi-square test) between the recombination-intense viral groups (38%) and the others (30%).
Regarding the bacterial host species of the recombination-intense viral groups (Table 1), many are recognized as part of the normal human oral flora, but sometimes cause disease. For example, some Atopobium species have been identified as agents of chronic periodontitis and bacteremia42,43. The viridans group streptococci (VGS), which consist of Streptococcus mitis, Streptococcus gordonii, and Streptococcus oligofermentans, can cause a wide range of infections in humans, including bacteremia, infective endocarditis, and moderate or severe clinical disease (e.g., VGS shock syndrome)44. Among others, associations with endocarditis have been reported for Actinomyces viscosus45, Gemella haemolysans46, and Leptotrichia goodfellowii47, while associations with bacteremia were reported for Leptotrichia goodfellowii47 and Capnocytophaga species48. In addition, Campylobacter concisus has been linked to prolonged diarrhea and inflammatory bowel disease49. Meanwhile, the phage lytic lifecycle has been shown to play a role in preventing outgrowth and dysbiosis by killing the bacterial host39. According to a recent review50, endogenous phages can play an important role in human oral health by limiting overgrowth of bacteria and maintaining the commensal microbiota at acceptable levels that can then be controlled by the human immune system. The evolution of recombination-intense phages could contribute to such maintenance function against oral host bacterial species during their coevolution.
Given these observations, the recombination-intense viral groups which are particularly present in oral viromes, could boost phage evolution and contribute to the maintenance of the commensal microbiota (eubiosis)39. This is the first systematic, quantitative study of recombination in phage genomes across >200 diverse viral groups, and the first study to explore viral-host relationships from a viewpoint of recombination and host immune systems.
Materials and Methods
Selection of viral groups and individual scaffolds
From 17,803 viral groups defined in the previous study11 as having ≥90% bidirectional average amino acid identity and ≥50% total alignment fraction, we extracted 211 groups consisting of at least four individuals (scaffolds) required for the examination of minimum number of recombination events and carrying information of host bacteria at the species level. The information was inferred in the previous study11 and is stored in IMG/VR database13 by either perfect matches of viral tRNAs, or matches of CRISPR-Cas spacers requiring at least 95% identity over the whole spacer length, and allowing only 1–2 SNPs at the 5′ end of the sequence.
Scaffold IDs for each individual in each viral group is listed in Table S3. We downloaded nucleotide sequences of the scaffolds through the ‘Expert Review’ version of IMG/M ER (https://img.jgi.doe.gov/mer/)10 datamart12. We made the nucleotide sequence data directly downloadable at https://figshare.com/articles/211viralgroups_fas_tgz/6223769.
Proteomic tree of the viral groups
A proteomic tree of the 211 viral groups was constructed using the Virus Classification and Tree Building Online Resource (VICTOR) method, publicly available at https://victor.dsmz.de10. From the three distinct trees generated by VICTOR, the one based on distance formula d4 was chosen because this formula is robust51 when using incomplete genomes and represented the most reasonable choice in view of the partially incomplete nature of the metagenomic samples. The tree was finally visualized and annotated using the iTOL web service52.
Pan-genome analysis of recombination and nucleotide diversity
The prediction of protein-coding genes and gene annotation was performed using Prokka53 software with the “-k Viruses” option. We conducted pan-genome analyses using the Roary54 pipeline with “-e --mafft -i 90 -z” options and obtained alignments of 12,319 orthologous genes. The minimum number of recombination events (rmin) was calculated for each orthologous gene using the four-gamete test55 that locates pairs of closest segregating sites within 4 haplotypes that are likely to be generated by recombination between them. We used the method implemented in the PGEToolbox56, which filters gaps in advance. Basic population genetic statistics (e.g. nucleotide diversity) were also calculated for each orthologous gene using DnaSAM57.
Annotation of host bacteria species
We downloaded nucleotide sequences of individual genomes and their annotations of the host bacterial species that are available in the MBGD database58 (http://mbgd.genome.ad.jp/htbin/getData?table=genome). For each host bacterial species, we checked the frequency of individuals that have either Type I, II, or III restriction enzymes, or CRISPR-arrays that were detected by CRISPRDetect59.
