ABSTRACT
Optogenetic switches permit accurate control of gene expression upon light stimulation. These synthetic switches have become a powerful tool for gene regulation, allowing modulation of customized phenotypes, overcoming the obstacles of chemical inducers, and replacing their use by an inexpensive resource: light. In this work, we implemented FUN-LOV, an optogenetic switch based on the photon-regulated interaction of WC-1 and VVD, two LOV (light-oxygen-voltage) blue-light photoreceptors from the fungus Neurospora crassa. When tested in yeast, FUN-LOV yields light-controlled gene expression with exquisite temporal resolution and a broad dynamic range of over 1,300-fold, as measured by a luciferase reporter. We also tested the FUN-LOV switch for heterologous protein expression in Saccharomyces cerevisiae, where Western blot analysis confirmed strong induction upon light stimulation, surpassing by 2.5 times the levels achieved with a classic GAL4/galactose chemical-inducible system. Additionally, we utilized FUN-LOV to control the ability of yeast cells to flocculate. Light-controlled expression of the flocculin-encoding gene FLO1, by the FUN-LOV switch, yielded flocculation in light (FIL), whereas the light-controlled expression of the corepressor TUP1 provided flocculation in darkness (FID). Altogether, the results reveal the potential of the FUN-LOV optogenetic switch to control two biotechnologically relevant phenotypes such as heterologous protein expression and flocculation, paving the road for the engineering of new yeast strains for industrial applications. Importantly, FUN-LOV’s ability to accurately manipulate gene expression, with a high temporal dynamic range, can be exploited in the analysis of diverse biological processes in various organisms.
KEYWORDS: LOV domain, filamentous fungi, flocculation, heterologous gene expression, optogenetics, yeasts
IMPORTANCE
Optogenetic switches are molecular devices which allow the control of different cellular processes by light, such as gene expression, providing a versatile alternative to chemical inducers. Here, we report a novel optogenetic switch (FUN-LOV) based on the LOV domain interaction of two blue-light photoreceptors (WC-1 and VVD) from the fungus N. crassa. In yeast cells, FUN-LOV allowed tight regulation of gene expression, with low background in darkness and a highly dynamic and potent control by light. We used FUN-LOV to optogenetically manipulate, in yeast, two biotechnologically relevant phenotypes, heterologous protein expression and flocculation, resulting in strains with potential industrial applications. Importantly, FUN-LOV can be implemented in diverse biological platforms to orthogonally control a multitude of cellular processes.
INTRODUCTION
Control of gene expression is an important tool for basic and applied research in biology. Chemical inducers, such as isopropyl-β-d-thiogalactopyranoside (IPTG), methanol, or galactose, have been profusely employed for inducible gene regulation despite their obvious limitations, including potential interference with metabolic processes, difficulty in removal from the culture medium once added, and insufficient temporal and spatial/dose resolution (1). In addition, the cost of chemical inducers can restrict their uses in some industrial applications, whereas temperature induction or constitutive gene expression is chosen based on cost efficiency regardless of its far-from-optimal characteristics (2). Light constitutes a promising alternative for the control of gene expression, considering its low cost, reduced toxic effects, adjustable levels, and high temporal and spatial resolution (3). In several organisms, light readily controls different processes, including gene expression, through photoreceptors with specialized domains which, under light stimulation, undergo a conformational change, passing to an active state (4). Such photoreceptors have allowed the definition of basic building blocks from which to develop optogenetic switches, which can be engineered into synthetic light-controlled orthogonal transcription factors (5). During the past several years, a nascent repertoire of optogenetic switches responding to light of different wavelengths, and assembled in different platforms, has become available (6–11).
Fungal photoreceptors have been an underexploited source of biological parts for the implementation of optogenetic switches. The Neurospora crassa blue-light photoreceptor VIVID (VVD) has the capacity to self-dimerize upon light stimulation through its light-oxygen-voltage (LOV) domain (12). This feature of VVD already led to the development of the optogenetic system named “LightOn,” which was successfully utilized for light-controlled expression of transgenes in mice and mammalian cells (13, 14). Notably, in Neurospora, VVD also interacts with the blue-light photoreceptor White Collar 1 (WC-1), through WC-1’s LOV domain, allowing N. crassa to photoadapt in the presence of continuous illumination (15–17).
The budding yeast Saccharomyces cerevisiae ranks among the most relevant and versatile microorganisms for biotechnology. The absence of photoreceptors in the yeast genome (18) has fueled the implementation of different optogenetic switches in this biological chassis, allowing the orthogonal control of diverse cellular processes by light (19–21). However, despite their obvious advantages, the use of optogenetic switches to control biotechnologically relevant phenotypes in yeast has been seldom implemented (22). For example, optogenetic switches for heterologous protein expression in yeast would replace the addition of chemical inducers, reducing the cost of industrial-scale bioprocesses, providing also a dynamic control of gene expression and the effective temporal control of the on and off states. Another biotechnologically relevant operation in yeast fermentation is flocculation, which allows a fast, accessible, and efficient way to remove remaining yeast cells after fermentation processes (23). Flocculation is controlled by the FLO genes, a family of subtelomeric genes which trigger cell aggregation upon nutrient starvation or under environmental stress conditions (24, 25). These features position flocculation as an ideal and biotechnologically relevant target for optogenetic control.
In this work, we present as proof of concept a new optogenetic switch that provides an ample dynamic range of expression, with low background in the off (dark) state. Its implementation in yeast provides evidence of tight regulation of gene expression by blue light and the control of biotechnologically relevant features and phenotypes, such as heterologous protein expression and flocculation.
RESULTS
FUN-LOV provides a dynamic range of gene expression.
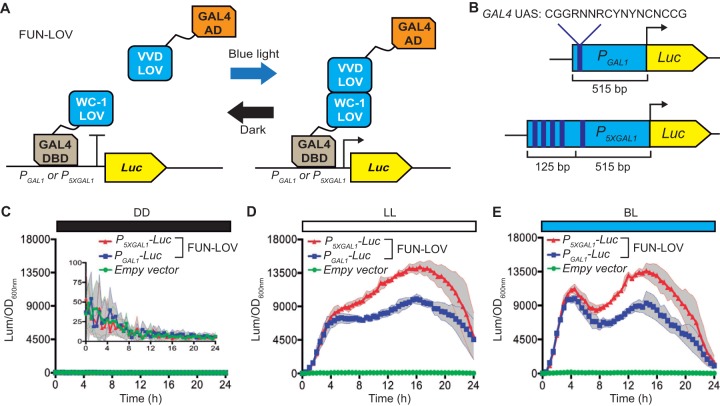
The new optogenetic switch, named FUN-LOV, was developed based on the pairing of WC-1 and VVD LOV domains, an interaction known to occur as part of the N. crassa photoadaptation process (17). The configuration of the FUN-LOV switch follows a yeast two-hybrid system design logic, where the LOV domain of WC-1 is bound to a Gal4 DNA binding domain (GAL4-DBD) and the full-length VVD (which contains a LOV domain) is tethered to the Gal4 transactivation domain (GAL4-AD). Thus, upon the light-stimulated interaction of this LOV domain pair, a functional transcription factor is reconstituted, activating transcription as evidenced by a destabilized firefly luciferase reporter (Luc) under the control of the GAL1 promoter (PGAL1) (Fig. 1A). Additionally, with the aim to increase gene expression induction even further, we designed a synthetic version of the GAL1 promoter (P5XGAL1), which included four additional GAL4-UAS DNA binding site sequences (Fig. 1B).
FIG 1 .
The FUN-LOV switch allows high levels of gene expression by light. (A) FUN-LOV is based on the light-controlled interaction of the LOV (light-oxygen-voltage) domains of WC-1 and VVD photoreceptors which are tethered to the Gal4 DNA binding domain (DBD) and Gal4 activation domain (AD), respectively. (B) Design of the synthetic promoter P5XGAL1, which contains four additional Gal4-UAS sequences upstream of the GAL1 (PGAL1) promoter sequence. Both promoters are controlling luciferase (Luc) expression, which serves as a reporter of FUN-LOV activity. (C to E) Luciferase expression under constant darkness (DD) (C), constant white light (LL) (D), and constant blue light (BL) (E). In panels C to E, standard deviations are represented as shadowed regions.
FUN-LOV revealed high levels of luciferase expression under constant white-light (LL) or constant blue-light (BL) conditions, with notably low levels of background expression in constant darkness (DD) (Fig. 1C, D, and E; see also Fig. S1 and S2 in the supplemental material). In our hands, the expression levels achieved by the FUN-LOV switch were superior to those obtained by applying classical galactose induction (Fig. S2). Furthermore, the maximum luciferase expression levels of the system were 1,218-fold induction for white light and 1,316-fold induction for blue light, utilizing the synthetic P5XGAL1 promoter (Fig. 2A, B, C, and D). The FUN-LOV system yielded similar results employing two different yeast genetic backgrounds (including the absence of endogenous Gal4/Gal80 proteins) and displayed robust luciferase expression either when maintained episomally or when integrated in the genome (Fig. 2, S1, and S2). As luminescence was being measured directly from living cells, we wanted to confirm that luciferase levels from protein extracts would reflect the same range of induction. Therefore, luciferase activity was measured in extracts from cells grown under constant light (LL) or darkness (DD) for 8 h, ratifying the strength of the FUN-LOV switch and revealing even higher levels of induction (~2,500-fold, Fig. 2E). Thus, the FUN-LOV switch provides remarkably high levels of gene expression, as measured by a luciferase reporter, under constant illumination with low background expression in darkness.
