ABSTRACT
Nontypeable Haemophilus influenzae (NTHi) is an important pathogen in individuals of all ages. The lipooligosaccharide (LOS) of NTHi has evolved a complex structure that can be attributed to a multiplicity of glycosyltransferases, the random switching of glycosyltransferase gene expression via phase variation, and the complex structure of its core region with multiple glycoform branch points. This article adds to that complexity by describing a multifunctional enzyme (LsgB) which optimally functions when the species is grown on a solid surface and which can add either a ketodeoxyoctanoate (KDO) or an N-acetylneuramic acid (Neu5Ac) moiety to a terminal N-acetyllactosamine structure of LOS. Our studies show that expression of lsgB is reduced four- to sixfold when NTHi is grown in broth. The substrate that the enzyme utilizes is dependent upon the concentration of free Neu5Ac (between 1 and 10 µg/ml) in the environment. In environments in which Neu5Ac is below that level, the enzyme utilizes endogenous CMP-KDO as the substrate. Our studies show that during in vivo growth in an NTHi biofilm, the KDO moiety is expressed by the organism. Monoclonal antibody 6E4, which binds KDO, is bactericidal for NTHi strains that express the KDO epitope at high levels. In a survey of 33 NTHi strains isolated from healthy and diseased individuals, the antibody was bactericidal (>90% kill) for 12 strains (36%). These studies open up the possibility of using a KDO-based glycoconjugate vaccine as part of a multicomponent vaccine against NTHi.
KEYWORDS: biofilm, ELISA, lipooligosaccharide, bactericidal activity, keto-deoxyoctanoate, nontypeable Haemophilus influenzae, sialic acid, vaccine
IMPORTANCE
Nontypeable Haemophilus influenzae is an important pathogen in middle ear infections in children, sinusitis in adults, and acute bronchitis in individuals with chronic obstructive lung disease. The organism is very well adapted to the human host environment, and this has hindered successful development of an effective vaccine. In this article, we describe a mechanism by which the bacteria decorates its surface lipooligosaccharide with a sugar unique to Gram-negative bacteria, ketodeoxyoctanoate (KDO). This sugar decoration is present during active infection and we have shown that an antibody directed against this sugar can result in killing of the organism. These data demonstrate that the lipooligosaccharide ketodeoxyoctanoate epitope may be a novel NTHi-specific candidate vaccine antigen.
INTRODUCTION
The lipooligosaccharides (LOS) of nontypeable Haemophilus influenzae (NTHi) are composed of multiple heterogeneous glycoforms (1). This is due in part to the process of phase variation by which a number of the LOS glycoforms vary due to changes in expression of transferases (2). In addition, among NTHi strains, the fact that glycoform chain extension can occur from any of three and, in some instances, four heptoses adds to the complexity of the structures formed (3). In the pathogenesis of NTHi, the LOS plays a number of roles in its ability to act as a colonizer and pathogen (4–8). LOS glycoforms of NTHi can be capped with N-acetyl-5-neuraminic acid (Neu5Ac) (9, 10). Many NTHi strains can utilize Neu5Ac as a nutrient source for carbon and nitrogen. This sugar has been shown to be important in pathogenesis, as it is needed to sustain an infection in the chinchilla otitis media model (11) and is a factor necessary for successful biofilm formation in vivo and in vitro (12). NTHi cannot synthesize Neu5Ac. It acquires free Neu5Ac via unique mechanisms which are composed of an ATP-independent transport system comprised of a binding protein, SiaP, a transmembrane transporter, SiaT, and a sensitive Neu5Ac-sensing system (13–15). The presence of Neu5Ac has also been shown to render NTHi resistant to killing by normal human serum (15–18) and is presumed to be critical to survival in the host. NTHi strains can have multiple distinct sialyltransferases (Lic3A, Lic3B, SiaA, and LsgB) (19–21). Both Lic3A and Lic3B are phase variable and sialylate terminal lactose moieties. Lic3B has also been shown to have transialidase activity and is capable of adding Neu5Ac to a terminal galactose (18). SiaA sialylates a terminal N-acetyllactosamine (17), and like LsgB, it is not phase variable.
The NTHi sialyltransferases, LsgB, is transcribed in a six-gene operon (22, 23). This operon contains genes that synthesize a glycoform containing hexoses, N-acetylhexosamines and Neu5Ac (24). Studies have shown that when this operon is cloned into an Escherichia coli K-12 strain lacking CMP-Neu5Ac synthase, the N-acetyllactosamine acceptor for Neu5Ac is capped by 2-keto-3-deoxyoctanoate (KDO), suggesting that this enzyme may be able to substitute KDO for Neu5Ac (22, 23, 25). When siaB is cloned into the E. coli background, LsgB places Neu5Ac on the terminal N-acetyllactosamine (26).
In the studies described in this article, we demonstrate that the sialyltransferase LsgB in NTHi can incorporate KDO on a terminal N-acetyllactosamine on their LOS and that this sialyltransferase can utilize endogenous CMP-KDO as a substrate in the absence of CMP-Neu5Ac.
RESULTS
Surface plasmon resonance analysis of MAb 6E4.
To confirm the specificity of monoclonal antibody (MAb) 6E4, surface plasmon resonance (SPR) was used to test the binding specificity of MAb 6E4 binding to KDO monosaccharide. MAb 6E4 was immobilized on a series S sensor chip CM5. Five sugars were tested, KDO, Neu5Ac, Neu5Gc, and the trisaccharides, α2-3 sialylactosamine (2-3SLN) containing either an acetyl or glycolyl group. As can be seen in Table 1 and Fig. 1, the binding affinity (KD) of MAb 6E4 to KDO was 11.5 ± 1.1 nM, which was 300-fold better binding than to Neu5Ac or Neu5Gc. No binding was detected with the trisaccharides tested.
TABLE 1 .
Surface plasmon resonance results of MAb 6E4 antibody with five sugars
| MAb | SPR result with the following sugara: |
||||
|---|---|---|---|---|---|
| KDO | Neu5Ac | Neu5Gc | Acα2-3SLN | Gcα2-3SLN | |
| 6E4 | 11.5 ± 1.1 nM | 3.45 ± 2.1 µM | 9.33 ± 2.2 µM | NCDI | NCDI |
SPR results of MAb 6E4 with free KDO, Neu5Ac, Neu5Gc, and α2-3 sialylactosamine (2-3SLN) containing either an acetyl group (Acα2-3SLN) or glycolyl group (Gcα2-3SLN). NCDI, no concentration-dependent interaction.
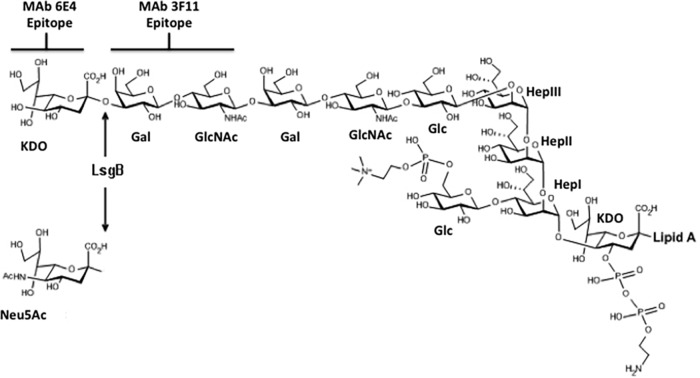
FIG 1 .
Structure of the KDO-containing glycoform of NTHi strain 2019 (23, 26, 40). The MAb 6E4 and 3F11 binding sites are demarcated.