A 16S rRNA gene sequence phylogeny was inferred by the GGDC web server51, available at https://ggdc.dsmz.de/, using the DSMZ phylogenomics pipeline60 adapted to single genes. A multiple sequence alignment was created with MUSCLE61. Maximum likelihood (ML) and maximum parsimony (MP) trees were inferred from the alignment with RAxML62 and TNT63, respectively. For ML, rapid bootstrapping in conjunction with the autoMRE bootstrapping criterion64, and subsequent search for the best tree was used; for MP, 1000 bootstrapping replicates were used in conjunction with tree-bisection-and-reconnection branch swapping and 10 random sequence addition replicates. The sequences were checked for compositional bias using the Chi-squared test as implemented in PAUP*65.
Identification and taxonomic assignments of recombination-intense viral groups
We calculated the sum of the rmin across the orthologous genes divided by the sum of their lengths, in base pairs, for each viral group. We also calculated nucleotide diversity of each orthologous gene using DnaSAM57, and its average across the orthologous genes for each viral group. We then conducted multiple linear regressions to capture the overall relationship between the rmin per nucleotide and nucleotide diversity after controlling for differences in the number of individuals in a viral group: where, for viral group i, yi is the minimum number of recombination events per nucleotide; x1,i is nucleotide diversity; x2,i is the number of individuals; β0 is the intercept; β1 and β2 are regression coefficients; and εi is error, which is normally distributed. We plotted the regression line in Fig. 2 given the parameter estimates, holding constant x2 as the average number of individuals among the viral groups. Using this relationship, we identified the top 25 viral groups having >0.009 deviation from the regression line as recombination-intense. We chose the cutoff by examining the empirical distribution (Fig. S2) and looking for approximately top 10 percentile. A caveat is frequent recombination among very closely related sequences might not be identified by this approach because such recombined sequences are expected to be almost the same as their parental sequences, and in principle be difficult to be detected by comparison of nucleotide sequences.
The recombination-intense viral groups were added to a recently published ICTV reference dataset10 which together was used for the inference of a proteome-based tree via the VICTOR method10. In particular, pairwise distances were inferred from the distance formula d4 because this formula is robust when using incomplete sequences10,51.
Identification and annotation of recombination-intense genes
For each recombination-intense viral group, we conducted multiple linear regressions, similar to above, but at the gene level rather than the viral group level: εi where, for gene i in a viral group, yi is the minimum number of recombination events per nucleotide; x1,i is nucleotide diversity; x2,i is the number of aligned sequences; β0 is the intercept; β1 and β2 are regression coefficients; and εi is error, which is normally distributed. We plotted the regression lines in Fig. 4 and Fig. S5 given the parameter estimates, holding constant x2 as the average number of aligned sequences in a gene. For each gene, we translated the alignment and conducted iterative protein searches using HHblits31, which represents both query and database sequences by profile hidden Markov models (i.e., condensed representation of multiple sequence alignments specifying, for each sequence position, the probability of observing each of the 20 amino acids) instead of single sequences for the detection of remote homology. We used the clustered uniprot20_2016_02 database (http://wwwuser.gwdg.de/~compbiol/data/hhsuite/databases/hhsuite_dbs/), which covers essentially all of the sequence universe by clustering the UniProt database66 from EBI/SIB/PIR and the non-redundant (nr) database from the NCBI. For all hits with >99% probability of being true positives, we individually examined each annotation, and the extent of deviation from the regression line, to identify notable recombination-intense genes.
Mapping reads back to genomes of recombination-intense viral groups
For each recombination-intense viral group carrying at least a notable recombination hot gene, we selected a representative with the longest possible scaffold, and mapping reads from their respective sample stored in back to it using Bowtie267. Raw reads were collected from the Sequence Read Archive (SRA) according to their run and sample identifiers (shown in Table S1) obtained via Genomes OnLine Database (GOLD)68. Visualization and statistics (mean coverage and standard deviation) were obtained using Geneious software (Biomatters Ltd., Auckland, New Zealand). Visualization of the viral contigs after gene calling was obtained from the Integrated Microbial Genomes with Metagenomes (IMG/M) platform12. Genes are colored in the maps according with their predicted function (based on clusters of orthologous groups; COGs).
Electronic supplementary material
Acknowledgements
Computational calculations were performed at the Human Genome Center at the Institute of Medical Science (the University of Tokyo), and at the National Institute of Genetics. This work was partly conducted by the US Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, under contract number DE-AC02-05CH11231. This research was supported by a Grant-in-Aid for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Science, Sports, and Technology (MEXT) of Japan (No. 16H06429, 16K21723, and 17H05826).
