This study raises two issues related to HIV-1-specific CTL responses. These are key immune responses that retard disease progression in infected persons that are highly relevant to immunotherapies and vaccines for HIV-1. First, we make the novel observation that these responses are promiscuous and that CTLs targeting one epitope may cross-recognize other, completely distinct epitopes in the virus. While these are low-avidity interactions that do not appear to contribute directly to the antiviral activity of CTLs, this raises interesting biologic implications regarding the purpose of the phenomenon, such as providing a stimulus for these responses to persist long term. Second, the data raise a technical caveat to detection of CTL responses against particular epitopes, suggesting that some methodologies may unintentionally detect cross-reactivity and overestimate responses against an epitope.
KEYWORDS: cross-reactivity, cytotoxic T lymphocyte, HIV
ABSTRACT
Although a high level of promiscuity for heterologous epitopes is believed to exist for cellular immunity, limited data explore this issue for human immunodeficiency virus type 1 (HIV-1)-specific CD8+ T lymphocyte (CTL) responses. Here, we found an unexpected degree of heterologous cross-reactivity against HIV-1 epitopes, in addition to the targeted index epitope. Most CTL clones screened cross-reacted against other known HIV-1 epitopes of the same major histocompatibility complex type I (MHC-I) restriction, up to 40% of tested nonindex epitopes in some cases. The observed cross-reactivity was universally lower avidity than recognition of the index epitope when examined for several A*02- and B*57-restricted CTL clones, demonstrating that the high concentrations of exogenous epitope typically used for screening of CTL responses are prone to detect such cross-reactivity spuriously. In agreement with this, we found that these cross-reactive responses do not appear to mediate CTL activity against HIV-1-infected cells. Overall, our data indicate that low-level cross-reactivity is remarkably common for HIV-1-specific CTLs. The role of this phenomenon is unclear, but low-avidity interactions have been shown to foster homeostatic proliferation of memory T cells.
IMPORTANCE This study raises two issues related to HIV-1-specific CTL responses. These are key immune responses that retard disease progression in infected persons that are highly relevant to immunotherapies and vaccines for HIV-1. First, we make the novel observation that these responses are promiscuous and that CTLs targeting one epitope may cross-recognize other, completely distinct epitopes in the virus. While these are low-avidity interactions that do not appear to contribute directly to the antiviral activity of CTLs, this raises interesting biologic implications regarding the purpose of the phenomenon, such as providing a stimulus for these responses to persist long term. Second, the data raise a technical caveat to detection of CTL responses against particular epitopes, suggesting that some methodologies may unintentionally detect cross-reactivity and overestimate responses against an epitope.
INTRODUCTION
T cell responses are generally considered to be specifically targeted by virtue of their recognizing an index epitope, but it is known that T cell receptors (TCRs) have a degree of heterologous promiscuity. Based on the empirical observation that mice can mount T cell responses against any peptide bound to mouse major histocompatibility complex (MHC), and the maximum number of T cells per mouse, it has been estimated that a given TCR may recognize an average of a million epitopes (1). Frequent examples have been identified where CD8+ T lymphocytes (CTLs) cross-target distinct epitopes from different pathogens, and indeed, exposure to one pathogen can cause immunologic priming that alters the immunodominance pattern to a second pathogen, supporting this concept (2).
While numerous studies have examined “cross-reactivity” of HIV-1-specific CTLs against target epitope variation within HIV-1, the degree to which they recognize truly heterologous epitopes is unclear. Limited data have suggested that there can be CTL cross-reactivity with microbial peptides (3), hepatitis C virus (4), and a self-protein (5). Here, we find unexpectedly that HIV-1-specific CTLs commonly exhibit cross-reactivity against epitopes from other regions of the HIV-1 proteome. This cross-reactivity is relatively low avidity, however, and the functional implications are yet to be defined.
RESULTS
HIV-1-specific CTL clones demonstrate cross-reactivity when screened against panels of other optimal HIV-1 epitopes.
