The rise in zoonotic infections of humans by emerging influenza viruses is a worldwide public health concern. The majority of recent zoonotic human influenza cases were caused by H7N9 and H5Nx viruses and were associated with high morbidity and mortality. In addition, seasonal influenza viruses are estimated to cause up to 650,000 deaths annually worldwide. Currently available antiviral treatment options include only neuraminidase inhibitors, but some influenza viruses are naturally resistant to these drugs, and others quickly develop resistance-conferring mutations. Alternative therapeutics are urgently needed. Broadly protective antibodies that target the conserved “stalk” domain of the hemagglutinin represent potential potent antiviral prophylactic and therapeutic agents that can assist pandemic preparedness. Here, we describe four human monoclonal antibodies that target conserved regions of influenza HA and characterize their binding spectrum as well as their protective capacity in prophylactic and therapeutic settings against a lethal challenge with a zoonotic influenza virus.
KEYWORDS: H5N1, HA stalk, hemagglutinin, hemagglutinin stalk, influenza, influenza virus, monoclonal antibody
ABSTRACT
Broadly cross-reactive antibodies (Abs) that recognize conserved epitopes within the influenza virus hemagglutinin (HA) stalk domain are of particular interest for their potential use as therapeutic and prophylactic agents against multiple influenza virus subtypes, including zoonotic virus strains. Here, we characterized four human HA stalk-reactive monoclonal antibodies (MAbs) for their binding breadth and affinity, in vitro neutralization capacity, and in vivo protective potential against an highly pathogenic avian influenza virus. The monoclonal antibodies were isolated from individuals shortly following infection with (70-1F02 and 1009-3B05) or vaccination against (05-2G02 and 09-3A01) A(H1N1)pdm09. Three of the MAbs bound HAs from multiple strains of group 1 viruses, and one MAb, 05-2G02, bound to both group 1 and group 2 influenza A virus HAs. All four antibodies prophylactically protected mice against a lethal challenge with the highly pathogenic A/Vietnam/1203/04 (H5N1) strain. Two MAbs, 70-1F02 and 09-3A01, were further tested for their therapeutic efficacy against the same strain and showed good efficacy in this setting as well. One MAb, 70-1F02, cocrystallized with H5 HA and showed heavy-chain-only interactions similar to those seen with the previously described CR6261 anti-stalk antibody. Finally, we show that antibodies that compete with these MAbs are prevalent in serum from an individual recently infected with the A(H1N1)pdm09 virus. The antibodies described here can be developed into broad-spectrum antiviral therapeutics that could be used to combat infections by zoonotic or emerging pandemic influenza viruses.
IMPORTANCE The rise in zoonotic infections of humans by emerging influenza viruses is a worldwide public health concern. The majority of recent zoonotic human influenza cases were caused by H7N9 and H5Nx viruses and were associated with high morbidity and mortality. In addition, seasonal influenza viruses are estimated to cause up to 650,000 deaths annually worldwide. Currently available antiviral treatment options include only neuraminidase inhibitors, but some influenza viruses are naturally resistant to these drugs, and others quickly develop resistance-conferring mutations. Alternative therapeutics are urgently needed. Broadly protective antibodies that target the conserved “stalk” domain of the hemagglutinin represent potential potent antiviral prophylactic and therapeutic agents that can assist pandemic preparedness. Here, we describe four human monoclonal antibodies that target conserved regions of influenza HA and characterize their binding spectrum as well as their protective capacity in prophylactic and therapeutic settings against a lethal challenge with a zoonotic influenza virus.
INTRODUCTION
Seasonal influenza virus infections cause acute respiratory illness in humans and are responsible for an estimated 290,000 to 650,000 deaths annually on a global scale (1). In addition, influenza pandemics occur at irregular intervals and are usually caused by viruses that emerge from reassortments between human viruses and viruses circulating in the animal reservoir (2). These pandemics are not predictable and can cause high numbers of fatalities (3). Furthermore, influenza viruses that circulate in avian or mammalian species can cause severe zoonotic infections. Since 2004, human infections with avian origin influenza A viruses of the H5N1 (4), H5N6 (5), H6N1 (6), H7N2 (7, 8), H7N3 (9–11), H7N7 (12), H7N9 (13), H9N2 (14), and H10N8 (15) subtypes have been documented. Most of these zoonotic infections were sporadic, but they have also resulted in epidemics, as in the case of A(H7N9) infections in China (13). Human infections with A(H7N9) viruses were first reported in 2013, and since then those viruses have triggered multiple waves of infection that overlapped the Northern Hemisphere seasonal influenza virus epidemics (13, 16).
Annual vaccination against seasonal influenza virus strains remains the most effective means to protect humans against influenza. Vaccines that elicit antibodies (Abs) specific for hemagglutinin (HA) are associated with providing the best protection (17). Seasonal vaccines are generally most efficacious when the vaccine is antigenically well matched to the circulating strains (18). However, many severe cases of seasonal influenza virus infections still occur due to poor vaccine coverage, vaccine mismatch, and weak immune responses to vaccination in some target populations (19). Vaccines against viruses with pandemic potential, such as H5 and H7 subtypes, have been prepared and tested in humans but are often poorly immunogenic (20). Furthermore, it is hard to predict which zoonotic virus would cause a future pandemic, and therefore it is unlikely that a matched vaccine would be available in sufficient quantities in time to provide effective protection to the population.
Current therapeutic treatment options for seasonal, pandemic, and zoonotic influenza virus infections are restricted to neuraminidase inhibitors (e.g., oseltamivir, zanamivir, peramivir, and laninamivir) (21). Therapeutic agents of a second type, M2 ion channel inhibitors (amantadine), show little or no efficacy against the 2009 pandemic H1N1 [A(H1N1)pdm09] strains or current seasonal H3N2 strains since most of these viruses harbor resistance mutations in their M2 gene (21). Neuraminidase inhibitors show efficacy against influenza virus infections when administered prophylactically or within 2 days following the onset of symptoms (22). However, when given at a later time point, these inhibitors lose effectiveness. In addition, viruses have been shown to develop resistance against neuraminidase inhibitors rapidly and often without loss of viral fitness in experimental settings (23) or in populations (24) and some zoonotic viruses (e.g., H7N9) already carry mutations conferring resistance to neuraminidase inhibitors (25). Therefore, novel treatment options are urgently needed.
Broadly cross-reactive antibodies that recognize conserved structures in the membrane-proximal “stalk” domain of the HA (anti-HA-stalk), comprising the HA1/HA2 interface, have been isolated from mice and from humans (26). These anti-HA stalk antibodies are typically not induced at high levels after regular seasonal influenza virus vaccination but have been shown to be induced at increased levels in humans following primary exposure to novel viruses such as A(H1N1)pdm09, H5N1, and H7N9 (27–33). Anti-HA stalk antibodies have been shown to be broadly cross-reactive with a variety of influenza A viruses and can provide protection against lethal viral challenge in animal models (34–38). Considerable efforts are under way to develop broad-spectrum therapeutics based on anti-HA stalk monoclonal antibodies (MAbs) that could provide therapeutic and/or prophylactic efficacy against future seasonal, zoonotic, and pandemic influenza virus infections (39–41). Anti-HA stalk MAbs could also be used prophylactically in at-risk populations during seasonal epidemics and in the event of a pandemic to protect health care personnel, public safety personnel, and essential community personnel.
Anti-stalk MAbs have been shown to bind a broad variety of HAs within a subtype (37) or different HA subtypes within a phylogenetic group (42–44) or, rarely, different HAs from different phylogenetic groups (i.e., group 1 and group 2 HAs [45, 46]). MAbs 70-1F02 and 1009-3B05 were derived from plasmablasts isolated from patients recently infected with A(H1N1)pdm09 virus (29). Both MAbs are encoded by the IGHV1-69 germ line, and both competed for binding with the previously reported C179 anti-HA stalk MAb (29, 47–49), neutralized a panel of H1N1 viruses, and protected mice from lethal challenge with A(H1N1)pdm09 in prophylactic and therapeutic settings. MAbs 05-2G02, 1009-3B05, and 09-3A01 were derived from plasmablasts isolated from healthy volunteers following vaccination with the monovalent A(H1N1)pdm09 vaccine (27). 05-2G02 and 09-3A01 are encoded by germ lines IGHV1-18 and IGHV4-39, respectively, bound to H1 and H5 HAs (27), competed for binding with 70-1F02, and also neutralized a panel of H1N1 viruses. In addition, 05-2G02 bound and neutralized an H3N2 virus, indicating a binding profile across both phylogenetic groups. The aim of this study was to further characterize the breadth and functionality of these antibodies, determine their protective potential in vivo against a highly pathogenic avian influenza virus, shed light on their molecular interactions with an avian influenza virus HA, and determine their prevalence in convalescent human sera.
RESULTS
MAbs 70-1F02, 1009-3B05, and 09-3A01 bind broadly to group 1 HAs whereas 05-2G02 shows a cross-group influenza A virus HA binding pattern.
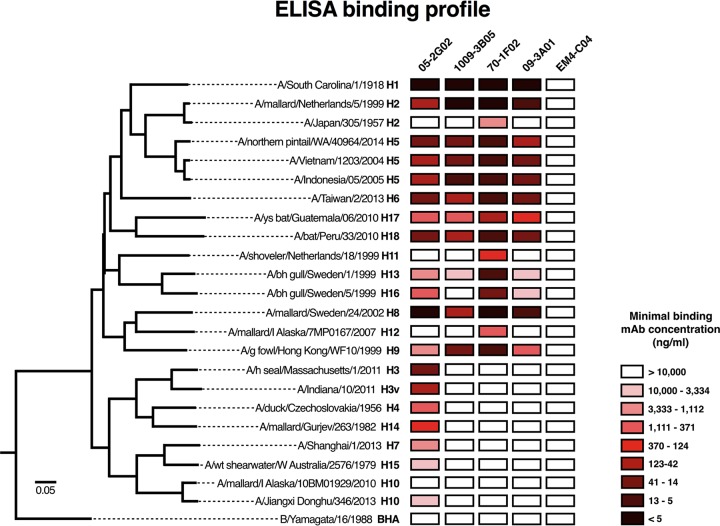
The binding spectrum of anti-influenza MAbs dictates their therapeutic and prophylactic potential. We therefore performed quantitative enzyme-linked immunosorbent assays (ELISAs) with the four MAbs as well as a control immunoglobulin G 1 (IgG1) MAb [EM4-C04; A(H1N1)pdm09 H1 specific] to determine their binding to a panel of HAs derived from all influenza virus subtypes, with a focus on novel subtypes and HAs from viruses with pandemic potential. MAbs 70-1F02, 1009-3B05, and 09-3A01 showed very similar binding patterns across group 1 HA subtypes (H1, H2, H5, H6, H8, H9, H11, H12, H13, H16, H17, and H18) (Fig. 1). Of note, all three MAbs bound to an H2 HA from a recent avian isolate and to an H6 HA from a recent human case in Taiwan as well as to H17 and H18, which have recently been isolated from bats. None of the three MAbs showed high-affinity binding to the group 2 HAs tested (H3, H4, H7, H10, H14, and H15) or to influenza B virus HA, and only 70-1F02 recognized the H2 HA from the 1957 pandemic H2N2 strain (group 1). Overall, MAb 70-1F02 gave the strongest binding signal whereas MAbs 1009-3B05 and 09-3A01 showed weaker or no binding to H11, H12, H13, and H16. MAb 05-2G02 showed broad cross-group influenza A virus HA binding, including binding to important group 2 HAs such as the H7 HA from H7N9, but did not bind influenza B virus HA.
