Abstract
Background
This meta‐analysis was conducted to investigate the diagnostic performance of P16 INK4a gene promoter methylation as a biomarker of non‐small cell lung cancer (NSCLC).
Methods
Two reviewers independently searched the Web of Science, PubMed, Cochrane, Embase, China National Knowledge Infrastructure, and Chinese Biomedical Literature databases. Publications relevant to P16 INK4a gene promoter methylation in serum or bronchoalveolar fluid/sputum were screened and included in this meta‐analysis. Pooled diagnostic sensitivity, specificity, and symmetric receiver operating characteristic curve were calculated.
Results
Twenty‐six publications with 1768 lung cancer cases and 1323 controls were included. The pooled sensitivity, specificity, positive and negative likelihood ratios, and diagnostic odds ratio were 0.46 (95% confidence interval [CI] 0.43–0.48), 0.90 (95% CI 0.88–0.91), 6.33 (95% CI 3.89–10.30), 0.57 (95% CI 0.50–0.65) and 10.72 (95% CI 6.94–16.56), respectively, for P16 INK4a gene promoter methylation as a biomarker for the diagnosis of NSCLC. The area under the symmetric receiver operating characteristic curve was 0.75 with a standard error of 0.004. No publication bias was detected via line regression test (t = 0.95; P = 0.35) and Begg's funnel plot.
Conclusion
P16 INK4a gene promoter methylation detection in serum or bronchoalveolar fluid/sputum may be a potential biomarker for NSCLC diagnosis; however, the sensitivity was relatively low, which is not suitable for NSCLC screening.
Keywords: Aberrant methylation, bronchoalveolar fluid, meta‐analysis, P16INK4a gene, serum
Introduction
Non‐small cell lung cancer (NSCLC) is one of the most common clinically diagnosed malignant carcinomas. It is estimated that 234 030 new cases and 154 050 deaths from NSCLC will occur in the United States in 2018.1 The general prognosis of NSCLC is poor, particularly in advanced‐stage patients, with an extremely low five‐year survival rate. One of the major reasons for this poor prognosis is the lack of effective lung cancer screening or early diagnostic methods.2 Several studies have evaluated lung cancer screening methods such as X‐ray,3, 4 sputum cytology, and chest computed tomography (CT);5 however, such methods yield low sensitivity or specificity and thus are not adequate to diagnose NSCLC at an early stage.
Promoter methylation of tumor suppressor genes is frequently detected in cancer tissue and body fluid in malignant carcinomas such as lung,6, 7 colorectal,8 and esophageal cancers. Previous studies have reported that methylation of the P16 INK4a gene promoter is common in lung cancer. The methylation frequency of P16 INK4a in serum or bronchoalveolar fluid (BAF)/sputum in lung cancer patients has been widely discussed; however, the exact diagnostic performance of P16 INK4a as a biomarker for NSCLC remains inconclusive. Therefore, we conducted this updated meta‐analysis to further evaluate the diagnostic performance of P16 INK4a as a biomarker for NSCLC.
Methods
Electronic database search strategy
Two reviewers independently searched the Web of Science, PubMed, Cochrane, Embase, China National Knowledge Infrastructure, and Chinese Biomedical Literature databases for studies relevant to P16 INK4a gene promoter methylation in serum or BAF/sputum. The following keywords were used: non‐small cell lung cancer; non‐small cell carcinoma, NSCLC, P16, P16 INK4a; cyclin‐dependent kinase inhibitor 2A, CDKN2A; CDK4 inhibitor; multiple tumor suppressor 1; TP16; methylation; and hypermethylation. Relevant studies were identified and duplicated publications or data were excluded. The title and abstract were then reviewed to locate relevant studies. All potentially suitable studies were reviewed in full‐text and all references of included publications were further screened to identify additional relevant publications. The publication search process is demonstrated in Figure 1.
Figure 1.