Author Contributions
K.Y. conceived and designed the study. K.Y., J.M.K. and D.P. analyzed the data. K.Y., J.M.K., J.U., H.Y. and D.P. contributed to the writing of the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-29272-2.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Rohwer F. Global phage diversity. Cell. 2003;113:141. doi: 10.1016/S0092-8674(03)00276-9. [DOI] [PubMed] [Google Scholar]
- 2.Grose JH, Casjens SR. Understanding the enormous diversity of bacteriophages: the tailed phages that infect the bacterial family Enterobacteriaceae. Virology. 2014;468-470:421–443. doi: 10.1016/j.virol.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suttle CA. Marine viruses–major players in the global ecosystem. Nat Rev Microbiol. 2007;5:801–812. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- 4.Williamson KE, Fuhrmann JJ, Wommack KE, Radosevich M. Viruses in Soil Ecosystems: An Unknown Quantity Within an Unexplored Territory. Annu Rev Virol. 2017;4:201–219. doi: 10.1146/annurev-virology-101416-041639. [DOI] [PubMed] [Google Scholar]
- 5.Clokie MR, Millard AD, Letarov AV, Heaphy S. Phages in nature. Bacteriophage. 2011;1:31–45. doi: 10.4161/bact.1.1.14942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Howard-Varona C, Hargreaves KR, Abedon ST, Sullivan MB. Lysogeny in nature: mechanisms, impact and ecology of temperate phages. ISME J. 2017;11:1511–1520. doi: 10.1038/ismej.2017.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manrique, P., Dills, M. & Young, M. J. The Human Gut Phage Community and Its Implications for Health and Disease. Viruses9, 10.3390/v9060141 (2017). [DOI] [PMC free article] [PubMed]
- 8.Wahida A, Ritter K, Horz HP. The Janus-Face of Bacteriophages across Human Body Habitats. PLoS Pathog. 2016;12:e1005634. doi: 10.1371/journal.ppat.1005634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simmonds P, et al. Consensus statement: Virus taxonomy in the age of metagenomics. Nat Rev Microbiol. 2017;15:161–168. doi: 10.1038/nrmicro.2016.177. [DOI] [PubMed] [Google Scholar]
- 10.Meier-Kolthoff JP, Goker M. VICTOR: genome-based phylogeny and classification of prokaryotic viruses. Bioinformatics. 2017;33:3396–3404. doi: 10.1093/bioinformatics/btx440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paez-Espino D, et al. Uncovering Earth’s virome. Nature. 2016;536:425–430. doi: 10.1038/nature19094. [DOI] [PubMed] [Google Scholar]
- 12.Chen IA, et al. IMG/M: integrated genome and metagenome comparative data analysis system. Nucleic Acids Res. 2017;45:D507–D516. doi: 10.1093/nar/gkw929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paez-Espino D, et al. IMG/VR: a database of cultured and uncultured DNA Viruses and retroviruses. Nucleic Acids Res. 2017;45:D457–D465. doi: 10.1093/nar/gkw1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao NL, et al. MVP: a microbe-phage interaction database. Nucleic Acids Res. 2017 doi: 10.1093/nar/gkx1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paez-Espino D, Pavlopoulos GA, Ivanova NN, Kyrpides NC. Nontargeted virus sequence discovery pipeline and virus clustering for metagenomic data. Nat Protoc. 2017;12:1673–1682. doi: 10.1038/nprot.2017.063. [DOI] [PubMed] [Google Scholar]
- 16.MaynardSmith, J. The Evolution of Sex. (Cambridge University Press, 1978).