FIG 1.
Heat map of antibody binding profile by ELISA. Antibodies were tested by ELISA against recombinantly produced HA proteins. A heat map was generated based on minimal binding concentration and plotted next to a phylogenetic tree (based on amino acid difference) to indicate the similarity of HAs that were tested. Each MAb was tested once in duplicate.
We next determined the binding affinity of the MAbs for a panel of HAs, including H1, H2, H5, H6, H8, H9, H11, H12, H13, and H16 HAs, by biolayer interferometry. All MAbs bound with nanomolar affinity to the H1 HA from A/California/07/2009 (Cal09) and with various levels of affinity to other HA subtypes (Table 1). Consistent with the ELISA data, 70-1F02, 1009-3B05, and 09-3A01 bound only to group 1 HA subtypes, whereas 05-2G02 bound to all group 1 and group 2 HAs tested. In summary, we characterized three pan-group 1 HA binding MAbs as well as one pan-influenza A virus HA binding MAb.
TABLE 1.
Affinity measurements
| Subtype | Strain name | Antibody affinitya |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 70-1F02 |
9-3A01 |
1009-3B05 |
05-2G02 |
||||||||||
| KD (M) | Kon | Koff | KD (M) | Kon | Koff | KD (M) | Kon | Koff | KD (M) | Kon | Koff | ||
| H1N1 | A/California/07/2009 | 1.03 × 10−10 | 2.59 × 105 | 2.86 × 10−5 | 7.93 × 10−11 | 2.14 × 105 | 1.69 × 10−5 | 2.18 × 10−11 | 1.65 × 105 | 3.59 × 10−6 | 1.63 × 10−10 | 2.41 × 105 | 3.94 × 10−5 |
| H1N1 | A/Solomon Islands/03/2006 | 7.49 × 10−12 | 1.83 × 105 | 1.29 × 10−6 | 1.79 × 10−10 | 1.87 × 105 | 3.34 × 10−5 | 1.45 × 10−11 | 1.37 × 105 | 1.98 × 10−6 | 3.52 × 10−11 | 1.58 × 105 | 5.55 × 10−6 |
| H1N1 | A/Brisbane/59/2007 | 8.84 × 10−11 | 2.91 × 105 | 2.57 × 10−5 | 3.34 × 10−10 | 2.79 × 105 | 9.3 × 10−5 | 6.30 × 10−11 | 2.03 × 105 | 1.28 × 10−5 | 2.55 × 10−10 | 2.28 × 105 | 5.81 × 10−5 |
| H1N1 | A/South Carolina/01/1918 | 7.07 × 10−12 | 3.39 × 105 | 2.54 × 10−6 | 2.63 × 10−11 | 3.13 × 105 | 8.23 × 10−6 | 2.52 × 10−10 | 2.4 × 105 | 6.05 × 10−5 | 6.16 × 10−12 | 2.73 × 105 | 1.68 × 10−6 |
| H1N1 | A/Swine/Iowa/15/1930 | 8.96 × 10−11 | 4.46 × 105 | 4.00 × 10−5 | 9.15 × 10−11 | 5.26 × 105 | 4.81 × 10−5 | 8.82 × 10−11 | 3.58 × 105 | 3.16 × 10−5 | 1.25 × 10−10 | 4.7 × 105 | 5.88 × 10−5 |
| H2N2 | A/Japan/305/1957 | 9.21 × 10−10 | 1.8 × 105 | 1.66 × 10−4 | 6.93 × 10−9 | 1.73 × 105 | 1.20 × 10−3 | 3.55 × 10−11 | 6.0 × 104 | 2.13 × 10−6 | 5.73 × 10−8 | 2.29 × 105 | 1.31 × 10−2 |
| H5N1 | A/Vietnam/1203/2004 | 6.88 × 10−10 | 7.72 × 104 | 5.31 × 10−5 | 6.62 × 10−9 | 8.58 × 104 | 5.68 × 10−4 | 2.65 × 10−10 | 5.3 × 104 | 1.41 × 10−5 | 1.37 × 10−8 | 8.37 × 104 | 1.15 × 10−3 |
| H5N1 | A/Hubei/05/2010 | 1.04 × 10−11 | 2.43 × 105 | 2.54 × 10−6 | 5.44 × 10−10 | 2.76 × 105 | 1.5 × 10−4 | 9.16 × 10−11 | 1.73 × 105 | 1.59 × 10−5 | 4.08 × 10−10 | 2.28 × 105 | 9.29 × 10−5 |
| H6N2 | A/chicken/NY/14677-13/1998 | 9.77 × 10−12 | 9.7 × 104 | 9.48 × 10−7 | 7.18 × 10−10 | 5.14 × 104 | 3.7 × 10−5 | 4.46 × 10−9 | 2.67 × 104 | 1.23 × 10−4 | 5.45 × 10−10 | 6.12 × 104 | 3.34 × 10−5 |
| H8N4 | A/Turkey/Ontario/1968 | 1.59 × 10−10 | 2.01 × 105 | 3.21 × 10−5 | 3.27 × 10−10 | 2.61 × 105 | 8.53 × 10−5 | 1.50 × 10−9 | 1.65 × 105 | 2.48 × 10−4 | 2.18 × 10−13 | 2.59 × 105 | 5.66 × 10−8 |
| H9N2 | A/HongKong/1999 | 3.08 × 10−10 | 8.39 × 105 | 2.58 × 10−4 | 1.51 × 10−9 | 7.9 × 105 | 1.19 × 10−3 | 1.17 × 10−9 | 7.09 × 105 | 8.25 × 10−4 | 4.59 × 10−9 | 1.81 × 106 | 6.29 × 10−3 |
| H11N9 | A/Duck/Memphis/1974 | 3.44 × 10−11 | 6.9 × 104 | 2.37 × 10−6 | 1.30 × 10−9 | 7.8 × 104 | 1.02 × 10−4 | 9.49 × 10−9 | 2.48 × 105 | 2.36 × 10−3 | 1.73 × 10−8 | 1.59 × 105 | 2.76 × 10−3 |
| H12N5 | A/Mallard/Alberta/1976 | 4.07 × 10−9 | 2.07 × 105 | 8.44 × 10−4 | 1.57 × 10−8 | 2.86 × 104 | 4.49 × 10−4 | 1.63 × 10−8 | 2.28 × 105 | 3.72 × 10−3 | 9.32 × 10−14 | 6.0 × 104 | 5.59 × 10−9 |
| H13N9 | A/Shorebird/DE/68/2004 | 3.39 × 10−9 | 3.55 × 104 | 1.20 × 10−4 | 3.28 × 10−8 | 8.96 × 104 | 2.93 × 10−3 | 4.78 × 10−8 | 3.73 × 104 | 1.78 × 10−3 | 3.12 × 10−8 | 8.94 × 104 | 2.78 × 10−3 |
| H16N3 | A/Shorebird/DE/172/2006 | 6.62 × 10−10 | 3.27 × 104 | 1.84 × 10−5 | 1.32 × 10−8 | 1.61 × 105 | 2.14 × 10−3 | NA | NA | NA | 2.31 × 10−8 | 2.0 × 105 | 4.62 × 10−3 |
| H3N2 | A/Brisbane/10/2007 | NA | NA | NA | NA | NA | NA | NA | NA | NA | 9.87 × 10−10 | 1.81 × 105 | 1.79 × 10−4 |
KD, equilibrium dissociation constant; Kon, rate constant for the association of the antibody-antigen complex; Koff, rate constant for the dissociation of the antibody-antigen complex; NA, not applicable.
MAbs 70-1F02, 09-3B05, 09-3A01, and 05-2G02 neutralize H5N1 in vitro.
Binding, as assessed above, is not a sufficient indicator for antiviral activity. To test if the four MAbs show antiviral activity, we assessed their neutralizing potency against a representative zoonotic virus, H5N1. Microneutralization assays with all four MAbs were performed against an H5N1 virus expressing the HA from A/Vietnam/1203/04 (low-pathogenicity version). We found that all MAbs potently neutralized H5N1 with slightly higher neutralization activity for 05-2G02 and 70-1F02 (50% inhibitory concentrations [IC50] of 0.244 μg/ml and 0.263 μg/ml, respectively) than for 09-3A01 and 1009-3B05 (IC50s of 0.395 μg/ml and 0.604 μg/ml, respectively) (Fig. 2). No neutralization was detected with the isotype control MAb.
FIG 2.
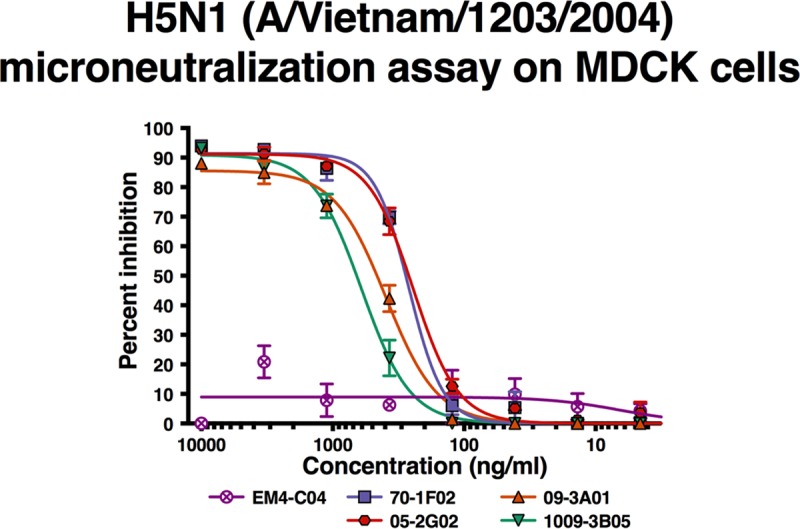
H5N1 (A/Vietnam/1203/2004) microneutralization assay on MDCK cells. Antibodies were tested in a neutralization assay against a low-pathogenicity H5 reassortant virus. The x axis shows the amount of monoclonal antibody tested, and the y axis indicates percent inhibition in the assay. Each point shows the mean of data from two replicates, and error bars indicate errors of the means. A nonlinear curve fit was performed for all antibody dilution series (colored lines). Each MAb was tested once in duplicate.
70-1F02 and 09-3A01 show therapeutic protective efficacy against highly pathogenic H5N1 challenge in mice.
Next, we measured the protective efficacy of this panel of broadly reactive MAbs in a mouse model of infection with a highly pathogenic avian influenza virus. One of the MAbs, 70-1F02, was previously shown to protect mice against infection with an H1N1 virus when administered as a prophylactic or a therapeutic agent (29). All four MAbs have been shown to neutralize a panel of H1N1 viruses (27, 29) and a low-pathogenicity version of H5N1 (see above). However, it was unclear if these MAbs would also show a protective effect in vivo against a stringent challenge with a highly pathogenic H5N1 virus, which represents a pandemic threat.
