Abstract
Rhizophora mucronata is an important ecosystem entity of the Malaysian mangrove forest. Since the species grows in a harsh environment, any organism that is isolated from this species would be of huge interest due to its potential in having novel bioactive compounds. In the present work, we isolated, identified and characterized, a total of 78 fungal isolates harboring inside the leaf tissues of R. mucronata. Molecular identification using the nuclear ribosomal DNA internal transcribe spacer (ITS) sequences returned with high similarity matches to known sequences in the GenBank. Maximum likelihood analysis revealed the phylogenetic relationship of all isolates from this study. Most of the dominating fungal endophytes were from the genera Pestalotiopsis, followed by Alternaria and Cladosporium. Six isolates representing the genera Alternaria, Fusarium, Nigrospora, Pestalotiopsis, Phoma, and Xylaria, were further screened for their antagonism activities. Dual culture test assay revealed their inhibition percentages against the phytopathogenic fungus Fusarium solani between 45–66%, and 0.8–23% when using non-volatile test assay. Of the six isolates, only Fusarium lateritium and Xylaria sp. showed antibacterial activities against the pathogenic bacteria, Bacillus subtilis, Escherichia coli, Pseudomonas aeruginosa, and Staphylococcus aureus, with the Minimum Inhibitory Concentration (MIC) and Minimum Bactericidal Concentration (MBC) ranging from 0.5 to 2 mg/mL. The DPPH radical scavenging assay recorded a high level of antioxidant activity in Xylaria sp., 3-fold above that of F. lateritium. We demonstrate for the first time, two members belonging to the endophytic fungal community in the tropical mangrove species that have potential use as antagonists and antibacterial agents for future biotechnological applications.
Keywords: fungal endophyte, ITS, dual culture, minimum inhibitory concentration, scavenging activity
Introduction
Mangroves of South and Southeast Asia comprise 41.4% of the global mangroves (Singh et al., 2014). There are ~70 species of true mangrove species, belonging to 17 different families. The largest mangrove family, Rhizophoraceae, comprises of four mangrove genera: Bruguiera, Ceriops, Kandelia, and Rhizophora (Tomlinson, 1986; Polidoro et al., 2010). Of these genera, Rhizophora has the widest distribution and is commonly found growing in the tropical and subtropical coastal areas (Duke, 2006). Recorded with a total of six true mangrove species and one hybrid names (The Plant List, 2013), three of these species: R. apiculata, R. mucronata, and R. stylosa, could be found populating the Indo-Malaya region (Duke et al., 2007). Rhizophora mucronata (Malay: bakau kurap) is characterized by its broadly elliptic to oblong leaves with yellowish stalked flowers (Qin and Boufford, 2007; Setyawan and Ulumuddin, 2012).
Different parts of R. mucronata such as the bark and leaves have been used traditionally as astringent and antiseptic, and have been shown to possess activities against bacteria, ulcers, and inflammation (Kokpol et al., 1990; Suganthy and Pandima Devi, 2016). Furthermore, pharmacological studies have shown that the leaf exhibits antiviral, anti-inflammatory, and anti-diabetic activities (Gaffar et al., 2011). The leaves also contain (+)-catechin, which has properties in treating Alzheimer disease (Suganthy and Pandima Devi, 2016). Apart from focusing on the plant itself, several studies have been conducted on microorganisms associated with R. mucronata, including fungal endophytes. Endophytic fungi from the genera Penicillium, Ampelomyces, and Fusarium, isolated from R. mucronata were found to be active against E. coli (Prihanto et al., 2011), while several others originating from R. apiculata, have led to the discovery of novel compounds including acropyrone, bicytosporone D, waol acid, and pestalotiopene (Klaiklay et al., 2012). Fungi from marine environments grow in habitats with unique conditions that attributed to the activation of metabolic pathways and the synthesis of distinct unknown molecules (de Souza Sebastianes et al., 2013). Production of these compounds aids in supporting the adaptation and survival of the fungi in marine ecosystems (Fox and Howlett, 2008).
Endophytic fungi live internally within various tissues of a host plant, asymptomatically, without causing any negative effect to the host plant (Aly et al., 2011). When a host plant harbors endophytes, their concurrence may help the host to adapt to biotic and abiotic stress factors (Hartley et al., 2015; Amin, 2016; Potshangbam et al., 2017). Tolerance to biotic stress as shown by some plants has been associated with fungal natural products (Rodriguez et al., 2008; Douanla-Meli et al., 2013). Despite endophytic fungi being regarded as new sources of novel active compounds, biological activity, and biotechnological developments; their true potential remains underexplored (Debbab et al., 2011).
In general, leaves have a more diverse fungal endophytes community when compared to other parts of the plant (Bayman et al., 1997; Shreelalitha and Sridhar, 2015). For endophytic fungi of mangrove leaf origin, significant biological activities have been reported from these fungi such as having antibiotic, anticancer, antimicrobial and insecticidal properties (Chaeprasert et al., 2010; Abraham et al., 2015; Thomas et al., 2016). The current study was designed to determine species diversity of the endophytic fungal community in the leaf tissue of R. mucronata, a tropical mangrove species commonly found at the Matang Mangrove Forest Reserve (MMFR), Perak, Malaysia, and to characterize biological activities of several selected species. These fungal endophytes might offer novel species or strains that possess valuable bioactive compounds, which demonstrates the significance of their study.
Materials and methods
Plant materials
Six marine mangrove trees of R. mucronata were identified from the Matang Mangrove Forest Reserve (MMFR) (coordinate E 100°3649.1 N 04°5049.1). The trees' diameter breast heights were about 12–14 cm. From each tree, ten mature (dark green) leaves were randomly sampled from the lowest branch, making a total of 60 leaves sampled. The leaves were kept in polyethene bags and brought back directly to the laboratory to be processed immediately. The leaves were washed thoroughly under running tap water to remove adherent debris on the surface of the leaves. Each leaf was cut into five 0.5 cm2 segments using a blade, accumulated to 300 leaf segments in total.
Endophytic fungi isolation and cultivation
Leaf segments were surface-sterilized according to the protocol suggested by Kjer et al. (2010). The segments were washed under running tap water, immersed in 70% ethanol for 60–120 s, followed by soaking in 4% NaOCl for 60 s, rinsed several times in sterile distilled water and then dried on a sterile filter paper. For control, the final sterile water rinse was plated and observed post-incubation period. Absence of fungal growth indiciate the leaf surface was sterile. The sterilized sample was excised 1–2 mm from the edge and the explant plated on a special-made agar medium, Medium A (Kjer et al., 2010). Medium A contained bacto agar (15 g/L), malt extract (15 g/L), artificial sea salt (10 g/L), chloramphenicol (0.2 g/L), and sterile distilled water (1 L), with the final pH adjusted from 7.4 to 7.8. The plates were incubated at 27°C for 7 days. Hyphae tips that grew out from the cultivated leaf segments were sub-cultured into fresh plates containing another special-made agar medium, Medium B (Kjer et al., 2010). Medium B contained similar ingredients to medium A, but without chloramphenicol. Pure cultures were grouped according to colony color, form, elevation and margin characteristics on Medium B. Based on the groupings, isolates with distinct morphology were selected for molecular identification. Cultures were maintained on Medium B for 7 days before being used for genomic DNA extraction and as inoculum for production of fungal extracts. For bioactivity assays, pure cultures were grown on Potato Dextrose Agar (PDA) (Friendemann Schmidt, Australia) at room temperature for 7 days.
DNA extraction and PCR amplification
Approximately 100 mg of fungal mycelia was used for fungal genomic DNA extraction. Fungal genomic DNA was extracted as previously described by Landum et al. (2016), in accordance to the manufacturer's instructions, using the DNeasy Plant Minikit (Qiagen, Germany). The nuclear ribosomal DNA internal transcribed spacer (ITS) of the fungal isolates were amplified using the forward primer, ITS1-F (5′-CTTGGTCATTTAGAGGAAGTAA-3′ and the reverse primer, ITS4 (5′-CTTGGTCATTTAGAGGAAGTAA-3′) (White et al., 1990). The final reaction volume was 25 μL, containing 12.5 μL of 2X PCRBio Taq Mix Red (PCR Biosystems, UK), 0.4 μM of forward and reverse primers, and 10 ng of genomic DNA template. For negative control, the DNA was replaced with distilled water to verify absence of contamination. PCR was carried out using MyCycler™ (Bio-Rad, USA), programmed for 1 min 95°C; 35 cycles for 15 s at 95°C, 15 s at 55°C, and 1 min at 72°C; and a final 10 min extension at 72°C. The PCR products were separated using 1% agarose gel in 1X TAE buffer (90 mM Tris-acetate and 2 nM EDTA, pH 8.0), stained with ethidium bromide (0.5 μg/mL) and documented using FluorChem™ (Alpha Innotech, USA). PCR products were sent for direct bi-directionally sequencing using ABI PRISM 3730 × 1 Genetic Analyzer (Applied Biosystems, USA) at the First BASE Laboratory Sdn. Bhd., Selangor, Malaysia.
Sequence and phylogenetic analysis
The resulting DNA sequences were aligned using MUSCLE software embedded in MEGA 7 (Kumar et al., 2016), manually trimmed and edited to obtain complete sequences. Homology searches were carried out using the BLASTn program against the NCBI GenBank database (https://blast.ncbi.nlm.nih.gov/Blast.cgi). DNA substitution models which were suitable for the ITS gene that were assessed using the “find best DNA/Protein Models (ML)” function embedded in the software MEGA 7 by implementing the maximum likelihood (ML) statistical method to test the goodness of fit to several models of evolution. According to the estimated values of all parameters for each model, the model best fitting to the dataset from the ITS sequences was general time reversible (GTR) model and gamma distributed (+G) with invariant sites (+I) (= GTR+G+I) model. ML tree was constructed using MEGA 7 with all positions containing gaps and missing data were included for analysis. Clade supports were calculated based on 1,000 bootstrap re-samplings. A soil fungus, Mortierella longigemmata (phyla Zygomycota, family Mortierellaceae) was included as out-group.
Fungal isolates for antagonism assays
Identified endophytic fungal isolates, which displayed distinct pigmentation and have been previously reported as having some biological activities (Hamzah, 2018) were selected for further analysis. They were Alternaria macrospora, Fusarium lateritium, Nigrospora oryzae, Pestalotiopsis sp., Phoma sp., and Xylaria sp. Fungi were cultivated on PDA plates at 26°C for 7 days. Antagonistic activity of the six fungal isolates were evaluated through the dual culture plate assay (Coşkuntuna and Özer, 2008), and the non-volatile compound test (Hajieghrari et al., 2008), against a known prevalent fungal pathogen, Fusarium solani. The phytopathogenic F. solani strain used in this study was isolated from a tropical tree, Aquilaria sp., (Faizal et al., 2017) and maintained at 27°C on PDA media at the Research, Development and Innovation Agency of the Indonesian Ministry of Environment and Forestry (FOERDIA), Indonesia. For all antagonism assays, culture of 7-day-old was used.
