Abstract
A long-term animal experiment involving inoculation with bovine coronavirus (BCoV) was conducted to verify its persistent infection in cattle. Three colostrum-deprived Holstein calves were housed separately in individual rooms of a high-containment facility and inoculated with the BCoV strain Kumamoto/1/07. Until the end of the experiment (1,085, 700 and 280 days, respectively), viral RNAs were detected sporadically by RT-PCR and nested PCR from plasma, nasal discharge, and feces. Seroconversion and titer changes were validated by hemagglutination inhibition tests and neutralization tests. Among the samples, nasal discharge showed a higher viral positivity than feces, which seemed to be associated with positive detection in the plasma. These data demonstrate the existence of persistent infection of BCoV in the respiratory tissues of cattle.
Keywords: bovine coronavirus, long-term animal experiment, persistent infection
Bovine coronavirus (BCoV) is a member of the order Nidovirales, family Coronaviridae, subfamily Coronavirinae, genus Betacoronavirus, species Betacoronavirus-1. This species also includes Human coronavirus OC43, Porcine hemagglutinating encephalomyelitis virus, Equine coronavirus, and Canine respiratory coronavirus [1].
BCoV infects the respiratory and digestive organs of cattle and causes neonatal calf diarrhea, bloody diarrhea in adult cattle (winter dysentery), and respiratory symptoms, including shipping fever in feedlot cattle [6, 12, 16]. BCoVs have spread widely across cattle farms all over the world, and thus, nearly all adult cattle have antibodies against the virus. Outbreaks typically occur during autumn and winter [2] and are associated with the housing period; however, several cases have also been reported during warmer seasons [5, 10, 13]. Although the mortality of this disease is low, it causes substantial economic losses owing to a reduction in milk production in dairy farms [14, 19] and meat production in beef farms [17, 18].
The main transmission route of BCoVs is horizontal infection, i.e., ingestion or inhalation of virus from feces or nasal discharges into the mouth or nasal cavity [7, 15]. There has been no report of vertical infection so far. BCoV antigens are detected in the feces of clinically healthy cattle [4], and studies on BCoV shedding in feces showed the virus to be detected over a long period on farms, even though the number of cattle shedding the virus decreased [3]. This suggests the existence of persistently infected cattle, which might also be the origin of transmission. To verify this hypothesis, we conducted a longitudinal animal experiment.
The BCoV used in this experimental study was obtained from a clarified suspension of feces from a cow on a farm where a BCoV case had been confirmed in 2007 by using HRT-18G cells (isolate Kumamoto/1/07) [11]. The culture supernatant of the isolate was inoculated into cattle by the oral route. Feces were collected when the cattle showed diarrhea. The isolate was passaged in colostrum-deprived Holstein calves twice, and feces of the infected calves were harvested as inoculum for this study (data not shown).
Three colostrum-deprived Holstein calves were used in this study. Each animal was maintained in a separate room of a high-containment facility at our institute. In this facility, the animal room area is separated from the preparation room by a shower room area. To enter the animal room area, the investigator undresses and puts on work clothes and boots in this area. The animal room area has 5 independent animal rooms and a corridor. Each animal room has a ventilator with HEPA filers to prevent the reintroduction of the virus from outside to the experimental cattle. During the animal experiment, we adhered to the strict high hygiene and biosecurity measures to prevent reinfection from other cattle. To enter the animal room, boots were removed at the corridor, and the “inside” boots and waterproof clothes kept in the animal room were worn. To exit the room after husbandry and sampling, put off the boots and waterproof clothes were taken off after disinfection with sodium hypochlorite using a spraying device.
The Animal Care and Use Committee of the National Institute of Animal Health approved all animal procedures prior to the initiation of this study (authorization number: 11-083).