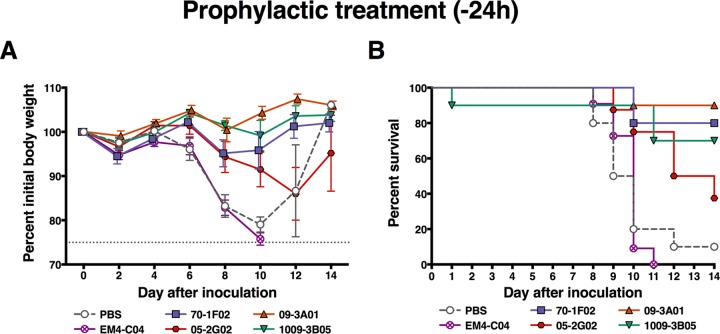
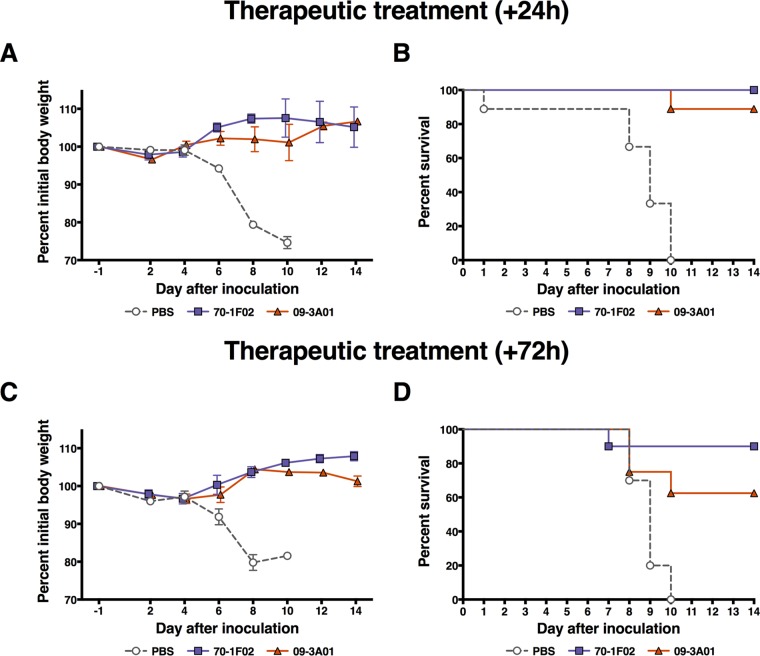
In a first experiment, the four MAbs were administered via intraperitoneal (i.p.) injection 24 h prior to infection at a dose of 20 mg/kg of body weight. This dose was chosen based on our experience with antibody prophylaxis and therapy in the mouse model. It is on the higher side of the range that we typically use because highly pathogenic H5N1 viruses are very lethal in this model and because the high containment level made it impracticable to test dose ranges (including lower doses). Animals were then challenged intranasally (i.n.) with 10× the 50% lethal dose (LD50) (6 PFU) of the highly pathogenic A/Vietnam/1203/04 (H5N1) virus. The H1 head-specific MAb EM4-C04 was included as a negative control. Of note, we did test the in vivo half-life of a subset of MAbs, including EM-C04 (control MAb), 70-1F02, and 1009-3B05 in mice and—unsurprisingly–detected very similar half-lives of 8.98, 8.14, and 7.01 days, respectively. MAbs 70-1F02, 1009-3B05, and 09-3A01 protected animals against infection-induced morbidity (Fig. 3A), and survival rates for these groups were 80%, 70%, and 90%, respectively (Fig. 3B). Animals that received 05-2G02 exhibited more weight loss than animals that received the other 3 cross-reactive MAbs and had a lower survival rate of 37.5% (Fig. 3). Mice that received the control MAb (EM4-C04, which binds to the H1 head domain [29]) or phosphate-buffered saline (PBS) were not protected and succumbed to infection, with the exception of a single mouse in the PBS control group. MAbs 70-1F02 and 09-3A01 were then chosen for follow-up experiments to test their protective potential in a therapeutic setting. The experimental design was similar to that of the previous experiment, but the groups were injected with the MAbs 24 or 72 h postinfection. In this experiment, therapeutic treatment after 24 h with 70-1F02 protected 100% of animals from mortality, and treatment after 72 h protected 90% of animals from mortality (Fig. 4). Results for 09-3A01 were similar, with 89% or 63% survival when the MAb was given 24 or 72 h postinfection, respectively (Fig. 4). In summary, all four MAbs increased survival after a lethal H5N1 challenge in mice, with 70-1F02, 1009-3B05, and 09-3A01 showing protection superior to that seen with the pan-influenza A virus HA antibody 05-2G02.
FIG 3.
In vivo challenge with highly pathogenic H5N1 after prophylactic treatment. Mice were intraperitoneally injected with antibodies or PBS (no-antibody control) 24 h before infection with highly pathogenic H5N1 virus. Survival was checked daily, and weight was recorded every other day. (A) Weight loss curves after challenge. Each point shows the average percentage of the initial body weight of the group, and error bars indicate the standard errors of the means. (B) Kaplan-Maier curves show the survival rate for each of the groups post-H5N1 challenge. Survival rates of all groups were compared to the rate determined for the group that received the isotype control EM4-C04 in a log rank (Mantel-Cox) test. MAbs 70-1F02 (P = 0.0001), 09-3A01 (P < 0.0001), 1009-3B05 (0.0005), and 05-2G02 (P = 0.0022) all conferred statistically significant protection compared to the isotype control, and no statistically significant difference was observed between mice receiving PBS and those receiving EM4-C04 (Bonferroni-corrected significance threshold = <0.01). Challenge studies were performed once with 8 to 10 mice per group.
FIG 4.
Therapeutic treatment postchallenge with highly pathogenic H5N1. Mice were intraperitoneally injected with 70-1F02, 09-3A01, or PBS (negative control) 24 h (A and B) or 72 h (C and D) postinfection with highly pathogenic H5N1 virus. Therapeutic treatment with both antibodies protected mice from weight loss (A and C) and mortality (B and D). As shown in panel B, all groups were compared to the PBS control group in a log rank (Mantel-Cox) test. MAbs 70-1F02 (P < 0.0001) and 09-3A01 (P < 0.0001) both conferred statistically significant protection compared to PBS (Bonferroni-corrected significance threshold = <0.025). In panel D, the same testing showed that MAbs 70-1F02 (P = 0.0002) and 09-3A01 (P = 0.0104) both conferred statistically significant protection compared to PBS (Bonferroni-corrected significance threshold = <0.025). Challenge studies were performed once with 8 to 10 mice per group.
Characterization of the 70-1F02 epitope.
Due to the high level of protection conferred by 70-1F02 in the challenge experiments, we selected this MAb for further characterization of its interaction with avian H5 HA. To comprehensively determine the 70-1F02 epitope, we crystallized the 70-1F02 Fab in complex with recombinant HA from the well-characterized A/Vietnam/1203/04 H5N1 (VN04) strain.
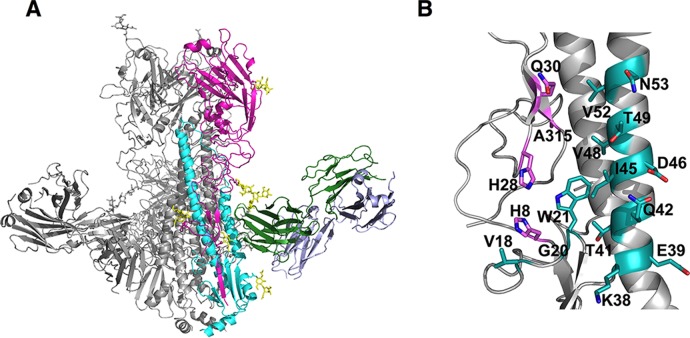
The crystallographic asymmetric unit contained two VN04 HA trimers and six 70-1F02 Fabs, with each FAb bound to a single HA1/HA2 heterodimer, as depicted in Fig. 5A. Data collection and refinement statistics are presented in Table 2. The overall structure of the VN04 HA trimer was equivalent to those described previously, with no significant conformational rearrangements upon 70-1F02 interaction. The 70-1F02 epitope within the membrane-proximal stalk region of VN04 HA was revealed at the junction of the HA1 and HA2 domains, such that the hydrophobic N-terminal strand of the HA2, which is involved in membrane fusion, formed a central component of the interface. Notably, the 70-1F02 binding site was restricted to a single HA1/HA2 monomer and did not span the HA trimerization interface. The footprint of the 70-1F02 Fab on the surface of H5 HA included 4 residues in the HA1 domain (His8, His28, Gln30, and Ala315) and 13 in the HA2 domain (Val18, Gly20, Trp21, Lys38, Glu39, Thr41, Gln42, Ile45, Asp46, Val48, Thr49, Val52, and Asn53). The relative locations of these residues are illustrated in Fig. 5B. Notably, the light chain of 70-1F02 did not contact VN04, and the interaction was mediated entirely by contacts with the heavy-chain subunit, reminiscent of previously described broadly neutralizing Abs that recognize epitopes in the HA stalk, such as CR6261 (42, 44). Superimposition of these Fab/HA complex structures with that of the 70-1F02-VN04 complex highlights the similarity of these epitopes (Fig. 6). For CR6261, 19 contact residues are described for VN04 H5 HA (44). Of these, 11 overlap the contact residues found for 70-1F02. In summary, the crystal structure revealed that 70-1F02 binds a membrane-proximal site of a single HA monomer with only the MAb's heavy-chain subunit, similarly to previously described HA stalk-specific antibodies.
FIG 5.
Crystal structure of the 701-F02 Fab/VN/04 HA complex. (A) Overview of the 70-1F02 FAb/H5 complex structure. Ribbons represent a single HA trimer bound to three 70-1F02 FAbs. For clarity, a single HA1 protomer and a single HA2 protomer are colored in magenta and cyan, respectively, with the 70-1F02 FAb heavy and light chains colored in green and blue, respectively. N-carbohydrates are indicated as yellow sticks. (B) The Ab 70-1F02 epitope on H5 HA. Residues on A/Vietnam/1203/2004 (H5N1) HA contacted by 70-1F02 are indicated. The primarily component of the epitope is the helical strand of the HA2 (11 residues). Residues within the 70-1F02 epitope are indicated as sticks on a ribbon representation of the H5 HA. Contact residues in the HA1 and HA2 subdomains are colored in magenta (HA1) and cyan (HA2), respectively. The epitope spans the two HA1 and HA2 subunits in a single HA trimer and does not contact the subunits of adjacent monomers. Residues are numbered consecutively, according to the mature protein.
TABLE 2.
Data collection and refinement statistics
| Parameter | Result(s) |
|---|---|
| Data collection | 70-1F02/H5 |
| Space group | P212121 |
| Cell dimensions | 174.6 Å, 205.4 Å, 222.3 Å; 90°, 90°, 90° |
| Resolution (Å) | 50–3.9 (4.0–3.9)a |
| Rsym (%) | 10.1 (94.4) |
| I/σ | 17.1 (1.8) |
| Completeness (%) | 98.8 (94.4) |
| Redundancy | 6.6 (5.9) |
| Refinement | |
| Resolution (Å) | 50.3–3.92 (4.03–3.92) |
| No. of reflections (total) | 64,141 |
| No. of reflections (test) | 3,422 |
| Rwork/Rfree | 28.6/32.9 |
| No. of atoms | 44,024 |
| RMSD; bond length (Å) | 0.0012 |
| RMSD; bond angle (°) | 1.607 |
| MolProbity scores | |
| Favored (%) | 90.5 |
| Allowed (%) | 99.0 |
| Outliers (%) (no. of outliers/total no. of residues) | 1.0 (55/5,458) |
Numbers in parentheses refer to the highest-resolution shell.