Dual culture plate assay
Fungal isolates were screened using in vitro dual culture assays for their ability to suppress the mycelial growth of F. solani. A fungal disc (5 mm) was taken from the test pathogen and placed 3 cm from the margin of the PDA plate (8 cm in diameter). A 5 mm disc of the endophytic fungus/antagonist was also placed in a similar manner but on the direct opposite of the pathogen disc. The plate was incubated at room temperature for 7 days. Plates inoculated with F. solani in the absence of antagonist fungus served as negative controls. The assay was replicated three times. Observations were carried out for 7 days, after which the mycelial radial growth of test pathogen on a control plate (r1) and in the direction of the antagonistic fungus (r2) were measured and the percentage inhibition (I%) in mycelial growth was calculated according to the formula (Hajieghrari et al., 2008): I% = [(r1–r2)/r1] × 100. Data in the form of I% were analyzed statistically with ANOVA using the SAS statistical package (SAS Institute, Cary, NC). For significant effect, the LSD was performed at P ≤ 0.05.
Clear inhibition zone (if any) was recorded, and the interactions between F. solani and fungal isolates were assessed using a classification system ranged from type A to F: “(1) Type A: mutual intermingling growth, where both fungi grew into each other without any macroscopic signs of interaction; (2) Type B: mutual inhibition on contact or space between colonies small (< 2 mm); (3) Type C: inhibition of one species on contact, the inhibited species continued to grow at a significantly reduced rate, while the inhibitor species grew at a slightly, reduced rate or unchanged; (4) Type D: mutual inhibition at a distance (>2 mm); (5) Type E: inhibition of one species on contact, the inhibitor species continuing to grow at a reduced rate through the inhibited colony; (6) Type F: inhibition of one species on contact or at a distance, the inhibitor species then continuing to grow at an unchanged rate through or over the inhibited colony” (Wheeler and Hocking, 1993).
Antifungal non-volatile compounds test
The effects of non-volatile metabolites produced by the selected six endophytic fungi/antagonists were determined using the method described by Hajieghrari et al. (2008). A fungal disc of 5 mm was inoculated into a 250 mL conical flask containing 100 mL of PDB and incubated at 27°C for 15 days, and then filtered through a Whatman filter paper. The filtrate was mixed into molten PDA at a final concentration of 20% (v/v) and poured into Petri dishes. Once solidified, a 5 mm disc of the test pathogen was placed at the center of the PDA plate. The plate was incubated at 27°C. Control plates were prepared without amending PDA with the culture filtrate. After 7 days, radial growth of the test pathogen on the culture filtrate-containing media and control plates were measured, and percentage inhibition in mycelial growth was calculated as explained in the dual culture assay.
In Vitro antibacterial assays
Four bacterial strains (test organisms) were used in this study including two gram-positive bacteria, Bacillus subtilis (UPMC 1175) and Staphylococcus aureus (ATCC 43300), and two gram-negative bacteria, Escherichia coli (ATCC E266) and Pseudomonas aeruginosa (ATCC 15442). They were obtained from the Institute of Biosciences (IBS), UPM. Bacterial cultures were grown in nutrient broth (NB) (Friendemann Schmidt, Australia) at 37°C for 24 h before being used in the assays.
Preliminary screening for antibacterial activity using plate assay
Antibacterial assay was performed according to Onn et al. (2016). Nutrient agar (NA) (Friendemann Schmidt, Australia) plate was prepared and a loopful of the test organism was then streaked onto the plate. A 1 cm disc (agar plug) was cut out from 1-week-old fungal culture, placed on the streaked agar and then incubated at 30°C for 24 h. The assay was replicated 3 times for each bacterium/antagonist combination. Streptomycin was used in place of the fungal agar plug to serve as positive control. The presence of a clear zone using naked eye observation indicates that the fungal isolate possess antibacterial activity, and thus was subjected to the Minimum Inhibitory Concentration (MIC) test. Only identified fungal isolates that showed inhibitory effects toward the bacterial strains in the preliminary screening were selected for subsequent tests.
Preparation of crude fungal extracts
Two fungal isolates, F. lateritium and Xylaria sp. were selected based on preliminary screenings against fungal pathogen F. solani using the dual culture test and the antibacterial plate assay. Actively growing fungal colony on a PDA plate was used as the inoculum. Five fungal discs were cut out from the plate and inoculated into a 500 mL Erlenmeyer flask containing 100 mL of Potato Dextrose Broth (PDB) (Friendemann Schmidt, Australia) at pH 6.5, and allowed to grow in a shaking incubator (120 rpm) at 26°C for 14 days. Then, the liquid culture was filtered using a Whatman filter paper and the filtrate was extracted twice with equal volume of ethyl acetate (R and M Chemicals, Malaysia). The extract was concentrated under reduced pressure at 40°C using a rotary vacuum evaporator and the crude extract was kept at −20°C until being used in the in vitro antibacterial and antioxidant assays.
Determination of minimum inhibitory concentration (MIC)
The MIC test was determined by micro-dilution method under aseptic condition in 96-well plate as described by Daouk et al. (1995). Crude extract of fungal isolates F. lateritium and Xylaria sp. were dissolved in ethyl acetate and working concentrations were prepared in a 2-fold serial dilutions with the highest concentration at 2 mg/mL. Starting with the lowest concentration, 100 uL of the extracts were pipetted into the first row of the plate, and then 50 uL of NB was added into each well, followed by 1 uL of the tested bacteria. For control, 10 mg/mL of Streptomycin (BioBasic Inc, Canada) was used, replacing the fungal crude extract. The plate was incubated overnight at 37°C. After 24 h, 30 μL of 0.02% (w/v) resazurin (Sigma-Aldrich, Germany) was added into each well and the plate was incubated for another 2 h and then subjected to naked eye observation. MIC is defined as the lowest concentration of the extract that inhibits the visible growth of the bacteria, which can be detected by changes in the color of the resazurin, from blue to pink.
Determination of minimum bactericidal concentration (MBC)
The MBC test was determined by micro-dilution method under aseptic condition in 96-well plate as described by Daouk et al. (1995). From the above mentioned MIC assay, contents from wells with concentrations higher than the MIC value were directly plated on NA (Friendemann Schmidt, Australia) plates. The MBC value was determined when there was no observable colony growth. In addition, the contents from wells with bacterial growth were serially diluted to quantify an end-point killing of the bacteria. Each experiment was performed in triplicates and repeated at three independent times.
Antioxidant activity
The antioxidant effect of the two fungal isolates F. lateritium and Xylaria sp. were monitored using the 1,1-diphenyl-2-picrylhydrazyl (DPPH) radical scavenging test according to the method described by Liu et al. (2007). Fungal crude extract was dissolved in methanol and working solutions were prepared in five different concentrations in a series of 2-fold dilutions starting at 1,000 μg/mL. 100 μL of the working solution was mixed with 2,900 μL of 120 μM DPPH (Friendemann Schmidt, Australia) in methanol. The mixture was vortexed and incubated at 37°C in the dark for 30 min. Absorbance was recorded at 517 nm using a UV spectrophotometer (GENESYS 10S UV-Vis, USA). Inhibition of free radical by DPPH in percentage (A%) was calculated using the following equation: A% = [(A0-Ai)/A0] × 100; where, A0 = absorbance of the control reaction (without test compound), and Ai = absorbance of the test compound. Ascorbic acid was used as positive control and all tests were carried out in triplicates. ANOVA was performed using the SAS statistical package (SAS Institute, Cary, NC). For significant effect, the LSD was performed at P ≤ 0.05.
Results
Endophytic fungi identification
A total of 350 endophytic fungi isolates (Supplementary Table S1) were successfully isolated from 300 leaf segments of R. mucronata trees growing in the mangrove forest. However, due to the large number of isolated fungi, they were grouped based on their morphological characteristics (colony form, mycelium color and reverse media color) to allow for a systematic selection of the isolates (Figure 1). The isolates were classified into five groups (A-E) according to their colony color: white, green, gray/black, brown, and others (orange, yellow, purple and violet), respectively. Other characteristics that were taken into account were colony form, colony margin, and pigmentation. Based on this grouping, 101 fungal isolates were chosen for molecular identification, of which only 78 were successfully amplified using primers ITS1 and ITS4. BLAST searches revealed their identities as members of 22 different genera (Table 1), with a majority (96.2%) belonging to the phylum Ascomycota, which includes two classes, Sordariomycetes and Dothideomycetes. On the other hand, members from the phylum Basidiomycota only comprised 3.8% of the total identified specimens. The dominating genera in this study were Pestalotiopsis (20.3%), Alternaria (10.1%), and Cladosporium (6.3%), whereas genera that were represented by a single isolate were Periconia, Pithomyces, and Xylaria. Other identified genera were Curvularia, Diaporthe, Epicoccum, Fusarium, Leptosphaeria, Lophiostoma, Nigrospora, Phaeosphaeriopsis, Phoma, Phomopsis, Schizophyllum, and Stagonosporopsis. The ITS sequences obtained through this study were deposited in the NCBI GenBank (Accession no. MH397067-MH397144) for future reference.
Figure 1.
Endophytic fungi isolated from the leaves of Rhizophora mucronata. Shown here are some representatives of the fungal isolates, grouped according to their color of colonies and morphological characteristics. All fungal strains were isolated from the inside leaf tissues of the tropical mangrove species and cultivated on conventional media (Medium B) for 7 days at 27°C. Details on the morphological characteristics of the 350 isolates are listed in Supplementary Table 1.
Table 1.
Maximum nucleotide identity match for 78 fungal isolates based on ITS sequences using BLAST analysis.