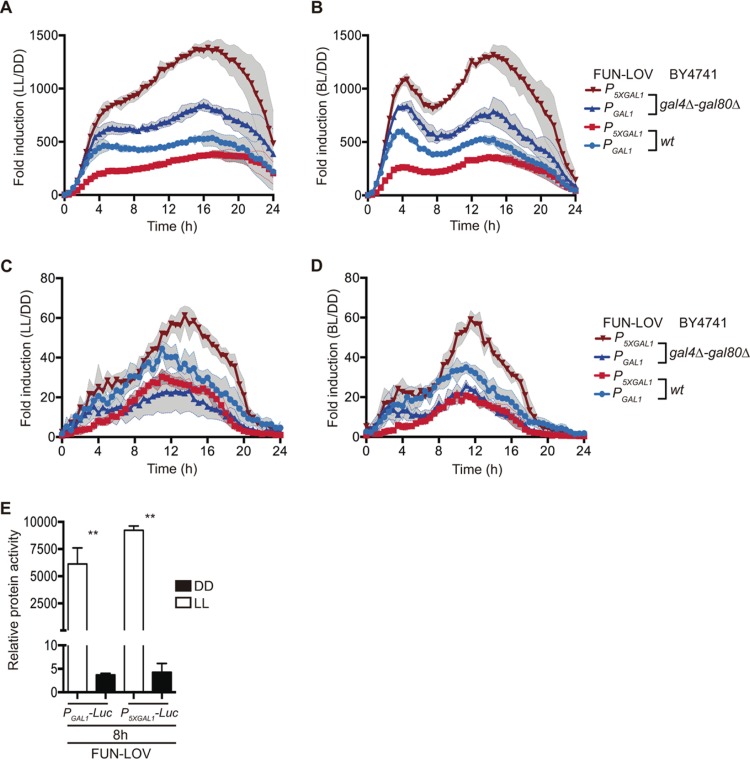
FIG 2 .
Fold induction achieved by the FUN-LOV system. Luciferase expression in constant white light (LL) (A and C) or constant blue light (BL) (B and D), with respect to the average background expression in constant darkness (DD). (A and B) Data from episomal luciferase reporters under the control of two different promoters (PGAL1 and P5XGAL1) and assayed in two different genetic backgrounds (BY4741 and BY4741 gal4Δ-gal80Δ). (C and D) Data from luciferase reporters inserted at the GAL3 locus under the control of PGAL1 and P5XGAL1 promoters, in two different genetic backgrounds (BY4741 wt and BY4741 gal4Δ-gal80Δ). (E) Luciferase activity from protein extracts of cells harvested at the 8-h time point. The double asterisk represents a significant statistical difference between LL and DD conditions (t test, P < 0.01). In panels A to D, the standard deviations are represented as shadowed regions.
FUN-LOV control of luciferase expression. Bioluminescence was monitored and normalized by OD600, as cultures were grown in constant darkness (DD), constant white light (LL), or constant blue light (BL), respectively. (A to F) Components of the FUN-LOV system and the reporter construct (PGAL1-luciferase or P5XGAL1-luciferase) were maintained episomally in a BY4741 wild-type strain (A to C) or in a BY4741 gal4Δ-gal80Δ strain (D to F). (G to L) BY4741 (G to I) or BY4741 gal4Δ-gal80Δ-derived strains (J to L), containing the FUN-LOV system episomally, but having the luciferase reporters integrated at the GAL3 locus. In panels A to L, standard deviations are represented as shadowed regions. Download FIG S1, PDF file, 0.8 MB (800.4KB, pdf) .
Copyright © 2018 Salinas et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Raw luciferase expression levels during active yeast growth. (A to C) The growth of yeast strains containing FUN-LOV and luciferase controlled by a PGAL1 or P5XGAL1 promoter was monitored as OD60 and bioluminescence levels, which were quantified in constant darkness (DD), constant white light (LL), or constant blue light (BL), respectively. (A to F) Components of the FUN-LOV switch and the reporter gene were maintained episomally in the BY4741 wild type (A to C) or BY4741 gal4Δ-gal80Δ strain (D to F). (G to L) Although the FUN-LOV switch was kept episomally, the luciferase reporter was chromosomally inserted at the GAL3 locus in BY4741 (G to I) or in the BY4741 gal4Δ-gal80Δ (J to L) background. (M and N) In addition, the behavior of the episomal Luc reporter in a BY4741 background, depending on the endogenous Gal4p, was evaluated in glucose (M) or galactose (N). When indicated, raw data for luciferase expression were acquired from two different types of promoters, PGAL1 and P5XGAL1. In panels A to N, standard deviations are represented as shadowed regions. Download FIG S2, PDF file, 1.3 MB (1.3MB, pdf) .
Copyright © 2018 Salinas et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
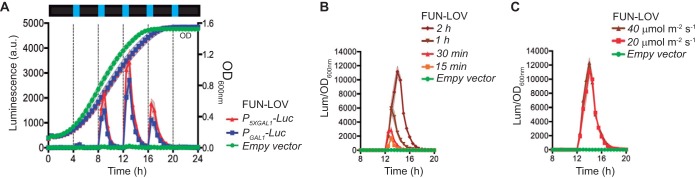
The feasibility of operating the FUN-LOV system as an on/off switch with dynamic and temporal resolution was also assayed. The FUN-LOV system yielded a distinct dynamic and temporal control of luciferase expression during the yeast exponential growth phase (Fig. 3A and S3). The system exhibited a decay in the stationary phase, probably resulting from the low transcriptional activity of the promoter (PADH1) controlling the expression of the FUN-LOV components, which appeared as a common feature of the constitutive promoters evaluated in this work (Fig. S4). This also suggests that the expression levels reached by the FUN-LOV system might be further incremented by expressing its components under stronger promoters, such as PTEF1 or PTDH3 (Fig. S4C and D), which in the future could further boost the relative levels of the FUN-LOV switch components, such as the GAL4-DBD moiety (Fig. S4E). We also found that longer exposure to a blue-light pulse increased the expression of the reporter gene for yeast cells growing in exponential phase. Thus, the system reached its maximum levels after 2 h of light exposure (Fig. 3B and S3E). However, an increase in the blue light-emitting diode (LED) light intensity (from 20 to 40 µmol m−2 s−1) did not further augment luciferase levels (Fig. 3C and S3F), probably reflecting a saturation of the system at 20 µmol m−2 s−1 based on the amount of existing photoreceptor molecules. Overall, the FUN-LOV system offers high expression levels of the luciferase reporter gene upon light, with great dynamic and temporal resolution, making it a suitable switch for the control of biotechnological phenotypes in yeast (26).
FIG 3 .
FUN-LOV allows dynamic control of gene expression. (A) Luciferase expression in yeast cells subjected to 30 min of blue-light pulses every 4 h. (B) Luciferase expression under blue-light pulses of different duration. (C) Luciferase expression using two different light intensities applied for 2 h. In panels B and C, data correspond to luciferase under the control of a P5XGAL1 promoter. In panels A to C, standard deviations are represented as shadowed regions.
Tuneable and dynamic control of luciferase expression by the FUN-LOV switch. Bioluminescence was actively monitored as yeast cells were grown in constant darkness and 30-min blue-light pulses were applied as depicted (A to D). PGAL1-Luc or P5XGAL1-Luc reporters were maintained episomally in a BY4741 (A) or BY4741 gal4Δ-gal80Δ (B) background or inserted at the GAL3 locus in BY4741 (C) or BY4741 gal4Δ-gal80Δ (D). The effect of the duration (E) or intensity (F) of a blue-light pulse was evaluated in a strain containing an episomally maintained PGAL1-Luc reporter. In panel F, the duration of the pulse was 2 h. In panels A to F, standard deviations are represented as shadowed regions. Download FIG S3, PDF file, 0.4 MB (417.5KB, pdf) .
Copyright © 2018 Salinas et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Behavior of the constitutive promoter controlling the FUN-LOV components. (A) Expression of the FUN-LOV switch components (also Fig. 1A) is regulated by the ADH1 promoter (PADH1) and ADH2 transcriptional terminator (ADH2ter). (B to D) The graphs depict luciferase expression regulated by the PADH1 (B), PTEF1 (C), and PTDH3 (D) promoters, as well as the OD600. (E) GAL4-DBD expression was measured by real-time PCR (qPCR) in two different genetic backgrounds (BY4741 and BY4741 gal4Δ-gal80Δ) and with/without the FUN-LOV system. In panels B to D, standard deviations are represented as shadowed regions. Download FIG S4, PDF file, 0.3 MB (263.2KB, pdf) .
Copyright © 2018 Salinas et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
FUN-LOV control of heterologous protein production.
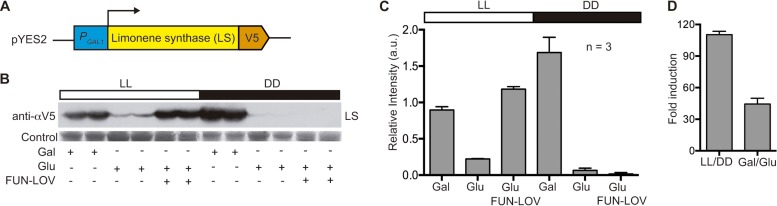
We then explored the capability of the FUN-LOV switch to regulate heterologous protein production upon light stimulation. Thus, the yeast codon-optimized version of the Cannabis sativa limonene synthase (LS) gene was cloned in a pYES2 plasmid, allowing PGAL1 promoter control and V5 tagging of the protein for Western blot analysis (Fig. 4A). This protein is involved in the production of limonene, a compound widely used by the food and household product industries to increase lemon scent in their products (27). Western blot analysis showed that yeast cells expressing the FUN-LOV system achieved an LS induction of 110-fold (LL/DD), which is 2.5 times higher than the average induction (44-fold) achieved, in our hands, by galactose. Notably, FUN-LOV yields lower background levels under DD conditions, particularly compared to the LS levels in the off state of the galactose/glucose systems (Fig. 4B, C, and D). Furthermore, when we evaluated the effect of LL on the temporal expression of proteins by exposing the cells to 2-h pulses of white light or blue light, high levels of protein expression were obtained, with no differences between the two light sources in two different genetic backgrounds (Fig. S5). Overall, FUN-LOV allows high levels of heterologous protein expression upon light stimulation, with low background expression in darkness, and reaching induction levels surpassing chemical induction approaches.