FIG 6.
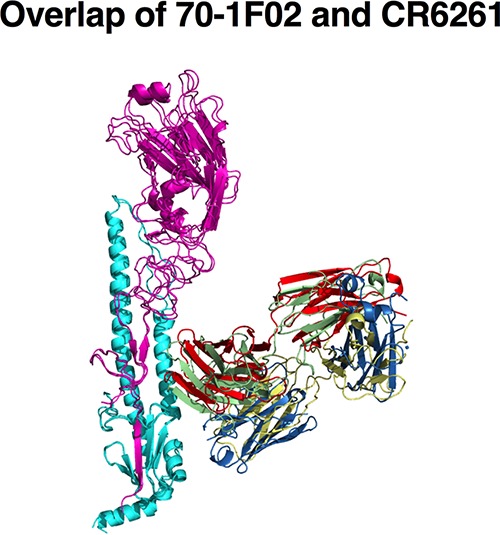
Overlap of 70-1F02 and CR6261 bound to an H5 monomer. An overlay of the previously crystallized human MAbs CR6261 and 70-1F02 reveals similar binding profiles of these antibodies.
Analysis of the epitope-specific component of IgG within sera from patients infected with pandemic H1N1.
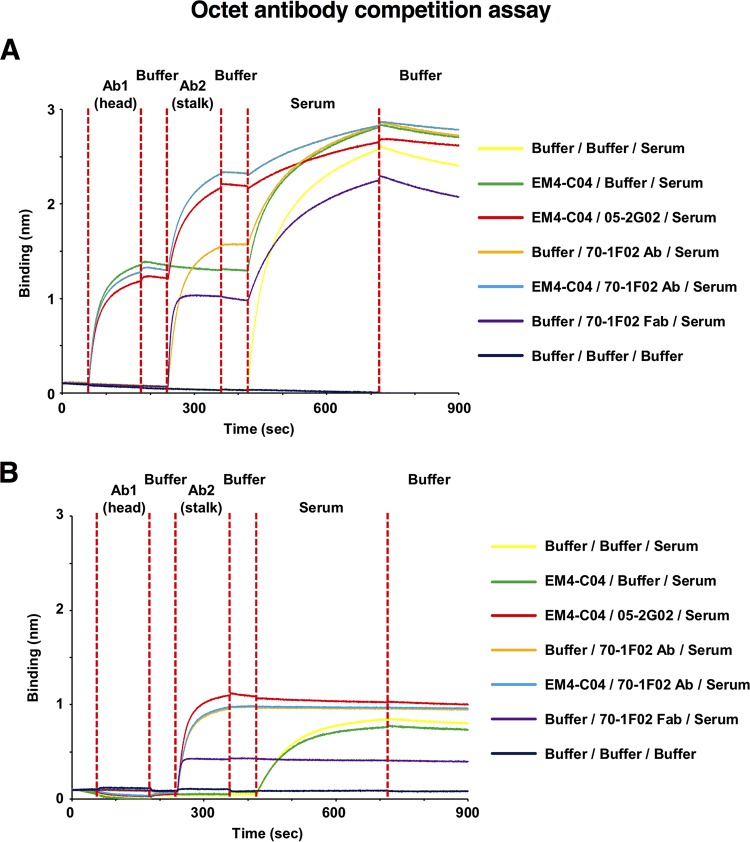
It has previously been reported that infection with A(H1N1)pdm09 virus in humans with preexisting immunity to seasonal H1N1 viruses leads to the preferential induction of stalk-specific antibodies (32). This is an interesting phenomenon, specifically because a high prevalence of anti-stalk antibodies could have a protective effect against subsequent infections with seasonal as well as emerging pandemic or zoonotic influenza viruses. Having established that multiple antibodies that are broadly cross-reactive to the HA stalk bind overlapping or similar epitopes, we designed a competition assay to measure reactivity to highly conserved epitopes within the HA stalk within human sera using biolayer interferometry. We used MAbs that bound either the globular head (MAb EM4-C04 [29]) or the stalk (MAb 70-1F02 and MAb 05-2G02) of H1 from Cal09, to block these respective epitopes, within serum from a patient infected with the A(H1N1)pdm09 virus. To specifically identify highly cross-reactive antibodies, we assessed the competition shown by serum antibodies binding to recombinant HA from both homologous pandemic H1N1 virus and the phylogenetically distinct VN04 H5N1 virus (Fig. 7) (Table 3). Biosensors coupled to HAs and IgGs were probed with serum diluted 1:80 in kinetics buffer (Table 3), and the relative (percent) levels of inhibition of binding to Abs within the sera were determined by comparison with the value observed for Ab binding to HA alone (Table 3). Of note, binding of either MAb 70-1F02 or MAb 05-2G02 to the stalk of Cal09 H1 did not prevent binding of MAb EM4-C04 to the globular head of HA (Fig. 7A). To confirm that the observed inhibition of Ab binding to the stalk was not due to steric blockade from the Fc (noninteracting) component of 70-1F02 IgG, we used the 70-1F02 Fab fragment and detected inhibition equivalent to that observed with the intact IgG, indicating that the Fc component did not significantly influence the results of this experiment (Table 3).
FIG 7.
Inhibition of binding to Abs within RDE-treated sera at 1/80 dilution. Recombinant Cal09 HA (A) and VN04 H5 HA (B) were bound, in parallel, to eight Octet Red biosensors (HA binding) and analyzed. Each curve represents a separate biosensor, bound to an equivalent quantity of HA. After washing was performed to remove excess HA, biosensors were incubated with EM4-C04 (see Ab1 [head]; red, blue, and green traces) and then washed with kinetics buffer prior to incubation with Ab2 (stalk), either Ab 70-1F02 (see blue and orange traces) or Ab 05-2G02 (red trace), or 70-1F02 MAb Fab (purple trace). Controls without antibody or serum (black trace) and serum only (yellow trace) were included as well. Biosensors were subsequently washed again in kinetics buffer to remove unbound Ab and then incubated in RDE-treated serum. Following incubation with serum, the biosensors were washed once more to remove unbound IgG prior to data analysis. The instrument detects binding to the biosensor tip, which results in a wavelength shift (measured in nanometers). This experiment was performed once with 5 to 7 different MAb concentrations.
TABLE 3.
Relative levels of inhibition of binding to Abs within RDE-treated sera with 1/80 dilution and purified IgG
| Antibody |
Response (nm) (% inhibition) |
|||
|---|---|---|---|---|
| 1 | 2 | Cal09 (H1) | VN04 (H5) | Cal09 (H1), pure IgG |
| None | None | 2.55 (0) | 0.84 (0) | 0.88 (0) |
| EM4-C04 | None | 1.54 (39.7) | 0.76 (9) | 0.45 (48.9) |
| None | 70-1F02 | 1.29 (49.9) | 0 (100) | 0.33 (62.5) |
| None | 05-2G02 | NA | 0.02 (99) | 0.32 (64) |
| None | 70-1F02 Fab | 1.3 (49.9) | 0.03 (99) | 0.33 (62.5) |
| EM4-C04 | 70-1F02 | 0.51 (80) | 0 (100) | 0.043 (95.1) |
| EM4-C04 | 05-2G02 | 0.54 (79.9) | 0 (100) | 0.034 (96.1) |
Under the conditions of this assay, significant, dose-dependent binding of polyclonal antibodies to Cal09 HA was observed. Approximately 50% of this serum response was blocked by 70-1F02 or 05-2G02 Ab, while 40% of the binding was blocked by MAb EM4-C04 (Fig. 7A) (Table 3). Importantly, a significant dose-dependent binding of polyclonal antibodies to H5 HA was also observed; however, 100% of this response was blocked by the 70-1F02 or 05-2G02 Ab, indicating the presence of broadly cross-reactive IgG against the HA stalk within the serum of this pandemic-H1N1-infected patient (Fig. 7B) (Table 3). Hence, a component of the HA-specific Ab pool within this human serum targeted the conserved stalk region. To assess the extent to which the observed blockade of immunoglobulin from sera was due to the inhibition of IgG, we repeated the competition analyses using IgG purified from sera. The results of the blockade experiment were highly similar to those obtained using whole sera (Table 3), indicating that the majority of the stalk-specific response was associated with serum IgG. Therefore, as indicated by our MAb competition data in a biolayer inferometry assay, we could confirm previous findings that patients, after exposure with pandemic H1N1, have high cross-reactive antibody titers mediated against the HA stalk domain.
DISCUSSION
Therapeutic options to treat influenza virus infections include neuraminidase and M2 ion channel inhibitors. M2 ion channel inhibitors are currently not in use due to widespread viral resistance. Neuraminidase inhibitors work optimally when given within a short window of time after infection/onset of disease, and their use can also lead to the quick emergence of viral escape mutants within the patient. Furthermore, some viruses—including H7N9 isolates—might be naturally resistant to these inhibitors. In recent years, efforts have been made to develop novel treatment options based on the use of broadly protective monoclonal antibodies that target conserved parts of the virus, including the M2 ectodomain and the stalk domain of the HA (39, 40, 50).
Here we characterized four human monoclonal antibodies that had been isolated from individuals infected with or vaccinated against pandemic H1N1 and provide novel insights into their binding breadth, protective efficacy against highly pathogenic avian influenza viruses in vivo, the molecular interactions of one of the MAbs with H5 HA, and the prevalence of these antibodies in human serum. The MAbs had shown broad neutralizing activity in initial studies (27, 29), which we have now shown extends to binding of three MAbs (70-1F02, 1009-3B05, and 09-3A01), to broad binding of group 1 HA subtypes, and, for a fourth MAb (05-2G02), to binding broadly to both group 1 and group 2 influenza A virus HAs. All antibodies showed neutralizing activity against H5N1 in vitro, and, in a stringent challenge experiment performed with a highly pathogenic H5N1 virus, three of the four MAbs showed excellent prophylactic activity. Two MAbs were also tested in a therapeutic setting and provided a high percentage of protection for the challenged mice, even when given 72 h after infection. While we did not extend the treatment window further, these two MAbs might show efficacy even at later time points.