| No. | Isolate ID (GenBank Accession) | Match identity (%) | E-value | Species | GenBank Accession | Phylum, Class, Family |
|---|---|---|---|---|---|---|
| 1 | RM1.18A.01 (MH397067) | 100 | 0.0 | Pestalotiopsis maculans | KX610327 | Ascomycota, Sordariomycetes, Pestalotiopsidaceae |
| 2 | RM1.18A.02 (MH397068) | 97 | 0.0 | Diaporthe phaseolorum | AF001017 | Ascomycota, Sordariomycetes, Diaporthaceae |
| 3 | RM1.18A.03 (MH397069) | 100 | 0.0 | Neopestalotiopsis egyptiaca | KP943747 | Ascomycota, Sordariomycetes, Pestalotiopsidaceae |
| 4 | RM1.18A.04 (MH397070) | 100 | 0.0 | Phaeosphaeriopsis musae | KR056291 | Ascomycota, Dothideomycetes, Phaeosphaeriaceae |
| 5 | RM1.18A.05 (MH397071) | 100 | 0.0 | Lophiostoma helminthicola | JQ026217 | Ascomycota, Dothideomycetes, Lophiostomataceae |
| 6 | RM1.18A.06 (MH397072) | 99 | 0.0 | Nigrospora oryzae | KM979813 | Ascomycota, Sordariomycetes, Apiosporaceae |
| 7 | RM1.18A.07 (MH397073) | 99 | 0.0 | Letendraea helminthicola | KU529827 | Ascomycota, Dothideomycetes, Tubeufiaceae |
| 8 | RM1.18A.08 (MH397074) | 97 | 0.0 | Xylaria sp. | KC507212 | Ascomycota, Sordariomycetes, Xylariaceae |
| 9 | RM1.18A.09 (MH397075) | 99 | 0.0 | Alternaria alternate | KY609180 | Ascomycota, Dothideomycetes, Pleosporaceae |
| 10 | RM1.18A.10 (MH397076) | 99 | 0.0 | Fusarium lateritium | AY266406 | Ascomycota, Sordariomycetes, Nectriaceae |
| 11 | RM1.18A.11 (MH397077) | 99 | 0.0 | Pestalotiopsis protearum | JX556231 | Ascomycota, Sordariomycetes, Pestalotiopsidaceae |
| 12 | RM1.18A.13 (MH397078) | 99 | 0.0 | Nigrospora oryzae | HQ608152 | Ascomycota, Sordariomycetes, Apiosporaceae |
| 13 | RM1.18A.14 (MH397079) | 99 | 0.0 | Nigrospora oryzae | HQ608152 | Ascomycota, Sordariomycetes, Apiosporaceae |
| 14 | RM2.18A.01 (MH397080) | 99 | 0.0 | Fusarium proliferatum | KR350649 | Ascomycota, Sordariomycetes, Nectriaceae |
| 15 | RM2.18A.02 (MH397081) | 100 | 0.0 | Neopestalotiopsis egyptiaca | KP943747 | Ascomycota, Sordariomycetes, Pestalotiopsidaceae |
| 16 | RM2.18A.03 (MH397082) | 100 | 0.0 | Neopestalotiopsis egyptiaca | KP943747 | Ascomycota, Sordariomycetes, Pestalotiopsidaceae |
| 17 | RM2.18A.06 (MH397083) | 98 | 0.0 | Fusarium verticillioides | KR183784 | Ascomycota, Sordariomycetes, Nectriaceae |
| 18 | RM2.18A.07 (MH397084) | 100 | 0.0 | Fusarium proliferatum | KR350649 | Ascomycota, Sordariomycetes, Nectriaceae |
| 19 | RM3.18A.01 (MH397085) | 99 | 0.0 | Pestalotiopsis protearum | JX556231 | Ascomycota, Sordariomycetes, Pestalotiopsidaceae |
| 20 | RM3.18A.02 (MH397086) | 99 | 0.0 | Pestalotiopsis protearum | JX556231 | Ascomycota, Sordariomycetes, Pestalotiopsidaceae |
| 21 | RM3.18A.03 (MH397087) | 99 | 0.0 | Neopestalotiopsis egyptiaca | KP943747 | Ascomycota, Sordariomycetes, Pestalotiopsidaceae |
| 22 | RM3.18A.04 (MH397088) | 99 | 0.0 | Neopestalotiopsis clavispora | KY319134 | Ascomycota, Sordariomycetes, Pestalotiopsidaceae |
| 23 | RM3.18A.05 (MH397089) | 99 | 0.0 | Schizophyllum commune | KP202299 | Basidiomycota, Agaricomycetes, Schizophyllaceae |
| 24 | RM3.18A.06 (MH397090) | 99 | 0.0 | Schizophyllum commune | AB369909 | Basidiomycota, Agaricomycetes, Schizophyllaceae |
| 25 | RM3.18A.07 (MH397091) | 99 | 0.0 | Phomopsis sp. | KX020566 | Ascomycota, Sordariomycetes, Diaporthaceae |
| 26 | RM3.18A.08 (MH397092) | 99 | 0.0 | Cladosporium perangustum | KP701968 | Ascomycota, Dothideomycetes, Cladosporiaceae |
| 27 | RM3.18A.09 (MH397093) | 100 | 0.0 | Cladosporium tenuissimum | KP701937 | Ascomycota, Dothideomycetes, Cladosporiaceae |
| 28 | RM3.18A.10 (MH397094) | 93 | 0.0 | Periconia pseudobyssoides | KC954161 | Ascomycota, Dothideomycetes |
| 29 | RM3.18A.11 (MH397095) | 100 | 0.0 | Arthrinium sp. | HQ022507 | Ascomycota, Sordariomycetes, Apiosporaceae |
| 30 | RM3.18A.13 (MH397096) | 100 | 0.0 | Phaeosphaeriopsis musae | KR056291 | Ascomycota, Dothideomycetes, Phaeosphaeriaceae |
| 31 | RM3.18A.14 (MH397097) | 99 | 0.0 | Curvularia lunata | DQ836800 | Ascomycota, Dothideomycetes, Pleosporaceae |
| 32 | RM3.18A.15 (MH397098) | 99 | 0.0 | Epicoccum nigrum | KM507768 | Ascomycota, Dothideomycetes, Pleosporaceae |
| 33 | RM3.18A.16 (MH397099) | 100 | 0.0 | Phoma sp. | KT150683 | Ascomycota, Dothideomycetes, Didymellaceae |
| 34 | RM3.18A.17 (MH397100) | 99 | 0.0 | Stagonosporopsis cucurbitacearum | KU059901 | Ascomycota, Dothideomycetes, Didymellaceae |
| 35 | RM3.18A.18 (MH397101) | 100 | 0.0 | Curvularia brachyspora | HG778983 | Ascomycota, Dothideomycetes, Pleosporaceae |
| 36 | RM3.18A.20 (MH397102) | 99 | 0.0 | Stagonosporopsis cucurbitacearum | KU059901 | Ascomycota, Dothideomycetes, Didymellaceae |
| 37 | RM3.18A.23 (MH397103) | 99 | 0.0 | Paraphaeosphaeria sp. | KX611058 | Ascomycota, Dothideomycetes, Didymosphaeriaceae |
| 38 | RM3.18A.24 (MH397104) | 99 | 0.0 | Pithomyces maydicus | HG933803 | Ascomycota, Dothideomycetes, Pleosporaceae |
| 39 | RM3.18A.25 (MH397105) | 100 | 0.0 | Arthrinium sp. | HQ022507 | Ascomycota, Sordariomycetes, Apiosporaceae |
| 40 | RM3.18A.26 (MH397106) | 100 | 0.0 | Phoma sp. | KT150683 | Ascomycota, Dothideomycetes, Didymellaceae |
| 41 | RM1.30.01 (MH397107) | 99 | 0.0 | Stagonosporopsis cucurbitacearum | KU059901 | Ascomycota, Dothideomycetes, Didymellaceae |
| 42 | RM1.30.02 (MH397108) | 99 | 0.0 | Epicoccum nigrum | KR094452 | Ascomycota, Dothideomycetes, Pleosporaceae |
| 43 | RM1.30.04 (MH397109) | 99 | 0.0 | Alternaria eichhorniae | KU593527 | Ascomycota, Dothideomycetes, Pleosporaceae |
| 44 | RM1.30.05 (MH397110) | 100 | 0.0 | Neopestalotiopsis egyptiaca | KP943747 | Ascomycota, Sordariomycetes, Pestalotiopsidaceae |
| 45 | RM1.30.07 (MH397111) | 100 | 0.0 | Alternaria eichhorniae | KU593527 | Ascomycota, Dothideomycetes, Pleosporaceae |
| 46 | RM1.30.08 (MH397112) | 99 | 0.0 | Epicoccum nigrum | HQ728258 | Ascomycota, Dothideomycetes, Pleosporaceae |
| 47 | RM1.30.09 (MH397113) | 99 | 0.0 | Epicoccum nigrum | HQ728258 | Ascomycota, Dothideomycetes, Pleosporaceae |
| 48 | RM1.30.11 (MH397114) | 99 | 0.0 | Neopestalotiopsis protearum | KT936426 | Ascomycota, Sordariomycetes, Pestalotiopsidaceae |
| 49 | RM1.30.12 (MH397115) | 100 | 0.0 | Neopestalotiopsis protearum | KX631739 | Ascomycota, Sordariomycetes, Pestalotiopsidaceae |
| 50 | RM1.30.13 (MH397116) | 99 | 0.0 | Neopestalotiopsis egyptiaca | KP943747 | Ascomycota, Sordariomycetes, Pestalotiopsidaceae |
| 51 | RM1.30.15 (MH397117) | 100 | 0.0 | Alternaria arborescens | KP942903 | Ascomycota, Dothideomycetes, Pleosporaceae |
| 52 | RM2.30.02 (MH397118) | 100 | 0.0 | Lophiostoma helminthicola | JN116664 | Ascomycota, Dothideomycetes, Lophiostomataceae |
| 53 | RM2.30.03 (MH397119) | 99 | 0.0 | Neopestalotiopsis foedans | KU593530 | Ascomycota, Sordariomycetes, Pestalotiopsidaceae |
| 54 | RM2.30.05 (MH397120) | 100 | 0.0 | Phoma leveillei | KT963795 | Ascomycota, Dothideomycetes, Didymellaceae |
| 55 | RM2.30.07 (MH397121) | 100 | 0.0 | Curvularia affinis | HG778982 | Ascomycota, Dothideomycetes, Pleosporaceae |
| 56 | RM2.30.11 (MH397122) | 100 | 0.0 | Schizophyllum commune | KP202299 | Basidiomycota, Agaricomycetes, Schizophyllaceae |
| 57 | RM2.30.12 (MH397123) | 100 | 0.0 | Cladosporium cladosporioides | JX230994 | Ascomycota, Dothideomycetes, Cladosporiaceae |
| 58 | RM2.30.15 (MH397124) | 100 | 0.0 | Phoma herbarum | MF120206 | Ascomycota, Dothideomycetes, Didymellaceae |