- 17.Yahara K, Didelot X, Ansari MA, Sheppard SK, Falush D. Efficient inference of recombination hot regions in bacterial genomes. Mol Biol Evol. 2014;31:1593–1605. doi: 10.1093/molbev/msu082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weller SK, Sawitzke JA. Recombination promoted by DNA viruses: phage lambda to herpes simplex virus. Annu Rev Microbiol. 2014;68:237–258. doi: 10.1146/annurev-micro-091313-103424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simon-Loriere E, Holmes EC. Why do RNA viruses recombine? Nat Rev Microbiol. 2011;9:617–626. doi: 10.1038/nrmicro2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joseph SB, Hanley KA, Chao L, Burch CL. Coinfection rates in Phi6 bacteriophage are enhanced by virus-induced changes in host cells. Evol Appl. 2009;2:24–31. doi: 10.1111/j.1752-4571.2008.00055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Paepe M, et al. Temperate phages acquire DNA from defective prophages by relaxed homologous recombination: the role of Rad52-like recombinases. PLoS Genet. 2014;10:e1004181. doi: 10.1371/journal.pgen.1004181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hatfull GF, Hendrix RW. Bacteriophages and their genomes. Curr Opin Virol. 2011;1:298–303. doi: 10.1016/j.coviro.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yahara K, Horie R, Kobayashi I, Sasaki A. Evolution of DNA double-strand break repair by gene conversion: coevolution between a phage and a restriction-modification system. Genetics. 2007;176:513–526. doi: 10.1534/genetics.106.056150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Otto SP, Lenormand T. Resolving the paradox of sex and recombination. Nat Rev Genet. 2002;3:252–261. doi: 10.1038/nrg761. [DOI] [PubMed] [Google Scholar]
- 25.Yahara K, et al. Genome-wide survey of mutual homologous recombination in a highly sexual bacterial species. Genome Biol Evol. 2012;4:628–640. doi: 10.1093/gbe/evs043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perry A, Lambert P. Propionibacterium acnes: infection beyond the skin. Expert Rev Anti Infect Ther. 2011;9:1149–1156. doi: 10.1586/eri.11.137. [DOI] [PubMed] [Google Scholar]
- 27.van Houte S, Buckling A, Westra ER. Evolutionary Ecology of Prokaryotic Immune Mechanisms. Microbiol Mol Biol Rev. 2016;80:745–763. doi: 10.1128/MMBR.00011-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pingoud A, Wilson GG, Wende W. Type II restriction endonucleases–a historical perspective and more. Nucleic Acids Res. 2014;42:7489–7527. doi: 10.1093/nar/gku447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amitai G, Sorek R. CRISPR-Cas adaptation: insights into the mechanism of action. Nat Rev Microbiol. 2016;14:67–76. doi: 10.1038/nrmicro.2015.14. [DOI] [PubMed] [Google Scholar]
- 30.Hatfull GF. Bacteriophage genomics. Curr Opin Microbiol. 2008;11:447–453. doi: 10.1016/j.mib.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Remmert M, Biegert A, Hauser A, Soding J. HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat Methods. 2011;9:173–175. doi: 10.1038/nmeth.1818. [DOI] [PubMed] [Google Scholar]
- 32.Roos WH, Ivanovska IL, Evilevitch A, Wuite GJ. Viral capsids: mechanical characteristics, genome packaging and delivery mechanisms. Cell Mol Life Sci. 2007;64:1484–1497. doi: 10.1007/s00018-007-6451-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Isidro A, Henriques AO, Tavares P. The portal protein plays essential roles at different steps of the SPP1 DNA packaging process. Virology. 2004;322:253–263. doi: 10.1016/j.virol.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 34.Shen X, et al. Functional identification of the DNA packaging terminase from Pseudomonas aeruginosa phage PaP3. Arch Virol. 2012;157:2133–2141. doi: 10.1007/s00705-012-1409-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Veesler, D. & Cambillau, C. A common evolutionary origin for tailed-bacteriophage functional modules and bacterial machineries. Microbiol Mol Biol Rev75, 423–433, first page of table of contents, 10.1128/MMBR.00014-11 (2011). [DOI] [PMC free article] [PubMed]
- 36.Li X, et al. An essential role for the baseplate protein Gp45 in phage adsorption to Staphylococcus aureus. Sci Rep. 2016;6:26455. doi: 10.1038/srep26455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Young R. Bacteriophage lysis: mechanism and regulation. Microbiol Rev. 1992;56:430–481. doi: 10.1128/mr.56.3.430-481.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lum AG, et al. Global transcription of CRISPR loci in the human oral cavity. BMC Genomics. 2015;16:401. doi: 10.1186/s12864-015-1615-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baker JL, Bor B, Agnello M, Shi W, He X. Ecology of the Oral Microbiome: Beyond Bacteria. Trends Microbiol. 2017;25:362–374. doi: 10.1016/j.tim.2016.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paez-Espino, D. et al. CRISPR immunity drives rapid phage genome evolution in Streptococcus thermophilus. MBio6, 10.1128/mBio.00262-15 (2015). [DOI] [PMC free article] [PubMed]
