Abstract
Proteolytic removal of membrane protein ectodomains (ectodomain shedding) is a post‐translational modification that controls levels and function of hundreds of membrane proteins. The contributing proteases, referred to as sheddases, act as important molecular switches in processes ranging from signaling to cell adhesion. When deregulated, ectodomain shedding is linked to pathologies such as inflammation and Alzheimer's disease. While proteases of the “a disintegrin and metalloprotease” (ADAM) and “beta‐site APP cleaving enzyme” (BACE) families are widely considered as sheddases, in recent years a much broader range of proteases, including intramembrane and soluble proteases, were shown to catalyze similar cleavage reactions. This review demonstrates that shedding is a fundamental process in cell biology and discusses the current understanding of sheddases and their substrates, molecular mechanisms and cellular localizations, as well as physiological functions of protein ectodomain shedding. Moreover, we provide an operational definition of shedding and highlight recent conceptual advances in the field. While new developments in proteomics facilitate substrate discovery, we expect that shedding is not a rare exception, but rather the rule for many membrane proteins, and that many more interesting shedding functions await discovery.
Keywords: matrix metalloproteases, meprin β, pro‐protein convertases, rhomboids, signal peptide peptidase‐like
Subject Categories: Membrane & Intracellular Transport; Post-translational Modifications, Proteolysis & Proteomics
Introduction
Membrane proteins are essential for health and disease and have a large variety of fundamental physiological functions. Levels of individual membrane proteins and their functions are tightly controlled through different mechanisms, including post‐translational modifications such as proteolytic ectodomain shedding (or briefly shedding). Shedding is a form of limited proteolysis and thus an irreversible post‐translational modification (Fig 1). During the shedding process, a protease (referred to as sheddase) cleaves a membrane protein substrate close to or within its transmembrane (TM) domain, resulting in release of the soluble extracellular domain (ectodomain) from the membrane and a fragment that remains bound to the membrane (Fig 1) (Kapeller et al, 1973; Black, 1980a; Ehlers & Riordan, 1991). Some sheddases are also referred to as secretases (Selkoe, 1990), as the cleaved substrate ectodomain may be secreted.
Figure 1. Sheddases trigger the release of a wide range of proteins from the membrane.
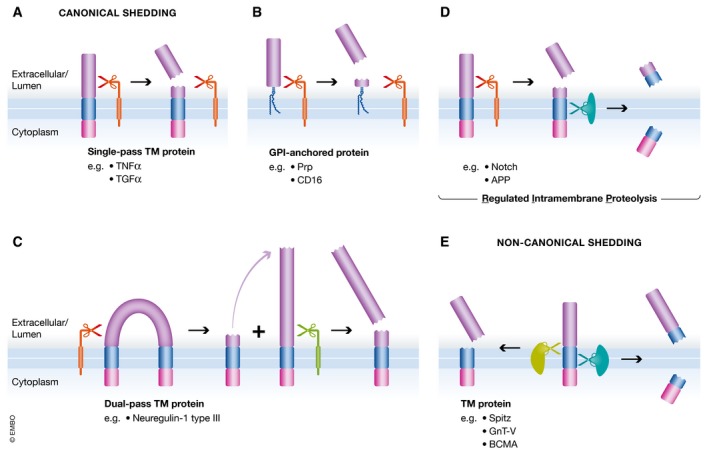
(A) Canonical sheddases cleave single‐pass TM membrane proteins in their luminal juxtamembrane region, thereby releasing ectodomains from their membrane‐integral domains. Ectodomain refers to that part of the protein that is found on the extracellular side of the membrane—in case that the protein localizes to the plasma membrane—or within the lumen of organelles of the secretory and endocytic pathway, which is topologically equivalent to the extracellular space. (B) GPI‐anchored proteins are separated from their lipid modification by cleavage within the C‐terminus of the protein. (C) Dual‐pass and polytopic membrane proteins (not shown) can be cleaved in loops and ectodomains (not shown). Neuregulin‐1 type III is cleaved at two sites in its loop domain, thereby releasing a bioactive peptide from its membrane anchors. (D) As a variation of canonical shedding, in regulated intramembrane proteolysis (RIP), the sheddase‐generated membrane‐integral fragment is further processed in the plane of the lipid bilayer, releasing an intracellular domain and a short extracellular peptide fragment. In this case, shedding is the first step of two subsequent proteolytic cleavages. (E) Non‐canonical sheddases cleave their substrate in or close to the TM domain without requiring any preceding cleavage. Depending on the site of cleavage, the intracellular fragment is released from the lipid bilayer or stays anchored by a slightly shortened TM domain.
Shedding is best understood in mammals, where it has emerged as a key cellular mechanism to control not only abundance, but also activation and inactivation of membrane proteins, for example, through release of membrane‐bound growth factors and cytokines or through degradation of surface receptors and cell adhesion proteins (e.g., Black et al, 1997; Moss et al, 1997; Peschon et al, 1998; Colombo et al, 2018). Given the large number of substrates, shedding influences many processes in development, physiology, and disease, such as connectivity in the nervous system (e.g., Hattori et al, 2000), cholesterol homeostasis (Sakai et al, 1996, 1998), Alzheimer's disease (e.g., Vassar et al, 1999), and inflammatory disorders (e.g., Black et al, 1997; Moss et al, 1997). Yet, for other membrane proteins, shedding may simply be a mechanism of protein turnover and may not be coupled to (patho)physiological consequences.
In the literature, the term shedding sometimes also refers to the non‐proteolytic release of membrane proteins and the release of vesicles from the plasma membrane (Black, 1980a,1980b), which are different molecular processes and are not covered here.
This review gives an overview of ectodomain shedding, starting with an operational definition of shedding, then highlighting the involved proteases and substrates and their regulation, and finally describing the functional consequences and medical implications of shedding. The aim of this review article is to use selected examples a) to demonstrate that shedding is a fundamental cell biological process, b) to illustrate general principles of shedding that emerge from the comparison of different sheddase families, and c) to highlight new trends and conceptual advances in the field.
Definition of ectodomain shedding
Shedding occurs for single‐span TM proteins (Fig 1A), GPI‐anchored proteins (Fig 1B), and proteins with two or more TM domains (Fig 1C). For several substrates, shedding is the first proteolytic cleavage and may be followed by additional proteolytic cleavage(s) within the TM segment. Both cleavages together are conceptually referred to as “regulated intramembrane proteolysis” (Fig 1D) (Brown et al, 2000; Lichtenthaler et al, 2011). In all cases, shedding refers to the release of a protein's ectodomain from the membrane.
Initially, the term ectodomain shedding was used in a narrow manner with regard to cellular localization (plasma membrane) (Kapeller et al, 1973; Black, 1980a; Arribas et al, 1996), the position of the cleavage sites within the substrates (lumenal juxtamembrane domain) and the number of proteases and substrates involved (Ehlers & Riordan, 1991; Massague & Pandiella, 1993). However, several key studies over the past years, which will be discussed in more detail below, demonstrated that shedding occurs in all cellular organelles of the secretory and endocytic pathway, happens both outside and even within the substrates’ TM domain (Fig 1E), and is mediated by many more proteases than previously thought, including membrane‐bound, intramembrane, and even soluble proteases. Moreover, it now has become clear that shedding impacts on many, if not all single‐span membrane proteins and numerous polytopic TM proteins at some stage during their lifetime. In order to reflect these new findings, we propose a broader definition of shedding. Ectodomain shedding is the proteolytic release of the bulk or even the entire ectodomain of a mature membrane protein into the luminal or extracellular space and often alters the substrate's function. Depending on the cellular compartment where shedding occurs, the ectodomain is released into the extracellular space (at the plasma membrane) or into the lumen of the organelles (e.g., Golgi or endosome), which is topologically equivalent to the extracellular space (Schatz & Dobberstein, 1996), and from where it may subsequently be secreted into the extracellular space. The proteolytic cut occurs within the extracellular or luminal juxtamembrane (membrane‐proximal) region or within the TM anchor of a membrane protein substrate. Cleavage sites within the juxtamembrane region are typically at a short distance of often 10 ‐ 35 amino acids from the TM segment, but more distant cleavage sites are possible and, in fact, the exact cleavage sites have only been determined for few shedding substrates (e.g., summarized for ADAMs and BACE1 in Caescu et al, 2009; Yan, 2017).
Several other proteolytic events in cells, such as removal of a signal peptide by signal peptidase (Blobel & Dobberstein, 1975) and proteolytic cleavages by mitochondrial AAA proteases (Levytskyy et al, 2017), formally share similarities to ectodomain shedding, but will not be discussed in this review, as they occur either during protein biosynthesis but not on the mature protein (signal peptidase), or do not occur in the secretory or endocytic pathway (mitochondria).
Hardware: canonical sheddases
The human genome contains nearly 600 protease‐encoding genes (Lopez‐Otin & Bond, 2008), and an increasing number of them are recognized to act as sheddases, with some having many shedding substrates and others so far having only a single substrate reported to undergo shedding, as will be discussed below. Proteases are commonly considered as sheddases if they cleave their substrates in the luminal juxtamembrane domain with a short distance to the membrane‐anchoring domain (Ehlers & Riordan, 1991). We refer to these proteases as canonical sheddases (Table 1) to distinguish them from the more recently described non‐canonical sheddases (described below) that cleave within a substrates’ TM domain or at the membrane boundary (Table 2). Canonical sheddases are typically themselves membrane‐bound, but more and more soluble proteases, such as matrix metalloproteases (MMPs), are also reported to mediate shedding, as will be discussed below. In Tables 1 and 2, we additionally distinguish between sheddases whose primary function is ectodomain release and proteases that mostly have non‐shedding functions, but can additionally act as secondary or “part‐time” sheddases. Some of the best‐characterized sheddases, such as “a disintegrin and metalloprotease 10” (ADAM10), ADAM17 (also known as TACE for TNFα‐converting enzyme), and “β‐site APP cleaving enzyme” (BACE1), have many shedding substrates and act as “full‐time” sheddases. In contrast, other proteases, such as matrix metalloproteases (MMPs) or pro‐protein convertases, have mostly non‐shedding functions, because they cleave soluble proteins (MMPs) or remove pro‐peptides (pro‐protein convertases) without shedding the whole ectodomain. As will be discussed below, such proteases are increasingly found to additionally act as sheddases on a few selected substrates. This qualifies them as “part‐time” sheddases.
Table 1.
List of canonical, mammalian sheddase families
| Sheddase type | Protease family | Protease type | Cellular localization | References |
|---|---|---|---|---|
| Full‐time sheddases |
ADAM proteases
(metalloproteases) ADAM8, ADAM9, ADAM10, ADAM12, ADAM15, ADAM17, ADAM19, ADAM20, ADAM21, ADAM28, ADAM30, ADAM33 |
Membrane‐anchored, type I | Late secretory pathway and plasma membrane | Pruessmeyer and Ludwig (2009), Saftig and Lichtenthaler (2015), Weber and Saftig (2012), Zunke and Rose‐John (2017) |
|
BACE proteases
(aspartyl proteases) BACE1, BACE2 |
Membrane‐anchored, type I | Trans‐Golgi network and endosomes | Barao et al (2016), Dislich and Lichtenthaler (2012), Vassar et al (2014), Yan (2017) | |
| Site‐1 protease (serine protease), also known as SKI‐1 or S1P | Membrane‐anchored, type I | Golgi | Seidah et al (2017), Seidah and Prat (2012) | |
| Part‐time sheddases |
Meprin β
(metalloprotease) |
Membrane‐anchored, type I | Broder and Becker‐Pauly (2013) | |
|
MT‐MMPs
(metalloproteases) MT1‐MMP, MT2‐MMP, MT3‐MMP, MT4‐MMP, MT5‐MMP, MT6‐MMP, also named MMP14‐MMP17, MMP24, MMP25 |
Membrane‐anchored, type I or GPI‐anchored | Late secretory pathway and plasma membrane | Hayashida et al (2010), Itoh (2015) | |
|
Pro‐protein convertases
(serine proteases) PCSK1/3, PCSK2, furin, PCSK4, PCSK5/6, PACE4, PCSK7 and PCSK9 |
Membrane‐anchored, type I or soluble | Late secretory pathway and plasma membrane | Seidah et al (2017), Seidah and Prat (2012) | |
|
Transmembrane serine proteases
Matriptase, Matriptase‐2, Matriptase‐3, Polyserase‐1, Corin, Hepsin, TMPRSS2, TMPRSS3, TMPRSS4, MSPL, Spinesin, Enteropeptidase, HAT, DESC1, TMPRSS11A, HAT‐like 4, HAT‐like 5 |
Membrane‐anchored, type II | Szabo and Bugge (2011), Tanabe and List (2017) | ||
|
Matrix metalloproteases (MMPs)
MMP1, MMP2, MMP3, MMP7, MMP8, MMP9, MMP10, MMP11, MMP12, MMP13, MMP19, MMP20, MMP21, MMP23, MMP26, MMP27, MMP28 |
Soluble | Extracellular space | Freitas‐Rodriguez et al (2017), Klein and Bischoff (2011), Peixoto et al (2012) | |
|
Legumain (δ‐secretase)
cysteine protease |
Soluble | Zhang et al (2016) | ||
|
Cathepsin S and L
(cysteine protease) |
Soluble | Extracellular space | Sobotic et al (2015) |
Family members with known shedding function are indicated in bold and italics. Selected review articles that typically describe the whole protease family are given. Some articles also contain lists of identified substrates. For proteases with few shedding substrates, the original study is cited. Site‐1 protease belongs to the family of pro‐protein convertases, but is listed separately to highlight that it acts as a full‐time sheddase in contrast to the other members of the same family.
Table 2.
List of non‐canonical, mammalian sheddase families
| Sheddase type | Protease family and members | Protease type | Cellular localization | References |
|---|---|---|---|---|
| Full‐time sheddases |
Rhomboid proteases
(serine proteases) RHBDL1, RHBDL2, RHBDL3, RHBDL4 |
Integral multi‐pass TM protein | Golgi (RHBDL1), plasma membrane (RHBDL2), endosomes (RHBDL3), ER (RHBDL4) | Freeman (2014), Lemberg (2013) |
|
SPP/SPPL family (aspartyl proteases) SPPL3 SPP, SPPL2a, SPPL2b, SPPL2c, (SPPL3 is a major sheddase; SPP acts as a sheddase only in exceptional cases) |
Integral multi‐pass TM protein | ER (SPP), lysosomes (SPPL2a), cell surface (SPPL2b), ER (SPPL2c), Golgi (SPPL3) | Kuhn et al (2015), Voss et al (2014) Boname et al (2014), Chen et al (2014) | |
| Part‐time sheddases | ||||
|
Presenilin/γ‐secretase
(aspartyl protease) Presenilin‐1, Presenilin‐2 (γ‐secretase acts as a sheddase only in exceptional cases) |
Integral multi‐pass TM protein, | Plasma membrane, endosomes | Laurent et al (2015), Schauenburg et al (2018) |
Family members with known shedding function are indicated in bold and italics. Selected review articles are given that typically describe the whole protease family. Some articles also contain lists of identified substrates. For proteases with few shedding substrates, the original study is cited.
The following paragraphs first describe the canonical sheddases, starting with membrane‐bound sheddases, followed by soluble ones. As some sheddases have more than 100 substrates, only selected substrates are listed, in particular those that have been validated under protease‐deficient conditions or through in vivo studies.
ADAM10 and ADAM17
The best‐characterized canonical sheddases, ADAM10 and ADAM17, are most likely active in the trans‐Golgi network (TGN), in later secretory pathway compartments, and at the plasma membrane (Fig 2). More than 100 substrates for ADAM10 and similar numbers for ADAM17 have been identified in different tissues and cells using candidate testing and advanced proteomics, although not all of them have been validated under physiological conditions and with in vitro assays (for detailed lists, see e.g., Pruessmeyer & Ludwig, 2009; Weber & Saftig, 2012; Kawahara et al, 2014; Saftig & Lichtenthaler, 2015; Kuhn et al, 2016; Zunke & Rose‐John, 2017). Selected substrates are highlighted in Table 3. Substrate cleavage by ADAM10 often happens constitutively under non‐stimulated conditions, whereas substrate shedding by ADAM17 is mostly observed, when cells are stimulated, either with physiological activators or phorbol esters such as PMA (phorbol‐12‐myristat‐13‐acetat, also known as TPA, 12‐O‐tetradecanoylphorbol‐13‐acetat).
Figure 2. Cellular localization of sheddases.
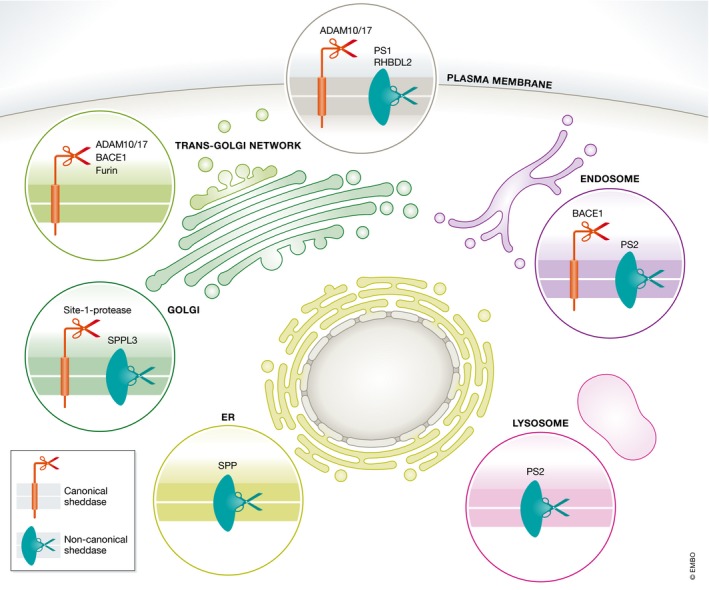
Catalytically active canonical and non‐canonical sheddases not only localize to the cell surface but also to different subcellular compartments. The localization of selected canonical (red) and non‐canonical (green) sheddases is indicated.
Table 3.
Examples of substrates of selected canonical and non‐canonical, mammalian sheddases.a
| Sheddase | Selected substrates | References |
|---|---|---|
| ADAM10 | Notch, APP, PrP, EGF, ephrin‐A5, N‐cadherin, DR6, CD23 | Altmeppen et al (2011), Colombo et al (2018), Hartmann et al (2002), Janes et al (2005), Jorissen et al (2010), Kuhn et al (2016, 2010), Pan and Rubin (1997), Postina et al (2004), Reiss et al (2005), Sahin et al (2004), Suh et al (2013), Vincent et al (2001), Weskamp et al (2006) |
| ADAM17 | TGFα, TNFα, IL6R, amphiregulin, epiregulin, heparin‐binding EGF‐like growth factor, L‐selectin, TNFR2 | Althoff et al (2000), Black et al (1997), Ludwig et al (2005), Moss et al (1997), Peschon et al (1998), Sahin et al (2004) |
| BACE1 | APP, NRG1, SEZ6, CHL1 | Dislich et al (2015), Esterhazy et al (2011), Hemming et al (2009), Kuhn et al (2012), Stutzer et al (2013), Zhou et al (2012) |
| BACE2 | TMEM27, PMEL17 | Esterhazy et al (2011), Rochin et al (2013) |
| Meprin β | CD99, APP | Arolas et al (2012), Bedau et al (2017), Jefferson et al (2011) |
| MT1‐MMP | CD44, syndecan, RANKL | Endo et al (2003), Hikita et al (2006), Kajita et al (2001), Tam et al (2004) |
| MT3‐MMP | NgR1 | Ferraro et al (2011), Sanz et al (2018) |
| MT5‐MMP | N‐cadherin, APP | Baranger et al (2016), Folgueras et al (2009), Porlan et al (2014), Willem et al (2015) |
| MMP9, MMP12 | N‐cadherin, NLG1 | Dwivedi et al (2009), Peixoto et al (2012) |
| PC7 | Transferrin receptor | Guillemot et al (2013), Wang and Pei (2001) |
| Site‐1 protease (S1P, SKI‐1) | SREBP, ATF6, GlcNAc‐1‐phosphotransferase | Marschner et al (2011), Sakai et al (1998), Seidah et al (2017), Ye et al (2000) |
| RHBDL2 | Thrombomodulin, EGF, BCAM, Spint‐1, CLCP1 | Adrain et al (2011), Cheng et al (2011), Johnson et al (2017), Lohi et al (2004) |
| RHBDL4 | ERAD substrates, APP | Fleig et al (2012), Johnson et al (2017), Paschkowsky et al (2016) |
| SPP | XBP1u | Chen et al (2014) |
| SPPL3 | GnT‐V and other glycan‐modifying enzymes | Kuhn et al (2015), Voss et al (2014) |
| γ‐secretase | BCMA | Laurent et al (2015) |
Sheddases with many substrates and examples of recent studies are listed.