Expression of the KDO epitope on NTHi is optimized on solid media and in the absence of Neu5Ac.
Studies in our laboratory have previously shown that MAb 6E4 derived from immunizing mice with NTHi 2019 binds to a terminal LOS epitope containing KDO (24) (Fig. 1). Our recent studies have indicated that expression of the KDO epitope is substantially reduced on bacteria grown in broth and in the presence of Neu5Ac. To address this, we tested strain NTHi 2019 grown either on supplemented RPMI 1640 (sRPMI) agar or in sRPMI broth in a whole-cell enzyme-linked immunosorbent assay (ELISA). The results shown in Fig. 2A demonstrate that there is a significant increase in epitope expression when the organisms are grown on plates. Figure 2B shows that as the concentration of Neu5Ac is increased to concentrations greater that 1 µg/µl (bars C and D) in the medium, the expression of the KDO epitope decreases significantly.
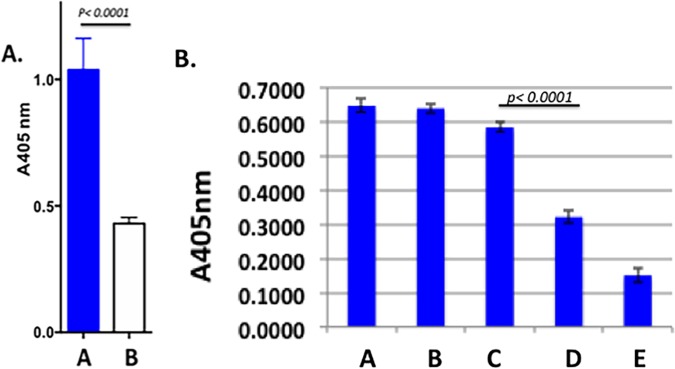
FIG 2 .
(A) Results of a MAb 6E4 whole-cell ELISA demonstrating the significantly different increase in expression of the MAb 6E4 epitope when NTHi strain 2019 is grown on solid medium (blue bar) compared to growth in broth (white bar). The antibody dilution is 1:40. A405 nm, absorbance at 405 nm. (B) Results of a whole-cell ELISA on increasing amounts of Neu5Ac in the solid medium. NTHi bacteria were grown on medium supplemented with 0, 0.1, 1, 10, and 100 μg/ml Neu5Ac, which is shown by bars A, B, C, D, and E, respectively. Significant loss of epitope expression occurs between the addition of 1 to 10 µg/ml Neu5Ac. All studies were performed in triplicate.
Neu5Ac and the KDO epitope.
Previous studies in our laboratory indicated that Neu5Ac was present on the NTHi 2019 LOS N-acetyllactosamine in the presence of 100 µM Neu5Ac (15). To define the structure upon which the KDO was being added, we performed Western blots of LOS preparations from NTHi strain 2019 grown in the absence of Neu5Ac and in 100 µM Neu5Ac before and after sialidase treatment (Fig. 3). In the presence of 100 µM Neu5Ac, neither MAb 3F11 nor 6E4 binds to the LOS (lane A). After sialidase treatment, only MAb 3F11 binds (lane B), while the LOS from NTHi strain 2019 grown in the absence of Neu5Ac shows binding by MAb 6E4 (lane C) and reduced binding by MAb 3F11. These data indicate that KDO and Neu5Ac compete for the N-acetyllactosamine terminal structure and combined with the data in Fig. 1B indicate that the sialyltransferase involved preferentially binds CMP-Neu5Ac but that in the absence of that sugar nucleotide, it will utilize endogenous CMP-KDO.
FIG 3 .
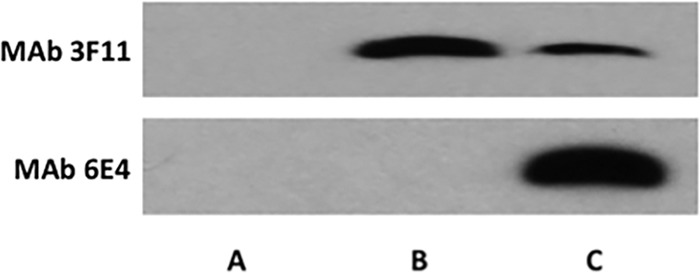
Western blots of proteinase K extracts of NTHi strain 2019 grown on sRMPI medium in the presence of 100μM Neu5Ac (lane A), the same extract treated with sialidase (lane B), and strain 2019 grown on sRMPI medium in the absence of Neu5Ac (lane C). As can be seen when the terminal N-acetyllactosamine is occupied by Neu5Ac, neither MAb 3F11 nor 6E4 binds NTHi LOS in the presence of Neu5Ac. Loss of the Neu5Ac after sialidase treatment exposes the terminal N-acetyllactosamine, resulting in MAb 3F11 binding, MAb 6E4 does not bind in this instance. When NTHi is grown in the absence of Neu5Ac, the majority of the acetyllactosamine is occupied by KDO, which is indicated by the strong MAb 6E4 binding and the weaker MAb 3F11 binding.
Figure S2 in the supplemental material shows this interaction.
Sensorgrams of single-cycle kinetics of interactions between 6E4 antibody and sugars. The panels show interactions between MAb 6E4 and KDO, Neu5Ac, Neu5Gc, α2-3 sialylactosamine (2-3SLN) containing either an acetyl group (2-3SLN-AC) or glycolyl group (2-3SLN-Gc). Download FIG S1, TIF file, 3.6 MB (3.7MB, tif) .
Copyright © 2018 Apicella et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Results of two Western blots of studies with proteinase K extracts of NTHi strain 2019 (lanes 1 to 3), ΔsiaA1 mutant (lanes 4 to 6), ΔsiaA1 Δlic3B mutant (lanes 7 to 9), ΔsiaA1 Δlic3A ΔlsgB mutant (lanes 10 to 12), and ΔsiaA1 Δlic3A ΔlsgB::lic3B mutant (lanes 13 to 15). Extracts in lanes 1, 2, 4, 5, 7, 8, 10, 11, 13, and 14 were from organisms grown in the presence of 100 μg Neu5Ac. Extracts in lane 2, 5, 8, 11, and 14 were from organisms treated with sialidase prior to electrophoresis. Extracts in lanes 3, 6, 9, 12, and 15 were from organisms grown in the absence of Neu5Ac. Lanes 16 contain LOS from N. gonorrhoeae PID 2 as a control in which a 4.5- and a 5.0-kDa band bind MAb 3F11. These blots demonstrate that MAb 6E4 did not react with any extracts derived from strains grown in the presence of Neu5Ac and that only extracts containing a functional LsgB reacted with Mab 6E4. In addition, the bottom blot shows that MAb 3F11 reactivity is reduced when 6E4 binding occurs. Finally, binding of both 6E4 and 3F11 MAbs does not occur to the LOS in extracts of organisms grown in the presence of Neu5Ac in the absence of sialidase treatment. Download FIG S2, TIF file, 3.1 MB (3.2MB, tif) .
Copyright © 2018 Apicella et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Analysis of the transferase involved in synthesis of the KDO epitope.