| 59 | RM2.30.17 (MH397125) | 99 | 0.0 | Pestalotiopsis sp. | KR085971 | Ascomycota, Sordariomycetes, Pestalotiopsidaceae |
| 60 | RM3.30.01 (MH397126) | 99 | 0.0 | Diaporthe yunnanensis | KY491542 | Ascomycota, Sordariomycetes, Diaporthaceae |
| 61 | RM3.30.02 (MH397127) | 99 | 0.0 | Leptosphaeria sp. | KM979814 | Ascomycota, Dothideomycetes, Leptosphaeriaceae |
| 62 | RM3.30.04 (MH397128) | 99 | 0.0 | Alternaria macrospora | AY154689 | Ascomycota, Dothideomycetes, Pleosporaceae |
| 63 | RM3.30.05 (MH397129) | 99 | 0.0 | Phaeosphaeriopsis sp. | KR012892 | Ascomycota, Dothideomycetes, Phaeosphaeriaceae |
| 64 | RM3.30.06 (MH397130) | 100 | 0.0 | Arthrinium sp. | HQ022507 | Ascomycota, Sordariomycetes, Apiosporaceae |
| 65 | RM3.30.07 (MH397131) | 100 | 0.0 | Cladosporium tenuissimum | KP701937 | Ascomycota, Dothideomycetes, Cladosporiaceae |
| 66 | RM3.30.08 (MH397132) | 100 | 0.0 | Cladosporium cladosporioides | KM980007 | Ascomycota, Dothideomycetes, Cladosporiaceae |
| 67 | RM3.30.09 (MH397133) | 99 | 0.0 | Leptosphaeria sp. | KM979814 | Ascomycota, Dothideomycetes, Leptosphaeriaceae |
| 68 | RM3.30.10 (MH397134) | 99 | 0.0 | Periconia byssoides | MF435088 | Ascomycota, Dothideomycetes |
| 69 | RM3.30.12 (MH397135) | 99 | 0.0 | Paraphaeosphaeria sp. | KX611058 | Ascomycota, Dothideomycetes, Didymosphaeriaceae |
| 70 | RM3.30.13 (MH397136) | 100 | 0.0 | Lophiostoma helminthicola | JN116664 | Ascomycota, Dothideomycetes, Lophiostomataceae |
| 71 | RM3.30.14 (MH397137) | 100 | 0.0 | Arthrinium sp. | HQ022507 | Ascomycota, Sordariomycetes, Apiosporaceae |
| 72 | RM3.30.15 (MH397138) | 99 | 0.0 | Leptosphaeria sp. | KM979814 | Ascomycota, Dothideomycetes, Leptosphaeriaceae |
| 73 | RM3.30.16 (MH397139) | 100 | 0.0 | Alternaria arborescens | KP942903 | Ascomycota, Dothideomycetes, Pleosporaceae |
| 74 | RM3.30.17 (MH397140) | 100 | 0.0 | Letendraea helminthicola | KP263123 | Ascomycota, Dothideomycetes, Tubeufiaceae |
| 75 | RM3.30.18 (MH397141) | 100 | 0.0 | Alternaria arborescens | KP942903 | Ascomycota, Dothideomycetes, Pleosporaceae |
| 76 | RM3.30.19 (MH397142) | 99 | 0.0 | Curvularia eragrostidis | HG778986 | Ascomycota; Dothideomycetes, Pleosporaceae |
| 77 | RM3.30.20 (MH397143) | 99 | 0.0 | Leptosphaeria sp. | KM979814 | Ascomycota, Dothideomycetes, Leptosphaeriaceae |
| 78 | RM3.30.22 (MH397144) | 100 | 0.0 | Curvularia lunata | HQ607991 | Ascomycota, Dothideomycetes, Pleosporaceae |
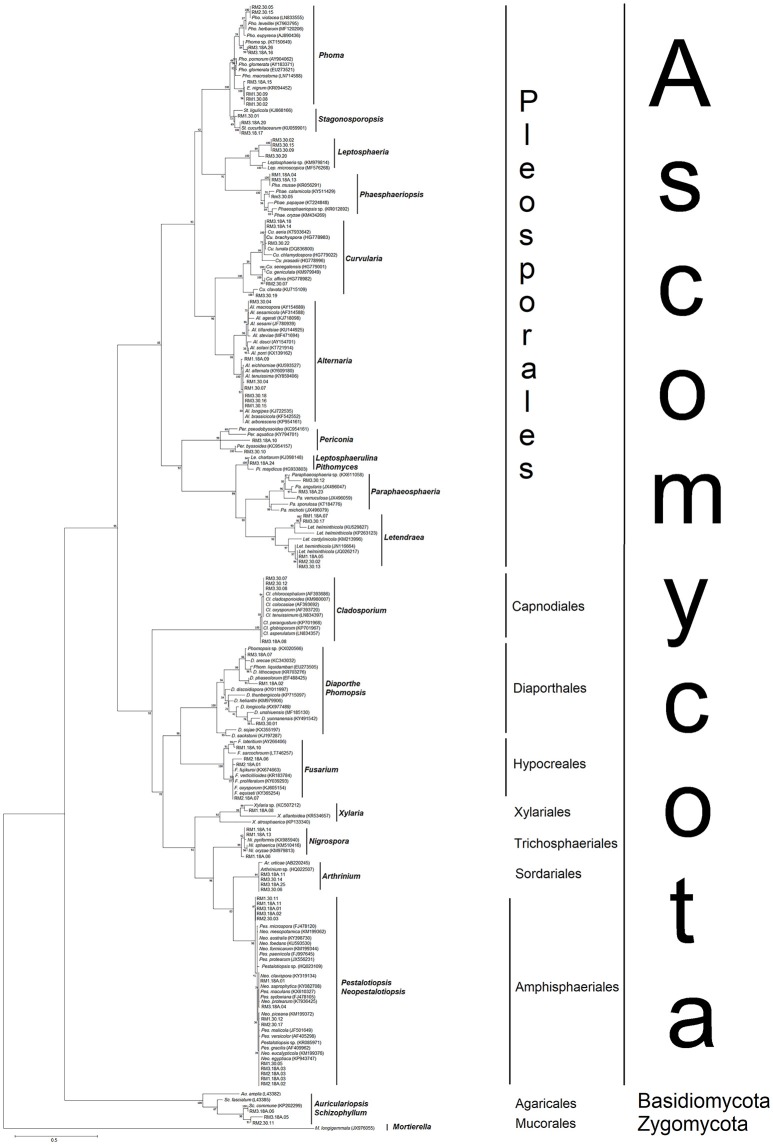
A total of 117 sequences of close relatives (Supplementary Table S2) were downloaded from the NCBI GenBank and combined with sequences of the 78 endophytes for phylogenetic tree construction (Figure 2). Nine different orders were observed, of which seven representing Ascomycota, and one order each representing Basidiomycota and the outgroup Zygomycota. Most of the fungal isolates clustered under the order Pleosporales, belonging to at least 12 different genera. Among the 78 isolates obtained from this study, eight (RM3.18A.15, RM3.18A.16, RM3.18A.26, RM1.30.02, RM1.30.08, RM1.30.09, RM2.30.05, and RM2.30.15) were grouped under the Phoma-Epicoccum clade with 100% bootstrap support (Figure 2). Similarly, other identified isolates clustered into clades of known sequences and received high bootstrap support (91–100%). For example, seven isolates (RM3.30.04, RM1.18A.09, RM1.30.04, RM1.30.07, RM3.30.18, RM3.30.16, and RM1.30.15) identified from their sequences as closely related to Alternaria sp., clustered into their predicted clade (99%). For the order Capnodiales, four isolates (RM3.30.07, RM2.30.12, RM3.30.08, and RM3.18A.08) clustered under Cladosporium (100%) and for Hypocreales, four isolates (RM1.18A.10, RM2.18A.06, RM2.18A.01, and RM2.18A.07) clustered with Fusarium (100%). For the order Xylariales, RM1.18A.08 was clustered under the Xylaria clade with 92% bootstrap support.
Figure 2.
Maximum likelihood (ML) phylogenetic tree based on rDNA ITS sequences of endophytic fungal isolates and fungal ITS sequences from the GenBank. ML tree was constructed using the substitution model general time reversible (GTR) model and gamma distributed (+G) with invariant sites (+I) (= GTR+G+I). All positions containing gaps and missing data were included for analysis. Clade supports were calculated based on 1000 bootstrap re-samplings. Accession numbers of the 78 sequences belonging to the endophytic fungi isolated in this study are listed in Table 1, while the reference sequences (117) from the GenBank are listed in Supplementary Table 2.
In Trichospaeriales, three isolates (RM1.18A.06, RM1.18A.13, and RM1.18A.14) clustered under the Nigrospora clade (99%). Four isolates (RM3.18A.11, RM3.18A.25, RM3.30.04, and RM3.30.06) clustered under Athrinium of Sordariales. The order with the most isolates obtained from this study was Amphisphaeriales. The cluster was constructed within a polytomy consisting Pestalotiopsis and Neopestalotiopsis (98%), recorded with 14 isolates (RM1.18A.01, RM1.18A.11, RM1.18A.03, RM2.18A.02, RM2.18A.03, RM3.18A.01, RM3.18A.02, RM3.18A.03, RM3.18A.04, RM1.30.05, RM1.30.11, RM1.30.12, RM2.30.03, and RM2.30.17). As for the phylum Basidiomycota, three isolates (RM2.30.11, RM3.18A.05 and RM3.18A.06) were clustered under the Auriculariopsis-Schizophyllum clade, showing sister relationship with S. commune.
Fungal interaction during dual culture assay
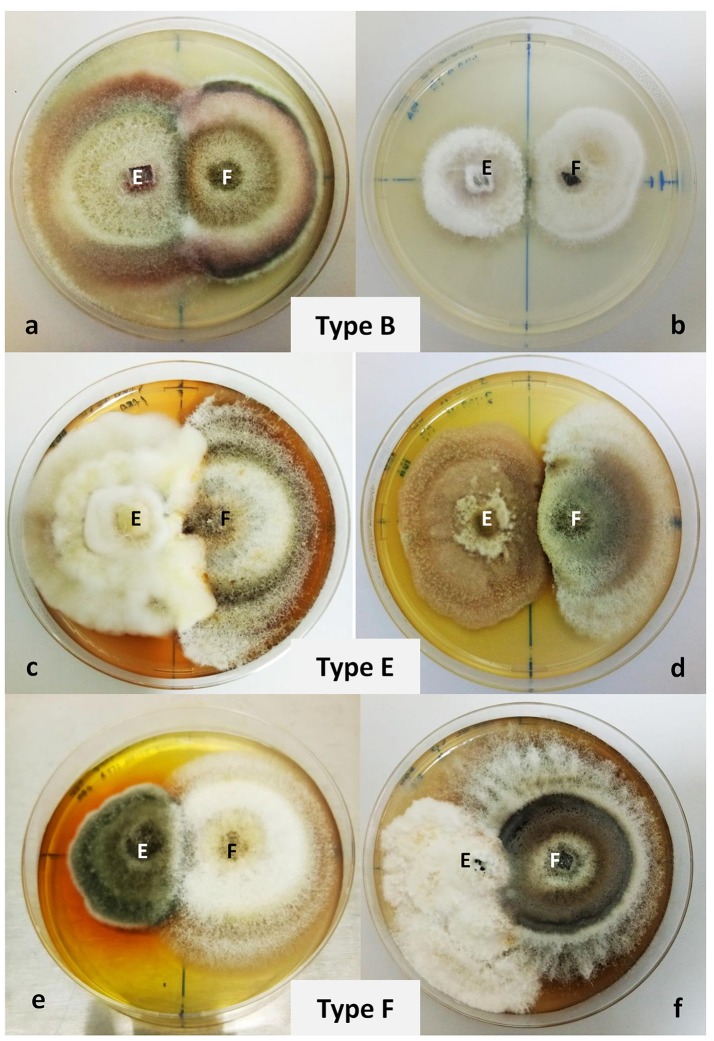
Six fungal isolates were selected for antagonism assay on the basis of their distinct morphology such as pigment production, indicating their potential in possessing significant bioactive compounds, and based on previous literature reporting on fungal genera with strong bioactivities against other microorganisms (Oliveira et al., 2012; Zhao et al., 2012a). The isolates were RM1.18A.08, RM1.18A.10, RM1.18A.13, RM3.18A.16, RM2.30.17, RM3.30.04, and identified through NCBI Blast match identity as Xylaria sp., Fusarium lateritium, Nigrospora oryzae, Phoma sp., Pestalotiopsis sp., and Alternaria macrospora, respectively. From the dual culture plates, we observed 3 different fungal interactions (Wheeler and Hocking, 1993) between the isolated fungal antagonists and the pathogen F. solani. Both F. lateritium and N. oryzae exhibited type B interaction when cultured with F. solani. The fungal endophytes and pathogen displayed a mutual inhibition on contact (Figures 3a,b). However, in the dual culture plate of N. oryzae and F. solani, a small clear zone (>2 mm) was recorded. In Xylaria sp./F. solani interaction, the mycelium of the antagonist had breached into the pathogen's colony. Yellowish pigmentation was first observed at the border of the two colonies, which later turned into rust after several days (Figure 3c). In Phoma sp./F. solani interaction, dark purple pigmentation was observed on the test pathogen. This combination exhibited type E interaction—growth of the test endophyte was unaffected while that of the pathogen was reduced (Figure 3d). For A. macrospora and Pestalotiopsis sp., they both showed type F interaction when co-cultivated with F. solani (Figures 3e,f). Growth of the endophytes were inhibited by the pathogen, which eventually overgrown the inhibited colony. The A. macrospora colony had been producing yellow pigments (data not shown) beginning on the third day after inoculation, which then turned into dark brown on the seventh day (Figure 3e).
Figure 3.