FIG 4 .
Heterologous protein expression controlled by the FUN-LOV switch. (A) The limonene synthase protein (LS) was expressed under the control of the PGAL1 promoter, including a C-terminal V5 tag for Western blot detection. (B and C) LS expression was analyzed by Western blotting (B) and normalized to membrane-stained proteins (C), after the yeast cells were grown in glucose (Glu) or galactose (Gal) under constant white-light (LL) or constant darkness (DD) conditions. (D) Comparison of the traditional Gal/Glu induction ratio in respect to LL/DD induction achieved with FUN-LOV. The average from three biological replicates (n = 3) with its standard deviation is shown in panels C and D.
FUN-LOV-controlled expression of a heterologous protein. (A to C) Expression of V5-tagged limonene synthase (LS) was analyzed by Western blotting (A) and quantified with respect to membrane-stained proteins (B) under three conditions: after 2 h of blue-light pulse (BLP), after 2 h of white-light pulse (WLP), and in constant darkness (DD). The fold induction for LS expression was calculated after the BLP and WLP with respect to DD (C). The experiments were carried out in the BY4741 wild-type strain. (D to F) Similar experiments as those in panels A to C performed in the BY4741 gal4Δ-gal80Δ genetic background. Download FIG S5, PDF file, 0.1 MB (109.5KB, pdf) .
Copyright © 2018 Salinas et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Modulation of yeast flocculation by light.
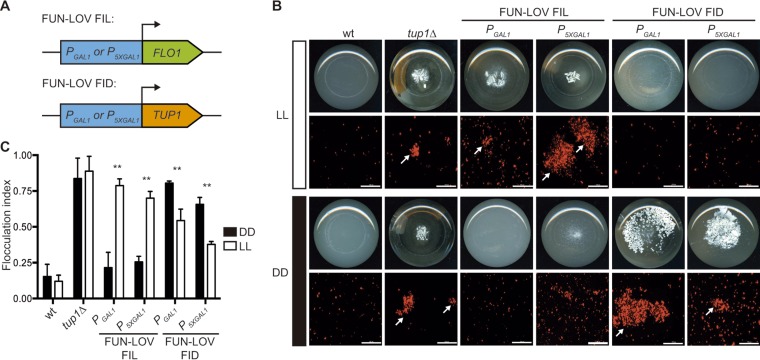
Finally, we sought to apply the FUN-LOV system to optogenetically modulate a biotechnologically relevant process such as flocculation. We rationalized that direct overexpression of the main genes responsible for flocculation (FLO1 or FLO11) would trigger such a phenotype, in agreement with previous reports for these genes (23, 28). On the other hand, low expression of the yeast transcriptional corepressor TUP1 should lead to upregulation of its targets (29), among which is FLO1, therefore triggering strong flocculation. Initially, we swapped the endogenous promoters of FLO1, FLO11, and TUP1 genes for PGAL1 and P5XGAL1 promoters (Fig. 5A). We confirmed the correct replacement of the endogenous promoters and analyzed the behavior of the resulting strains by growing them under conditions of galactose and glucose as carbon sources. Using bright-field microscopy, we observed strong cell aggregation in strains carrying PGAL1-FLO1 in galactose but not in glucose but moderate aggregation in the ones bearing PGAL1-FLO11 (Fig. S6). As expected, strains carrying PGAL1-TUP1 showed strong cell aggregation in glucose but not in galactose, confirming the flocculation phenotype caused by the lack of TUP1 expression (Fig. S6). Then, we episomally incorporated the FUN-LOV system into the strains carrying the swapped promoters. The strains with PGAL1-FLO1 and carrying the FUN-LOV system demonstrated high levels of cell aggregation in LL and low or no aggregation in DD (Fig. S7). We designated this FUN-LOV-controlled phenotype flocculation in light (FIL). Compared to the FUN-LOV control of FLO1-driven flocculation, only a modest FIL phenotype was observed in LL for strains with PGAL1-FLO11. These phenotypes were confirmed by macroscopic observation, as well as by fluorescence microscopy of yeast cells constitutively expressing mCherry, and by calculating the flocculation index of each strain (Fig. 5B and C and S8). On the other hand, strains with FUN-LOV control of PGAL1-TUP1 revealed high levels of cell aggregation in DD but not in LL (Fig. 5B and C and S7). This phenotype was designated flocculation in darkness (FID).
FIG 5 .
The FUN-LOV switch allows precise control of flocculation by light conditions. (A) FUN-LOV-dependent expression of FLO1 or TUP1 genes allows flocculation to occur in light (FIL) or in darkness (FID). (B) FIL and FID phenotypes observed as macroscopic cell aggregation in culture flasks (bottom view) and fluorescence microscopy of cells expressing mCherry under the PTDH3 constitutive promoter. Experiments were performed under constant white-light (LL) or constant darkness (DD) conditions. The BY4741 wild-type (wt) and BY4741 tup1Δ strains were utilized as negative and positive controls of flocculation, respectively. Cell aggregates are indicated by arrows; bar, 100 µm. (C) Quantification of the FIL and FID phenotypes observed in panel B by calculating the flocculation index. Statistically significant differences between LL and DD conditions are indicated as ** (t test, P < 0.01).
Galactose and glucose control of the flocculation phenotype. (A and B) Strains with FLO1, FLO11, and TUP1 promoter swapping (PGAL1 and P5XGAL1) were grown in galactose and glucose as carbon sources, and the flocculation phenotype was assayed by bright-field microscopy (A) and macroscopic analyses of the culture flasks (bottom view) (B). BY4741 and BY4741 flo1Δ strains were used as negative controls of flocculation, whereas the BY4741 tup1Δ strain was used as a positive control of flocculation. Bars, 100 µm at ×20 magnification and 20 µm at ×60 magnification. Download FIG S6, PDF file, 1.1 MB (1.2MB, pdf) .
Copyright © 2018 Salinas et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
FUN-LOV control of flocculation in light (FIL) and flocculation in darkness (FID) phenotypes. (A and B) Strains with FLO1, FLO11, and TUP1 promoter swapping (PGAL1 and P5XGAL1) and carrying the FUN-LOV system were grown under constant light (LL) and constant darkness (DD) conditions, and the flocculation phenotype was assayed by bright-field microscopy (A) and pictures of the culture flasks (bottom view) (B). The BY4741 wt and BY4741 flo1Δ strains were used as negative controls of flocculation, whereas the BY4741 tup1Δ strain was used as a positive control of flocculation. Bars, 100 µm at ×20 magnification and 20 µm at ×60 magnification. Download FIG S7, PDF file, 1.2 MB (1.3MB, pdf) .
Copyright © 2018 Salinas et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Quantification of light-regulated flocculation by FUN-LOV. (A) Flocculation in light (FIL) and flocculation in darkness (FID) were monitored as macroscopic phenotypes by observing cell aggregation in culture flasks (bottom view), as well as by fluorescence microscopy of cells expressing mCherry under a constitutive promoter (PTDH3). Experiments were performed under constant white light (LL) and constant darkness (DD). wt yeast cells (BY4741) as well as flo1Δ and flo11Δ strains were utilized as negative controls, whereas the tup1Δ strain was used as a positive control of flocculation. Cellular aggregations are highlighted with arrows. Bar, 100 µm. (B) Quantification of the flocculation index of the FIL and FID phenotypes depicted in panel A, where the symbol ** represents a significant statistical difference between LL and DD conditions (t test; **, P < 0.01). Download FIG S8, PDF file, 0.5 MB (522.5KB, pdf) .