Only such substrates are listed that have been validated, preferentially under sheddase‐inactivating conditions or through in vivo experiments.
ADAM10 is essential for ligand‐dependent shedding of the Notch1 receptor and its subsequent signaling (Pan & Rubin, 1997; Bozkulak & Weinmaster, 2009; van Tetering et al, 2009), which is required for embryonic development but also in several adult tissues (Sato et al, 2012; reviewed in Alabi et al, 2018). Additionally, it acts as α‐secretase for the amyloid precursor protein (APP) thereby preventing the generation of the neurotoxic amyloid‐β peptide (Lammich et al, 1999; Postina et al, 2004; Jorissen et al, 2010; Kuhn et al, 2010; Suh et al, 2013), and is, thus, considered a drug target for Alzheimer's disease. Numerous phenotypes have been identified in ADAM10‐deficient mice, for example, in the nervous system (Prox et al, 2013), but the many substrates still need to be assigned to the individual phenotypes and functions. It is also possible that some phenotypes are not just caused by the loss of cleavage of a single, but of multiple substrates simultaneously.
ADAM17 has a key function in tissue homeostasis through cleavage of several members of the epidermal growth factor (EGF) receptor (EGFR) ligand family, including TGFα (Peschon et al, 1998), and may be a drug target for EGFR‐dependent tumors (e.g., Schmidt et al, 2018). Additionally, ADAM17 has a fundamental role in inflammation by being the major sheddase for the cytokine tumor necrosis factor α (TNFα). Thus, ADAM17 is considered a major drug target for inflammatory diseases such as sepsis, rheumatoid arthritis, and inflammatory bowel disease (reviewed in Rose‐John, 2013).
Besides ADAM10 and ADAM17, the ADAM family has ten more members with proven or assumed proteolytic activity (Weber & Saftig, 2012), but only few (or in some cases no) physiological substrates for them have been identified to date (Table 1).
BACE1 and BACE2
Another class of sheddases in the endomembrane system are BACE1 and BACE2 (Fig 2), which were initially identified as APP sheddases (Hussain et al, 1999; Sinha et al, 1999; Vassar et al, 1999; Yan et al, 1999, 2001; Lin et al, 2000; Fluhrer et al, 2002). Candidate approaches and, more recently, proteomic studies identified more than 40 substrates and substrate candidates each for BACE1 and BACE2 (see Table 3 for selected substrates) (Hemming et al, 2009; Esterhazy et al, 2011; Kuhn et al, 2012; Zhou et al, 2012; Stutzer et al, 2013; Dislich et al, 2015), but many of them have not yet been validated under physiological conditions. Since BACE1 acts as the major β‐secretase for APP and catalyzes formation of the pathogenic amyloid‐β peptide, several inhibitors targeting BACE1 are currently in advanced clinical trials for Alzheimer's disease. However, BACE1 has additional functions in neurobiology, including in myelination, muscle spindle formation and maintenance, synapse formation, and axon targeting (Hu et al, 2006; Willem et al, 2006; Rajapaksha et al, 2011; Cao et al, 2012; Hitt et al, 2012; Cheret et al, 2013; Barao et al, 2015; Zhu et al, 2018).
For most substrates, the functional consequences of their cleavage by BACE1 have not yet been investigated, largely for lack of tools such as antibodies targeted to the substrates’ ectodomains or intracellular domains, or because little is known about the substrates.
While BACE1 is highly expressed in the nervous system, its homolog BACE2 is strongly expressed in pancreas (Vassar et al, 1999). In vivo experiments using BACE2‐deficient mice revealed that BACE2 regulates pancreatic β‐cell function and mass through cleavage of “transmembrane protein 27” (TMEM27) (Esterhazy et al, 2011), making BACE2 a potential drug target for diabetes, which needs to be further evaluated. Another BACE2 substrate is PMEL17, the cleavage of which is required for pigment production in melanocytes and thus, for pigmentation of hair, skin, and mucosa, at least in rodents (Rochin et al, 2013).
Meprin β
Meprin β is a homodimeric TM metalloprotease. Soluble and TM substrates as well as substrate candidates were identified proteomically from cell lines overexpressing or exposed to soluble meprin β (Bien et al, 2012; Jefferson et al, 2013). Several TM proteins were cleaved within their ectodomain at a large distance from the membrane, which is not seen as a shedding event, as a large part of the ectodomain remains. However, meprin β also acts as a sheddase and cleaves close to the TM domain in CD99 to promote transendothelial cell migration, and in APP, for which it acts as an alternative β‐secretase (Jefferson et al, 2011; Arolas et al, 2012; Bedau et al, 2017). To what extent meprin β may contribute to Alzheimer's disease still needs to be explored in more detail.
Membrane‐type matrix metalloproteases (MT‐MMPs)
The six MT‐MMPs are a subgroup of the larger MMP family. They are assumed to be active at the plasma membrane and are mostly known for their cleavage of soluble substrates (see Table 1), in particular extracellular matrix proteins, such as collagens and fibronectin (Itoh, 2015). Increasingly, they are reported to also act as canonical sheddases for TM proteins (see Table 3 and reviewed in Hayashida et al, 2010; Itoh, 2015), and more shedding substrates are likely to be identified in the future. For example, MT1‐MMP sheds RANKL (Receptor activator of NF‐kappaB ligand) and negatively regulates osteoclastogenesis (Hikita et al, 2006). MT3‐MMP was recently shown to shed the GPI‐anchored Nogo receptor 1, which promotes excitatory synapse formation in vitro and in vivo (Sanz et al, 2018). MT5‐MMP sheds N‐cadherin and controls peripheral thermal nociception, presumably through modulation of cell adhesion between mast cells and sensory fibers (Folgueras et al, 2009). MT5‐MMP shedding of N‐cadherin also controls adhesion of neuronal stem cells to ependymocytes and thereby stem cell quiescence versus proliferation (Folgueras et al, 2009; Porlan et al, 2014). MT5‐MMP was recently furthermore identified as the APP η‐secretase, and its inactivation reduced inflammation and amyloid pathology in an Alzheimer's disease mouse model (Willem et al, 2015; Baranger et al, 2016). However, the APP η‐secretase cleavage is more distant (~120 amino acids) from the membrane than most other shedding events. It is not yet clear for all examples mentioned above how exactly the MT‐MMPs contribute to the indicated (patho)physiological processes, and it is likely that more shedding functions of MT‐MMPs will be discovered.
Pro‐protein convertases, including site‐1‐protease (S1P)
Pro‐protein convertases are a family of nine soluble and membrane‐bound serine proteases that are commonly found in the TGN and later compartments of the secretory pathway (Fig 2). Several of them, such as furin, remove pro‐peptides from soluble or membrane‐bound inactive protein precursors (reviewed in Seidah & Prat, 2012), including ADAM and BACE proteases. These pro‐peptide cleavages are not considered as shedding event, since they often occur several hundred amino acids distant from the substrates’ TM domains and, therefore, do not remove the majority of the substrates’ ectodomains. Yet, pro‐protein convertases are increasingly reported to additionally act as sheddases for selected substrates. For example, PCSK7 sheds the transferrin receptor (Guillemot et al, 2013), whereas furin or another pro‐protein convertase sheds MT5‐MMP (Wang & Pei, 2001), with both cleavages occurring < 25 amino acids away from the substrates’ TM domains. Thus, while the functional consequences of these shedding events are not yet fully understood, pro‐protein convertases can act as “part‐time” sheddases.
One family member, site‐1 protease (S1P), also known as subtilins/kexin‐isozyme 1, stands out from the other family members in that it functions primarily as a sheddase (reviewed in Seidah et al, 2017). Known substrates of this Golgi‐resident protease include viral proteins as well as the latent transcription factors SREBP, involved in cholesterol homeostasis, and ATF6, which is proteolytically activated during the endoplasmic reticulum (ER) unfolded protein response, as well as the inactive α/β‐subunit precursor of GlcNAc‐1‐phosphotransferase, where proteolysis is required for lysosomal homeostasis (Sakai et al, 1998; Ye et al, 2000; Marschner et al, 2011). Both SREBP and ATF6 are shed at a distance of < 30 amino acids from the membrane and are further processed by site‐2 protease in the paradigm of regulated intramembrane proteolysis (Brown et al, 2000).
Transmembrane serine proteases (TTSPs)
Matriptase‐2 is a member of the type II transmembrane serine proteases (TTSPs), an understudied group of 17 membrane‐bound serine proteases (Szabo & Bugge, 2011), and has been reported to shed APP within the amyloid β domain, at least in transfected cells or in vitro (Beckmann et al, 2016). Other TTSPs appear to cleave predominantly soluble proteins or activate membrane‐bound proteins, but without shedding them in their juxtamembrane domains (Jackle et al, 2015; Murray et al, 2016). Thus, at present TTSPs belong to the group of “part‐time” sheddases. However, it is well possible that the future will reveal more TTSP shedding substrates.
Soluble sheddases
Canonical sheddases are typically single‐span TM or GPI‐anchored proteins (Table 1). Yet, several soluble proteases, which typically cleave non‐membrane‐bound substrates, are increasingly reported to also act as “part‐time” sheddases by cleaving within the substrate's juxtamembrane domain. For example, MMP9, a soluble MMP, sheds neuroligin 1 in the nervous system (Peixoto et al, 2012). Another example, the cysteine protease legumain (also known as asparagine endopeptidase), was recently shown to act as APP δ‐secretase (Zhang et al, 2015, 2017). This cleavage occurs in vicinity to the BACE1 cleavage site in APP and enhances amyloid‐β generation, which makes legumain a potential drug target in Alzheimer's disease. Likewise, the soluble protease cathepsin S sheds the cell adhesion proteins ALCAM and CD44 in vivo and, at least in vitro, numerous additional cell surface proteins (Sobotic et al, 2015). Taken together, these examples demonstrate that membrane attachment is not required for a protease to act as a sheddase. However, a membrane‐anchor offers the advantage to position the active site close to the membrane surface, facilitating rapid accessibility to the substrate's cleavage site.
Hardware: non‐canonical sheddases
Based on the position of their cleavage sites outside the substrates’ TM region, canonical sheddases were long thought to be clearly different from intramembrane proteases, which cleave their substrates within the TM domain. The best‐investigated intramembrane protease, γ‐secretase, cleaves its first‐identified substrates, which typically have a long ectodomain, only when their ectodomain is truncated by a canonical sheddase. Consequently, γ‐secretase and other intramembrane proteases initially were assumed to not directly shed membrane proteins, but only act secondary to a primary shedding event. However, recent studies demonstrated that some intramembrane proteases, in particular rhomboids and SPPL3, act as bona fide sheddases and that other intramembrane proteases, such as SPP and γ‐secretase, can—at least occasionally—also shed membrane protein ectodomains directly and thus act as “part‐time” sheddases. Thus, proteases cleaving in juxtamembrane and TM regions share more functional properties than previously expected.
Rhomboids
Rhomboids are intramembrane serine proteases first discovered in Drosophila, where they act as key activators of EGFR signaling (Lee et al, 2001; Urban et al, 2001). Drosophila Rhomboid‐1 is a Golgi‐resident protease that triggers secretion of the EGFR‐receptor ligand Spitz by cleaving within its TM domain, paralleling the physiological function of ADAM proteases in mammals (Fig 3A). Rhomboids are universally conserved, and different functions ranging from protein degradation to cleavage of cell adhesion molecules during parasite invasion have been described (for recent reviews see Lemberg, 2013; Freeman, 2014; Urban, 2016). The crystal structures of the E. coli rhomboid protease GlpG revealed a conserved six‐TM helix‐bundle forming a rhomboid active site cavity that opens to the periplasmic (luminal) side of the membrane (Wang et al, 2006). Consistent with this, rhomboid proteases have been shown to also cleave substrates within their ectodomains and within loops of multi‐TM domain proteins (Erez & Bibi, 2009; Fleig et al, 2012). Interestingly, in contrast to most other intramembrane proteases, rhomboids can directly act as sheddases on full‐length proteins with long ectodomains and do not require the substrate's ectodomain to be trimmed by a preceding canonical shedding event (Fig 3A). In mammals, four secretory pathway rhomboid proteases are known, which are referred to as RHBDL1 to 4 (Fig 2 and Table 2). The first known physiological substrate of a mammalian rhomboid protease was thrombomodulin, which is cleaved by RHBDL2 at the plasma membrane (Lohi et al, 2004; Cheng et al, 2011). RHBDL2 also cleaves EGF, but likely only to modulate its signaling, a process that may nevertheless be deregulated in certain cancer cells (Adrain et al, 2011). More recently, substrate proteomics identified several additional proteins to be cleaved by RHBDL2 (Table 3) (Johnson et al, 2017). RHBDL4 has been linked to the ER‐associated degradation (ERAD) pathway (Fleig et al, 2012) and has been suggested to act as a non‐canonical sheddase of APP (Paschkowsky et al, 2016).
Figure 3. Non‐canonical sheddases. One representative substrate per non‐canonical sheddase is given.
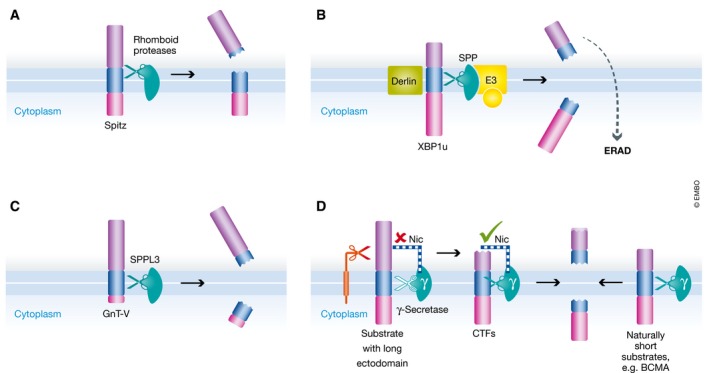
(A) Rhomboid proteases cleave the TM domain of their substrates in the luminal membrane leaflet, thereby triggering release of the ectodomain. (B) SPP assembles with the rhomboid pseudoprotease Derlin1, and ERAD E3 ubiquitin ligases TRC8 and MARCH6 to form a proteolytic ERAD complex that recognizes membrane proteins without preceding cleavage. In a concerted action, fragments are released to both sides of the membrane and degraded by further components of the ERAD pathway. (C) SPPL3 cleaves glycan‐modifying enzymes at the luminal border of their TM domains, releasing the active site containing ectodomain. (D) Membrane proteins with large ectodomains need shedding to truncate their ectodomain before their C‐terminal fragment (CTF) can be further processed by γ‐secretase. In contrast, substrates with a naturally short ectodomain are directly shed by γ‐secretase leading to secretion of their entire ectodomains. Nicastrin (Nic) serves as a molecular ruler accepting only membrane proteins with a short ectodomain.
Signal peptide peptidase (SPP)
Signal peptide peptidase (SPP) is a member of the heterogenous group of GxGD intramembrane aspartyl proteases (Ponting et al, 2002; Weihofen et al, 2002). A recent crystal structure of the archaeal GxGD protease MCMJR1 revealed the catalytic aspartate residues forming an aqueous active site 8 Å below the membrane surface (Li et al, 2013). SPP was first characterized as the activity that clears signal peptides from the ER following their removal from nascent secretory proteins by signal peptidase (Weihofen et al, 2000; Lemberg & Martoglio, 2002). More recently, SPP has also been recognized as an ERAD factor that may under certain circumstances act as a non‐canonical sheddase, where—different to its common role in regulated intramembrane proteolysis of signal peptides—it does not require initial activatory cleavage by signal peptidase (Boname et al, 2014; Chen et al, 2014; Hsu et al, 2015). Consistent with this dual role, SPP assembles as a homo‐tetramer that processes signal peptides (Schrul et al, 2010) and—for its sheddase function—as higher molecular weight assembly with the ERAD factor Derlin1 and ERAD E3 ubiquitin ligases (Fig 3B) (Stagg et al, 2009; Chen et al, 2014; Stefanovic‐Barrett et al, 2018). While SPP had initially been hypothesized to contribute to non‐proteolytic dislocation of certain ERAD substrates (Loureiro et al, 2006; Lee et al, 2010), heme oxigenase‐1 and XBP1u were recently shown to be shed by SPP, leading to their rapid degradation by the proteasome (Boname et al, 2014; Chen et al, 2014). Although the C‐terminal, luminal portion of SPP‐dependent ERAD substrates is not secreted (Fig 3B), cleavage without preceding substrate processing formally ranks SPP as a “part‐time” non‐canonical sheddase.
SPP‐like proteases (SPPL)
Besides SPP four SPP homologs, the SPP‐like (SPPL) proteases, SPPL2a, SPPL2b, SPPL2c, and SPPL3, have been identified in mammals (Grigorenko et al, 2002; Ponting et al, 2002; Weihofen et al, 2002). While all known SPPL2b substrates require processing by a canonical sheddase before SPPL2b processing can occur within the TM segment (Fluhrer et al, 2006; Martin et al, 2008, 2009; Zahn et al, 2013), SPPL3 was recently found to act as a non‐canonical sheddase independently of the substrates’ ectodomain length (Fig 3C; Voss et al, 2012). Proteomic approaches have identified many substrate candidates, in particular with functions in regulation of cellular N‐glycosylation (Voss et al, 2014; Kuhn et al, 2015). By shedding of various glycosyltransferases and glycosidases, SPPL3 removes the catalytic domain of these enzymes in the Golgi. Subsequent secretion of these domains results in inactivation of the glycan‐modifying enzymes. Consequently, increased expression of SPPL3 leads to protein hypoglycosylation, while reduced SPPL3 expression induces hyperglycosylation of cellular proteins. Thus, shedding mediated by SPPL3 may serve as a potent cellular switch that allows adaption of a cell's glycan pattern to environmental changes (Voss et al, 2014). SPPL2a is involved in processing of CD74, the invariant chain of the Major Histocompatibility Complex II (MHCII) (Beisner et al, 2013; Bergmann et al, 2013; Schneppenheim et al, 2013). Under physiological conditions, the type II‐oriented CD74 molecule is sequentially processed by several serine and cysteine proteases generating a stable membrane‐bound CD74 fragment that is subject to SPPL2a cleavage. Thus, CD74 processing reflects a classical cascade of regulated intramembrane proteolysis. Whether SPPL2a and its presently still orphan sister protease SPPL2c can also act as a non‐canonical sheddases remains to be elucidated.