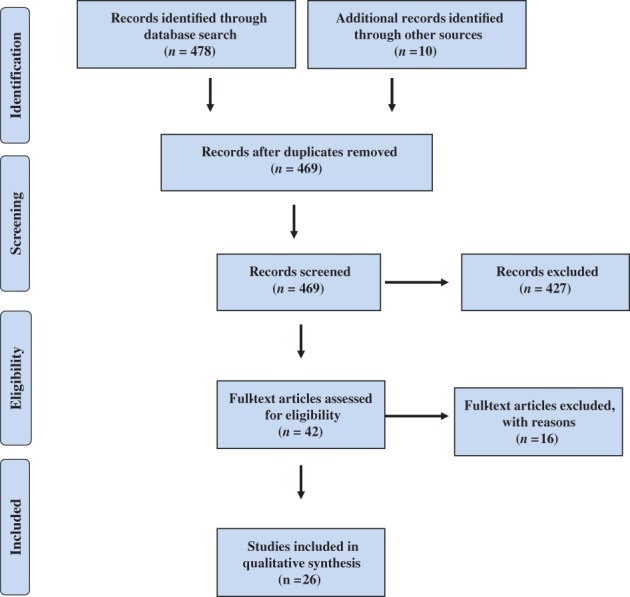
Publication search process.
Inclusion and exclusion criteria
The identified studies were further reviewed to assess whether the inclusion criteria were fulfilled: (i) diagnostic studies relevant to P16 INK4a promoter methylation and NSCLC; (ii) NSCLC diagnosis confirmed by pathology or cytology; (iii) P16 INK4a gene promoter methylation was detected by methylation‐specific PCR (MSP), real‐time MSP (RT‐MSP), or quantitative MSP (q‐MSP); (iv) P16 INK4a gene methylation status in serum or BAF/sputum in NSCLC and control subjects was available for each included study. The exclusion criteria were: (i) case reports or literature reviews; (ii) P16 INK4a gene methylation status detected in other specimens, not in serum or BAF/sputum; (iii) studies published in languages other than English or Chinese; and (iv) insufficient data to calculate sensitivity and specificity.
Statistical analysis
The diagnostic sensitivity, specificity, and symmetric receiver operating characteristic (SROC) curve were pooled by fixed or random effects method according to the statistical heterogeneity across the included studies. Diagnostic sensitivity and specificity were calculated using the following equations: sensitivity = true positive/(true positive + false negative); specificity = true negative/(true negative + false positive). Publication bias was evaluated by Egger's line regression test and Begger's funnel plot. P < 0.05 was considered to indicate significant statistical difference.
Results
Study characteristics
Initially, 488 relevant publications were identified. After reviewing the title, abstract, and full text, 26 studies relevant to P16 INK4a gene promoter methylation as a biomarker for the diagnosis of NSCLC were included for quantitative analysis.9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34 Sixteen publications evaluated P16 INK4a gene promoter methylation in serum and 10 in BAF/sputum. The general characteristics of the 26 studies are shown in Table 1.
Table 1.
Study characteristics
| Distribution | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Study | Year | Area | NSCLC | Control | Tp | Fp | Fn | Tn | Specimen |
| Kersting et al.9 | 2000 | US | 31 | 25 | 18 | 7 | 13 | 18 | Serum |
| Bearzatto et al.10 | 2002 | Italy | 30 | 15 | 12 | 0 | 18 | 15 | Serum |
| Wu et al.11 | 2002 | China | 14 | 26 | 4 | 0 | 10 | 26 | Serum |
| Cai et al.12 | 2003 | China | 49 | 55 | 15 | 1 | 34 | 54 | Serum |
| Kim et al.13 | 2004 | Korea | 85 | 127 | 14 | 8 | 71 | 119 | Serum |