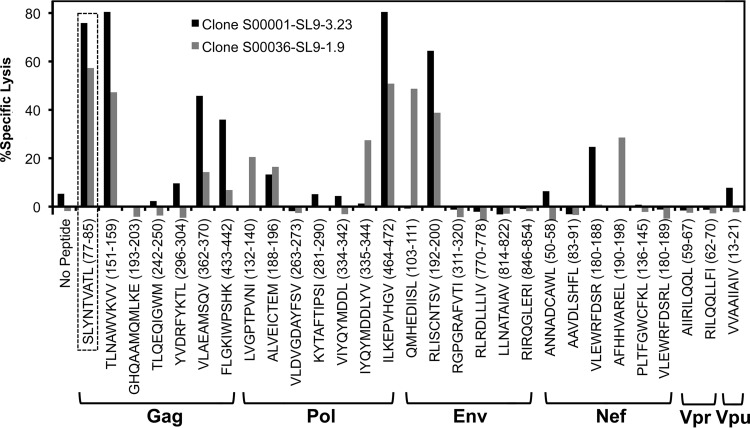
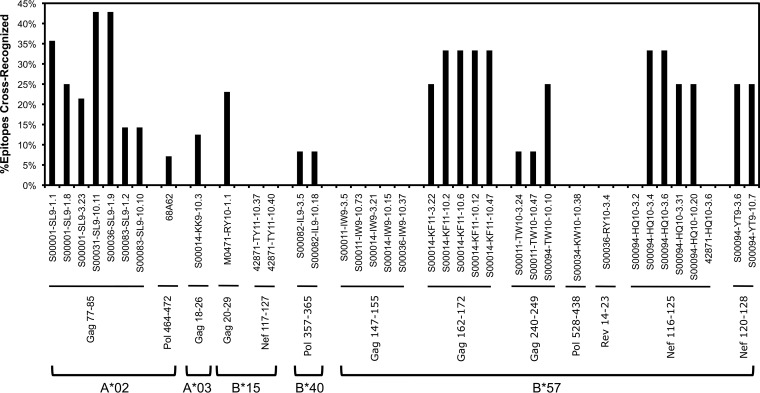
Panels of HIV-1-specific CTL clones derived from persons with chronic infection (Table 1) were screened against other well-defined HIV-1 optimal epitopes of the same human leukocyte antigen class I (HLA-I) restriction (Table 2) in standard chromium release killing assays (Fig. 1). Unexpectedly, many of the clones demonstrated cross-recognition of other epitopes, defined as ≥20% killing activity compared to the index epitope. For example, screening of seven clones from four different persons recognizing the A*02-restricted epitope SLYNTVATL (SL9; Gag 77–85) against 28 other known HIV-1 A*02 epitopes revealed cross-recognition of 4 to 12 (14.3 to 42.9%) other epitopes (Fig. 2 and 3). Although some peptides appeared to be more frequently cross-recognized (e.g., the TV9 epitope), there was no appreciable pattern of similarity to the index epitopes and shared amino acids to explain this finding. While similarity of cross-recognition between some clones derived from the same person (e.g., S00001-SL9 clones) may have been explained by identical T cell receptors, frequent cross-reactivity of some epitopes was shared even between clones derived from different persons (e.g., the TV9 epitope).
TABLE 1.
CTL clones utilized in this study
| HLA | Epitope location | Clonea |
|---|---|---|
| A*02 | Gag 77–85 | S00001-SL9-1.1 |
| S00001-SL9-1.8 | ||
| S00001-SL9-3.23 | ||
| S00031-SL9-10.11 | ||
| S00036-SL9-1.9 | ||
| S00083-SL9-1.2 | ||
| S00083-SL9-10.10 | ||
| Pol 464–472 | 68A62 (IV9) | |
| A*03 | Gag 18–26 | S00014-KK9-10.3 |
| B*15 | Gag 20–29 | M0471-RY10-1.1 |
| Nef 117–127 | 42871-TY11-10.37 | |
| 42871-TY11-10.40 | ||
| B*40 | Pol 357–365 | S00082-IL-9-3.5 |
| S00082-IL-9-10.18 | ||
| B*57 | Gag 147–155 | S00011-IW9-3.5 |
| S00011-IW9-10.73 | ||
| S00014-IW9-3.21 | ||
| S00014-IW9-10.15 | ||
| S00036-IW9-10.37 | ||
| Gag 162–172 | S00014-KF11-3.22 | |
| S00014-KF11-10.2 | ||
| S00014-KF11-10.6 | ||
| S00014-KF11-10.12 | ||
| S00014-KF11-10.47 | ||
| Gag 240–249 | S00011-TW10-3.24 | |
| S00011-TW10-10.47 | ||
| S00094-TW10-10.10 | ||
| Pol 528–438 | S00034-KW10-10.38 | |
| Rev 14–23 | S00036-RY10-3.4 | |
| Nef 116–125 | S00094-HQ10-3.2 | |
| S00094-HQ10-3.4 | ||
| S00094-HQ10-3.6 | ||
| S00094-HQ10-3.31 | ||
| S00094-HQ10-10.20 | ||
| 42871-HQ10-3.6 | ||
| Nef 120–128 | S00094-YT9-3.6 | |
| S00094-YT9-10.7 |
CTL clones are named according to the scheme X-Y-Z, where X is the individual from whom they are derived, Y is the recognized index epitope, and Z is an identifier number (with the exception of clone 68A62, a gift from Bruce D. Walker).
TABLE 2.