We also characterized the interactions of MAb 70-1F02 with H5 HA in detail. The crystallographically determined footprint of 70-1F02 on the surface of H5 HA revealed interfacing residues predominantly within the invariant HA2 subunit of HA, with conservation in the sizes and charges of residues at structurally equivalent positions among HAs of the H1, H2, and H5 subtypes, providing the basis for broad cross-reactivity of 70-1F02. Highly conserved, solvent-accessible residues in the HA stalk that comprise the potential epitopes for broadly cross-reactive antibodies represent a small proportion of the total surface area of the HA trimer. Crystal structures of other broadly cross-reactive antibodies, derived from the IGHV1-69 genetic lineage as well as from different genetic lineages, illustrate the close distribution of these epitopes within the spatially restrictive region of the HA stalk (42, 44). We therefore analyzed the percentage of stalk-reactive antibodies in serum from a pandemic-H1N1-infected patient and found that a large proportion (approximately 50%) of the serum response against H1 competed with stalk MAbs. This suggests a high prevalence of anti-stalk antibodies in sera of pandemic-H1N1-infected individuals, a finding that is corroborated by other studies (32). The patients in this study were all adults and had therefore likely been preexposed to influenza viruses. The antibody response to influenza viruses is primarily directed against the immunodominant HA head region. However, responses against the HA stalk are induced as well. When individuals who have been primed are then exposed to a novel virus with a different HA head domain but a similar HA stalk, B cells that recognize these conserved epitopes are preferentially boosted (28, 30–32, 51, 52). Of note, while all antibodies in this study showed neutralizing activity, it had been previously shown that some broadly binding MAbs do not have neutralizing activity and mediate their protective effect through Fc-dependent effective functions (53, 54). It is therefore possible that a proportion of the stalk response measured here in polyclonal sera consists of nonneutralizing antibodies that might nevertheless be useful for protection against infection and/or disease.
Given the rise of emerging influenza A viruses with pandemic potential and the increasing number of zoonotic infections (5, 13, 55, 56), the described MAbs could be important future tools for therapeutic treatment (41). These MAbs might also be useful in treating severe cases of seasonal influenza virus infection or could be used to prophylactically protect health care personnel, public safety personnel, and essential community personnel. Importantly, combinations of multiple MAbs could increase the protective effect and breadth of antibodies as has been recently shown against Ebola virus (57).
MATERIALS AND METHODS
Cells, viruses, and monoclonal antibodies.
The initial isolation of monoclonal antibodies 70-1F02, 9-3A01, 1009-3B05, 05-2G02, and EM4-C04 was described elsewhere (27, 29). MAbs were expressed in 293A cells and purified using a protein A Sepharose column (27). Madin-Darby canine kidney (MDCK) cells (sourced from the American Type Culture Collection) were grown in complete Dulbecco's modified Eagle medium (Life Technologies) supplemented with antibiotics (100 U/ml penicillin–100 μg/ml streptomycin [Pen-Strep]; Gibco) and 10% fetal bovine serum (HyClone).
Virus nomenclature is as follows: A/California/07/2009 [A(H1N1)pdm09, Cal09], A/Solomon Islands/03/06 (prepandemic seasonal H1N1, SI06), A/Brisbane/59/07 (prepandemic seasonal H1N1, Bris07), A/South Carolina/01/18 (1918 pandemic H1N1, SC18), A/swine/Iowa/15/30 (H1N1, Iowa30), A/Japan/305/57 (1957 pandemic H2N2, JA57), A/mallard/Netherlands/5/99 (H2N9, mallNL99) A/Brisbane/10/2007 (H3N2, Bris10), A/harbor seal/Massachusetts/1/11 (H3N8, hsMass11), A/Indiana/10/11 (human H3N2 variant isolate, Ind10), A/duck/Czechoslovakia/56 (H4N6, dCZ56), A/Vietnam/1203/04 (human highly pathogenic H5N1 isolate, VN04), A/Taiwan/2/13 (human H6N1 isolate, TW13), A/chicken/NY/14677-13/98 (H6N2, ckNY98), A/Shanghai/1/13 (human H7N9 isolate, SH13), A/mallard/Sweden/24/02 (H8N4, mallSwe02), A/turkey/Ontario/6118/68 (H8N4, tkOn68), A/guinea fowl/Hong Kong/WF10/99 (H9N2, gfHK99), A/Hong Kong/1073/99 (H9N2, HK'99), A/Northern pintail/Alaska/44161-018/06 (H10N7, npAla06), A/Jiangxi Donghu/346/13 (human H10N8 isolate, JD13), A/shoveler/Netherlands/18/99 (H11N7 sNeth99), A/duck/Memphis/546/74 (H11N9, duck74), A/mallard/Interior Alaska/7MP0167/07 (H12N5, mallIA07), A/mallard/Alberta/60/76 (H12N5, mallAlb76), A/black-headed gull/Sweden/1/99 (H13N6, bhgSwe99), A/shorebird/DE/68/04 (H13N9, sbD04), A/mallard/Gurjev/263/82 (H14N5, mallGur82), A/wedge-tailed shearwater/Western Australia/2576/79 (H15N9, wtsWA79), A/black-headed gull/Sweden/5/99 (H16N3, bhgSwe99), A/shorebird/DE/172/06 (H16N3, sbDE06), A/yellow-shouldered bat/Guatemala/060/10 (H17N10, ysbGua10), A/bat/Peru/33/10 (H18N11, batPeru10), and B/Yamagata/16/88 (B Yam88).
Highly pathogenic A/Vietnam/1203/2004 (H5N1) influenza virus was grown in the allantoic cavity of embryonated chicken eggs for 48 h at 35°C under biosafety level 3E (BSL-3E) conditions. Infectious titers were measured by titration on monolayers of MDCK cells as PFU per milliliter. The 50% lethal dose (LD50) in mice was determined by inoculation of naive BALB/c mice with 10-fold serial dilutions of virus and calculated using the Reed and Muench method (58) and was determined to be 6 PFU.
Human sera.
Deidentified human sera from individuals infected with H1N1 virus and from individuals vaccinated with H1N1 virus were collected with the approval of the Internal Review Boards (IRB) of Emory University (principal investigator, Rafi Ahmed; IRB approval numbers 22371 and 555-2000) and of the University of Chicago (principal investigator, Patrick Wilson; IRB approval number 16851E), respectively. All work with samples from infected patients was performed in a designated BSL2+ facility. Plasma samples were stored at −80°C. All subjects provided informed consent.
Mouse challenge experiments.
All animal experiments were conducted according to the guidelines of the UGA Institutional Animal Care and Use Committee (IACUC; approval number A2011 06-001). Challenge experiments were performed in a BSL3+ facility. Female 6-to-8-week-old BALB/c mice (Charles River) were randomly assigned into groups of 8 to 10 mice. Initially, all MAbs were tested in a prophylactic setting. Mice were dosed with MAb at 20 mg/kg of body weight (given i.p. in 600 μl in sterile PBS) 24 h prior to infection. Control mice received PBS or control MAb EM4-C04 (specific to pandemic H1N1). After transfer of the mice to the BSL-3E facility, the mice were anesthetized using 2,2,2-tribromoethyl alcohol and challenged i.n. with 10 LD50s (6 PFU) of highly pathogenic VN04 H5N1 virus in a volume of 50 μl. Following challenge, weights were monitored every 2 days and humane endpoints (>25% weight loss or signs of paresis) assessed daily. A follow-up experiment examined the therapeutic efficacy of MAbs 70-1F02 and 09-3A01. One group of mice was dosed with MAb at 20 mg/kg (given in 600 μl i.p.) 24 h before challenge (prophylactically), another group was dosed 24 h postchallenge (therapeutically), and a third group was dosed 72 h postchallenge (therapeutically). For each of the groups, a control group was added to the experiment. Animals in the control groups received phosphate-buffered saline (PBS). For the challenge experiments, animals were anesthetized using 2,2,2-tribromoethyl alcohol and infected i.n. with 10 LD50s (6 PFU) of highly pathogenic VN04 H5N1 virus in a volume of 50 μl. Mice were monitored twice daily postchallenge for humane endpoints, and weights were recorded every other day.
Recombinant hemagglutinins.
Production of proteins for ELISAs was performed as follows. High Five insect cells (Trichoplusia ni; BTI-TN-5B1-4 Vienna Institute of Biotechnology subclone; 59) for use in expression of protein for ELISAs were grown in SFX serum-free insect cell medium supplemented with antibiotics (penicillin-streptomycin mix [100 units/ml of penicillin and 100 μg/ml of streptomycin]; Gibco). Residues comprising the HA ectodomains from H1 SC18, H2 JA57, H2 mallNL99, H3 hsMass11, H3 Ind10, H4 dCZ56, H5 VN04, H6 TW13, H7 SH13, H8 mallSwe02, H9 gfHK99, H11 sNeth99, H10 npAla06, H10 JD13, H12 mallIA07, H13 bhgSwe99, H13 sbD04, H14 mallGur82, H15 wtsWA79, H16 bhgSwe99, H17 ysbGua10, H18 batPeru10, and B Yam88 were cloned into modified pFastBac transfer plasmids and expressed as described previously (60). Purification was performed in a manner similar to that described above following an established protocol. However, to guarantee trimer stability in ELISAs (60, 61), the foldon sequence (and the hexahistidine tag) was not removed.
For production of proteins for affinity measurements, High Five insect cells (T. ni; BTI-TN-5B1-4 cells) were grown in HyClone SFX-Insect cell culture media. Residues comprising the mature HA ectodomains from H1 Cal09, H1 SI06, H1 Bris07, H1 SC18, H1 Iowa30, H2 JA57, H3 Bris10, H5 VN04, H6 ckNY98, H8 tkOn68, H9 HK99, H11 duck74, H12 mallAlb76, H13 sbD04, and H16 sbDE06 were cloned into baculovirus shuttle vector pAcGP67-A (BD Pharmingen) and expressed as described previously (62–65). Briefly, High Five cells (Invitrogen) were infected with recombinant baculovirus at a multiplicity of infection of 5 to 10 at 28°C for 72 h. The secreted recombinant HA protein was purified from tissue culture supernatant by metal affinity chromatography and subsequent size exclusion gel filtration chromatography (Superdex 200 16/60 column; GE Healthcare). For crystallization, the C-terminal foldon sequence/histidine tag was removed from VN04 HA by thrombin treatment using 3 units of enzyme per mg HA overnight at 4°C.
Biolayer interferometry.
An Octet Red instrument (Fortebio, Inc., Menlo Park, CA) was used for all binding studies. Data were analyzed using the system software and exported as a Microsoft Excel file for analysis and presentation in other software packages. For antibody kinetics and binding assays, recombinant HA protein was immobilized on anti-hexahistidine biosensors at 50 to 100 μg/ml in kinetics buffer (PBS [pH 7.4], 0.01% bovine serum albumin [BSA], 0.002% Tween 20). For subtype specificity studies, biosensors were probed with MAb at a concentration of 10 μg/ml in each well. For comparisons of levels of MAb binding to recombinant HAs, 5 to 7 different concentrations of each MAb species were used, with the highest concentration being 21 nM. Baseline and dissociation steps were carried out in 1× kinetics buffer. For serum-antibody epitope blockade assays, recombinant HA protein from the Cal09 pH1N1 virus bearing a C-terminal hexahistidine tag was immobilized on anti-hexahistidine biosensors at 50 to 100 μg/ml in kinetics buffer (PBS [pH 7.4], 0.01% BSA, 0.002% Tween 20). Biosensors were probed sequentially with either one or two MAbs or Fabs at a concentration of 10 μg/ml in each well. Clarified sera from patient EM4-C04 was pretreated with receptor-destroying enzyme (RDE) overnight, to reduce nonspecific HA binding.
ELISA.
