Abstract
Background
The antisense of the OIP5‐AS1 gene is a long non‐coding RNA (lncRNA) that is reported to be upregulated and promotes cell proliferation in multiple human cancers; however, its function in lung cancer is unknown. We investigated the regulatory function and underlying mechanisms of OIP5‐AS1 in lung cancer.
Methods
OIP5‐AS1 and microRNA (miR)‐378a‐3p expression were assayed by quantitative real‐time PCR, and proliferation‐related protein expression was measured by Western blotting. Cell viability was detected using methyl thiazolyl tetrazolium assay. Luciferase reporter assay and RNA immunoprecipitation were used to detect the direct regulation of miR‐378a‐3p by OIP5‐AS1. Nude mice were used to test the function of OIP5‐AS1 in vivo.
Results
OIP5‐AS1 was highly expressed in lung cancer tissues and was correlated with tumor size and tumor growth speed. OIP5‐AS1 overexpression increased lung cancer cell proliferation in vitro. Further investigation revealed that OIP5‐AS1 functions as a competing endogenous RNA of miR‐378a‐3p. MiR‐378a‐3p overexpression inhibited cell proliferation and caused proliferation‐associated proteins CDK4 and CDK6 to decrease in A549 cells. Overexpression of wild type OIP5‐AS1 led to strong CDK4 and CDK6 expression; however, these two proteins did not change when mutated OIP5‐AS1 was upregulated. Finally, in vivo assay showed that the speed of tumor growth was increased and decreased when OIP5‐AS1 was upregulated and downregulated, respectively.
Conclusion
Our results revealed that OIP5‐AS1 acts as a growth‐promoting lncRNA in lung cancer by suppressing miR‐378a‐3p function. OIP5‐AS1 and miR‐378a‐3p interaction may provide a potential target for lung cancer treatment.
Keywords: Long non‐coding RNA, lung cancer, miR‐378a‐3p, OIP5‐AS1, proliferation
Introduction
Lung cancer is the most common cause of cancer‐related death in the world.1 Despite advances in therapy, the overall survival rate of patients with advanced lung cancer is low. Long non‐coding RNAs (lncRNAs) are transcripts that are longer than 200 nucleotides and have no protein coding function.2 Some lncRNAs are known to be involved in complex mechanisms underlying the development of malignancies, including carcinogenesis, progression, and metastasis, but most of the underlying molecular events are unknown.3, 4, 5
The human lncRNA OIP5‐AS1, which is transcribed in the antisense (AS) of OIP5, suppresses cell proliferation in multiple malignancies.6 OIP5‐AS1 is reported to be associated with HuR in Hela cells, reducing its availability to target messenger RNAs of cyclins A and D1 (CCNA2 and CCND1) and SIRT1. Accordingly, these proteins are more highly expressed when OIP5‐AS1 levels decline and are suppressed when OIP5‐AS1 is overexpressed.7 It is also regarded as a competing endogenous RNA (ceRNA) to interact with microRNAs (miRNAs, miRs) in some cancer cells.8 However, the function of OIP5‐AS1 in lung cancer has never been examined.
The hsa‐miR‐378 precursor gives rise to two mature strands: hsa‐miR‐378 (hsa‐miR‐378a‐3p) and hsamiR‐378* (hsa‐miR‐378a‐5p).9 According to previous studies, miR‐378 promotes cell survival and tumor growth of glioblastoma by targeting two tumor suppressors: Sufu and Fus‐1.10 Its interaction with HO‐1 protein in A549 adenocarcinoma cells inhibits growth, vascularization, and metastasis.11 MiR‐378 has also been identified as a novel target of the c‐Myc oncoprotein that is able to cooperate with activated Ras or HER2 in tumor development.12
Many lncRNAs regulate gene expression as ceRNAs to repress miRNA functions.13 In this study, we found that OIP5‐AS1 acts as a ceRNA to inhibit miR‐378a. Upregulation of OIP5‐AS1 in lung cancer cells and tissues was associated with increased lung cancer cell viability and promotion of colony formation. This study supplements our understanding of lung cancer pathogenesis.
Methods
Tissue collection
Eighty pairs of lung cancer tissues and adjacent nonmalignant lung tissue samples were obtained from patients at the Affiliated Hospital of Qingdao University between December 2015 and March 2016. Follow‐up was conducted each month. Specimens were immediately snap‐frozen in liquid nitrogen and stored at −80°C until processing. The basic clinical data of the patients are listed in Table 1. None of the patients underwent preoperative radiotherapy or chemotherapy, and none had distant metastases. Complete data of clinicopathologic features and follow‐up were available. Patients lost to follow‐up, who had received preoperative radiotherapy or chemotherapy, and those with recurrence and distant metastasis were excluded. Tumor growth speed was analyzed according to Ki67 expression, which was obtained from pathological reports.
Table 1.