Panel of HIV–1 epitopes
| HLA | Peptide (location) | Sequence |
|---|---|---|
| A*02 | SL9 (Gag 77–85) | SLYNTVATL |
| TV9 (Gag 151–159) | TLNAWVKVV | |
| GE11 (Gag 193–203) | GHQAAMQMLKE | |
| TM9 (Gag 242–250) | TLQEQIGWM | |
| YL9 (Gag 296–304) | YVDRFYKTL | |
| VV9 (Gag 362–370) | VLAEAMSQV | |
| FK10 (Gag 433–442) | FLGKIWPSHK | |
| LI9 (Pol 132–140) | LVGPTPVNI | |
| AM9 (Pol 188–196) | ALVEICTEM | |
| VV11 (Pol 263–273) | VLDVGDAYFSV | |
| KI10 (Pol 281–290) | KYTAFTIPSI | |
| VL9 (Pol 334–342) | VIYQYMDDL | |
| IV10 (Pol 335–344) | IYQYMDDLYV | |
| IV9 (Pol 464–472) | ILKEPVHGV | |
| RI9 (Env 846–854) | RIRQGLERI | |
| QL9 (Env 103–111) | QMHEDIISL | |
| RV9 (Env 192–200) | RLISCNTSV | |
| RI10 (Env 311–320) | RGPGRAFVTI | |
| RV9 (Env 770–778) | RLRDLLLIV | |
| LV9 (Env 814–822) | LLNATAIAV | |
| AL9 (Nef 50–58) | ANNADCAWL | |
| AL9 (Nef 83–91) | AAVDLSHFL | |
| VR9 (Nef 180–188) | VLEWRFDSR | |
| AL9 (Nef 190–198) | AFHHVAREL | |
| PL10 (Nef 136–145) | PLTFGWCYKL | |
| VL10 (Nef 180–189) | VLEWRFDSRL | |
| AL9 (Vpr 59–67) | AIIRILQQL | |
| RI9 (Vpr 62–70) | RILQQLLFI | |
| VV9 (Vpu 13–21) | VVAAIIAIV | |
| A*03 | KK9 (Gag 18–26) | KIRLRPGGK |
| AK11 (Pol 188–198) | ALVEICTEMEK | |
| AK9 (Pol 313–321) | AIFQSSMTK | |
| KA9 (Pol 685–693) | KVYLAWVPA | |
| RK10 (Vif 17–26) | RIRTWKSLVK | |
| HK9 (Vif 28–36) | HMYISKKAK | |
| KK11 (Vif 158–168) | KTKPPLPSVKK | |
| RR9 (Rev 58–66) | RILSTYLGR | |
| RR11 (Env 770–780) | RLRDLLLIVTR | |
| B*15 | RY10 (Gag 20–29) | RLRPGGKKKY |
| SL9 (Gag 77–85) | SLYNTVATL | |
| GL9 (Gag 193–201) | GHQAAMQML | |
| GY9 (Gag 269–277) | GLNKIVRMY | |
| IY10 (Pol 464–473) | IKLEPVHGVY | |
| VI10 (Pol 651–660) | VTDSQYALGI | |
| FY10 (Pol 900–909) | FKRKGGIGGY | |
| RY9 (Pol 978–986) | RKAKIIRDY | |
| HI8 (Vif 80–87) | HLGQGVSI | |
| FY10 (Tat 38–47) | FTTKGLGISY | |
| RA9 (Nef 19–27) | RMRRAEPAA | |
| AL8 (Nef 84–91) | AVDLSHFL | |
| TY11 (Nef 117–127) | TQGYFPDWQNY | |
| WF9 (Nef 183–191) | WRFDSRLAF | |
| B*40 | GI9 (Gag 11–19) | GELDKWEKI |
| IL–10 (Gag 92–101) | IEIKDTKEAL | |
| SL9 (Gag 176–184) | SEGATPQDL | |
| KA9 (Gag 202–210) | KETINEEAA | |
| AV9 (Gag 210–218) | AEWDRVHPV | |
| TL8 (Gag 427–434) | TERQANFL | |
| KL9 (Gag 481–489) | KELYPLASL | |
| IL–8 (Pol 160–167) | IETVPVKL | |
| IL–9 (Pol 357–365) | IEELRQHLL | |
| RL9 (Vpr 12–20) | REPHNEWTL | |
| QL10 (Env 805–814) | QELKNSAVSL | |
| LS9 (Nef 37–45) | LEKHGAITS | |
| KL9 (Nef 92–100) | KEKGGLEGL | |
| B*57 | GP10 (Gag 140–149) | GQMVHQAISP |
| IW9 (Gag 147–155) | ISPRTLNAW | |
| KF11 (Gag 162–172) | KAFSPEVIPMF | |
| TW10 (Gag 240–249) | TSTLQEQIGW | |
| QW9 (Gag 308–316) | QASQEVKNW | |
| IW9 (Pol 399–407) | IVLPEKDSW | |
| KW10 (Pol 529–538) | KIATESIVIW | |
| KF9 (Pol 888–896) | KTAVQMAVF | |
| AW9 (Vpr 30–38) | AVRHFPRIW | |
| IF9 (Vif 31–39) | ISRKAKDWF | |
| RY10 (Rev 14–23) | RTVRLIKLLY | |
| HQ10 (Nef 116–125) | HTQGYFPDWQ | |
| YT9 (Nef 120–128) | YFPDWQNYT |
FIG 1.
Example of testing two HIV-1-specific CTL clones for cross-reactivity against other HIV-1 epitopes. HIV-1-specific clones S00001-SL9-3.23 and S00036-SL9-1.9, both A*02 restricted and targeting the minimal epitope SLYNTVATL (Gag 77–85; SL9) (dashed box), were screened against a panel of other known A*02-restricted HIV-1 minimal epitopes (Table 2) in standard 4-h chromium release assays of peptide-labeled (1 μg/ml) T1 cells at an effector-to-target cell ratio of 5:1. Specific lysis is indicated for each epitope.
FIG 2.
Cross-reactivity of HIV-1-specific CTL clones against other HIV-1 epitopes. HIV-1-specific CTL clones were tested against panels of known HIV-1 epitopes of the same HLA-I restriction, as shown in Fig. 1. Cross-reactivity was defined as observed specific lysis at ≥20% of specific lysis against the index epitope. The CTL clones and peptides are grouped according to HLA-I restriction. Black squares indicate the index epitope, gray squares indicate cross-reactivity, and white squares indicate lack of cross-reactivity.
