Abstract
Kinesins are widely expressed, microtubule‐dependent motors that play vital roles in microtubule‐associated cellular activities, such as cell division and intracellular transport. Eg5, also known as kinesin‐5 or kinesin spindle protein, is a member of the kinesin family that contributes to the formation and maintenance of the bipolar mitotic spindle during cell division. Small‐molecule compounds that inhibit Eg5 activity have been shown to impair spindle assembly, block mitotic progression, and possess anti‐cancer activity. Recent studies focusing on the localization and functions of Eg5 in plants have demonstrated that in addition to spindle organization, this motor protein has non‐canonical functions, such as chromosome segregation and cytokinesis, that have not been observed in animals. In this review, we discuss the structure, function, and localization of Eg5 in various organisms, highlighting the specific role of this protein in plants. We also propose directions for the future studies of novel Eg5 functions based on the lessons learned from plants.
Keywords: Eg5, kinesin, localization, microtubule, mitotic spindle
Introduction
Mitosis is an essential cellular process in eukaryotic organisms. To ensure that mitosis is carried out with accuracy and at the appropriate frequency, eukaryotic cells have evolved complex and finely tuned mechanisms. At the core of these regulatory mechanisms is the mitotic spindle, a complex structure comprising microtubules; microtubule‐dependent motor proteins, such as dynein and kinesin family proteins; and non‐motor microtubule‐binding proteins.1, 2 Microtubules, as the key structural component of the bipolar mitotic spindle, are essential for its assembly and maintenance.3, 4 Microtubule‐binding proteins, especially motor proteins, are also essential for spindle integrity.5, 6, 7
A subset of kinesins have been shown to play vital roles in the process of cell division, including the establishment of spindle bipolarity, chromosome alignment, and cytokinesis; these processes all rely on the synergistic effects of kinesins.8 Based on sequence similarity, kinesin proteins have been divided into more than a dozen subfamilies.9, 10 Eg5, also known as kinesin‐5 or kinesin spindle protein, is a member of the kinesin family and has been shown to play a critical role in the establishment of spindle bipolarity.11 Recent studies in plants have found that Eg5 is also involved in spindle organization, chromosome segregation, and cytokinesis. Additionally, Eg5 possesses a post‐anaphase function in plants that has not been observed in animals.12, 13, 14, 15, 16 In this review, we discuss the structure, localization, and function of Eg5, with a focus on its unique localization and function in plants.
Eg5 structure and function
The amino acid sequences and structures of Eg5 are conserved across multiple species (Fig 1).10, 11 Most Eg5 proteins contain a motor domain, an internal stalk domain, and a tail domain. Four Eg5 proteins form a bipolar homotetrameric complex via interactions between the stalk domains, resulting in the positioning of two motor domains at each end of the tetramer (Fig 2a).11 The tetramers can simultaneously move toward the plus ends of two anti‐parallel microtubules, a movement that pushes anti‐parallel microtubules in opposite directions (Fig 2b).17, 18 Both ATPase activity and the microtubule‐binding property are carried out by the motor domain of Eg5.19 However, the non‐motor stalk and tail domains are also required for the protein to crosslink microtubules and slide apart anti‐parallel microtubules.20, 21 In addition, the tail domain of Eg5 contributes to its localization during mitosis and enhances its binding to microtubules.22
Figure 1.
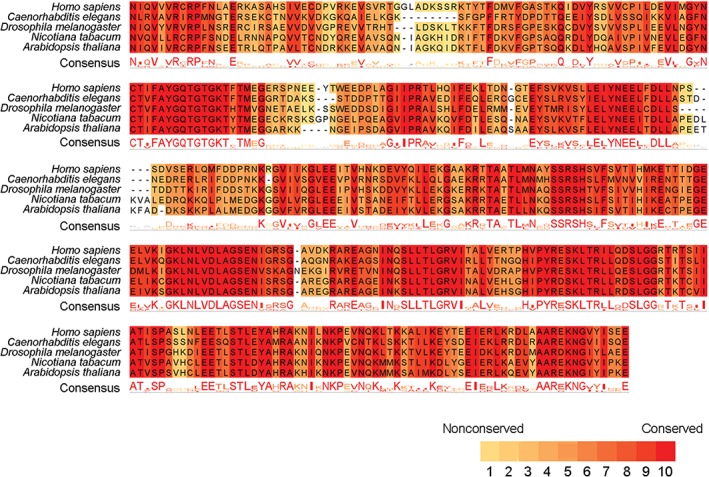
Multiple sequence alignment of conserved regions in the motor domain of Eg5 from five different species. The consistency of amino acids is classified from 0 (nonconserved) to 10 (conserved).
