Influenza control strategies rely on annual immunization and require frequent updates of the vaccine, which is not always a foolproof process. Furthermore, the current antivirals are also losing effectiveness as new viral strains are often refractory to conventional treatments. Thus, there is an urgent need to find new antiviral mechanisms and develop therapeutic drugs based on these mechanisms. Targeting the virus-host interface is an emerging new strategy because host factors controlling viral replication activity will be ideal candidates, and cellular proteins are less likely to mutate under drug-mediated selective pressure. Here, we show that the ubiquitin E3 ligase TRIM41 is an intrinsic host restriction factor to IAV. TRIM41 directly binds the viral nucleoprotein and targets it for ubiquitination and proteasomal degradation, thereby limiting viral infection. Exploitation of this natural defense pathway may open new avenues to develop antiviral drugs targeting the influenza virus.
KEYWORDS: intrinsic immunity, host defense, flu, antiviral, ubiquitin E3 ligase, proteasomal degradation
ABSTRACT
Influenza A virus (IAV) is a highly transmissible respiratory pathogen and a major cause of morbidity and mortality around the world. Nucleoprotein (NP) is an abundant IAV protein essential for multiple steps of the viral life cycle. Our recent proteomic study of the IAV-host interaction network found that TRIM41 (tripartite motif-containing 41), a ubiquitin E3 ligase, interacted with NP. However, the role of TRIM41 in IAV infection is unknown. Here, we report that TRIM41 interacts with NP through its SPRY domain. Furthermore, TRIM41 is constitutively expressed in lung epithelial cells, and overexpression of TRIM41 inhibits IAV infection. Conversely, RNA interference (RNAi) and knockout of TRIM41 increase host susceptibility to IAV infection. As a ubiquitin E3 ligase, TRIM41 ubiquitinates NP in vitro and in cells. The TRIM41 mutant lacking E3 ligase activity fails to inhibit IAV infection, suggesting that the E3 ligase activity is indispensable for TRIM41 antiviral function. Mechanistic analysis further revealed that the polyubiquitination leads to NP protein degradation and viral inhibition. Taking these observations together, TRIM41 is a constitutively expressed intrinsic IAV restriction factor that targets NP for ubiquitination and protein degradation.
IMPORTANCE Influenza control strategies rely on annual immunization and require frequent updates of the vaccine, which is not always a foolproof process. Furthermore, the current antivirals are also losing effectiveness as new viral strains are often refractory to conventional treatments. Thus, there is an urgent need to find new antiviral mechanisms and develop therapeutic drugs based on these mechanisms. Targeting the virus-host interface is an emerging new strategy because host factors controlling viral replication activity will be ideal candidates, and cellular proteins are less likely to mutate under drug-mediated selective pressure. Here, we show that the ubiquitin E3 ligase TRIM41 is an intrinsic host restriction factor to IAV. TRIM41 directly binds the viral nucleoprotein and targets it for ubiquitination and proteasomal degradation, thereby limiting viral infection. Exploitation of this natural defense pathway may open new avenues to develop antiviral drugs targeting the influenza virus.
INTRODUCTION
Influenza A virus (IAV) is a member of the Orthomyxoviridae family and a human respiratory pathogen that causes seasonal epidemics and occasional global pandemics with considerable economic and social impact (1, 2). The virus engages with the host cellular protein interaction network during infection. The engagement either facilitates virus hijacking of the host molecular machinery to fulfill the viral life cycle or triggers the host immune defense to eliminate the virus. In recent years, host intrinsic restriction factors have gained increasing importance in IAV inhibition (3). Host intrinsic restriction factors usually limit viral infection by direct interaction with viral proteins. For example, plakophilin 2 (PKP2) competes with PB2 for PB1 binding, thus disrupting IAV polymerase complex and inhibiting viral replication (4). The therapeutics targeting intrinsic immunity factors are more promising because cellular proteins are less likely to mutate under drug-mediated selective pressure.
The tripartite motif (TRIM) family members have been increasingly recognized as intrinsic immunity factors that inhibit viral infection. For example, TRIM5α is well known for species-specific retroviral restriction by binding to the viral capsid and inducing premature uncoating (5). TRIM79α restricts tick-borne encephalitis virus (6). TRIM28, also known as KAP1, restricts murine leukemia virus as well as facilitating the establishment of viral latency (7, 8). Recently, TRIM52 has been found to interact with the NS2A protein of Japanese encephalitis virus and target NS2A for proteasome-mediated destruction (9).
Several TRIM proteins have been found to inhibit IAV infection. For example, TRIM32 ubiquitinates PB1 and subsequently degrades PB1, thereby limiting viral infection (10). TRIM19 (also known as PML), TRIM22, and TRIM56 display broad intrinsic antiviral activity and inhibit multiple viruses, including IAV (11–13). In contrast, IAV evolves to subvert host immunity by targeting TRIM proteins (14). TRIM25 ubiquitinates and activates RIG-I-mediated innate immunity (15). The NS1 of IAV impairs the interferon (IFN)-dependent innate immune response by impeding TRIM25 multimerization and activation of RIG-I (16, 17).
Our recent study on the IAV-host protein interaction network found that TRIM41 interacted with the nucleoprotein (NP) (4). TRIM41, also known as the RING finger-interacting protein with C kinase (RINCK), regulates PKC kinase signaling (18). TRIM41 is also found to interact with the nucleotide binding oligomerization domain-containing 2 (NOD2) protein, but how TRIM41 regulates NOD2 signaling is not clear (19). Recently, a screening of TRIM proteins found that TRIM41 along with seven other TRIM proteins inhibited hepatitis B virus (HBV) transcription (20). However, the role of TRIM41 in IAV infection is unknown. Here, we characterized the physical interaction between TRIM41 and NP. Furthermore, overexpression of TRIM41 inhibits IAV infection while depletion of TRIM41 increases host susceptibility to viral infection. As a ubiquitin E3 ligase, TRIM41 ubiquitinates NP, and the ubiquitination leads to NP protein degradation. Thus, our study establishes the role of TRIM41 as a new host restriction factor in IAV infection.
RESULTS
TRIM41 interacts and colocalizes with NP.