FIG 3.
Amounts of cross-reactivity of HIV-1-specific CTL clones. For each CTL clone, the observed percentage of cross-reactivity (excluding the index epitope) in Fig. 2 is plotted.
CTL clones against another epitope appeared to be less cross-reactive; five clones from three different persons recognizing the B*57-restricted epitope ISPRTLNAW (IW9; Gag 147–155) screened against 12 other known HIV-1 B*57 epitopes showed no cross-recognition (Fig. 2 and 3). This did not appear to be HLA specific, as cross-reactivity was observed for other B*57-restricted clones. Overall, of the CTL clones recognizing 13 index epitopes screened, clones targeting 9 of those epitopes demonstrated cross-reactivity.
HIV-1-specific CTL clone cross-reactivity is lower avidity than recognition of the index epitope.
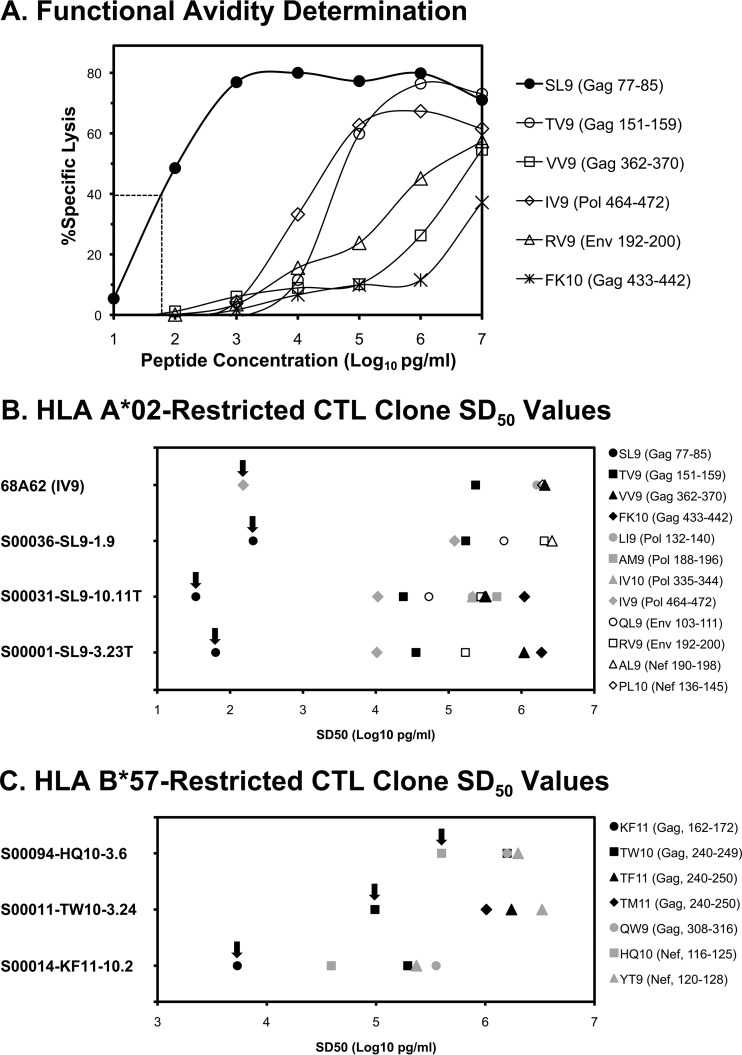
To evaluate the efficiency of cross-reactivity as reflected by the sensitizing dose for 50% maximal killing (SD50), CTL clones were tested for their functional avidity against index versus cross-reacting epitopes by peptide titration killing assays (Fig. 4). For cross-reactive clones recognizing two epitopes restricted by A*02 (Fig. 4B), avidity for the index epitope was consistently more than 100-fold lower (SD50) than the most avid cross-reactive epitopes. For cross-reactive clones recognizing three epitopes restricted by B*57 (Fig. 4C), avidity for the index epitope was also higher, but by a smaller margin. These data suggested that while cross-reactivity occurs, it is limited to relatively low-avidity interactions compared to the index epitope.
FIG 4.
Functional avidity values of CTL clones against index and cross-reactive epitopes. Functional avidity was defined as the sensitizing dose of peptide yielding 50% maximal lysis in chromium release assays (SD50). (A) Example of clone S00001-SL9-3.23 recognizing the Gag epitope SLYNTVATL (SL9; Gag 77–85), demonstrating titrations of the index peptide versus five cross-reactive peptides. The dashed line indicates the SD50 value for the index peptide. (B and C) Functional avidities of four A*02-restricted CTL clones (B) and three B*57-restricted CTL clones (C) against their index peptides and cross-reactive peptides. The arrows indicate the values for index peptides.