One calf (11 days old, referred to as “calf 1”) was inoculated orally using a catheter that delivered a sample of the centrifuged supernatant at 1,500 × g for 10 min from a mixture of 10 g feces as mentioned above and 50 ml phosphate-buffered saline (PBS) containing 100 µg/ml gentamicin into the esophagus. Samples of nasal discharge, feces, plasma, and sera were collected daily until 10 days post inoculation (dpi), followed by weekly collection until 141 dpi and then twice-weekly collection until the end of the experiment (1,085 dpi). The nasal discharge samples were collected by inserting cotton into the nasal cavity for a few min and extracting the discharge using sterilized disposable syringes. Feces were collected directly from the rectum and prepared as a 10% suspension in PBS containing 100 µg/ml gentamicin and 1 µg/ml trypsin for virus isolation and RNA extraction. The same experiment was carried out on another calf (8 days old, “calf 2”) using the centrifuged supernatant from a mixture of 50 ml PBS and 10 g feces of calf 1 collected at 3 dpi (after it showed diarrhea). Samples of nasal discharge, feces, plasma, and sera were collected daily until 11 dpi, followed by weekly collection until 78 dpi and twice-weekly collection until the end of the experiment (700 dpi). The same experiment was carried out on yet another calf (4 days old, “calf 3”) using centrifuged supernatant from a mixture of 50 ml PBS and 20 g feces of calf 2 collected at 5 dpi, when it showed diarrhea. Nasal discharge, feces, plasma, and serum samples were collected daily until 8 dpi, followed by weekly collection until 33 dpi and twice-weekly collection until the end of the experiment (280 dpi).
Viral RNA was extracted from plasma, nasal discharge, and 10% fecal suspensions in PBS using the High Pure Viral RNA Kit (Roche Diagnostics, Tokyo, Japan) according to the manufacturer’s instructions. BCoV-specific genes were detected from the extracted RNA by the RT-PCR assay using a Titan One Tube RT-PCR Kit (Roche Diagnostics), with the following thermal cycling profile: 30 min at 50°C; 2 min at 94°C; 35 cycles of denaturation at 94°C for 30 sec, annealing at 55°C for 30 sec, and extension at 68°C for 45 sec; and completion of amplification with a 7-min extension step at 68°C [20]. The oligonucleotide primers used for nested PCR were designed from the nucleotide sequence of the Kakegawa strain (GenBank accession no. AB354579) in this study. The primers were as follows: BCoV-nu, 5ʹ-TGCTACTTCTCAGCAACCATCAG-3ʹ (nt 29,540–29,562, sense primer), and BCoV-nd, 5ʹ-TTGGCATGCGGTCCTGTTCCAAG-3ʹ (nt 29,778–29,800, antisense primer). The size of the amplification fragment is 261 bp. Nested PCR was performed by using TaKaRa Ex Taq (Takara-Bio., Kusatsu, Japan), and the thermal cycling profile was as follows: 1 min at 98°C; 30 cycles of denaturation at 98°C for 10 sec, annealing at 55°C for 30 sec, and extension at 72°C for 45 sec; and completion of amplification with a 7-min extension step at 72°C. To prevent laboratory contamination in RT-PCR and nested PCR, we set up mixtures for these reactions in a laminar flow cabinet equipped with an UV lamp, wore fresh gloves at each step, used pipette tips with aerosol filters. Negative controls were also included in every PCR experiment to detection of contamination. The RT-PCR and nested PCR products were visualized on 1.8% agarose gels stained with ethidium bromide.
The hemagglutination inhibition (HI) test was conducted by the microtiter method [9]. Sera were treated with kaolin and chicken erythrocytes and heat inactivated. As antigen for the HI test, the Kakegawa strain was used in this assay because the hemagglutinating (HA) activity of the Kumamoto/1/07 strain was low and unstable with chicken and mouse erythrocytes, similar to recent Japanese isolates (Kanno et al., unpublished data), whereas the HA activity of the Kakegawa strain was sufficient. Briefly, twofold dilutions of serum were prepared in duplicate and reacted with the virus. The HI titers were expressed as the reciprocal of the highest serum dilution that completely inhibited the HA activity of 4 HA units of the virus.
Serum samples were also tested in a virus neutralization test by preparing serial twofold dilutions of serum were reacted in duplicate with 100 of 50% tissue culture infective doses (TCID50) of Kumamoto/1/07, followed by incubation for 1 hr at 37°C. After incubation, the serum–virus mixture was transferred onto monolayered human rectal tumor cells (HRT-18G) cultured in microplates and incubated for 5 days at 37°C. The neutralization antibody titers were expressed as the reciprocal of the highest serum dilution that completely inhibited the cytopathic effect (CPE).