| Fujiwara et al.14 | 2005 | US | 111 | 80 | 14 | 3 | 97 | 77 | Serum |
| Kong et al.15 | 2007 | China | 64 | 46 | 19 | 0 | 45 | 46 | Serum |
| Hsu et al.16 | 2007 | 51 | 33 | 21 | 3 | 30 | 30 | Serum | |
| Zhang et al.17 | 2008 | China | 95 | 22 | 52 | 2 | 43 | 20 | Serum |
| Ma et al.18 | 2009 | China | 62 | 19 | 32 | 0 | 30 | 19 | Serum |
| Hu et al.19 | 2009 | China | 46 | 21 | 22 | 1 | 24 | 20 | Serum |
| Chen et al.20 | 2010 | China | 159 | 81 | 39 | 0 | 120 | 81 | Serum |
| Wang et al.21 | 2016 | China | 50 | 50 | 22 | 0 | 28 | 50 | Serum |
| Wan et al.22 | 2017 | China | 98 | 60 | 69 | 9 | 29 | 51 | Serum |
| Liu et al.23 | 2017 | China | 120 | 46 | 45 | 3 | 75 | 43 | Serum |
| Destro et al.24 | 2004 | Italy | 24 | 100 | 16 | 4 | 8 | 96 | BAF/sputum |
| Konno et al.25 | 2004 | Japan | 78 | 94 | 44 | 20 | 34 | 74 | BAF/sputum |
| Wang et al.26 | 2004 | China | 34 | 21 | 11 | 0 | 23 | 21 | BAF/sputum |
| Georgiou et al.27 | 2007 | Greece | 80 | 40 | 55 | 9 | 25 | 31 | BAF/sputum |
| Liu et al.28 | 2008 | China | 58 | 107 | 41 | 55 | 17 | 52 | BAF/sputum |
| Zhang et al.29 | 2004 | China | 44 | 20 | 27 | 3 | 17 | 17 | BAF/sputum |
| Guo et al.30 | 2008 | China | 100 | 50 | 61 | 0 | 39 | 50 | BAF/sputum |
| Hu et al.31 | 2009 | China | 42 | 25 | 20 | 0 | 22 | 25 | BAF/sputum |
| Peng et al.32 | 2010 | China | 82 | 25 | 60 | 0 | 22 | 25 | BAF/sputum |
| Zhang et al.33 | 2012 | China | 41 | 15 | 21 | 2 | 20 | 13 | BAF/sputum |
| Sun et al.34 | 2012 | China | 120 | 120 | 56 | 6 | 64 | 114 | BAF/sputum |
BAF, bronchoalveolar fluid; fn, false negative; fp, false positive; NSCLC, non‐small cell lung cancer; tn, true negative; tp, true positive; US, United States.
Pooled sensitivity and specificity
Because of significant statistical heterogeneity, the diagnostic sensitivity and specificity were pooled using the random effects method. The pooled sensitivity and specificity were 0.46 (95% confidence interval [CI] 0.43–0.48) (Fig 2) and 0.90 (95% CI 0.88–0.91) (Fig 3), respectively, for P16 INK4a gene promoter methylation as a biomarker for the diagnosis of NSCLC.
Figure 2.
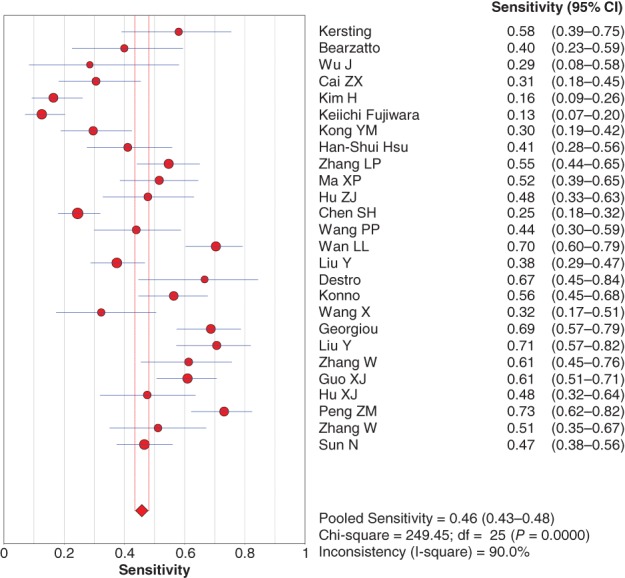
Forest plot if the sensitivity of P16 INK4a gene promoter methylation as a biomarker for the diagnosis of non‐small cell lung cancer. CI, confidence interval.
Figure 3.
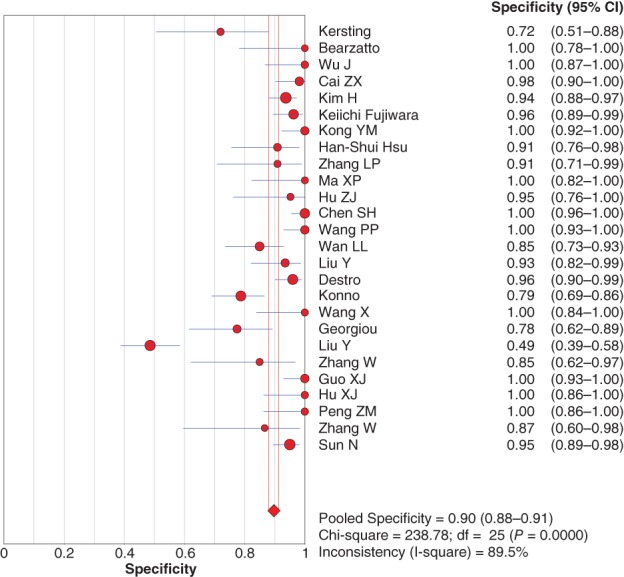
Forest plot for specificity of P16 INK4a gene promoter methylation as a biomarker for the diagnosis of non‐small cell lung cancer. CI, confidence interval.