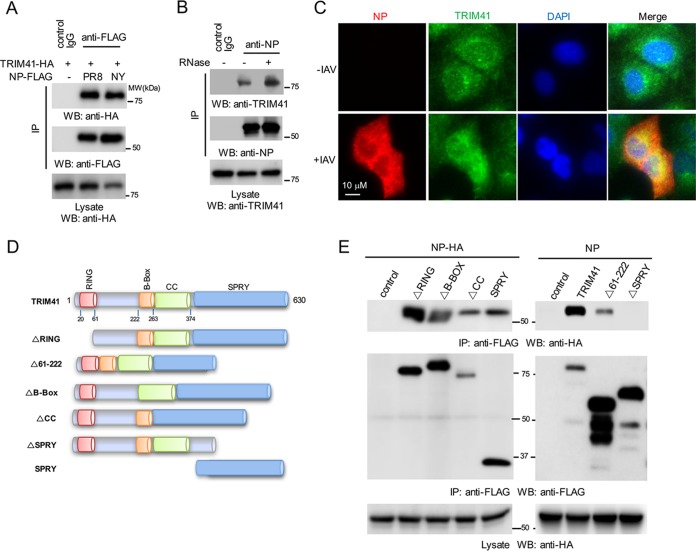
Our previous proteomic study showed that TRIM41 associated with NP (4), but the interaction is not well defined. Thus, we first validated the protein interaction between TRIM41 and NP. FLAG-tagged NP from influenza A/Puerto Rico/8/1934 (PR8) or A/New York/1682/2009 (NY) was cotransfected with hemagglutinin (HA)-tagged TRIM41 into HEK293 cells. Coimmunoprecipitation confirmed the interaction between TRIM41 and the two NP proteins (Fig. 1A). Next, we examined the TRIM41-NP protein interaction during viral infection. A549 cells were infected at a multiplicity of infection (MOI) of 1 with PR8 IAV for 12 h, and then cell lysates were immunoprecipitated with anti-NP antibody or control IgG. As shown in Fig. 1B, viral NP interacted with endogenous TRIM41. Furthermore, RNase treatment had little effect on the interaction (Fig. 1B), suggesting that the TRIM41-NP interaction is independent of RNA. We also examined the colocalization between TRIM41 and NP in A549 lung epithelial cells. Consistent with a previous report (21), endogenous TRIM41 was expressed in the cytoplasm and nucleus (Fig. 1C). Following IAV infection, NP was expressed and colocalized with TRIM41 in the cytoplasm and nucleus (Fig. 1C). Finally, to determine which domain of TRIM41 is required for NP interaction, we generated a series of TRIM41 truncates by mutagenesis (Fig. 1D). As shown in Fig. 1E, deletion of RING (ΔRING), the region of amino acids 61 to 222 (Δ61–222), the B-box (ΔB-Box), or the coiled coil (ΔCC) had marginal effects on TRIM41-NP interaction while deletion of SPRY (ΔSPRY) abolished the interaction (Fig. 1E), suggesting that the C-terminal SPRY domain is required. Indeed, the SPRY domain alone is sufficient for NP interaction (Fig. 1E). Thus, TRIM41 interacts with NP through the SPRY domain on the C terminus.
FIG 1.
TRIM41 colocalizes and interacts with NP. (A) TRIM41-HA (1.25 μg) was cotransfected with 1.25 μg of FLAG-tagged NP from the IAV PR8 or NY strain into HEK293 cells. Cell lysates were immunoprecipitated (IP) with anti-FLAG antibody or control IgG and then blotted as indicated. Molecular weights are indicated. (B) A549 cells were infected at an MOI of 1 with PR8 IAV for 12 h. Cell lysates were treated with or without RNase A and then immunoprecipitated with anti-NP antibody or control IgG. (C) A549 cells were mock infected or infected with PR8 IAV. After 12 h, cells were fixed with cold methanol and incubated with anti-NP and anti-TRIM41 antibodies. DAPI, 4′,6′-diamidino-2-phenylindole. (D) The schematics of TRIM41 mutants. RING, really interesting new gene; B-Box, B-box type zinc finger; CC, coiled coil; SPRY, SPla and the RYanodine receptor. (E) HA-tagged NP (NP-HA; 1.25 μg) was cotransfected with 1.25 μg of the indicated FLAG-tagged TRIM41 mutants into HEK293 cells. Cell lysates were immunoprecipitated with anti-FLAG antibody and blotted with the indicated antibodies. WB, Western blotting.
TRIM41 expression is not induced by type I IFN.
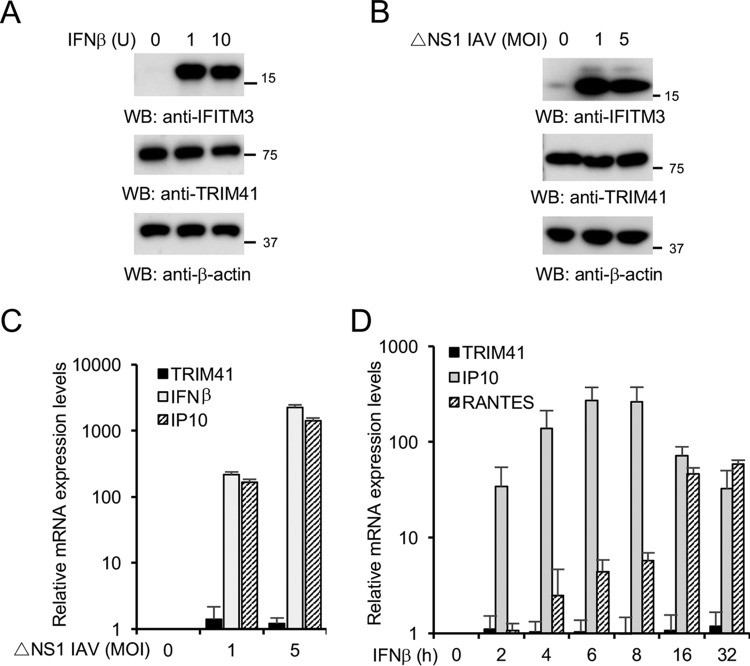
As IAV is intrinsically sensitive to the antiviral action of IFN, we speculated that type I IFN might regulate TRIM41 expression. Therefore, we treated the cells with IFN-β or infected A549 lung epithelial cells with a PR8 IAV mutant with an NS1 gene deletion (PR8 ΔNS1) (22). TRIM41 mRNA levels and protein expression were then examined. However, IFN-β stimulation or IAV infection marginally modulated TRIM41 protein and mRNA levels (Fig. 2A to D), suggesting that TRIM41 is constitutively expressed in A549 human lung epithelial cells.
FIG 2.
TRIM41 expression is not modulated by IFN and IAV. (A) A549 cells were treated with the designated units (U) of IFN-β for 12 h. Cell lysates were blotted with the indicated antibodies. The interferon-induced transmembrane protein 3 (IFITM3) was included as a positive control. (B) A549 cells were infected at the indicated MOI with PR8 ΔNS1 IAV for 12 h. Cell lysates were blotted with the indicated antibodies. IFITM3 was included as a positive control. (C) A549 cells were infected at the indicated MOI with the ΔNS1 mutant virus for 4 h. mRNA was extracted, and real-time PCR was performed to determine the relative levels of the indicated genes. IFN-β and IFN-γ-inducible protein 10 (IP10) are the positive-control genes that are known to be induced by viral infection. (D) A549 cells were treated with 1 unit of IFN-β for the designated times. mRNA was extracted, and real-time PCR was performed to determine the relative levels of the indicated genes. All experiments were biologically repeated three times. RANTES (regulated on activation, normal T cell expressed and secreted) and IP10 were the positive controls.
TRIM41 limits IAV infection.