To further characterize binding of MAbs to all 18 HA subtypes, including HAs from viruses of concern and viruses with pandemic potential, we performed quantitative ELISAs (52) with the minimum binding concentration as the readout. EM4-C04 was used as the isotype control. Briefly, Immulon 4HBX flat-bottom 96-well microtiter plates (Thermo Fisher Scientific) were coated for approximately 16 h with 2 μg/ml of recombinant protein in coating buffer (0.1 M Na2CO3, NaHCO3 [pH 9.4]; 50 μl/well) at 4°C. Then, the coating buffer was discarded and plates were blocked using 3% nonfat dry milk powder–PBS containing 0.1% Tween 20 (PBST). After 1 h at room temperature the blocking buffer was removed and was replaced with dilution buffer (1% nonfat dry milk powder–Tris-PBS [TPBS]). MAbs were diluted in 1:3 steps in the plate with a starting concentration of 30 μg/ml. After 1 h of incubation, the MAb dilutions were removed, plates were washed three times with TBPS, and secondary antibody (anti-human IgG; Sigma catalog no. A0293) was added in dilution buffer. After another 1-h incubation step, the secondary antibody was discarded and the plates were washed three times with TPBS and were developed using o-phenylenediamine dihydrochloride (SigmaFast OPD; Sigma) as the substrate. After 10 min of incubation, the reaction was stopped using 3 M HCl and the plates were read at 490 nm on a Synergy H1 plate reader (BioTek). The minimal binding concentration was calculated using a cutoff corresponding to the average blank values plus 3 standard deviations. The procedure followed a standard protocol to guarantee maximum comparability between ELISA HA substrates.
Microneutralization assays.
Incubations were performed with 96-well cell culture plates containing 100 μl of cell culture media with 1.5 × 107/ml to 1.8 × 107/ml Madin Darby canine kidney (MDCK) cells at 37°C overnight. Antibodies (starting concentration of 200 μg/ml; EM4-C04 served as the isotype control) were 3-fold serially diluted in minimum essential media containing tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK) trypsin at a concentration of 1:1,000 (T-MEM) and incubated at 50 μl each with H5N1 (7 + 1 reassortant) virus at a concentration of 200,000 PFU per ml at room temperature to allow antibody binding. After 1 h, 100 μl of the serum and virus mix was transferred onto the plates containing MDCK cells and incubated for 1 h at 37°C. Cells were then washed and incubated with sera serially diluted in T-MEM without virus for at 37°C. After 24 h, the cells were washed and fixed with ice-cold 70% acetone and refrigerated at −20°C for at least 1 h. Acetone was removed, cells were washed 3 times, and 100 μl of 3% hydrogen peroxide was added. After 30 min, the hydrogen peroxide was removed and 250 μl of blocking solution was added. After 30 min, the blocking solution was removed, 50 μl of blocking solution containing mouse anti-nucleoprotein antibody conjugated to biotin (Millipore) antibody at a concentration of 1:2,000 was added, and the cells were incubated at room temperature. After 1 h, the plates were washed 3 times, 50 μl of blocking solution containing streptavidin antibody conjugated to horseradish peroxidase (HRP) (Millipore) at a concentration of 1:5,000 was added, and the cells were incubated at room temperature. The plates were washed 3 times and developed with 100 μl of SigmaFast OPD (Sigma) per well. The reaction was stopped after 30 min with 50 μl of 3 M HCl and read at an absorbance of 490 nm with a Synergy H1 plate reader (BioTek).
Formation and purification of the 70-1F02 complexes.
Purified 70-1F02 IgG was digested to Fab and Fc fragments by exposure to 40 μg of activated papain protease (Sigma) per mg IgG, in a solution containing 10 mM cysteine, 100 mM sodium acetate (pH 5.5), and 125 μM EDTA, for 2 to 4 h at 37°C. Fab fragments were subsequently purified from Fc by affinity chromatography using a protein A HiTrap column (GE Healthcare) and purified to homogeneity by size exclusion gel filtration chromatography (Superdex 75 10/30; GE Healthcare). To ensure saturation of the HA trimer with Fab, Fab 70-1F02 was mixed with purified, hexahistidine-tag-depleted, recombinant VN04 HA trimers at a molar ratio of 5 parts FAb to 1 part HA. The resulting 70-1F02 Fab-VN04 HA (70/H5) complexes were purified from unbound substrates by size exclusion gel filtration chromatography (Superdex 200 10/30 column; GE Healthcare) in a buffer comprising 50 mM Tris-HCl (pH 8.0) and 150 mM NaCl. The 70-1F02/H5 complexes eluted as single peaks between the 158-kDa and 670-kDa molecular weight markers and were concentrated to 15 mg/ml.
Crystallization and structure determination of the 70-1F02/H5 complex.
To ensure saturation with Fab, 70-1F02 Fab was mixed with purified, hexahistidine-tag-depleted, recombinant VN04 HA trimers at a molar ratio of 5 parts Fab to 1 part HA. The resulting 70-1F02 Fab-VN04 HA (70/H5) complex was purified from unbound substrates by size exclusion gel filtration chromatography and crystallized. Subsequently, the complex structure was determined by the molecular replacement (MR) method using Phaser (66). A strong solution was obtained for the HA component of the complex using a single monomer of VN04 HA (chains A and B of PDB 2FK0 [62]). An initial search for 70-1F02 Fab determined the position of the variable (Fv) domains using the Fv component of a mouse IgG1 Fab (PDB 1WEJ [67]) as a search model. A subsequent search using the constant (Fc) domains of a mouse IgG2a Fcb (PDB 3F09 [68]) located the Fc region.
Initial sparse-matrix crystallization screening was carried out using a Topaz Free Interface Diffusion (FID) Crystallizer system (Fluidigm Corporation, San Francisco, CA). Hits were obtained after 24 h under several conditions containing the precipitant polyethylene glycol (PEG) 4000. Following optimization, diffraction quality crystals were obtained at 23°C by the use of the vapor batch diffusion method under an oil layer (comprising 6 parts paraffin oil and 4 parts silicon oil) in 1.0-μl drops containing 70-1F02/HA, 25% PEG 3350, 200 mM NaCl, and 100 mM Bis/Tris-HCl (pH 5.5). The 70-1F02 Fab-HA complex data set was collected from a single crystal at 3.9-Å resolution at the Advanced Photon Source (APS) Southeast Regional Collaborative Access Team (Ser-CAT) 22-ID beamline. 70-1F02 Fab-HA crystallized in orthorhombic space group p212121. Data were processed and scaled using HKL2000 and Denzo software (69). Rigid body refinement and restrained refinement of the molecular replacement solution were carried out using REFMAC v5.5 (70), and model building was performed in Coot v0.8.8 (71). The modest resolution of the data limited analysis of the VN04 srHA/70-1F02 Fab interaction for determination of the relative positions of the molecules within the complex. Nevertheless, the model may be interpreted with confidence, given the high resolution of the HA (2.9-Å) and Fab (2.8-Å) structures used as search models for molecular replacement (MR). 2Fo-Fc electron density was well defined throughout the model. Notably, residues 79 to 85 of HA1 adopted a conformation different from those in the apo-H5 HA structure. Additional positive electron density was observed in the region of 27 potential N-linked glycosylation sites. Sixty-six carbohydrate moieties were built into this density at these positions. Restrained refinement of the structure was completed in REFMAC5. Residues were numbered in the H5 HA of both complexes according to the mature H5 HA. Residues in the 70-1F02 Fab were numbered according to the Wu and Kabat numbering system (72). Structural validation was carried out using MolProbity (73) and Procheck software as well as the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB) validation server. The connectivity and nomenclature of carbohydrate moieties were validated using PDBCARE (Glycosciences.de). Model manipulations, root-mean-square-deviation (RMSD) calculations, and distance measurements were carried out using Coot and Pymol (74). Solvent-accessible surface area calculations were carried out using Proteins, Interfaces, Structures, Assemblies (PISA) (http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html) (75) and Protorp applications.
Immunoglobulin G purification.
Human sera were diluted 1:10 in PBS (pH 7.4) and were filtered through a 0.22-μm-pore-size filter unit. Immunoglobulin G (IgG) was then purified as described before (14) using 4 FastFlow Sepharose G resin (GE Healthcare) on a gravity flow column (Qiagen). Briefly, diluted and filtered serum was passed over the column three times; the resin was then washed with four column volumes of PBS, and IgG was eluted using a 0.1 M glycine/HCl buffer at pH 2.7. The eluate was immediately neutralized with 2 M Tris/HCl buffer at pH 10. Purified IgG was then concentrated and buffer exchanged against PBS using Amicon Ultra centrifugal filter units (Millipore) with a 30-kDa molecular weight cutoff in a swinging bucket rotor. Protein concentration was measured with a NanoDrop 2000 spectrophotometer using the A280 method.
Statistical analysis.
Animal survival data were analyzed using a log rank (Mantel-Cox) test in GraphPad Prism, and the significance levels were adjusted using the Bonferroni method.
Accession number(s).
The atomic coordinates and structure factors for the 70-1F02/H5 complex are available from the Research Collaboratory for Structural Bioinformatics (RCSB) PDB under accession no. 6B3M.
ACKNOWLEDGMENTS
We thank Ariana Hirsh for excellent technical support and protein expression. We also thank Arvind Rajabhathor, Jackson Turner, and Aaron Schmitz for their great technical assistance.
This work was supported by NIAID grant U19 AI109946 (F.K.), grant R21 AI139813 (A.H.E.) and by grants from the NIAID-funded Centers of Excellence in Influenza Research and Surveillance (CEIRS; HHSN266200700006C and HHSN272201400004C [R.A.] and HHSN272201400008C [F.K. and A.H.E.]).