Pooled positive and negative likelihood ratios
The diagnostic positive likelihood ratio (+LR) and negative likelihood ratio (−LR) were also pooled by random effect method because of significant heterogeneity. The pooled +LR and −LR were 6.33 (95% CI 3.89–10.30) (Fig 4) and 0.57 (95% CI 0.50–0.65) (Fig 5), respectively, for P16 INK4a gene promoter methylation as a biomarker for the diagnosis of NSCLC.
Figure 4.
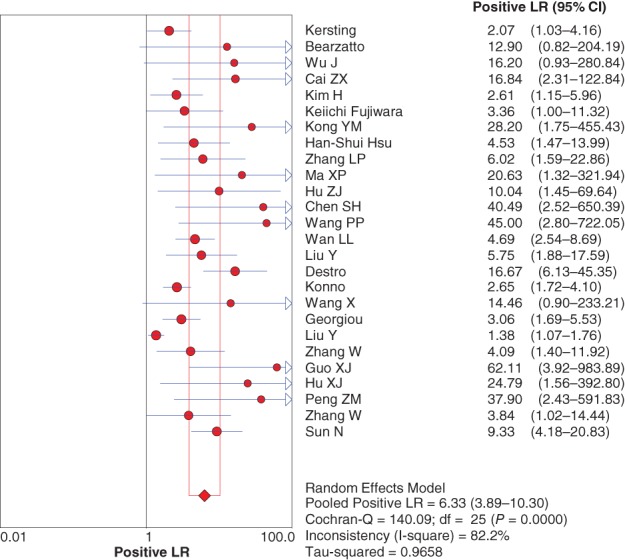
Forest plot of the negative likelihood ratio (LR). CI, confidence interval.
Figure 5.
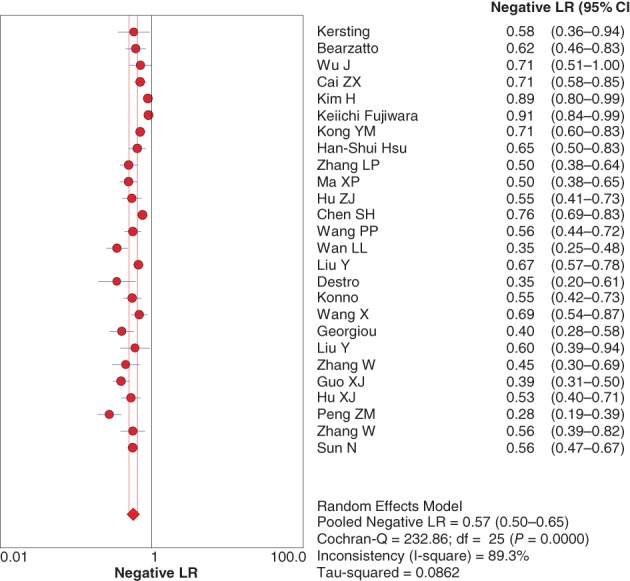
Forest plot of the positive likelihood ratio (LR). CI, confidence interval.
Pooled diagnostic odds ratio
The pooled diagnostic odds ratio (DOR) was 10.72 (95% CI 6.94–16.56) for P16 INK4a gene promoter methylation as a biomarker for the diagnosis of NSCLC (Fig 6).
Figure 6.
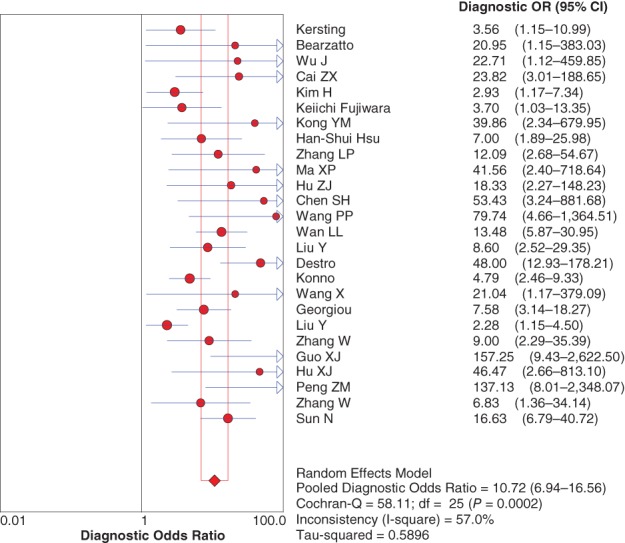
Forest plot of the diagnostic odds ratio (OR).