Figure 2.
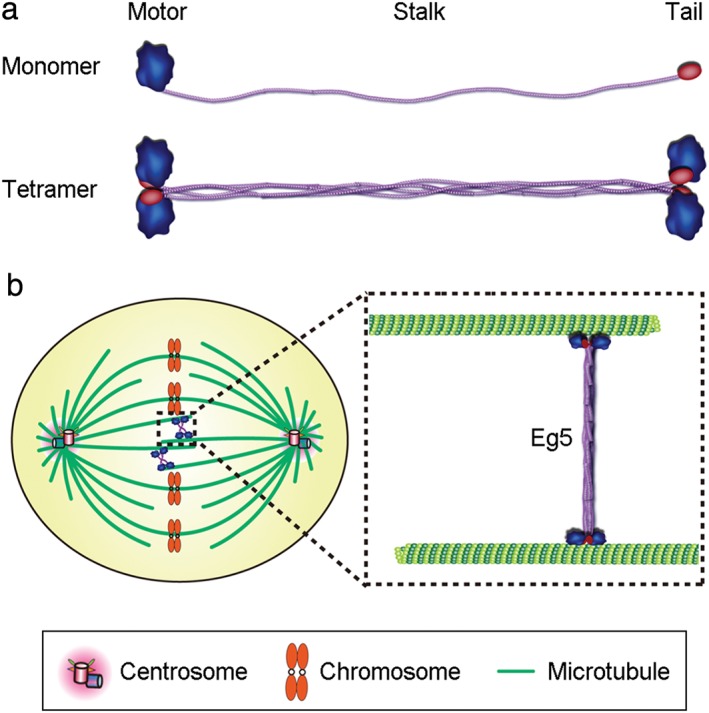
Eg5 structure and function. (a) An Eg5 monomer contains a motor domain, a stalk domain, and a tail domain (top). Four Eg5 monomers form a homotetramer via interactions between the stalk domains (bottom). (b) Schematic model showing that the Eg5 tetramer crosslinks and slides apart anti‐parallel microtubules, contributing to bipolar spindle formation and maintenance.
In multiple eukaryotic organisms, the core functions of Eg5 are similar. In animals, fungi, and plants, Eg5 monomers form tetramers that play an essential role in the establishment of spindle bipolarity. In addition, stable Eg5 dimers have been reported to promote microtubule polymerization in vitro.23 A requirement for Eg5 during mitosis has been demonstrated experimentally in cells from multiple species. Deletion of Eg5, reduction of its expression, or inhibition of its activity results in the formation of a monopolar spindle (Fig 3), leading to the activation of the spindle checkpoint and a subsequent block in cell division.11, 24, 25, 26 The formation of a monopolar spindle upon the impairment of Eg5 supports the general consensus that the core function of Eg5 is to slide apart anti‐parallel microtubules during mitosis.
Figure 3.
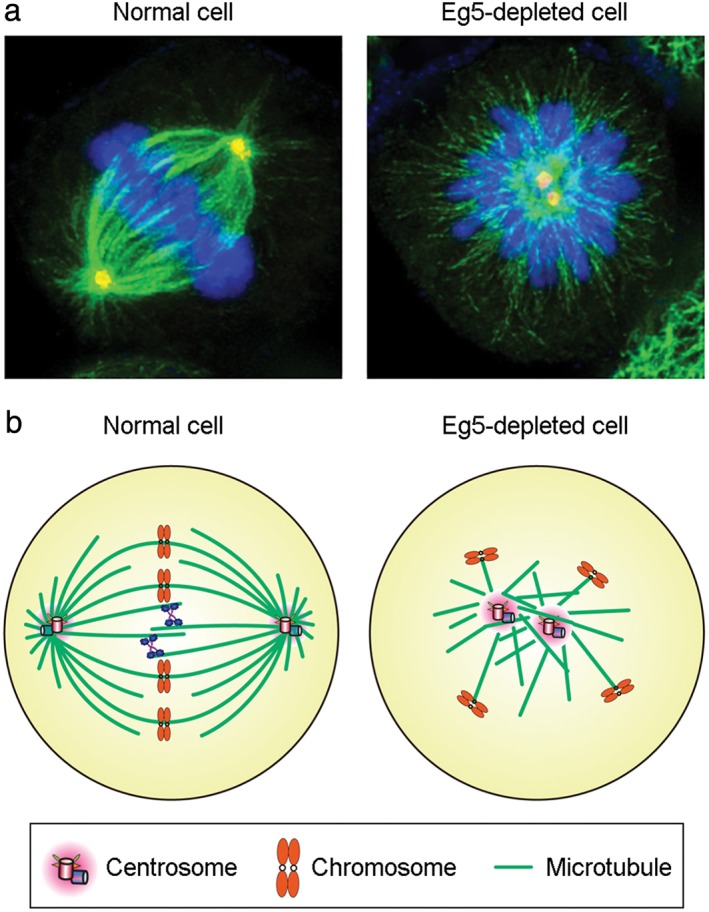
Depletion of Eg5 impairs mitotic spindle formation. (a) Immunofluorescence microscopy of microtubules (green), γ‐tubulin (red), and chromosomes (blue) in normal metaphase or Eg5‐depleted HeLa cells. (b) Schematic model illustrating a bipolar spindle in the normal metaphase cell and a monopolar spindle in the Eg5‐depleted cell.