We constructed a series of deletion mutant combinations in all four sialyltransferases and in all combinations of the sialyltransferases in strain NTHi 2019 (see Table S1 in the supplemental material). Western blot analysis shown in Fig. 4 using MAb 6E4 demonstrated that reactivity occurred only with extracts derived from NTHi strain 2019 grown in the absence of Neu5Ac and containing a functional copy of lsgB. Figure S2 shows Western blots containing additional NTHi 2019 sialyltransferase mutant combinations with MAbs 3F11 and 6E4. Figure S2 again demonstrates that MAb 6E4 did not react with any extracts derived from strains grown in the presence of Neu5Ac and only extracts derived from strains that contained a functional lsgB gene reacted with MAb 6E4. In addition, MAb 3F11 reactivity is reduced when MAb 6E4 binding occurs. These data indicate that the LsgB sialyltransferase has the capacity to utilize CMP-KDO as a substrate in the absence of CMP-Neu5Ac. In addition, LsgB can place KDO onto the terminal LOS acetyllactosamine of NTHi strains when the organisms are grown in the absence of Neu5Ac.
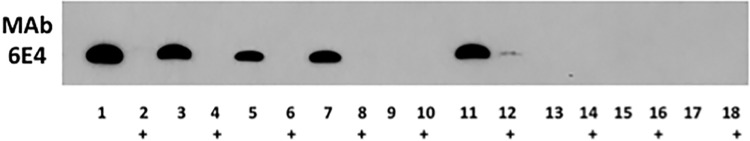
FIG 4 .
MAb 6E4 Western blot with proteinase K extracts of NTHi strains (wild type and mutants). Wild-type strain 2019 (lanes 1 and 2), ΔsiaA mutant (lanes 3 and 4), Δlic3 mutant (lanes 5 and 6), Δlic3B mutant (lanes 7 and 8), ΔlsgB mutant (lanes 9 and 10), ΔsiaA Δlic3 Δlic3B mutant (lanes 11 and 12), ΔsiaA Δlic3A ΔlsgB mutant (lanes 13 and 14), ΔsiaA Δlic3B ΔlsgB mutant (lanes 15 and 16), and ΔsiaB Δlic3A ΔlsgB mutant (lanes 17 and 18) were tested. Extracts in the even-numbered rows were from strains grown in the presence (+) of 100 μM Neu5Ac. As can be seen, MAb 6E4 did not react with any extracts derived from strains grown in the presence of 100 μM Neu5Ac, and reactivity with MAb 6E4 occurred only when the LsgB sialyltransferase was active in NTHi strain 2019. The slight band in lane 12 is due to spillover from lane 11.
Strains used in this study. Download TABLE S1, DOCX file, 0.01 MB (15.9KB, docx) .
Copyright © 2018 Apicella et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Expression of lsgB on solid and liquid media and in the presence of Neu5Ac.
Studies of lsgB gene expression were undertaken using quantitative real-time PCR (RT-PCR) on solid and liquid media and in the presence and absence of Neu5Ac. NTHi strain 2019 was studied after 12 h of growth on sRPMI agarose plates or in sRPMI broth. RNA was isolated, and the results are shown in Fig. 5A. As can be seen, expression of lsgB is increased approximately sevenfold when the organisms are grown on solid medium compared to organisms grown in broth (Fig. 5A). Similar studies were performed on lsgB expression in NTHi 2019siaR (Fig. 5B). No differences were seen in gene expression in this NTHi 2019 background on the different types of media. This suggests that siaR may be involved in the regulation of expression of lsgB.
FIG 5 .
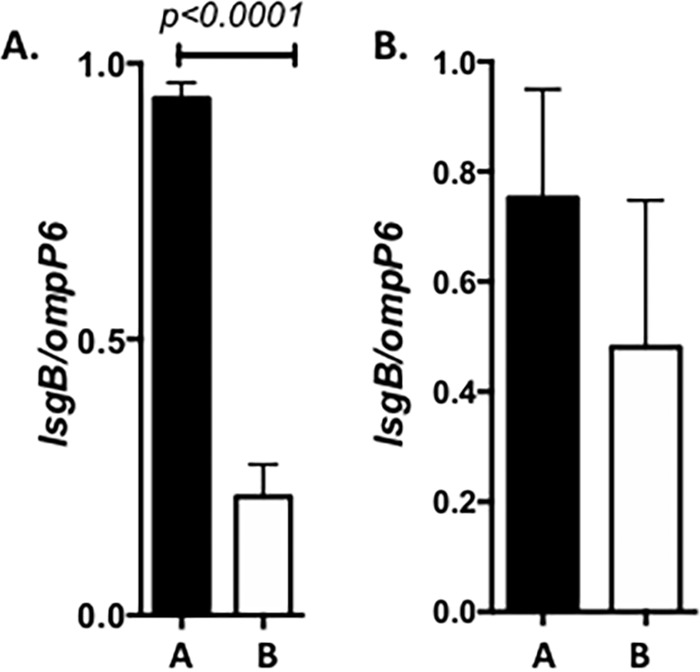
(A) lsgB expression in strain 2019 background when the organism is grown on an agarose surface (bar A) or in broth (bar B). As can be seen, there is a significant approximately fourfold increase in expression of lsgB when NTHi strain 2019 is grown on solid medium (P < 0.0001). (B) No significant differences in lsgB expression in NTHi 2019siaR strain comparing growth on solid medium (bar A) to growth in broth (bar B). The lack of difference in expression of the lsgB gene under these different growth conditions suggests that SiaR may play a role in the regulation of lsgB expression. All studies were performed in triplicate and measured in triplicate in the assay.
Prevalence of the KDO epitope among NTHi strains.
To address the frequency of the expression of the KDO epitope among NTHi strains, we performed MAb 6E4 whole-cell ELISA analysis on 33 NTHi plate-grown strains isolated from patients with different clinical conditions (Fig. 6). Relative to the NTHi strain 2019 used to generate MAb 6E4, 12 (36%) of the strains reacted strongly (>60% of the level of strain 2019), while the remaining 21 strains (64%) showed reactivity to MAb 6E4 from just over 40% to less than 10% of the reactivity of NTHi strain 2019. The mutant strain 2019lsgB serves as the negative control in these experiments.
FIG 6 .
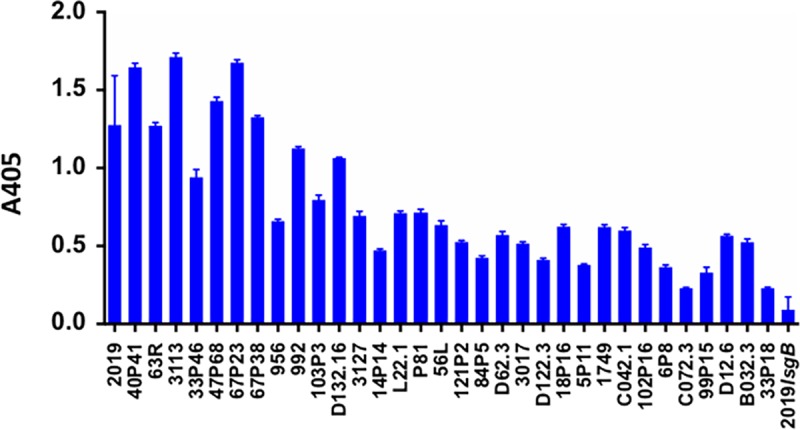
Results of whole-cell ELISA studies of 33 NTHi strains from different clinical situations and isolated from healthy carriers. The NTHi strains are shown on the x axis. These studies were performed in triplicate, and the error bars indicate the standard deviations from the means. An absorbance of 0.25 at a wavelength of 600 nm equals three times the background of this ELISA in this assay. Thirty-one of the 33 strains showed reactivity at least three times the background level of the assay (absorbance at 600 nm of 0.24). Two strains were either below or at that level. These results suggest that the majority of NTHi strains express the KDO epitope. All studies were performed in triplicate.