Clinical data of lung cancer patients (n = 80)
| Features | Total |
|---|---|
| Age (year) | |
| Mean | 61.12 ± 8.25 |
| > 50 | 36 |
| ≤ 50 | 44 |
| Gender | |
| Male | 45 |
| Female | 35 |
| Clinical Stage | |
| I | 43 |
| II | 29 |
| III | 8 |
| Tumor size (mm) | |
| Mean | 22.5 ± 12.51 |
| > 20 | 31 |
| ≤ 20 | 49 |
| Lymph node metastasis | |
| Yes | 25 |
| No | 55 |
| Lung adenocarcinoma | 62 |
| Squamous cell lung cancer | 18 |
Data are presented as mean ± standard deviation. Clinical stages and tumor extent were evaluated according to American Joint Committee on Cancer Tumor Node Metastasis classification. Tumor size and lymph node metastasis were determined according to pathological reports.
The Research Ethics Committee of the Affiliated Hospital of Qingdao University approved the study. Informed consent was obtained from all patients.
Cell culture
A549 and H520 human lung cancer cell lines were obtained from the Chinese Institute of Biochemistry and Cell Biology (Shanghai, China). Cells were cultured in RPMI 1640 medium (Invitrogen, Carlsbad, CA, USA) containing 10% fetal bovine serum, 100 U/mL penicillin, and 100 U/mL streptomycin in culture flasks at 37°C with 5% CO2.
Quantitative real‐time PCR
Total RNA was extracted from cells and tissue samples using TRIzol reagent (Invitrogen) following the manufacturer's instructions. Complementary (c)DNA synthesis was performed using a Prime Script Reverse Transcriptase Reagent Kit (TaKaRa, Dalian, China) according to the manufacturer's instructions; RNA was reverse transcribed to cDNA with a Reverse Transcription Kit (TaKaRa); and OIP5‐AS1, miR‐378a‐3p, CDK4, and CDK6 expression were assayed by quantitative real‐time (qRT) PCR using SYBR Premix Ex Taq (TaKaRa). The PCR primer sequences used were: OIP5‐AS1: 5′‐TGCGAAGATGGCGGAGTAAG ‐3′ and 5′‐TAGTTCCTCTCCTCTGGCCG‐3′; CDK4: forward, 5′‐ GGTGACAAGTGGTGGAACAG‐3′; reverse, 5′‐GCCCAATCAGGTCAAAGATT‐3′; CDK6: forward, 5′‐CGAATGCGTGGCGGAGATC‐3′; reverse, 5′‐CCACTGAGGTTAGAGCCATC‐3′. miR‐378a‐3p: 5′‐ACUGGACUUGGAGUCAGAAGG‐3′. LncRNA and miRNA expression were normalized against human glyceraldehyde 3‐phosphate dehydrogenase (GAPDH) and U6 small nuclear RNA (snRNA). Fold‐changes in expression were calculated using the relative quantification (2 − ΔΔCt) method. Assays were repeated three times for each sample.
Lentivirus infection and establishment of stable cell lines
Lentivirus expressing OIP5‐AS1, short hairpin (sh)RNA targeting OIP5‐AS1, and their corresponding controls were purchased from GenePharma (Shanghai, China). The overexpression vector and its control were named LV‐OIP5‐AS1 and LV‐NC; the silencing vector and its control were named LV‐shOIP5‐AS1 and LV‐shNC. The sequences were: shOIP5‐AS1, 5′‐CCGGGCTCCTAGGATTCCAGTTATCCTCGAGGCAGAAGGCTGAGTTTCATTTTTTTTG‐3′; shNC, 5′‐CACCGTTCTCCGAACGTGTCACGTCAAGAGATTACGTG ACACGTTCGGAGAATTTTTTG‐3′. MiRNA inhibitor and mimics were purchased from GenePharma. MiR‐378‐3p inhibitor: 5′‐ACUGGACUUGGAGUCAGAAGG‐3′; miR‐378a‐3p mimics: 5′‐ACUGGACUUGGAGUCAGAAGG‐3′. A549 cells were transduced with the recombinant lentivirus and stable cell lines were established. MiR‐378a‐3p mimics were also obtained from GenePharma. Transfections were repeated three times.
Western blotting
Total cell or tissue proteins were extracted using a Total Protein Extraction Kit (Jiancheng Bioengineering Institute, Nanjing, China). Proteins were electrophoresed in 12% polyacrylamide gels. Nonspecific binding was blocked with 5% skim milk for two hours at 37°C. Anti‐CDK4 polyclonal antibody (1:1000, ab68266), anti‐CDK6 (1:1000, ab131469), and anti‐Ki67 (1:1000, ab92742; Abcam, Cambridge, UK) were then used as the first antibodies; β‐actin antibody (1:1500 diluted, ab8227; Abcam) was used as a control. Assays were repeated three times.
Methyl thiazolyl tetrazolium assay
Cell proliferation was measured by 3‐(4, 5‐dimethylthiazole‐2‐yl)‐2, 5‐diphenyltetrazolium bromide (MTT) assay. LV‐infected cells were seeded into 96‐well plates at a density of 2000 cells/well and cultured for 24, 48, 72, or 96 hours. The spectrophotometric absorbance of each well was measured at 490 nm at different time points using a microplate reader absorbance test plate (Molecular Devices, Sunnyvale, CA, USA). Assays were repeated three times. Cell survival was calculated with optical density (OD) value, and the NC group in 0 hour was regarded as 100%.