In mammalian cells, overexpression of Eg5 leads to abnormal cell division and genomic instability, anomalies associated with tumorigenesis.27 For example, overexpression of Eg5 in pancreatic cancer cells causes the formation of multipolar spindles.28 Given that Eg5 plays an essential role during cell division and that it is highly expressed in many human cancer types,28, 29, 30, 31, 32, 33, 34 this protein is considered a promising target for cancer therapy. Numerous small‐molecule compounds that inhibit Eg5 activity have been shown to inhibit cancer cell division, induce apoptosis, and effectively block tumor growth in mice; in addition, several Eg5 inhibitors are already in clinical trials as potential anti‐cancer agents.25, 35
Subcellular localization of Eg5
The subcellular localization of Eg5 varies between species. For example, in Drosophila melanogaster cells, Eg5 localizes uniformly along spindle microtubules. In Xenopus laevis cells, Eg5 is enriched near the spindle poles, a localization pattern that results from dynein‐dependent transport.36, 37 In Caenorhabditis elegans, Eg5 is mainly expressed in the hermaphrodite germline of fertilized embryos; after fertilization, the subcellular localization changes during the cell cycle.38 In prophase, Eg5 begins to interact with microtubules, and in metaphase, Eg5 is enriched on the spindle poles and kinetochore microtubules. During anaphase, Eg5 is positioned at the central spindle, while in telophase, Eg5 localizes to the midbody. The localization of Eg5 in prophase has recently been shown to depend on TPX2, a microtubule nuclear factor.39, 40
In plants, Eg5 localization shows substantial divergence from the localization patterns observed in animals and fungi. In Arabidopsis thaliana, Eg5 is distributed along microtubules throughout the cell cycle, irrespective of whether cells are in interphase or mitosis.14, 41, 42, 43 In Nicotiana tabacum, the distribution of microtubules changes with the progression of the cell cycle, as does the localization of Eg5.13, 44 During the S phase of the cell cycle, Eg5 distributes along cortical microtubules. In pre‐mitotic cells, it localizes along microtubules in the pre‐prophase band and along perinuclear microtubules. During mitosis, Eg5 is distributed along spindle microtubules and on the equatorial plate, while in cytokinesis, Eg5 localizes to phragmoplast microtubules.13 In Physcomitrella patens, Eg5 localizes to cytoplasmic microtubules in prophase and to phragmoplast microtubules in prometaphase.15 Surprisingly, however, there is little localization of Eg5 to the equatorial plate, an area where anti‐parallel microtubules are enriched.
Non‐canonical functions of Eg5 in plants
The unique localization of Eg5 in plants suggests that this protein may possess novel functions in plants. These non‐canonical activities of Eg5 also suggest the possibility that additional roles for Eg5 and other kinesins may exist in animal cells. In Nicotiana tabacum, Eg5 is reportedly involved in separating anti‐parallel microtubules in the phragmoplast.13, 45, 46 Inhibition of Eg5 using a peptide that targets its motor domain blocks the translocation of phragmoplast microtubules, suggesting that the motor activity of Eg5 is required for the organization of phragmoplast microtubules. Additionally, this phenotype indicates a vital role for Eg5 in microtubule translocation, an event that is critical for the formation and maintenance of the bipolar structure of the phragmoplast.
