Abstract
Alterations in transcript‐specific translation are emerging as a driver of cellular transformation and cancer etiology. A new study provides evidence for enhanced codon‐dependent translation of hypoxia‐inducible factor 1α in promoting glycolytic metabolism and drug resistance in melanoma cells. This specialized translation reprogramming relies, in part, on mTORC2‐mediated phosphorylation of enzymes modifying the wobble position of the transfer RNA anticodon.
Subject Categories: Cancer, Molecular Biology of Disease
Cancer cells rapidly adapt to extreme environmental conditions by changes in specific metabolic pathways (Ward & Thompson, 2012). The mechanisms that control this metabolic reprogramming by immediate changes in specific gene expression programs remain less clear. Now, an exciting new study by Rapino et al (2018) provides evidence for an unexpected role of enzymes that modify transfer RNA (tRNA) in this process. Specifically, they show that enzymes modifying uridine 34 (U34) of tRNA are important for the maintenance and therapeutic resistance of an aggressive skin cancer known as melanoma. This tRNA modification is linked to codon‐mediated translation regulation of hypoxia‐inducible factor 1α (HIF1α), a transcription factor known for its role in metabolic adaptation to unfavorable growth conditions. From a clinical perspective, these exciting findings pave the way for targeting RNA‐modifying enzymes in drug‐resistant cancers. Of note, codon‐mediated translation regulation of the KRAS and HRAS oncogenes has recently been implicated in tumorigenesis and drug resistance (Lampson et al, 2013; Ali et al, 2017) suggesting that alternative codon usage may be a more widespread feature of human cancers than previously thought.
Codon usage, tRNA abundance and modifications, and codon‐anticodon base pairing are crucial for efficient and accurate translation of messenger RNAs (mRNAs) into proteins (Agris, 2004). How these basic decoding mechanisms are altered in cancer cells remains poorly understood. While U34 tRNA‐modifying enzymes have been implicated in metabolism, DNA damage response, and exocytosis (Esberg et al, 2006; Begley et al, 2007; Laxman et al, 2013), their role in cancer has not been extensively studied until now. By employing a combination of approaches and patient samples, Rapino et al (2018) demonstrate that key enzymes required for U34 tRNA modifications are increased in melanomas exhibiting highly glycolytic phenotypes that are often associated with mutations in BRAF (one of the most commonly mutated genes in melanoma patients). Rapino and colleagues provide compelling evidence that BRAFV600E melanomas are “addicted” to high levels of U34 tRNA‐modifying enzymes. Together with an assortment of additional proteins, these enzymes catalyze a rather unconventional chemical modification involving methoxycarbonylmethyl formation at position 5 (termed mcm5U) mediated by the multi‐subunit Elongator (ELP) complex (Huang et al, 2005) and a subsequent thiolation at position 2 (termed s2U) by cytosolic thiouridylases and the ubiquitin‐related modifier 1 (URM1) pathway (Leidel et al, 2009) to generate 5‐methoxycarbonylmethyl‐2‐thiouridine (termed mcm5s2U) at U34 of the tRNA anticodon loop. This study further shows that resistance to targeted BRAF inhibition (vemurafenib) is overcome by a reduction in Elongator proteins ELP1 and ELP3 and the cytosolic thiouridylase subunit 1 and 2 (CTU1/CTU2).