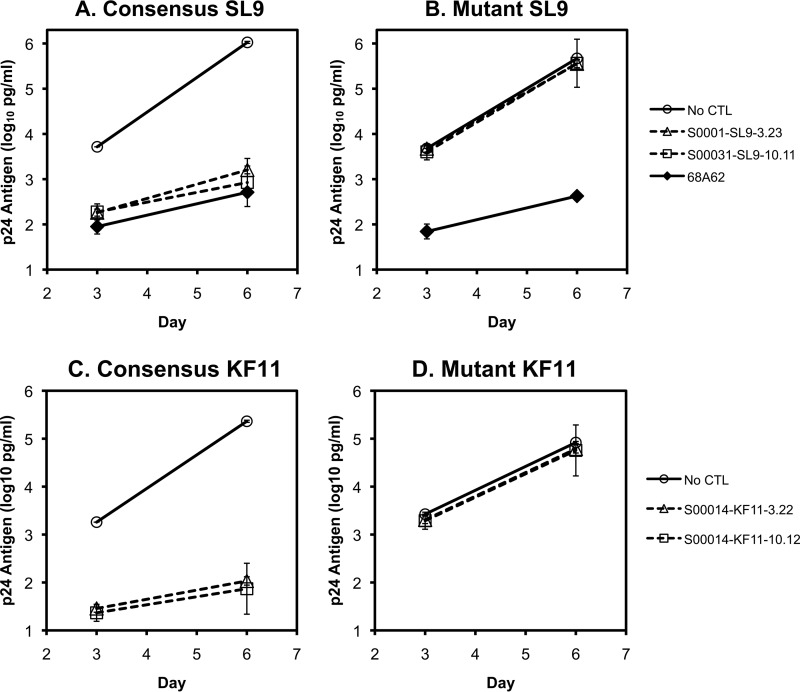
Cross-reactivity of HIV-1-specific CTL clones does not appreciably contribute to their antiviral activity.
To examine whether cross-reactivity contributes to the antiviral activity of HIV-1-specific CTLs, they were tested against HIV-1 containing escape mutations in the index epitope (Fig. 5). Three A*02-restricted clones targeting the SL9 epitope (Gag 77–85), and a control A*02-restricted clone not targeting SL9 (targeting IV9; Pol 464–472) were screened for suppression of viral replication against HIV-1 NL4-3 engineered to contain the consensus SL9 sequence SLYNTVATL or a nonrecognized sequence, SLYNLVAVL, as previously described. HIV-1 with the consensus SL9 epitope was efficiently suppressed by all clones (Fig. 5A), but despite the observed cross-recognition of multiple A*02 HIV-1 epitopes (Fig. 1 to 3), viral suppression was completely ablated by the nonrecognized SL9 epitope (Fig. 5B), while the virus remained sensitive to the control clone recognizing the IV9 epitope. Similarly, two clones targeting the B*57-restricted epitope KF11 (Gag 162–172) efficiently suppressed HIV-1 with the consensus epitope (Fig. 5C), but not a nonrecognized variant (Fig. 5D), despite observed cross-reactivity against other B*57-restricted HIV-1 epitopes (Fig. 2 and 3). These findings indicated that virus suppression is dependent on CTL recognition of the index epitope and that the lower-avidity cross-reactivity against other epitopes in HIV-1 does not contribute.
FIG 5.
Dependence of virus suppression on recognition of the index epitope. (A and B) HIV-1-infected T1 cells were cocultured with either of two CTL clones recognizing the SL9 index epitope (Gag 77–85) or a control clone recognizing the IV9 index epitope, and viral replication was monitored by quantitative p24 ELISA. (A) The infecting virus was a variant of NL4-3 altered to contain the consensus SL9 epitope sequence SLYNTVATL (and a substitution in Nef, M20A, to ablate HLA downregulation). (B) The infecting virus was identical except for mutation (underlined) of the SL9 epitope to the nonrecognized variant sequence SLYNLVAVL. The error bars represent standard deviations of triplicates. The data are representative of five independent experiments. (C and D) HIV-1-infected 1cc4.14 cells (expressing B*57) were cocultured with either of two CTL clones recognizing the KF11 index epitope (Gag 162–172), and viral replication was monitored by quantitative p24 ELISA. (C) The infecting virus was NL4-3, containing the consensus KF11 sequence KAFSPEVIPMF (and a substitution in Nef, M20A, to ablate HLA downregulation). (D) The infecting virus was identical except for mutation of the KF11 epitope to the nonrecognized variant sequence KGFNPEVIPMF. The error bars represent standard deviations of triplicates. The data are representative of nine independent experiments.
Cross-reactivity is detected by standard gamma interferon ELISpot assay.