In Arabidopsis thaliana, disruption of Eg5 activity leads to disorganized intracellular microtubules during interphase and disrupted spindle microtubules.14, 47 These changes affect the formation of the bipolar spindle, suggesting that the function of Eg5 in Arabidopsis thaliana may be similar to that in animals. However, in mammalian epithelial cells depleted of Eg5, expression of Arabidopsis thaliana Eg5 does not rescue the formation of bipolar spindles, despite the localization of exogenous Eg5 to spindle microtubules.14, 48 In Physcomitrella patens, Eg5 plays a significant role in spindle organization and chromosome segregation.15 Depletion of Eg5 induces the formation of multinucleated cells, a phenotype resulting from aberrant chromosome segregation. Additionally, spindle microtubules are disrupted in Eg5‐depleted cells, with metaphase spindles appearing longer and slackened and phragmoplast microtubules forming later and failing to properly align.15 However, depletion of Eg5 in Physcomitrella patens does not affect spindle bipolarity, an observation inconsistent with the classical function of Eg5 in the formation and maintenance of bipolar spindles.
Conclusions and perspectives
Given the vital role Eg5 plays during cell division, characterization of the regulation of Eg5 structure and function may promote a deeper understanding of the molecular mechanisms that underlie cell division. In animals, exploring the regulation of Eg5 may provide new knowledge regarding the pathogenesis of cancer and other diseases, leading to improved diagnosis and treatments.49, 50, 51, 52, 53, 54, 55, 56 In plants, exploring the function, localization, and motor activity of Eg5 may provide important insights into its roles in plant growth and development; furthermore, these studies will provide new knowledge for Eg5 research in animals.7, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66 It is worth noting that the localization of kinesins during mitosis has been systematically analyzed in Physcomitrella patens.15 Forty‐two kinesins are found in microtubule‐based structures, such as the kinetochores, spindles, and phragmoplasts. Only one kinesin shows the same localization pattern as its animal homologs, and many kinesins are enriched at unexpected locations.15 Therefore, studying the localization and function of Eg5 and other kinesins in plant cells could contribute to the discovery of novel Eg5 functions that may be difficult to uncover in animals.
Accumulating evidence indicates that Eg5 localization, motor activity, and function are modulated by post‐translational modifications (Fig 4).67, 68, 69, 70, 71, 72, 73, 74, 75 For example, the threonine at position 926 in the tail domain of human Eg5 can be phosphorylated by Cdk1; this modification is important for the interaction of Eg5 with microtubules and its localization to the spindle.76 In addition, the serine at position 1033, also in the tail domain of human Eg5, is phosphorylated by Nek6/7, a modification that contributes to its localization to the spindle pole.77 In contrast to the Eg5 tail domain, it is unclear whether the motor domain of Eg5 is phosphorylated, and whether such phosphorylation is involved in the modulation of Eg5 localization, motor activity, or function in spindle assembly and maintenance. Recent studies have suggested acetylation of Eg5 at lysine 146, which is located in the α2 helix of the motor domain, enhances its mechanochemical coupling, and alters its mitotic function.75 In addition, Src is reported to phosphorylate three tyrosines in the motor domain of human Eg578 however, the molecular mechanisms regulating this modification and its functional significance remain unknown. It also remains to be determined whether the phosphorylation of Eg5 affects its role in plant growth and development. Additional studies identifying other forms of post‐translational modifications, and determining how these modifications affect Eg5 localization and function may lead to the discovery of novel non‐canonical activities for Eg5.
Figure 4.
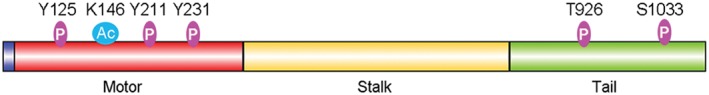
Schematic illustration of phosphorylated (P) and acetylated (Ac) amino acid residues in Eg5.
Disclosure
No authors report any conflict of interest.
Acknowledgment
This study was supported by grants from the National Natural Science Foundation of China (31671403 and 31741039).