- 41.Price, V. J., Huo, W., Sharifi, A. & Palmer, K. L. CRISPR-Cas and Restriction-Modification Act Additively against Conjugative Antibiotic Resistance Plasmid Transfer in Enterococcus faecalis. mSphere1, 10.1128/mSphere.00064-16 (2016). [DOI] [PMC free article] [PubMed]
- 42.Angelakis E, Roux V, Raoult D, Drancourt M. Human case of Atopobium rimae bacteremia. Emerg Infect Dis. 2009;15:354–355. doi: 10.3201/eid1502.071399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumar PS, et al. New bacterial species associated with chronic periodontitis. J Dent Res. 2003;82:338–344. doi: 10.1177/154405910308200503. [DOI] [PubMed] [Google Scholar]
- 44.Shelburne SA, et al. Streptococcus mitis strains causing severe clinical disease in cancer patients. Emerg Infect Dis. 2014;20:762–771. doi: 10.3201/eid2005.130953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mardis JS, Many WJ., Jr. Endocarditis due to Actinomyces viscosus. South Med J. 2001;94:240–243. doi: 10.1097/00007611-200194020-00014. [DOI] [PubMed] [Google Scholar]
- 46.Liu D, Bateman T, Carr E, Foster P. Endocarditis due to Gemella haemolysans in a newly diagnosed multiple myeloma patient. J Community Hosp Intern Med Perspect. 2016;6:32357. doi: 10.3402/jchimp.v6.32357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lim YK, Kweon OJ, Kim HR, Lee MK. Leptotrichia goodfellowii Infection: Case Report and Literature Review. Ann Clin Lab Sci. 2016;46:83–86. [PubMed] [Google Scholar]
- 48.Martino R, et al. Bacteremia caused by Capnocytophaga species in patients with neutropenia and cancer: results of a multicenter study. Clin Infect Dis. 2001;33:E20–22. doi: 10.1086/322649. [DOI] [PubMed] [Google Scholar]
- 49.Kirk KF, Nielsen HL, Thorlacius-Ussing O, Nielsen H. Optimized cultivation of Campylobacter concisus from gut mucosal biopsies in inflammatory bowel disease. Gut Pathog. 2016;8:27. doi: 10.1186/s13099-016-0111-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pinto G, Silva MD, Peddey M, Sillankorva S, Azeredo J. The role of bacteriophages in periodontal health and disease. Future Microbiol. 2016;11:1359–1369. doi: 10.2217/fmb-2016-0081. [DOI] [PubMed] [Google Scholar]
- 51.Meier-Kolthoff JP, Auch AF, Klenk HP, Goker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics. 2013;14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Letunic I, Bork P. Interactive tree of life (iTOL)v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 54.Page AJ, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hudson RR, Kaplan NL. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics. 1985;111:147–164. doi: 10.1093/genetics/111.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cai JJ. PGEToolbox: A Matlab toolbox for population genetics and evolution. J Hered. 2008;99:438–440. doi: 10.1093/jhered/esm127. [DOI] [PubMed] [Google Scholar]
- 57.Eckert AJ, Liechty JD, Tearse BR, Pande B, Neale DB. DnaSAM: Software to perform neutrality testing for large datasets with complex null models. Mol Ecol Resour. 2010;10:542–545. doi: 10.1111/j.1755-0998.2009.02768.x. [DOI] [PubMed] [Google Scholar]
- 58.Uchiyama I. MBGD: microbial genome database for comparative analysis. Nucleic Acids Res. 2003;31:58–62. doi: 10.1093/nar/gkg109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Biswas A, Staals RH, Morales SE, Fineran PC, Brown CM. CRISPRDetect: A flexible algorithm to define CRISPR arrays. BMC Genomics. 2016;17:356. doi: 10.1186/s12864-016-2627-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Meier-Kolthoff JP, et al. Complete genome sequence of DSM 30083(T), the type strain (U5/41(T)) of Escherichia coli, and a proposal for delineating subspecies in microbial taxonomy. Stand Genomic Sci. 2014;9:2. doi: 10.1186/1944-3277-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Goloboff PA, Farris JS, Nixon KC. TNT, a free program for phylogenetic analysis. Cladistics. 2008;24:774–786. doi: 10.1111/j.1096-0031.2008.00217.x. [DOI] [Google Scholar]
- 64.Pattengale ND, Alipour M, Bininda-Emonds OR, Moret BM, Stamatakis A. How many bootstrap replicates are necessary? J Comput Biol. 2010;17:337–354. doi: 10.1089/cmb.2009.0179. [DOI] [PubMed] [Google Scholar]
- 65.PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4.0 b10 (Sinauer Associates, Sunderland, 2002).
- 66.UniProt, C. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res 38, D142–148, 10.1093/nar/gkp846 (2010). [DOI] [PMC free article] [PubMed]
- 67.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mukherjee S, et al. Genomes OnLine Database (GOLD) v.6: data updates and feature enhancements. Nucleic Acids Res. 2017;45:D446–D456. doi: 10.1093/nar/gkw992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Farris, J. S. Estimating phylogenetic trees from distance matrices. The American Naturalist (1972).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.