Copyright © 2018 Salinas et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
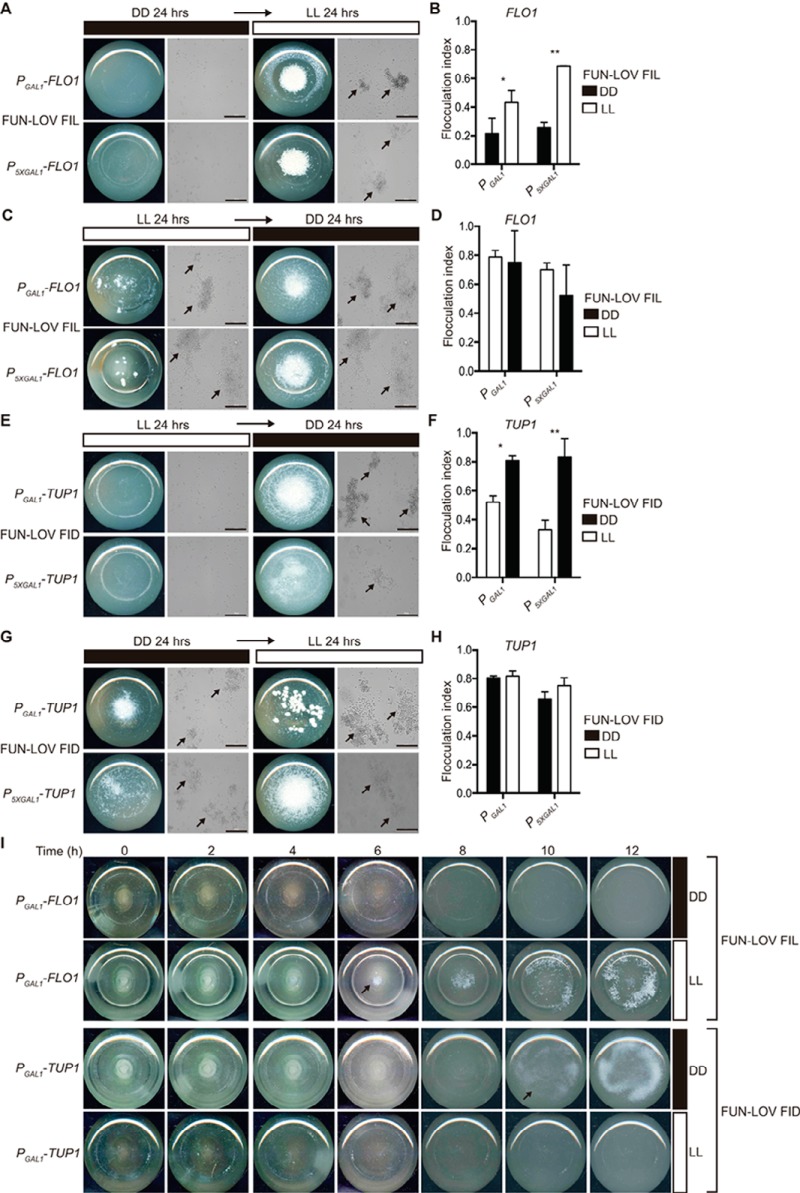
The reversibility of the flocculation phenotype was evaluated under different conditions of LL and DD. Growth of FLO1 FIL strains during 24 h in DD followed by 24 h in LL resulted in strong flocculation (Fig. 6A and B), whereas the opposite treatment—24 h in LL followed by 24 h in DD—revealed strong flocculation in LL without reversion of the phenotype after the transfer to DD (Fig. 6C and D). In the case of TUP1 FID strains, there was no flocculation in LL during the initial 24 h of incubation, followed by strong flocculation after the transfer to DD (Fig. 6E and F). As expected, no reversion was observed for the phenotype when cells were incubated for 24 h in DD and then transferred to LL (Fig. 6G and H). Finally, we evaluated the time necessary to induce the FIL and FID phenotypes in yeast cultures. The results showed that the FIL phenotype becomes readily visible after 6 h of light stimulation, whereas the FID phenotype can be observed after 10 h of incubation in the dark (Fig. 6I). Overall, depending on the genetic wiring of FUN-LOV, yeast flocculation can be triggered by light (FIL phenotype) or by its absence (FID phenotype). These phenotypes showed no reversibility, and the time of appearance exhibited different temporalities.
FIG 6 .
Reversibility and temporality of the different flocculation phenotypes. (A to D) The reversion of the FIL phenotype for strains with FLO1 promoter swapping and carrying the FUN-LOV system was assayed after 24 h of incubation in constant darkness (DD) and then transfer for 24 h to constant light (LL) (A and B); the opposite regimen was also assayed, with 24 h in LL and 24 h in DD (C and D). Cellular aggregation was recorded with pictures of the bottom view of the culture flasks and bright-field microscopy (A and C). Yeast aggregates are highlighted with arrows; bar, 100 µm. The flocculation index was assayed for the same set of strains and culture conditions (B and D). Single and double asterisks represent statistically significant differences between LL and DD conditions (t test; *, P < 0.05, and **, P < 0.01). (E to H) Reversion of the FID phenotype for strains with TUP1 promoter swapping and carrying the FUN-LOV system was assayed under the same experimental conditions as in panels A to D, starting from the nonflocculating condition (LL) to the flocculating condition (DD) (E); the opposite regimen was also assayed (G). The flocculation index was assayed for the same set of strains and culture conditions (F and H). (I) Time course of the FIL and FID phenotypes for the strains with FLO1 and TUP1 promoter swapping (by PGAL1) and carrying the FUN-LOV system. The cellular aggregation was recorded with pictures of the bottom view of the culture flasks every 2 h under DD and LL conditions. The arrows highlight the time point where the macroscopic flocculation phenotype started to be visible.
DISCUSSION
Based on the LOV domains of two blue-light photoreceptors from the fungus N. crassa (proteins WC-1 and VVD), we developed a new optogenetic switch named FUN-LOV. We set up FUN-LOV in yeast cells, since this microorganism is a model eukaryotic system for genetics and molecular biology studies and a powerful platform for biotechnology, including applications such as production of high-value metabolites and proteins (30, 31). Using yeast as a biological chassis, FUN-LOV showed three singular features: (i) high levels of gene expression with a broad dynamic range and temporal resolution (Fig. 1, 2 and 3); (ii) low background expression under dark conditions (Fig. 1C); and (iii) the control of biotechnologically relevant phenotypes in yeast, such as heterologous protein expression (Fig. 4) and flocculation (Fig. 5 and 6). Those characteristics achieved by FUN-LOV increase its potential uses under industrial or high-volume bioprocess conditions.
Light has been positioned in previous years as a promising tool to control gene expression due to its reduced toxic effects on cells, low costs compared to chemical inducers, and the ability to confer spatiotemporal modulation of biological processes (3). In this work, we selected yeast as a biological platform for the implementation of FUN-LOV, ensuring the orthogonal effect of light over gene expression. Importantly, it has been described that extremely high intensities of blue, green, and white lights can affect yeast growth, producing impaired respiration, increased levels of reactive oxygen species, and upregulation of the stress response (32). In this sense, FUN-LOV reached the maximum level of gene expression at 20 µmol m−2 s−1 of blue-light intensity, consistent with previous reports where between 2 and 24 µmol m−2 s−1 of blue light is enough for VVD photocycle activation (33). Utilizing rather low light levels could be considered an advantage of our system, as it reduces the possibility of undesirable effects of high light on yeast physiology and metabolism. Nevertheless, it is interesting that the dynamics of the luciferase reporter under blue light show a drop not seen in white light (i.e., Fig. 1D and E), a feature that is not observed in the luciferase data prior to normalization by optical density (OD) (see Fig. S2 in the supplemental material).
The first optogenetic switch developed and implemented in yeast was based on the red-light-dependent interactions of the photoreceptor phytochrome B (PhyB) and its interacting protein PIF3 from Arabidopsis thaliana (6). Ever since, this switch has been adapted in yeast for different purposes such as reconstitution of protein activities (7), subcellular protein localization and control of cell polarity and budding phenotypes (34, 35), and reporter gene expression (yellow fluorescent protein [YFP] or green fluorescent protein [GFP]) (19, 36). Recently, a new red-light optogenetic switch also based on PhyB-PIF3 interaction (named PhiReX), which overcomes the obstacle of external addition of the chromophore phycocyanobilin (PCB), has been implemented and tested in yeast (36). These types of toggle switches can be quite powerful as they provide activation with a given wavelength (red) to then be turned off by another one (far-red). Yet, in some cases that contrasts with the simplicity of blue-light switches, which can be readily activated by blue or white light, while they can be turned off with fast kinetics upon transfer to the dark, without the need of extra specific wavelengths.
In general, multiple blue-light optogenetic switches have been implemented in several biological platforms, utilizing a wide repertoire of biological parts and resulting in diverse expression levels. The latter has been monitored by different reporter genes, a fact that complicates the direct comparison between systems (22). In yeast, one of the most widely used optogenetic switches is based on the blue-light-dependent interaction of the photoreceptor cryptochrome 2 (CRY2) and its interacting protein CIB1 from A. thaliana, which has allowed light-controlled reporter gene expression (20), heterologous expression of YFP or mCherry (37, 38), and cell cycle modulation (19). Besides cryptochromes, blue-light photoreceptors containing LOV domains have been also used in yeast optogenetics. The LOV2 domain from Avena sativa phototropin 1 (AsLOV2) was utilized for light-controlled caging of peptides and β-galactosidase expression in yeast (39). Similarly, the LOV2 domain from A. thaliana phototropin 1 (AtLOV2) has been also used for blue-light control of the cell cycle progression and expression of conditional essential genes (40, 41).
Fungal blue-light photoreceptors containing LOV domains have been seldom utilized in optogenetics, and their uses have not been exploited in yeast, regardless of the functional conservation between fungal and plant LOV domains (42). Only the protein VIVID (VVD) from the fungus N. crassa has been used in optogenetics switches, permitting light-controlled expression of transgenes in mice and mammalian cells (13, 14). The latter optogenetic switch, named GAVPO and commonly known as the “LightOn” system, is based on the homodimerization of VVD LOV domains upon light stimulation and also includes components such as a GAL4 DNA binding domain (GAL4-DBD) and a p65 transactivation domain, reaching 200- to 300-fold induction, as measured by a luciferase reporter in mammalian cells (14). Thus, the fact that “LightOn” shares several components with FUN-LOV (VVD and GAL4-DBD) is a positive feature that supports future adaptability of FUN-LOV beyond yeast, including mammalian systems.
Optogenetic systems based on the homodimerization of LOV domains have shown proper functionality for light-controlled gene expression. However, they have limited applications in light-activated control of subcellular protein localization, due to the possibility of homodimerization of one of the target proteins. This problem has been nicely solved using the “Magnets” system, where the VVD LOV domain has been engineered to recognize a VVD partner with the opposite electrostatic charge, showing successful subcellular protein localization (43) when tested in mammalian cell lines (44). In this sense, it is worth mentioning that VVD/VVD or WC-1/WC-1 self-dimerization could be decreasing the induction levels achieved by FUN-LOV, as it would reduce the proportion of WC-1/VVD pairing, which is the one needed to reconstruct a chimeric transcription factor that allows expression of a target gene. Therefore, future efforts will seek to implement the “Magnets” strategy in our system to avoid the formation of nonfunctional homodimers, boosting induction to even higher levels.