MAb 6E4 bactericidal activity.
Experiments in our laboratory indicated that as little as 0.1 µg purified MAb 6E4 can result in killing of 75% of NTHi strain 2019 in our bactericidal assay (Fig. 7). MAb 6E4 bactericidal activity can be inhibited by 10 µg of Neu5Ac in the media (Fig. S3). At the same time, we performed the ELISAs (Fig. 6) and bactericidal studies were done with protein G-purified MAb 6E4 on the same population of 33 NTHi strains. Figure 8 also shows the results of bactericidal studies in these strains using 1 µg protein G-purified MAb 6E4. As with the ELISA studies, Fig. 8 demonstrates a comparison to the bactericidal results in NTHi 2019 (100% killed in the assay) with each of the strains in the assay. The strains listed on the x axis are in identical positions in Fig. 6 and 8. These studies demonstrate that MAb 6E4 is bactericidal for NTHi strains if there is sufficient expression of the KDO epitope and these studies would suggest that more than one-third of NTHi strains would be susceptible to antibody directed at the KDO epitope.
FIG 7 .
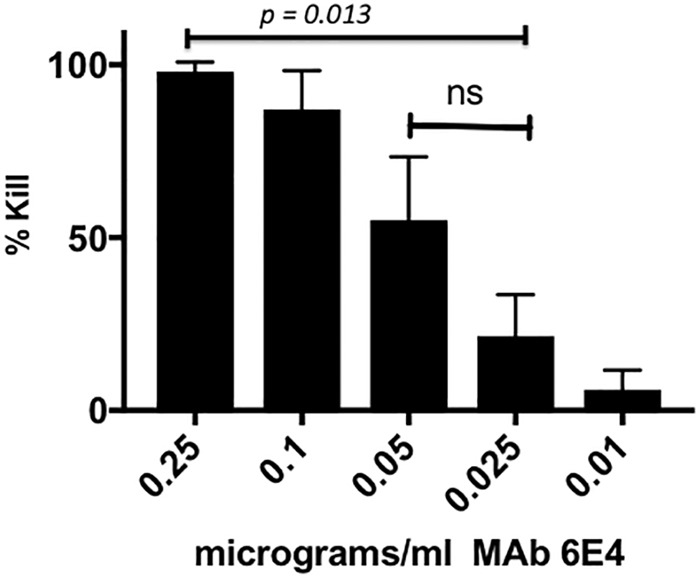
Bactericidal activity of protein G affinity-purified MAb 6E4 against NTHi strain 2019 as a function of the concentration of the antibody. The amounts (in micrograms per milliliter) of MAb 6E4 are shown below the bars. These studies show that the MAb 6E4 has significant bactericidal activity between 0.1 and 0.05 μg/ml MAb. All studies were performed in triplicate. The values that are significantly different are indicated by a bar. The values that are not significantly different (ns) are indicated by a bar.
FIG 8 .
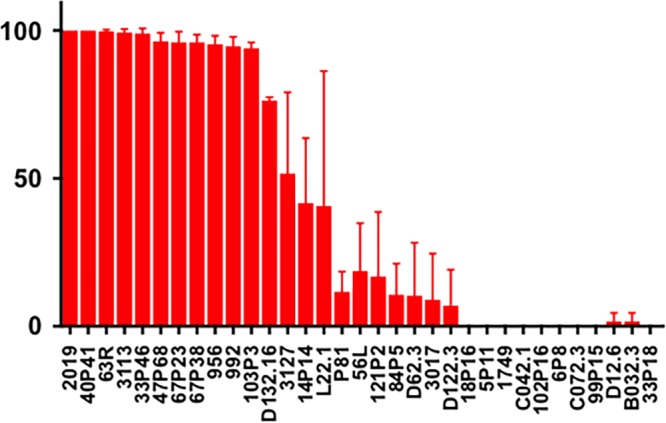
Bactericidal response for the same 33 NTHi strains shown in Fig. 6. MAb 6E4 was used at a concentration of 1 μg/ml. AGS at a 1:20 dilution was used as the complement source. The strains are arranged in the same order as in Fig. 6, demonstrating the concordance of KDO surface epitope expression with bactericidal activity. All studies were performed in triplicate.
MAb 6E4 bactericidal effect is directly related to the presence of Neu5Ac in the NTHi strain 2019. Neu5Ac was sequentially increased in the growth medium from 0.1 to 100 µg/ml. Growth of NTHi in the presence of greater than 1 µg of Neu5Ac significantly inhibits (P < 0.05) the bactericidal effect of MAb 6E4. Download FIG S3, TIF file, 3.7 MB (3.8MB, tif) .
Copyright © 2018 Apicella et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The KDO epitope is present during in vivo growth of NTHi.
This raised the question of whether there is sufficient free Neu5Ac available in clinically infected tissue to inhibit deposition of the KDO epitope. To address this question of whether NTHi grown in vivo incorporates the KDO epitope onto its LOS, we performed confocal microscopy using MAb 6E4 on biofilms frozen in archival samples of chinchilla middle ear infections. Figure 9 demonstrates the presence of the KDO epitope on NTHi strain 2019 within the biofilm during growth in the chinchilla middle ear. This suggests that within the middle ear environment during clinical infection, there may not be sufficient free Neu5Ac to inhibit deposition of KDO on the LPS by LsgB.
FIG 9 .
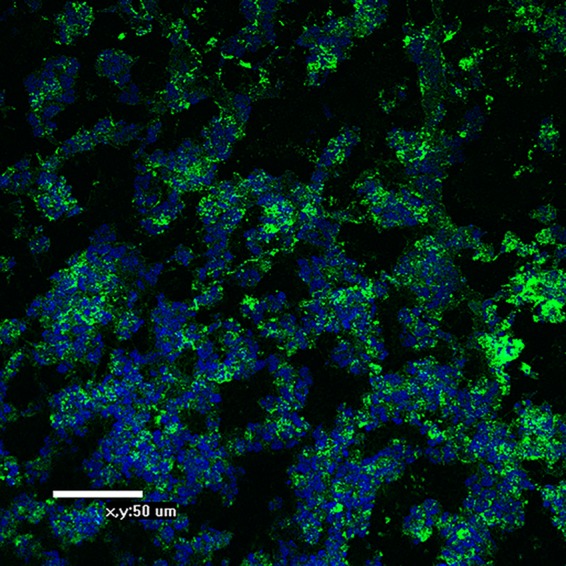
Confocal micrograph of a biofilm formed in a chinchilla middle ear by NTHi strain 2019 stained with MAb 6E4. The antibody was counterstained with a goat anti-mouse anti IgG antibody conjugated to fluorescein. The DNA in the biofilm was counterstained with DAPI and fluoresces blue. Organisms can be seen throughout the biofilm stained with MAb 6E4, indicating the presence of the KDO epitope in vivo.
DISCUSSION
NTHi LOS is an extremely complex structure. This paper documents this complexity by describing an enzyme, LsgB, which has been shown to be an LOS sialyltransferase (19), that can also substitute KDO on a terminal N-acetyllactosamine when Neu5Ac is present at levels below 10 µg/ml in the growth media. Genome analysis indicates that this enzyme is present in almost all of the NTHi genomes that have been sequenced. lsgB is a part of an operon containing genes involved in the biosynthesis of the LOS N-acetyllactosamine-containing glycoform (26). More than 90% (31/33) of the NTHi strains we tested with MAb 6E4 expressed the KDO epitope on their LOS, indicating its widespread prevalence among the species.