References
- 1. Prosser SL, Pelletier L. Mitotic spindle assembly in animal cells: A fine balancing act. Nat Rev Mol Cell Biol 2017; 18: 187–201. [DOI] [PubMed] [Google Scholar]
- 2. Yamada M, Goshima G. Mitotic spindle assembly in land plants: Molecules and mechanisms. Biology (Basel) 2017; 6: pii:E6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Xie S, Zhou J. Harnessing plant biodiversity for the discovery of novel anticancer drugs targeting microtubules. Front Plant Sci 2017; 8: 720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Masoud K, Herzog E, Chabouté ME, Schmit AC. Microtubule nucleation and establishment of the mitotic spindle in vascular plant cells. Plant J 2013; 75: 245–57. [DOI] [PubMed] [Google Scholar]
- 5. Xie S, Ogden A, Aneja R, Zhou J. Microtubule‐binding proteins as promising biomarkers of paclitaxel sensitivity in cancer chemotherapy. Med Res Rev 2016; 36: 300–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. He X, Liu Z, He Q et al Identification of novel microtubule‐binding proteins by taxol‐mediated microtubule stabilization and mass spectrometry analysis. Thorac Cancer 2015; 6: 649–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu W, Ji SX, Fang XL et al Protein kinase LTRPK1 influences cold adaptation and microtubule stability in rice. J Plant Growth Regul 2013; 32: 483–90. [Google Scholar]
- 8. Lu W, Gelfand VI. Moonlighting motors: Kinesin, dynein, and cell polarity. Trends Cell Biol 2017; 27: 505–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang W, Cao L, Wang C, Gigant B, Knossow M. Kinesin, 30 years later: Recent insights from structural studies. Protein Sci 2015; 24: 1047–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hirokawa N, Tanaka Y. Kinesin superfamily proteins (KIFs): Various functions and their relevance for important phenomena in life and diseases. Exp Cell Res 2015; 334: 16–25. [DOI] [PubMed] [Google Scholar]
- 11. Waitzman JS, Rice SE. Mechanism and regulation of kinesin‐5, an essential motor for the mitotic spindle. Biol Cell 2014; 106: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gillmor CS, Roeder AH, Sieber P, Somerville C, Lukowitz W. A genetic screen for mutations affecting cell division in the Aarabidopsis thaliana embryo identifies seven loci required for cytokinesis. PLoS One 2016; 11: e0146492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Asada T, Kuriyama R, Shibaoka H. TKRP125, a kinesin‐related protein involved in the centrosome‐independent organization of the cytokinetic apparatus in tobacco BY‐2 cells. J Cell Sci 1997; 110: 179–89. [DOI] [PubMed] [Google Scholar]
- 14. Bannigan A, Scheible WR, Lukowitz W et al A conserved role for kinesin‐5 in plant mitosis. J Cell Sci 2007; 120: 2819–27. [DOI] [PubMed] [Google Scholar]
- 15. Miki T, Naito H, Nishina M, Goshima G. Endogenous localizome identifies 43 mitotic kinesins in a plant cell. Proc Natl Acad Sci U S A 2014; 111: E1053–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lee YR, Qiu W, Liu B. Kinesin motors in plants: From subcellular dynamics to motility regulation. Curr Opin Plant Biol 2015; 28: 120–6. [DOI] [PubMed] [Google Scholar]
- 17. Kapitein LC, Kwok BH, Weinger JS, Schmidt CF, Kapoor TM, Peterman EJ. Microtubule cross‐linking triggers the directional motility of kinesin‐5. J Cell Biol 2008; 182: 421–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shimamoto Y, Forth S, Kapoor TM. Measuring pushing and braking forces generated by ensembles of kinesin‐5 crosslinking two microtubules. Dev Cell 2015; 34: 669–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen GY, Mickolajczyk KJ, Hancock WO. The kinesin‐5 chemomechanical cycle is dominated by a two‐heads‐bound state. J Biol Chem 2016; 291: 20283–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Acar S, Carlson DB, Budamagunta MS et al The bipolar assembly domain of the mitotic motor kinesin‐5. Nat Commun 2013; 4: 1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Muretta JM, Jun Y, Gross SP, Major J, Thomas DD, Rosenfeld SS. The structural kinetics of switch‐1 and the neck linker explain the functions of kinesin‐1 and Eg5. Proc Natl Acad Sci U S A 2015; 112: E6606–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Weinger JS, Qiu M, Yang G, Kapoor TM. A nonmotor microtubule binding site in kinesin‐5 is required for filament crosslinking and sliding. Curr Biol 2011; 21: 154–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen Y, Hancock WO. Kinesin‐5 is a microtubule polymerase. Nat Commun 2015; 6: 8160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu M, Li D, Sun L et al Modulation of Eg5 activity contributes to mitotic spindle checkpoint activation and Tat‐mediated apoptosis in CD4‐positive T‐lymphocytes. J Pathol 2014; 233: 138–47. [DOI] [PubMed] [Google Scholar]