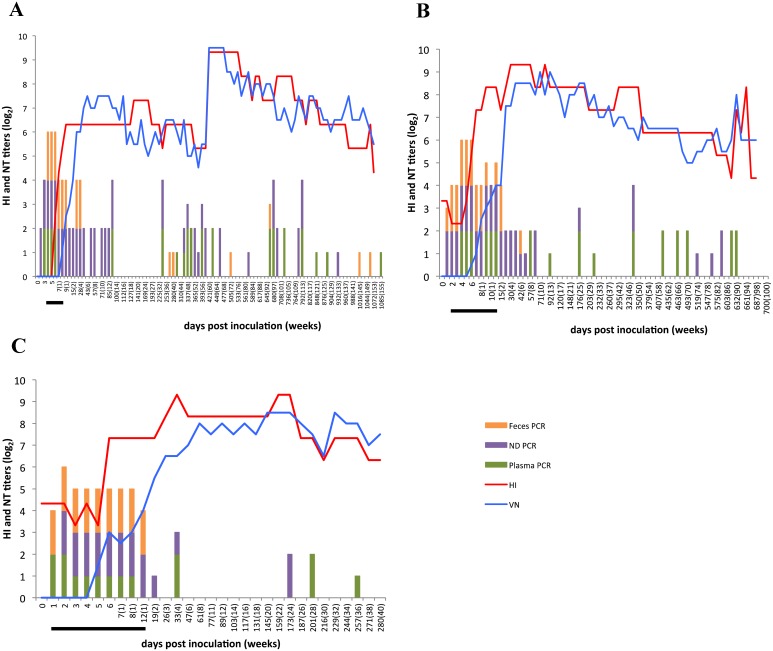
All 3 calves showed clinical signs, such as loose or diarrheal stool, up to 8–12 dpi. Viral RNA was detected from the plasma during the period of onset (Fig. 1A–C). After recovery from the disease, viral RNA was sporadically detected by RT-PCR and nested PCR from plasma. Final detection days (last day when a sample showed a positive result) by RT-PCR were 792, 632, and 201 dpi, and by nested PCR, 1,085, 632, and 257 dpi, respectively, in the 3 calves. As with the plasma, viral RNA was detected from nasal discharge on the basis of viral titers, and was detected sporadically by PCR-based experiments. Final detection days by RT-PCR were 792, 589 and 173 dpi, and by nested PCR, 932, 589 and 173 dpi, respectively, in the 3 calves. Viral RNA was not detected from the feces after the disease onset in calves 2 and 3. However, nested PCR was positive for the virus at 42 dpi in calf 2. In calf 1, viral RNA from feces was detected sporadically by the PCR-based experiments. In this calf, the final detection day was 28 dpi by RT-PCR and 1,058 dpi by nested PCR.
Fig. 1.
Detection of viral genes from clinical samples by RT-PCR and nested PCR assays and of antibodies by the hemagglutination inhibition (HI) test and neutralization test (NT) in calves 1 (A), 2 (B) and 3 (C). The days when the viral RNA was detected from the plasma, nasal discharge, and feces are indicated by green, violet, and orange bars, respectively. The numbers in parentheses indicate weeks post inoculation. The half-size bar showed that the viral RNA was detected only by nested PCR. The antibody titers measured in the HI and VN tests are on the left y-axes. The period of disease onset is indicated by the black bar.
Virus isolation was conducted from the samples showing positive results by RT-PCR and nested PCR. The samples were inoculated into HRT-18G monolayer cells in 12-well plates and incubated for 5 days at 37°C and 5% CO2. If a CPE was not observed in the incubation period, passaging into fresh cells was conducted twice using the inoculum after freeze-thawing the cultures thrice. However, the virus was not isolated, except in the samples from the period of onset.