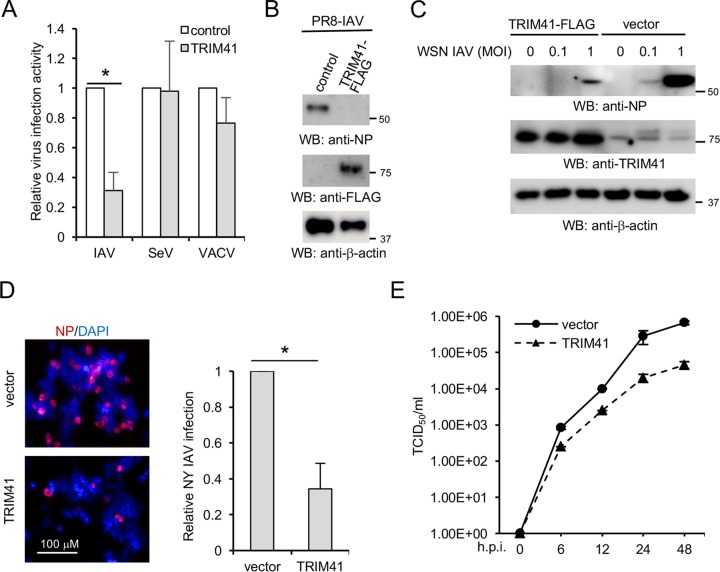
These initial observations led to our working hypothesis that TRIM41 modulates IAV infection. To test this hypothesis, four assays (reporter assay, Western blotting, and immunofluorescence and 50% tissue infective dose [TCID50] assays) were adopted to evaluate the biological effects of TRIM41 overexpression on IAV infection. First, FLAG-tagged TRIM41 or vector was transfected into HEK293 cells, followed by infection with a PR8-Gaussia luciferase reporter virus (PR8-Gluc) (23). The reporter assay showed that TRIM41 overexpression inhibited IAV replication activity (Fig. 3A). We also examined whether TRIM41 restricted other viruses. HEK293 cells expressing TRIM41-FLAG were also infected with two other reporter viruses, Sendai virus with a firefly luciferase gene (SeV-Luc) or vaccinia virus with a firefly luciferase gene (VACV-Luc). TRIM41 selectively restricted IAV replication (Fig. 3A). We then determined the effect of TRIM41 overexpression on viral infection by examining NP expression of PR8 IAV and A/WSN/1933 (WSN). TRIM41 overexpression impaired the replication of both IAV strains, as indicated by decreased levels of NP protein (Fig. 3B and C). An immunofluorescence assay (IFA) was also performed to visualize viral restriction. Ectopic expression of TRIM41 restricted NY IAV infection in HEK293 cells, as indicated by the number of infected cells (Fig. 3D). A TCID50 assay was used to determine the effect of TRIM41 on the production of infectious IAV particles. Overexpression of TRIM41 consistently reduced viral titers of WSN IAV (Fig. 3E). Taken together, these findings indicate that TRIM41 is an IAV restriction factor.
FIG 3.
Ectopic expression of TRIM41 inhibits IAV infection. (A) HEK293 cells transfected with 0.5 μg of TRIM41-FLAG or pCMV-3Tag-8 vector were infected at an MOI of 0.1 with IAV-Gluc, SeV-Luc, and VACV-Luc for 12 h. Relative luciferase activities were examined. Data represent means ± standard deviations of three independent experiments. The P value was calculated (two-tailed Student's t test) by comparison of results with those with the vector control (*, P < 0.05). (B) HEK293 cells were transfected with 0.5 μg of pCMV-3Tag-8 vector or TRIM41-FLAG for 24 h and then infected at an MOI of 1 with PR8 IAV for 12 h. Cell lysates were blotted using the indicated antibodies. (C) HEK293 cells were transfected with 0.5 μg of pCMV-3Tag-8 vector or TRIM41-FLAG for 24 h and then infected at the indicated MOI with WSN IAV for 12 h. Cell lysates were blotted using the indicated antibodies. (D) HEK293 cells were transfected with 0.5 μg of pCMV-3Tag-8 vector or TRIM41-FLAG for 24 h and then infected at an MOI of 1 with NY IAV for 12 h. Fixed cells were stained with anti-NP antibody. The percentage of stained cells is summarized in the graph. *, P < 0.05. (E) HEK293 cells were transfected with 0.5 μg of pCMV-3Tag-8 vector or TRIM41-FLAG. After 24 h, cells were infected at an MOI of 0.001 with WSN/33 IAV. After the designated hour postinfection (h.p.i.), virus titers were determined by TCID50 assay in MDCK cells. All experiments were biologically repeated three times.
TRIM41 deficiency increases host susceptibility to IAV.
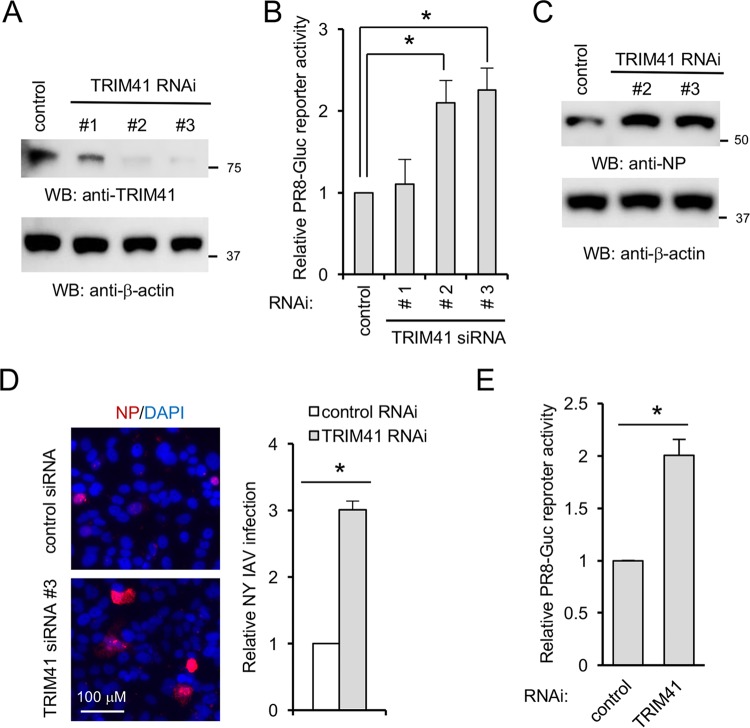
To complement the above gain of function, we first depleted TRIM41 using a small interfering RNA (siRNA). Three pairs of siRNA duplexes against TRIM41 were individually transfected into A549 cells. After 48 h, cells were infected with PR8-Gluc IAV for 12 h. Reduced TRIM41 protein expression was correlated with increased IAV reporter activity in A549 lung cells (Fig. 4A and B). Silencing TRIM41 also enhanced IAV propagation in A549 cells, as detected by Western blotting and IFA (Fig. 4C and D). Furthermore, we knocked down TRIM41 in human primary tracheal cells and then infected these cells with PR8-Gluc IAV. Luciferase assays showed that RNA interference (RNAi) depletion of TRIM41 increased IAV infection in primary tracheal cells (Fig. 4E).