- 25. Pérez‐Melero C. KSP inhibitors as antimitotic agents. Curr Top Med Chem 2014; 14: 2286–311. [DOI] [PubMed] [Google Scholar]
- 26. Chen H, Connell M, Mei L, Reid GSD, Maxwell CA. The nonmotor adaptor HMMR dampens Eg5‐mediated forces to preserve the kinetics and integrity of chromosome segregation. Mol Biol Cell 2018; 29: 786–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Castillo A, Morse HC 3rd, Godfrey VL, Naeem R, Justice MJ. Overexpression of Eg5 causes genomic instability and tumor formation in mice. Cancer Res 2007; 67: 10138–47. [DOI] [PubMed] [Google Scholar]
- 28. Liu M, Wang X, Yang Y et al Ectopic expression of the microtubule‐dependent motor protein Eg5 promotes pancreatic tumourigenesis. J Pathol 2010; 221: 221–8. [DOI] [PubMed] [Google Scholar]
- 29. Ding S, Xing N, Lu J et al Overexpression of Eg5 predicts unfavorable prognosis in non‐muscle invasive bladder urothelial carcinoma. Int J Urol 2011; 18: 432–8. [DOI] [PubMed] [Google Scholar]
- 30. Liu C, Zhou N, Li J, Kong J, Guan X, Wang X. Eg5 overexpression is predictive of poor prognosis in hepatocellular carcinoma patients. Dis Markers 2017; 2017: 2176460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu L, Liu X, Mare M et al Overexpression of Eg5 correlates with high grade astrocytic neoplasm. J Neurooncol 2016; 126: 77–80. [DOI] [PubMed] [Google Scholar]
- 32. Liu M, Aneja R, Sun X et al Parkin regulates Eg5 expression by Hsp70 ubiquitination‐dependent inactivation of c‐Jun NH2‐terminal kinase. J Biol Chem 2008; 283: 35783–8. [DOI] [PubMed] [Google Scholar]
- 33. Liu M, Aneja R, Liu C et al Inhibition of the mitotic kinesin Eg5 up‐regulates Hsp70 through the phosphatidylinositol 3‐kinase/Akt pathway in multiple myeloma cells. J Biol Chem 2006; 281: 18090–7. [DOI] [PubMed] [Google Scholar]
- 34. Liu M, Yu H, Huo L, Liu J, Li M, Zhou J. Validating the mitotic kinesin Eg5 as a therapeutic target in pancreatic cancer cells and tumor xenografts using a specific inhibitor. Biochem Pharmacol 2008; 76: 169–78. [DOI] [PubMed] [Google Scholar]
- 35. Myers SM, Collins I. Recent findings and future directions for interpolar mitotic kinesin inhibitors in cancer therapy. Future Med Chem 2016; 8: 463–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sawin KE, Mitchison TJ. Mutations in the kinesin‐like protein Eg5 disrupting localization to the mitotic spindle. Proc Natl Acad Sci U S A 1995; 92: 4289–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fernández JP, Agüero TH, Vega López GA, Marranzino G, Cerrizuela S, Aybar MJ. Developmental expression and role of kinesin Eg5 during Xenopus laevis embryogenesis. Dev Dyn 2014; 243: 527–40. [DOI] [PubMed] [Google Scholar]
- 38. Bishop JD, Han Z, Schumacher JM. The Caenorhabditis elegans Aurora B kinase AIR‐2 phosphorylates and is required for the localization of a BimC kinesin to meiotic and mitotic spindles. Mol Biol Cell 2005; 16: 742–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Eibes S, Gallisà‐Suñe N, Rosas‐Salvans M, Martínez‐Delgado P, Vernos I, Roig J. Nek9 phosphorylation defines a new role for TPX2 in Eg5‐dependent centrosome separation before nuclear envelope breakdown. Curr Biol 2018; 28: 121–9.e4. [DOI] [PubMed] [Google Scholar]
- 40. Meng XQ, Dai YY, Jing LD et al Subcellular localization of proline‐rich tyrosine kinase 2 during oocyte fertilization and early‐embryo development in mice. J Reprod Dev 2016; 62: 351–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ren XL, Qi GN, Feng HQ et al Calcineurin B‐like protein CBL10 directly interacts with AKT1 and modulates K+ homeostasis in Arabidopsis. Plant J 2013; 74: 258–66. [DOI] [PubMed] [Google Scholar]