REFERENCES
- 1.World Health Organization. 2009. WHO fact sheet no. 211: Influenza. http://www.who.int/mediacentre/factsheets/fs211/en/index.html. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Palese P. 2004. Influenza: old and new threats. Nat Med 10:S82–S87. doi: 10.1038/nm1141. [DOI] [PubMed] [Google Scholar]
- 3.Taubenberger JK. 2006. The origin and virulence of the 1918 “Spanish” influenza virus. Proc Am Philos Soc 150:86–112. [PMC free article] [PubMed] [Google Scholar]
- 4.Harfoot R, Webby RJ. 2017. H5 influenza, a global update. J Microbiol 55:196–203. doi: 10.1007/s12275-017-7062-7. [DOI] [PubMed] [Google Scholar]
- 5.Jiang H, Wu P, Uyeki TM, He J, Deng Z, Xu W, Lv Q, Zhang J, Wu Y, Tsang TK, Kang M, Zheng J, Wang L, Yang B, Qin Y, Feng L, Fang VJ, Gao GF, Leung GM, Yu H, Cowling BJ. 1 August 2017. Preliminary epidemiologic assessment of human infections with highly pathogenic avian influenza A(H5N6) virus, China. Clin Infect Dis doi: 10.1093/cid/cix334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuan J, Zhang L, Kan X, Jiang L, Yang J, Guo Z, Ren Q. 2013. Origin and molecular characteristics of a novel 2013 avian influenza A(H6N1) virus causing human infection in Taiwan. Clin Infect Dis 57:1367–1368. doi: 10.1093/cid/cit479. [DOI] [PubMed] [Google Scholar]
- 7.Belser JA, Pulit-Penaloza JA, Sun X, Brock N, Pappas C, Creager HM, Zeng H, Tumpey TM, Maines TR. 12 July 2017. A novel A(H7N2) influenza virus isolated from a veterinarian caring for cats in a New York City animal shelter causes mild disease and transmits poorly in the ferret model. J Virol doi: 10.1128/JVI.00672-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belser JA, Blixt O, Chen LM, Pappas C, Maines TR, Van Hoeven N, Donis R, Busch J, McBride R, Paulson JC, Katz JM, Tumpey TM. 2008. Contemporary North American influenza H7 viruses possess human receptor specificity: implications for virus transmissibility. Proc Natl Acad Sci U S A 105:7558–7563. doi: 10.1073/pnas.0801259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Centers for Disease Control and Prevention (CDC). 2012. Notes from the field: highly pathogenic avian influenza A (H7N3) virus infection in two poultry workers—Jalisco, Mexico, July 2012. MMWR Morb Mortal Wkly Rep 61:726–727. [PubMed] [Google Scholar]
- 10.Tweed SA, Skowronski DM, David ST, Larder A, Petric M, Lees W, Li Y, Katz J, Krajden M, Tellier R, Halpert C, Hirst M, Astell C, Lawrence D, Mak A. 2004. Human illness from avian influenza H7N3, British Columbia. Emerg Infect Dis 10:2196–2199. doi: 10.3201/eid1012.040961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lopez-Martinez I, Balish A, Barrera-Badillo G, Jones J, Nuñez-García TE, Jang Y, Aparicio-Antonio R, Azziz-Baumgartner E, Belser JA, Ramirez-Gonzalez JE, Pedersen JC, Ortiz-Alcantara J, Gonzalez-Duran E, Shu B, Emery SL, Poh MK, Reyes-Teran G, Vazquez-Perez JA, Avila-Rios S, Uyeki T, Lindstrom S, Villanueva J, Tokars J, Ruiz-Matus C, Gonzalez-Roldan JF, Schmitt B, Klimov A, Cox N, Kuri-Morales P, Davis CT, Diaz-Quiñonez JA. 2013. Highly pathogenic avian influenza A(H7N3) virus in poultry workers, Mexico, 2012. Emerg Infect Dis 19:1531–1534. doi: 10.3201/eid1909.130087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fouchier RA, Schneeberger PM, Rozendaal FW, Broekman JM, Kemink SA, Munster V, Kuiken T, Rimmelzwaan GF, Schutten M, Van Doornum GJ, Koch G, Bosman A, Koopmans M, Osterhaus AD. 2004. Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proc Natl Acad Sci U S A 101:1356–1361. doi: 10.1073/pnas.0308352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iuliano AD, Jang Y, Jones J, Davis CT, Wentworth DE, Uyeki TM, Roguski K, Thompson MG, Gubareva L, Fry AM, Burns E, Trock S, Zhou S, Katz JM, Jernigan DB. 2017. Increase in human infections with avian influenza A(H7N9) virus during the fifth epidemic—China, October 2016–February 2017 MMWR Morb Mortal Wkly Rep 66:254–255. doi: 10.15585/mmwr.mm6609e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang Y, Li X, Zhang H, Chen B, Jiang Y, Yang L, Zhu W, Hu S, Zhou S, Tang Y, Xiang X, Li F, Li W, Gao L. 2015. Human infection with an avian influenza A (H9N2) virus in the middle region of China. J Med Virol 87:1641–1648. doi: 10.1002/jmv.24231. [DOI] [PubMed] [Google Scholar]
- 15.Chen H, Yuan H, Gao R, Zhang J, Wang D, Xiong Y, Fan G, Yang F, Li X, Zhou J, Zou S, Yang L, Chen T, Dong L, Bo H, Zhao X, Zhang Y, Lan Y, Bai T, Dong J, Li Q, Wang S, Li H, Gong T, Shi Y, Ni X, Li J, Fan J, Wu J, Zhou X, Hu M, Wan J, Yang W, Li D, Wu G, Feng Z, Gao GF, Wang Y, Jin Q, Liu M, Shu Y. 2014. Clinical and epidemiological characteristics of a fatal case of avian influenza A H10N8 virus infection: a descriptive study. Lancet 383:714–721. doi: 10.1016/S0140-6736(14)60111-2. [DOI] [PubMed] [Google Scholar]
- 16.Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, Xu X, Lu H, Zhu W, Gao Z, Xiang N, Shen Y, He Z, Gu Y, Zhang Z, Yang Y, Zhao X, Zhou L, Li X, Zou S, Zhang Y, Yang L, Guo J, Dong J, Li Q, Dong L, Zhu Y, Bai T, Wang S, Hao P, Yang W, Han J, Yu H, Li D, Gao GF, Wu G, Wang Y, Yuan Z, Shu Y. 2013. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med 368:1888–1897. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- 17.Krammer F, Palese P. 2015. Advances in the development of influenza virus vaccines. Nat Rev Drug Discov 14:167–182. doi: 10.1038/nrd4529. [DOI] [PubMed] [Google Scholar]
- 18.Tricco AC, Chit A, Soobiah C, Hallett D, Meier G, Chen MH, Tashkandi M, Bauch CT, Loeb M. 2013. Comparing influenza vaccine efficacy against mismatched and matched strains: a systematic review and meta-analysis. BMC Med 11:153. doi: 10.1186/1741-7015-11-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Centers for Disease Control and Prevention (CDC). 2010. Estimates of deaths associated with seasonal influenza—United States, 1976–2007. MMWR Morb Mortal Wkly Rep 59:1057–1062. [PubMed] [Google Scholar]
- 20.Couch RB, Decker WK, Utama B, Atmar RL, Niño D, Feng JQ, Halpert MM, Air GM. 2012. Evaluations for in vitro correlates of immunogenicity of inactivated influenza a H5, H7 and H9 vaccines in humans. PLoS One 7:e50830. doi: 10.1371/journal.pone.0050830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hussain M, Galvin HD, Haw TY, Nutsford AN, Husain M. 2017. Drug resistance in influenza A virus: the epidemiology and management. Infect Drug Resist 10:121–134. doi: 10.2147/IDR.S105473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen-Van-Tam JS, Venkatesan S, Muthuri SG, Myles PR. 2015. Neuraminidase inhibitors: who, when, where? Clin Microbiol Infect 21:222–225. doi: 10.1016/j.cmi.2014.11.020. [DOI] [PubMed] [Google Scholar]
- 23.Seibert CW, Rahmat S, Krammer F, Palese P, Bouvier NM. 2012. Efficient transmission of pandemic H1N1 influenza viruses with high-level oseltamivir resistance. J Virol 86:5386–5389. doi: 10.1128/JVI.00151-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meijer A, Lackenby A, Hungnes O, Lina B, van-der-Werf S, Schweiger B, Opp M, Paget J, van-de-Kassteele J, Hay A, Zambon M; European Influenza Surveillance Scheme. 2009. Oseltamivir-resistant influenza virus A (H1N1), Europe, 2007–08 season. Emerg Infect Dis 15:552–560. doi: 10.3201/eid1504.181280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hai R, Schmolke M, Leyva-Grado VH, Thangavel RR, Margine I, Jaffe EL, Krammer F, Solórzano A, García-Sastre A, Palese P, Bouvier NM. 2013. Influenza A(H7N9) virus gains neuraminidase inhibitor resistance without loss of in vivo virulence or transmissibility. Nat Commun 4:2854. doi: 10.1038/ncomms3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krammer F, Palese P. 2013. Influenza virus hemagglutinin stalk-based antibodies and vaccines. Curr Opin Virol 3:521–530. doi: 10.1016/j.coviro.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li GM, Chiu C, Wrammert J, McCausland M, Andrews SF, Zheng NY, Lee JH, Huang M, Qu X, Edupuganti S, Mulligan M, Das SR, Yewdell JW, Mehta AK, Wilson PC, Ahmed R. 2012. Pandemic H1N1 influenza vaccine induces a recall response in humans that favors broadly cross-reactive memory B cells. Proc Natl Acad Sci U S A 109:9047–9052. doi: 10.1073/pnas.1118979109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ellebedy AH, Krammer F, Li GM, Miller MS, Chiu C, Wrammert J, Chang CY, Davis CW, McCausland M, Elbein R, Edupuganti S, Spearman P, Andrews SF, Wilson PC, García-Sastre A, Mulligan MJ, Mehta AK, Palese P, Ahmed R. 2014. Induction of broadly cross-reactive antibody responses to the influenza HA stem region following H5N1 vaccination in humans. Proc Natl Acad Sci U S A 111:13133–13138. doi: 10.1073/pnas.1414070111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wrammert J, Koutsonanos D, Li GM, Edupuganti S, Sui J, Morrissey M, McCausland M, Skountzou I, Hornig M, Lipkin WI, Mehta A, Razavi B, Del Rio C, Zheng NY, Lee JH, Huang M, Ali Z, Kaur K, Andrews S, Amara RR, Wang Y, Das SR, O'Donnell CD, Yewdell JW, Subbarao K, Marasco WA, Mulligan MJ, Compans R, Ahmed R, Wilson PC. 2011. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J Exp Med 208:181–193. doi: 10.1084/jem.20101352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nachbagauer R, Wohlbold TJ, Hirsh A, Hai R, Sjursen H, Palese P, Cox RJ, Krammer F. 2014. Induction of broadly reactive anti-hemagglutinin stalk antibodies by an H5N1 vaccine in humans. J Virol 88:13260–13268. doi: 10.1128/JVI.02133-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu L, Nachbagauer R, Zhu L, Huang Y, Xie X, Jin S, Zhang A, Wan Y, Hirsh A, Tian D, Shi X, Dong Z, Yuan S, Hu Y, Krammer F, Zhang X, Xu J. 2017. Induction of broadly cross-reactive stalk-specific antibody responses to influenza group 1 and group 2 hemagglutinins by natural H7N9 virus infection in humans. J Infect Dis 215:518–528. doi: 10.1093/infdis/jiw608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pica N, Hai R, Krammer F, Wang TT, Maamary J, Eggink D, Tan GS, Krause JC, Moran T, Stein CR, Banach D, Wrammert J, Belshe RB, García-Sastre A, Palese P. 2012. Hemagglutinin stalk antibodies elicited by the 2009 pandemic influenza virus as a mechanism for the extinction of seasonal H1N1 viruses. Proc Natl Acad Sci U S A 109:2573–2578. doi: 10.1073/pnas.1200039109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomson CA, Wang Y, Jackson LM, Olson M, Wang W, Liavonchanka A, Keleta L, Silva V, Diederich S, Jones RB, Gubbay J, Pasick J, Petric M, Jean F, Allen VG, Brown EG, Rini JM, Schrader JW. 2012. Pandemic H1N1 influenza infection and vaccination in humans induces cross-protective antibodies that target the hemagglutinin stem. Front Immunol 3:87. doi: 10.3389/fimmu.2012.