NTHi cannot synthesize Neu5Ac and must acquire it from its environment (15). Previously, we studied the questions of uptake of Neu5Ac and its regulation (13, 15, 17, 27). NTHi has a unique Neu5Ac ATP-independent two-component uptake system, consisting of two proteins, SiaT, the transporter, and SiaP, the binding protein, which facilitates transport. NTHi can metabolize Neu5Ac, and the operon adjacent to the operon containing SiaP and SiaT genes contain genes for enzymes, which may be involved in that degradation. Within this operon is siaR, which with crp, is involved in the intricate regulation of the uptake of Neu5Ac (Neu5Ac can be toxic to NTHi when uptake is unregulated) (13, 17, 27). Once Neu5Ac is acquired, it can be activated by SiaB to form CMP-Neu5Ac, which can be utilized by the organism to allow incorporation into the LOS structures. Studies presented in this article show that when less than 1 µg/ml of Neu5Ac is present in the growth media, LsgB utilizes CMP-KDO as the nucleotide sugar for incorporation into the LOS structure. Our studies with chinchilla biofilms formed by NTHi strain 2019 indicate that the majority, if not all, of the organisms within the biofilm express the KDO epitope. This would suggest that the level of free Neu5Ac within the biofilm is at least less than 1 µg/ml. Because of the amount of glycoproteins expressing Neu5Ac in the human host, it is difficult to measure free Neu5Ac levels. Older methodologies employed assays using strong acids which could cleave bound Neu5Ac, and determining free Neu5Ac from bound Neu5Ac has been especially difficult with the exception of certain pathological states. A recent study using derivatization of free Neu5Ac to a quinoxalinone and quantitation by gas chromatography (GC)/mass spectrometry (MS) has shown that levels of free Neu5Ac in healthy people ranges from 280 to 350 ng/ml, well below the level necessary for sialylation of the LOS by LsgB (28). NTHi does not carry a gene for a sialidase, and it would appear that a major source of Neu5Ac for the organism may be the trans-sialidase activity of NTHi sialyltransferase, Lic3B, described by Fox and coworkers (20).
It is interesting that lsgB activity is suppressed when the organism is grown in liquid media with an ~7-fold difference in expression comparing growth in solid media growth to growth in broth. siaR appears to play a role in this difference, as it disappears when siaR is mutated. The biological significance of this is not clear, but the in vivo studies showing expression of the KDO epitope in infected tissue may suggest that plate growth better simulates the in vivo environment than in vitro growth in broth.
Our studies have shown that the KDO epitope can be a target for bactericidal antibodies. Studies with purified MAb 6E4 demonstrate that significant killing of NTHi strain 2019 can occur in the presence of 0.1 µg/ml of the antibody. Studies of 33 strains isolated from healthy carriers and individuals with different diseases show that 12 (36%) of the strains showed significant killing after exposure to MAb 6E4. Given the difficulties in developing a successful vaccine to NTHi, using a glycoconjugate composed of either multiple KDOs or a KDO-N-acetyllactosamine conjugated to an immunogenic protein may be an approach to develop a component of a multicomponent vaccine similar to Bexsero or Trumenba used for protection against the serogroup B meningococcus. Additionally, the KDO epitope offers the ability to potentially use a sugar-based vaccine for NTHi without concern about cross-reactivity with human tissues, since KDO is found only in Gram-negative bacteria. Also, its effect on commensal Gram-negative bacteria would not be a concern, since KDO is buried in the deep LPS core and not available to be bound by MAb 6E4 (22, 23).
These studies further demonstrate the unique biology of NTHi LOS. The observation that the LsgB sialyltransferase is responsible for placing a terminal KDO residue on a N-acetyllactosamine glycoform in vivo and the possibility that this may be a target for bactericidal antibody opens the possibility of developing a multicomponent vaccine to NTHi.
MATERIALS AND METHODS
Strains, plasmids, and culture conditions.
The mutant strains used in this study are listed in Table S1 in the supplemental material. NTHi was grown at 37°C in the presence of 5% CO2. RPMI 1640 (Sigma-Aldrich, Saint Louis, MO) was used as a sialic acid-free chemically defined medium. Supplemented RPMI 1640 (sRPMI) was prepared with protoporphyrin IX (0.5 µg/ml), β-NAD (10 µg/ml), hypoxanthine (0.1 mg/ml), uracil (0.1 mg/ml), and sodium pyruvate (0.8 mM). Neu5Ac was added as indicated. In addition, 34 NTHi strains from different clinical conditions were studied in the whole-cell ELISA assay. These 33 strains have been previously described (29). These strains were confirmed to be H. influenzae based on hybridization to an iga gene probe (30).
Construction of mutants and selection of constitutively phase-on lic3A and lic3B in NTHi strain 2019.
Deletions of siaA1, lic3B, lic3A, and lsgB were performed as described elsewhere (31). To produce constitutively phase-on lic3A and lic3B in NTHi strain 2019, the following method was used. Single colonies of the strain of interest were selected. Genomic DNA was isolated using the MasterPure DNA purification kit from EpiCentre Biotechnologies (Madison, WI). PCR was performed using primers obtained from Integrated DNA Technologies (Iowa City, IA). Primers were labeled with 6-carboxyfluorescein. A modified version of the capillary electrophoresis fragment length analysis developed by Fox et al. (2) was used to determine the length of the tandem repeats in PCR products for lic3A and siaA2, using Peak Scanner software (Applied Biosystems International). Colonies in which the gene of interest was predominantly 90% phase on based on capillary electrophoresis studies were selected for further study.
Monoclonal antibodies.
Monoclonal antibody (MAb) 6E4 is a murine IgG3 monoclonal antibody that has been previously described (22, 23, 25). This antibody recognizes a terminal 2-keto-3-deoxyoctanoate epitope (25). MAb 3F11 is a murine IgM antibody that recognizes a terminal N-acetyllactosamine epitope (32). MAb 2C3 is a murine IgG1 antibody that binds to the 14.6-kDa H.8 protein of pathogenic Neisseria (33, 34). For bactericidal studies, MAb 6E4 was purified over a protein G column using standard elution techniques (35).
Surface plasmon resonance of 6E4 antibody and sugars.
The 6E4 antibody was immobilized onto a series S CM5 sensor chip via amine coupling using the amine coupling wizard template on a Biacore T200 system. Flow cell 1 was made as a blank cell (ethanolamine-blocked surface) to allow for double reference subtraction. Sugars were initially flowed over the 6E4 antibody from 0.01 to 100 µM, and the concentration for each sample was adjusted on the basis of observed binding as previously described (36). KDO was run at a maximum concentration of 500 nM, Neu5Ac was run at a maximum concentration of 10 µM, and N-glycolylneuraminic acid (Neu5Gc) was reduced to 50 µM. Analysis was carried out using the Biacore T200 evaluation software.
Whole-cell ELISA.
A whole-cell enzyme-linked immunosorbent assay was performed using monoclonal antibodies 6E4 and 3F11 by the method of Abdillahi and Poolman (37). Bacteria were grown on sRPMI plates, which were supplemented with Neu5Ac where indicated. Cells were harvested and resuspended in phosphate-buffered saline (PBS) to an optical density at 600 nm of 0.10. Microtiter plate wells were filled with 100 µl of the bacterial suspension and dried at 40°C overnight. The wells were washed, and ELISA was performed with MAb 6E4 at a dilution of 1:100. Secondary antibody (alkaline phosphatase-conjugated goat anti-mouse IgG; Jackson ImmunoResearch, West Grove, PA) was used at a dilution of 1:10,000. The substrate p-nitrophenylphosphate was applied at a concentration of 1.0 mg/ml. Absorbance was read in a Tecan Model Infinite 200 Pro plate reader at 405 nm.