Symmetric receiver operating characteristic curve
The area under the SROC curve was 0.75 with a standard error of 0.004 for P16 INK4a gene promoter methylation as a biomarker for the diagnosis of lung cancer (Fig 7).
Figure 7.
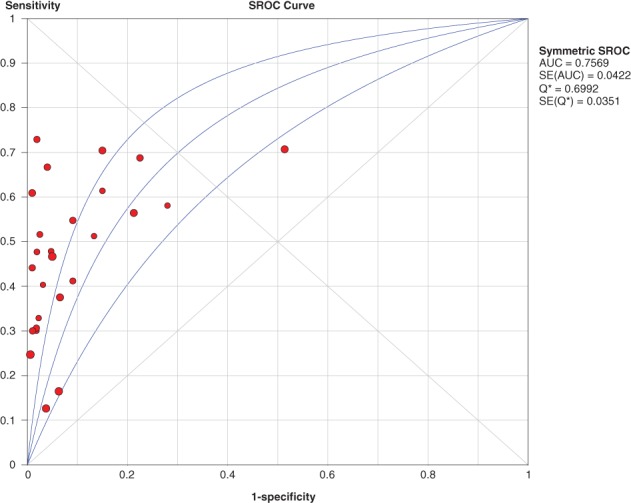
The pooled symmetric receiver operating characteristic (SROC) curve for P16 gene promoter methylation for the diagnosis of non‐small cell lung cancer. AUC, area under the curve; SE, standard error.
Subgroup analysis
We also conducted subgroup analysis, detecting P16 INK4a gene promoter methylation in serum or BAF/sputum. The pooled diagnostic performances in serum and BAF/sputum are shown in Table 2.
Table 2.
Diagnostic performance in subgroup analysis
| Serum | BAF/sputum | |||||
|---|---|---|---|---|---|---|
| Diagnostic index | Point estimate | 95% CI | I2 | Point estimate | 95% CI | I2 |
| Sensitivity | 0.37 | 0.34–0.40 | 89.8% | 0.59 | 0.55–0.62 | 71.7% |
| Specificity | 0.95 | 0.93–0.96 | 72.5% | 0.84 | 0.81–0.87 | 93.1% |
| +LR | 5.46 | 3.43–8.69 | 39.6% | 6.15 | 2.81–13.46 | 89.6% |
| −LR | 0.64 | 0.57–0.73 | 86.9% | 0.49 | 0.51–0.57 | 64.2% |
| DOR | 9.41 | 5.67–15.62 | 32.5% | 12.32 | 5.86–25.94 | 73.0% |
| AUC | 0.78 | 0.74–0.82 | — | 0.71 | 0.69–0.77 | — |
+LR, positive likelihood ratio; ‐LR, negative likelihood ratio; AUC, area under the curve; BAF, bronchoalveolar fluid; CI, confidence interval; DOR, diagnostic odds ratio.
Evaluation of publication bias
Publication bias was evaluated by Egger's line regression test and Begg's funnel plot. No publication bias was detected by line regression test (t = 0.95; P = 0.35) or Begg's funnel plot (Fig 8).
Figure 8.