Dual culture plate assay between six endophytic fungal isolates against the pathogen Fusarium solani. Fusarium solani was grown in PDA plates together with (a) Fusarium lateritium, (b) Nigrospora oryzae, (c) Xylaria sp., (d) Phoma sp., (e) Alternaria macrospora, and (f) Pestalotiopsis sp. The plates were cultivated for 7 days at 27°C. Radial growths were measured and types of interaction were observed. Type B interaction was observed for F. lateritium and N. oryzae, type E for Xylaria sp. and Phoma sp., and type F for A. macrospora and Pestalotiopsis sp.
Antagonistic activity of the fungal isolates against Fusarium solani
Dual culture assay revealed the inhibition percentage (I%) of each endophytic fungus against the pathogen F. solani (Table 2). Phoma sp. recorded the highest I% at 69.64%. Xylaria sp. the second highest I% (65.11%), followed by N. oryzae (64.03%), and F. lateritium (61.89%). Mycelium development of F. solani in dual culture plates was significantly inhibited (F test = 56.31, p < 0.0001). There was significant differences between means of inhibition by Phoma sp. when compared to the other five species, while no differences were observed between Xylaria sp. and N. oryzae, and N. oryzae with F. lateritium. Their antagonistic ability to produce non-volatile compounds to inhibit the test pathogen was further confirmed. Radial growth of F. solani mycelia in plates containing the endophytic fungal filtrates showed significant I% for Xylaria sp. (25.07%), Phoma sp. (23.36%), and F. lateritium (21.19%) (F = 63.71, p < 0.0001) (Table 2).
Table 2.
In vitro antagonism of six selected endophytic fungal isolates against the pathogenic fungus, Fusarium solani, using two types of assays.
| Isolate name | Percent inhibition growth over control (I%) ± SD | |
|---|---|---|
| Dual culture assayA | Non-volatile compound assayB | |
| Phoma sp. | 69.64a ± 1.76 | 23.26a, c ± 3.31 |
| Xylaria sp. | 65.11b ± 0.91 | 25.07a, b ± 1.61 |
| Nigrospora oryzae | 64.03b, c ± 0.40 | 2.07d ± 3.58 |
| Fusarium lateritium | 61.89c ± 2.05 | 21.19b, c ± 0.45 |
| Alternaria macrospora | 59.05d ± 1.61 | 1.29d ± 1.19 |
| Pestalotiopsis sp. | 49.40e ± 1.61 | 11.63e ± 2.05 |
Data are means of percentage of growth inhibition ± standard deviation (SD). Means followed by the same letter in each group are not significantly different at α = 0.05 according to Tukey test (LSD = 2.83 and 4.11, for A and B assays, respectively).
Antibacterial properties
From the preliminary qualitative antibacterial screening of the six fungal isolates, F. lateritium showed inhibition to all four bacterial pathogens tested, while Xylaria sp. to three (Table 3). However, the other four endophytic isolates including the high performer from the in vitro antagonistic assays, Phoma sp., gave no inhibition effect to any of the tested bacteria. Ethyl acetate-extracts of the fungi were used to determine the minimum concentration required to inhibit bacterial growths. For these two fungi, crude extract at 0.50 mg/mL was sufficient to inhibit E. coli and P. aeruginosa; at 1.00 mg/mL was needed to inhibit S. aureus, and the highest concentration at 2.00 mg/mL was required for B. subtilis (Table 4).
Table 3.
Preliminary screening for antibacterial activities in six selected endophytic fungal isolates against Bacillus subtilis, Escherichia coli, Pseudomonas aeruginosa, and Staphylococcus aureus using plate assay.
| Isolates | Staphylococcus aureus ATCC 43300 | Escherichia coli ATCC 25922 | Pseudomonas aeruginosa ATCC 15542 | Bacillussubtilis B29 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| i | ii | iii | i | ii | iii | i | ii | iii | i | ii | iii | |
| Alternaria macrospora | – | – | – | – | – | – | – | – | – | – | – | – |
| Fusarium lateritium | + | – | + | + | + | – | + | – | – | + | + | – |
| Nigrospora oryzae | – | – | – | – | – | – | – | – | – | – | – | – |
| Pestalotiopsis sp. | – | – | – | – | – | – | – | – | – | – | – | – |
| Phoma sp. | – | – | – | – | – | – | – | – | – | – | – | – |
| Xylaria sp. | – | – | – | + | + | – | + | + | + | + | + | – |
| Streptomycin | 35 | 35 | 35 | 30 | 30 | 30 | 35 | 35 | 35 | 30 | 30 | 30 |
“+” indicates the fungal isolate inhibited bacterial growth, and “–” indicates no inhibition. Streptomycin was used as positive control.
Table 4.
Determination of the Minimum Inhibitory Concentration (MIC) and Minimum Bactericidal Concentration (MBC) of ethyl-acetate extracts of two endophytic fungal isolates against several pathogenic bacteria using the microdilution method.
| Isolates | MIC / MBC concentration (mg/mL) | |||
|---|---|---|---|---|
| Bacillus subtilis | Escherichia coli | Pseudomonas aeruginosa | Staphylococcus aureus | |
| Fusarium lateritium | 2.00/2.00 | 0.50/0.50 | 0.50/0.50 | 1.00/1.00 |
| Xylaria sp. | 2.00/2.00 | 0.50/0.50 | 0.50/0.50 | 1.00/1.00 |
Antioxidant activity
For antioxidant activity, methanol-extracts of F. lateritium and Xylaria sp. were evaluated by DPPH radical scavenging assay. The extracts had significant radical scavenging capacity with significant F test (F = 57.05, p < 0.0001). The methanol-extract from Xylaria sp. notably reduced the absorbance of DPPH free radical (Table 5). It recorded significantly high scavenging activity (A%) of over 90% at the two highest extract concentrations (500 and 1,000 μg/mL) (Table 5). The A% was reduced to almost half at 250 μg/mL, and further lowered with decreasing concentrations. In contrast, the methanol-extract of F. lateritium had free radicals inhibition activity of only 29.95% at the highest concentration (1,000 μg/mL) and 6.61% at the lowest concentration (62.5 μg/mL), which were significantly lower when compared to Xylaria sp. and the positive control, ascorbic acid.
Table 5.
Antioxidant activity of Fusarium lateritium and Xylaria sp. as determined from their free radical-scavenging capacity measured by DPPH assay.
| Extract concentration (μg/mL) | Inhibition of free radical by DPPH in percentage (A%) ± SD (%) | ||
|---|---|---|---|
| Fusarium lateritium | Xylaria sp. | Ascorbic acid | |
| 1,000 | 29.95c±0.373 | 95.82a±0.079 | 61.16b±0.518 |
| 500 | 14.03c±0.879 | 91.89a±0.925 | 56.82b±4.57 |
| 250 | 8.03c±2.567 | 51.32b±0.356 | 53.03a±7.906 |
| 125 | 6.74c±1.59 | 30.32b±1.060 | 47.45a±9.733 |
| 62.5 | 6.61c±0.273 | 18.76b±1.292 | 39.61a±2.739 |
The fungal extract was prepared in a series of 2-fold dilutions. Ascorbic acid served as positive control. Data are means of inhibition of free radicals by DPPH. Means followed by the same letter across each concentration are not significantly different at α = 0.05 according to Tukey test (LSD = 9.07).
Discussion
Endophytic fungal diversity
From this study, ITS sequences identified 75 fungal isolates representing the phylum Ascomycota, and three from Basidiomycota. The phylum Ascomycota is reportedly the most common representative of endophytic fungi community when isolated using standard isolation protocol (Crozier et al., 2006; He et al., 2012; Koukol et al., 2012), and the same finding was reflected in our study. Fungi from the phylum Basidiomycota have been reported to be culture-method dependent (Crozier et al., 2006), which explains the small number of Basidiomycota isolates in this study. Comparative studies have also indicated that only a small portion of the microorganisms in nature was amenable to cultivation using conventional microbiological techniques (Amann et al., 1995). There are many unfavorable factors that could affect the ability to grow microorganisms in laboratory conditions. Among them are lack of knowledge about their nutritional requirements and demanding nature of the microorganisms specifically when they originate from environmental samples.
Based on the total fungal isolates identified using the DNA method, the genus Pestalotiopsis was the most frequent, consisting 20.25% of total fungal isolates obtained from this study. Pestalotiopsis as a dominant fungal endophyte has been reported in other plants such as Carapa guianensis, Catostemma fragrans, Chlorocardium rodiei, and Eperua falcata (Cannon and Simmons, 2002). Pestalotiopsis is a beneficial member of the foliar endophytic community because it is capable to switch its nutritional-mode, allowing itself to stay as an endophyte or become a saprobe when necessary (Maharachchikumbura et al., 2012; Douanla-Meli et al., 2013). Besides Pestalotiopsis, other fungal genera such as Alternaria, Cladosporium, Curvularia, and Fusarium were also reportedly predominant mangrove endophytic fungi (Udayanga et al., 2011) and are widespread in different mangrove species (Cheng et al., 2009). Meanwhile, the single isolate genus Xylaria is a common endophytic fungal genus found in trees (Suryanarayanan et al., 2002). Although it is widely available in most tropical plants investigated in past studies, we recorded a low isolation frequency for this genus (Chen et al., 2013; Douanla-Meli et al., 2013). Our finding also revealed lesser-known fungal genera, namely Periconia, Phaeosphaeriopsis, Pithomyces, and Stagonosporopsis as community members in the leaf of R. mucronata.
Potential bioactivities
Antagonism properties
Antagonism has been defined as “to include any activity of one organism which in some way adversely affects another growing in association with it” (Khara and Hadwan, 1990). There are several types of antagonism that fungal antagonists could exhibit, such as competition, mycoparasitism, and the production of extracellular metabolites (Siameto et al., 2010). These metabolites, namely antibiotics and lytic enzymes, have been widely applied in various fields such as crop-pathogen controls. Endophytic microorganisms isolated from plants were also reported to be capable of producing a variety of novel bioactive metabolites that are source for novel secondary metabolites (Firáková et al., 2007; Ramasamy et al., 2010). Therefore, attention can be given to the bioactive compounds produced by plants, microbes, and marine organisms for the discovery of compounds useful for the development of new drugs.
Fusarium solani is a common tree pathogen and has been isolated from diseased mangrove plants (Umechuruba, 2005). We showed that endophytes F. lateritium, N. oryzae, Phoma sp., and Xylaria sp., successfully inhibited the pathogenic F. solani in the dual culture assay (Figure 3d). The ability to out-grow the pathogenic fungus suggested that Phoma sp. inhibited growth of F. solani by competing for space and nutrients. Spreading of F. solani was further restricted when mycelium of the two fungi came into contact. Biological agents with antifungal properties are known to secrete certain enzymes, which are responsible for the breaking down of the pathogen's cell wall, thus restricting its growth (Sharon et al., 2001). Antagonist mechanism displayed by Xylaria sp. was more aggressive when compared to Phoma sp. as it advances and further colonized over the surface of the pathogen (Figure 3c). This phenomenon could be attributed to the production of lytic enzymes by Xylaria sp., which degrade the pathogen's cell wall and allows Xylaria sp. to colonize the surface of the pathogen's colony, similar to what has been reported in Streptomyces griseus (Anitha and Rabeeth, 2010). The antagonistic activity displayed by Phoma sp. and Xylaria sp. could be explained by their secreting secondary metabolites into the agar, which are detrimental to the pathogen's growth. Arguably, such inhibition could also be due to nutrient depletion in the media (Robinson et al., 2014). Variations in fungal interactions might also be influenced by antibiotics produced from the isolates, which could be fungicidal to a single fungus, but fungistatic to other fungi (Dharmaputra, 2003). Meanwhile, a small inhibition zone >2 mm, was recorded in the dual culture plate of N. oryzae/F. solani (Figure 3b). It could be due to the production of hydrolytic enzymes by certain fungal isolates, or the production of antibiotics by the antagonists (Kamala and Indira, 2011). The various types of interactions we observed are dependent on the specific fungal species combination, because of differences in compounds of the volatile mixture, substrates, or abilities to detoxify the volatile metabolites such as enzymatic activity (Kai et al., 2007). Compound isolation from the endophytic Nigrospora sp., has revealed griseofulvin responsible for its strong antifungal activity against a plant pathogenic fungus, Botrytis cinerea (Zhao et al., 2012a). Griseofulvin has been widely used to treat pathogenic fungi causal diseases, acting as antibiotics (Zhao et al., 2012a). Many fungi including Xylaria sp. have been reported to be typical griseofulvin-producing fungi (Park et al., 2005).