γ‐secretase
The last member of the GxGD‐type aspartyl proteases is γ‐secretase, which has presenilin‐1 or ‐2 as the catalytic subunit and which acts as a sheddase on full‐length proteins only in exceptional cases. Initially identified because of its link to APP processing in Alzheimer's disease (Sherrington et al, 1995; De Strooper et al, 1998; Wolfe et al, 1999), γ‐secretase currently has more than 100 known substrates (for an overview see Haapasalo & Kovacs, 2011). Interestingly, presenilins are distant homologs of the SPP/SPPL family members, but have opposite membrane topology and therefore only cleave type I membrane protein substrates, whereas SPP/SPPL proteases selectively cleave type II‐oriented TM segments (Weihofen et al, 2002). Besides presenilin, γ‐secretase consists of three additional proteins (Aph‐1, nicastrin, and PEN2) (Fig 3D), which are essential for γ‐secretase maturation and activity (Edbauer et al, 2003; Kimberly et al, 2003; Takasugi et al, 2003). Cryo‐electron microscopy shows that the nicastrin subunit has a compactly folded ectodomain that forms a lid on top of the active site of γ‐secretase (Bai et al, 2015), making nicastrin a molecular ruler that prevents membrane proteins with long ectodomains from getting cleaved by γ‐secretase (Bolduc et al, 2016). As a consequence, γ‐secretase substrates with long ectodomains require prior shedding by canonical sheddases, reducing ectodomain length to < ~50 amino acids and allowing subsequent intramembrane proteolysis by γ‐secretase (Struhl & Adachi, 2000). Thus, γ‐secretase generally does not act as a sheddase on intact membrane proteins, with the striking exception of the recently described γ‐secretase shedding of the B cell maturation antigen (BCMA) that alters its function as a B cell surface receptor required for NFκB signaling and maintenance of long‐lived plasma cells (Laurent et al, 2015). The cleavage takes place within the BCMA TM domain, but—in contrast to other γ‐secretase substrates—does not require prior ectodomain shedding, as the ectodomain is naturally short enough (54 amino acids) for direct non‐canonical shedding by γ‐secretase. The mammalian proteome contains additional type I membrane proteins with naturally short ectodomains, and BCMA may therefore be the founding member of a new class of naturally short γ‐secretase shedding substrates. Interestingly, a recent study reported that also the APP‐homolog APLP1, which has a large ectodomain of several hundred amino acids, may be directly shed by γ‐secretase, at least to a small extent, in addition to its usual shedding by ADAM10 and BACE1 (Schauenburg et al, 2018). However, it is in this case not yet clear how the long APLP1 ectodomain could mechanistically bypass the strict, short ectodomain length requirement imposed by nicastrin.
Taken together, intramembrane proteases can act as non‐canonical sheddases and thus directly influence the physiological functions of their substrates. While some intramembrane proteases, like rhomboids and SPPL3, primarily act as sheddases and have multiple substrates, others including SPP and γ‐secretase appear to have exceptional “part‐time” sheddase functions only on selected targets under specific conditions, and otherwise mostly act as intramembrane proteases requiring a prior shedding event.
Hardware: higher order assembly and non‐proteolytic subunits
Most sheddases are assumed to act as monomers or, as observed for BACE1, homodimers (Schmechel et al, 2004; Westmeyer et al, 2004), but there is increasing evidence that certain sheddases may assemble into higher order complexes, as has been described for ADAM10 that interacts with γ‐secretase (Chen et al, 2015). While this may allow efficient coupling of shedding and subsequent intramembrane proteolysis, it is unknown whether all ADAM10 substrates are further processed by γ‐secretase, and it remains to be determined which fraction of both proteases is found in the complex. In addition, ADAM10 was also reported to associate with certain members of another class of multi‐pass TM proteins, the tetraspanins (reviewed in Matthews et al, 2017). These non‐proteolytic partners have been attributed functions in ADAM10 maturation (Arduise et al, 2008; Dornier et al, 2012; Haining et al, 2012; Prox et al, 2012), but their exact impact on activity, regulation, and substrate specificity of ADAM10 remains to be determined. Similarly, larger complexes have been observed for ADAM17, which associates with the catalytically inactive rhomboid‐family proteins iRhom1 or ‐2 (Adrain et al, 2012; McIlwain et al, 2012; Christova et al, 2013; Maretzky et al, 2013; Cavadas et al, 2017; Grieve et al, 2017), and for SPP, which assembles with Derlin1 (Chen et al, 2014). iRhoms and derlins can be seen as part of the sheddase hardware and serve as substrate adaptors or trafficking regulators for their active protease partners ADAM17 or SPP (Maretzky et al, 2013). This is reminiscent of γ‐secretase with its three non‐proteolytic subunits required for maturation, trafficking, and activity of the whole protease complex (Edbauer et al, 2003; Kimberly et al, 2003; Takasugi et al, 2003).
Hardware: substrates
To date, the number of shedding substrates is unknown, but given the large numbers of canonical sheddase substrates mentioned above (see substrates and review articles cited in Table 3), it is clear that shedding affects at least a few hundred different mammalian membrane proteins. Moreover, a systematic screening of published reports led to a database (SheddomeDB) listing over 400, mostly human, shed proteins (Tien et al, 2017). Furthermore, several recent proteomic studies detected hundreds of membrane proteins in conditioned medium of various cell lines and in body fluids (e.g., Faca et al, 2008; Kuhn et al, 2012; Meissner et al, 2013; Kim et al, 2014; Wilhelm et al, 2014; Dislich et al, 2015). These proteins may well constitute cleavage products of shedding substrates, although not all of them have been validated by independent methods. Interestingly, sets of shed proteins differed significantly between different cell types or even between distinct cancer cell lines (Faca et al, 2008; Kuhn et al, 2012). Given the large variety of cell types in mammals, it thus seems reasonable to estimate that the total number of shed proteins in a given organism may exceed 1,000. Potentially, all of the more than 2,000 human single‐span membrane proteins listed in UniProt may undergo shedding, at least at some point during their life cycle. In fact, it has proven difficult to find proteins that are not shed at all, in particular when proteins are overexpressed in cell lines. Thus, it appears possible that shedding substrates fall into two distinct categories. One of them includes substrates for which shedding is coupled to physiological consequences, as discussed in the next paragraph. The other category comprises membrane proteins where shedding does not lead to major functional changes but may be a mechanism of protein turnover. Yet, more functional studies with a larger number of shedding substrates are required to firmly distinguish between both categories.
A future establishment of a more comprehensive catalog of sheddase substrates appears possible and is facilitated by the recent development of new proteomic methods for in vitro and in vivo substrate identification (Gevaert et al, 2003; Kleifeld et al, 2010; Eichelbaum et al, 2012; Kuhn et al, 2012; Dislich et al, 2015; reviewed in Muller et al, 2016). However, before concluding that all proteins identified in a given proteomic study or from overexpression studies are new sheddase substrates, careful validation by independent methods must be executed. This includes in vitro assays to test the direct cleavage and the demonstration of physiological relevance, in particular when substrates have been identified upon protease overexpression or exogenous addition of recombinant protease in in vitro assays.
How does shedding alter membrane protein function?
As for all other enzymes, the function of sheddases is determined by their substrates. Given the large number and diversity of their substrates, it is clear that shedding affects numerous physiological processes. In the following, we will illustrate three fundamental ways in which the shedding process can alter a substrate protein's function.
First, where the full‐length membrane‐bound form of the substrate displays its physiological function, shedding provides a mechanism to terminate the function of a full‐length membrane protein (Fig 4A). Examples are cell adhesion proteins (e.g., selectins), glycosyltransferases (e.g., GnT‐V), and cell surface receptors (e.g., TNFRs). This is not just a mechanism leading to membrane protein degradation, but the shed ectodomain can even further block the physiological function of the remaining full‐length proteins. For example, the shed ectodomain of the B cell maturation antigen (BCMA) acts as a decoy receptor that sequesters the cognate ligand and thereby further attenuates cell signaling in addition to shedding of the full‐length BCMA receptor (Laurent et al, 2015).
Figure 4. Functional consequences of shedding.
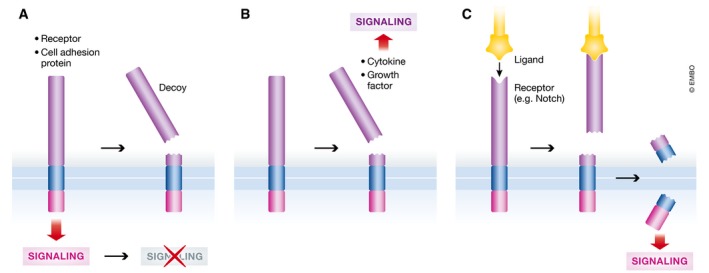
(A) By proteolytic processing of TM proteins that display a physiological function, like cell adhesion or receptor‐mediated signaling, sheddases terminate these functions. (B) Shedding generates biologically active signaling molecules from membrane‐anchored precursors, e.g., cytokines or growth factors that act on neighboring or far distant cells. (C) As part of regulated intramembrane proteolysis, sheddases induce a proteolytic cascade ultimately activating signaling molecules, like the Notch intracellular domain, that act within the same cell. Shedding may be ligand‐induced and the ligand may even be a membrane‐anchored protein itself, as in case of Notch.
Second, if cleavage releases the biologically active ectodomain of the membrane protein, shedding can activate a membrane protein (Fig 4B). Examples are growth factors (e.g., TGFα) and cytokines (e.g., TNFα). In these cases, shedding is a mechanism to timely and spatially control release and activity of the biomolecule. For example, shed TNFα acts in a paracrine manner to activate proinflammatory TNFR1 signaling, whereas membrane‐bound TNFα activates anti‐inflammatory TNFR2 (Grell et al, 1995), but can only do this upon direct cell–cell contact. Interestingly, also the membrane‐bound fragment remaining after shedding can be biologically active. This is observed for the BACE1‐generated C‐terminal fragment of CHL1, which functions in growth cone collapse during axon guidance in the nervous system (Barao et al, 2015).
Third, shedding can induce subsequent processing by an intramembrane protease. A key example is activation of the Notch receptor, which is induced by a membrane‐anchored ligand on another cell. Ligand‐binding triggers endocytosis of both the ligand and of Notch into the opposing cell. This exposes the membrane‐proximal ADAM10 cleavage site. ADAM10 cleavage is followed by γ‐secretase‐mediated processing to release the intracellular domain, which then acts as a transcription activator (Pan & Rubin, 1997; De Strooper et al, 1999; Tiyanont et al, 2011; Fig 4C).
Taken together, shedding is a versatile mechanism to control the activity of membrane proteins. Yet, for most shedding substrates, it has not yet been explored how shedding controls or alters their function. As a result, more functional consequences in addition to the three categories described above may be discovered in the future. Likewise, it appears possible that—for some substrates—the shedding process is simply contributing to protein turnover and may not be coupled to a major functional consequence, for example, in cell signaling.
Cellular localization of ectodomain shedding
Initially, the term shedding referred to cleavages occurring at or very close to the plasma membrane (Arribas et al, 1996), where the cleaved, soluble ectodomain was released from cells into conditioned medium or into body fluids. It is now clear that shedding additionally takes place in multiple cellular compartments, including all organelles of the secretory and endocytic pathway (Fig 2). Related proteases may be active in different cellular compartments, as seen for SPP cleaving in the ER (Weihofen et al, 2002) and SPPL3 being active in the Golgi (Voss et al, 2014), or presenilin‐1‐containing γ‐secretase being more active at the plasma membrane while γ‐secretase complexes containing the presenilin‐2 paralog are predominantly active in endo‐ and lysosomes, but potentially also in the trans‐Golgi network (Meckler & Checler, 2016; Sannerud et al, 2016). In which cellular compartment a given sheddase cleaves its substrate is largely determined by where the substrate meets the active enzyme. Many sheddases, including ADAMs, BACEs, and MT‐MMPs, require removal of their pro‐peptide by furin or related pro‐protein convertases for gaining their full proteolytic activity (e.g., Lopez‐Perez et al, 1999, 2001; Bennett et al, 2000; Capell et al, 2000; Huse et al, 2000; Schlondorff et al, 2000; Creemers et al, 2001). Pro‐peptide removal often occurs in the trans‐Golgi compartment and, thus, efficiently prevents premature substrate cleavage in the ER and the Golgi compartment. Activity of other sheddases is pH‐dependent. For instance, BACE1 has an acidic pH optimum (Vassar et al, 1999; Shimizu et al, 2005), thus only cleaving in acidic cellular compartments. Therefore, depending on the substrates’ localization, some BACE1 substrates are predominantly cleaved in endosomes, whereas others are mostly cleaved in the TGN. These different cellular localizations of BACE1 activity are even exploited for the development of substrate‐specific BACE1 inhibitors, which selectively target BACE1 in endosomes (Rajendran et al, 2008; Mitterreiter et al, 2010; Ben Halima et al, 2016). On the other hand, partly due to lack of suitable reagents, the exact cleavage compartment—assumed to be late in the secretory pathway or at the plasma membrane—has been identified for only few substrates of ADAM10. For instance, that ADAM10 cleavage of the Eph‐receptor ligand ephrin‐A5 takes place at the plasma membrane has been inferred mainly because this cleavage happens in trans with ephrin‐A5 being expressed on the surface of one cell and ADAM10 on the surface of another cell (Janes et al, 2005). Otherwise, shedding events known to date happen in cis, with substrate and sheddase expressed within the same cell.
Substrate recognition of sheddases
The increasing number of substrates that are assigned to sheddases allows to highlight two major requirements governing substrate recognition by sheddases: (i) substrate sequence and structure and (ii) vicinity of the cleavage site close to or within the membrane.
Sheddases recognize amino acid motifs and/or secondary structures in their substrates
Sheddases often have preferences for certain amino acid motifs, which is seen in in vitro assays (e.g., Gruninger‐Leitch et al, 2002; Caescu et al, 2009), by mutational analyses (e.g., Sisodia, 1992) and by sheddase structure determinations (e.g., Hong et al, 2000; Seegar et al, 2017). However, this requirement is less pronounced compared to many soluble proteases, such as trypsin and caspases. As a consequence, point mutations in the substrates close to the cleavage site rarely fully abolish cleavage, as for example shown for APP (Sisodia, 1992; Citron et al, 1995). Additionally, it is possible that mutations simply shift the cleavage site by a few amino acids to an alternative, cryptic cleavage site, or that other proteases cleave at a site close by. This may be overlooked in typical cellular shedding experiments, where levels of one of the cleavage products are measured, but where the exact cleavage sites have mostly not been determined. Not only the amino acid sequence, but also secondary structure elements around the substrate's cleavage site may contribute to specificity of the shedding event. For example, cleavage sites for rhomboids are within or at the border of substrates’ TM domains and are part of helical structures, which need to be unfolded before cleavage (Urban & Freeman, 2003; Strisovsky et al, 2009). Consequently, point mutations increasing or decreasing the propensity to unfold the helical structure increased or decreased the extent of substrate cleavage, respectively (Strisovsky et al, 2009; Moin & Urban, 2012; Strisovsky, 2016). Overall, substrate specificity of sheddases is not only determined by a binary interaction between sequence surrounding the scissile peptide bond and protease active site, but additional protein interactions to so‐called exosites located either on the sheddase or adaptor proteins. For example, it has been suggested that iRhoms and tetraspanins present substrates to ADAM17 and ADAM10 (Maretzky et al, 2013; Matthews et al, 2017), respectively, proposing an interaction of the substrate with both the adapter and the protease. Similarly, SPP is primed toward the full‐length type II membrane protein XBP1u by the rhomboid pseudoprotease Derlin1 (Chen et al, 2014). Likewise, substrates are thought to interact with different subunits of the four‐member γ‐secretase complex before they reach the active site (Fukumori & Steiner, 2016).
Location of the cleavage site with respect to the membrane
A second major determinant of substrate recognition by sheddases is the location of the substrate's cleavage site. The cleavage site typically localizes in the luminal juxtamembrane domains (canonical sheddases) and TM domains (non‐canonical sheddases) of the substrate. Thus, a “perfect” sequence motif for a membrane‐bound sheddase may still not be sufficient for cleavage if the sequence motif in the substrate is too far away from the membrane. Likewise, once the membrane is removed, e.g., in in vitro sheddase assays, the substrate specificity may be different compared to cellular and in vivo experiments, as the active site may gain access to potential cleavage sites that would not be reachable and therefore never get cleaved under physiological conditions (e.g., Schlondorff et al, 2000; Brummer et al, 2018). Conversely, protease cleavage specificities determined in vitro using peptide libraries may not necessarily be relevant in full‐length substrates in cellular membranes. Thus, it is difficult to predict sheddase substrates simply based on sequence analysis, and experimental substrate identification is required.
Taken together, substrate recognition of sheddases requires a permissive sequence and structure around the cleavage site of the substrate and the correct positioning of the sheddase's active site toward the substrate's scissile peptide bond.
Substrate repertoire of sheddases
For most known shedding substrates, the protease has not yet been identified. And conversely, only for few sheddases, a comprehensive list of substrates is known. In the following, we will summarize key lessons about shedding substrates learned from the comparative study of different sheddases with multiple substrates.
Developmental stage matters
Numerous substrates are known for ADAM10 and ADAM17. Yet, only one or at most few of them were assumed to be of major physiological relevance. This conclusion was largely based on the most obvious phenotype(s) of the corresponding sheddase‐deficient mice. For example, ADAM10‐deficient mice die at embryonic day 9.5, because the ADAM10 substrate Notch is no longer cleaved, thus preventing Notch signaling during embryonic development (Hartmann et al, 2002). However, conditional postnatal ADAM10 deletion circumventing embryonic lethality demonstrated additional phenotypes, particularly in the brain, that are not related to Notch but to other substrates; for example, defects in synaptic connectivity were caused by defective processing of NrCAM (Jorissen et al, 2010; Prox et al, 2013; Kuhn et al, 2016). Thus, the most pertinent physiological function of a sheddase may be mediated by different substrates at distinct developmental or adult stages.
Tissue‐dependent shedding
A substrate may be cleaved by different sheddases in a tissue‐dependent manner, depending on the expression pattern of substrate and sheddases. For example, BACE1 is highly expressed in the brain, but at low levels in most other tissues (Vassar et al, 1999). As a result, APP and the cell adhesion protein L1 are mostly shed by BACE1 in the brain, but predominantly by ADAM10 in peripheral cells and tissues (Gutwein et al, 2003; Kuhn et al, 2012, 2016; Colombo et al, 2013). Likewise, the surface protein SEZ6L is mostly cleaved by BACE1 in the brain, but by BACE2 in the pancreas (Stutzer et al, 2013; Pigoni et al, 2016).
Subcellular localization
The substrate spectrum may also depend on the subcellular localization of protease and substrate, in particular in polarized cells such as neurons. For example, neuronal ADAM10 predominantly localizes to the somatodendritic compartment (Marcello et al, 2007), whereas BACE1 is found more in axons (Kandalepas et al, 2013). Similar differences are seen in polarized epithelial cells (Capell et al, 2002; Wild‐Bode et al, 2006). SPPL3 mainly localizes to the Golgi and preferentially sheds glycan‐modifying enzymes in this compartment, while glycosyltransferases localizing to the ER are not affected by SPPL3 (Voss et al, 2014; Kuhn et al, 2015). Thus, it is likely that these proteases mostly cleave substrates that localize to the same subcellular compartment.
One substrate may be cleaved by multiple sheddases within one cell type
A major insight from recent quantitative proteomic studies for substrate identification is that some targets are predominantly shed by a single protease within one cell type, whereas other substrates within the same cell type may be cleaved by more than one protease. For instance, in neurons, SEZ6 is only cleaved by BACE1 and MMP17 is only shed by ADAM10, whereas the cell adhesion protein CHL1 is cleaved to more than 40% by each BACE1 and ADAM10 (Kuhn et al, 2012, 2016) and to some extent by ADAM8 (Naus et al, 2004). For most substrates with multiple sheddases, it remains unclear which additional sheddase(s) contribute and whether the cleavage sites of the different proteases are identical or different. For example, numerous glycan‐modifying enzymes are shed by SPPL3, but even upon loss of the protease some substrates still undergo significant shedding (Kuhn et al, 2015). Additionally, compensatory effects need to be considered, when one sheddase is inactivated. For example, blocking BACE1‐medidated shedding of the Alzheimer's disease‐linked APP protein in neurons leads to a compensatory increase in APP shedding by ADAM10, as observed in neurons and even in humans in an Alzheimer clinical trial (May et al, 2011; Colombo et al, 2013). A key challenge for the future will be to understand whether the different proteases that cleave a single protein have redundant functions or whether the corresponding cleavages lead to different functional consequences, as in the case of APP and neuregulin (Hu et al, 2006; Willem et al, 2006; Ring et al, 2007; Li et al, 2010; La Marca et al, 2011).