Luciferase reporter assay
OIP5‐AS1 fragments containing the putative binding sequence of miR‐378a‐3p and its mutant sequence were cloned into a pGL3‐control vector (Promega, Madison, WI, USA). The vectors were sequenced and named OIP5‐AS1‐WT and OIP5‐AS1‐Mut. A549 cells were co‐transfected with the appropriate reporter plasmid, miRNA, or pRL–TK Renilla plasmid (Promega) using Lipofectamine 2000 (Invitrogen). Luciferase activity was measured 48 hours post‐transfection using a dual‐luciferase reporter assay system (Promega) following the manufacturer's instructions. Assays were repeated three times.
RNA immunoprecipitation (RIP)
RNA immunoprecipitation (RIP) was performed using an EZ‐Magna RIP RNA‐binding protein immunoprecipitation kit (Millipore, Billerica, MA, USA) following the manufacturer's instructions. A549 and H520 cell lysates containing OIP5‐AS1 and miRNAs were prepared and incubated with RIP buffer containing magnetic beads conjugated to human anti‐argonaute2 (Ago2) antibody (Millipore). Normal mouse immunoglobulin G (IgG; Millipore) was used as a negative control. OIP5‐AS1 and miRNAs present in the precipitates were assayed by qRT‐PCR. Assays were repeated three times.
Tumor growth in nude mice
Twelve four‐week‐old female nude mice were used in our study. Nude mice were purchased from Vital River Laboratories (VRL, Beijing, China) and bred at the Medical College of Qingdao University animal center. Cells were washed with phosphate buffered saline and harvested with 1640 culture medium, and then re‐suspended in 1640 culture medium. Equal amounts of matrix gum were mixed in single cell suspension. Subsequently, 2 × 106 cells (0.1 mL) were injected subcutaneously into the two lower limbs of the mice. Six mice were injected with LV‐OIP5‐AS1 ‐A549 cells in the right and LV‐NC‐A549 cells in the left; the other six mice were injected with LV‐shOIP5‐AS1‐A549 cells in the right and LV‐shNC‐A549 cells in the left. The mice were sacrificed in the fourth week. The Animal Ethics Committee of the hospital research department approved all animal experiments.
Statistical analysis
Data were reported as mean ± standard deviation (SD) and analyzed using SPSS version 17.0 (SPSS Inc., Chicago, IL, USA). Between‐group differences were tested for significance using Student's t‐test and one‐way analysis of variance. Pearson correlation coefficients were calculated to determine the significance of the relationship between OIP5‐AS1 and miR‐378a‐3p expression. P values of < 0.05 were considered significant. *P < 0.05; **P < 0.01; ***P < 0.005. BiBiServ2 (https://bibiserv.cebitec) was used to predict potential lncRNA–miRNA interactions.
Results
Elevated OIP5‐AS1 in human lung cancer tissue is correlated with tumor size and poor prognosis
The lncRNA OIP5‐AS1 expression level was first examined in lung cancer tissues and matched adjacent normal tissue samples. As shown in Figure 1a, OIP5‐AS1 expression was significantly higher in lung cancer tissues than in controls (P = 0.010). All patients were sequenced according to OIP5‐AS1 level, and then further divided into OIP5‐AS1high and OIP5‐AS1low groups (OIP5‐AS1high n = 40; OIP5‐AS1low n = 40). We then analyzed the correlation between OIP5‐AS1 and tumor size (Fig 1b). Tumor sizes in the OIP5‐AS1high group were significantly larger than in the OIP5‐AS1low (P = 0.012). The association between OIP5‐AS1 and Ki67 expression in tumor tissues was also analyzed according to patients’ pathological data (Fig 1c). The OIP5‐AS1high group showed a higher Ki67 expression rate than the OIP5‐AS1low (P = 0.045). Finally, we analyzed the correlation between OIP5‐AS1 expression and lymph node metastasis; however, there was no significant difference in OIP5‐AS1 expression between lymph node positive and negative patients (Fig 1d).
Figure 1.