Over time during the dual culture assay, we observed several fungi had produced pigmentation, including Xylaria sp., A. macrospora, and the pathogen F. solani (Figure 3). Fungi are known to produce a range of bioactive compounds that hold significant roles in the diversification and adaptability of these micro-organisms in various ecological niches (Fox and Howlett, 2008; da Costa Souza et al., 2016). Among the compounds induced by fungi, pigments have been accentuated as the contributor to biological activities exhibited by the fungi (dos Reis Celestino et al., 2014). There are a variety of pigments produced by fungi including carotenoids, melanins, flavins, phenazines, and quinos (Mapari et al., 2010; Dufossé et al., 2014). These pigments are attributed to crucial activities such as antibacterial, antifungal, and herbicidal (Geweely, 2011; Premalatha et al., 2012; Teixeira et al., 2012). This was shown in the present study, where yellow pigmentation was produced by Xylaria sp. in the dual culture plate against the pathogen F. solani (Figure 3c). The same condition was observed for isolate A. macrospora, where it started producing yellow pigmentation since day three of the dual culture assay. The yellowish pigmentation turned darker, eventually into brown, after 7 days. While, in the case of Phoma sp./F. solani, the pathogen started producing dark purple pigmentation when their mycelia got in contact. Many fungi produce extracellular pigments in response to adverse conditions, such as low moisture, pH, and UV light, or as a way to protect their resources from other fungi (Score et al., 1997; Tudor et al., 2013). Under natural conditions, when the supplies of vital nutrients become depleted, parts of the fungal mycelium will switch their biochemical activity to the pathways of secondary metabolism (Isaac, 1994). Rather than producing new fungal building material, these fungi start to produce metabolites manifested as pigmentation. The production of the pigments might have resulted from the defense mechanism of the fungi to protect their mycelia from being hydrolyzed by the enzymes produced by other microbes (Isaac, 1994).
Production of antimicrobial compounds is one of the main mechanisms employed by antagonists for pathogen control. The antimicrobial compounds may be produced in the form of volatiles and non-volatiles (Cazar et al., 2005). One of the simplest and most effective method to identify the potential antagonists against pathogen is by the non-volatile compound test. In this study, we examined six fungal isolates for their production of non-volatile compounds that could inhibit the growth of F. solani in vitro. Based on the results, it is evident that the non-volatile compounds suppressed the pathogen's growth, causing mycelial growth reduction in F. solani when compared to the control plates. The culture filtrates from these isolates displayed inhibitory effects on the radial growth of the pathogen, and the mycelia accumulation observed in this study indicated the presence of non-volatile antibiotics in the filtrate. Antibiotic inhibitions have been reportedly useful in reducing plant diseases in previous studies (Dubey and Suresh, 2006).
Antibacterial activities
Since the last three decades, pathogenic microbes have become more resistant to modern antibiotics (Cui et al., 2015). It was reported that the antibacterial activities exhibited by leaf extracts of several mangrove species such as Acanthus ilicifolius, Excoecaria agallocha, R. mucronata, and Sonneratia caseolaris, showed significant inhibition on the growth of multidrug resistance pathogenic bacteria Staphylococcus aureus (Prihanto et al., 2011). We observed similar findings through this study. The enormous diversity of endophytic fungi of the mangrove species was believed to be the major factor. Previously, it was reported that Xylaria sp. contained 7-amino-4-methylcoumarin compound, which was proven to exhibit strong inhibitory activities against 13 microorganisms, including E. coli and S. aureus (Liu et al., 2008). Other genera which also showed effectiveness in inhibition of microbial growth were Aspergillus, Botryosphaeria, Eutypella, Fusarium, Guignardia, Penicillium and Phomopsis (Phongpaichit et al., 2006; Bernardi-Wenzel et al., 2010; Rhoden et al., 2012). Therefore, it could be suggested that the antibacterial activities exhibited in mangrove leaf extracts were probably contributed by the endophytic fungi living in them. This claim was supported by the findings of a group of Chinese researchers who isolated 130 endophytic fungi from several Chinese medicinal plants, and further tested the antitumor and antifungal activities from their extracts. As a result, 9.2% of the extracts were positive in antitumour activity, with 30% of the endophytes extracted from the plants exhibited antifungal activity, thus indicating presence in fungal compounds that could be associated with the host plants (Li et al., 2005). In addition, it was also reported that endophytic fungi could produce bioactive compounds associated with their host plants (Kusari et al., 2013).
In the recent study, F. lateritium was shown to successfully inhibit all four pathogenic bacteria, with low MIC values. Endophytic Fusarium species of medicinal plants are well-known sources of bioactive compounds including against microbial activity (Liang et al., 2012). Several compounds that were proven to exhibit antimicrobial activities include pentaketide (2-methylbutyraldehyde-substituted-α-pyrone) isolated from Fusarium sp. of Selaginella pallescens, which showed strong activity against Candida albicans (Brady and Clardy, 2000); beauvericin from F. oxysporum, isolated from the bark of Cinnamomum kanehirae, which significantly suppressed the growth of methicillin-resistant bacteria, B. subtillis and S. aureus (Wang et al., 2011); and subglutinol A and B, from Fusarium sp. of a medicinal plant Tripterygium wilfordii (Lee et al., 1995). Another compound equisetin, extracted from Fusarium sp., was not only responsible for the antimicrobial activity, but also aided the plant to successfully withstand stressful environmental conditions (Ratnaweera et al., 2015).
Antioxidant activity
The DPPH test showed that the methanol extract of F. lateritium had a lower antioxidant activity (6.6–29.9) in contrast to that of Xylaria sp. (18.7–95.8%) (Table 5). At the concentration of 1,000 μg/mL, Xylaria sp. displayed higher scavenging activity even above that of the positive control, ascorbic acid (61.6%). The activity was observed to be extract concentration-dependent, with scavenging activity directly proportional to the concentration. This is noticeable by the color change in the solution. The big difference between these two isolates could be due to the high compound content in Xylaria sp. An isolate of Xylaria sp. from Gingko biloba had several phenolic compounds present in its methanolic extract such as 2-hydrazino-8-hydroxy-4-phenylquinoline, 3,4-dimethoxy-phenol, 2,4-bis(1,1-dimethylethyl)-phenol, 3,4-dihydro-8-hydroxy-3-methyl-isocoumarin and ferruginol. These compounds are believed to be contributors to the strong antioxidant activities exhibited by Xylaria sp. (Liu et al., 2007). In contrast to Xylaria sp., F. lateritium, did not display distinct scavenging activity. This result however is not as observed by previous studies, which regarded Fusarium sp. as a potent source of antioxidant based on their significant antioxidant activity recorded (Vasundhara et al., 2016). Such example include F. oxysporum, an endophytic fungus from the rhizomes of Dioscorea zingiberensis (Li et al., 2012) and Fusarium proliferatum, from pigeon pea (Zhao et al., 2012b).
Data availability
The dataset generated in this study has been deposited in the GenBank, accession codes MH397067 to MH397144.
Author contributions
TH and RM designed the study. TH performed the experiments and analyzed the data with help from SL. AH helped with antagonistic analyses. TH and SL led the writing with contributions from RT. IF-H helped in site identification and logistics. RM reviewed and edited the manuscript. All authors reviewed and approved the final manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank the Forestry Department of Peninsular Malaysia (FDPM) for granting permission and assisting in sample collection at the Matang Mangrove Forest Reserve, Perak, Malaysia. This work was supported by the Universiti Putra Malaysia Research University Grant Scheme (GP-IBT/2013/9418400 and GP-IPB/2014/9441200).