Substrates can have major and minor sheddases
Another future challenge will be to detect (patho)physiologically relevant, but minor proteolytic cleavage events in a given substrate. For example, APP in neurons is mostly shed by the α‐secretase ADAM10 and the β‐secretase BACE1. Yet, to an apparently smaller extent, APP is also shed by additional proteases, including meprin β and BACE2 (Farzan et al, 2000; Yan et al, 2001; Fluhrer et al, 2002; Jefferson et al, 2011) and the recently identified δ‐ and η‐secretases legumain and MT5‐MMP, respectively, which may be relevant for Alzheimer's disease (Willem et al, 2015; Zhang et al, 2015). APP in non‐neuronal cells may further be cleaved by RHBDL4 (Paschkowsky et al, 2016), but the pathophysiological relevance of this cleavage still needs to be demonstrated. Other examples are the Nogo‐66 receptor (NgR1), which is predominantly shed by MT3‐MMP in neurons (Sanz et al, 2018), and the transferrin receptor that is mostly shed by PC7 in hepatocytes (Guillemot et al, 2013), with presumed minor sheddases in these cases yet to be identified. The glycosylation enzyme ST6GalI is a shedding substrate for both SPPL3 (Kuhn et al, 2015) and BACE1 (Kitazume et al, 2001).
Establishment of minor cleavage events is difficult, because the major sheddase would still cut a substrate protein even when the minor sheddase is inactivated. However, proteomic methods specifically determining the neo‐N‐/C‐termini of cleavage products can determine even such minor cleavage sites (reviewed in Muller et al, 2016). Another difficulty in detecting minor or even major cleavage events is that the shedding by a specific protease may only take place under activated cellular conditions, such as inflammation, infection, or cell stress. Thus, as long as the stimulus is unknown, the cleavage event will not be detected.
Taken together, sheddases can have different substrates and substrates can have different sheddases. As a last expansion of the complexity, a protease that cleaves a given substrate may differ between various organisms. This still appears as an exception, but during evolution different proteases have evolved in distinct organisms to cleave similar types of substrates. One prominent example is EGFR ligands, which are cleaved by ADAM10 and ADAM17 in mammals but by rhomboid proteases in Drosophila, despite both classes of sheddases being conserved between the two species (Lee et al, 2001). Interestingly, mammalian rhomboid proteases still retain the ability to cleave EGFR ligands, but they appear to be less prominent, as ADAM17 has taken the lead during mammalian evolution. Hence, EGFR ligand cleavage by rhomboids only becomes visible upon ADAM17 inhibition or deficiency (Adrain et al, 2011). Over all, we can expect that the substrate spectrum in common model organisms will be more comprehensively defined so that we shall learn more about the fascinating evolution of canonical and non‐canonical sheddases.
Regulation of shedding
Shedding is frequently regulated by mechanisms ranging from trafficking control to natural protein inhibitors and activators. Given the number of sheddases, substrates, and mechanisms, there is a wealth of studies on this topic (reviewed e.g., in Hayashida et al, 2010; Lichtenthaler, 2012; Adrain & Freeman, 2014; Clark, 2014). In the following, we will illustrate key principles with selected examples and highlight new developments in shedding regulation.
Trafficking introduces a major layer of control
Protein trafficking is arguably one of the most important regulatory mechanisms for ectodomain shedding. While a soluble protease may meet its substrate through diffusion, membrane‐bound sheddases and their TM substrates need to be transported to the same organelle for cleavage to occur. This was first shown for the pathway regulating shedding and activation of SREBP. When cellular cholesterol levels drop, ER‐localized SREBP translocates to the Golgi, where it gets shed by S1P and subsequently cleaved by site‐2‐protease within its TM domain (Rawson et al, 1997; Sakai et al, 1998). This dual cleavage results in release and activation of the cytoplasmic SREBP domain, which induces transcription of genes involved in cholesterol biosynthesis (Sakai et al, 1996). Regulated trafficking also happens for other shedding substrates such as APP, where endocytic trafficking controls APP cleavage by either the α‐secretase ADAM10 or the β‐secretase BACE1 (e.g., Haass et al, 1993; Koo & Squazzo, 1994; Chyung & Selkoe, 2003; Carey et al, 2005; Schobel et al, 2008) (and reviewed in Lichtenthaler, 2012). Trafficking also controls the activity of sheddases. For example, upon activation of NMDA receptors, the cytoplasmic adaptor protein Sap97 binds the cytoplasmic tail of ADAM10, thereby promoting its trafficking and activation (Marcello et al, 2007). ADAM10 trafficking is also controlled by tetraspanins (Dornier et al, 2012). Likewise, iRhoms have been implicated in trafficking and regulation of ADAM17 (Adrain et al, 2012; McIlwain et al, 2012).
Abundance control is key
More sheddase or more substrate typically results in more cleavage, and levels of both enzyme and substrates are typically controlled through transcription, translation, and protein degradation. For example, LPS stimulation of immune cells induces TNFα transcription followed by increased TNFα shedding through ADAM17 (Black et al, 1997; Moss et al, 1997). Translational repression has been intensively studied for BACE1 (De Pietri Tonelli et al, 2004; Lammich et al, 2004; Rogers et al, 2004; Zhou & Song, 2006; Mihailovich et al, 2007; Faghihi et al, 2008; Hebert et al, 2008; Wang et al, 2008), and this repression may be relieved upon cellular stress or during disease (e.g., O'Connor et al, 2008). Lysosomal protein degradation is an additional mechanism to control levels of sheddases and substrates. For instance, binding of BACE1 to the adaptor protein GGA promotes degradation (Tesco et al, 2007) and this is blocked by a specific sugar modification, bisecting N‐acetylglucosamine (Kizuka et al, 2015). As an alternative to lysosomal degradation, classical sheddases may be shed themselves, e.g., ADAM10, BACE1, and meprin β (Hussain et al, 2003; Tousseyn et al, 2009), which may be considered a mechanism for inactivating a sheddase (Wichert et al, 2017).
Integration of signaling at the level of sheddase and regulatory subunits
Besides stimulation through regulation of trafficking and protein abundance, signaling pathways can also acutely stimulate shedding within minutes, allowing cells to quickly respond to external stimuli without the need for time‐consuming protein biosynthesis. A prime example is the fast activation of ADAM17 by the phorbol ester PMA (Peschon et al, 1998; Doedens et al, 2003; Sahin et al, 2004). This occurs through phosphorylation of the ADAM17‐associated iRhom protein independently of the cytoplasmic tail of ADAM17 and appears to induce a fast structural change in ADAM17 (Doedens et al, 2003; Le Gall et al, 2010; Cavadas et al, 2017; Grieve et al, 2017). This fast ADAM17 activation also occurs for a physiological process called transactivation, in which agonists of G protein‐coupled receptors indirectly activate the EGFR (Prenzel et al, 1999; Maretzky et al, 2011). Although this fast activation appears to be independent of phosphorylation of ADAM17 itself, other sheddases can be regulated by direct phosphorylation as observed for the δ‐secretase legumain (Wang et al, 2017). Another mechanism that is increasingly linked to the shedding control is calcium signaling, as observed for human meprin β (Arnold et al, 2015) or certain rhomboid proteases (Baker & Urban, 2015).
Inhibitors and activators of protein shedding
In addition to direct activation of the enzymes, various mechanisms tune access of sheddases to their substrates. One prominent example is the soluble tissue inhibitors of metalloproteases that (to various extent) block ADAM10, ADAM17, and MT‐MMPs (e.g., Amour et al, 1998, 2000). Conversely, other ligands are known to induce shedding as observed for Notch, RGMa, BAFFR, and DDR1 (Pan & Rubin, 1997; Bozkulak & Weinmaster, 2009; van Tetering et al, 2009; van Erp et al, 2015; Shitomi et al, 2015; Smulski et al, 2017). An emerging layer of regulation is post‐translational modification at the substrate level. For instance, O‐glycosylation at amino acids close to the cleavage site controls substrate shedding by ADAM proteases (Goth et al, 2015) and in context of the ER, substrate ubiquitination has been shown to trigger intramembrane proteolysis (Fleig et al, 2012).
Modulation by lipids
Since sheddases and their substrates are mostly membrane proteins, they are in direct contact with membrane lipids. It is now becoming clear that lipids are not only bystanders, but can directly control proteolytic activity, as is clearly seen for intramembrane sheddases, in particular γ‐secretase and rhomboids (Urban & Wolfe, 2005; Bondar et al, 2009; Holmes et al, 2012; Winkler et al, 2012). Yet, even the activity of canonical sheddases can be affected by lipids, for example, BACE1 by cholesterol (Ehehalt et al, 2003; Kalvodova et al, 2005) or ADAM17 by phosphatidylserine (Sommer et al, 2016). However, the exact mechanisms by which lipids regulate sheddase activity await further clarification.
Disease association and shedding‐based drugs
Deregulation of shedding may result in diseases caused by too much or too little of the substrate or the sheddase or the cleavage activity. Sheddases and the relevant substrates are consequently considered as drug targets. A key example is an excessive level of shed TNFα, which is implicated in numerous inflammatory diseases, including sepsis, rheumatoid arthritis, and lupus. Clinical treatment blocks the function of shed TNFα by neutralizing it with antibodies or antibody‐like proteins, which are among the best‐selling drugs worldwide (Udalova et al, 2016). Other approaches were less successful, such as the use of small‐molecule inhibitors of the TNFα sheddase ADAM17, because of the broad ADAM17 substrate spectrum and the cross‐reactivity of inhibitors with ADAM17‐related metalloproteases. Besides small‐molecule drugs and antibodies, shed substrate ectodomains may be employed as decoy receptors. For example, recombinant soluble gp130, corresponding to the shed gp130 ectodomain, is used to block excessive IL‐6 signaling in inflammatory conditions and is currently being tested in a phase II clinical trial (reviewed in Rose‐John, 2017). Another important example of a shedding‐related condition is Alzheimer's disease, where pathogenic amyloid‐β peptide results from shedding of APP by BACE1 followed by γ‐secretase‐mediated intramembrane proteolysis. Naturally occurring mutations at the BACE1 cleavage site in APP result in enhanced APP shedding by BACE1 and increased amyloid‐β levels, thus causing an inherited form of the disease (Citron et al, 1992). For some diseases, increased or reduced levels of sheddases have been reported, but it is not always clear whether this is a cause or a consequence of the disease pathogenesis and whether it might be therapeutically exploited, for example, in cancer (reviewed in Murphy, 2008). Selected examples of shedding‐related diseases and potential drugs are listed in Table 4. The shed substrate ectodomains may also serve as potential companion diagnostics to monitor drug responses in patients upon sheddase inhibition or activation. For example, the BACE1‐cleaved APP ectodomain serves as a marker to monitor BACE1 activity in clinical trials with BACE1 inhibitors against AD (May et al, 2011).
Table 4.
Shedding‐related diseases and drugs
| Sheddase | Substrate | Disease | Therapeutic strategy and stage of development | References (review articles or original study) |
|---|---|---|---|---|
| ADAM8 | Unknown | Breast cancer | Inhibition of ADAM8 (mouse study) | Romagnoli et al (2014) |
| Unknown | Pancreatic cancer | Inhibition of ADAM8 (mouse study) | Schlomann et al (2015) | |
| ADAM9 | EGF, FGFR2iiib | Prostate cancer | Inhibition of ADAM9 (mouse study) | Peduto et al (2005) |
| ADAM10 | APP | Alzheimer's disease | Activation of ADAM10 (phase II clinical trial terminated), | Endres et al (2014), Suh et al (2013) |
| CD23 | Asthma | Inhibition of ADAM10 (mouse study) | Mathews et al (2011), Weskamp et al (2006) | |
| Ephrin‐B2 | Lung fibrosis | Inhibition of ADAM10 (mouse study) | Lagares et al (2017) | |
| PrP | Prion diseases | Activation of ADAM10 (mouse study) | Altmeppen et al (2015), Endres et al (2009) | |
| ADAM17 | TNFα |
Inflammatory diseases Sepsis Rheumatoid arthritis Crohn's disease Psoriasis Lupus nephritis |
Inhibition of TNFα, (approved drugs), blocking ADAM17 through exosite inhibitors, soluble prodomain or iRhoms (mouse study) | Adrain et al (2012), Horiuchi et al (2007), Issuree et al (2013), McIlwain et al (2012), Qing et al (2018), Udalova et al (2016), Wong et al (2016) |
| IL6 receptor |
Inflammatory diseases Intestinal inflammation, intestinal cancer, Rheumatoid arthritis, Lupus erythematosus, Asthma, Sepsis, Nephrotoxic nephritis, Arterosclerosis, Lung emphysema and others |
Inhibition of IL6 signaling through soluble gp130 (clinical trials ongoing) | Rose‐John (2017), Schmidt et al (2018) | |
| BACE1 | APP | Alzheimer's disease | Inhibition of BACE1 with small‐molecule drugs (clinical trials ongoing) | Barao et al (2016), Vassar et al (2014) |
| δ‐secretase | APP | Alzheimer's disease | Inhibition (mouse study) | Zhang et al (2017, 2015) |
The table lists selected examples where the pathological role of a sheddase or its substrate to a disease has been established in animal models and through human genetics. The many instances where altered sheddase expression only correlates with disease are not listed.
Conclusion and outlook
ADAM10 and ADAM17, the first known proteases with sheddase activity, were identified 21 years ago (Black et al, 1997; Moss et al, 1997; Pan & Rubin, 1997). Initially considered as a process affecting selected membrane protein substrates only (Ehlers & Riordan, 1991; Massague & Pandiella, 1993; Arribas et al, 1996), ectodomain shedding is now a fundamental process in cell biology. It controls the communication between cells and their environment and impacts on many areas in life sciences and medicine. It is becoming increasingly clear that proteolysis of membrane proteins is not the exception, but rather the rule for many membrane proteins. Despite the tremendous progress over the past years, there are many central open questions and challenges that lie ahead. Given the large number of known and still to be identified shedding substrates and the increasing number of sheddases, a major challenge for the future will be to assign individual substrates to proteases and determine how the proteolytic cleavage alters the substrates’ function. This is particularly important as sheddases are used as drug targets. Their inhibition or activation may not only interfere in the desired way with the function of the disease‐linked protein, but may affect the function of multiple other substrates of the same protease as well. Yet, these hurdles may be overcome by developing substrate‐selective inhibitors or by designing drugs targeting protease exosites where only a subset of substrates binds. This will require a better understanding of the molecular mechanisms underlying the sheddases’ substrate specificity. It will be equally important to understand the spatial organization of proteolysis, for example, within cells or even whole tissues, as well as the timing, kinetics, and regulation of shedding, including the potential identification of more non‐proteolytic subunits of sheddases. Taken together, it is a fascinating time to study ectodomain shedding and we can stay tuned for more major discoveries over the years to come.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgements
We are grateful to Carl Blobel, Paul Saftig, and Harald Steiner for helpful comments on this manuscript. This work was supported by funds from the Deutsche Forschungsgemeinschaft within the research unit FOR2290. We apologize to the colleagues that only selected examples of the vast shedding literature could be cited in this review article.
The EMBO Journal (2018) 37: e99456
Contributor Information
Stefan F Lichtenthaler, Email: stefan.lichtenthaler@dzne.de.
Marius K Lemberg, Email: m.lemberg@zmbh.uni-heidelberg.de.
Regina Fluhrer, Email: regina.fluhrer@mail03.med.uni-muenchen.de.