An exciting question arises as to why a reduction in U34 tRNA‐modifying enzymes elucidates such a profound growth inhibition phenotype in highly metabolic melanomas. Rapino et al (2018) employ an elegant combination of proteomics and ribosome profiling experiments to identify a role for specific codons in rewiring translation mediated by U34 tRNA‐modifying enzymes in skin cancer cells. The authors show that the abundance of proteins enriched in codons, known to require thiolated tRNAs for decoding, such as Gln, Glu, and Lys codons, are significantly increased in BRAFV600E‐driven melanoma. Moreover, ELP3 depletion enhanced ribosome occupancy of mRNAs biased for codons decoded by U34‐modified tRNAs, without globally impacting ribosome footprinting density. Having identified that ELP3 influences decoding of specific codons during translation in melanoma, the authors next focus on HIF1α whose mRNA transcript was significantly enriched in codons that require U34 tRNA modifications and whose protein, but not mRNA, was strongly decreased upon depletion of ELP3 or CTU1/2 (Fig 1). Mechanistically, the authors demonstrate that upon ELP3 reduction, ribosomes accumulate on HIF1α mRNA indicating a decrease in decoding during translation elongation. Rapino and colleagues identify that replacing codons that rely on U34 tRNA‐modifying enzymes with synonymous codons rescues HIF1α protein expression, defects in cell viability, and glucose metabolism observed upon ELP3 or CTU1 depletion. Consistent with previous reports (Nedialkova & Leidel, 2015), it was further shown that deficient decoding due to loss of U34 tRNA‐modifying enzymes results in accumulation of protein aggregates. Together, the findings presented in this study clearly demonstrate that in melanoma, accurate HIF1α mRNA translation requires U34 tRNA‐modifying enzymes (Fig 1). While the authors unquestionably show the importance of the U34 tRNA‐modifying enzymes in this process, they do not determine a direct change in tRNA modifications in their study. An exciting avenue for future research involves understanding how tRNA modifications within the anticodon loop interphase with the growing repertoire of RNA modifications (coined the epitranscriptome) to modulate translation elongation.
Figure 1. Codon‐dependent translation regulation of HIF1α couples glycolytic metabolism to drug resistance in melanoma.
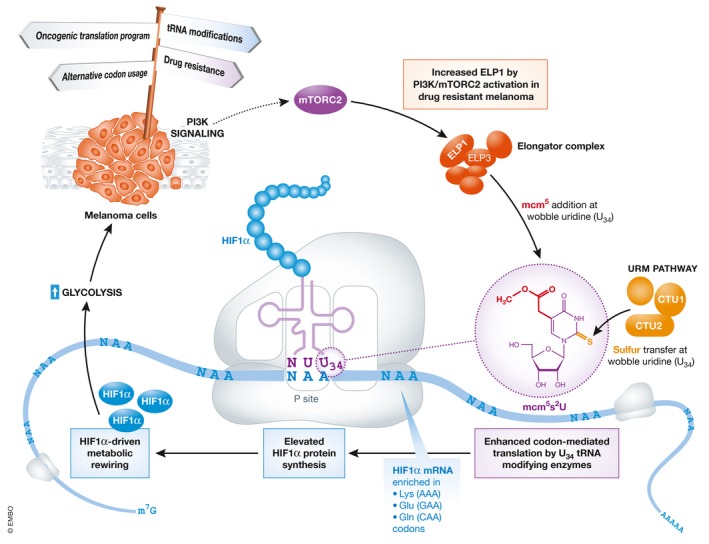
Cartoon depicts the proposed mechanism by which increased activation of U34 tRNA‐modifying enzymes promotes HIF1α protein synthesis, leading to enhanced glycolysis and conferring mTORC2‐dependent resistance toward targeted BRAF inhibition.
Given the observation that HIF1α is a determinant of response to BRAF inhibition in melanoma, the authors next probed the role of U34 tRNA‐modifying enzymes in acquired resistance to targeted BRAF therapy. This is an extremely relevant question as acquired resistance to BRAF inhibitors such as vemurafenib or dabrafenib is a characteristic of BRAFV600E‐driven melanoma. Interestingly, the authors demonstrate that reducing U34 tRNA‐modifying enzymes re‐sensitized resistant BRAFV600E melanoma cells to vemurafenib and significantly reduced melanoma growth in vivo, which appears to rely on HIF1α. Finally, the authors show that increased levels of U34 tRNA‐modifying enzymes in resistant BRAFV600E melanoma are associated with elevated PI3K/mTOR signaling. Mechanistically, they illustrate that mTORC2 activity, but not mTORC1, enhances ELP1 phosphorylation at S1174, leading to elevated ELP1 levels and is crucial for the survival of HIF1α‐dependent glycolytic melanoma cells. Altogether, these exciting findings provide a strong rationale for targeting RNA‐modifying enzymes in drug‐resistant melanoma and open a new portal into investigating the role of tRNA modifications and codon‐mediated translation regulation in cancer pathogenesis.