Fluorescence analysis of archival chinchilla biofilm samples with MAb 6E4.
To preserve the architecture of biofilms that had formed in vivo, we embedded the biofilm sample in an OCT compound (Fisher Scientific, Pittsburgh, PA) as previously described (38). Serial sections (4-µm thickness) were cut on a Leica CM3050S cryotome (Leica Microsystems, Inc., Bannockburn, IL). The sections were placed on Superfrost slides (Fisher Scientific) and stored at −80°C. Prior to staining, sections were fixed in 4% (wt/vol) paraformaldehyde (in 0.1 M phosphate buffer [pH 7.4]). The DNA matrix in the specimens was stained with DAPI (4′,6-diamidino-2-phenylindole). The bacteria were stained with MAb 6E4 and secondary antibody goat anti-mouse IgG conjugated to fluorescein isothiocyanate (FITC) (Jackson ImmunoResearch).
LOS preparation and Western blotting.
Cells were grown on sRPMI solid medium in the presence or absence of 100 µM Neu5Ac as indicated. Bacteria were collected by scraping and suspended in a solution of 60 mM Tris, 10 mM EDTA, and 2% (wt/vol) sodium dodecyl sulfate, pH 6.8. Proteinase K was added to a final concentration of 50 µg/ml, and samples were incubated overnight at 37°C. After three ethanol precipitations, samples were treated with DNase I and RNase A. Samples were then phenol extracted, ethanol precipitated three more times, and centrifuged at 120,000 × g for 75 min. The pellets were resuspended in water, frozen, and then lyophilized. Neuraminidase treatments were carried out by incubating 2.5 µg LOS with 0.25 mU of neuraminidase purified from Vibrio cholerae (Roche Diagnostics, Wilmington, MA) in neuraminidase buffer (0.15 M NaCl, 4 mM CaCl2 [pH 5.5]) at 37°C for 2 h. Western blotting was performed by the method of Blake et al. (39).
RNA isolation and quantitative real-time PCR.
Cell samples were homogenized by pipetting with 1 ml of TRIZOL total RNA isolation reagent (Life Technologies, Grand Island, NY). Lysates were transferred to Lysing Matrix B tubes (MP Biomedicals, Santa Ana, CA) and vortexed for 60 s. The lysates were extracted with 200 µl of chloroform and centrifuged for 15 min. The supernatant was precipitated with 1 volume of isopropanol for 10 min at 25°C (room temperature). RNA was pelleted and washed with 75% ethanol, and the pellet was dried at room temperature. RNA was reconstituted in ultrapure water and cleaned with a Qiagen RNeasy minikit (Valencia, CA). The RNA was then treated with reagents in the TURBO DNA-free kit (Life Technologies, Carlsbad, CA) for 30 min at 37°C and purified using a phenol-chloroform-isoamyl alcohol (25:24:1; Roche, Indianapolis, IN) extraction, followed by an ethanol precipitation. One microgram of RNA was reverse transcribed using SuperScript II reverse transcriptase (Life Technologies). SYBR green quantitative real-time PCR (qRT-PCR) was used to measure gene expression. For each target, qRT-PCR primer sets were selected using Primer Express software (Agilent Technologies, Santa Clara, CA) and obtained from Integrated DNA Technologies (Coralville, IA). Relative RNA quantities were determined by standard curve (fivefold dilutions of purified genomic DNA [gDNA] ranging from 100 to 0.032 ng/µl), and all values were normalized to the amount of outer membrane protein 6 (OmpP6) RNA in each sample. OmpP6 is considered constitutively expressed in NTHi. Our data on more than 60 NTHi mRNA samples confirmed this fact. We performed 20-µl reactions in triplicate in 1× Kapa SYBR Fast qPCR master mix (Kapa Biosystems, Wilmington, MA) with 4 ng of template or gDNA standard and a final concentration of 250 nM for each primer. qRT-PCR was performed on an ABI Prism 7900 sequence detection system (Life Technologies). The data were normalized against NTHi P6 and standardized against genomic DNA from wild-type NTHi 2019 strain.
Statistical analysis.
Statistical analysis was performed with Prism 7 software (GraphPad Software, Inc., La Jolla, CA). Student’s t tests were used to compare differences in ELISA, bactericidal, and RT-PCR results. Values that met a P value cutoff of <0.05 were considered statistically different.
ACKNOWLEDGMENTS
This work was supported by NIH grants AI024616 (M.A.A.), DC015688 (M.P.J.), and NHMRC Program grant (APP1071659) and an NHMRC Principal research fellowship (APP1138466) to M.P.J. M.P.J. was supported by a University of Iowa Helen C. Levitt Visiting Professorship.
Footnotes
Citation Apicella MA, Coffin J, Ketterer M, Post DMB, Day CJ, Jen FE-C, Jennings MP. 2018. Nontypeable Haemophilus influenzae lipooligosaccharide expresses a terminal ketodeoxyoctanoate in vivo, which can be used as a target for bactericidal antibody. mBio 9:e01401-18. https://doi.org/10.1128/mBio.01401-18.