FIG 4.
Knockdown of TRIM41 increases IAV infection in A549 cells. (A) A549 cells were transfected with 5 pmol of a scrambled control siRNA or three different siRNA duplexes against TRIM41. After 48 h, cell lysates were blotted with anti-TRIM41 or anti-β-actin antibody. (B) A549 cells were transfected with 5 pmol of the control siRNA or the indicated siRNA duplex against TRIM41. After 48 h, the cells were infected at an MOI of 0.1 with IAV PR8-Gluc for 12 h. Relative luciferase activities were examined. All experiments were biologically repeated three times. Data represent means ± standard deviations of three independent experiments. The P value was calculated (two-tailed Student's t test) by comparison of results with those of the siRNA control in each cell group (*, P < 0.05). (C) A549 cells were transfected with 5 pmol of the control siRNA or the TRIM41 siRNA 2 or 3 duplex for 48 h and then infected at an MOI of 1 with WSN IAV for 12 h. Cell lysates were blotted using the indicated antibodies. (D) A549 cells were transfected with 5 pmol of the control siRNA or the TRIM41 siRNA 3 duplex for 48 h and then infected at an MOI of 0.1 with NY IAV for 12 h. Fixed cells were stained with anti-NP antibody. The percentage of stained cells is summarized in the graph. *, P < 0.05. DAPI, 4′,6′-diamidino-2-phenylindole. (E) Primary human tracheal epithelial cells were transfected with 5 pmol of the control siRNA or the TRIM41 siRNA 3 duplex. After 48 h, the cells were infected at an MOI of 0.1 with PR8-Gluc for 16 h. The relative luciferase activity was examined. Data represent means ± standard deviations of three independent experiments. *, P < 0.05.
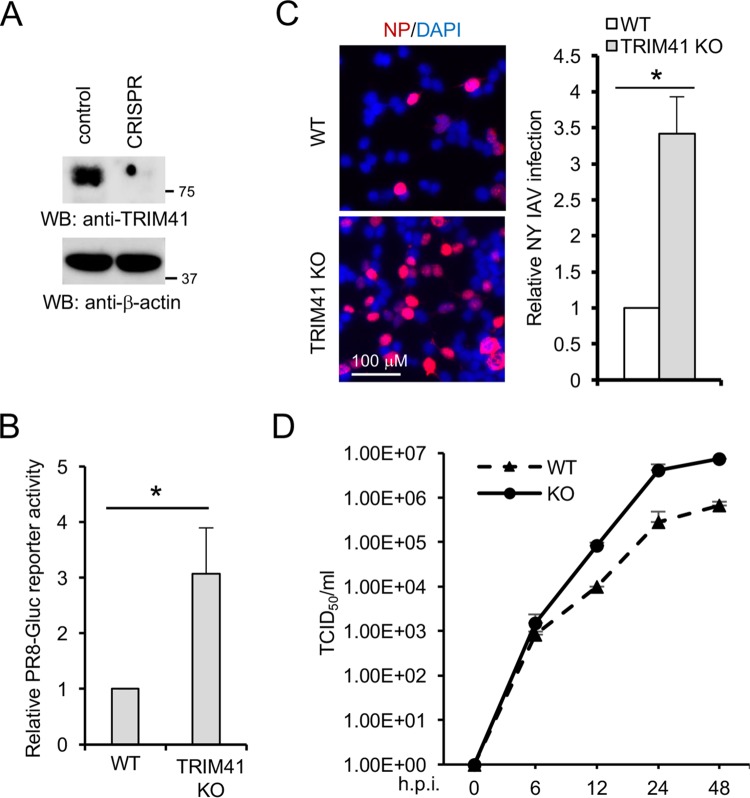
We next examined the effects of TRIM41 knockout on IAV infection. In this regard, a single guide RNA (sgRNA) was cloned into the lentiCRISPR v2 containing Cas9 (24) and transfected into HEK293 cells. After 48 h, cells were selected using puromycin. Single clones were picked and expanded for knockout confirmation by Western blotting. TRIM41 expression was abolished in CRIPSR knockout clone 4 as shown by Western blotting (Fig. 5A). DNA sequencing found a 1-bp insertion in the DNA genome of knockout clone 4 (data not shown). An increase in PR8-GLuc reporter activity was noted in TRIM41 knockout cells compared to that in the wild-type controls (Fig. 5B). In line with these results, significantly more TRIM41 knockout cells were stained with anti-NP antibody (Fig. 5C). We also evaluated the impact of TRIM41 deficiency on viral propagation using a TCID50 assay. As predicted, TRIM41 knockout cells produced more infectious viral particles than control HEK293 cells (Fig. 5D). Taken together, these data suggest that endogenous TRIM41 is essential for host restriction to IAV.
FIG 5.
Knockout of TRIM41 increases host susceptibility to IAV infection. (A) CRISPR knockout of TRIM41 in HEK293 cells. Cell lysates were blotted as indicated. (B) Wild-type (WT) HEK293 and TRIM41 knockout (KO) cells were infected at an MOI of 0.1 with IAV PR8-Gluc for 12 h. Relative luciferase activities were examined. All experiments were biologically repeated three times. *, P < 0.05. (C) Wild-type and TRIM41 knockout cells were infected at an MOI of 1 with NY IAV for 12 h. Cells were fixed and stained with anti-NP antibody. The percentage of stained cells is summarized in the graph. *, P < 0.05. (D) Wild-type and TRIM41 knockout cells were infected at an MOI of 0.001 with WSN/33 IAV. After the designated hour postinfection, virus titers were determined by TCID50 assay in MDCK cells. All experiments were biologically repeated three times.
Ubiquitin E3 ligase activity is required for TRIM41 antiviral function.
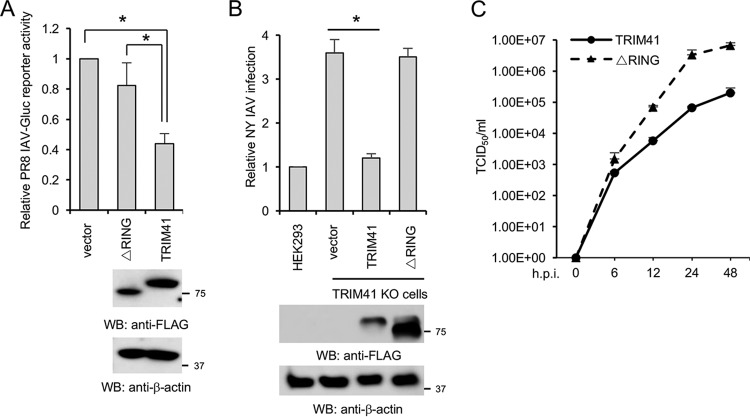
The RING domain is critical for the E3 ligase activity of TRIM proteins. Thus, we evaluated the role of the RING domain in TRIM41-mediated viral restriction. TRIM41 or the RING deletion (ΔRING) mutant was transfected into HEK293 cells, followed by IAV infection. Ectopic expression of wild-type TRIM41 attenuated infection of the IAV reporter virus while the ΔRING mutant failed to defend against IAV infection (Fig. 6A). Furthermore, wild-type TRIM41, but not the mutant, restored antiviral activity in the TRIM41 knockout cells (Fig. 6B). Similarly, the mutant failed to inhibit IAV infection in the TRIM41 knockout cells as determined by TCID50 assay (Fig. 6C). Thus, we concluded that E3 ligase activity is critical for the antiviral function of TRIM41.