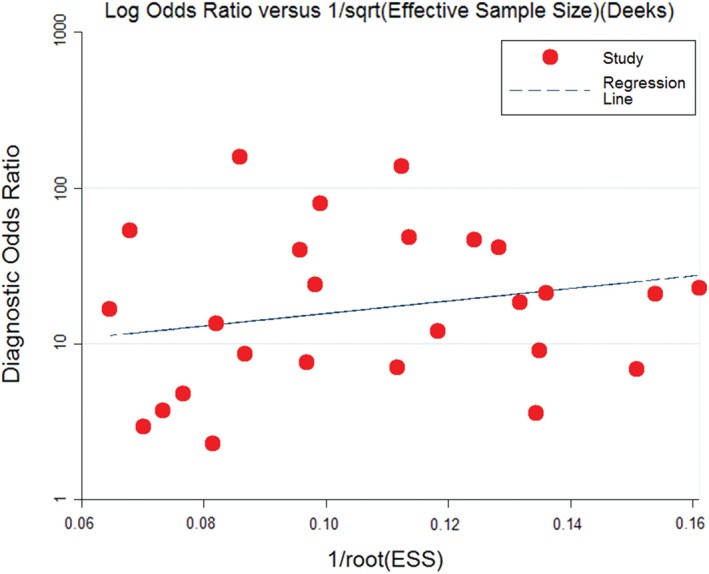
A funnel plot of publication evaluation. ESS, effective sample size. ( ) Study and (
) Study and ( ) Regression lines.
) Regression lines.
Discussion
In China, lung cancer is the most commonly diagnosed malignant carcinoma and the leading cause of cancer mortality in both men and women, particularly in men aged ≥ 75 years. As most NSCLC patients are only diagnosed at locally advanced‐stage or after remote metastasis has occurred, they are ineligible for surgery. Prognosis is poor, with an extremely low five‐year survival rate, because of the lack of effective methods for lung cancer screening or early diagnosis.
Promoter methylation of tumor suppressor genes is common in body fluid and can be used as a lung cancer diagnosis method or biomarker. Several studies have evaluated its clinical application with acceptable diagnostic performance and high specificity.35, 36, 37, 38
The P16 INK4a gene, also known as the CDKN2A gene, is located on chromosome 9 (9p21.3) and plays an important role in regulating the cell cycle.39 The promoter region of P16 ink4a is usually hypermethylated in cancer cells of NSCLC patients. Studies have found that P16 INK4a methylation can be detected in body fluid, such as serum and sputum,23, 24 indicating that detection of P16 INK4a methylation status may be used as an important tool for lung cancer diagnosis, screening, or the monitoring of recurrence.
Two previous meta‐analyses evaluated P16 INK4a methylation in serum and sputum as a biomarker for lung cancer diagnosis and concluded that detection of P16 INK4a promoter methylation via these methods was a useful tool for lung cancer diagnosis.40, 41 However, several recently published relevant studies were not included in these meta‐analyses. Therefore, we performed an updated meta‐analysis, including recently published relevant publications and further evaluated the clinical value of P16 INK4a methylation as a biomarker for NSCLC diagnosis. We confirmed that P16 INK4a gene promoter methylation detection in serum or BAF/sputum may be a potential biomarker for NSCLC diagnosis; however, the sensitivity was relatively low and was thus not suitable for NSCLC screening.
Although our results indicate that P16 INK4a gene promoter methylation represents a promising method for NSCLC diagnosis, there was significant statistical heterogeneity in the process of data merging, which inevitably affected our results. Furthermore, the sample sizes of the included studies were relatively small, which can reduce the statistical power of each included study. Large‐scale prospective diagnostic tests should be conducted by multiple health centers to further evaluate the clinical application value of P16 INK4a gene promoter methylation as an NSCLC diagnostic method.
Disclosure
No authors report any conflict of interest.