The HI and neutralization titers in the 3 calves increased several days after disease onset and peaked at 19, 4 and 4 weeks post inoculation, and at 6, 10 and 20 weeks post inoculation, respectively, in the 3 calves. Thereafter, the titers gradually decreased. However, both titers showed a rapid rise at 421 dpi in calf 1 (640 and 724, respectively) (Fig. 1A). Ten days before the sampling, calf 1 showed diarrhea at 411 and 412 dpi (data not shown). Therefore, we surmised that the virus was reactivated in the cattle, probably in the respiratory tissues, causing diarrhea followed by this booster effect. The virus in the digestive tract might have been quickly inactivated and excreted; therefore, viral RNAs were not detected from nasal discharge and feces at 421 dpi, 10 days after the onset. Unfortunately, feces samples had not been collected at the onset, so no virus was isolated. A rapid rise in these titers was also found in calf 2 at 632 dpi, just after the detection of viral RNA from the plasma at 617 dpi by both RT-PCR and nested PCR (Fig. 1B). This finding might also be attributed to virus reactivation in respiratory tissues and quick inactivation by host immunity without clinical signs.
This study showed that the BCoV RNA was long-lasting, having been detected from the nasal discharge of cattle that had been maintained in an isolated room of a high-containment facility to prevent virus intrusion from outside. This indicated the existence of persistent infection of BCoV in the respiratory tissues of cattle, although further investigations are warranted because the virus was not isolated. Other coronaviruses, such as Transmissible gastroenteritis virus, Canine coronavirus, and Feline coronavirus, have also been reported to cause long-term infection [8, 21, 22]. Thus, coronavirus tends to cause persistent infection in host animals. Often, calf diarrhea caused by BCoV repeatedly occurs at the same farm every year. This might be explained by the presence of persistently infected cattle.
CONFLICTS OF INTEREST
The authors have no potential conflicts of interest to declare with respect to the research, authorship, and/or publication of this article.
Acknowledgments
We are grateful to Mr. Keiji Itoh, Mr. Kazuhiko Takase, Mr. Mitsutoshi Noi, Mr. Yoshihiro Himoro, and Mr. Kiyoshi Tanaka for their skilled handling of the animals at the Institute of Animal Health, Hokkaido Research Center.
REFERENCES
- 1.Carstens E. B.2010. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2009). Arch. Virol. 155: 133–146. doi: 10.1007/s00705-009-0547-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clark M. A.1993. Bovine coronavirus. Br. Vet. J. 149: 51–70. doi: 10.1016/S0007-1935(05)80210-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collins J. K., Riegel C. A., Olson J. D., Fountain A.1987. Shedding of enteric coronavirus in adult cattle. Am. J. Vet. Res. 48: 361–365. [PubMed] [Google Scholar]
- 4.Crouch C. F., Raybould T. J., Acres S. D.1984. Monoclonal antibody capture enzyme-linked immunosorbent assay for detection of bovine enteric coronavirus. J. Clin. Microbiol. 19: 388–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Decaro N., Mari V., Desario C., Campolo M., Elia G., Martella V., Greco G., Cirone F., Colaianni M. L., Cordioli P., Buonavoglia C.2008. Severe outbreak of bovine coronavirus infection in dairy cattle during the warmer season. Vet. Microbiol. 126: 30–39. doi: 10.1016/j.vetmic.2007.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hasoksuz M., Lathrop S. L., Gadfield K. L., Saif L. J.1999. Isolation of bovine respiratory coronaviruses from feedlot cattle and comparison of their biological and antigenic properties with bovine enteric coronaviruses. Am. J. Vet. Res. 60: 1227–1233. [PubMed] [Google Scholar]