FIG 6.
E3 ligase activity is required for TRIM41 antiviral function. (A) HEK293 cells were transfected with 0.5 μg of pCMV-3Tag-8 vector, FLAG-tagged TRIM41, or the ΔRING mutant. After 24 h, cells were infected at an MOI of 0.1 with PR8-Gluc for 12 h. Relative luciferase activities were examined. Data represent means ± standard deviations of three independent experiments. The P value was calculated by two-tailed Student's t test (*, P < 0.05). (B) TRIM41 knockout cells were transfected with 0.5 μg of pCMV-3Tag-8 vector, TRIM41-FLAG, or the ΔRING mutant for 24 h and then infected at an MOI of 1 with NY IAV for 12 h. HEK293 cells were included as the control. IFA was performed, and the percentage of stained cells was used to determine the infection activity. *, P < 0.05. (C) TRIM41 knockout cells were transfected with 0.5 μg of pCMV-3Tag-8 vector, TRIM41-FLAG, or the ΔRING mutant. After 24 h, cells were infected at an MOI of 0.001 with WSN/33 IAV. After the designated hour postinfection, virus titers were determined by TCID50 assay in MDCK cells. All experiments were biologically repeated three times.
TRIM41 ubiquitinates and degrades NP.
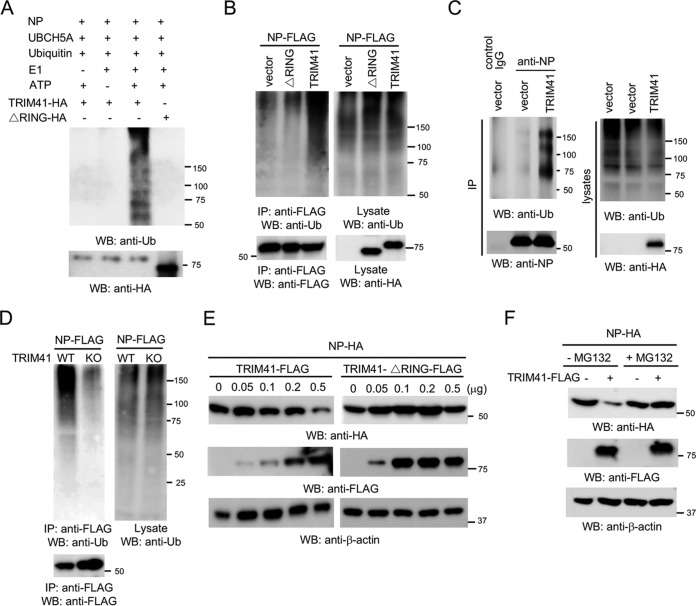
The requirement of E3 ligase activity indicates that TRIM41 may ubiquitinate NP. To determine whether TRIM41 can couple ubiquitin onto NP, we first performed an in vitro ubiquitination assay. TRIM41 is able to heavily conjugate ubiquitin onto NP in vitro, but the ΔRING mutant failed (Fig. 7A). We next examined NP ubiquitination in cells. NP was transfected with vector, TRIM41, or the ΔRING mutant into HEK293 cells. As shown in Fig. 7B, TRIM41 increased the levels of NP ubiquitination. However, ectopic expression of the ΔRING mutant showed little or no effect on NP ubiquitination (Fig. 7B). We also examined whether overexpression of TRIM41 increases NP ubiquitination during IAV infection. HEK293 cells were transfected with vector or TRIM41 and then infected at an MOI of 1 with PR8 IAV for 12 h. As shown in Fig. 7C, TRIM41 increased the levels of NP ubiquitination during IAV infection. Consistently, ubiquitinated NP was barely detected in TRIM41 knockout cells (Fig. 7D).
FIG 7.
TRIM41 ubiquitinates and degrades NP. (A) In vitro ubiquitination of NP by TRIM41. FLAG-tagged NP, HA-tagged TRIM41, and the RING deletion mutant plus E1, E2 (UBCH5A), ATP, and ubiquitin were added as indicated and incubated at 30°C for 2 h. Samples were blotted with the indicated antibodies. Ub, ubiquitin. (B) NP-FLAG (0.5 μg) was cotransfected with 2 μg of vector, TRIM41-HA, or the HA-tagged ΔRING mutant into HEK293 cells. Cells were treated with 1 μM MG132 for 12 h to prevent NP protein degradation. Cell lysates were immunoprecipitated with anti-FLAG antibody and blotted with the indicated antibodies. (C) HEK293 cells were transfected with 2.5 μg of vector or TRIM41. After 24 h, cells were infected at an MOI of 1 with PR8 IAV and treated with 1 μM MG132 for 12 h. Cell lysates were immunoprecipitated with anti-NP antibody and blotted with the indicated antibodies. (D) Wild-type and TRIM41 knockout cells were transfected with 0.5 μg of NP-FLAG. Cell lysates were immunoprecipitated with anti-FLAG antibody and blotted with the indicated antibodies. (E) NP-HA (0.25 μg) was cotransfected with the designated amounts of FLAG-tagged TRIM41 or the ΔRING mutant into HEK293 cells. Cell lysates were blotted as indicated. (F) NP-HA (0.25 μg) was cotransfected with 0.25 μg of vector or TRIM41-FLAG into HEK293 cells. After 24 h, cells were treated with dimethyl sulfoxide or 1 μM MG132 for 12 h. Cell lysates were then blotted with the indicated antibodies.
Proteins modified with polyubiquitin are classic targets for proteasomal degradation. To determine the role of TRIM41 in NP protein turnover, NP was transfected with wild-type TRIM41 or the ΔRING mutant into HEK293 cells. Wild-type TRIM41, but not the mutant, reduced NP protein expression (Fig. 7E), indicating that TRIM41 promotes NP destabilization. To corroborate the role of TRIM41 in NP degradation, HEK293 cells were transfected with NP along with TRIM41 or vector. After 24 h, cells were treated with the protease inhibitor MG132 for 12 h. Western blotting found that MG132 prevented TRIM41-mediated NP degradation (Fig. 7F). Taken together, the combined data indicate that TRIM41 mediates ubiquitination of NP, which leads to NP degradation and viral inhibition.