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018; 68: 7–30. [DOI] [PubMed] [Google Scholar]
- 2. Woodard GA, Jones KD, Jablons DM. Lung cancer staging and prognosis. Cancer Treat Res 2016; 170: 47–75. [DOI] [PubMed] [Google Scholar]
- 3. Pertile P, Poli A, Dominioni L et al Is chest X‐ray screening for lung cancer in smokers cost‐effective? Evidence from a population‐based study in Italy. Cost Eff Resour Alloc 2015; 13: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Strauss GM, Dominioni L. Chest X‐ray screening for lung cancer: Overdiagnosis, endpoints, and randomized population trials. J Surg Oncol 2013; 108: 294–300. [DOI] [PubMed] [Google Scholar]
- 5. Hoffman RM, Sanchez R. Lung cancer screening. Med Clin North Am 2017; 101: 769–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hua F, Fang N, Li X, Zhu S, Zhang W, Gu J. A meta‐analysis of the relationship between RARβ gene promoter methylation and non‐small cell lung cancer. PLoS One 2014; 9: e96163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gu J, Wen Y, Zhu S et al Association between P(16INK4a) promoter methylation and non‐small cell lung cancer: A meta‐analysis. PLoS One 2013; 8: e60107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hu H, Li B, Zhou C et al Diagnostic value of WIF1 methylation for colorectal cancer: A meta‐analysis. Oncotarget 2018; 9: 5378–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kersting M, Friedl C, Kraus A, Behn M, Pankow W, Schuermann M. Differential frequencies of p16(INK4a) promoter hypermethylation, p53 mutation, and K‐ras mutation in exfoliative material mark the development of lung cancer in symptomatic chronic smokers. J Clin Oncol 2000; 18: 3221–9. [DOI] [PubMed] [Google Scholar]
- 10. Bearzatto A, Conte D, Frattini M et al p16(INK4A) Hypermethylation detected by fluorescent methylation‐specific PCR in plasmas from non‐small cell lung cancer. Clin Cancer Res 2002; 8: 3782–7. [PubMed] [Google Scholar]
- 11. Wu Jun LB, He Jianqiu ZH, Zhigang W. [Promoter methylation of P16 AND DAP gen in serum of NSCLC.] Chin J Lung Cancer 2002; 5: 188–90 (In Chinese.) [DOI] [PubMed] [Google Scholar]
- 12. Cai ZX, Liang QZ, Liao SX, Zhang XX, Wang X. [Methylation of p16 gene promoter in serum of lung cancer]. China Cancer 2003; 12: 42–4 (In Chinese.) [Google Scholar]
- 13. Kim H, Kwon YM, Kim JS et al Tumor‐specific methylation in bronchial lavage for the early detection of non‐small‐cell lung cancer. J Clin Oncol 2004; 22: 2363–70. [DOI] [PubMed] [Google Scholar]
- 14. Fujiwara K, Fujimoto N, Tabata M et al Identification of epigenetic aberrant promoter methylation in serum DNA is useful for early detection of lung cancer. Clin Cancer Res 2005; 11: 1219–25. [PubMed] [Google Scholar]
- 15. Kong JY, Jin YT, Xue SL et al [Detection and significance of promoter hypermethylation of p16 and MGMT gene methylation in plasma from NSCLC patients.] Tumor 2007; 27: 715–8 (In Chinese.) [Google Scholar]
- 16. Hsu HS, Chen TP, Hung CH et al Characterization of a multiple epigenetic marker panel for lung cancer detection and risk assessment in plasma. Cancer 2007; 110: 2019–26. [DOI] [PubMed] [Google Scholar]
- 17. Zhang LP, Yao QF, Ning Y. [Significance of p16 and MGMT gene promoter methylation for early diagnosis of lung cancer.] Chin J Public Health 2008; 24: 52–3 (In Chinese.) [Google Scholar]
- 18. Ma Xinping WY. The clinical value of p16 gene promoter methylation detection in diagnosis of lung cancer. Pract Prev Med 2009; 16: 76–8. [Google Scholar]
- 19. Hu ZJ, Hu HB, Liu DY, Chen ZP. [Clinical significance of p16 gene methylation in early diagnosis of lung cancer.] Chin J Pathophysiol 2009; 1941–5 (In Chinese.) [Google Scholar]
- 20. Chen SH, Xue SL, Jin YT et al [Methylation of p16 gene in plasma and tissues from non‐small cell lung cancer patients.] Chin J Lab Diagn 2010; 14: 1035–8 (In Chinese.) [Google Scholar]
- 21. Wang PP, Zhang CY. [Methylation of RASSF 1A,P16,DAPK AND RUNX 3 gene and its role in Inner Mongolia non‐small‐cell lung cancer.] J Inner Mongolia Med Univ 2016; 38: 98–102 (In Chinese.) [Google Scholar]