REFERENCES
- 1.Preston A, Mandrell RE, Gibson BW, Apicella MA. 1996. The lipooligosaccharides of pathogenic Gram-negative bacteria. Crit Rev Microbiol 22:139–180. doi: 10.3109/10408419609106458. [DOI] [PubMed] [Google Scholar]
- 2.Fox KL, Atack JM, Srikhanta YN, Eckert A, Novotny LA, Bakaletz LO, Jennings MP. 2014. Selection for phase variation of LOS biosynthetic genes frequently occurs in progression of non-typeable Haemophilus influenzae infection from the nasopharynx to the middle ear of human patients. PLoS One 9:e90505. doi: 10.1371/journal.pone.0090505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morey P, Viadas C, Euba B, Hood DW, Barberán M, Gil C, Grilló MJ, Bengoechea JA, Garmendia J. 2013. Relative contributions of lipooligosaccharide inner and outer core modifications to nontypeable Haemophilus influenzae pathogenesis. Infect Immun 81:4100–4111. doi: 10.1128/IAI.00492-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swords WE, Ketterer MR, Shao J, Campbell CA, Weiser JN, Apicella MA. 2001. Binding of the non-typeable Haemophilus influenzae lipooligosaccharide to the PAF receptor initiates host cell signalling. Cell Microbiol 3:525–536. doi: 10.1046/j.1462-5822.2001.00132.x. [DOI] [PubMed] [Google Scholar]
- 5.Harvey HA, Swords WE, Apicella MA. 2001. The mimicry of human glycolipids and glycosphingolipids by the lipooligosaccharides of pathogenic Neisseria and Haemophilus. J Autoimmun 16:257–262. doi: 10.1006/jaut.2000.0477. [DOI] [PubMed] [Google Scholar]
- 6.Edwards JL, Apicella MA. 2002. The role of lipooligosaccharide in Neisseria gonorrhoeae pathogenesis of cervical epithelia: lipid A serves as a C3 acceptor molecule. Cell Microbiol 4:585–598. doi: 10.1046/j.1462-5822.2002.00212.x. [DOI] [PubMed] [Google Scholar]
- 7.Edwards JL, Brown EJ, Ault KA, Apicella MA. 2001. The role of complement receptor 3 (CR3) in Neisseria gonorrhoeae infection of human cervical epithelia. Cell Microbiol 3:611–622. doi: 10.1046/j.1462-5822.2001.00140.x. [DOI] [PubMed] [Google Scholar]
- 8.Harvey HA, Jennings MP, Campbell CA, Williams R, Apicella MA. 2001. Receptor-mediated endocytosis of Neisseria gonorrhoeae into primary human urethral epithelial cells: the role of the asialoglycoprotein receptor. Mol Microbiol 42:659–672. doi: 10.1046/j.1365-2958.2001.02666.x. [DOI] [PubMed] [Google Scholar]
- 9.Mandrell RE, Apicella MA. 1993. Lipo-oligosaccharides (LOS) of mucosal pathogens: molecular mimicry and host-modification of LOS. Immunobiology 187:382–402. doi: 10.1016/S0171-2985(11)80352-9. [DOI] [PubMed] [Google Scholar]
- 10.Mandrell RE, McLaughlin R, Aba Kwaik Y, Lesse A, Yamasaki R, Gibson B, Spinola SM, Apicella MA. 1992. Lipooligosaccharides (LOS) of some Haemophilus species mimic human glycosphingolipids, and some LOS are sialylated. Infect Immun 60:1322–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bouchet V, Hood DW, Li J, Brisson JR, Randle GA, Martin A, Li Z, Goldstein R, Schweda EK, Pelton SI, Richards JC, Moxon ER. 2003. Host-derived sialic acid is incorporated into Haemophilus influenzae lipopolysaccharide and is a major virulence factor in experimental otitis media. Proc Natl Acad Sci U S A 100:8898–8903. doi: 10.1073/pnas.1432026100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jurcisek J, Greiner L, Watanabe H, Zaleski A, Apicella MA, Bakaletz LO. 2005. Role of sialic acid and complex carbohydrate biosynthesis in biofilm formation by nontypeable Haemophilus influenzae in the chinchilla middle ear. Infect Immun 73:3210–3218. doi: 10.1128/IAI.73.6.3210-3218.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnston JW, Apicella MA. 2008. Sialic acid metabolism and regulation by Haemophilus influenzae: potential novel antimicrobial therapies. Curr Infect Dis Rep 10:83–84. doi: 10.1007/s11908-008-0014-y. [DOI] [PubMed] [Google Scholar]
- 14.Johnston JW, Zaleski A, Allen S, Mootz JM, Armbruster D, Gibson BW, Apicella MA, Munson RS. 2007. Regulation of sialic acid transport and catabolism in Haemophilus influenzae. Mol Microbiol 66:26–39. doi: 10.1111/j.1365-2958.2007.05890.x. [DOI] [PubMed] [Google Scholar]
- 15.Allen S, Zaleski A, Johnston JW, Gibson BW, Apicella MA. 2005. Novel sialic acid transporter of Haemophilus influenzae. Infect Immun 73:5291–5300. doi: 10.1128/IAI.73.9.5291-5300.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ward ME, Watt PJ, Glynn AA. 1970. Gonococci in urethral exudates possess a virulence factor lost on subculture. Nature 227:382–384. doi: 10.1038/227382a0. [DOI] [PubMed] [Google Scholar]
- 17.Johnston JW, Coussens NP, Allen S, Houtman JC, Turner KH, Zaleski A, Ramaswamy S, Gibson BW, Apicella MA. 2008. Characterization of the N-acetyl-5-neuraminic acid-binding site of the extracytoplasmic solute receptor (SiaP) of nontypeable Haemophilus influenzae strain 2019. J Biol Chem 283:855–865. doi: 10.1074/jbc.M706603200. [DOI] [PubMed] [Google Scholar]
- 18.Parsons NJ, Andrade JR, Patel PV, Cole JA, Smith H. 1989. Sialylation of lipopolysaccharide and loss of absorption of bactericidal antibody during conversion of gonococci to serum resistance by cytidine 5′-monophospho-N-acetyl neuraminic acid. Microb Pathog 7:63–72. doi: 10.1016/0882-4010(89)90112-5. [DOI] [PubMed] [Google Scholar]
- 19.Jones PA, Samuels NM, Phillips NJ, Munson RS, Bozue JA, Arseneau JA, Nichols WA, Zaleski A, Gibson BW, Apicella MA. 2002. Haemophilus influenzae type b strain A2 has multiple sialyltransferases involved in lipooligosaccharide sialylation. J Biol Chem 277:14598–14611. doi: 10.1074/jbc.M110986200. [DOI] [PubMed] [Google Scholar]
- 20.Fox KL, Cox AD, Gilbert M, Wakarchuk WW, Li J, Makepeace K, Richards JC, Moxon ER, Hood DW. 2006. Identification of a bifunctional lipopolysaccharide sialyltransferase in Haemophilus influenzae: incorporation of disialic acid. J Biol Chem 281:40024–40032. doi: 10.1074/jbc.M602314200. [DOI] [PubMed] [Google Scholar]
- 21.Hood DW, Cox AD, Gilbert M, Makepeace K, Walsh S, Deadman ME, Cody A, Martin A, Månsson M, Schweda EK, Brisson JR, Richards JC, Moxon ER, Wakarchuk WW. 2001. Identification of a lipopolysaccharide alpha-2,3-sialyltransferase from Haemophilus influenzae. Mol Microbiol 39:341–350. doi: 10.1046/j.1365-2958.2001.02204.x. [DOI] [PubMed] [Google Scholar]
- 22.Abul Kwaik Y, McLaughlin RE, Apicella MA, Spinola SM. 1992. Cloning and analysis of lipooligosaccharide synthesis genes of Haemophilus influenzae type b that assemble or expose a 2-keto-3-deoxyoctulosonic acid epitope in Escherichia coli. J Infect Dis 165(Suppl 1):S195–S196. [DOI] [PubMed] [Google Scholar]
- 23.Abu Kwaik Y, McLaughlin RE, Apicella MA, Spinola SM. 1991. Analysis of Haemophilus influenzae type b lipooligosaccharide-synthesis genes that assemble or expose a 2-keto-3-deoxyoctulosonic acid epitope. Mol Microbiol 5:2475–2480. doi: 10.1111/j.1365-2958.1991.tb02092.x. [DOI] [PubMed] [Google Scholar]