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01707/full#supplementary-material
References
- Abraham S., Basukriadi A., Pawiroharsono S., Sjamsuridzal W. (2015). Insecticidal activity of ethyl acetate extracts from culture filtrates of mangrove fungal endophytes. Mycobiology 43, 137–149. 10.5941/MYCO.2015.43.2.137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aly A. H., Debbab A., Proksch P. (2011). Fungal endophytes: unique plant inhabitants with great promises. Appl. Microbiol. Biotechnol. 90, 1829–1845. 10.1007/s00253-011-3270-y [DOI] [PubMed] [Google Scholar]
- Amann R. I., Ludwig W., Schleifer K. H. (1995). Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Mol. Biol. Rev. 59, 143–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin N. (2016). Endophytic fungi to control of cocoa pod borer (Conopomorpha cramerella) on Cocoa plantation. Res. J. Pharm. Biol. Chem. Sci. 6:1496–1501 [Google Scholar]
- Anitha A., Rabeeth M. (2010). Degradation of fungal cell walls of phytopathogenic fungi by lytic enzyme of Streptomyces griseus. Afr. J. Plant Sci. 4, 061–066. [Google Scholar]
- Bayman P., Lebron L. L., Tremblay R. L., Lodge D. J. (1997). Variation in endophytic fungi from roots and leaves of Lepanthes (Orchidaceae). New Phytol. 135, 143–149. 10.1046/j.1469-8137.1997.00618.x [DOI] [PubMed] [Google Scholar]
- Bernardi-Wenzel J., Garcia A., Rubim Filho C. J., Prioli A. J., Pamphile J. A. (2010). Evaluation of foliar fungal endophyte diversity and colonization of medicinal plant Luehea divaricata (Martius et Zuccarini). Bio. Res. 43, 375–384. 10.4067/S0716-97602010000400001 [DOI] [PubMed] [Google Scholar]
- Brady S. F., Clardy J. (2000). CR377, a new pentaketide antifungal agent isolated from an endophytic fungus. J. Nat. Prod. 63, 1447–1448. 10.1021/np990568p [DOI] [PubMed] [Google Scholar]
- Cannon P. F., Simmons C. M. (2002). Diversity and host preference of leaf endophytic fungi in the Iwokrama Forest Reserve, Guyana. Mycologia 94, 210–220. 10.1080/15572536.2003.11833226 [DOI] [PubMed] [Google Scholar]
- Cazar M. E., Schmeda-Hirschmann G., Astudillo L. (2005). Antimicrobial butyrolactone I derivatives from the Ecuadorian soil fungus Aspergillus terreus Thorn. var terreus. World J. Microbiol. Biotechnol. 21, 1067–1075. 10.1007/s11274-004-8150-526811110 [DOI] [Google Scholar]
- Chaeprasert S., Piapukiew J., Whalley A. J., Sihanonth P. (2010). Endophytic fungi from mangrove plant species of Thailand: their antimicrobial and anticancer potentials. Bot. Mar. 53, 555–564. 10.1515/bot.2010.074 [DOI] [Google Scholar]
- Chen J., Zhang L., Xing Y., Wang Y., Xing X., Zhang D., et al. (2013). Diversity and taxonomy of endophytic xylariaceous fungi from medicinal plants of Dendrobium (Orchidaceae). PLoS ONE 8:58268. 10.1371/journal.pone.0058268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z., Pan J., Tang W., Chen Q., Lin Y. (2009). Biodiversity and biotechnological potential of mangrove-associated fungi. J. For. Res. 20, 63–71. 10.1007/s11676-009-0012-426811110 [DOI] [Google Scholar]
- Coşkuntuna A., Özer N. (2008). Biological control of onion basal rot disease using Trichoderma harzianum and induction of antifungal compounds in onion set following seed treatment. Crop Protect. 27, 330–336. 10.1016/j.cropro.2007.06.002 [DOI] [Google Scholar]
- Crozier J., Thomas S. E., Aime M. C., Evans H. C., Holmes K. A. (2006). Molecular characterization of fungal endophytic morphospecies isolated from stems and pods of Theobroma cacao. Plant Pathol. 55, 783–791. 10.1111/j.1365-3059.2006.01446.x24843434 [DOI] [Google Scholar]
- Cui J., Guo T., Ren Z., Zhang N., Wang M. (2015). Diversity and antioxidant activity of culturable endophytic fungi from alpine plants of Rhodiola crenulata, R. angusta, and R. sachalinensis. PLoS ONE 10:0118204. 10.1371/journal.pone.0118204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Costa Souza P. N., Grigoletto T. L., de Moraes L. A., Abreu L. M., Guimarães L. H., Santos C., et al. (2016). Production and chemical characterization of pigments in filamentous fungi. Microbiology 162, 12–22. 10.1099/mic.0.000168 [DOI] [PubMed] [Google Scholar]
- Daouk R. K., Dagher S. M., Sattout E. J. (1995). Antifungal activity of the essential oil of Origanum syriacum L. J. Food Prot. 58, 1147–1149. [DOI] [PubMed] [Google Scholar]
- Debbab A., Aly A. H., Proksch P. (2011). Bioactive secondary metabolites from endophytes and associated marine derived fungi. Fungal Divers. 49, 1–12. 10.1007/s13225-011-0114-026811110 [DOI] [Google Scholar]
- de Souza Sebastianes F. L., Romao-Dumaresq A. S., Lacava P. T., Harakava R., Azevedo J. L., de Melo I. S., et al. (2013). Species diversity of culturable endophytic fungi from Brazilian mangrove forests. Curr. Genet. 59, 153–166. 10.1007/s00294-013-0396-8 [DOI] [PubMed] [Google Scholar]
- Dharmaputra O. S. (2003). Control of aflatoxigenic Aspergillus flavus in Peanuts Using Nonaflatoxigenic A. flavus, A. niger and Trichoderma harzianum. Biotropia 21, 32–44. 10.11598/BTB.2003.0.21.187 [DOI] [Google Scholar]
- Douanla-Meli C., Langer E., Mouafo F. T. (2013). Fungal endophyte diversity and community patterns in healthy and yellowing leaves of Citrus limon. Fungal Ecol. 6, 212–222. 10.1016/j.funeco.2013.01.004 [DOI] [Google Scholar]
- Dubey S. C., Suresh M. (2006). Randomly amplified polymorphic DNA markers for Trichoderma species and antagonism against Fusarium oxysporum f. sp. ciceris causing chickpea wilt. J. Phytopathol. 154, 663–669. 10.1111/j.1439-0434.2006.01167.x24843434 [DOI] [Google Scholar]
- Dufossé L., Fouillaud M., Caro Y., Mapari S. A., Sutthiwong N. (2014). Filamentous fungi are large-scale producers of pigments and colorants for the food industry. Curr. Opin. Biotechnol. 26, 56–61. 10.1016/j.copbio.2013.09.007 [DOI] [PubMed] [Google Scholar]
- Duke N. C. (2006). Australia's Mangroves: the Authoritative Guide to Australia's Mangrove Plants. Brisbane: University of Queensland. [Google Scholar]
- Duke N. C., Meynecke J. O., Dittmann S., Ellison A. M., Anger K., Berger U., et al. (2007). A world without mangroves? Science 317, 41–42. 10.1126/science.317.5834.41b [DOI] [PubMed] [Google Scholar]
- Faizal A., Esyanti R. R., Aulianisa E. N., Santoso E., Turjaman M. (2017). Formation of agarwood from Aquilaria malaccensis in response to inoculation of local strains of Fusarium solani. Trees 31, 189–197. 10.1007/s00468-016-1471-926811110 [DOI] [Google Scholar]
- Firáková S., Proksa B., Šturdíková M. (2007). Biosynthesis and biological activity of enniatins. Pharmazie 62, 563–568. 10.1691/ph.2007.8.7600 [DOI] [PubMed] [Google Scholar]
- Fox E. M., Howlett B. J. (2008). Biosynthetic gene clusters for epipolythiodioxopiperazines in filamentous fungi. Mycol. Res. 112, 162–169. 10.1016/j.mycres.2007.08.017 [DOI] [PubMed] [Google Scholar]
- Gaffar M. U., Morshed M. A., Uddin A., Roy S., Hannan J. M. A. (2011). Study the efficacy of Rhizophora mucornata Poir. leaves for diabetes therapy in Long Evans rats. Int. J. Biomol. Biomed. 1, 20–26. [Google Scholar]
- Geweely N. S. (2011). Investigation of the optimum condition and antimicrobial activities of pigments from four potent pigment-producing fungal species. J. Life Sci. 5, 697–711. 10.4172/2153-2435.10000S4 [DOI] [Google Scholar]
- Hajieghrari B., Torabi-Giglou M., Mohammadi M. R., Davari M. (2008). Biological potantial of some Iranian Trichoderma isolates in the control of soil borne plant pathogenic fungi. Afr. J. Biotechnol. 7, 967–972. [Google Scholar]
- Hamzah T. N. T. (2018). Assessment on the Diversity and Bioactive Compounds Present in Endophytic Fungi Isolated from Rhizophora mucronata in Matang Mangrove Forest Reserve, Perak. Master's thesis, Universiti Putra Malaysia, Serdang. [Google Scholar]
- Hartley S. E., Eschen R., Horwood J. M., Gange A. C., Hill E. M. (2015). Infection by a foliar endophyte elicits novel arabidopside-based reactions in its host, Cirsium arvense. New Phytol. 205, 816–827. 10.1111/nph.13067 [DOI] [PubMed] [Google Scholar]
- He X., Han G., Lin Y., Tian X., Xiang C., Tian Q., et al. (2012). Diversity and decomposition potential of endophytes in leaves of a Cinnamomum camphora plantation in China. Ecol. Res. 27, 273–284. 10.1007/s11284-011-0898-026811110 [DOI] [Google Scholar]
- Isaac S. (1994). Mycology answers: many fungi are brightly coloured; does pigmentation provide any advantage to those species? Mycologist 8, 178–179. [Google Scholar]
- Kai M., Effmert U., Berg G., Piechulla B. (2007). Volatiles of bacterial antagonists inhibit mycelial growth of the plant pathogen Rhizoctonia solani. Arch. Microbiol. 187, 351–360. 10.1007/s00203-006-0199-0 [DOI] [PubMed] [Google Scholar]
- Kamala T., Indira S. (2011). Evaluation of indigenous Trichoderma isolates from Manipur as biocontrol agent against Pythium aphanidermatum on common beans. 3 Biotech. 1, 217–225. 10.1007/s13205-011-0027-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khara H. S., Hadwan H. A. (1990). In vitro studies on antagonism of Trichoderma spp. against Rhizoctonia solani the causal agent of damping off of tomato. Plant Dis. Res. 5, 144–147. [Google Scholar]
- Kjer J., Debbab A., Aly A. H., Proksch P. (2010). Methods for isolation of marine-derived endophytic fungi and their bioactive secondary products. Nat. Prot. 5, 479–490. 10.1038/nprot.2009.233 [DOI] [PubMed] [Google Scholar]
- Klaiklay S., Rukachaisirikul V., Phongpaichit S., Pakawatchai C., Saithong S., Buatong J., et al. (2012). Anthraquinone derivatives from the mangrove-derived fungus Phomopsis sp. PSU-MA214. Phytochem. Lett. 5, 738–742. 10.1016/j.phytol.2012.08.003 [DOI] [Google Scholar]
- Kokpol U., Thebpatiphat S., Boonyaratavej S., Chedchuskulcai V., Ni C. Z., Clardy J., et al. (1990). Structure of trigonostemone, a new phenanthrenone from the Thai plant Trigonostemon reidioides. J. Nat. Prod. 53, 1148–1151. [Google Scholar]
- Koukol O., Kolarík M., Kolárová Z., Baldrian P. (2012). Diversity of foliar endophytes in wind-fallen Picea abies trees. Fungal Divers. 54, 69–77. 10.1007/s13225-011-0112-226811110 [DOI] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusari S., Pandey S. P., Spiteller M. (2013). Untapped mutualistic paradigms linking host plant and endophytic fungal production of similar bioactive secondary metabolites. Phytochem. 91, 81–87. 10.1016/j.phytochem.2012.07.021 [DOI] [PubMed] [Google Scholar]
- Landum M. C., do Rosário Félix M., Alho J., Garcia R., Cabrita M. J., Rei F., et al. (2016). Antagonistic activity of fungi of Olea europaea L. against Colletotrichum acutatum. Microbiol. Res. 183, 100–108. 10.1016/j.micres.2015.12.001 [DOI] [PubMed] [Google Scholar]