- 42. Li SP, Chen M, Yu DL et al EXO70A1‐mediated vesicle trafficking is critical for tracheary element development in Arabidopsis. Plant Cell 2013; 25: 1774–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhao Y, Zhao SS, Mao TL et al The plant‐specific actin binding protein SCAB1 stabilizes actin filaments and regulates stomatal movement in Arabidopsis. Plant Cell 2011; 23: 2314–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tang GY, Shao FX, Xu PL, Shan L, Liu ZJ. Overexpression of a peanut NAC gene, AhNAC4, confers enhanced drought tolerance in tobacco. Russ J Plant Physiol 2017; 64: 525–35. [Google Scholar]
- 45. Li S, van Os GM, Ren S et al Expression and functional analyses of EXO70 genes in Arabidopsis implicate their roles in regulating cell type‐specific exocytosis. Plant Physiol 2010; 154: 1819–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tang GY, Wei LQ, Liu ZJ, Bi YP, Shan L. Ectopic expression of peanut acyl carrier protein in tobacco alters fatty acid composition in the leaf and resistance to cold stress. Biol Plantarum 2012; 56: 493–501. [Google Scholar]
- 47. Zhang LY, Zhang XJ, Fan SJ. Meta‐analysis of salt‐related gene expression profiles identifies common signatures of salt stress responses in Arabidopsis. Plant Syst Evol 2017; 303: 757–74. [Google Scholar]
- 48. Zheng Y, Liao C, Zhao S, Wang C, Guo Y. The glycosyltransferase QUA1 regulates chloroplast‐associated calcium signaling during salt and drought stress in Arabidopsis. Plant Cell Physiol 2017; 58: 329–41. [DOI] [PubMed] [Google Scholar]
- 49. Liu M, Xie S, Zhou J. Use of animal models for the imaging and quantification of angiogenesis. Exp Anim 2018; 67: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang W, Song Z, Zhang Y. Response to crizotinib in a squamous cell lung carcinoma patient harbouring echinoderm microtubule‐associated protein‐like 4‐anaplastic lymphoma translocation: A case report. Thorac Cancer 2016; 7: 355–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Xie W, Zhou J. Aberrant regulation of autophagy in mammalian diseases. Biol Lett 2018; 14: pii:: 20170540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yang Y, Mu T, Li T et al Effects of FSTL1 on the proliferation and motility of breast cancer cells and vascular endothelial cells. Thorac Cancer 2017; 8: 606–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li H, Yang GW, Ma F et al Molecular characterization of a fish‐specific toll‐like receptor 22 (TLR22) gene from common carp (Cyprinus carpio L.): Evolutionary relationship and induced expression upon immune stimulants. Fish Shellfish Immunol 2017; 63: 74–86. [DOI] [PubMed] [Google Scholar]
- 54. Guo T, Zhang L, Cheng D et al Low‐density lipoprotein receptor affects the fertility of female mice. Reprod Fertil Dev 2015; 27: 1222–32. [DOI] [PubMed] [Google Scholar]
- 55. Du X, Zhou J. Application of biosensors to detection of epidemic diseases in animals. Res Vet Sci 2018; 118: 444–8. [DOI] [PubMed] [Google Scholar]
- 56. Yang HT, Zou SS, Zhai LJ et al Pathogen invasion changes the intestinal microbiota composition and induces innate immune responses in the zebrafish intestine. Fish Shellfish Immunol 2017; 71: 35–42. [DOI] [PubMed] [Google Scholar]
- 57. Huang J, Li Z, Biener G et al Carbonic anhydrases function in anther cell differentiation downstream of the receptor‐like kinase EMS1. Plant Cell 2017; 29: 1335–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhao S, Jiang Y, Zhao Y et al Casein kinase1‐like protein 2 regulates actin filament stability and stomatal closure via phosphorylation of actin depolymerizing factor. Plant Cell 2016; 28: 1422–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhao BT, Dai AH, Wei HC et al Arabidopsis KLU homologue GmCYP78A72 regulates seed size in soybean. Plant Mol Biol 2016; 90: 33–47. [DOI] [PubMed] [Google Scholar]
- 60. Yuan F, Lyu MJA, Leng BY, Zhu XG, Wang BS. The transcriptome of NaCl‐treated Limonium bicolor leaves reveals the genes controlling salt secretion of salt gland. Plant Mol Biol 2016; 91: 241–56. [DOI] [PubMed] [Google Scholar]