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marjuki H, Mishin VP, Chai N, Tan MW, Newton EM, Tegeris J, Erlandson K, Willis M, Jones J, Davis T, Stevens J, Gubareva LV. 2016. Human monoclonal antibody 81.39a effectively neutralizes emerging influenza A viruses of group 1 and 2 hemagglutinins. J Virol 90:10446–10458. doi: 10.1128/JVI.01284-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Friesen RH, Koudstaal W, Koldijk MH, Weverling GJ, Brakenhoff JP, Lenting PJ, Stittelaar KJ, Osterhaus AD, Kompier R, Goudsmit J. 2010. New class of monoclonal antibodies against severe influenza: prophylactic and therapeutic efficacy in ferrets. PLoS One 5:e9106. doi: 10.1371/journal.pone.0009106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baranovich T, Jones JC, Russier M, Vogel P, Szretter KJ, Sloan SE, Seiler P, Trevejo JM, Webby RJ, Govorkova EA. 2016. The hemagglutinin stem-binding monoclonal antibody VIS410 controls influenza virus-induced acute respiratory distress syndrome. Antimicrob Agents Chemother 60:2118–2131. doi: 10.1128/AAC.02457-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tan GS, Krammer F, Eggink D, Kongchanagul A, Moran TM, Palese P. 2012. A pan-h1 anti-hemagglutinin monoclonal antibody with potent broad-spectrum efficacy in vivo. J Virol 86:6179–6188. doi: 10.1128/JVI.00469-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tan GS, Lee PS, Hoffman RM, Mazel-Sanchez B, Krammer F, Leon PE, Ward AB, Wilson IA, Palese P. 2014. Characterization of a broadly neutralizing monoclonal antibody that targets the fusion domain of group 2 influenza a virus hemagglutinin. J Virol 88:13580–13592. doi: 10.1128/JVI.02289-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wollacott AM, Boni MF, Szretter KJ, Sloan SE, Yousofshahi M, Viswanathan K, Bedard S, Hay CA, Smith PF, Shriver Z, Trevejo JM. 2016. Safety and upper respiratory pharmacokinetics of the hemagglutinin stalk-binding antibody VIS410 support treatment and prophylaxis based on population modeling of seasonal influenza A outbreaks. EBioMedicine 5:147–155. doi: 10.1016/j.ebiom.2016.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nachbagauer R, Krammer F. 2017. Universal influenza virus vaccines and therapeutic antibodies. Clin Microbiol Infect 23:222–228. doi: 10.1016/j.cmi.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marston HD, Paules CI, Fauci AS. 7 March 2018. Monoclonal antibodies for emerging infectious diseases—borrowing from history. N Engl J Med doi: 10.1056/NEJMp1802256. [DOI] [PubMed] [Google Scholar]
- 42.Sui J, Hwang WC, Perez S, Wei G, Aird D, Chen LM, Santelli E, Stec B, Cadwell G, Ali M, Wan H, Murakami A, Yammanuru A, Han T, Cox NJ, Bankston LA, Donis RO, Liddington RC, Marasco WA. 2009. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol 16:265–273. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ekiert DC, Friesen RH, Bhabha G, Kwaks T, Jongeneelen M, Yu W, Ophorst C, Cox F, Korse HJ, Brandenburg B, Vogels R, Brakenhoff JP, Kompier R, Koldijk MH, Cornelissen LA, Poon LL, Peiris M, Koudstaal W, Wilson IA, Goudsmit J. 2011. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science 333:843–850. doi: 10.1126/science.1204839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ekiert DC, Bhabha G, Elsliger MA, Friesen RH, Jongeneelen M, Throsby M, Goudsmit J, Wilson IA. 2009. Antibody recognition of a highly conserved influenza virus epitope. Science 324:246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dreyfus C, Laursen NS, Kwaks T, Zuijdgeest D, Khayat R, Ekiert DC, Lee JH, Metlagel Z, Bujny MV, Jongeneelen M, van der Vlugt R, Lamrani M, Korse HJ, Geelen E, Sahin Ö Sieuwerts M, Brakenhoff JP, Vogels R, Li OT, Poon LL, Peiris M, Koudstaal W, Ward AB, Wilson IA, Goudsmit J, Friesen RH. 2012. Highly conserved protective epitopes on influenza B viruses. Science 337:1343–1348. doi: 10.1126/science.1222908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Corti D, Voss J, Gamblin SJ, Codoni G, Macagno A, Jarrossay D, Vachieri SG, Pinna D, Minola A, Vanzetta F, Silacci C, Fernandez-Rodriguez BM, Agatic G, Bianchi S, Giacchetto-Sasselli I, Calder L, Sallusto F, Collins P, Haire LF, Temperton N, Langedijk JP, Skehel JJ, Lanzavecchia A. 2011. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 333:850–856. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- 47.Okuno Y, Isegawa Y, Sasao F, Ueda S. 1993. A common neutralizing epitope conserved between the hemagglutinins of influenza A virus H1 and H2 strains. J Virol 67:2552–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Okuno Y, Matsumoto K, Isegawa Y, Ueda S. 1994. Protection against the mouse-adapted A/FM/1/47 strain of influenza A virus in mice by a monoclonal antibody with cross-neutralizing activity among H1 and H2 strains. J Virol 68:517–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Avnir Y, Watson CT, Glanville J, Peterson EC, Tallarico AS, Bennett AS, Qin K, Fu Y, Huang CY, Beigel JH, Breden F, Zhu Q, Marasco WA. 2016. IGHV1-69 polymorphism modulates anti-influenza antibody repertoires, correlates with IGHV utilization shifts and varies by ethnicity. Sci Rep 6:20842. doi: 10.1038/srep20842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramos EL, Mitcham JL, Koller TD, Bonavia A, Usner DW, Balaratnam G, Fredlund P, Swiderek KM. 2015. Efficacy and safety of treatment with an anti-m2e monoclonal antibody in experimental human influenza. J Infect Dis 211:1038–1044. doi: 10.1093/infdis/jiu539. [DOI] [PubMed] [Google Scholar]
- 51.Andrews SF, Huang Y, Kaur K, Popova LI, Ho IY, Pauli NT, Henry Dunand CJ, Taylor WM, Lim S, Huang M, Qu X, Lee JH, Salgado-Ferrer M, Krammer F, Palese P, Wrammert J, Ahmed R, Wilson PC. 2015. Immune history profoundly affects broadly protective B cell responses to influenza. Sci Transl Med 7:316ra192. doi: 10.1126/scitranslmed.aad0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nachbagauer R, Choi A, Hirsh A, Margine I, Iida S, Barrera A, Ferres M, Albrecht RA, García-Sastre A, Bouvier NM, Ito K, Medina RA, Palese P, Krammer F. 2017. Defining the antibody cross-reactome directed against the influenza virus surface glycoproteins. Nat Immunol 18:464–473. doi: 10.1038/ni.3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.DiLillo DJ, Tan GS, Palese P, Ravetch JV. 2014. Broadly neutralizing hemagglutinin stalk-specific antibodies require FcγR interactions for protection against influenza virus in vivo. Nat Med 20:143–151. doi: 10.1038/nm.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Henry Dunand CJ, Leon PE, Huang M, Choi A, Chromikova V, Ho IY, Tan GS, Cruz J, Hirsh A, Zheng NY, Mullarkey CE, Ennis FA, Terajima M, Treanor JJ, Topham DJ, Subbarao K, Palese P, Krammer F, Wilson PC. 2016. Both neutralizing and non-neutralizing human H7N9 influenza vaccine-induced monoclonal antibodies confer protection. Cell Host Microbe 19:800–813. doi: 10.1016/j.chom.2016.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang F, Bi Y, Wang J, Wong G, Shi W, Hu F, Yang Y, Yang L, Deng X, Jiang S, He X, Liu Y, Yin C, Zhong N, Gao GF. 6 April 2017. Human infections with recently-emerging highly pathogenic H7N9 avian influenza in China. J Infect doi: 10.1016/j.jinf.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 56.Krammer F. 2015. Emerging influenza viruses and the prospect of a universal influenza virus vaccine. Biotechnol J 10:690–701. doi: 10.1002/biot.201400393. [DOI] [PubMed] [Google Scholar]
- 57.Qiu X, Wong G, Audet J, Bello A, Fernando L, Alimonti JB, Fausther-Bovendo H, Wei H, Aviles J, Hiatt E, Johnson A, Morton J, Swope K, Bohorov O, Bohorova N, Goodman C, Kim D, Pauly MH, Velasco J, Pettitt J, Olinger GG, Whaley K, Xu B, Strong JE, Zeitlin L, Kobinger GP. 2014. Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature 514:47–53. doi: 10.1038/nature13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am J Epidemiol 27:493–497. doi: 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
- 59.Krammer F, Schinko T, Palmberger D, Tauer C, Messner P, Grabherr R. 2010. Trichoplusia ni cells (High Five) are highly efficient for the production of influenza A virus-like particles: a comparison of two insect cell lines as production platforms for influenza vaccines. Mol Biotechnol 45:226–234. doi: 10.1007/s12033-010-9268-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Margine I, Palese P, Krammer F. 6 November 2013. Expression of functional recombinant hemagglutinin and neuraminidase proteins from the novel H7N9 influenza virus using the baculovirus expression system. J Vis Exp doi: 10.3791/51112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Krammer F, Margine I, Tan GS, Pica N, Krause JC, Palese P. 2012. A carboxy-terminal trimerization domain stabilizes conformational epitopes on the stalk domain of soluble recombinant hemagglutinin substrates. PLoS One 7:e43603. doi: 10.1371/journal.pone.0043603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stevens J, Blixt O, Tumpey TM, Taubenberger JK, Paulson JC, Wilson IA. 2006. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science 312:404–410. doi: 10.1126/science.1124513. [DOI] [PubMed] [Google Scholar]
- 63.Carney PJ, Lipatov AS, Monto AS, Donis RO, Stevens J. 2010. Flexible label-free quantitative assay for antibodies to influenza virus hemagglutinins. Clin Vaccine Immunol 17:1407–1416. doi: 10.1128/CVI.00509-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang H, Carney P, Stevens J. 2010. Structure and receptor binding properties of a pandemic H1N1 virus hemagglutinin. PLoS Curr 2:RRN1152. doi: 10.1371/currents.RRN1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang H, Chen LM, Carney PJ, Donis RO, Stevens J. 2010. Structures of receptor complexes of a North American H7N2 influenza hemagglutinin with a loop deletion in the receptor binding site. PLoS Pathog 6:e1001081. doi: 10.1371/journal.ppat.1001081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mylvaganam SE, Paterson Y, Getzoff ED. 1998. Structural basis for the binding of an anti-cytochrome c antibody to its antigen: crystal structures of FabE8-cytochrome c complex to 1.8 A resolution and FabE8 to 2.26 A resolution. J Mol Biol 281:301–322. doi: 10.1006/jmbi.1998.1942. [DOI] [PubMed] [Google Scholar]
- 68.Zhu X, Tanaka F, Lerner RA, Barbas CF III, Wilson IA. 2009. Direct observation of an enamine intermediate in amine catalysis. J Am Chem Soc 131:18206–18207. doi: 10.1021/ja907271a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Otwinowski Z, Minor W. 1997. Processing of X ray diffraction data collected in oscillation mode. Methods Enzymol 276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 70.Murshudov GN, Vagin AA, Dodson EJ. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 71.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu TT, Kabat EA. 1970. An analysis of the sequences of the variable regions of Bence Jones proteins and myeloma light chains and their implications for antibody complementarity. J Exp Med 132:211–250. doi: 10.1084/jem.132.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB III, Snoeyink J, Richardson JS, Richardson DC. 2007. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res 35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Delano WL. 2002. The PyMOL molecular graphics system. https://pymol.org/2/ Schrödinger, New York, NY. [Google Scholar]
- 75.Krissinel E, Henrick K. 2007. Inference of macromolecular assemblies from crystalline state. J Mol Biol 372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]