Indeed, in N. crassa not only has the protein VVD the capacity to self-dimerize upon light stimulation, but additionally VVD interacts with the protein WC-1, a LOV-LOV interaction that allows photoadaptation to different light intensities (17, 45). Importantly, WC-1 is a GATA transcription factor and also a blue-light photoreceptor containing a LOV domain, participating (with WC-2 protein) in the White-Collar complex (WCC), which acts as a positive element in the circadian clock of N. crassa (46). Therefore, when it comes to LOV, finding the right partner may improve the performance of the system, as evidenced by the VVD/WC-1 combination. This naturally occurring LOV-LOV interaction of WC-1 and VVD opens the door for the development of novel optogenetic switches based on different LOV pairs, such as WC-1/WC-1 from either Neurospora or other fungi; considering the diversity of such sequences in different fungal genomes, such LOV domains are a promising source for novel optogenetic switches such as the one presented here. Moreover, the ability to easily assess functional interactions between full-length (as utilized in FUN-LOV) or N-terminally truncated VVD (as in GAPVO) and homo- or hetero-partners in yeast can help advance structure-function studies allowing evaluation of the effect of different mutations in LOV-LOV interactions.
In conclusion, in this work we reported the implementation in yeast of a novel optogenetic switch, FUN-LOV, which provides accurate and strong light-controlled gene expression, exemplified also by the regulation of two phenotypes of biotechnological relevance, such as heterologous protein expression and flocculation. The current challenge is to successfully scale up this technology for industrial bioprocesses, under conditions where high culture densities could preclude efficient light delivery to all yeast cells. Importantly, in addition to its applicability to producing high-value metabolites or heterologous proteins, its low background and broad dynamic range make FUN-LOV a powerful tool to exquisitely regulate the expression of any gene of interest and to probe complex biological phenomena. In addition, its modular design can help the implementation of different optologic gates, by combining it with optogenetic switches for other light wavelengths (22), in order to rewire, build, or perturb complex gene regulatory networks in yeast. Notably, as FUN-LOV utilizes a Gal4-DBD, it could be readily domesticated in other systems such as mammals, or Drosophila, where Gal4 orthogonal control of gene expression has proven extremely successful.
MATERIALS AND METHODS
Yeast strains, medium, and culture conditions.
Saccharomyces cerevisiae strains BY4741 wild type (wt) (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) and BY4741 with GAL4 and GAL80 deletions (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 gal4Δ::NatMx gal80Δ::HphMx) were used as genetic background for gene deletions and promoter swapping. The strains used and generated in this work were maintained in YDPA medium (2% glucose, 2% peptone, 1% yeast extract, and 2% agar) at 30°C, and their genotypes are listed in Table 1. Strains carrying plasmids with auxotrophic markers were maintained in synthetic complete (SC) medium (0.67% yeast nitrogen base without amino acids, 2% glucose, 0.2% dropout mix, and 2% agar) minus the corresponding amino acid (dropout mix). Growth of yeast cultures under different white-light (LL) and darkness (DD) conditions was conducted at 30°C, using 100 µmol m−2 s−1 of white-light intensity, which was the light intensity provided by the Percival incubators (Percival Scientific, USA). In general, cells were grown in 50 ml of SC medium at 30°C with 130 rpm of shaking in flasks or in 200 µl of SC medium using 96-well plates at 30°C.
TABLE 1 .
Strains of Saccharomyces cerevisiae used and generated in this work
| Strain | Genotype | Source |
|---|---|---|
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Euroscarf |
| gal4Δ-gal80Δ | BY4741; gal4Δ::NatMx gal80Δ::HphMx | This work |
| flo1Δ | BY4741; flo1Δ::URA3 | This work |
| flo11Δ | BY4741; flo11Δ::URA3 | This work |
| tup1Δ | BY4741; tup1Δ::KanMx | Euroscarf |
| PGAL1-FLO1 | BY4741; PFLO1Δ::KanMxRV-PGAL1 | This work |
| P5XGAL1-FLO1 | BY4741; PFLO1Δ::KanMxRV-P5XGAL1 | This work |
| PGAL1-FLO11 | BY4741; PFLO11Δ::KanMxRV-PGAL1 | This work |
| P5XGAL1-FLO11 | BY4741; PFLO11Δ::KanMxRV-PGAL1 | This work |
| PGAL1-TUP1 | BY4741; PTUP1Δ::KanMxRV-PGAL1 | This work |
| P5XGAL1-TUP1 | BY4741; PTUP1Δ::KanMxRV-P5XGAL1 | This work |
| gal3Δ::PGAL1-Luc | BY4741; gal3Δ::KanMxRV-PGAL1-Luc | This work |
| gal3Δ::P5XGAL1-Luc | BY4741; gal3Δ::KanMxRV-P5XGAL1-Luc | This work |
| gal4Δ-gal80Δ PGAL1-FLO1 | BY4741; gal4Δ-gal80Δ PFLO1Δ::KanMxRV-PGAL1 | This work |
| gal4Δ-gal80Δ PGAL1-FLO11 | BY4741; gal4Δ-gal80Δ PFLO11Δ::KanMxRV-PGAL1 | This work |
| gal4Δ-gal80Δ PGAL1-TUP1 | BY4741; gal4Δ-gal80Δ PTUP1Δ::KanMxRV-PGAL1 | This work |
| gal4Δ-gal80Δ gal3Δ::PGAL1-Luc | BY4741; gal4Δ-gal80Δ gal3Δ::KanMxRV-PGAL1-Luc | This work |
| gal4Δ-gal80Δ gal3Δ::P5XGAL1-Luc | BY4741; gal4Δ-gal80Δ gal3Δ::KanMxRV-P5XGAL1-Luc | This work |
Illumination conditions.
Blue-light (BL) experiments under microcultivation conditions were carried out using an LED lamp (growth LED, model i5038, 2 W of potency, and 38 LED pieces) at 20 µmol m−2 s−1 of light intensity, except in the data presented in Fig. 3C, where 40 µmol m−2 s−1 was utilized. In the case of white light (LL), experiments were performed with an LED lamp (General Electric; type A60, 10 W of potency) at 100 µmol m−2 s−1 of light intensity. Light intensity was measured using a quantum light meter, model LightScout (Spectrum Technologies Inc., USA), which measures the photosynthetically active radiation (PAR) light between 400 and 700 nm. Additionally, light intensity was regulated by modifying the distance between the 96-well plate and the light source. The spectrum of the white and blue LED lamps was determined using an HR2000 high-resolution spectrometer (Ocean Optics, USA), showing that the two light sources have similar peaks at 460 nm. The heterologous protein expression and flocculation experiments were conducted in a Percival incubator, which delivered 100 µmol m−2 s−1 of light intensity. Manipulation of strains in the “dark” was conducted in a light-tight room equipped with red safety lights.
Plasmid construct and strain generation.
Plasmids pBM1 and pBM2 carrying the LOV domains of WC-1 (WC-1 LOV-BD) and VVD (VVD-AD, which actually contains the full-length VVD and not just VVD-36) were generously provided by the Brunner lab (17). The plasmids used and generated in this work are shown in Table 2. The FUN-LOV components were cloned into pRS423 (WC-1 LOV plus GAL4-DBD) and pRS425 (VVD plus GAL4-AD) plasmids for HIS3 and LEU2 auxotrophic selection, respectively. All the cloning experiments were performed and genetic constructs were generated using yeast recombinational cloning in vivo assembly (47, 48).
TABLE 2 .
Plasmids assembled in this work using yeast recombinational cloning
| Plasmid | Construct |
|---|---|
| pRS423-WC-1 | PADH1-WC-1 LOV-GAL4 DBD-ADH2ter |
| pRS425-VVD | PADH1-VVD-GAL4 AD-ADH2ter |
| pRS426-PGAL1-Luc | KanMxRV-PGAL1-Luc-CYC1ter |
| pRS426-P5XGAL1-Luc | KanMxRV-P5XGAL1-Luc-CYC1ter |
| pRS426-PGAL1 | KanMxRV-PGAL1 |
| pRS426-P5XGAL1 | KanMxRV-P5XGAL1 |
| pRS426-mCherry | PTDH3-mCherry-CYC1ter-HphMx |
| pRS426-PADH1−Luc | PADH1-Luc-CYC1ter |
| pRS426-PTEF1-Luc | PTEF1-Luc-CYC1ter |
| pRS426-PTDH3-Luc | PTDH3-Luc-CYC1ter |
The synthetic version of the PGAL1 promoter, called P5XGAL1, carrying five DNA binding sites for the Gal4 transcription factor (GAL4-UAS), was synthesized using the Genewiz gene synthesis service. In the construct assembly, the PGAL1 and P5XGAL1 promoters were amplified by PCR using Phusion Flash high-fidelity PCR master mix (Thermo Scientific, USA). Additionally, the kanamycin (KanMx) antibiotic resistance cassette was added in the reverse direction upstream of each promoter (KanMxRv-PGAL1 or KanMxRv-P5XGAL1) using yeast recombinational cloning (48). The complete genetic construct (KanMxRv-PGAL1 or KanMxRv-P5XGAL1) was used to transform the BY4741 wt and BY4741 gal4Δ-gal80Δ strains. The complete genetic constructs (KanMxRv-PGAL1 or KanMxRv-P5XGAL1) were amplified by PCR using a Phusion Flash high-fidelity PCR master mix (Thermo Scientific, USA) and 70-bp primers for direct homologous recombination on the target locus, allowing the swapping of the endogenous promoter region. The promoter swapping of FLO1, FLO11, and TUP1 was confirmed by PCR under standard conditions; primers used for plasmid assembly, promoter swapping, and promoter swapping confirmations are shown in Table S1 in the supplemental material. The same assembly procedure was followed to construct different versions of the luciferase reporter gene under the control of PGAL1 and P5XGAL1 promoters. The deletion of GAL4 or GAL80 or the integration of the luciferase reporter at the GAL3 locus was carried out using one-step PCR deletion by recombination (49). Primers used for gene deletion and reporter gene integration in the genome and its confirmation are listed in Table S1.