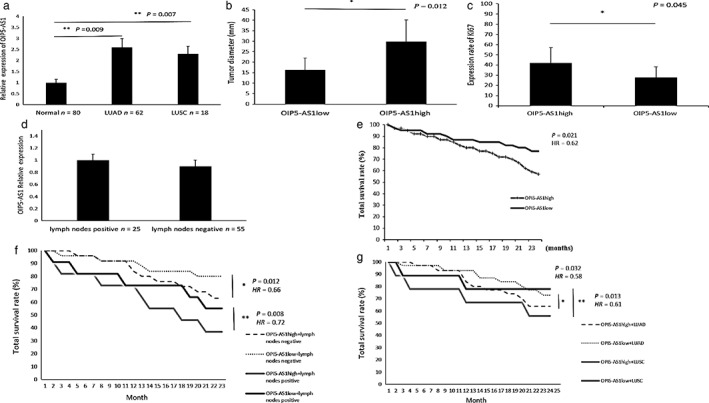
OIP5‐AS1 was overexpressed in lung cancer tissues and was associated with tumor growth and prognosis. (a) OIP5‐AS1 was upregulated in lung adenocarcinoma (LUAD) and lung squamous cell cancer (LUSC) compared to adjacent normal tissue (P < 0.01). (b) OIP5‐AS1 expression and tumor size (40 vs. 40). (c) OIP5‐AS1 and Ki67 expression rates (40 vs. 40). (d) No difference in OIP5‐AS1 expression between cells with and without lymph node metastasis was observed. (e) OIP5‐AS1 expression level is associated with poor lung cancer prognosis and (f) lower total survival rate in patients positive and negative for lymph node metastasis (negative: OIP5‐AS1high n = 27, OIP5‐AS1low n = 27; positive: OIP5‐AS1high n = 11, OIP5‐AS1low n = 11). (g) High OIP5‐AS1 expression leads to lower total survival rate both in LUAD and LUSC patients (LUAD: OIP5‐AS1high n = 31, OIP5‐AS1low n = 31; LUSC: OIP5‐AS1high n = 9, OIP5‐AS1low n = 9). HR, hazard ratio.
Kaplan–Meier plotter survival analysis was performed to study the relationship between OIP5‐AS1 and prognosis. We compared the survival curves in the OIP5‐AS1high and OIP5‐AS1low groups. Patients in the OIP5‐AS1low group achieved higher total survival than in the OIP5‐AS1high (hazard ratio [HR] 0.62, 95% confidence interval [CI] 0.51–0.69; P = 0.021) (Fig 1e). In order to exclude confounding factors, we divided the 80 patients into lymph node metastasis positive (n = 55) and negative (n = 25) groups. Patients were again then divided in OIP5‐AS1high and OIP5‐AS1low groups and the survival curves were analyzed. Both lymph node positive and negative patients showed higher total survival in the OIP5‐AS1low than in the OIP5‐AS1high group (positive: HR 0.72, 95% CI 0.612–0.920, P = 0.008; negative: HR 0.66, 95% CI 0.581–0.734, P = 0.012) (Fig 1f). We analyzed lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) patient survival using the same method and the results showed that both LUAD and LUSC patients in the OIP5‐AS1low group achieved higher total survival than the OIP5‐AS1high (LUAD: HR 0.58, 95% CI 0.422–0.720, P = 0.032; LUSC: HR 0.61, 95% CI 0.525–0.716, P = 0.013) (Fig 1g).
Collectively, the results indicated that upregulated OIP5‐AS1 in lung cancer promoted tumor growth. High OIP5‐AS1 expression indicated poor prognosis in lung cancer.
OIP5‐AS1 promoted cell proliferation in lung cancer cells
We performed in vitro gain and loss of function analyses to determine whether OIP5‐AS1 regulates lung cancer cell proliferation. Transfection of overexpression and silencing of OIP5‐AS1 were performed in A549 and H520 lung cancer cells (Fig 2a,b). MTT assay showed that OIP5‐AS1 overexpression significantly promoted lung cancer cell viability compared to the negative control (Fig 2c). Cell viability was significantly inhibited when OIP5‐AS1 was silenced with LV‐shOIP5‐AS1 (Fig 2d). In order to further test the cell promoting function of OIP5‐AS1, we detected Ki67 protein expression. The results showed that OIP5‐AS1 promoted Ki67 protein expression in lung cancer cells (Fig 2e,f). When OIP5‐AS1 was overexpressed, cell proliferation was activated. These results confirmed that OIP5‐AS1 functions as a proliferation‐promoter in lung cancer cells.
Figure 2.
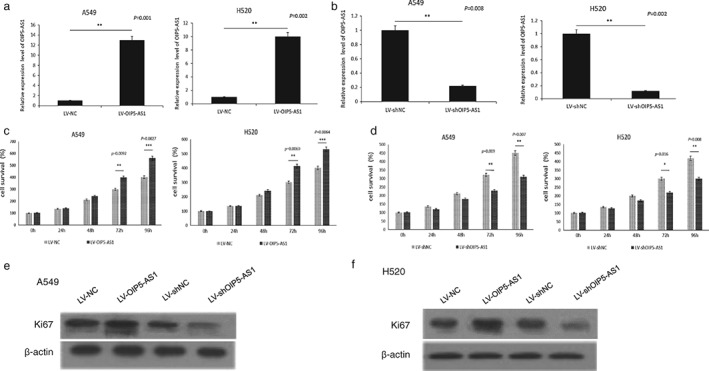
OIP5‐AS1 promoted cell proliferation in lung cancer cells. Transfection efficiency of (a) LV‐OIP5‐AS1l and (b) LV‐shOIP5‐AS1l in A549 and H520 cells. Cell proliferation activity was tested using methyl thiazolyl tetrazolium assay after transfection in (c) A549 and (d) H520 cells. High OIP5‐AS1 expression improved Ki67 protein expression in both (e) A549 and (f) H520 cells.