References
- Adrain C, Strisovsky K, Zettl M, Hu L, Lemberg MK, Freeman M (2011) Mammalian EGF receptor activation by the rhomboid protease RHBDL2. EMBO Rep 12: 421–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adrain C, Zettl M, Christova Y, Taylor N, Freeman M (2012) Tumor necrosis factor signaling requires iRhom2 to promote trafficking and activation of TACE. Science 335: 225–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adrain C, Freeman M (2014) Regulation of receptor tyrosine kinase ligand processing. Cold Spring Harb Perspect Biol 6: a008995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabi RO, Farber G, Blobel CP (2018) Intriguing roles for endothelial ADAM10/Notch signaling in the development of organ‐specific vascular beds. Physiol Rev in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- Althoff K, Reddy P, Voltz N, Rose‐John S, Mullberg J (2000) Shedding of interleukin‐6 receptor and tumor necrosis factor alpha. Contribution of the stalk sequence to the cleavage pattern of transmembrane proteins. Eur J Biochem 267: 2624–2631 [DOI] [PubMed] [Google Scholar]
- Altmeppen HC, Prox J, Puig B, Kluth MA, Bernreuther C, Thurm D, Jorissen E, Petrowitz B, Bartsch U, De Strooper B, Saftig P, Glatzel M (2011) Lack of a‐disintegrin‐and‐metalloproteinase ADAM10 leads to intracellular accumulation and loss of shedding of the cellular prion protein in vivo . Mol Neurodegener 6: 36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmeppen HC, Prox J, Krasemann S, Puig B, Kruszewski K, Dohler F, Bernreuther C, Hoxha A, Linsenmeier L, Sikorska B, Liberski PP, Bartsch U, Saftig P, Glatzel M (2015) The sheddase ADAM10 is a potent modulator of prion disease. Elife 4: e04260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amour A, Slocombe PM, Webster A, Butler M, Knight CG, Smith BJ, Stephens PE, Shelley C, Hutton M, Knauper V, Docherty AJ, Murphy G (1998) TNF‐alpha converting enzyme (TACE) is inhibited by TIMP‐3. FEBS Lett 435: 39–44 [DOI] [PubMed] [Google Scholar]
- Amour A, Knight CG, Webster A, Slocombe PM, Stephens PE, Knauper V, Docherty AJ, Murphy G (2000) The in vitro activity of ADAM‐10 is inhibited by TIMP‐1 and TIMP‐3. FEBS Lett 473: 275–279 [DOI] [PubMed] [Google Scholar]
- Arduise C, Abache T, Li L, Billard M, Chabanon A, Ludwig A, Mauduit P, Boucheix C, Rubinstein E, Le Naour F (2008) Tetraspanins regulate ADAM10‐mediated cleavage of TNF‐alpha and epidermal growth factor. J Immunol 181: 7002–7013 [DOI] [PubMed] [Google Scholar]
- Arnold P, Schmidt F, Prox J, Zunke F, Pietrzik C, Lucius R, Becker‐Pauly C (2015) Calcium negatively regulates meprin beta activity and attenuates substrate cleavage. FASEB J 29: 3549–3557 [DOI] [PubMed] [Google Scholar]
- Arolas JL, Broder C, Jefferson T, Guevara T, Sterchi EE, Bode W, Stocker W, Becker‐Pauly C, Gomis‐Ruth FX (2012) Structural basis for the sheddase function of human meprin beta metalloproteinase at the plasma membrane. Proc Natl Acad Sci USA 109: 16131–16136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribas J, Coodly L, Vollmer P, Kishimoto TK, Rose JS, Massague J (1996) Diverse cell surface protein ectodomains are shed by a system sensitive to metalloprotease inhibitors. J Biol Chem 271: 11376–11382 [DOI] [PubMed] [Google Scholar]
- Bai XC, Yan C, Yang G, Lu P, Ma D, Sun L, Zhou R, Scheres SHW, Shi Y (2015) An atomic structure of human gamma‐secretase. Nature 525: 212–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker RP, Urban S (2015) Cytosolic extensions directly regulate a rhomboid protease by modulating substrate gating. Nature 523: 101–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranger K, Marchalant Y, Bonnet AE, Crouzin N, Carrete A, Paumier JM, Py NA, Bernard A, Bauer C, Charrat E, Moschke K, Seiki M, Vignes M, Lichtenthaler SF, Checler F, Khrestchatisky M, Rivera S (2016) MT5‐MMP is a new pro‐amyloidogenic proteinase that promotes amyloid pathology and cognitive decline in a transgenic mouse model of Alzheimer's disease. Cell Mol Life Sci 73: 217–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barao S, Gartner A, Leyva‐Diaz E, Demyanenko G, Munck S, Vanhoutvin T, Zhou L, Schachner M, Lopez‐Bendito G, Maness PF, De Strooper B (2015) Antagonistic effects of BACE1 and APH1B‐gamma‐secretase control axonal guidance by regulating growth cone collapse. Cell Rep 12: 1367–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barao S, Moechars D, Lichtenthaler SF, De Strooper B (2016) BACE1 physiological functions may limit its use as therapeutic target for Alzheimer's disease. Trends Neurosci 39: 158–169 [DOI] [PubMed] [Google Scholar]
- Beckmann AM, Glebov K, Walter J, Merkel O, Mangold M, Schmidt F, Becker‐Pauly C, Gutschow M, Stirnberg M (2016) The intact Kunitz domain protects the amyloid precursor protein from being processed by matriptase‐2. Biol Chem 397: 777–790 [DOI] [PubMed] [Google Scholar]
- Bedau T, Peters F, Prox J, Arnold P, Schmidt F, Finkernagel M, Kollmann S, Wichert R, Otte A, Ohler A, Stirnberg M, Lucius R, Koudelka T, Tholey A, Biasin V, Pietrzik CU, Kwapiszewska G, Becker‐Pauly C (2017) Ectodomain shedding of CD99 within highly conserved regions is mediated by the metalloprotease meprin beta and promotes transendothelial cell migration. FASEB J 31: 1226–1237 [DOI] [PubMed] [Google Scholar]
- Beisner DR, Langerak P, Parker AE, Dahlberg C, Otero FJ, Sutton SE, Poirot L, Barnes W, Young MA, Niessen S, Wiltshire T, Bodendorf U, Martoglio B, Cravatt B, Cooke MP (2013) The intramembrane protease Sppl2a is required for B cell and DC development and survival via cleavage of the invariant chain. J Exp Med 210: 23–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Halima S, Mishra S, Raja KMP, Willem M, Baici A, Simons K, Brustle O, Koch P, Haass C, Caflisch A, Rajendran L (2016) Specific inhibition of beta‐secretase processing of the Alzheimer disease amyloid precursor protein. Cell Rep 14: 2127–2141 [DOI] [PubMed] [Google Scholar]
- Bennett BD, Denis P, Haniu M, Teplow DB, Kahn S, Louis JC, Citron M, Vassar R (2000) A furin‐like convertase mediates propeptide cleavage of BACE, the Alzheimer's beta ‐secretase. J Biol Chem 275: 37712–37717 [DOI] [PubMed] [Google Scholar]
- Bergmann H, Yabas M, Short A, Miosge L, Barthel N, Teh CE, Roots CM, Bull KR, Jeelall Y, Horikawa K, Whittle B, Balakishnan B, Sjollema G, Bertram EM, Mackay F, Rimmer AJ, Cornall RJ, Field MA, Andrews TD, Goodnow CC et al (2013) B cell survival, surface BCR and BAFFR expression, CD74 metabolism, and CD8‐ dendritic cells require the intramembrane endopeptidase SPPL2A. J Exp Med 210: 31–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bien J, Jefferson T, Causevic M, Jumpertz T, Munter L, Multhaup G, Weggen S, Becker‐Pauly C, Pietrzik CU (2012) The metalloprotease meprin beta generates amino terminal‐truncated amyloid beta peptide species. J Biol Chem 287: 33304–33313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black PH (1980a) Shedding from normal and cancer‐cell surfaces. N Engl J Med 303: 1415–1416 [DOI] [PubMed] [Google Scholar]
- Black PH (1980b) Shedding from the cell surface of normal and cancer cells. Adv Cancer Res 32: 75–199 [DOI] [PubMed] [Google Scholar]
- Black RA, Rauch CT, Kozlosky CJ, Peschon JJ, Slack JL, Wolfson MF, Castner BJ, Stocking KL, Reddy P, Srinivasan S, Nelson N, Boiani N, Schooley KA, Gerhart M, Davis R, Fitzner JN, Johnson RS, Paxton RJ, March CJ, Cerretti DP (1997) A metalloproteinase disintegrin that releases tumour‐necrosis factor‐alpha from cells. Nature 385: 729–733 [DOI] [PubMed] [Google Scholar]
- Blobel G, Dobberstein B (1975) Transfer of proteins across membranes. I. Presence of proteolytically processed and unprocessed nascent immunoglobulin light chains on membrane‐bound ribosomes of murine myeloma. J Cell Biol 67: 835–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolduc DM, Montagna DR, Gu Y, Selkoe DJ, Wolfe MS (2016) Nicastrin functions to sterically hinder gamma‐secretase‐substrate interactions driven by substrate transmembrane domain. Proc Natl Acad Sci USA 113: E509–E518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boname JM, Bloor S, Wandel MP, Nathan JA, Antrobus R, Dingwell KS, Thurston TL, Smith DL, Smith JC, Randow F, Lehner PJ (2014) Cleavage by signal peptide peptidase is required for the degradation of selected tail‐anchored proteins. J Cell Biol 205: 847–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondar AN, del Val C, White SH (2009) Rhomboid protease dynamics and lipid interactions. Structure 17: 395–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozkulak EC, Weinmaster G (2009) Selective use of ADAM10 and ADAM17 in activation of Notch1 signaling. Mol Cell Biol 29: 5679–5695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broder C, Becker‐Pauly C (2013) The metalloproteases meprin alpha and meprin beta: unique enzymes in inflammation, neurodegeneration, cancer and fibrosis. Biochem J 450: 253–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MS, Ye J, Rawson RB, Goldstein JL (2000) Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell 100: 391–398 [DOI] [PubMed] [Google Scholar]
- Brummer T, Pigoni M, Rossello A, Wang H, Noy PJ, Tomlinson MG, Blobel CP, Lichtenthaler SF (2018) The metalloprotease ADAM10 (a disintegrin and metalloprotease 10) undergoes rapid, postlysis autocatalytic degradation. FASEB J 32: 3560–3573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caescu CI, Jeschke GR, Turk BE (2009) Active‐site determinants of substrate recognition by the metalloproteinases TACE and ADAM10. Biochem J 424: 79–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Rickenbacher GT, Rodriguez S, Moulia TW, Albers MW (2012) The precision of axon targeting of mouse olfactory sensory neurons requires the BACE1 protease. Sci Rep 2: 231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capell A, Steiner H, Willem M, Kaiser H, Meyer C, Walter J, Lammich S, Multhaup G, Haass C (2000) Maturation and pro‐peptide cleavage of beta‐secretase. J Biol Chem 275: 30849–30854 [DOI] [PubMed] [Google Scholar]
- Capell A, Meyn L, Fluhrer R, Teplow DB, Walter J, Haass C (2002) Apical sorting of beta‐secretase limits amyloid beta‐peptide production. J Biol Chem 277: 5637–5643 [DOI] [PubMed] [Google Scholar]
- Carey RM, Balcz BA, Lopez‐Coviella I, Slack BE (2005) Inhibition of dynamin‐dependent endocytosis increases shedding of the amyloid precursor protein ectodomain and reduces generation of amyloid beta protein. BMC Cell Biol 6: 30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavadas M, Oikonomidi I, Gaspar CJ, Burbridge E, Badenes M, Felix I, Bolado A, Hu T, Bileck A, Gerner C, Domingos PM, von Kriegsheim A, Adrain C (2017) Phosphorylation of iRhom2 controls stimulated proteolytic shedding by the metalloprotease ADAM17/TACE. Cell Rep 21: 745–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Malchus NS, Hehn B, Stelzer W, Avci D, Langosch D, Lemberg MK (2014) Signal peptide peptidase functions in ERAD to cleave the unfolded protein response regulator XBP1u. EMBO J 33: 2492–2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AC, Kim S, Shepardson N, Patel S, Hong S, Selkoe DJ (2015) Physical and functional interaction between the alpha‐ and gamma‐secretases: a new model of regulated intramembrane proteolysis. J Cell Biol 211: 1157–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng TL, Wu YT, Lin HY, Hsu FC, Liu SK, Chang BI, Chen WS, Lai CH, Shi GY, Wu HL (2011) Functions of rhomboid family protease RHBDL2 and thrombomodulin in wound healing. J Invest Dermatol 131: 2486–2494 [DOI] [PubMed] [Google Scholar]
- Cheret C, Willem M, Fricker FR, Wende H, Wulf‐Goldenberg A, Tahirovic S, Nave KA, Saftig P, Haass C, Garratt AN, Bennett DL, Birchmeier C (2013) Bace1 and Neuregulin‐1 cooperate to control formation and maintenance of muscle spindles. EMBO J 32: 2015–2028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christova Y, Adrain C, Bambrough P, Ibrahim A, Freeman M (2013) Mammalian iRhoms have distinct physiological functions including an essential role in TACE regulation. EMBO Rep 14: 884–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chyung JH, Selkoe DJ (2003) Inhibition of receptor mediated endocytosis demonstrates generation of amyloid beta ‐protein at the cell surface. J Biol Chem 278: 51035–51043 [DOI] [PubMed] [Google Scholar]
- Citron M, Oltersdorf T, Haass C, McConlogue L, Hung AY, Seubert P, Vigo‐Pelfrey C, Lieberburg I, Selkoe DJ (1992) Mutation of the beta‐amyloid precursor protein in familial Alzheimer's disease increases beta‐protein production. Nature 360: 672–674 [DOI] [PubMed] [Google Scholar]
- Citron M, Teplow DB, Selkoe DJ (1995) Generation of amyloid beta protein from its precursor is sequence specific. Neuron 14: 661–670 [DOI] [PubMed] [Google Scholar]
- Clark P (2014) Protease‐mediated ectodomain shedding. Thorax 69: 682–684 [DOI] [PubMed] [Google Scholar]
- Colombo A, Wang H, Kuhn PH, Page R, Kremmer E, Dempsey PJ, Crawford HC, Lichtenthaler SF (2013) Constitutive alpha‐ and beta‐secretase cleavages of the amyloid precursor protein are partially coupled in neurons, but not in frequently used cell lines. Neurobiol Dis 49: 137–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo A, Hsia HE, Wang M, Kuhn PH, Brill MS, Canevazzi P, Feederle R, Taveggia C, Misgeld T, Lichtenthaler SF (2018) Non‐cell‐autonomous function of DR6 in Schwann cell proliferation. EMBO J 37: e97390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creemers JW, Ines Dominguez D, Plets E, Serneels L, Taylor NA, Multhaup G, Craessaerts K, Annaert W, De Strooper B (2001) Processing of beta‐secretase by furin and other members of the proprotein convertase family. J Biol Chem 276: 4211–4217 [DOI] [PubMed] [Google Scholar]
- De Pietri Tonelli D, Mihailovich M, Di Cesare A, Codazzi F, Grohovaz F, Zacchetti D (2004) Translational regulation of BACE‐1 expression in neuronal and non‐neuronal cells. Nucleic Acids Res 32: 1808–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Strooper B, Saftig P, Craessaerts K, Vanderstichele H, Guhde G, Annaert W, Von Figura K, Van Leuven F (1998) Deficiency of presenilin‐1 inhibits the normal cleavage of amyloid precursor protein. Nature 391: 387–390 [DOI] [PubMed] [Google Scholar]
- De Strooper B, Annaert W, Cupers P, Saftig P, Craessaerts K, Mumm JS, Schroeter EH, Schrijvers V, Wolfe MS, Ray WJ, Goate A, Kopan R (1999) A presenilin‐1‐dependent gamma‐secretase‐like protease mediates release of Notch intracellular domain. Nature 398: 518–522 [DOI] [PubMed] [Google Scholar]
- Dislich B, Lichtenthaler SF (2012) The membrane‐bound aspartyl protease BACE1: molecular and functional properties in Alzheimer's disease and beyond. Front Physiol 3: 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dislich B, Wohlrab F, Bachhuber T, Muller SA, Kuhn PH, Hogl S, Meyer‐Luehmann M, Lichtenthaler SF (2015) Label‐free quantitative proteomics of mouse cerebrospinal fluid detects beta‐Site APP cleaving enzyme (BACE1) protease substrates in vivo . Mol Cell Proteomics 14: 2550–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doedens JR, Mahimkar RM, Black RA (2003) TACE/ADAM‐17 enzymatic activity is increased in response to cellular stimulation. Biochem Biophys Res Commun 308: 331–338 [DOI] [PubMed] [Google Scholar]
- Dornier E, Coumailleau F, Ottavi JF, Moretti J, Boucheix C, Mauduit P, Schweisguth F, Rubinstein E (2012) TspanC8 tetraspanins regulate ADAM10/Kuzbanian trafficking and promote Notch activation in flies and mammals. J Cell Biol 199: 481–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi A, Slater SC, George SJ (2009) MMP‐9 and ‐12 cause N‐cadherin shedding and thereby beta‐catenin signalling and vascular smooth muscle cell proliferation. Cardiovasc Res 81: 178–186 [DOI] [PubMed] [Google Scholar]
- Edbauer D, Winkler E, Regula JT, Pesold B, Steiner H, Haass C (2003) Reconstitution of gamma‐secretase activity. Nat Cell Biol 5: 486–488 [DOI] [PubMed] [Google Scholar]
- Ehehalt R, Keller P, Haass C, Thiele C, Simons K (2003) Amyloidogenic processing of the Alzheimer beta‐amyloid precursor protein depends on lipid rafts. J Cell Biol 160: 113–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers MR, Riordan JF (1991) Membrane proteins with soluble counterparts: role of proteolysis in the release of transmembrane proteins. Biochemistry 30: 10065–10074 [DOI] [PubMed] [Google Scholar]
- Eichelbaum K, Winter M, Berriel Diaz M, Herzig S, Krijgsveld J (2012) Selective enrichment of newly synthesized proteins for quantitative secretome analysis. Nat Biotechnol 30: 984–990 [DOI] [PubMed] [Google Scholar]
- Endo K, Takino T, Miyamori H, Kinsen H, Yoshizaki T, Furukawa M, Sato H (2003) Cleavage of syndecan‐1 by membrane type matrix metalloproteinase‐1 stimulates cell migration. J Biol Chem 278: 40764–40770 [DOI] [PubMed] [Google Scholar]
- Endres K, Mitteregger G, Kojro E, Kretzschmar H, Fahrenholz F (2009) Influence of ADAM10 on prion protein processing and scrapie infectiosity in vivo . Neurobiol Dis 36: 233–241 [DOI] [PubMed] [Google Scholar]
- Endres K, Fahrenholz F, Lotz J, Hiemke C, Teipel S, Lieb K, Tuscher O, Fellgiebel A (2014) Increased CSF APPs‐alpha levels in patients with Alzheimer disease treated with acitretin. Neurology 83: 1930–1935 [DOI] [PubMed] [Google Scholar]
- Erez E, Bibi E (2009) Cleavage of a multispanning membrane protein by an intramembrane serine protease. Biochemistry 48: 12314–12322 [DOI] [PubMed] [Google Scholar]
- van Erp S, van den Heuvel DM, Fujita Y, Robinson RA, Hellemons AJ, Adolfs Y, Van Battum EY, Blokhuis AM, Kuijpers M, Demmers JA, Hedman H, Hoogenraad CC, Siebold C, Yamashita T, Pasterkamp RJ (2015) Lrig2 negatively regulates ectodomain shedding of axon guidance receptors by ADAM proteases. Dev Cell 35: 537–552 [DOI] [PubMed] [Google Scholar]
- Esterhazy D, Stutzer I, Wang H, Rechsteiner MP, Beauchamp J, Dobeli H, Hilpert H, Matile H, Prummer M, Schmidt A, Lieske N, Boehm B, Marselli L, Bosco D, Kerr‐Conte J, Aebersold R, Spinas GA, Moch H, Migliorini C, Stoffel M (2011) Bace2 is a beta cell‐enriched protease that regulates pancreatic beta cell function and mass. Cell Metab 14: 365–377 [DOI] [PubMed] [Google Scholar]