DISCUSSION
The innate immune system uses pattern recognition receptors (PRRs) in different cellular compartments to discriminate microbial components that mark invading viruses. Many of these PRRs have been characterized, including the Toll-like receptors (TLRs) and RIG-I-like receptors (RLRs) (25, 26). TLRs and RLRs sense viral nucleic acids and indirectly inhibit viral infection by eliciting signaling cascades that activate the expression of type I IFN. The type I IFN acts as a master cytokine that activates hundreds of interferon-stimulated genes, which in turn inhibits viral infection. For example, interferon-induced transmembrane proteins (IFITMs) recruit and coordinate with the zinc metalloprotease ste24-like protein (ZMPSTE24) to block influenza virus entry on endosomes (27, 28). In contrast, intrinsic immunity impairs viral infection by direct interaction between host factors and virus (3). Several TRIM proteins have been reported as intrinsic restriction factors, such as TRIM5α (5), TRIM32 (10), and TRIM22 (12). Similarly, our study demonstrates that TRIM41 directly restricts viral infection through ubiquitination of NP. Thus, TRIM41 is a new intrinsic immune factor that inhibits IAV infection.
TRIM proteins are ubiquitin E3 ligases that share a conserved N-terminal tripartite motif (comprising RING, B-box, and coiled-coil domains) and a versatile C-terminal domain (29–32). TRIM41 belongs to the subgroup of the TRIM family that possesses a SPRY domain on the C terminus. Our data showed that the SPRY domain of TRIM41 is responsible for the interaction with NP. More importantly, the RING domain of TRIM proteins confers the ubiquitin E3 ligase activity. Our study found a requirement of E3 ligase activity for TRIM41 antiviral activity. Interestingly, a recent study reported that the E3 ligase activity of TRIM41 is also required for HBV inhibition (20), but the antiviral mechanism is unknown. However, our study further revealed that TRIM41 mediates the ubiquitination of NP, which leads to NP protein degradation and viral inhibition.
NP has an essential role in the IAV life cycle, including nuclear transport of viral RNP (vRNP) and transcription and replication of the viral genome (3). Several host factors have been shown to inhibit IAV infection by interacting with NP. For example, myxovirus resistance protein 1 interacts with NP and suppresses influenza virus transcription (33). RuvB-like protein 2 disrupts vRNP assembly by interfering with NP oligomerization (34). Interferon-stimulated gene 20 interacts with NP and inhibits viral protein expression (35). Similarly, our study showed that TRIM41 modulates IAV infection by interaction with NP, which allows NP ubiquitination and subsequent protein degradation.
TRIM22 was reported to ubiquitinate NP and target it for protein degradation (12). Unlike TRIM41, TRIM22 is an IFN-stimulated gene, and a previous study showed that the protein expression of TRIM22 is very low or undetectable in cells without IFN stimulation (12). Thus, TRIM22 might not be present in the cells during the early stage of viral infection before the host IFN response is elicited. In contrast, TRIM41 is constitutively expressed, and its expression is not modulated by IFN and IAV. Therefore, it is possible that TRIM41 and TRIM22 cooperate to limit NP expression at different stages of the IAV life cycle. Future experiments will investigate the distinct roles of TRIM41 and TRIM22 in IAV restriction. Furthermore, CNOT4 is also reported to ubiquitinate NP (36). However, CNOT4-mediated ubiquitination promotes viral RNA replication by monoubiquitination of NP (36).
In conclusion, we demonstrate that TRIM41 inhibits IAV by targeting NP for proteasomal degradation and provides intrinsic cellular restriction against IAV infection. Upregulation of TRIM41 E3 ligase activity may boost host immunity to limit viral infection. Thus, future investigation of the regulation of TRIM41 activity might provide the insights and opportunities needed for therapeutic development.
MATERIALS AND METHODS
Cells and viruses.
HEK293 cells (ATCC CRL-1573) and MDCK cells (ATCC CCL-34) were maintained in Dulbecco's modified Eagle medium (Life Technologies) containing antibiotics (Life Technologies) and 10% fetal bovine serum (Life Technologies). A549 cells (ATCC CCL-185) were cultured in RPMI 1640 medium (Life Technologies) plus 10% fetal bovine serum and 1× minimal essential medium (MEM) supplemented with nonessential amino acids (MEM Non-Essential Amino Acids Solution; Life Technologies). Primary human tracheal epithelial cells and supporting medium were purchased from Lifeline Cell Technology (FC-0035 and LL-0023, respectively; Frederick, MD).
Influenza A/Puerto Rico/8/34 (H1N1) (catalog number 10100374; Charles River Laboratories), A/WSN/33 (H1N1) (a kind gift of Peter Palese, Mount Sinai School of Medicine, NY), and A/New York/18/2009 (H1N1) pdm09 (BEI Resources) were used. Influenza PR8-GLuc virus was a generous gift from Peter Palese and features a Gaussia luciferase (Gluc) gene inserted downstream of PB2 (23). IAV propagation in specific-pathogen-free fertilized eggs and a TCID50 assay were performed as described by Szretter et al. (37). Sendai virus with a luciferase gene (SeV-Luc) was a kind gift from Charles Russell (St. Jude's Hospital, Memphis, TN). VACV with a firefly luciferase gene (VACV-Luc) was a gift from D. L. Bartlett (University of Pittsburg, Pittsburg, PA).
Plasmids and antibodies.
TRIM41-HA was a generous gift from Adolfo Garcia-Sastre (Mount Sinai School of Medicine, NY) (30). TRIM41 was cloned into pCMV-3Tag-8 (Stratagene) to generate TRIM41-FLAG. Deletion mutants of TRIM41-FLAG were constructed using a Q5 site-directed mutagenesis kit (New England BioLabs).
Antibodies included the following: anti-β-actin (ab8227; Abcam), anti-FLAG (F3165; Sigma), anti-IFITM3 (GTX63349; GeneTex), anti-ubiquitin (sc-8017; Santa Cruz Biotechnology), anti-TRIM41 (ARP34763_P050; Aviva Systems Biology), anti-HA epitope (3724; Cell Signaling Technology), anti-NP (NR-4282; BEI Resources), anti-NP (MAB8800; EMD Millipore), and anti-NP (A01506-40; GenScript). Goat anti-mouse IgG-horseradish peroxidase (HRP) (sc-2055; Santa Cruz Biotechnology), goat anti-rabbit IgG-HRP (sc-2030; Santa Cruz Biotechnology), Alexa Fluor 594-goat anti-mouse IgG(H+L) (A11005; Life Technologies), and Alexa Fluor 488-goat anti-rabbit IgG(H+L) (A11034; Life Technologies) were also used.
Sample preparation, Western blotting, and immunoprecipitation.
Approximately 1 × 106 cells were lysed in 500 μl of tandem affinity purification (TAP) lysis buffer (50 mM Tris-HCl [pH 7.5], 10 mM MgCl2, 100 mM NaCl, 0.5% Nonidet P40, 10% glycerol, Complete EDTA-free protease inhibitor cocktail tablets [Roche]) for 30 min at 4°C. The lysates were then centrifuged for 30 min at 15,000 × g. Supernatants were collected and mixed with 1× Lane Marker Reducing Sample Buffer (Thermo Fisher Scientific). Western blotting and immunoprecipitation were performed as described in a previous study (4).