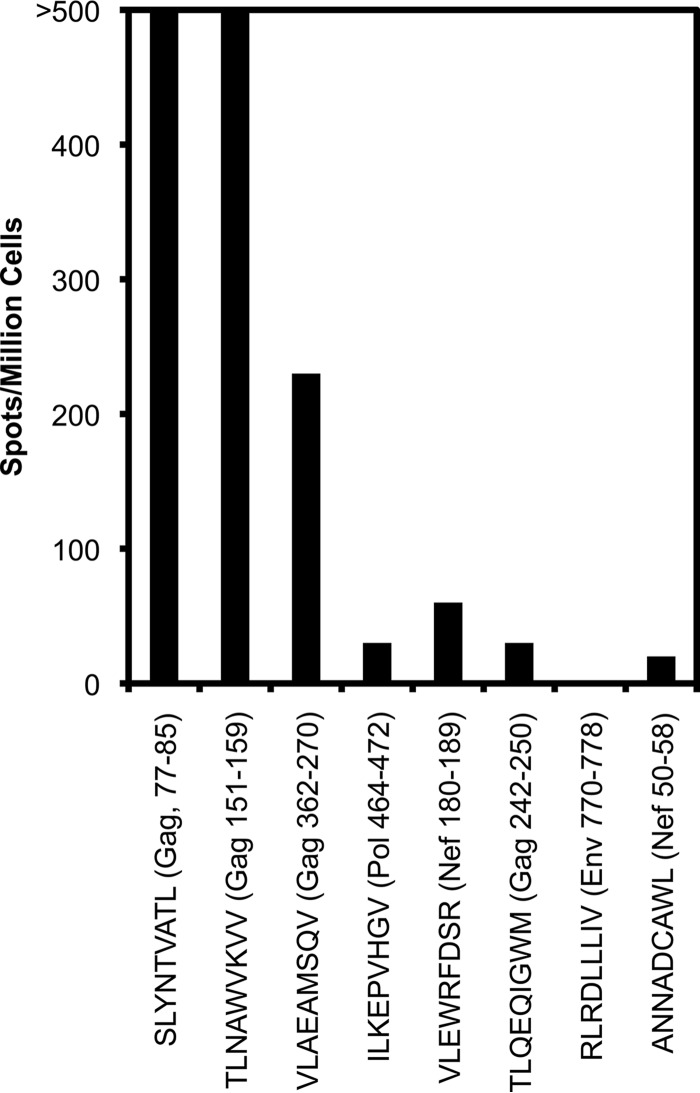
To assess whether cross-reactivity is also observed by another assay, a CTL clone recognizing the A*02-restricted Gag 77–85 epitope was exposed to the index epitope or other selected A*02-restricted epitopes and assessed for reactivity by gamma interferon enzyme-linked immunospot (ELISpot) assay. As observed above in killing assays (Fig. 1), reactivity against other nonindex epitopes was demonstrated (Fig. 6). In particular, the most strongly cross-reactive response against the TV9 epitope (Gag 151–159) in killing assays also showed high cross-reactivity by this assay. This result confirmed the potential for cross-reactive detection of CTL responses by ELISpot assay, as well as killing assays.
FIG 6.
Detection of cross-reactivity by gamma interferon ELISpot assay. The CTL clone S00001-SL9-3.23 recognizing the A*02-restricted minimal epitope SLYNTVATL (Gag 77–85:, SL9) was screened against seven other known A*02-restricted HIV-1 minimal epitopes (Table 2) in a standard ELISpot assay. Background-subtracted counts are plotted. The data are representative of two independent experiments.
DISCUSSION
Defining cross-reactivity as significant killing of target cells labeled with excess optimal epitopes (1 μg/ml), our data suggest that cross-reactivity within the HIV-1 proteome is unexpectedly frequent among HIV-1-specific CTLs. If it is roughly assumed that TCRs bind the central 6 amino acids between the MHC class I-binding anchor residues at positions 2 and 9 of a typical 9-amino-acid CTL epitope, it would suggest that there are about 206, or 64,000,000, possible MHC-binding variants. The estimate of a million recognized epitopes per TCR (1) would suggest that <2% of these variants should be recognized, on average, including minor variants of the index epitope that were not tested in this study. However, we observed several CTL clones that recognize multiple HIV-1 variants that are highly distinct from their index epitopes. It is unclear whether this reflects the fact that HIV-1-specific CTLs are highly promiscuous in general, or whether this promiscuity is limited to other HIV-1 epitopes. Although we did not systematically test non-HIV epitopes for cross-reactivity to distinguish these possibilities, we found that three A*02-restricted CTL clones (recognizing SL9 Gag 77–85 and IV9 Pol 464–472) cross-recognized the A*02-restricted immunodominant influenza virus matrix epitope GILGFVFTL (data not shown), suggesting a general phenomenon. This also agrees with a study where SL9-specific CTL clones from HIV-1-infected persons were screened against combinatorial peptide libraries, identifying frequent recognition of highly substituted epitope variants (e.g., sharing only 3 of 6 amino acids between the anchor residues), including some corresponding to epitopes from other, unrelated viruses (6). Why HIV-1-specific CTLs would be generally promiscuous is unclear; selection by a rapidly evolving quasispecies could be a mechanism, as proposed for broadly neutralizing antibodies.
Our data also highlight the somewhat ambiguous nature of defining and detecting cross-reactivity. While cross-reactivity is seen at relatively high concentrations of peptides, the functional avidity for these responses is relatively low compared to the index peptide. We have previously demonstrated that exogenously added epitope peptides at typical excess concentrations used to screen for CTL responses are supraphysiologic compared to the levels expressed on HIV-1-infected cells, which can lead to detection of responses that does not reflect recognition of infected cells (7, 8). Given our finding that the observed cross-reactivity does not mediate antiviral activity, it is a reflection of this phenomenon of detection that does not necessarily correspond to a physiologic function due to narrow avidity thresholds that must be exceeded for CTL recognition of HIV-1-infected cells (8). Given the limited sampling of HIV-1-specific CTL clones and HIV-1 epitopes, our data do not exclude the possibility that there are functionally significant instances of cross-reactivity against multiple HIV-1 epitopes that we did not detect. Another mechanistic possibility behind the observed cross-reactivity is that these low-avidity interactions could be a mechanism for persistence of HIV-1-specific CTLs, particularly after escape mutation in the index epitope, as low-level TCR interactions are believed to help drive homeostatic proliferation of memory cells (9).