- Lee J. C., Lobkovsky E., Pliam N. B., Strobel G., Clardy J. (1995). Subglutinols A and B: immunosuppressive compounds from the endophytic fungus Fusarium subglutinans. J. Org. Chem. 60, 7076–7707. 10.1021/jo00127a001 [DOI] [Google Scholar]
- Li H., Qing C., Zhang Y., Zhao Z. (2005). Screening for endophytic fungi with antitumour and antifungal activities from Chinese medicinal plants. World J. Microbiol. Biotechnol. 21, 1515–1519. 10.1007/s11274-005-7381-4 [DOI] [Google Scholar]
- Li P., Lou J., Mou Y., Sun W., Shan T., Zhou L. (2012). Effects of oligosaccharide elicitors from endophyitc Fusarium oxysporum Dzf17 on diosgenin accumulation in Dioscorea zingiberensis seedling cultures. J. Med. Plants Res. 6, 5128–5134. 10.5897/JMPR12.120 [DOI] [Google Scholar]
- Liang H., Xing Y., Chen J., Zhang D., Guo S., Wang C. (2012). Antimicrobial activities of endophytic fungi isolated from Ophiopogon japonicus (Liliaceae). BMC Compl. Alt. Med. 12:238. 10.1186/1472-6882-12-238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Dong M., Chen X., Jiang M., Lv X., Yan G. (2007). Antioxidant activity and phenolics of an endophytic Xylaria sp. from Ginkgo biloba. Food Chem. 105, 548–554. 10.1016/j.foodchem.2007.04.008 [DOI] [Google Scholar]
- Liu X., Dong M., Chen X., Jiang M., Lv X., Zhou J. (2008). Antimicrobial activity of an endophytic Xylaria sp. YX-28 and identification of its antimicrobial compound 7-amino-4-methylcoumarin. Appl. Microbiol. Biotechnol. 78, 241–247. 10.1007/s00253-007-1305-1 [DOI] [PubMed] [Google Scholar]
- Maharachchikumbura S. S., Guo L. D., Cai L., Chukeatirote E., Wu W. P., Sun X., et al. (2012). A multi-locus backbone tree for Pestalotiopsis, with a polyphasic characterization of 14 new species. Fungal Divers. 56, 95–129. 10.1007/s13225-012-0198-126811110 [DOI] [Google Scholar]
- Mapari S. A., Thrane U., Meyer A. S. (2010). Fungal polyketide azaphilone pigments as future natural food colorants? Trends Biotechnol. 28, 300–307. 10.1016/j.tibtech.2010.03.004 [DOI] [PubMed] [Google Scholar]
- Oliveira A. L. L. D., Felício R. D., Debonsi H. M. (2012). Marine natural products: chemical and biological potential of seaweeds and their endophytic fungi. Braz. J. Pharm. 22, 906–920. 10.1590/S0102-695X2012005000083 [DOI] [Google Scholar]
- Onn M. L., Lim P. T., Mujahid A., Proksch P., Müller M. (2016). Initial screening of mangrove endophytic fungi for antimicrobial compounds and heavy metal biosorption potential. Sains Malaysiana, 45, 1063–1071. [Google Scholar]
- Park J. H., Choi G. J., Lee H. B., Kim K. M., Jung H. S., Lee S. W., et al. (2005). Griseofulvin from Xylaria sp. strain F0010, an endophytic fungus of Abies holophylla and its antifungal activity against plant pathogenic fungi. J. Microbiol. Biotechnol. 15, 112–117. [Google Scholar]
- Phongpaichit S., Rungjindamai N., Rukachaisirikul V., Sakayaroj J. (2006). Antimicrobial activity in cultures of endophytic fungi isolated from Garcinia species. FEMS Immun. Med. Microbiol. 48, 367–372. 10.1111/j.1574-695X.2006.00155.x [DOI] [PubMed] [Google Scholar]
- The Plant List (2013). Version 1.1. Published on the Internet. Available Online at: http://www.theplantlist.org/
- Polidoro B. A., Carpenter K. E., Collins L., Duke N. C., Ellison A. M., Ellison J. C., et al. (2010). The loss of species: mangrove extinction risk and geographic areas of global concern. PLoS ONE 5:e10095. 10.1371/journal.pone.0010095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potshangbam M., Devi S. I., Sahoo D., Strobel G. A. (2017). Functional characterization of endophytic fungal community associated with Oryza sativa L. and Zea mays L. Front. Microbiol. 8:325. 10.3389/fmicb.2017.00325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premalatha B., Pradeep F. S., Pradeep B. V., Palaniswamy M. (2012). Production and characterization of naphthoquinone pigment from Fusarium moniliforme MTCC6985. World. J. Pharm. Res. 1, 1126–1142. [Google Scholar]
- Prihanto A. A., Firdaus M., Nurdiani R. (2011). Endophytic fungi isolated from mangrove (Rhyzopora mucronata) and its antibacterial activity on Staphylococcus aureus and Escherichia coli. J. Food Sci. Eng. 1, 386–389. [Google Scholar]
- Qin H., Boufford D. E. (2007). Rhizophoraceae, in Flora of China, Vol. 13, eds Wu Z., Raven P. H., Hong D. (Beijing; St. Louis, MO: Science Press; Missouri Botanical Garden Press; ), 295–299. [Google Scholar]
- Ramasamy K., Lim S. M., Bakar H. A., Ismail N., Ismail M. S., Ali M. F., et al. (2010). Antimicrobial and cytotoxic activities of Malaysian endophytes. Phytoth. Res. 24, 640–643. 10.1002/ptr.2891 [DOI] [PubMed] [Google Scholar]
- Ratnaweera P. B., de Silva E. D., Williams D. E., Andersen R. J. (2015). Antimicrobial activities of endophytic fungi obtained from the arid zone invasive plant Opuntia dillenii and the isolation of equisetin, from endophytic Fusarium sp. BMC Compl. Alt. Med. 15:220. 10.1186/s12906-015-0722-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Reis Celestino J., de Carvalho L. E., da Paz Lima M., Lima A. M., Ogusku M. M., de Souza J. V. B. (2014). Bioprospecting of Amazon soil fungi with the potential for pigment production. Proc. Biochem. 49, 569–575. 10.1016/j.procbio.2014.01.018 [DOI] [Google Scholar]
- Rhoden S. A., Garcia A., Bongiorno V. A., Azevedo J. L., Pamphile J. A. (2012). Antimicrobial activity of crude extracts of endophytic fungi isolated from medicinal plant Trichilia elegans A. Juss. J. App. Pharm. Sci. 02, 57–59. 10.7324/JAPS.2012.2807 [DOI] [Google Scholar]
- Robinson J. G., Nedergaard B. S., Rogers W. J., Fialkow J., Neutel J. M., Ramstad D., et al. (2014). Effect of evolocumab or ezetimibe added to moderate-or high-intensity statin therapy on LDL-C lowering in patients with hypercholesterolemia: the LAPLACE-2 randomized clinical trial. JAMA. 311, 1870–1883. 10.1001/jama.2014.4030 [DOI] [PubMed] [Google Scholar]
- Rodriguez R. J., Henson J., Van V., Hoy M., Wright L., Beckwith F., et al. (2008). Stress tolerance in plants via habitat-adapted symbiosis. Int. Soc. Microbiol. Ecol. J. 2, 404–416. 10.1038/ismej.2007.106 [DOI] [PubMed] [Google Scholar]
- Score A. J., Palfreyman J. W., White N. A. (1997). Extracellular phenoloxidase and peroxidase enzyme production during interspecific fungal interactions. Int. Biodeter. Biodegrad. 39, 225–233. [Google Scholar]
- Setyawan A. D., Ulumuddin Y. I. (2012). Species diversity of Rhizophora in Tambelan Islands, Natuna Sea, Indonesia. Biodiversitas 13, 172–177. 10.13057/biodiv/d130402 [DOI] [Google Scholar]
- Sharon E., Bar-Eyal M., Chet I., Herrera-Estrella A., Kleifeld O., Spiegel Y. (2001). Biological control of the root-knot nematode Meloidogyne javanica by Trichoderma harzianum. Phytopathol. 91, 687–693. 10.1016/j.soilbio.2008.03.011 [DOI] [PubMed] [Google Scholar]
- Shreelalitha S. J., Sridhar K. R. (2015). Endophytic fungi of wild legume Sesbania bispinosa in coastal sand dunes and mangroves of the Southwest coast of India. J. For. Res. 26, 1003–1011. 10.1007/s11676-015-0103-326811110 [DOI] [Google Scholar]
- Siameto E. N., Okoth S., Amugune N. O., Chege N. C. (2010). Antagonism of Trichoderma farzianum isolates on soil borne plant pathogenic fungi from Embu District, Kenya. J. Yeast Fungal Res. 1, 47–54. [Google Scholar]
- Singh S. K., Srivastava P. K., Gupta M., Thakur J. K., Mukherjee S. (2014). Appraisal of land use/land cover of mangrove forest ecosystem using support vector machine. Env. Earth Sci. 71, 2245–2255. 10.1007/s12665-013-2628-026811110 [DOI] [Google Scholar]
- Suganthy N., Pandima Devi K. (2016). In vitro antioxidant and anti-cholinesterase activities of Rhizophora mucronata. Pharm. Biol. 54, 118–129. 10.3109/13880209.2015.1017886 [DOI] [PubMed] [Google Scholar]
- Suryanarayanan T. S., Murali T. S., Venkatesan G. (2002). Occurrence and distribution of fungal endophytes in tropical forests across a rainfall gradient. Can. J. Bot. 80, 818–826. 10.1139/b02-069 [DOI] [Google Scholar]
- Teixeira M. F., Martins M. S., Da Silva J. C., Kirsch L. S., Fernandes O. C., Carneiro A. L., et al. (2012). Amazonian biodiversity: pigments from Aspergillus and Penicillium-characterizations, antibacterial activities and their toxicities. Curr. Trends in Biotech. Pharm. 6, 300–311. [Google Scholar]
- Thomas S. A. L., Fleming R., Shaw L. N., Baker B. J. (2016). Isolation of bioactive secondary metabolites from mangrove fungal endophytes using epigenetic regulation. Planta Med. 82, S1–S381. 10.1055/s-0036-159673726606158 [DOI] [Google Scholar]
- Tomlinson P. B. (1986). The Botany of Mangroves. Cambridge Tropical Biology Series. New York, NY: Cambridge University Press. [Google Scholar]
- Tudor D., Robinson S. C., Cooper P. A. (2013). The influence of pH on pigment formation by lignicolous fungi. Int. Biodet. Biodegrad. 80, 22–28. 10.1016/j.ibiod.2012.09.013 [DOI] [Google Scholar]
- Udayanga D., Liu X., McKenzie E. H., Chukeatirote E., Bahkali A. H., Hyde K. D. (2011). The genus Phomopsis: biology, applications, species concepts and names of common phytopathogens. Fungal Divers. 50, 189–225. 10.1007/s13225-011-0126-926811110 [DOI] [Google Scholar]
- Umechuruba C. I. (2005). Health impact assessment of mangrove vegetation in an oil spilled site at the Bodo West field in Rivers State, Nigeria. J. Appl. Sci. Environ. Manag. 9, 69–73. [Google Scholar]
- Vasundhara M., Baranwal M., Kumar A. (2016). Fusarium tricinctum, an endophytic fungus exhibits cell growth inhibition and antioxidant activity. Ind. J. Microbiol. 56, 433–438. 10.1007/s12088-016-0600-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Li S., Zhao F., Dai H., Bao L., Ding R., et al. (2011). Chemical constituents from endophytic fungus Fusarium oxysporum. Fitoterapia 85, 777–781. 10.1016/j.fitote.2011.04.002 [DOI] [PubMed] [Google Scholar]
- Wheeler K. A., Hocking A. D. (1993). Interactions among xerophilic fungi associated with dried salted fish. J. Appl. Microbiol. 74, 164–169. [DOI] [PubMed] [Google Scholar]
- White T. J., Bruns T., Lee S. J. W. T., Taylor J. L. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: a guide to methods and applications 18, 315–322. 10.1016/B978-0-12-372180-8.50042-1 [DOI] [Google Scholar]
- Zhao J., Fu Y., Luo M., Zu Y., Wang W., Zhao C., et al. (2012a). Endophytic fungi from pigeon pea [Cajanus cajan (L.) Millsp.] produce antioxidant cajaninstilbene acid. J. Agr. Food Chem. 60, 4314–4319. 10.1021/jf205097y [DOI] [PubMed] [Google Scholar]
- Zhao Y., Wei T., Yin K., Chen Z., Gu H., Qu L., et al. (2012b). Arabidopsis RAP2. 2 plays an important role in plant resistance to Botrytis cinerea and ethylene responses. New Phytologist 195, 450–460. 10.1111/j.1469-8137.2012.04160.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The dataset generated in this study has been deposited in the GenBank, accession codes MH397067 to MH397144.