- 7.Heckert R. A., Saif L. J., Hoblet K. H., Agnes A. G.1990. A longitudinal study of bovine coronavirus enteric and respiratory infections in dairy calves in two herds in Ohio. Vet. Microbiol. 22: 187–201. doi: 10.1016/0378-1135(90)90106-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoshino Y., Scott F. W.1980. Coronavirus-like particles in the feces of normal cats. Arch. Virol. 63: 147–152. doi: 10.1007/BF01320772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inaba Y., Sato K., Takahashi E., Kurogi H., Satoda K.1977. Hemagglutination with Nebraska calf diarrhea virus. Microbiol. Immunol. 21: 531–534. doi: 10.1111/j.1348-0421.1977.tb00319.x [DOI] [PubMed] [Google Scholar]
- 10.Kanno T., Hatama S., Ishihara R., Uchida I.2007. Molecular analysis of the S glycoprotein gene of bovine coronaviruses isolated in Japan from 1999 to 2006. J. Gen. Virol. 88: 1218–1224. doi: 10.1099/vir.0.82635-0 [DOI] [PubMed] [Google Scholar]
- 11.Kanno T., Kamiyoshi T., Ishihara R., Hatama S., Uchida I.2009. Phylogenetic studies of bovine coronaviruses isolated in Japan. J. Vet. Med. Sci. 71: 83–86. doi: 10.1292/jvms.71.83 [DOI] [PubMed] [Google Scholar]
- 12.Lathrop S. L., Wittum T. E., Brock K. V., Loerch S. C., Perino L. J., Bingham H. R., McCollum F. T., Saif L. J.2000. Association between infection of the respiratory tract attributable to bovine coronavirus and health and growth performance of cattle in feedlots. Am. J. Vet. Res. 61: 1062–1066. doi: 10.2460/ajvr.2000.61.1062 [DOI] [PubMed] [Google Scholar]
- 13.Park S. J., Jeong C., Yoon S. S., Choy H. E., Saif L. J., Park S. H., Kim Y. J., Jeong J. H., Park S. I., Kim H. H., Lee B. J., Cho H. S., Kim S. K., Kang M. I., Cho K. O.2006. Detection and characterization of bovine coronaviruses in fecal specimens of adult cattle with diarrhea during the warmer seasons. J. Clin. Microbiol. 44: 3178–3188. doi: 10.1128/JCM.02667-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saif L. J.2010. Bovine respiratory coronavirus. Vet. Clin. North Am. Food Anim. Pract. 26: 349–364. doi: 10.1016/j.cvfa.2010.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saif L. J., Redman D. R., Moorhead P. D., Theil K. W.1986. Experimentally induced coronavirus infections in calves: viral replication in the respiratory and intestinal tracts. Am. J. Vet. Res. 47: 1426–1432. [PubMed] [Google Scholar]
- 16.Storz J., Stine L., Liem A., Anderson G. A.1996. Coronavirus isolation from nasal swab samples in cattle with signs of respiratory tract disease after shipping. J. Am. Vet. Med. Assoc. 208: 1452–1455. [PubMed] [Google Scholar]
- 17.Svensson C., Lundborg K., Emanuelson U., Olsson S. O.2003. Morbidity in Swedish dairy calves from birth to 90 days of age and individual calf-level risk factors for infectious diseases. Prev. Vet. Med. 58: 179–197. doi: 10.1016/S0167-5877(03)00046-1 [DOI] [PubMed] [Google Scholar]
- 18.Torsein M., Lindberg A., Sandgren C. H., Waller K. P., Törnquist M., Svensson C.2011. Risk factors for calf mortality in large Swedish dairy herds. Prev. Vet. Med. 99: 136–147. doi: 10.1016/j.prevetmed.2010.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tråvén M., Näslund K., Linde N., Linde B., Silván A., Fossum C., Hedlund K. O., Larsson B.2001. Experimental reproduction of winter dysentery in lactating cows using BCV — comparison with BCV infection in milk-fed calves. Vet. Microbiol. 81: 127–151. doi: 10.1016/S0378-1135(01)00337-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsunemitsu H., Smith D. R., Saif L. J.1999. Experimental inoculation of adult dairy cows with bovine coronavirus and detection of coronavirus in feces by RT-PCR. Arch. Virol. 144: 167–175. doi: 10.1007/s007050050493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Underdahl N. R., Mebus C. A., Torres-Medina A.1975. Recovery of transmissible gastroenteritis virus from chronically infected experimental pigs. Am. J. Vet. Res. 36: 1473–1476. [PubMed] [Google Scholar]
- 22.Williams F. P., Jr.1980. Astrovirus-like, coronavirus-like, and parvovirus-like particles detected in the diarrheal stools of beagle pups. Arch. Virol. 66: 215–226. doi: 10.1007/BF01314735 [DOI] [PMC free article] [PubMed] [Google Scholar]