A question raised by our data is whether the difference between the functional avidities of index versus cross-recognized epitopes for A*02 (consistently ∼100-fold or more) versus B*57 (consistently ∼3- to 10-fold) reflects a true difference between A*02 and B*57, which would be consistent with a prior report hypothesizing greater promiscuity for B*57-restricted HIV-1-specific CTLs (10). Counter to this would be the observation that some B*57-restricted CTLs (recognizing the epitope IW9, Gag 147–155) did not appear to be cross-reactive. These data and generalized conclusions are limited by the technical inability to screen against all possible epitopes with a large number of clones.
Finally, our data raise a caveat to quantifying CTL responses using minimal epitope peptides at typical concentrations used for screening in killing or ELISpot assays, which are excess. At these supraphysiologic concentrations, low-avidity cross-reactivity against other HIV-1 epitopes is frequently detected. It is therefore likely that responses against a particular epitope would be overestimated, including purely false-positive detection. However, the use of nonoptimal longer peptides, such as the overlapping 15-mer peptides offered by the NIH AIDS Reagent Repository, appears not to detect such cross-reactivity in ELISpot assays (data not shown), likely due to the relatively poor efficiency of such peptides compared to optimal minimal epitopes.
MATERIALS AND METHODS
HIV-1-permissive cell lines.
Cell lines included T1 cells (11) and 1cc4.14 cells generated by fusion of T1 cells with primary CD4+ T lymphocytes (12). They were maintained as previously described in RPMI 1640 medium supplemented with heat-inactivated fetal calf serum, penicillin-streptomycin, and l-glutamine (13, 14).
CTL clones.
HIV-1-specific CTL clones were derived by limiting-dilution cloning from peripheral blood mononuclear cells (PBMC) of persons with chronic HIV-1 infection, as previously described (13, 14). Clone 68A62 was the generous gift of Bruce D. Walker.
HIV-1.
HIV-1 molecular clone NL4-3 was modified to have the M20A substitution mutation in Nef (which ablates MHC class I downregulation) as previously described (15). This strain was further modified to have the specified mutations in the Gag epitope SL9 (Gag 77–85) or KF11 (Gag 162–172), and virus stocks whose titers were determined were produced by plasmid transfection of HEK 293T cells as previously described (12, 16).
Chromium release assays.
HIV-1-infected cells were assayed for susceptibility in chromium release assays as previously described (8, 13). In brief, target cells were 51Cr labeled (in the presence of synthetic epitope peptide [Sigma] as appropriate) and incubated with or without CTLs for 4 h at an effector-to-target cell ratio of 5:1 in 96-well U-bottom plates. Peptide labeling was performed at a concentration of 1 μg/ml for qualitative screening of cross-reactivity or titrated ranging from 10 μg/ml to 1 pg/ml for functional-avidity (SD50) determinations as previously described (8, 17). Supernatants were harvested for measurement of released 51Cr by microscintillation counting. Specific lysis was calculated by subtracting the control spontaneous release from the test release and dividing that number by the difference of the control maximum release minus the spontaneous release: specific lysis = (observed chromium release − spontaneous chromium release)/(maximal chromium release − spontaneous chromium release).
Virus suppression assays.
Virus suppression assays were performed in a 96-well plate format version (8, 15) of a previously described assay (14). Briefly, target cells were acutely infected with HIV-1 using a multiplicity of infection of 10−2 infectious dose/cell and then cocultured with CTL clones at an effector-to-target ratio of 0.25:1 (1.25 × 104 CTLs with 5 × 104 target cells per well) in triplicate wells. Viral replication was monitored by quantitative p24 enzyme-linked immunosorbent assay (ELISA) measurement of supernatant.
Gamma interferon ELISpot assays.
Gamma interferon ELISpot assays were performed as previously described (18) with the following modifications. In brief, each indicated peptide was added at a final concentration of 1 μg/ml to 105 CTL clones per well in precoated 96-well filter plates for overnight incubation. Final counts were background subtracted using means of negative-control wells with no peptide.
ACKNOWLEDGMENTS
We thank Mary Ann Hausner for technical assistance. Recombinant human interleukin-2 was provided by the NIH AIDS Reagent Repository.
This work was primarily supported by a grant from the AIDS Healthcare Foundation (O.O.Y.), with additional support from PHS grants DE025166 (O.O.Y.) and AI028697 (UCLA AIDS Institute and Center for AIDS Research), the James B. Pendleton Trust, and the McCarthy Foundation. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We declare that we have no financial conflicts of interest related to this work.