Immunofluorescence assay.
Cells were cultured in a Lab-Tek II CC2 chamber slide system (four-well; Thermo Fisher Scientific). After IAV infection, the cells were fixed and permeabilized in cold methanol for 10 min at −20 °C. Then the slides were washed with 1× phosphate-buffered saline (PBS) for 10 min and blocked with Odyssey blocking buffer (Li-Cor Biosciences) for 1 h. The slides were incubated in Odyssey blocking buffer with appropriately diluted primary antibodies at 4 °C for 12 h. Images were captured and analyzed using an iRiS digital cell imaging system (Logos Biosystems).
Real-time PCR.
Total RNA was prepared using RNeasy columns (Qiagen). A1-μg quantity of RNA was transcribed into cDNA using a QuantiTect reverse transcription kit (Qiagen). For one real-time reaction, 10 μl of SYBR green PCR mix (Eurogentec) including a 1/10 volume of the synthesized cDNA plus an appropriate oligonucleotide primer pair was analyzed on a 7500 Fast real-time PCR system (Applied Biosystems). The comparative threshold cycle (CT) method was used to determine the relative mRNA expression levels of genes normalized by the level of the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) housekeeping gene. The primer sequences were as follows: for IP10, forward primer 5′-TTCAAGGAGTACCTCTCTCTAG-3′ and reverse primer 5′-CTGGATTCAGACATCTCTTCTC-3′; for IFN-β, forward primer 5′-CAGCAATTTTCAGTGTCAGAAGC-3′ and reverse primer 5′-TCATCCTGTCCTTGAGGCAGT-3′; for TRIM41, forward primer 5′-AGAATCCAGGAGCCACAAAC-3′ and reverse primer 5′-TCTTCAGCCAGAAACCGTG-3′; for GAPDH, forward primer 5′-AGGTGAAGGTCGGAGTCA-3′ and reverse primer 5′-GGTCATTGATGGCAACAA-3′.
Plasmid transfection.
HEK293 cells were transfected using Lipofectamine 2000 or Lipofectamine 3000 transfection reagent (Life Technologies) according to the manufacturer's protocol. For coimmunoprecipitation experiments, a total of 2.5 μg of plasmids was transfected into approximately 1.2 × 106 cells. For other experiments, a total of 0.5 μg of plasmids was transfected into approximately 0.2 × 106 cells.
RNAi depletion.
RNAi target sequences (sense strand) were as follows: TRIM41 siRNA 1, GAGGCGAGTGACAGAACTGAA; TRIM41 siRNA 2, AAGGCGTGCTGTGGAAATAAA; TRIM41 siRNA 3, TTCAATAGGTGTGAAGAGGTA. An siGENOME nontargeting control siRNA (D-001210-02-05; Dharmacon) was used as the control siRNA. siRNA duplexes (5 pmol) were transfected into A549 cells using Lipofectamine RNAiMAX transfection reagent (Life Technologies) according to the manufacturer's protocol.
Cell viability.
Cell viability was assessed by using a CellTiter-Glo luminescent cell viability assay (Promega) according to the manufacturer's instructions.
CRISPR/Cas9.
The single guide RNA (sgRNA) sequence targeting human TRIM41 is GTAGTCTTCATCCCGCATGG. The sgRNA was cloned into lentiCRISPR v2 (24) (Addgene). Lentiviral construct (0.5 μg) was transfected into HEK293 cells using Lipofectamine 2000. Cells were selected with 10 μg/ml puromycin for 14 days. Single clones were expanded for knockout confirmation by Western blotting and DNA sequencing.
In vitro ubiquitination.
An in vitro ubiquitination assay was performed according to the manufacturer's instructions (Boston Biochem). Ubiquitin (3 μg), E1 (200 ng), UBCH5A (300 ng; Boston Biochem), NP-FLAG (0.5 μg) bound to the anti-FLAG resin (Sigma), and TRIM41-HA or ΔRING-HA (0.5 μg) were incubated at 30°C in ubiquitin assay buffer (20 mM Tris-HCl, pH 7.5, plus 1× energy regeneration solution [ERS] buffer) for 2 h. Two hours later, the anti-FLAG resin was washed with 1 M urea for 15 min to exclude potential binding of unanchored polyubiquitin. Then the resin was incubated with 40 μl of 0.2 μg/ml FLAG peptide to elute NP protein. The eluates were subsequently analyzed by SDS-PAGE, followed by Western blotting.
Statistical analysis.
The sample size was sufficient for data analysis using paired two-tailed Student's t tests. For all statistical analysis, differences were considered to be statistically significant at a P value of <0.05.
ACKNOWLEDGMENTS
We thank Peter Palese (Mount Sinai School of Medicine, New York, NY), Adolfo García-Sastre (Mount Sinai School of Medicine, New York, NY), Charles Russell (St. Jude Children's Research Hospital, Memphis, TN), and D. L. Bartlett (University of Pittsburg, Pittsburg, PA) for reagents.
This work was supported by NIH grants R15AI126360 (S.L.), R21AI137750 (S.L.), and P20GM103648 (S.L.), OCAST grant HR17-045 (S.L.), and RAC Fund (S.L.).