- 24.Johansen EB, Szoka FC, Zaleski A, Apicella MA, Gibson BW. 2010. Utilizing the O-antigen lipopolysaccharide biosynthesis pathway in Escherichia coli to interrogate the substrate specificities of exogenous glycosyltransferase genes in a combinatorial approach. Glycobiology 20:763–774. doi: 10.1093/glycob/cwq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spinola SM, Kwaik YA, Lesse AJ, Campagnari AA, Apicella MA. 1990. Cloning and expression in Escherichia coli of a Haemophilus influenzae type b lipooligosaccharide synthesis gene(s) that encodes a 2-keto-3-deoxyoctulosonic acid epitope. Infect Immun 58:1558–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phillips NJ, Miller TJ, Engstrom JJ, Melaugh W, McLaughlin R, Apicella MA, Gibson BW. 2000. Characterization of chimeric lipopolysaccharides from Escherichia coli strain JM109 transformed with lipooligosaccharide synthesis genes (lsg) from Haemophilus influenzae. J Biol Chem 275:4747–4758. doi: 10.1074/jbc.275.7.4747. [DOI] [PubMed] [Google Scholar]
- 27.Johnston JW, Shamsulddin H, Miller AF, Apicella MA. 2010. Sialic acid transport and catabolism are cooperatively regulated by SiaR and CRP in nontypeable Haemophilus influenzae. BMC Microbiol 10:240. doi: 10.1186/1471-2180-10-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang D, Zhou X, Wang L, Wang S, Sun XL. 2014. Quantification of free sialic acid in human plasma through a robust quinoxalinone derivatization and LC-MS/MS using isotope-labeled standard calibration. J Chromatogr B Analyt Technol Biomed Life Sci 944:75–81. doi: 10.1016/j.jchromb.2013.11.016. [DOI] [PubMed] [Google Scholar]
- 29.Post DM, Ketterer MR, Coffin JE, Reinders LM, Munson RS, Bair T, Murphy TF, Foster ED, Gibson BW, Apicella MA. 2016. Comparative analyses of the lipooligosaccharides from nontypeable Haemophilus influenzae and Haemophilus haemolyticus show differences in sialic acid and phosphorylcholine modifications. Infect Immun 84:765–774. doi: 10.1128/IAI.01185-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCrea KW, Xie J, LaCross N, Patel M, Mukundan D, Murphy TF, Marrs CF, Gilsdorf JR. 2008. Relationships of nontypeable Haemophilus influenzae strains to hemolytic and nonhemolytic Haemophilus haemolyticus strains. J Clin Microbiol 46:406–416. doi: 10.1128/JCM.01832-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McLaughlin R, Spinola SM, Apicella MA. 1992. Generation of lipooligosaccharide mutants of Haemophilus influenzae type b. J Bacteriol 174:6455–6459. doi: 10.1128/jb.174.20.6455-6459.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mandrell RE, Griffiss JM, Macher BA. 1988. Lipooligosaccharides (LOS) of Neisseria gonorrhoeae and Neisseria meningitidis have components that are immunochemically similar to precursors of human blood group antigens. Carbohydrate sequence specificity of the mouse monoclonal antibodies that recognize crossreacting antigens on LOS and human erythrocytes. J Exp Med 168:107–126. doi: 10.1084/jem.168.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Apicella MA, Mandrell RE, Shero M, Wilson ME, Griffiss JM, Brooks GF, Lammel C, Breen JF, Rice PA. 1990. Modification by sialic acid of Neisseria gonorrhoeae lipooligosaccharide epitope expression in human urethral exudates: an immunoelectron microscopic analysis. J Infect Dis 162:506–512. doi: 10.1093/infdis/162.2.506. [DOI] [PubMed] [Google Scholar]
- 34.Mandrell RE, Lesse AJ, Sugai JV, Shero M, Griffiss JM, Cole JA, Parsons NJ, Smith H, Morse SA, Apicella MA. 1990. In vitro and in vivo modification of Neisseria gonorrhoeae lipooligosaccharide epitope structure by sialylation. J Exp Med 171:1649–1664. doi: 10.1084/jem.171.5.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J, Zhou H, Ji Z, Regnier F. 1998. Monoclonal antibody production with on-line harvesting and process monitoring. J Chromatogr B Biomed Sci Appl 707:257–265. doi: 10.1016/S0378-4347(97)00472-6. [DOI] [PubMed] [Google Scholar]
- 36.Day CJ, Paton AW, Higgins MA, Shewell LK, Jen FE, Schulz BL, Herdman BP, Paton JC, Jennings MP. 2017. Structure aided design of a Neu5Gc specific lectin. Sci Rep 7:1495. doi: 10.1038/s41598-017-01522-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abdillahi H, Poolman JT. 1988. Typing of group-B Neisseria meningitidis with monoclonal antibodies in the whole-cell ELISA. J Med Microbiol 26:177–180. [PubMed] [Google Scholar]
- 38.Cho C, Chande A, Gakhar L, Bakaletz LO, Jurcisek JA, Ketterer M, Shao J, Gotoh K, Foster E, Hunt J, O’Brien E, Apicella MA. 2015. Role of the nuclease of nontypeable Haemophilus influenzae in dispersal of organisms from biofilms. Infect Immun 83:950–957. doi: 10.1128/IAI.02601-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blake MS, Johnston KH, Russell-Jones GJ, Gotschlich EC. 1984. A rapid, sensitive method for detection of alkaline phosphatase-conjugated anti-antibody on Western blots. Anal Biochem 136:175–179. doi: 10.1016/0003-2697(84)90320-8. [DOI] [PubMed] [Google Scholar]
- 40.Nichols WA, Gibson BW, Melaugh W, Lee NG, Sunshine M, Apicella MA. 1997. Identification of the ADP-l-glycero-d-manno-heptose-6-epimerase (rfaD) and heptosyltransferase II (rfaF) biosynthesis genes from nontypeable Haemophilus influenzae 2019. Infect Immun 65:1377–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sensorgrams of single-cycle kinetics of interactions between 6E4 antibody and sugars. The panels show interactions between MAb 6E4 and KDO, Neu5Ac, Neu5Gc, α2-3 sialylactosamine (2-3SLN) containing either an acetyl group (2-3SLN-AC) or glycolyl group (2-3SLN-Gc). Download FIG S1, TIF file, 3.6 MB (3.7MB, tif) .
Copyright © 2018 Apicella et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Results of two Western blots of studies with proteinase K extracts of NTHi strain 2019 (lanes 1 to 3), ΔsiaA1 mutant (lanes 4 to 6), ΔsiaA1 Δlic3B mutant (lanes 7 to 9), ΔsiaA1 Δlic3A ΔlsgB mutant (lanes 10 to 12), and ΔsiaA1 Δlic3A ΔlsgB::lic3B mutant (lanes 13 to 15). Extracts in lanes 1, 2, 4, 5, 7, 8, 10, 11, 13, and 14 were from organisms grown in the presence of 100 μg Neu5Ac. Extracts in lane 2, 5, 8, 11, and 14 were from organisms treated with sialidase prior to electrophoresis. Extracts in lanes 3, 6, 9, 12, and 15 were from organisms grown in the absence of Neu5Ac. Lanes 16 contain LOS from N. gonorrhoeae PID 2 as a control in which a 4.5- and a 5.0-kDa band bind MAb 3F11. These blots demonstrate that MAb 6E4 did not react with any extracts derived from strains grown in the presence of Neu5Ac and that only extracts containing a functional LsgB reacted with Mab 6E4. In addition, the bottom blot shows that MAb 3F11 reactivity is reduced when 6E4 binding occurs. Finally, binding of both 6E4 and 3F11 MAbs does not occur to the LOS in extracts of organisms grown in the presence of Neu5Ac in the absence of sialidase treatment. Download FIG S2, TIF file, 3.1 MB (3.2MB, tif) .
Copyright © 2018 Apicella et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Strains used in this study. Download TABLE S1, DOCX file, 0.01 MB (15.9KB, docx) .
Copyright © 2018 Apicella et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
MAb 6E4 bactericidal effect is directly related to the presence of Neu5Ac in the NTHi strain 2019. Neu5Ac was sequentially increased in the growth medium from 0.1 to 100 µg/ml. Growth of NTHi in the presence of greater than 1 µg of Neu5Ac significantly inhibits (P < 0.05) the bactericidal effect of MAb 6E4. Download FIG S3, TIF file, 3.7 MB (3.8MB, tif) .
Copyright © 2018 Apicella et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.