REFERENCES
- 1.Mason D. 1998. A very high level of crossreactivity is an essential feature of the T-cell receptor. Immunol Today 19:395–404. doi: 10.1016/S0167-5699(98)01299-7. [DOI] [PubMed] [Google Scholar]
- 2.Cornberg M, Clute SC, Watkin LB, Saccoccio FM, Kim SK, Naumov YN, Brehm MA, Aslan N, Welsh RM, Selin LK. 2010. CD8 T cell cross-reactivity networks mediate heterologous immunity in human EBV and murine vaccinia virus infections. J Immunol 184:2825–2838. doi: 10.4049/jimmunol.0902168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pohlmeyer CW, Laskey SB, Beck SE, Xu DC, Capoferri AA, Garliss CC, May ME, Livingston A, Lichmira W, Moore RD, Leffell MS, Butler NJ, Thorne JE, Flynn JA, Siliciano RF, Blankson JN. 2018. Cross-reactive microbial peptides can modulate HIV-specific CD8+ T cell responses. PLoS One 13:e0192098. doi: 10.1371/journal.pone.0192098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vali B, Tohn R, Cohen MJ, Sakhdari A, Sheth PM, Yue FY, Wong D, Kovacs C, Kaul R, Ostrowski MA. 2011. Characterization of cross-reactive CD8+ T-cell recognition of HLA-A2-restricted HIV-Gag (SLYNTVATL) and HCV-NS5b (ALYDVVSKL) epitopes in individuals infected with human immunodeficiency and hepatitis C viruses. J Virol 85:254–263. doi: 10.1128/JVI.01743-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mason RD, Bowmer MI, Howley CM, Grant MD. 2005. Cross-reactive cytotoxic T lymphocytes against human immunodeficiency virus type 1 protease and gamma interferon-inducible protein 30. J Virol 79:5529–5536. doi: 10.1128/JVI.79.9.5529-5536.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boggiano C, Moya R, Pinilla C, Bihl F, Brander C, Sidney J, Sette A, Blondelle SE. 2005. Discovery and characterization of highly immunogenic and broadly recognized mimics of the HIV-1 CTL epitope Gag77-85. Eur J Immunol 35:1428–1437. doi: 10.1002/eji.200425903. [DOI] [PubMed] [Google Scholar]
- 7.Bennett MS, Ng HL, Ali A, Yang OO. 2008. Cross-clade detection of HIV-1-specific cytotoxic T lymphocytes does not reflect cross-clade antiviral activity. J Infect Dis 197:390–397. doi: 10.1086/525281. [DOI] [PubMed] [Google Scholar]
- 8.Bennett MS, Ng HL, Dagarag M, Ali A, Yang OO. 2007. Epitope-dependent avidity thresholds for cytotoxic T-lymphocyte clearance of virus-infected cells. J Virol 81:4973–4980. doi: 10.1128/JVI.02362-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morris GP, Allen PM. 2012. How the TCR balances sensitivity and specificity for the recognition of self and pathogens. Nat Immunol 13:121–128. doi: 10.1038/ni.2190. [DOI] [PubMed] [Google Scholar]
- 10.Kosmrlj A, Read EL, Qi Y, Allen TM, Altfeld M, Deeks SG, Pereyra F, Carrington M, Walker BD, Chakraborty AK. 2010. Effects of thymic selection of the T-cell repertoire on HLA class I-associated control of HIV infection. Nature 465:350–354. doi: 10.1038/nature08997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salter RD, Howell DN, Cresswell P. 1985. Genes regulating HLA class I antigen expression in T-B lymphoblast hybrids. Immunogenetics 21:235–246. doi: 10.1007/BF00375376. [DOI] [PubMed] [Google Scholar]
- 12.Balamurugan A, Ali A, Boucau J, Le Gall S, Ng HL, Yang OO. 2013. HIV-1 gag cytotoxic T lymphocyte epitopes vary in presentation kinetics relative to HLA class I downregulation. J Virol 87:8726–8734. doi: 10.1128/JVI.01040-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang OO, Kalams SA, Rosenzweig M, Trocha A, Jones N, Koziel M, Walker BD, Johnson RP. 1996. Efficient lysis of human immunodeficiency virus type 1-infected cells by cytotoxic T lymphocytes. J Virol 70:5799–5806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang OO, Kalams SA, Trocha A, Cao H, Luster A, Johnson RP, Walker BD. 1997. Suppression of human immunodeficiency virus type 1 replication by CD8+ cells: evidence for HLA class I-restricted triggering of cytolytic and noncytolytic mechanisms. J Virol 71:3120–3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adnan S, Balamurugan A, Trocha A, Bennett MS, Ng HL, Ali A, Brander C, Yang OO. 2006. Nef interference with HIV-1-specific CTL antiviral activity is epitope specific. Blood 108:3414–3419. doi: 10.1182/blood-2006-06-030668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bennett MS, Joseph A, Ng HL, Goldstein H, Yang OO. 2010. Fine-tuning of T-cell receptor avidity to increase HIV epitope variant recognition by cytotoxic T lymphocytes. AIDS 24:2619–2628. doi: 10.1097/QAD.0b013e32833f7b22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang OO, Sarkis PT, Trocha A, Kalams SA, Johnson RP, Walker BD. 2003. Impacts of avidity and specificity on the antiviral efficiency of HIV-1-specific CTL. J Immunol 171:3718–3724. doi: 10.4049/jimmunol.171.7.3718. [DOI] [PubMed] [Google Scholar]
- 18.Ibarrondo FJ, Anton PA, Fuerst M, Ng HL, Wong JT, Matud J, Elliott J, Shih R, Hausner MA, Price C, Hultin LE, Hultin PM, Jamieson BD, Yang OO. 2005. Parallel human immunodeficiency virus type 1-specific CD8+ T-lymphocyte responses in blood and mucosa during chronic infection. J Virol 79:4289–4297. doi: 10.1128/JVI.79.7.4289-4297.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]