Primers used in this work. Download TABLE S1, PDF file, 0.1 MB (55.6KB, pdf) .
Copyright © 2018 Salinas et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Luciferase in vivo expression and Western blot assay.
We used a previously described destabilized version of firefly luciferase for real-time monitoring of gene expression in yeast (50). We cloned into plasmid pRS426 the luciferase reporter gene under the control of the PGAL1 and P5XGAL1 promoters using yeast recombinational cloning as we described above. Real-time luciferase expression was measured under DD, LL, and BL conditions using a Cytation 3 microplate reader (BioTek, USA), which allows the measurement of both optical density at 600 nm (OD600) and luminescence of the cell cultures over time. Briefly, the yeast strains were grown overnight in a 96-well plate with 200 µl of SC medium at 30°C under the DD condition, and 10 µl of these cultures was used to inoculate a new 96-well plate containing 290 µl (30-fold dilution) of fresh medium plus 1 mM luciferin. The OD600 and the luminescence were acquired every 30 min using a Cytation 3 microplate reader, running a discontinuous kinetic protocol with 30 s of shaking (285 cycles/min) before each measurement. This protocol also allowed illumination of the 96-well plate between each measurement, keeping the plate outside the equipment and exposing it to the light source. Luciferase expression was normalized by OD600 of the yeast cultures, and all experiments were performed in six biological replicates (51). Normalization of real-time luciferase levels was necessary as yeast biomass rapidly changed over the length of the experiment, something that may not be necessary when monitoring luciferase from filamentous fungi (52).
Limonene synthase (LS) enzyme cDNA sequence from Cannabis sativa was codon optimized for S. cerevisiae and cloned into pYES2 expression vector. The strain BY4741 wt was cotransformed with pYES2-LS plasmid and the components of the FUN-LOV system. Strains were grown under DD and LL conditions until the OD600 was 1, and protein extractions of the yeast cellular pellets were carried out under standard conditions (53). We used 25 µg of total protein from each sample for Western blot assays; detection of the LS was performed using anti-α-V5 primary antibody (Invitrogen, USA) and goat anti-mouse IgG(H+L)-horseradish peroxidase (HRP)-conjugated antibody (Bio-Rad, USA) as secondary antibody (52, 54). All the Western blot experiments were performed in three biological replicates (n = 3).
Gene expression by qPCR.
The gene expression levels for the FUN-LOV component (GAL4-DBD) was measured by real-time PCR (qPCR) in the BY4741 genetic background under the LL condition. Briefly, the yeast strains were grown for 8 h under the LL condition at 30°C in SC medium for RNA extractions. RNA extractions were carried out using Trizol reagent (Thermo Fisher Scientific, USA); the quality and integrity of the total RNA were confirmed by 1% agarose gel and NanoDrop (Thermo Fisher Scientific, USA) quantification. We used 100 ng of total RNA for reverse transcription using Superscript III transcriptase (Invitrogen, USA) under standard conditions. The cDNA was amplified using 2× SensiMix SYBR Hi-ROX kit (Bioline, USA) and using StepOnePlus real-time PCR equipment (Thermo Fisher Scientific, USA). The relative gene expression was calculated using the threshold cycle (2−ΔΔCT) method and utilizing two different reference genes (HEM2 and TAF10) (55, 56). All the qPCR experiments were performed in three biological replicates (n = 3), and primers used for qPCR amplifications are listed in Table S1.
Luciferase enzymatic activity.
The luciferase activity assays were carried out in three biological replicates (n = 3). The luciferase protein activity was assayed using the luciferase assay system kit (catalog no. E1500; Promega, USA) with modifications. Briefly, the yeast strains were grown for 8 h at 30°C in SC medium and harvested by centrifugation for 5 min at 4,000 × g. The cell pellet was disrupted using 200 µl of 2× lysis buffer reagent (Promega, USA) and 200 µl of glass beads in a TissueLyser II equipment (Qiagen, USA) for 3 min. Cells were centrifuged for 5 min at maximum speed, and the supernatant containing the protein extract was recovered and quantified by the Bradford standard method. The luciferase activity was assayed combining 5 µl of the total extracted proteins plus 100 µl of luciferase assay reagent (Promega, USA). The luminescence was immediately recorded in a Cytation 3 microplate reader (BioTek, USA), and it was normalized using the total protein concentration of each sample.
Flocculation phenotypes.
Strains with promoter swapping in FLO1, FLO11, and TUP1 and carrying the FUN-LOV system were evaluated under DD and LL conditions as we previously described. Scans of the flocculation phenotype were taken after 24 h of growth in culture flasks under DD or LL conditions. Additionally, the strains were transformed with a pRS426 plasmid carrying mCherry controlled by the PTDH3 promoter. Pictures of yeast cells were taken under bright-field and fluorescence microscopy using a Cytation 3 in microscope mode (BioTek, USA). In the time course experiments, scanning of the culture flask was performed every 2 h.
The flocculation of each strain was quantified by calculating the flocculation index of each strain at OD600 (57, 58). The flocculation index was calculated as the OD difference in a yeast culture after 30 min of static incubation: 1 − (finalOD/initialOD). All the flocculation experiments were conducted in three biological replicates (n = 3).
Statistical analysis.
All the statistical analyses were carried out using GraphPad (Prism) software version 6.0.
ACKNOWLEDGMENTS
We thank Consuelo Olivares, Rodrigo Santibañez, Cristobal Mena, and Patricio Romero for technical help during this work and members of the Larrondo lab for their constructive comments on the manuscript.
This research was funded by MIISSB Iniciativa Científica Milenio-MINECON, CONICYT/FONDEQUIP EQM130158, and CONICYT/FONDECYT 1171151 and 1170745 to L.F.L. and E.A., respectively. F.S. was supported by CONICYT/FONDECYT (grants no. 3150156 and 11170158), and V.R. and V.D. were supported by CONICYT/PhD scholarships 21170331 and 6313018, respectively. L.F.L. is an International Research Scholar of the Howard Hughes Medical Institute.
F.S., V.R., V.D., E.A., and L.F.L. designed the research; F.S., V.R., V.D., and J.L. performed lab experiments and analyzed the experimental data; F.S. and L.F.L. wrote the paper with insight from all the authors.
Footnotes
Citation Salinas F, Rojas V, Delgado V, López J, Agosin E, Larrondo LF. 2018. Fungal light-oxygen-voltage domains for optogenetic control of gene expression and flocculation in yeast. mBio 9:e00626-18. https://doi.org/10.1128/mBio.00626-18.