Several questions arise, including whether the change in tRNA modification machinery identified in this study may be a harbinger of a more global effect of human oncogenes in controlling tRNA function to regulate specific gene expression programs to sustain malignant properties. Even though not addressed in this study, the tight interplay between U34 tRNA‐modifying enzymes and glycolysis in melanoma poses the question as to whether feedback loops exist whereby the products of metabolic pathways can, in turn, directly influence tRNA modifications to drive tumorigenesis. Notably, various RNA‐modifying enzymes require co‐factors such as ATP, vitamins, and metal ions to modulate their activity (Helm & Alfonzo, 2014), all of which are often considered by‐products of cancer cell metabolism. This study will no doubt open several exciting avenues for future research and ignite the design of novel therapeutic strategies for “designer tRNAs” optimized for suppression of gene‐specific translation downstream of oncogenic activation.
The EMBO Journal (2018) 37: e99978
See also: https://doi.org/10.1038/s41586-018-0243-7 (June 2018)
Contributor Information
Mary McMahon, Email: mary.mcmahon@ucsf.edu.
Davide Ruggero, Email: davide.ruggero@ucsf.edu.
References
- Agris PF (2004) Decoding the genome: a modified view. Nucleic Acids Res 32: 223–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali M, Kaltenbrun E, Anderson GR, Stephens SJ, Arena S, Bardelli A, Counter CM, Wood KC (2017) Codon bias imposes a targetable limitation on KRAS‐driven therapeutic resistance. Nat Commun 8: 15617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begley U, Dyavaiah M, Patil A, Rooney JP, DiRenzo D, Young CM, Conklin DS, Zitomer RS, Begley TJ (2007) Trm9‐catalyzed tRNA modifications link translation to the DNA damage response. Mol Cell 28: 860–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esberg A, Huang B, Johansson MJO, Bystrom AS (2006) Elevated levels of two tRNA species bypass the requirement for elongator complex in transcription and exocytosis. Mol Cell 24: 139–148 [DOI] [PubMed] [Google Scholar]
- Helm M, Alfonzo JD (2014) Posttranscriptional RNA modifications: playing metabolic games in a cell's chemical legoland. Chem Biol 21: 174–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Johansson MJO, Bystrom AS (2005) An early step in wobble uridine tRNA modification requires the elongator complex. RNA 11: 424–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson BL, Pershing NLK, Prinz JA, Lacsina JR, Marzluff WF, Nicchitta CV, MacAlpine DM, Counter CM (2013) Rare codons regulate KRas oncogenesis. Curr Biol 23: 70–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laxman S, Sutter BM, Wu X, Kumar S, Guo XF, Trudgian DC, Mirzaei H, Tu BP (2013) Sulfur amino acids regulate translational capacity and metabolic homeostasis through modulation of tRNA thiolation. Cell 154: 416–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidel S, Pedrioli PGA, Bucher T, Brost R, Costanzo M, Schmidt A, Aebersold R, Boone C, Hofmann K, Peter M (2009) Ubiquitin‐related modifier Urm1 acts as a sulphur carrier in thiolation of eukaryotic transfer RNA. Nature 458: 228–232 [DOI] [PubMed] [Google Scholar]
- Nedialkova DD, Leidel SA (2015) Optimization of codon translation rates via tRNA modifications maintains proteome integrity. Cell 161: 1606–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapino F, Delaunay S, Rambow F, Zhou Z, Tharun L, De Tullio P, Sin O, Shostak K, Schmitz S, Piepers J, Ghesquière B, Karim L, Charloteaux B, Jamart D, Florin A, Lambert C, Rorive A, Jerusalem G, Leucci E, Dewaele M et al (2018) Codon‐specific translation reprogramming promotes resistance to targeted therapy. Nature 558: 605–609 [DOI] [PubMed] [Google Scholar]
- Ward PS, Thompson CB (2012) Metabolic reprogramming: a cancer hallmark even Warburg did not anticipate. Cancer Cell 21: 297–308 [DOI] [PMC free article] [PubMed] [Google Scholar]