- Faca VM, Ventura AP, Fitzgibbon MP, Pereira‐Faca SR, Pitteri SJ, Green AE, Ireton RC, Zhang Q, Wang H, O'Briant KC, Drescher CW, Schummer M, McIntosh MW, Knudsen BS, Hanash SM (2008) Proteomic analysis of ovarian cancer cells reveals dynamic processes of protein secretion and shedding of extra‐cellular domains. PLoS ONE 3: e2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G III, Kenny PJ, Wahlestedt C (2008) Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed‐forward regulation of beta‐secretase. Nat Med 14: 723–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farzan M, Schnitzler CE, Vasilieva N, Leung D, Choe H (2000) BACE2, a beta ‐secretase homolog, cleaves at the beta site and within the amyloid‐beta region of the amyloid‐beta precursor protein. Proc Natl Acad Sci USA 97: 9712–9717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraro GB, Morrison CJ, Overall CM, Strittmatter SM, Fournier AE (2011) Membrane‐type matrix metalloproteinase‐3 regulates neuronal responsiveness to myelin through Nogo‐66 receptor 1 cleavage. J Biol Chem 286: 31418–31424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleig L, Bergbold N, Sahasrabudhe P, Geiger B, Kaltak L, Lemberg MK (2012) Ubiquitin‐dependent intramembrane rhomboid protease promotes ERAD of membrane proteins. Mol Cell 47: 558–569 [DOI] [PubMed] [Google Scholar]
- Fluhrer R, Capell A, Westmeyer G, Willem M, Hartung B, Condron MM, Teplow DB, Haass C, Walter J (2002) A non‐amyloidogenic function of BACE‐2 in the secretory pathway. J Neurochem 81: 1011–1020 [DOI] [PubMed] [Google Scholar]
- Fluhrer R, Grammer G, Israel L, Condron MM, Haffner C, Friedmann E, Bohland C, Imhof A, Martoglio B, Teplow DB, Haass C (2006) A gamma‐secretase‐like intramembrane cleavage of TNFalpha by the GxGD aspartyl protease SPPL2b. Nat Cell Biol 8: 894–896 [DOI] [PubMed] [Google Scholar]
- Folgueras AR, Valdes‐Sanchez T, Llano E, Menendez L, Baamonde A, Denlinger BL, Belmonte C, Juarez L, Lastra A, Garcia‐Suarez O, Astudillo A, Kirstein M, Pendas AM, Farinas I, Lopez‐Otin C (2009) Metalloproteinase MT5‐MMP is an essential modulator of neuro‐immune interactions in thermal pain stimulation. Proc Natl Acad Sci USA 106: 16451–16456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman M (2014) The rhomboid‐like superfamily: molecular mechanisms and biological roles. Annu Rev Cell Dev Biol 30: 235–254 [DOI] [PubMed] [Google Scholar]
- Freitas‐Rodriguez S, Folgueras AR, Lopez‐Otin C (2017) The role of matrix metalloproteinases in aging: tissue remodeling and beyond. Biochim Biophys Acta 1864: 2015–2025 [DOI] [PubMed] [Google Scholar]
- Fukumori A, Steiner H (2016) Substrate recruitment of gamma‐secretase and mechanism of clinical presenilin mutations revealed by photoaffinity mapping. EMBO J 35: 1628–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevaert K, Goethals M, Martens L, Van Damme J, Staes A, Thomas GR, Vandekerckhove J (2003) Exploring proteomes and analyzing protein processing by mass spectrometric identification of sorted N‐terminal peptides. Nat Biotechnol 21: 566–569 [DOI] [PubMed] [Google Scholar]
- Goth CK, Halim A, Khetarpal SA, Rader DJ, Clausen H, Schjoldager KT (2015) A systematic study of modulation of ADAM‐mediated ectodomain shedding by site‐specific O‐glycosylation. Proc Natl Acad Sci USA 112: 14623–14628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grell M, Douni E, Wajant H, Lohden M, Clauss M, Maxeiner B, Georgopoulos S, Lesslauer W, Kollias G, Pfizenmaier K, Scheurich P (1995) The transmembrane form of tumor necrosis factor is the prime activating ligand of the 80 kDa tumor necrosis factor receptor. Cell 83: 793–802 [DOI] [PubMed] [Google Scholar]
- Grieve AG, Xu H, Kunzel U, Bambrough P, Sieber B, Freeman M (2017) Phosphorylation of iRhom2 at the plasma membrane controls mammalian TACE‐dependent inflammatory and growth factor signalling. Elife 6: e23968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigorenko AP, Moliaka YK, Korovaitseva GI, Rogaev EI (2002) Novel class of polytopic proteins with domains associated with putative protease activity. Biochemistry 67: 826–835 [DOI] [PubMed] [Google Scholar]
- Gruninger‐Leitch F, Schlatter D, Kung E, Nelbock P, Dobeli H (2002) Substrate and inhibitor profile of BACE (beta‐secretase) and comparison with other mammalian aspartic proteases. J Biol Chem 277: 4687–4693 [DOI] [PubMed] [Google Scholar]
- Guillemot J, Canuel M, Essalmani R, Prat A, Seidah NG (2013) Implication of the proprotein convertases in iron homeostasis: proprotein convertase 7 sheds human transferrin receptor 1 and furin activates hepcidin. Hepatology 57: 2514–2524 [DOI] [PubMed] [Google Scholar]
- Gutwein P, Mechtersheimer S, Riedle S, Stoeck A, Gast D, Joumaa S, Zentgraf H, Fogel M, Altevogt DP (2003) ADAM10‐mediated cleavage of L1 adhesion molecule at the cell surface and in released membrane vesicles. FASEB J 17: 292–294 [DOI] [PubMed] [Google Scholar]
- Haapasalo A, Kovacs DM (2011) The many substrates of presenilin/gamma‐secretase. J Alzheimers Dis 25: 3–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haass C, Hung AY, Schlossmacher MG, Oltersdorf T, Teplow DB, Selkoe DJ (1993) b‐Amyloid peptide and a 3‐kDa fragment are derived by distinct cellular mechanisms. J Biol Chem 268: 3021–3024 [PubMed] [Google Scholar]
- Haining EJ, Yang J, Bailey RL, Khan K, Collier R, Tsai S, Watson SP, Frampton J, Garcia P, Tomlinson MG (2012) The TspanC8 subgroup of tetraspanins interacts with A disintegrin and metalloprotease 10 (ADAM10) and regulates its maturation and cell surface expression. J Biol Chem 287: 39753–39765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann D, de Strooper B, Serneels L, Craessaerts K, Herreman A, Annaert W, Umans L, Lubke T, Lena Illert A, von Figura K, Saftig P (2002) The disintegrin/metalloprotease ADAM 10 is essential for Notch signalling but not for alpha‐secretase activity in fibroblasts. Hum Mol Genet 11: 2615–2624 [DOI] [PubMed] [Google Scholar]
- Hattori M, Osterfield M, Flanagan JG (2000) Regulated cleavage of a contact‐mediated axon repellent. Science 289: 1360–1365 [DOI] [PubMed] [Google Scholar]
- Hayashida K, Bartlett AH, Chen Y, Park PW (2010) Molecular and cellular mechanisms of ectodomain shedding. Anat Rec 293: 925–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert SS, Horre K, Nicolai L, Papadopoulou AS, Mandemakers W, Silahtaroglu AN, Kauppinen S, Delacourte A, De Strooper B (2008) Loss of microRNA cluster miR‐29a/b‐1 in sporadic Alzheimer's disease correlates with increased BACE1/beta‐secretase expression. Proc Natl Acad Sci USA 105: 6415–6420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemming ML, Elias JE, Gygi SP, Selkoe DJ (2009) Identification of beta‐secretase (BACE1) substrates using quantitative proteomics. PLoS ONE 4: e8477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikita A, Yana I, Wakeyama H, Nakamura M, Kadono Y, Oshima Y, Nakamura K, Seiki M, Tanaka S (2006) Negative regulation of osteoclastogenesis by ectodomain shedding of receptor activator of NF‐kappaB ligand. J Biol Chem 281: 36846–36855 [DOI] [PubMed] [Google Scholar]
- Hitt B, Riordan SM, Kukreja L, Eimer WA, Rajapaksha TW, Vassar R (2012) beta‐Site amyloid precursor protein (APP)‐cleaving enzyme 1 (BACE1)‐deficient mice exhibit a close homolog of L1 (CHL1) loss‐of‐function phenotype involving axon guidance defects. J Biol Chem 287: 38408–38425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes O, Paturi S, Ye W, Wolfe MS, Selkoe DJ (2012) Effects of membrane lipids on the activity and processivity of purified gamma‐secretase. Biochemistry 51: 3565–3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong L, Koelsch G, Lin X, Wu S, Terzyan S, Ghosh AK, Zhang XC, Tang J (2000) Structure of the protease domain of memapsin 2 (beta‐secretase) complexed with inhibitor. Science 290: 150–153 [DOI] [PubMed] [Google Scholar]
- Horiuchi K, Kimura T, Miyamoto T, Takaishi H, Okada Y, Toyama Y, Blobel CP (2007) Cutting edge: TNF‐alpha‐converting enzyme (TACE/ADAM17) inactivation in mouse myeloid cells prevents lethality from endotoxin shock. J Immunol 179: 2686–2689 [DOI] [PubMed] [Google Scholar]
- Hsu FF, Yeh CT, Sun YJ, Chiang MT, Lan WM, Li FA, Lee WH, Chau LY (2015) Signal peptide peptidase‐mediated nuclear localization of heme oxygenase‐1 promotes cancer cell proliferation and invasion independent of its enzymatic activity. Oncogene 34: 2360–2370 [DOI] [PubMed] [Google Scholar]
- Hu X, Hicks CW, He W, Wong P, Macklin WB, Trapp BD, Yan R (2006) Bace1 modulates myelination in the central and peripheral nervous system. Nat Neurosci 9: 1520–1525 [DOI] [PubMed] [Google Scholar]
- Huse JT, Pijak DS, Leslie GJ, Lee VM, Doms RW (2000) Maturation and endosomal targeting of beta‐site amyloid precursor protein‐cleaving enzyme. The Alzheimer's disease beta‐secretase. J Biol Chem 275: 33729–33737 [DOI] [PubMed] [Google Scholar]
- Hussain I, Powell D, Howlett DR, Tew DG, Meek TD, Chapman C, Gloger IS, Murphy KE, Southan CD, Ryan DM, Smith TS, Simmons DL, Walsh FS, Dingwall C, Christie G (1999) Identification of a novel aspartic protease (Asp 2) as beta‐secretase. Mol Cell Neurosci 14: 419–427 [DOI] [PubMed] [Google Scholar]
- Hussain I, Hawkins J, Shikotra A, Riddell DR, Faller A, Dingwall C (2003) Characterization of the ectodomain shedding of the beta‐site amyloid precursor protein‐cleaving enzyme 1 (BACE1). J Biol Chem 278: 36264–36268 [DOI] [PubMed] [Google Scholar]
- Issuree PD, Maretzky T, McIlwain DR, Monette S, Qing X, Lang PA, Swendeman SL, Park‐Min KH, Binder N, Kalliolias GD, Yarilina A, Horiuchi K, Ivashkiv LB, Mak TW, Salmon JE, Blobel CP (2013) iRHOM2 is a critical pathogenic mediator of inflammatory arthritis. J Clin Invest 123: 928–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y (2015) Membrane‐type matrix metalloproteinases: their functions and regulations. Matrix Biol 44–46: 207–223 [DOI] [PubMed] [Google Scholar]
- Jackle F, Schmidt F, Wichert R, Arnold P, Prox J, Mangold M, Ohler A, Pietrzik CU, Koudelka T, Tholey A, Gutschow M, Stirnberg M, Becker‐Pauly C (2015) Metalloprotease meprin beta is activated by transmembrane serine protease matriptase‐2 at the cell surface thereby enhancing APP shedding. Biochem J 470: 91–103 [DOI] [PubMed] [Google Scholar]
- Janes PW, Saha N, Barton WA, Kolev MV, Wimmer‐Kleikamp SH, Nievergall E, Blobel CP, Himanen JP, Lackmann M, Nikolov DB (2005) Adam meets Eph: an ADAM substrate recognition module acts as a molecular switch for ephrin cleavage in trans. Cell 123: 291–304 [DOI] [PubMed] [Google Scholar]
- Jefferson T, Causevic M, auf dem Keller U, Schilling O, Isbert S, Geyer R, Maier W, Tschickardt S, Jumpertz T, Weggen S, Bond JS, Overall CM, Pietrzik CU, Becker‐Pauly C (2011) Metalloprotease meprin beta generates nontoxic N‐terminal amyloid precursor protein fragments in vivo . J Biol Chem 286: 27741–27750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson T, Auf dem Keller U, Bellac C, Metz VV, Broder C, Hedrich J, Ohler A, Maier W, Magdolen V, Sterchi E, Bond JS, Jayakumar A, Traupe H, Chalaris A, Rose‐John S, Pietrzik CU, Postina R, Overall CM, Becker‐Pauly C (2013) The substrate degradome of meprin metalloproteases reveals an unexpected proteolytic link between meprin beta and ADAM10. Cell Mol Life Sci 70: 309–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson N, Brezinova J, Stephens E, Burbridge E, Freeman M, Adrain C, Strisovsky K (2017) Quantitative proteomics screen identifies a substrate repertoire of rhomboid protease RHBDL2 in human cells and implicates it in epithelial homeostasis. Sci Rep 7: 7283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorissen E, Prox J, Bernreuther C, Weber S, Schwanbeck R, Serneels L, Snellinx A, Craessaerts K, Thathiah A, Tesseur I, Bartsch U, Weskamp G, Blobel CP, Glatzel M, De Strooper B, Saftig P (2010) The disintegrin/metalloproteinase ADAM10 is essential for the establishment of the brain cortex. J Neurosci 30: 4833–4844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajita M, Itoh Y, Chiba T, Mori H, Okada A, Kinoh H, Seiki M (2001) Membrane‐type 1 matrix metalloproteinase cleaves CD44 and promotes cell migration. J Cell Biol 153: 893–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalvodova L, Kahya N, Schwille P, Ehehalt R, Verkade P, Drechsel D, Simons K (2005) Lipids as modulators of proteolytic activity of BACE: involvement of cholesterol, glycosphingolipids, and anionic phospholipids in vitro . J Biol Chem 280: 36815–36823 [DOI] [PubMed] [Google Scholar]
- Kandalepas PC, Sadleir KR, Eimer WA, Zhao J, Nicholson DA, Vassar R (2013) The Alzheimer's beta‐secretase BACE1 localizes to normal presynaptic terminals and to dystrophic presynaptic terminals surrounding amyloid plaques. Acta Neuropathol 126: 329–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapeller M, Gal‐Oz R, Grover NB, Doljanski F (1973) Natural shedding of carbohydrate‐containing macromolecules from cell surfaces. Exp Cell Res 79: 152–158 [PubMed] [Google Scholar]
- Kawahara R, Lima RN, Domingues RR, Pauletti BA, Meirelles GV, Assis M, Figueira AC, Paes Leme AF (2014) Deciphering the role of the ADAM17‐dependent secretome in cell signaling. J Proteome Res 13: 2080–2093 [DOI] [PubMed] [Google Scholar]
- Kim MS, Pinto SM, Getnet D, Nirujogi RS, Manda SS, Chaerkady R, Madugundu AK, Kelkar DS, Isserlin R, Jain S, Thomas JK, Muthusamy B, Leal‐Rojas P, Kumar P, Sahasrabuddhe NA, Balakrishnan L, Advani J, George B, Renuse S, Selvan LD et al (2014) A draft map of the human proteome. Nature 509: 575–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimberly WT, LaVoie MJ, Ostaszewski BL, Ye W, Wolfe MS, Selkoe DJ (2003) Gamma‐secretase is a membrane protein complex comprised of presenilin, nicastrin, Aph‐1, and Pen‐2. Proc Natl Acad Sci USA 100: 6382–6387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitazume S, Tachida Y, Oka R, Shirotani K, Saido TC, Hashimoto Y (2001) Alzheimer's beta‐secretase, beta‐site amyloid precursor protein‐ cleaving enzyme, is responsible for cleavage secretion of a Golgi‐ resident sialyltransferase. Proc Natl Acad Sci USA 98: 13554–13559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kizuka Y, Kitazume S, Fujinawa R, Saito T, Iwata N, Saido TC, Nakano M, Yamaguchi Y, Hashimoto Y, Staufenbiel M, Hatsuta H, Murayama S, Manya H, Endo T, Taniguchi N (2015) An aberrant sugar modification of BACE1 blocks its lysosomal targeting in Alzheimer's disease. EMBO Mol Med 7: 175–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleifeld O, Doucet A, auf dem Keller U, Prudova A, Schilling O, Kainthan RK, Starr AE, Foster LJ, Kizhakkedathu JN, Overall CM (2010) Isotopic labeling of terminal amines in complex samples identifies protein N‐termini and protease cleavage products. Nat Biotechnol 28: 281–288 [DOI] [PubMed] [Google Scholar]
- Klein T, Bischoff R (2011) Physiology and pathophysiology of matrix metalloproteases. Amino Acids 41: 271–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo EH, Squazzo SL (1994) Evidence that production and release of amyloid beta‐protein involves the endocytic pathway. J Biol Chem 269: 17386–17389 [PubMed] [Google Scholar]
- Kuhn PH, Wang H, Dislich B, Colombo A, Zeitschel U, Ellwart JW, Kremmer E, Rossner S, Lichtenthaler SF (2010) ADAM10 is the physiologically relevant, constitutive alpha‐secretase of the amyloid precursor protein in primary neurons. EMBO J 29: 3020–3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn PH, Koroniak K, Hogl S, Colombo A, Zeitschel U, Willem M, Volbracht C, Schepers U, Imhof A, Hoffmeister A, Haass C, Rossner S, Brase S, Lichtenthaler SF (2012) Secretome protein enrichment identifies physiological BACE1 protease substrates in neurons. EMBO J 31: 3157–3168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn PH, Voss M, Haug‐Kroper M, Schroder B, Schepers U, Brase S, Haass C, Lichtenthaler SF, Fluhrer R (2015) Secretome analysis identifies novel signal Peptide peptidase‐like 3 (sppl3) substrates and reveals a role of sppl3 in multiple Golgi glycosylation pathways. Mol Cell Proteomics 14: 1584–1598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn PH, Colombo AV, Schusser B, Dreymueller D, Wetzel S, Schepers U, Herber J, Ludwig A, Kremmer E, Montag D, Muller U, Schweizer M, Saftig P, Brase S, Lichtenthaler SF (2016) Systematic substrate identification indicates a central role for the metalloprotease ADAM10 in axon targeting and synapse function. Elife 5: e12748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Marca R, Cerri F, Horiuchi K, Bachi A, Feltri ML, Wrabetz L, Blobel CP, Quattrini A, Salzer JL, Taveggia C (2011) TACE (ADAM17) inhibits Schwann cell myelination. Nat Neurosci 14: 857–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagares D, Ghassemi‐Kakroodi P, Tremblay C, Santos A, Probst CK, Franklin A, Santos DM, Grasberger P, Ahluwalia N, Montesi SB, Shea BS, Black KE, Knipe R, Blati M, Baron M, Wu B, Fahmi H, Gandhi R, Pardo A, Selman M et al (2017) ADAM10‐mediated ephrin‐B2 shedding promotes myofibroblast activation and organ fibrosis. Nat Med 23: 1405–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammich S, Kojro E, Postina R, Gilbert S, Pfeiffer R, Jasionowski M, Haass C, Fahrenholz F (1999) Constitutive and regulated alpha‐secretase cleavage of Alzheimer's amyloid precursor protein by a disintegrin metalloprotease. Proc Natl Acad Sci USA 96: 3922–3927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammich S, Schobel S, Zimmer AK, Lichtenthaler SF, Haass C (2004) Expression of the Alzheimer protease BACE1 is suppressed via its 5′‐untranslated region. EMBO Rep 5: 620–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent SA, Hoffmann FS, Kuhn PH, Cheng Q, Chu Y, Schmidt‐Supprian M, Hauck SM, Schuh E, Krumbholz M, Rubsamen H, Wanngren J, Khademi M, Olsson T, Alexander T, Hiepe F, Pfister HW, Weber F, Jenne D, Wekerle H, Hohlfeld R et al (2015) gamma‐Secretase directly sheds the survival receptor BCMA from plasma cells. Nat Commun 6: 7333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gall SM, Maretzky T, Issuree PD, Niu XD, Reiss K, Saftig P, Khokha R, Lundell D, Blobel CP (2010) ADAM17 is regulated by a rapid and reversible mechanism that controls access to its catalytic site. J Cell Sci 123: 3913–3922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JR, Urban S, Garvey CF, Freeman M (2001) Regulated intracellular ligand transport and proteolysis control EGF signal activation in Drosophila . Cell 107: 161–171 [DOI] [PubMed] [Google Scholar]
- Lee SO, Cho K, Cho S, Kim I, Oh C, Ahn K (2010) Protein disulphide isomerase is required for signal peptide peptidase‐mediated protein degradation. EMBO J 29: 363–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemberg MK, Martoglio B (2002) Requirements for signal peptide peptidase‐catalyzed intramembrane proteolysis. Mol Cell 10: 735–744 [DOI] [PubMed] [Google Scholar]