REFERENCES
- 1.Tscherne DM, Garcia-Sastre A. 2011. Virulence determinants of pandemic influenza viruses. J Clin Invest 121:6–13. doi: 10.1172/JCI44947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fukuyama S, Kawaoka Y. 2011. The pathogenesis of influenza virus infections: the contributions of virus and host factors. Curr Opin Immunol 23:481–486. doi: 10.1016/j.coi.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao M, Wang L, Li S. 2017. Influenza A virus-host protein interactions control viral pathogenesis. Int J Mol Sci 18:E1673. doi: 10.3390/ijms18081673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang L, Fu B, Li W, Patil G, Liu L, Dorf ME, Li S. 2017. Comparative influenza protein interactomes identify the role of plakophilin 2 in virus restriction. Nat Commun 8:13876. doi: 10.1038/ncomms13876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. 2004. The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature 427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 6.Taylor RT, Lubick KJ, Robertson SJ, Broughton JP, Bloom ME, Bresnahan WA, Best SM. 2011. TRIM79α, an interferon-stimulated gene product, restricts tick-borne encephalitis virus replication by degrading the viral RNA polymerase. Cell Host Microbe 10:185–196. doi: 10.1016/j.chom.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wolf D, Goff SP. 2007. TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell 131:46–57. doi: 10.1016/j.cell.2007.07.026. [DOI] [PubMed] [Google Scholar]
- 8.Rauwel B, Jang SM, Cassano M, Kapopoulou A, Barde I, Trono D. 2015. Release of human cytomegalovirus from latency by a KAP1/TRIM28 phosphorylation switch. Elife 4:e06068. doi: 10.7554/eLife.06068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan W, Wu M, Qian S, Zhou Y, Chen H, Li X, Qian P. 2016. TRIM52 inhibits Japanese encephalitis virus replication by degrading the viral NS2A. Sci Rep 6:33698. doi: 10.1038/srep33698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu B, Wang L, Ding H, Schwamborn JC, Li S, Dorf ME. 2015. TRIM32 senses and restricts influenza A virus by ubiquitination of PB1 polymerase. PLoS Pathog 11:e1004960. doi: 10.1371/journal.ppat.1004960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu B, Li NL, Shen Y, Bao X, Fabrizio T, Elbahesh H, Webby RJ, Li K. 2016. The C-terminal tail of TRIM56 dictates antiviral restriction of influenza A and B viruses by impeding viral RNA synthesis. J Virol 90:4369–4382. doi: 10.1128/JVI.03172-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Pietro A, Kajaste-Rudnitski A, Oteiza A, Nicora L, Towers GJ, Mechti N, Vicenzi E. 2013. TRIM22 inhibits influenza A virus infection by targeting the viral nucleoprotein for degradation. J Virol 87:4523–4533. doi: 10.1128/JVI.02548-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chelbi-Alix MK, Quignon F, Pelicano L, Koken MH, de The H. 1998. Resistance to virus infection conferred by the interferon-induced promyelocytic leukemia protein. J Virol 72:1043–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Tol S, Hage A, Giraldo MI, Bharaj P, Rajsbaum R. 2017. The TRIMendous role of TRIMs in virus-host interactions. Vaccines (Basel) 5:E23. doi: 10.3390/vaccines5030023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, Jung JU. 2007. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 16.Rajsbaum R, Albrecht RA, Wang MK, Maharaj NP, Versteeg GA, Nistal-Villan E, Garcia-Sastre A, Gack MU. 2012. Species-specific inhibition of RIG-I ubiquitination and IFN induction by the influenza A virus NS1 protein. PLoS Pathog 8:e1003059. doi: 10.1371/journal.ppat.1003059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanchez-Aparicio MT, Ayllon J, Leo-Macias A, Wolff T, Garcia-Sastre A. 2017. Subcellular localizations of RIG-I, TRIM25, and MAVS complexes. J Virol 91:e01155-16. doi: 10.1128/JVI.01155-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen D, Gould C, Garza R, Gao T, Hampton RY, Newton AC. 2007. Amplitude control of protein kinase C by RINCK, a novel E3 ubiquitin ligase. J Biol Chem 282:33776–33787. doi: 10.1074/jbc.M703320200. [DOI] [PubMed] [Google Scholar]
- 19.Thiebaut R, Esmiol S, Lecine P, Mahfouz B, Hermant A, Nicoletti C, Parnis S, Perroy J, Borg JP, Pascoe L, Hugot JP, Ollendorff V. 2016. Characterization and genetic analyses of new genes coding for NOD2 interacting proteins. PLoS One 11:e0165420. doi: 10.1371/journal.pone.0165420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang S, Guo JT, Wu JZ, Yang G. 2013. Identification and characterization of multiple TRIM proteins that inhibit hepatitis B virus transcription. PLoS One 8:e70001. doi: 10.1371/journal.pone.0070001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanaka M, Fukuda Y, Mashima K, Hanai R. 2005. Intracellular localization and domain organization of human TRIM41 proteins. Mol Biol Rep 32:87–93. doi: 10.1007/s11033-004-6613-2. [DOI] [PubMed] [Google Scholar]
- 22.Garcia-Sastre A, Egorov A, Matassov D, Brandt S, Levy DE, Durbin JE, Palese P, Muster T. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- 23.Heaton NS, Leyva-Grado VH, Tan GS, Eggink D, Hai R, Palese P. 2013. In vivo bioluminescent imaging of influenza A virus infection and characterization of novel cross-protective monoclonal antibodies. J Virol 87:8272–8281. doi: 10.1128/JVI.00969-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanjana NE, Shalem O, Zhang F. 2014. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takeuchi O, Akira S. 2010. Pattern recognition receptors and inflammation. Cell 140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 26.Wilkins C, Gale M Jr. 2010. Recognition of viruses by cytoplasmic sensors. Curr Opin Immunol 22:41–47. doi: 10.1016/j.coi.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, Feeley EM, Ryan BJ, Weyer JL, van der Weyden L, Fikrig E, Adams DJ, Xavier RJ, Farzan M, Elledge SJ. 2009. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139:1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fu B, Wang L, Li S, Dorf ME. 2017. ZMPSTE24 defends against influenza and other pathogenic viruses. J Exp Med 214:919–929. doi: 10.1084/jem.20161270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hatakeyama S. 2017. TRIM family proteins: roles in autophagy, immunity, and carcinogenesis. Trends Biochem Sci 42:297–311. doi: 10.1016/j.tibs.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 30.Versteeg GA, Rajsbaum R, Sanchez-Aparicio MT, Maestre AM, Valdiviezo J, Shi M, Inn KS, Fernandez-Sesma A, Jung J, Garcia-Sastre A. 2013. The E3-ligase TRIM family of proteins regulates signaling pathways triggered by innate immune pattern-recognition receptors. Immunity 38:384–398. doi: 10.1016/j.immuni.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ozato K, Shin DM, Chang TH, Morse HC III. 2008. TRIM family proteins and their emerging roles in innate immunity. Nat Rev Immunol 8:849–860. doi: 10.1038/nri2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawai T, Akira S. 2011. Regulation of innate immune signalling pathways by the tripartite motif (TRIM) family proteins. EMBO Mol Med 3:513–527. doi: 10.1002/emmm.201100160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turan K, Mibayashi M, Sugiyama K, Saito S, Numajiri A, Nagata K. 2004. Nuclear MxA proteins form a complex with influenza virus NP and inhibit the transcription of the engineered influenza virus genome. Nucleic Acids Res 32:643–652. doi: 10.1093/nar/gkh192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kakugawa S, Shimojima M, Neumann G, Goto H, Kawaoka Y. 2009. RuvB-like protein 2 is a suppressor of influenza A virus polymerases. J Virol 83:6429–6434. doi: 10.1128/JVI.00293-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qu H, Li J, Yang L, Sun L, Liu W, He H. 2016. Influenza A Virus-induced expression of ISG20 inhibits viral replication by interacting with nucleoprotein. Virus Genes 52:759–767. doi: 10.1007/s11262-016-1366-2. [DOI] [PubMed] [Google Scholar]
- 36.Lin YC, Jeng KS, Lai MMC. 2017. CNOT4-mediated ubiquitination of influenza A virus nucleoprotein promotes viral RNA replication. mBio 8:e00597-17. doi: 10.1128/mBio.00597-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szretter KJ, Balish AL, Katz JM. 2006. Influenza: propagation, quantification, and storage. Curr Protoc Microbiol Chapter. 15:Unit 15G.1. doi: 10.1002/0471729256.mc15g01s3. [DOI] [PubMed] [Google Scholar]