REFERENCES
- 1.Deiters A. 2010. Principles and applications of the photochemical control of cellular processes. Chembiochem 11:47–53. doi: 10.1002/cbic.200900529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mokdad-Gargouri R, Abdelmoula-Soussi S, Hadiji-Abbès N, Amor IY, Borchani-Chabchoub I, Gargouri A. 2012. Yeasts as a tool for heterologous gene expression. Methods Mol Biol 824:359–370. doi: 10.1007/978-1-61779-433-9_18. [DOI] [PubMed] [Google Scholar]
- 3.Drepper T, Krauss U, Meyer zu Berstenhorst S, Pietruszka J, Jaeger KE. 2011. Lights on and action! Controlling microbial gene expression by light. Appl Microbiol Biotechnol 90:23–40. doi: 10.1007/s00253-011-3141-6. [DOI] [PubMed] [Google Scholar]
- 4.Schmidt D, Cho YK. 2015. Natural photoreceptors and their application to synthetic biology. Trends Biotechnol 33:80–91. doi: 10.1016/j.tibtech.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 5.Deisseroth K. 2011. Optogenetics. Nat Methods 8:26–29. doi: 10.1038/nmeth.f.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shimizu-Sato S, Huq E, Tepperman JM, Quail PH. 2002. A light-switchable gene promoter system. Nat Biotechnol 20:1041–1044. doi: 10.1038/nbt734. [DOI] [PubMed] [Google Scholar]
- 7.Tyszkiewicz AB, Muir TW. 2008. Activation of protein splicing with light in yeast. Nat Methods 5:303–305. doi: 10.1038/nmeth.1189. [DOI] [PubMed] [Google Scholar]
- 8.Levskaya A, Weiner OD, Lim WA, Voigt CA. 2009. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature 461:997–1001. doi: 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kennedy MJ, Hughes RM, Peteya LA, Schwartz JW, Ehlers MD, Tucker CL. 2010. Rapid blue-light-mediated induction of protein interactions in living cells. Nat Methods 7:973–975. doi: 10.1038/nmeth.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yazawa M, Sadaghiani AM, Hsueh B, Dolmetsch RE. 2009. Induction of protein-protein interactions in live cells using light. Nat Biotechnol 27:941–945. doi: 10.1038/nbt.1569. [DOI] [PubMed] [Google Scholar]
- 11.Müller K, Engesser R, Schulz S, Steinberg T, Tomakidi P, Weber CC, Ulm R, Timmer J, Zurbriggen MD, Weber W. 2013. Multi-chromatic control of mammalian gene expression and signaling. Nucleic Acids Res 41:e124. doi: 10.1093/nar/gkt340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zoltowski BD, Schwerdtfeger C, Widom J, Loros JJ, Bilwes AM, Dunlap JC, Crane BR. 2007. Conformational switching in the fungal light sensor Vivid. Science 316:1054–1057. doi: 10.1126/science.1137128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma Z, Du Z, Chen X, Wang X, Yang Y. 2013. Fine tuning the LightOn light-switchable transgene expression system. Biochem Biophys Res Commun 440:419–423. doi: 10.1016/j.bbrc.2013.09.092. [DOI] [PubMed] [Google Scholar]
- 14.Wang X, Chen X, Yang Y. 2012. Spatiotemporal control of gene expression by a light-switchable transgene system. Nat Methods 9:266–269. doi: 10.1038/nmeth.1892. [DOI] [PubMed] [Google Scholar]
- 15.Chen CH, DeMay BS, Gladfelter AS, Dunlap JC, Loros JJ. 2010. Physical interaction between VIVID and white collar complex regulates photoadaptation in Neurospora. Proc Natl Acad Sci U S A 107:16715–16720. doi: 10.1073/pnas.1011190107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hunt SM, Thompson S, Elvin M, Heintzen C. 2010. VIVID interacts with the WHITE COLLAR complex and FREQUENCY-interacting RNA helicase to alter light and clock responses in Neurospora. Proc Natl Acad Sci U S A 107:16709–16714. doi: 10.1073/pnas.1009474107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malzahn E, Ciprianidis S, Káldi K, Schafmeier T, Brunner M. 2010. Photoadaptation in Neurospora by competitive interaction of activating and inhibitory LOV domains. Cell 142:762–772. doi: 10.1016/j.cell.2010.08.010. [DOI] [PubMed] [Google Scholar]
- 18.Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, Louis EJ, Mewes HW, Murakami Y, Philippsen P, Tettelin H, Oliver SG. 1996. Life with 6000 genes. Science 274:546–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 19.Hughes RM, Bolger S, Tapadia H, Tucker CL. 2012. Light-mediated control of DNA transcription in yeast. Methods 58:385–391. doi: 10.1016/j.ymeth.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pathak GP, Strickland D, Vrana JD, Tucker CL. 2014. Benchmarking of optical dimerizer systems. ACS Synth Biol 3:832–838. doi: 10.1021/sb500291r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sorokina O, Kapus A, Terecskei K, Dixon LE, Kozma-Bognar L, Nagy F, Millar AJ. 2009. A switchable light-input, light-output system modelled and constructed in yeast. J Biol Eng 3:15. doi: 10.1186/1754-1611-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salinas F, Rojas V, Delgado V, Agosin E, Larrondo LF. 2017. Optogenetic switches for light-controlled gene expression in yeast. Appl Microbiol Biotechnol 101:2629–2640. doi: 10.1007/s00253-017-8178-8. [DOI] [PubMed] [Google Scholar]
- 23.Govender P, Domingo JL, Bester MC, Pretorius IS, Bauer FF. 2008. Controlled expression of the dominant flocculation genes FLO1, FLO5, and FLO11 in Saccharomyces cerevisiae. Appl Environ Microbiol 74:6041–6052. doi: 10.1128/AEM.00394-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soares EV. 2011. Flocculation in Saccharomyces cerevisiae: a review. J Appl Microbiol 110:1–18. doi: 10.1111/j.1365-2672.2010.04897.x. [DOI] [PubMed] [Google Scholar]
- 25.Teunissen AW, Steensma HY. 1995. The dominant flocculation genes of Saccharomyces cerevisiae constitute a new subtelomeric gene family. Yeast 11:1001–1013. [DOI] [PubMed] [Google Scholar]
- 26.Stewart CJ, McClean MN. 2017. Design and implementation of an automated illuminating, culturing, and sampling system for microbial optogenetic applications. J Vis Exp doi: 10.3791/54894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jongedijk E, Cankar K, Buchhaupt M, Schrader J, Bouwmeester H, Beekwilder J. 2016. Biotechnological production of limonene in microorganisms. Appl Microbiol Biotechnol 100:2927–2938. doi: 10.1007/s00253-016-7337-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smukalla S, Caldara M, Pochet N, Beauvais A, Guadagnini S, Yan C, Vinces MD, Jansen A, Prevost MC, Latgé JP, Fink GR, Foster KR, Verstrepen KJ. 2008. FLO1 is a variable green beard gene that drives biofilm-like cooperation in budding yeast. Cell 135:726–737. doi: 10.1016/j.cell.2008.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fleming AB, Beggs S, Church M, Tsukihashi Y, Pennings S. 2014. The yeast Cyc8-Tup1 complex cooperates with Hda1p and Rpd3p histone deacetylases to robustly repress transcription of the subtelomeric FLO1 gene. Biochim Biophys Acta 1839:1242–1255. doi: 10.1016/j.bbagrm.2014.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amano K, Chiba Y, Kasahara Y, Kato Y, Kaneko MK, Kuno A, Ito H, Kobayashi K, Hirabayashi J, Jigami Y, Narimatsu H. 2008. Engineering of mucin-type human glycoproteins in yeast cells. Proc Natl Acad Sci U S A 105:3232–3237. doi: 10.1073/pnas.0710412105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paddon CJ, Westfall PJ, Pitera DJ, Benjamin K, Fisher K, McPhee D, Leavell MD, Tai A, Main A, Eng D, Polichuk DR, Teoh KH, Reed DW, Treynor T, Lenihan J, Fleck M, Bajad S, Dang G, Dengrove D, Diola D, Dorin G, Ellens KW, Fickes S, Galazzo J, Gaucher SP, Geistlinger T, Henry R, Hepp M, Horning T, Iqbal T, Jiang H, Kizer L, Lieu B, Melis D, Moss N, Regentin R, Secrest S, Tsuruta H, Vazquez R, Westblade LF, Xu L, Yu M, Zhang Y, Zhao L, Lievense J, Covello PS, Keasling JD, Reiling KK, Renninger NS, Newman JD. 2013. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 496:528–532. doi: 10.1038/nature12051. [DOI] [PubMed] [Google Scholar]
- 32.Robertson JB, Davis CR, Johnson CH. 2013. Visible light alters yeast metabolic rhythms by inhibiting respiration. Proc Natl Acad Sci U S A 110:21130–21135. doi: 10.1073/pnas.1313369110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dasgupta A, Chen CH, Lee C, Gladfelter AS, Dunlap JC, Loros JJ. 2015. Biological significance of photoreceptor photocycle length: VIVID photocycle governs the dynamic VIVID-white collar complex pool mediating photo-adaptation and response to changes in light intensity. PLoS Genet 11:e1005215. doi: 10.1371/journal.pgen.1005215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jost AP, Weiner OD. 2015. Probing yeast polarity with acute, reversible, optogenetic inhibition of protein function. Synth Biol 4:1077–1085. doi: 10.1021/acssynbio.5b00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang X, Jost AP, Weiner OD, Tang C. 2013. A light-inducible organelle-targeting system for dynamically activating and inactivating signaling in budding yeast. Mol Biol Cell 24:2419–2430. doi: 10.1091/mbc.E13-03-0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hochrein L, Machens F, Messerschmidt K, Mueller-Roeber B. 2017. PhiReX: a programmable and red light-regulated protein expression switch for yeast. Nucleic Acids Res 45:9193–9205. doi: 10.1093/nar/gkx610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Melendez J, Patel M, Oakes BL, Xu P, Morton P, McClean MN. 2014. Real-time optogenetic control of intracellular protein concentration in microbial cell cultures. Integr Biol 6:366–372. doi: 10.1039/c3ib40102b. [DOI] [PubMed] [Google Scholar]
- 38.Gerhardt KP, Olson EJ, Castillo-Hair SM, Hartsough LA, Landry BP, Ekness F, Yokoo R, Gomez EJ, Ramakrishnan P, Suh J, Savage DF, Tabor JJ. 2016. An open-hardware platform for optogenetics and photobiology. Sci Rep 6:35363. doi: 10.1038/srep35363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lungu OI, Hallett RA, Choi EJ, Aiken MJ, Hahn KM, Kuhlman B. 2012. Designing photoswitchable peptides using the AsLOV2 domain. Chem Biol 19:507–517. doi: 10.1016/j.chembiol.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Renicke C, Schuster D, Usherenko S, Essen LO, Taxis C. 2013. A LOV2 domain-based optogenetic tool to control protein degradation and cellular function. Chem Biol 20:619–626. doi: 10.1016/j.chembiol.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 41.Usherenko S, Stibbe H, Muscò M, Essen LO, Kostina EA, Taxis C. 2014. Photo-sensitive degron variants for tuning protein stability by light. BMC Syst Biol 8:128. doi: 10.1186/s12918-014-0128-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheng P, He Q, Yang Y, Wang L, Liu Y. 2003. Functional conservation of light, oxygen, or voltage domains in light sensing. Proc Natl Acad Sci U S A 100:5938–5943. doi: 10.1073/pnas.1031791100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kawano F, Suzuki H, Furuya A, Sato M. 2015. Engineered pairs of distinct photoswitches for optogenetic control of cellular proteins. Nat Commun 6:6256. doi: 10.1038/ncomms7256. [DOI] [PubMed] [Google Scholar]