- Lemberg MK (2013) Sampling the membrane: function of rhomboid‐family proteins. Trends Cell Biol 23: 210–217 [DOI] [PubMed] [Google Scholar]
- Levytskyy RM, Bohovych I, Khalimonchuk O (2017) Metalloproteases of the inner mitochondrial membrane. Biochemistry 56: 4737–4746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Wang B, Wang Z, Guo Q, Tabuchi K, Hammer RE, Sudhof TC, Zheng H (2010) Soluble amyloid precursor protein (APP) regulates transthyretin and Klotho gene expression without rescuing the essential function of APP. Proc Natl Acad Sci USA 107: 17362–17367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Dang S, Yan C, Gong X, Wang J, Shi Y (2013) Structure of a presenilin family intramembrane aspartate protease. Nature 493: 56–61 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler SF, Haass C, Steiner H (2011) Regulated intramembrane proteolysis–lessons from amyloid precursor protein processing. J Neurochem 117: 779–796 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler SF (2012) Alpha‐secretase cleavage of the amyloid precursor protein: proteolysis regulated by signaling pathways and protein trafficking. Curr Alzheimer Res 9: 165–177 [DOI] [PubMed] [Google Scholar]
- Lin X, Koelsch G, Wu S, Downs D, Dashti A, Tang J (2000) Human aspartic protease memapsin 2 cleaves the beta‐secretase site of beta‐amyloid precursor protein. Proc Natl Acad Sci USA 97: 1456–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohi O, Urban S, Freeman M (2004) Diverse substrate recognition mechanisms for rhomboids; thrombomodulin is cleaved by Mammalian rhomboids. Curr Biol 14: 236–241 [DOI] [PubMed] [Google Scholar]
- Lopez‐Otin C, Bond JS (2008) Proteases: multifunctional enzymes in life and disease. J Biol Chem 283: 30433–30437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez‐Perez E, Seidah NG, Checler F (1999) Proprotein convertase activity contributes to the processing of the Alzheimer's beta‐amyloid precursor protein in human cells: evidence for a role of the prohormone convertase PC7 in the constitutive alpha‐secretase pathway. J Neurochem 73: 2056–2062 [PubMed] [Google Scholar]
- Lopez‐Perez E, Zhang Y, Frank SJ, Creemers J, Seidah N, Checler F (2001) Constitutive alpha‐secretase cleavage of the beta‐amyloid precursor protein in the furin‐deficient LoVo cell line: involvement of the pro‐hormone convertase 7 and the disintegrin metalloprotease ADAM10. J Neurochem 76: 1532–1539 [DOI] [PubMed] [Google Scholar]
- Loureiro J, Lilley BN, Spooner E, Noriega V, Tortorella D, Ploegh HL (2006) Signal peptide peptidase is required for dislocation from the endoplasmic reticulum. Nature 441: 894–897 [DOI] [PubMed] [Google Scholar]
- Ludwig A, Hundhausen C, Lambert MH, Broadway N, Andrews RC, Bickett DM, Leesnitzer MA, Becherer JD (2005) Metalloproteinase inhibitors for the disintegrin‐like metalloproteinases ADAM10 and ADAM17 that differentially block constitutive and phorbol ester‐inducible shedding of cell surface molecules. Comb Chem High Throughput Screen 8: 161–171 [DOI] [PubMed] [Google Scholar]
- Marcello E, Gardoni F, Mauceri D, Romorini S, Jeromin A, Epis R, Borroni B, Cattabeni F, Sala C, Padovani A, Di Luca M (2007) Synapse‐associated protein‐97 mediates alpha‐secretase ADAM10 trafficking and promotes its activity. J Neurosci 27: 1682–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maretzky T, Evers A, Zhou W, Swendeman SL, Wong PM, Rafii S, Reiss K, Blobel CP (2011) Migration of growth factor‐stimulated epithelial and endothelial cells depends on EGFR transactivation by ADAM17. Nat Commun 2: 229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maretzky T, McIlwain DR, Issuree PD, Li X, Malapeira J, Amin S, Lang PA, Mak TW, Blobel CP (2013) iRhom2 controls the substrate selectivity of stimulated ADAM17‐dependent ectodomain shedding. Proc Natl Acad Sci USA 110: 11433–11438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschner K, Kollmann K, Schweizer M, Braulke T, Pohl S (2011) A key enzyme in the biogenesis of lysosomes is a protease that regulates cholesterol metabolism. Science 333: 87–90 [DOI] [PubMed] [Google Scholar]
- Martin L, Fluhrer R, Reiss K, Kremmer E, Saftig P, Haass C (2008) Regulated intramembrane proteolysis of Bri2 (Itm2b) by ADAM10 and SPPL2a/SPPL2b. J Biol Chem 283: 1644–1652 [DOI] [PubMed] [Google Scholar]
- Martin L, Fluhrer R, Haass C (2009) Substrate requirements for SPPL2b‐dependent regulated intramembrane proteolysis. J Biol Chem 284: 5662–5670 [DOI] [PubMed] [Google Scholar]
- Massague J, Pandiella A (1993) Membrane‐anchored growth factors. Annu Rev Biochem 62: 515–541 [DOI] [PubMed] [Google Scholar]
- Mathews JA, Ford J, Norton S, Kang D, Dellinger A, Gibb DR, Ford AQ, Massay H, Kepley CL, Scherle P, Keegan AD, Conrad DH (2011) A potential new target for asthma therapy: a disintegrin and metalloprotease 10 (ADAM10) involvement in murine experimental asthma. Allergy 66: 1193–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews AL, Noy PJ, Reyat JS, Tomlinson MG (2017) Regulation of A disintegrin and metalloproteinase (ADAM) family sheddases ADAM10 and ADAM17: the emerging role of tetraspanins and rhomboids. Platelets 28: 333–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- May PC, Dean RA, Lowe SL, Martenyi F, Sheehan SM, Boggs LN, Monk SA, Mathes BM, Mergott DJ, Watson BM, Stout SL, Timm DE, Smith Labell E, Gonzales CR, Nakano M, Jhee SS, Yen M, Ereshefsky L, Lindstrom TD, Calligaro DO et al (2011) Robust central reduction of amyloid‐beta in humans with an orally available, non‐peptidic beta‐secretase inhibitor. J Neurosci 31: 16507–16516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIlwain DR, Lang PA, Maretzky T, Hamada K, Ohishi K, Maney SK, Berger T, Murthy A, Duncan G, Xu HC, Lang KS, Haussinger D, Wakeham A, Itie‐Youten A, Khokha R, Ohashi PS, Blobel CP, Mak TW (2012) iRhom2 regulation of TACE controls TNF‐mediated protection against Listeria and responses to LPS. Science 335: 229–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meckler X, Checler F (2016) Presenilin 1 and presenilin 2 target gamma‐secretase complexes to distinct cellular compartments. J Biol Chem 291: 12821–12837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner F, Scheltema RA, Mollenkopf HJ, Mann M (2013) Direct proteomic quantification of the secretome of activated immune cells. Science 340: 475–478 [DOI] [PubMed] [Google Scholar]
- Mihailovich M, Thermann R, Grohovaz F, Hentze MW, Zacchetti D (2007) Complex translational regulation of BACE1 involves upstream AUGs and stimulatory elements within the 5′ untranslated region. Nucleic Acids Res 35: 2975–2985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitterreiter S, Page RM, Kamp F, Hopson J, Winkler E, Ha HR, Hamid R, Herms J, Mayer TU, Nelson DJ, Steiner H, Stahl T, Zeitschel U, Rossner S, Haass C, Lichtenthaler SF (2010) Bepridil and amiodarone simultaneously target the Alzheimer's disease beta‐ and gamma‐secretase via distinct mechanisms. J Neurosci 30: 8974–8983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moin SM, Urban S (2012) Membrane immersion allows rhomboid proteases to achieve specificity by reading transmembrane segment dynamics. Elife 1: e00173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss ML, Jin SL, Milla ME, Bickett DM, Burkhart W, Carter HL, Chen WJ, Clay WC, Didsbury JR, Hassler D, Hoffman CR, Kost TA, Lambert MH, Leesnitzer MA, McCauley P, McGeehan G, Mitchell J, Moyer M, Pahel G, Rocque W et al (1997) Cloning of a disintegrin metalloproteinase that processes precursor tumour‐necrosis factor‐alpha. Nature 385: 733–736 [DOI] [PubMed] [Google Scholar]
- Muller SA, Scilabra SD, Lichtenthaler SF (2016) Proteomic substrate identification for membrane proteases in the brain. Front Mol Neurosci 9: 96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy G (2008) The ADAMs: signalling scissors in the tumour microenvironment. Nat Rev Cancer 8: 929–941 [DOI] [PubMed] [Google Scholar]
- Murray B, Pearson CS, Aranjo A, Cherupalla D, Belfort G (2016) Mechanism of four de novo designed antimicrobial peptides. J Biol Chem 291: 25706–25715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naus S, Richter M, Wildeboer D, Moss M, Schachner M, Bartsch JW (2004) Ectodomain shedding of the neural recognition molecule CHL1 by the metalloprotease‐disintegrin ADAM8 promotes neurite outgrowth and suppresses neuronal cell death. J Biol Chem 279: 16083–16090 [DOI] [PubMed] [Google Scholar]
- O'Connor T, Sadleir KR, Maus E, Velliquette RA, Zhao J, Cole SL, Eimer WA, Hitt B, Bembinster LA, Lammich S, Lichtenthaler SF, Hebert SS, De Strooper B, Haass C, Bennett DA, Vassar R (2008) Phosphorylation of the translation initiation factor eIF2alpha increases BACE1 levels and promotes amyloidogenesis. Neuron 60: 988–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D, Rubin GM (1997) Kuzbanian controls proteolytic processing of Notch and mediates lateral inhibition during Drosophila and vertebrate neurogenesis. Cell 90: 271–280 [DOI] [PubMed] [Google Scholar]
- Paschkowsky S, Hamze M, Oestereich F, Munter LM (2016) Alternative processing of the amyloid precursor protein family by rhomboid protease RHBDL4. J Biol Chem 291: 21903–21912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peduto L, Reuter VE, Shaffer DR, Scher HI, Blobel CP (2005) Critical function for ADAM9 in mouse prostate cancer. Cancer Res 65: 9312–9319 [DOI] [PubMed] [Google Scholar]
- Peixoto RT, Kunz PA, Kwon H, Mabb AM, Sabatini BL, Philpot BD, Ehlers MD (2012) Transsynaptic signaling by activity‐dependent cleavage of neuroligin‐1. Neuron 76: 396–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschon JJ, Slack JL, Reddy P, Stocking KL, Sunnarborg S, Lee DC, Russell WE, Castner BJ, Johnson RS, Fitzner JN, Boyce RW, Nelson N, Kozlosky CJ, Wolfson MF, Rauch CT, Cerretti DP, Paxton RJ, March CJ, Black RA (1998) An essential role for ectodomain shedding in mammalian development. Science 282: 1281–1284 [DOI] [PubMed] [Google Scholar]
- Pigoni M, Wanngren J, Kuhn PH, Munro KM, Gunnersen JM, Takeshima H, Feederle R, Voytyuk I, De Strooper B, Levasseur MD, Hrupka BJ, Muller SA, Lichtenthaler SF (2016) Seizure protein 6 and its homolog seizure 6‐like protein are physiological substrates of BACE1 in neurons. Mol Neurodegener 11: 67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Hutton M, Nyborg A, Baker M, Jansen K, Golde TE (2002) Identification of a novel family of presenilin homologues. Hum Mol Genet 11: 1037–1044 [DOI] [PubMed] [Google Scholar]
- Porlan E, Marti‐Prado B, Morante‐Redolat JM, Consiglio A, Delgado AC, Kypta R, Lopez‐Otin C, Kirstein M, Farinas I (2014) MT5‐MMP regulates adult neural stem cell functional quiescence through the cleavage of N‐cadherin. Nat Cell Biol 16: 629–638 [DOI] [PubMed] [Google Scholar]
- Postina R, Schroeder A, Dewachter I, Bohl J, Schmitt U, Kojro E, Prinzen C, Endres K, Hiemke C, Blessing M, Flamez P, Dequenne A, Godaux E, van Leuven F, Fahrenholz F (2004) A disintegrin‐metalloproteinase prevents amyloid plaque formation and hippocampal defects in an Alzheimer disease mouse model. J Clin Invest 113: 1456–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prenzel N, Zwick E, Daub H, Leserer M, Abraham R, Wallasch C, Ullrich A (1999) EGF receptor transactivation by G‐protein‐coupled receptors requires metalloproteinase cleavage of proHB‐EGF. Nature 402: 884–888 [DOI] [PubMed] [Google Scholar]
- Prox J, Willenbrock M, Weber S, Lehmann T, Schmidt‐Arras D, Schwanbeck R, Saftig P, Schwake M (2012) Tetraspanin15 regulates cellular trafficking and activity of the ectodomain sheddase ADAM10. Cell Mol Life Sci 69: 2919–2932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prox J, Bernreuther C, Altmeppen H, Grendel J, Glatzel M, D'Hooge R, Stroobants S, Ahmed T, Balschun D, Willem M, Lammich S, Isbrandt D, Schweizer M, Horre K, De Strooper B, Saftig P (2013) Postnatal disruption of the disintegrin/metalloproteinase ADAM10 in brain causes epileptic seizures, learning deficits, altered spine morphology, and defective synaptic functions. J Neurosci 33: 12915–12928, 12928a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruessmeyer J, Ludwig A (2009) The good, the bad and the ugly substrates for ADAM10 and ADAM17 in brain pathology, inflammation and cancer. Semin Cell Dev Biol 20: 164–174 [DOI] [PubMed] [Google Scholar]
- Qing X, Chinenov Y, Redecha P, Madaio M, Roelofs JJ, Farber G, Issuree PD, Donlin L, McIlwain DR, Mak TW, Blobel CP, Salmon JE (2018) iRhom2 promotes lupus nephritis through TNF‐alpha and EGFR signaling. J Clin Invest 128: 1397–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajapaksha TW, Eimer WA, Bozza TC, Vassar R (2011) The Alzheimer's beta‐secretase enzyme BACE1 is required for accurate axon guidance of olfactory sensory neurons and normal glomerulus formation in the olfactory bulb. Mol Neurodegener 6: 88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendran L, Schneider A, Schlechtingen G, Weidlich S, Ries J, Braxmeier T, Schwille P, Schulz JB, Schroeder C, Simons M, Jennings G, Knolker HJ, Simons K (2008) Efficient inhibition of the Alzheimer's disease beta‐secretase by membrane targeting. Science 320: 520–523 [DOI] [PubMed] [Google Scholar]
- Rawson RB, Zelenski NG, Nijhawan D, Ye J, Sakai J, Hasan MT, Chang TY, Brown MS, Goldstein JL (1997) Complementation cloning of S2P, a gene encoding a putative metalloprotease required for intramembrane cleavage of SREBPs. Mol Cell 1: 47–57 [DOI] [PubMed] [Google Scholar]
- Reiss K, Maretzky T, Ludwig A, Tousseyn T, de Strooper B, Hartmann D, Saftig P (2005) ADAM10 cleavage of N‐cadherin and regulation of cell‐cell adhesion and beta‐catenin nuclear signalling. EMBO J 24: 742–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ring S, Weyer SW, Kilian SB, Waldron E, Pietrzik CU, Filippov MA, Herms J, Buchholz C, Eckman CB, Korte M, Wolfer DP, Muller UC (2007) The secreted beta‐amyloid precursor protein ectodomain APPs alpha is sufficient to rescue the anatomical, behavioral, and electrophysiological abnormalities of APP‐deficient mice. J Neurosci 27: 7817–7826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochin L, Hurbain I, Serneels L, Fort C, Watt B, Leblanc P, Marks MS, De Strooper B, Raposo G, van Niel G (2013) BACE2 processes PMEL to form the melanosome amyloid matrix in pigment cells. Proc Natl Acad Sci USA 110: 10658–10663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers GW Jr, Edelman GM, Mauro VP (2004) Differential utilization of upstream AUGs in the beta‐secretase mRNA suggests that a shunting mechanism regulates translation. Proc Natl Acad Sci USA 101: 2794–2799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romagnoli M, Mineva ND, Polmear M, Conrad C, Srinivasan S, Loussouarn D, Barille‐Nion S, Georgakoudi I, Dagg A, McDermott EW, Duffy MJ, McGowan PM, Schlomann U, Parsons M, Bartsch JW, Sonenshein GE (2014) ADAM8 expression in invasive breast cancer promotes tumor dissemination and metastasis. EMBO Mol Med 6: 278–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose‐John S (2013) ADAM17, shedding, TACE as therapeutic targets. Pharmacol Res 71: 19–22 [DOI] [PubMed] [Google Scholar]
- Rose‐John S (2017) The soluble interleukin 6 receptor: advanced therapeutic options in inflammation. Clin Pharmacol Ther 102: 591–598 [DOI] [PubMed] [Google Scholar]
- Saftig P, Lichtenthaler SF (2015) The alpha secretase ADAM10: a metalloprotease with multiple functions in the brain. Prog Neurobiol 135: 1–20 [DOI] [PubMed] [Google Scholar]
- Sahin U, Weskamp G, Kelly K, Zhou HM, Higashiyama S, Peschon J, Hartmann D, Saftig P, Blobel CP (2004) Distinct roles for ADAM10 and ADAM17 in ectodomain shedding of six EGFR ligands. J Cell Biol 164: 769–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai J, Duncan EA, Rawson RB, Hua X, Brown MS, Goldstein JL (1996) Sterol‐regulated release of SREBP‐2 from cell membranes requires two sequential cleavages, one within a transmembrane segment. Cell 85: 1037–1046 [DOI] [PubMed] [Google Scholar]
- Sakai J, Rawson RB, Espenshade PJ, Cheng D, Seegmiller AC, Goldstein JL, Brown MS (1998) Molecular identification of the sterol‐regulated luminal protease that cleaves SREBPs and controls lipid composition of animal cells. Mol Cell 2: 505–514 [DOI] [PubMed] [Google Scholar]
- Sannerud R, Esselens C, Ejsmont P, Mattera R, Rochin L, Tharkeshwar AK, De Baets G, De Wever V, Habets R, Baert V, Vermeire W, Michiels C, Groot AJ, Wouters R, Dillen K, Vints K, Baatsen P, Munck S, Derua R, Waelkens E et al (2016) Restricted location of PSEN2/gamma‐secretase determines substrate specificity and generates an intracellular abeta pool. Cell 166: 193–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz RL, Ferraro GB, Kacervosky J, Salesse C, Gowing E, Hua L, Rambaldi I, Beaubien F, Holmbeck K, Cloutier JF, Levesque M, Murai K, Fournier AE (2018) MT3‐MMP promotes excitatory synapse formation by promoting nogo‐66 receptor ectodomain shedding. J Neurosci 38: 518–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato C, Zhao G, Ilagan MX (2012) An overview of notch signaling in adult tissue renewal and maintenance. Curr Alzheimer Res 9: 227–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz G, Dobberstein B (1996) Common principles of protein translocation across membranes. Science 271: 1519–1526 [DOI] [PubMed] [Google Scholar]
- Schauenburg L, Liebsch F, Eravci M, Mayer MC, Weise C, Multhaup G (2018) APLP1 is endoproteolytically cleaved by gamma‐secretase without previous ectodomain shedding. Sci Rep 8: 1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlomann U, Koller G, Conrad C, Ferdous T, Golfi P, Garcia AM, Hofling S, Parsons M, Costa P, Soper R, Bossard M, Hagemann T, Roshani R, Sewald N, Ketchem RR, Moss ML, Rasmussen FH, Miller MA, Lauffenburger DA, Tuveson DA et al (2015) ADAM8 as a drug target in pancreatic cancer. Nat Commun 6: 6175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlondorff J, Becherer JD, Blobel CP (2000) Intracellular maturation and localization of the tumour necrosis factor alpha convertase (TACE). Biochem J 347(Pt 1): 131–138 [PMC free article] [PubMed] [Google Scholar]
- Schmechel A, Strauss M, Schlicksupp A, Pipkorn R, Haass C, Bayer TA, Multhaup G (2004) Human BACE forms dimers and colocalizes with APP. J Biol Chem 279: 39710–39717 [DOI] [PubMed] [Google Scholar]