- 61. Wang SS, Wang F, Tan SJ, Wang MX, Sui N, Zhang XS. Transcript profiles of maize embryo sacs and preliminary identification of genes involved in the embryo sac‐pollen tube interaction. Front Plant Sci 2014; 5: 702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sui N, Tian SS, Wang WQ, Wang MJ, Fan H. Overexpression of glycerol‐3‐phosphate acyltransferase from Suaeda salsa improves salt tolerance in Arabidopsis. Front Plant Sci 2017; 8: 1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sui N, Wang Y, Liu SS, Yang Z, Wang F, Wan SB. Transcriptomic and physiological evidence for the relationship between unsaturated fatty acid and salt stress in peanut. Front Plant Sci 2018; 9:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Liu F, Yang YJ, Gao JW, Ma CL, Bi YP. A comparative transcriptome analysis of a wild purple potato and its red mutant provides insight into the mechanism of anthocyanin transformation. PLoS One 2018; 13:e019406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Guo JR, Li YD, Han GL, Song J, Wang BS. NaCl markedly improved the reproductive capacity of the euhalophyte Suaeda salsa . Funct Plant Biol 2018; 45: 350–61. [DOI] [PubMed] [Google Scholar]
- 66. Zhao CZ, Qiu JJ, Agarwal G et al Genome‐wide discovery of microsatellite markers from diploid progenitor species, Arachis duranensis and A. ipaensis, and their application in cultivated peanut (A. hypogaea). Front Plant Sci 2017; 8: 1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Duan Y, Huo D, Gao J et al Ubiquitin ligase RNF20/40 facilitates spindle assembly and promotes breast carcinogenesis through stabilizing motor protein Eg5. Nat Commun 2016; 7: 12648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. He J, Zhang Z, Ouyang M et al PTEN regulates EG5 to control spindle architecture and chromosome congression during mitosis. Nat Commun 2016; 7: 12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Liu Y, Wang Y, Du Z, Yan X, Zheng P, Liu Y. Fbxo30 regulates mammopoiesis by targeting the bipolar mitotic kinesin Eg5. Cell Rep 2016; 15: 1111–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Nalawansha DA, Gomes ID, Wambua MK, Pflum MKH. HDAC inhibitor‐induced mitotic arrest is mediated by Eg5/KIF11 acetylation. Cell Chem Biol 2017; 24: 481–92.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. van Ree JH, Nam HJ, Jeganathan KB, Kanakkanthara A, van Deursen JM. Pten regulates spindle pole movement through Dlg1‐mediated recruitment of Eg5 to centrosomes. Nat Cell Biol 2016; 18: 814–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Xie W, Yang Y, Gao S et al The tumor suppressor CYLD controls epithelial morphogenesis and homeostasis by regulating mitotic spindle behavior and adherens junction assembly. J Genet Genomics 2017; 44: 343–53. [DOI] [PubMed] [Google Scholar]
- 73. Luo Y, Ran J, Xie S et al ASK1 controls spindle orientation and positioning by phosphorylating EB1 and stabilizing astral microtubules. Cell Discov 2016; 2: 16033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yang Y, Liu M, Li D et al CYLD regulates spindle orientation by stabilizing astral microtubules and promoting dishevelled‐NuMA‐dynein/dynactin complex formation. Proc Natl Acad Sci U S A 2014; 111: 2158–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Muretta JM, Reddy BJN, Scarabelli G et al A posttranslational modification of the mitotic kinesin Eg5 that enhances its mechanochemical coupling and alters its mitotic function. Proc Natl Acad Sci U S A 2018; 115: E1779–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Blangy A, Lane HA, d'Hérin P, Harper M, Kress M, Nigg EA. Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin‐related motor essential for bipolar spindle formation in vivo. Cell 1995; 83: 1159–69. [DOI] [PubMed] [Google Scholar]
- 77. Rapley J, Nicolàs M, Groen A et al The NIMA‐family kinase Nek6 phosphorylates the kinesin Eg5 at a novel site necessary for mitotic spindle formation. J Cell Sci 2008; 121: 3912–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Bickel KG, Mann BJ, Waitzman JS, Poor TA, Rice SE, Wadsworth P. Src family kinase phosphorylation of the motor domain of the human kinesin‐5, Eg5. Cytoskeleton 2017; 74: 317–30. [DOI] [PMC free article] [PubMed] [Google Scholar]