OIP5‐AS1 acts as a molecular sponge of miR‐378a‐3p
To determine whether OIP5‐AS1 acts as a ceRNA to bind some miRNAs, we used BiBiServ2 (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid) to predict potential lncRNA–miRNA interactions. Among several miRNAs, we found that miR‐378a‐3p had the highest possibility of binding OIP5‐AS1 (Fig 3a). MiR‐378a‐3p expression was detected in lung cancer tissues and was downregulated in tumors compared to adjacent normal tissues (P = 0.029) (Fig 3b). Moreover, an RIP assay was performed in A549 and H520 cells to test whether OIP5‐AS1 is associated with an miR‐378a‐3p‐component RNA‐induced silencing complex (RISC).14 We examined miR‐378a‐3p and OIP5‐AS1 on magnetic beads conjugated to anti‐Ago2 antibody and found that miR‐378a‐3p was highly enriched (Fig 3c). This result suggested that OIP5‐AS1 and miR‐378a‐3p can both combine with AGO2 and format an RISC and that miR‐378a‐3p binds to OIP5‐AS1 in lung cancer cells. In addition, a luciferase assay was performed in A549 and H520 cells to test the binding site of miR‐378a‐3p in OIP5‐AS1. The activity of luciferase reporters containing the theoretical binding site of OIP5‐AS1 in miR‐378a‐3p was significantly decreased in OIP5‐AS1‐WT constructs, while there was no affect in OIP5‐AS1‐Mut constructs (Fig 3d,e). Collectively, these results suggested that OIP5‐AS1 acts as a molecular sponge of miR‐378a‐3p.
Figure 3.
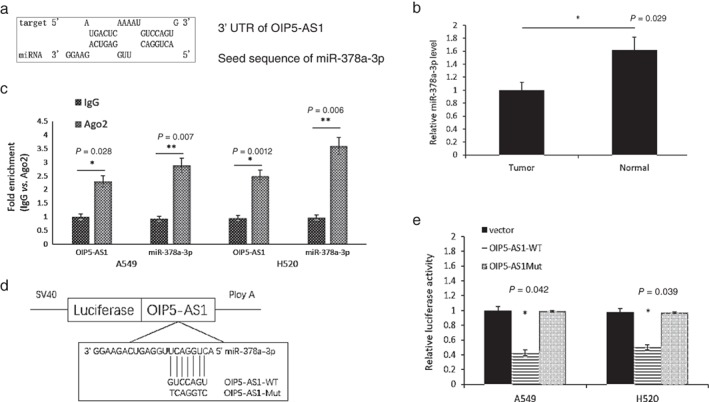
OIP5‐AS1 acted as a competing endogenous RNA (ceRNA) of miR‐378a‐3p. (a) Analysis of miR‐378a‐3p‐binding sequence in OIP5‐AS1. (b) MiR‐378a‐3p expression was examined in lung cancer tissues by quantitative real‐time PCR. (c) Associations between OIP5‐AS1, miR‐378a‐3p, and AGO2 were confirmed by RNA immunoprecipitation with an AGO2 antibody. OIP5‐AS1 and miR‐378a‐3p were both highly expressed in the precipitate of AGO2 in A549 and H520 cells, indicating that OIP5‐AS1 and miR‐378a‐3p can both combine with AGO2 and form an RNA‐induced silencing complex. (d) Putative miR‐378a‐3p‐binding sequence of OIP5‐AS1 and design of OIP5‐AS1‐Mut vector. (e) Relative luciferase activities were measured in A549 and H520 cells. Cells were transfected with OIP5‐AS1‐WT or OIP5‐AS1‐Mut vectors.
MiR‐378a‐3p inhibits lung cancer cell proliferation
According to previous studies, miR‐378 functions as an inhibitor in lung adenocarcinoma.15 To study the function of miR‐378a‐3p in lung cancer, miR‐378a‐3p mimics and inhibitors were transfected in A549 and H520 cells (Fig 4a). MTT assay and Ki67 expression showed that miR‐378a‐3p overexpression significantly inhibited the proliferation of lung cancer cells compared to untreated cells. The proliferation of lung cancer cells was significantly upregulated when miR‐378a‐3p was silenced (Fig 4b,c). CDK4 and CDK6 have been reported as important proliferation‐associated proteins in multiple human cancers.16 To confirm whether miR‐378a‐3p inhibited lung cancer cell proliferation, we examined changes in CDK4 and CDK6 expression in transfected A549 and H520 cells. CDK4 and CDK6 protein expression was significantly enhanced after being transfected with miR‐378a‐3p inhibitor. When A549 and H520 cells were transfected with miR‐378a‐3p mimics, CDK4 and CDK6 proteins were significantly reduced compared to cells that were not treated (Fig 4d). These results showed that miR‐378a‐3p functions as an inhibitor of lung cancer proliferation and the underlying mechanisms are related to CDK4 and CDK6 proteins. Further experiments are required to confirm the relationship between OIP5‐AS1, miR‐378a‐3p, and lung cancer proliferation.