- Schmidt S, Schumacher N, Schwarz J, Tangermann S, Kenner L, Schlederer M, Sibilia M, Linder M, Altendorf‐Hofmann A, Knosel T, Gruber ES, Oberhuber G, Bolik J, Rehman A, Sinha A, Lokau J, Arnold P, Cabron AS, Zunke F, Becker‐Pauly C et al (2018) ADAM17 is required for EGF‐R‐induced intestinal tumors via IL‐6 trans‐signaling. J Exp Med 215: 1205–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneppenheim J, Dressel R, Huttl S, Lullmann‐Rauch R, Engelke M, Dittmann K, Wienands J, Eskelinen EL, Hermans‐Borgmeyer I, Fluhrer R, Saftig P, Schroder B (2013) The intramembrane protease SPPL2a promotes B cell development and controls endosomal traffic by cleavage of the invariant chain. J Exp Med 210: 41–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schobel S, Neumann S, Hertweck M, Dislich B, Kuhn PH, Kremmer E, Seed B, Baumeister R, Haass C, Lichtenthaler SF (2008) A novel sorting nexin modulates endocytic trafficking and alpha‐secretase cleavage of the amyloid precursor protein. J Biol Chem 283: 14257–14268 [DOI] [PubMed] [Google Scholar]
- Schrul B, Kapp K, Sinning I, Dobberstein B (2010) Signal peptide peptidase (SPP) assembles with substrates and misfolded membrane proteins into distinct oligomeric complexes. Biochem J 427: 523–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seegar TCM, Killingsworth LB, Saha N, Meyer PA, Patra D, Zimmerman B, Janes PW, Rubinstein E, Nikolov DB, Skiniotis G, Kruse AC, Blacklow SC (2017) Structural basis for regulated proteolysis by the alpha‐secretase ADAM10. Cell 171: 1638–1648.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidah NG, Prat A (2012) The biology and therapeutic targeting of the proprotein convertases. Nat Rev Drug Discov 11: 367–383 [DOI] [PubMed] [Google Scholar]
- Seidah NG, Abifadel M, Prost S, Boileau C, Prat A (2017) The proprotein convertases in hypercholesterolemia and cardiovascular diseases: emphasis on proprotein convertase subtilisin/kexin 9. Pharmacol Rev 69: 33–52 [DOI] [PubMed] [Google Scholar]
- Selkoe DJ (1990) Deciphering Alzheimer's disease: the amyloid precursor protein yields new clues. Science 248: 1058–1060 [DOI] [PubMed] [Google Scholar]
- Sherrington R, Rogaev EI, Liang Y, Rogaeva EA, Levesque G, Ikeda M, Chi H, Lin C, Li G, Holman K, Tsuda T, Mar L, Foncin JF, Bruni AC, Montesi MP, Sorbi S, Rainero I, Pinessi L, Nee L, Chumakov I et al (1995) Cloning of a gene bearing missense mutations in early‐onset familial Alzheimer's disease. Nature 375: 754–760 [DOI] [PubMed] [Google Scholar]
- Shimizu T, Oishi T, Omori A, Sugiura A, Hirota K, Aoyama H, Saito T, Sugaya T, Kon Y, Engel JD, Fukamizu A, Tanimoto K (2005) Identification of cis‐regulatory sequences in the human angiotensinogen gene by transgene coplacement and site‐specific recombination. Mol Cell Biol 25: 2938–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shitomi Y, Thogersen IB, Ito N, Leitinger B, Enghild JJ, Itoh Y (2015) ADAM10 controls collagen signaling and cell migration on collagen by shedding the ectodomain of discoidin domain receptor 1 (DDR1). Mol Biol Cell 26: 659–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S, Anderson JP, Barbour R, Basi GS, Caccavello R, Davis D, Doan M, Dovey HF, Frigon N, Hong J, Jacobson‐Croak K, Jewett N, Keim P, Knops J, Lieberburg I, Power M, Tan H, Tatsuno G, Tung J, Schenk D et al (1999) Purification and cloning of amyloid precursor protein beta‐secretase from human brain. Nature 402: 537–540 [DOI] [PubMed] [Google Scholar]
- Sisodia SS (1992) Beta‐amyloid precursor protein cleavage by a membrane‐bound protease. Proc Natl Acad Sci USA 89: 6075–6079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smulski CR, Kury P, Seidel LM, Staiger HS, Edinger AK, Willen L, Seidl M, Hess H, Salzer U, Rolink AG, Rizzi M, Schneider P, Eibel H (2017) BAFF‐ and TACI‐dependent processing of BAFFR by ADAM proteases regulates the survival of B cells. Cell Rep 18: 2189–2202 [DOI] [PubMed] [Google Scholar]
- Sobotic B, Vizovisek M, Vidmar R, Van Damme P, Gocheva V, Joyce JA, Gevaert K, Turk V, Turk B, Fonovic M (2015) Proteomic identification of cysteine cathepsin substrates shed from the surface of cancer cells. Mol Cell Proteomics 14: 2213–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer A, Kordowski F, Buch J, Maretzky T, Evers A, Andra J, Dusterhoft S, Michalek M, Lorenzen I, Somasundaram P, Tholey A, Sonnichsen FD, Kunzelmann K, Heinbockel L, Nehls C, Gutsmann T, Grotzinger J, Bhakdi S, Reiss K (2016) Phosphatidylserine exposure is required for ADAM17 sheddase function. Nat Commun 7: 11523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagg HR, Thomas M, van den Boomen D, Wiertz EJ, Drabkin HA, Gemmill RM, Lehner PJ (2009) The TRC8 E3 ligase ubiquitinates MHC class I molecules before dislocation from the ER. J Cell Biol 186: 685–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovic‐Barrett S, Dickson AS, Burr SP, Williamson JC, Lobb IT, van den Boomen DJ, Lehner PJ, Nathan JA (2018) MARCH6 and TRC8 facilitate the quality control of cytosolic and tail‐anchored proteins. EMBO Rep 19: e45603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strisovsky K, Sharpe HJ, Freeman M (2009) Sequence‐specific intramembrane proteolysis: identification of a recognition motif in rhomboid substrates. Mol Cell 36: 1048–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strisovsky K (2016) Why cells need intramembrane proteases ‐ a mechanistic perspective. FEBS J 283: 1837–1845 [DOI] [PubMed] [Google Scholar]
- Struhl G, Adachi A (2000) Requirements for presenilin‐dependent cleavage of notch and other transmembrane proteins. Mol Cell 6: 625–636 [DOI] [PubMed] [Google Scholar]
- Stutzer I, Selevsek N, Esterhazy D, Schmidt A, Aebersold R, Stoffel M (2013) Systematic proteomic analysis identifies beta‐site amyloid precursor protein cleaving enzyme 2 and 1 (BACE2 and BACE1) substrates in pancreatic beta‐cells. J Biol Chem 288: 10536–10547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh J, Choi SH, Romano DM, Gannon MA, Lesinski AN, Kim DY, Tanzi RE (2013) ADAM10 missense mutations potentiate beta‐amyloid accumulation by impairing prodomain chaperone function. Neuron 80: 385–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo R, Bugge TH (2011) Membrane‐anchored serine proteases in vertebrate cell and developmental biology. Annu Rev Cell Dev Biol 27: 213–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasugi N, Tomita T, Hayashi I, Tsuruoka M, Niimura M, Takahashi Y, Thinakaran G, Iwatsubo T (2003) The role of presenilin cofactors in the gamma‐secretase complex. Nature 422: 438–441 [DOI] [PubMed] [Google Scholar]
- Tam EM, Morrison CJ, Wu YI, Stack MS, Overall CM (2004) Membrane protease proteomics: isotope‐coded affinity tag MS identification of undescribed MT1‐matrix metalloproteinase substrates. Proc Natl Acad Sci USA 101: 6917–6922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe LM, List K (2017) The role of type II transmembrane serine protease‐mediated signaling in cancer. FEBS J 284: 1421–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesco G, Koh YH, Kang EL, Cameron AN, Das S, Sena‐Esteves M, Hiltunen M, Yang SH, Zhong Z, Shen Y, Simpkins JW, Tanzi RE (2007) Depletion of GGA3 stabilizes BACE and enhances beta‐secretase activity. Neuron 54: 721–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Tetering G, van Diest P, Verlaan I, van der Wall E, Kopan R, Vooijs M (2009) Metalloprotease ADAM10 is required for Notch1 site 2 cleavage. J Biol Chem 284: 31018–31027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tien WS, Chen JH, Wu KP (2017) SheddomeDB: the ectodomain shedding database for membrane‐bound shed markers. BMC Bioinformatics 18: 42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiyanont K, Wales TE, Aste‐Amezaga M, Aster JC, Engen JR, Blacklow SC (2011) Evidence for increased exposure of the Notch1 metalloprotease cleavage site upon conversion to an activated conformation. Structure 19: 546–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tousseyn T, Thathiah A, Jorissen E, Raemaekers T, Konietzko U, Reiss K, Maes E, Snellinx A, Serneels L, Nyabi O, Annaert W, Saftig P, Hartmann D, De Strooper B (2009) ADAM10, the rate‐limiting protease of regulated intramembrane proteolysis of Notch and other proteins, is processed by ADAMS‐9, ADAMS‐15, and the gamma‐secretase. J Biol Chem 284: 11738–11747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udalova I, Monaco C, Nanchahal J, Feldmann M (2016) Anti‐TNF therapy. Microbiol Spectr 4: MCHD‐0022‐2015 [DOI] [PubMed] [Google Scholar]
- Urban S, Lee JR, Freeman M (2001) Drosophila rhomboid‐1 defines a family of putative intramembrane serine proteases. Cell 107: 173–182 [DOI] [PubMed] [Google Scholar]
- Urban S, Freeman M (2003) Substrate specificity of rhomboid intramembrane proteases is governed by helix‐breaking residues in the substrate transmembrane domain. Mol Cell 11: 1425–1434 [DOI] [PubMed] [Google Scholar]
- Urban S, Wolfe MS (2005) Reconstitution of intramembrane proteolysis in vitro reveals that pure rhomboid is sufficient for catalysis and specificity. Proc Natl Acad Sci USA 102: 1883–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban S (2016) A guide to the rhomboid protein superfamily in development and disease. Semin Cell Dev Biol 60: 1–4 [DOI] [PubMed] [Google Scholar]
- Vassar R, Bennett BD, Babu‐Khan S, Kahn S, Mendiaz EA, Denis P, Teplow DB, Ross S, Amarante P, Loeloff R, Luo Y, Fisher S, Fuller J, Edenson S, Lile J, Jarosinski MA, Biere AL, Curran E, Burgess T, Louis JC et al (1999) Beta‐secretase cleavage of Alzheimer's amyloid precursor protein by the transmembrane aspartic protease BACE. Science 286: 735–741 [DOI] [PubMed] [Google Scholar]
- Vassar R, Kuhn PH, Haass C, Kennedy ME, Rajendran L, Wong PC, Lichtenthaler SF (2014) Function, therapeutic potential and cell biology of BACE proteases: current status and future prospects. J Neurochem 130: 4–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent B, Paitel E, Saftig P, Frobert Y, Hartmann D, De Strooper B, Grassi J, Lopez‐Perez E, Checler F (2001) The disintegrins ADAM10 and TACE contribute to the constitutive and phorbol ester‐regulated normal cleavage of the cellular prion protein. J Biol Chem 276: 37743–37746 [DOI] [PubMed] [Google Scholar]
- Voss M, Fukumori A, Kuhn PH, Kunzel U, Klier B, Grammer G, Haug‐Kroper M, Kremmer E, Lichtenthaler SF, Steiner H, Schroder B, Haass C, Fluhrer R (2012) Foamy virus envelope protein is a substrate for signal peptide peptidase‐like 3 (SPPL3). J Biol Chem 287: 43401–43409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss M, Kunzel U, Higel F, Kuhn PH, Colombo A, Fukumori A, Haug‐Kroper M, Klier B, Grammer G, Seidl A, Schroder B, Obst R, Steiner H, Lichtenthaler SF, Haass C, Fluhrer R (2014) Shedding of glycan‐modifying enzymes by signal peptide peptidase‐like 3 (SPPL3) regulates cellular N‐glycosylation. EMBO J 33: 2890–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Pei D (2001) Shedding of membrane type matrix metalloproteinase 5 by a furin‐type convertase: a potential mechanism for down‐regulation. J Biol Chem 276: 35953–35960 [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhang Y, Ha Y (2006) Crystal structure of a rhomboid family intramembrane protease. Nature 444: 179–180 [DOI] [PubMed] [Google Scholar]
- Wang WX, Rajeev BW, Stromberg AJ, Ren N, Tang G, Huang Q, Rigoutsos I, Nelson PT (2008) The expression of microRNA miR‐107 decreases early in Alzheimer's disease and may accelerate disease progression through regulation of beta‐site amyloid precursor protein‐cleaving enzyme 1. J Neurosci 28: 1213–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZH, Liu P, Liu X, Manfredsson FP, Sandoval IM, Yu SP, Wang JZ, Ye K (2017) Delta‐secretase phosphorylation by SRPK2 enhances its enzymatic activity, provoking pathogenesis in Alzheimer's disease. Mol Cell 67: 812–825 e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber S, Saftig P (2012) Ectodomain shedding and ADAMs in development. Development 139: 3693–3709 [DOI] [PubMed] [Google Scholar]
- Weihofen A, Lemberg MK, Ploegh HL, Bogyo M, Martoglio B (2000) Release of signal peptide fragments into the cytosol requires cleavage in the transmembrane region by a protease activity that is specifically blocked by a novel cysteine protease inhibitor. J Biol Chem 275: 30951–30956 [DOI] [PubMed] [Google Scholar]
- Weihofen A, Binns K, Lemberg MK, Ashman K, Martoglio B (2002) Identification of signal peptide peptidase, a presenilin‐type aspartic protease. Science 296: 2215–2218 [DOI] [PubMed] [Google Scholar]
- Weskamp G, Ford JW, Sturgill J, Martin S, Docherty AJ, Swendeman S, Broadway N, Hartmann D, Saftig P, Umland S, Sehara‐Fujisawa A, Black RA, Ludwig A, Becherer JD, Conrad DH, Blobel CP (2006) ADAM10 is a principal ‘sheddase’ of the low‐affinity immunoglobulin E receptor CD23. Nat Immunol 7: 1293–1298 [DOI] [PubMed] [Google Scholar]
- Westmeyer GG, Willem M, Lichtenthaler SF, Lurman G, Multhaup G, Assfalg‐Machleidt I, Reiss K, Saftig P, Haass C (2004) Dimerization of beta‐site beta‐amyloid precursor protein‐cleaving enzyme. J Biol Chem 279: 53205–53212 [DOI] [PubMed] [Google Scholar]
- Wichert R, Ermund A, Schmidt S, Schweinlin M, Ksiazek M, Arnold P, Knittler K, Wilkens F, Potempa B, Rabe B, Stirnberg M, Lucius R, Bartsch JW, Nikolaus S, Falk‐Paulsen M, Rosenstiel P, Metzger M, Rose‐John S, Potempa J, Hansson GC et al (2017) Mucus detachment by host metalloprotease meprin beta requires shedding of its inactive pro‐form, which is abrogated by the pathogenic protease RgpB. Cell Rep 21: 2090–2103 [DOI] [PubMed] [Google Scholar]
- Wild‐Bode C, Fellerer K, Kugler J, Haass C, Capell A (2006) A basolateral sorting signal directs ADAM10 to adherens junctions and is required for its function in cell migration. J Biol Chem 281: 23824–23829 [DOI] [PubMed] [Google Scholar]
- Wilhelm M, Schlegl J, Hahne H, Moghaddas Gholami A, Lieberenz M, Savitski MM, Ziegler E, Butzmann L, Gessulat S, Marx H, Mathieson T, Lemeer S, Schnatbaum K, Reimer U, Wenschuh H, Mollenhauer M, Slotta‐Huspenina J, Boese JH, Bantscheff M, Gerstmair A et al (2014) Mass‐spectrometry‐based draft of the human proteome. Nature 509: 582–587 [DOI] [PubMed] [Google Scholar]
- Willem M, Garratt AN, Novak B, Citron M, Kaufmann S, Rittger A, DeStrooper B, Saftig P, Birchmeier C, Haass C (2006) Control of peripheral nerve myelination by the beta‐secretase BACE1. Science 314: 664–666 [DOI] [PubMed] [Google Scholar]
- Willem M, Tahirovic S, Busche MA, Ovsepian SV, Chafai M, Kootar S, Hornburg D, Evans LD, Moore S, Daria A, Hampel H, Muller V, Giudici C, Nuscher B, Wenninger‐Weinzierl A, Kremmer E, Heneka MT, Thal DR, Giedraitis V, Lannfelt L et al (2015) eta‐Secretase processing of APP inhibits neuronal activity in the hippocampus. Nature 526: 443–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler E, Kamp F, Scheuring J, Ebke A, Fukumori A, Steiner H (2012) Generation of Alzheimer disease‐associated amyloid beta42/43 peptide by gamma‐secretase can be inhibited directly by modulation of membrane thickness. J Biol Chem 287: 21326–21334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe MS, Xia W, Ostaszewski BL, Diehl TS, Kimberly WT, Selkoe DJ (1999) Two transmembrane aspartates in presenilin‐1 required for presenilin endoproteolysis and gamma‐secretase activity. Nature 398: 513–517 [DOI] [PubMed] [Google Scholar]
- Wong E, Cohen T, Romi E, Levin M, Peleg Y, Arad U, Yaron A, Milla ME, Sagi I (2016) Harnessing the natural inhibitory domain to control TNFalpha Converting Enzyme (TACE) activity in vivo . Sci Rep 6: 35598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R, Bienkowski MJ, Shuck ME, Miao H, Tory MC, Pauley AM, Brashier JR, Stratman NC, Mathews WR, Buhl AE, Carter DB, Tomasselli AG, Parodi LA, Heinrikson RL, Gurney ME (1999) Membrane‐anchored aspartyl protease with Alzheimer's disease beta‐ secretase activity. Nature 402: 533–537 [DOI] [PubMed] [Google Scholar]
- Yan R, Munzner JB, Shuck ME, Bienkowski MJ (2001) BACE2 functions as an alternative alpha‐secretase in cells. J Biol Chem 276: 34019–34027 [DOI] [PubMed] [Google Scholar]
- Yan R (2017) Physiological functions of the beta‐site amyloid precursor protein cleaving enzyme 1 and 2. Front Mol Neurosci 10: 97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Rawson RB, Komuro R, Chen X, Dave UP, Prywes R, Brown MS, Goldstein JL (2000) ER stress induces cleavage of membrane‐bound ATF6 by the same proteases that process SREBPs. Mol Cell 6: 1355–1364 [DOI] [PubMed] [Google Scholar]
- Zahn C, Kaup M, Fluhrer R, Fuchs H (2013) The transferrin receptor‐1 membrane stub undergoes intramembrane proteolysis by signal peptide peptidase‐like 2b. FEBS J 280: 1653–1663 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Song M, Liu X, Su Kang S, Duong DM, Seyfried NT, Cao X, Cheng L, Sun YE, Ping Y, Jia J, Levey AI, Ye K (2015) Delta‐secretase cleaves amyloid precursor protein and regulates the pathogenesis in Alzheimer's disease. Nat Commun 6: 8762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Xie M, Ye K (2016) Asparagine endopeptidase is an innovative therapeutic target for neurodegenerative diseases. Expert Opin Ther Targets 20: 1237–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Obianyo O, Dall E, Du Y, Fu H, Liu X, Kang SS, Song M, Yu SP, Cabrele C, Schubert M, Li X, Wang JZ, Brandstetter H, Ye K (2017) Inhibition of delta‐secretase improves cognitive functions in mouse models of Alzheimer's disease. Nat Commun 8: 14740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Song W (2006) Leaky scanning and reinitiation regulate BACE1 gene expression. Mol Cell Biol 26: 3353–3364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Barao S, Laga M, Bockstael K, Borgers M, Gijsen H, Annaert W, Moechars D, Mercken M, Gevaer K, De Strooper B (2012) The neural cell adhesion molecules L1 and CHL1 are cleaved by BACE1 protease in vivo . J Biol Chem 287: 25927–25940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu K, Xiang X, Filser S, Marinkovic P, Dorostkar MM, Crux S, Neumann U, Shimshek DR, Rammes G, Haass C, Lichtenthaler SF, Gunnersen JM, Herms J (2018) Beta‐site amyloid precursor protein cleaving enzyme 1 inhibition impairs synaptic plasticity via seizure protein 6. Biol Psychiatry 83: 428–437 [DOI] [PubMed] [Google Scholar]
- Zunke F, Rose‐John S (2017) The shedding protease ADAM17: physiology and pathophysiology. Biochim Biophys Acta 1864: 2059–2070 [DOI] [PubMed] [Google Scholar]