- 44.Kawano F, Okazaki R, Yazawa M, Sato M. 2016. A photoactivatable Cre-loxP recombination system for optogenetic genome engineering. Nat Chem Biol 12:1059–1064. doi: 10.1038/nchembio.2205. [DOI] [PubMed] [Google Scholar]
- 45.Heintzen C, Loros JJ, Dunlap JC. 2001. The PAS protein VIVID defines a clock-associated feedback loop that represses light input, modulates gating, and regulates clock resetting. Cell 104:453–464. doi: 10.1016/S0092-8674(01)00232-X. [DOI] [PubMed] [Google Scholar]
- 46.Montenegro-Montero A, Canessa P, Larrondo LF. 2015. Around the fungal clock: recent advances in the molecular study of circadian clocks in Neurospora and other fungi. Adv Genet 92:107–184. doi: 10.1016/bs.adgen.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 47.Gibson DG, Benders GA, Axelrod KC, Zaveri J, Algire MA, Moodie M, Montague MG, Venter JC, Smith HO, Hutchison CA III. 2008. One-step assembly in yeast of 25 overlapping DNA fragments to form a complete synthetic Mycoplasma genitalium genome. Proc Natl Acad Sci U S A 105:20404–20409. doi: 10.1073/pnas.0811011106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oldenburg KR, Vo KT, Michaelis S, Paddon C. 1997. Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res 25:451–452. doi: 10.1093/nar/25.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Longtine MS, McKenzie A III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953–961. doi:. [DOI] [PubMed] [Google Scholar]
- 50.Rienzo A, Pascual-Ahuir A, Proft M. 2012. The use of a real-time luciferase assay to quantify gene expression dynamics in the living yeast cell. Yeast 29:219–231. doi: 10.1002/yea.2905. [DOI] [PubMed] [Google Scholar]
- 51.Salinas F, de Boer CG, Abarca V, García V, Cuevas M, Araos S, Larrondo LF, Martínez C, Cubillos FA. 2016. Natural variation in non-coding regions underlying phenotypic diversity in budding yeast. Sci Rep 6:21849. doi: 10.1038/srep21849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Larrondo LF, Olivares-Yañez C, Baker CL, Loros JJ, Dunlap JC. 2015. Decoupling circadian clock protein turnover from circadian period determination. Science 347:1257277. doi: 10.1126/science.1257277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kushnirov VV. 2000. Rapid and reliable protein extraction from yeast. Yeast 16:857–860. doi:. [DOI] [PubMed] [Google Scholar]
- 54.Baker CL, Kettenbach AN, Loros JJ, Gerber SA, Dunlap JC. 2009. Quantitative proteomics reveals a dynamic interactome and phase-specific phosphorylation in the Neurospora circadian clock. Mol Cell 34:354–363. doi: 10.1016/j.molcel.2009.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 57.Liu N, Wang D, Wang ZY, He XP, Zhang B. 2007. Genetic basis of flocculation phenotype conversion in Saccharomyces cerevisiae. FEMS Yeast Res 7:1362–1370. doi: 10.1111/j.1567-1364.2007.00294.x. [DOI] [PubMed] [Google Scholar]
- 58.Oud B, Guadalupe-Medina V, Nijkamp JF, de Ridder D, Pronk JT, van Maris AJ, Daran JM. 2013. Genome duplication and mutations in ACE2 cause multicellular, fast-sedimenting phenotypes in evolved Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 110:E4223–E4231. doi: 10.1073/pnas.1305949110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FUN-LOV control of luciferase expression. Bioluminescence was monitored and normalized by OD600, as cultures were grown in constant darkness (DD), constant white light (LL), or constant blue light (BL), respectively. (A to F) Components of the FUN-LOV system and the reporter construct (PGAL1-luciferase or P5XGAL1-luciferase) were maintained episomally in a BY4741 wild-type strain (A to C) or in a BY4741 gal4Δ-gal80Δ strain (D to F). (G to L) BY4741 (G to I) or BY4741 gal4Δ-gal80Δ-derived strains (J to L), containing the FUN-LOV system episomally, but having the luciferase reporters integrated at the GAL3 locus. In panels A to L, standard deviations are represented as shadowed regions. Download FIG S1, PDF file, 0.8 MB (800.4KB, pdf) .
Copyright © 2018 Salinas et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Raw luciferase expression levels during active yeast growth. (A to C) The growth of yeast strains containing FUN-LOV and luciferase controlled by a PGAL1 or P5XGAL1 promoter was monitored as OD60 and bioluminescence levels, which were quantified in constant darkness (DD), constant white light (LL), or constant blue light (BL), respectively. (A to F) Components of the FUN-LOV switch and the reporter gene were maintained episomally in the BY4741 wild type (A to C) or BY4741 gal4Δ-gal80Δ strain (D to F). (G to L) Although the FUN-LOV switch was kept episomally, the luciferase reporter was chromosomally inserted at the GAL3 locus in BY4741 (G to I) or in the BY4741 gal4Δ-gal80Δ (J to L) background. (M and N) In addition, the behavior of the episomal Luc reporter in a BY4741 background, depending on the endogenous Gal4p, was evaluated in glucose (M) or galactose (N). When indicated, raw data for luciferase expression were acquired from two different types of promoters, PGAL1 and P5XGAL1. In panels A to N, standard deviations are represented as shadowed regions. Download FIG S2, PDF file, 1.3 MB (1.3MB, pdf) .
Copyright © 2018 Salinas et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Tuneable and dynamic control of luciferase expression by the FUN-LOV switch. Bioluminescence was actively monitored as yeast cells were grown in constant darkness and 30-min blue-light pulses were applied as depicted (A to D). PGAL1-Luc or P5XGAL1-Luc reporters were maintained episomally in a BY4741 (A) or BY4741 gal4Δ-gal80Δ (B) background or inserted at the GAL3 locus in BY4741 (C) or BY4741 gal4Δ-gal80Δ (D). The effect of the duration (E) or intensity (F) of a blue-light pulse was evaluated in a strain containing an episomally maintained PGAL1-Luc reporter. In panel F, the duration of the pulse was 2 h. In panels A to F, standard deviations are represented as shadowed regions. Download FIG S3, PDF file, 0.4 MB (417.5KB, pdf) .
Copyright © 2018 Salinas et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Behavior of the constitutive promoter controlling the FUN-LOV components. (A) Expression of the FUN-LOV switch components (also Fig. 1A) is regulated by the ADH1 promoter (PADH1) and ADH2 transcriptional terminator (ADH2ter). (B to D) The graphs depict luciferase expression regulated by the PADH1 (B), PTEF1 (C), and PTDH3 (D) promoters, as well as the OD600. (E) GAL4-DBD expression was measured by real-time PCR (qPCR) in two different genetic backgrounds (BY4741 and BY4741 gal4Δ-gal80Δ) and with/without the FUN-LOV system. In panels B to D, standard deviations are represented as shadowed regions. Download FIG S4, PDF file, 0.3 MB (263.2KB, pdf) .
Copyright © 2018 Salinas et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
FUN-LOV-controlled expression of a heterologous protein. (A to C) Expression of V5-tagged limonene synthase (LS) was analyzed by Western blotting (A) and quantified with respect to membrane-stained proteins (B) under three conditions: after 2 h of blue-light pulse (BLP), after 2 h of white-light pulse (WLP), and in constant darkness (DD). The fold induction for LS expression was calculated after the BLP and WLP with respect to DD (C). The experiments were carried out in the BY4741 wild-type strain. (D to F) Similar experiments as those in panels A to C performed in the BY4741 gal4Δ-gal80Δ genetic background. Download FIG S5, PDF file, 0.1 MB (109.5KB, pdf) .
Copyright © 2018 Salinas et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Galactose and glucose control of the flocculation phenotype. (A and B) Strains with FLO1, FLO11, and TUP1 promoter swapping (PGAL1 and P5XGAL1) were grown in galactose and glucose as carbon sources, and the flocculation phenotype was assayed by bright-field microscopy (A) and macroscopic analyses of the culture flasks (bottom view) (B). BY4741 and BY4741 flo1Δ strains were used as negative controls of flocculation, whereas the BY4741 tup1Δ strain was used as a positive control of flocculation. Bars, 100 µm at ×20 magnification and 20 µm at ×60 magnification. Download FIG S6, PDF file, 1.1 MB (1.2MB, pdf) .
Copyright © 2018 Salinas et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
FUN-LOV control of flocculation in light (FIL) and flocculation in darkness (FID) phenotypes. (A and B) Strains with FLO1, FLO11, and TUP1 promoter swapping (PGAL1 and P5XGAL1) and carrying the FUN-LOV system were grown under constant light (LL) and constant darkness (DD) conditions, and the flocculation phenotype was assayed by bright-field microscopy (A) and pictures of the culture flasks (bottom view) (B). The BY4741 wt and BY4741 flo1Δ strains were used as negative controls of flocculation, whereas the BY4741 tup1Δ strain was used as a positive control of flocculation. Bars, 100 µm at ×20 magnification and 20 µm at ×60 magnification. Download FIG S7, PDF file, 1.2 MB (1.3MB, pdf) .
Copyright © 2018 Salinas et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Quantification of light-regulated flocculation by FUN-LOV. (A) Flocculation in light (FIL) and flocculation in darkness (FID) were monitored as macroscopic phenotypes by observing cell aggregation in culture flasks (bottom view), as well as by fluorescence microscopy of cells expressing mCherry under a constitutive promoter (PTDH3). Experiments were performed under constant white light (LL) and constant darkness (DD). wt yeast cells (BY4741) as well as flo1Δ and flo11Δ strains were utilized as negative controls, whereas the tup1Δ strain was used as a positive control of flocculation. Cellular aggregations are highlighted with arrows. Bar, 100 µm. (B) Quantification of the flocculation index of the FIL and FID phenotypes depicted in panel A, where the symbol ** represents a significant statistical difference between LL and DD conditions (t test; **, P < 0.01). Download FIG S8, PDF file, 0.5 MB (522.5KB, pdf) .
Copyright © 2018 Salinas et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used in this work. Download TABLE S1, PDF file, 0.1 MB (55.6KB, pdf) .
Copyright © 2018 Salinas et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.