Figure 4.
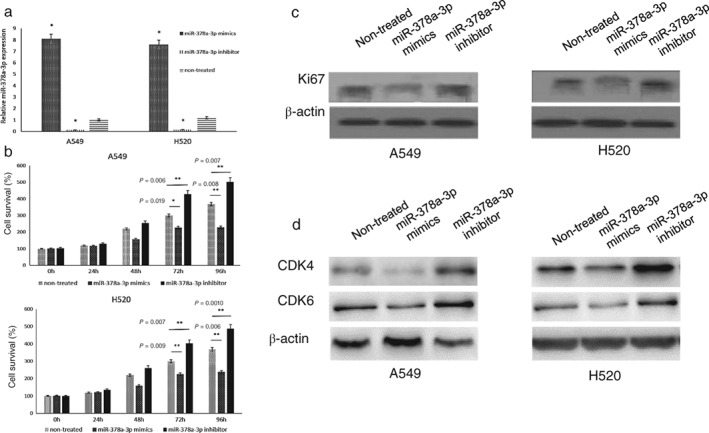
MiR‐378a‐3p inhibits lung cancer cell proliferation. (a) Transfection efficiency of miR‐378a‐3p mimics and inhibitor in A549 and H520 cells. (b) Cell proliferation activity of A549 and H520 cells tested with methyl thiazolyl tetrazolium assay. Differences were significant at 72 hours. (c) Ki67 protein expression was blocked by miR‐378a‐3p in both A549 and H520 cells. (d) CDK4 and CDK6 protein expression in A549 and H520 cell lines transfected with miR‐378a‐3p mimics or inhibitor. Non‐treated refers to cells that were not transfected.
OIP5‐AS1 reverses the inhibiting effects of miR‐378a‐3p in lung cancer cells
To investigate whether the proliferation‐inhibiting effects of miR‐378a‐3p were suppressed by OIP5‐AS1, we co‐transfected A549 and H520 cells with miR‐378a‐3p mimics and OIP5‐AS1‐WT expression vector. MTT assays showed higher cell proliferation activity in the control group transfected with miR‐378a‐3p mimics only. When we co‐transfected A549 cells with miR‐378a‐3p mimics and an OIP5‐AS1‐Mut expression vector, the proliferation‐inhibiting function of miR‐378a‐3p barely differed from the control (Fig 5a). Ki67, CDK4, and CDK6 expression were also increased after treatment with the OIP5‐AS1‐WT vector, while OIP5‐AS1‐Mut exerted no function in lung cancer cells (Fig 5b). These observations suggest that OIP5‐AS1 promoted tumor cell growth in part by competitively binding miR‐378a‐3p.
Figure 5.
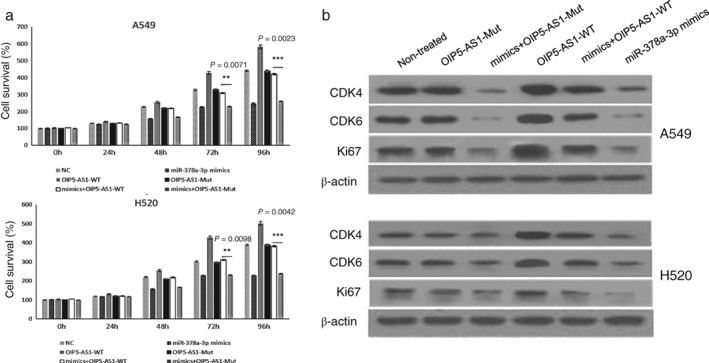
OIP5‐AS1 promoted A549 and H520 cell proliferation by competitively binding miR‐378a‐3p. Binding‐site‐mutated OIP5‐AS1 did not exert the proliferation promoting function. (a) Methyl thiazolyl tetrazolium assay was conducted in A549 and H520 cells after transfection with OIP5‐AS1‐WT or OIP5‐AS1‐Mut vectors and miR‐378a‐3p mimics. (b) Ki67, CDK4, and CDK6 protein expression in lung cancer cells after transfection with OIP5‐AS1‐WT or OIP5‐AS1‐Mut vectors and microRNA mimics. Non‐treated A549 and H520 cells were used as the control.
OIP5‐AS1 inhibits tumor growth in vivo
The function of OIP5‐AS1 in vivo was investigated with nude mice. Six mice were used to perform OIP5‐AS1‐overexpression assay. We injected LV‐OIP5‐AS1‐A549 cells into the right limbs of the mice and LV‐NC‐A549 cells into the left. Four weeks later, we found that the LV‐OIP5‐AS1 side tumors were significantly larger than in the LV‐NC side (Fig 6a). In addition, another six mice were used to conduct OIP5‐AS1‐knockdown assay. The right limbs of the mice were injected with LV‐shOIP5‐AS1‐A549 cells, and left with LV‐shNC‐A549 cells. The results showed that tumors in the LV‐shOIP5‐AS1 side were significantly smaller than in the LV‐shNC side (Fig 6b). After excision, all of the tumors were weighed. The average weight of LV‐OIP5‐AS1 tumors was significantly higher than LV‐NC tumors (P = 0.005), and the average weight of LV‐shOIP5‐AS1 tumors was lower than LV‐shNC (P = 0.023) (Fig 6c). These results demonstrated that lncRNA OIP5‐AS1 suppresses lung tumor growth in vivo.