- 22. Wan L, Liang Y, He Y, Mi X, Song G, Pan J. [Methylation of p16 gene and p53 antibody as tumor marker in the diagnosis of non‐small cell lung cancer.] J Chin Oncol 2017; 23: 372–6 (In Chinese.) [Google Scholar]
- 23. Liu Y, Wang Y, Jia W et al [Determination of p16, FHIT and APC gene methylation in serum from patients with lung cancer.] J Zhengzhou Univ (Med Sci) 2017; 52: 51–4 (In Chinese.) [Google Scholar]
- 24. Destro A, Bianchi P, Alloisio M et al K‐ras and p16(INK4A)alterations in sputum of NSCLC patients and in heavy asymptomatic chronic smokers. Lung Cancer 2004; 44: 23–32. [DOI] [PubMed] [Google Scholar]
- 25. Konno S, Morishita Y, Fukasawa M et al Anthracotic index and DNA methylation status of sputum contents can be used for identifying the population at risk of lung carcinoma. Cancer 2004; 102: 348–54. [DOI] [PubMed] [Google Scholar]
- 26. Wang X, Cao A, Peng M et al The value of chest CT scan and tumor markers detection in sputum for early diagnosis of peripheral lung cancer. Chin J Lung Cancer 2004; 7: 58–63. [DOI] [PubMed] [Google Scholar]
- 27. Georgiou E, Valeri R, Tzimagiorgis G et al Aberrant p16 promoter methylation among Greek lung cancer patients and smokers: Correlation with smoking. Eur J Cancer Prev 2007; 16: 396–402. [DOI] [PubMed] [Google Scholar]
- 28. Liu Y, Lan Q, Shen M et al Aberrant gene promoter methylation in sputum from individuals exposed to smoky coal emissions. Anticancer Res 2008; 28: 2061–6. [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang W, Sun Y, Lu G. [The diagnostic value of determination of p16 methylation of sputum exfoliated cells for peripheral lung cancer.] Chin J Lung Cancer 2004; 7: 46–9 (In Chinese.) [DOI] [PubMed] [Google Scholar]
- 30. Guo X, Shen L. Diagnostic value of combinative detection of hypermethylation of p16 gene and cytokeratin 19 fragment and CA15‐3 antigen for non‐small cell lung cancer patients. Clin Focus 2008; 23: 1067–70. [Google Scholar]
- 31. Hu X, Kang J, Hou X, Yu R. [Expression of tumor suppressor gene pRb and p16 in pulmonary adenocarcinoma: Its clinical significance.] Chin J Pract Intern Med 2002; 22: 410–1 (In Chinese.) [Google Scholar]
- 32. Peng Z, Shan C, Wang H. [Value of promoter methylation of RASSF1A, p16, and DAPK genes in induced sputum in diagnosing lung cancer.] J Central South Univ (Med Sci) 2010; 35: 247–53 (In Chinese.) [DOI] [PubMed] [Google Scholar]
- 33. Zhang W, Yu C, Xia H, Cai Q, Liu Y. [Diagnostic value of determination of p16 methylation and k‐ras mutation of sputum exfoliated cells for peripheral lung cancer.] China J Mod Med 2012; 22(8): 9–13 (In Chinese.) [Google Scholar]
- 34. Sun N, Zhang L, Liu YY, Zheng S, Zhao X. [Methylation of P16 and RASSF1A genes in sputum samples associated with peripheral non‐small cell lung cancer.] Prog Mod Biomed 2012; 12: 2503–10 (In Chinese.) [Google Scholar]
- 35. Zhao QT, Guo T, Wang HE et al Diagnostic value of SHOX2 DNA methylation in lung cancer: A meta‐analysis. Onco Targets Ther 2015; 8: 3433–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gu C, Lu J, Cui T et al Association between MGMT promoter methylation and non‐small cell lung cancer: A meta‐analysis. PLoS One 2013; 8: e72633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Song L, Yu H, Li Y. Diagnosis of lung cancer by SHOX2 gene methylation assay. Mol Diagn Ther 2015; 19: 159–67. [DOI] [PubMed] [Google Scholar]
- 38. Schmidt B, Liebenberg V, Dietrich D et al SHOX2 DNA methylation is a biomarker for the diagnosis of lung cancer based on bronchial aspirates. BMC Cancer 2010; 10: 600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stone S, Jiang P, Dayananth P et al Complex structure and regulation of the P16 (MTS1) locus. Cancer Res 1995; 55: 2988–94. [PubMed] [Google Scholar]
- 40. Yifan D, Qun L, Yingshuang H et al Bronchial lavage P 16INK4A gene promoter methylation and lung cancer diagnosis: A meta‐analysis. Indian J Cancer 2015; 52 (Suppl 2): e96–8. [DOI] [PubMed] [Google Scholar]
- 41. Yan R, Chi L, Zheng X, Sun R, You J, Ye X. A meta‐analysis of serum p16 gene promoter methylation for diagnosis of nonsmall cell lung cancer. Indian J Cancer 2015; 52 (Suppl 2): e116–8. [DOI] [PubMed] [Google Scholar]