Figure 6.
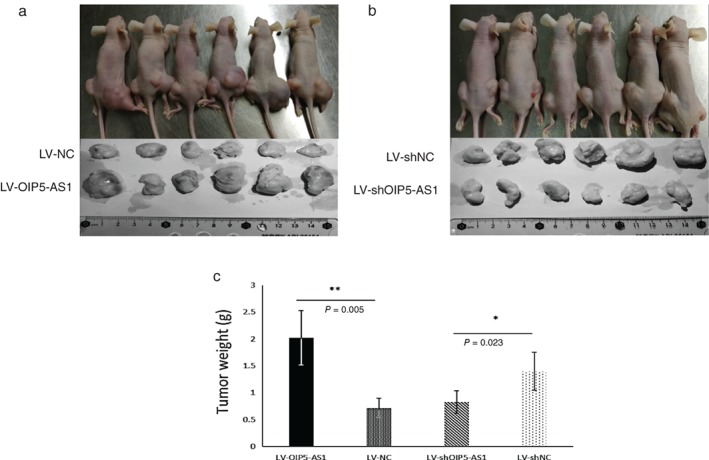
OIP5‐AS1 promotes tumor growth in vivo. Nude mice were inoculated with (a) LV‐OIP5‐AS1‐A549 cells in the right lower limb and LV‐NC‐A549 cells in the left and (b) LV‐shOIP5‐AS1‐A549 cells in the right and LV‐shNC‐A549 cells in the left lower limb. Tumors were resected in the fourth week. (c) Comparison of tumor weight among the different groups.
Discussion
Study of miRNAs has dominated the field of noncoding RNA regulation in the past decade.17, 18, 19 Recently, more and more studies have indicated that lncRNAs provide a growth advantage to malignant cells, resulting in progressive and uncontrolled tumor growth.20, 21 This provides us with new directions for cancer treatment.
OIP5‐AS1 activity in lung cancer has not previously been described. In this study, we found that OIP5‐AS1 is upregulated in lung cancer cells and tissues, and overexpression significantly promotes cell proliferation and colony formation, whereas OIP5‐AS1 knockdown negatively regulates cell growth. The findings show that OIP5‐AS1 plays an important role in the modulation of lung cancer progression. Whether OIP5‐AS1 can be a therapeutic target in lung cancer warrants further research.
Increasing studies have revealed the existence of a widespread interaction network involving ceRNAs, where ncRNAs could regulate modulatory RNA by binding and titrating them off their binding sites on protein coding messengers. We hypothesized that lncRNA OIP5‐AS1 functioned as a ceRNA to promote lung cancer progression and investigated potential interactions with miRNAs. We used bioinformatics analysis and luciferase assays to verify the direct binding of predicted miRNA response elements on the OIP5‐AS1 transcript. The results show that miR‐378a‐3p could form complementary base pairing with OIP5‐AS1 and reduce pGL3‐OIP5‐AS1 reporter gene expression. In RIP assay, OIP5‐AS1 expression immunoprecipitated with Ago2 was higher than when immunoprecipitated with immunoglobulin G, indicating the reciprocal repression of OIP5‐AS1 and miR‐378a‐3p caused by RISC. We also show that miR‐378a‐3p overexpression arrested lung cancer cell growth and OIP5‐AS1 can inhibit its function. These findings demonstrate that OIP5‐AS1 interacts with miR‐378a‐3p in lung cancer pathogenesis.
MiR‐378a‐3p is known to inhibit tumor proliferation in multiple cancers.22, 23 Our study showed that miR‐378a‐3p is downregulated in human lung cancer tissues, which indicates that the expression and functions of this molecule are suppressed by some mechanisms in lung cancer. In addition, we found that miR‐378a‐3p negatively regulates the expression of proliferation‐associated proteins, CDK4 and CDK6.16 These two proteins are significantly increased again by upregulating OIP5‐AS1. These results indicate an OIP5‐AS1/miR‐378a‐3p/CDK4/CDK6 axis that regulates cell proliferation in lung cancer.
In summary, we showed that the lncRNA OIP5‐AS1 promotes lung cancer cell proliferation by competitively binding miR‐378a‐3p. We describe a novel OIP5‐AS1/miR‐378a‐3p/CDK4/CDK6 axis with a regulatory function in lung cancer. Further studies should focus on the mechanisms between CDK4/CDK6 and OIP5‐AS1/miR‐378a‐3p. Our findings show that OIP5‐AS1 may be a target for lung cancer therapy, with crosstalk between miR‐378a‐3p and OIP5‐AS1 shedding new light on the potential treatment of lung cancer.
Disclosure
No authors report any conflict of interest.
Acknowledgment
This study was funded by the Natural Science Foundation of Shandong Province (Grant Number: ZR2016HM58).
References
- 1. Wei S, Tian J, Song X, Wu B, Liu L. Causes of death and competing risk analysis of the associated factors for non‐small cell lung cancer using the Surveillance, Epidemiology, and End Results database. J Cancer Res Clin Oncol 2018; 144: 145–55. [DOI] [PubMed] [Google Scholar]
- 2. Khandelwal A, Bacolla A, Vasquez KM et al Long non‐coding RNA: A new paradigm for lung cancer. Mol Carcinog 2015; 54: 1235–51. [DOI] [PubMed] [Google Scholar]
- 3. Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell 2018; 172: 393–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Atianand MK, Caffrey DR, Fitzgerald KA. Immunobiology of long noncoding RNAs. Annu Rev Immunol 2017; 35: 177–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sullenger BA, Nair S. From the RNA world to the clinic. Science 2016; 352: 1417–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Naemura M, Kuroki M, Tsunoda T et al The long noncoding RNA OIP5‐AS1 is involved in the regulation of cell proliferation. Anticancer Res 2018; 38: 77–81. [DOI] [PubMed] [Google Scholar]
- 7. Kim J, Abdelmohsen K, Yang X et al LncRNA OIP5‐AS1/cyrano sponges RNA‐binding protein HuR. Nucleic Acids Res 2016; 44: 2378–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu X, Zheng J, Xue Y et al PIWIL3/OIP5‐AS1/miR‐367‐3p/CEBPA feedback loop regulates the biological behavior of glioma cells. Theranostics 2018; 8: 1084–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Velazquez‐Torres G, Shoshan E, Ivan C et al A‐to‐I miR‐378a‐3p editing can prevent melanoma progression via regulation of PARVA expression. Nat Commun 2018; 9: 461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tang X, Zhou J, Zhang J et al Low expression of FUS1 is negatively correlated with miR‐378 and may predict adverse prognoses in acute myeloid leukemia. Acta Haematol 2018; 139: 89–95. [DOI] [PubMed] [Google Scholar]
- 11. Skrzypek K, Tertil M, Golda S et al Interplay between heme oxygenase‐1 and miR‐378 affects non‐small cell lung carcinoma growth, vascularization, and metastasis. Antioxid Redox Signal 2013; 19: 644–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Feng M, Li Z, Aau M et al Myc/miR‐378/TOB2/cyclin D1 functional module regulates oncogenic transformation. Oncogene 2011; 30: 2242–51. [DOI] [PubMed] [Google Scholar]
- 13. Li MJ, Zhang J, Liang Q et al Exploring genetic associations with ceRNA regulation in the human genome. Nucleic Acids Res 2017; 45: 5653–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yi T. Identifying RISC components using Ago2 immunoprecipitation and mass spectrometry. Methods Mol Biol 2018; 1720: 149–59. [DOI] [PubMed] [Google Scholar]
- 15. Ho CS, Noor SM, Nagoor NH. MiR‐378 and mir‐1827 regulate tumor invasion, migration and angiogenesis in human lung adenocarcinoma by targeting RBX1 and CRKL, respectively. J Cancer 2018; 9: 331–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kozar K, Sicinski P. Cell cycle progression without cyclin D‐CDK4 and cyclin D‐CDK6 complexes. Cell Cycle 2005; 4: 388–91. [DOI] [PubMed] [Google Scholar]
- 17. Mirzaei H, Masoudifar A, Sahebkar A et al MicroRNA: A novel target of curcumin in cancer therapy. J Cell Physiol 2018; 233: 3004–15. [DOI] [PubMed] [Google Scholar]
- 18. Kumar R, Xi Y. MicroRNA, epigenetic machinery and lung cancer. Thorac Cancer 2011; 2: 35–44. [DOI] [PubMed] [Google Scholar]
- 19. Pu Q, Huang Y, Lu Y et al Tissue‐specific and plasma microRNA profiles could be promising biomarkers of histological classification and TNM stage in non‐small cell lung cancer. Thorac Cancer 2016; 7: 348–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu S J, Horlbeck M A, Cho S W et al CRISPRi‐based genome‐scale identification of functional long noncoding RNA loci in human cells. Science 2017; 355: pii: aah7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li S, Sun X, Miao S, Liu J, Jiao W et al Differential protein‐coding gene and long noncoding RNA expression in smoking‐related lung squamous cell carcinoma. Thorac Cancer 2017; 8: 672–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zeng M, Zhu L, Li L et al miR‐378 suppresses the proliferation, migration and invasion of colon cancer cells by inhibiting SDAD1. Cell Mol Biol Lett 2017; 22: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen X, Jiang Y, Huang Z et al miRNA‐378 reverses chemoresistance to cisplatin in lung adenocarcinoma cells by targeting secreted clusterin. Sci Rep 2016; 6: 19455. [DOI] [PMC free article] [PubMed] [Google Scholar]


