A better understanding of the immune correlates of protection against HIV might facilitate the development of a prophylactic vaccine. Therefore, we investigated simian immunodeficiency virus (SIV) infection outcomes in rhesus macaques expressing the major histocompatibility complex class I allele Mamu-B*17. Approximately 21% of Mamu-B*17+ macaques spontaneously controlled chronic phase viremia after SIV infection, an effect that may involve CD8+ T cells targeting Mamu-B*17-restricted SIV epitopes. We vaccinated Mamu-B*17+ macaques with genes encoding immunodominant epitopes in Vif and Nef alone (group 1) or together with env (group 2). Although neither vaccine regimen prevented SIV infection, 5/8 group 2 vaccinees controlled viremia to below detection limits shortly after infection. This outcome, which was not observed in group 1, was associated with vaccine-induced, nonneutralizing Env-binding antibodies. Together, these findings suggest a limited contribution of Vif- and Nef-specific CD8+ T cells for virologic control in Mamu-B*17+ macaques and implicate anti-Env antibodies in containment of SIV infection.
KEYWORDS: AIDS, human immunodeficiency virus, simian immunodeficiency virus, vaccines
ABSTRACT
Certain major histocompatibility complex class I (MHC-I) alleles are associated with spontaneous control of viral replication in human immunodeficiency virus (HIV)-infected people and simian immunodeficiency virus (SIV)-infected rhesus macaques (RMs). These cases of “elite” control of HIV/SIV replication are often immune-mediated, thereby providing a framework for studying anti-lentiviral immunity. In this study, we examined how vaccination impacts SIV replication in RMs expressing the MHC-I allele Mamu-B*17. Approximately 21% of Mamu-B*17+ and 50% of Mamu-B*08+ RMs control chronic-phase viremia after SIVmac239 infection. Because CD8+ T cells targeting Mamu-B*08-restricted SIV epitopes have been implicated in virologic suppression in Mamu-B*08+ RMs, we investigated whether this might also be true for Mamu-B*17+ RMs. Two groups of Mamu-B*17+ RMs were vaccinated with genes encoding Mamu-B*17-restricted epitopes in Vif and Nef. These genes were delivered by themselves (group 1) or together with env (group 2). Group 3 included MHC-I-matched RMs and served as the control group. Surprisingly, the group 1 vaccine regimen had little effect on viral replication compared to group 3, suggesting that unlike Mamu-B*08+ RMs, preexisting SIV-specific CD8+ T cells alone do not facilitate long-term virologic suppression in Mamu-B*17+ RMs. Remarkably, however, 5/8 group 2 vaccinees controlled viremia to <15 viral RNA copies/ml soon after infection. No serological neutralizing activity against SIVmac239 was detected in group 2, although vaccine-elicited gp140-binding antibodies correlated inversely with nadir viral loads. Collectively, these data shed new light on the unique mechanism of elite control in Mamu-B*17+ RMs and implicate vaccine-induced, nonneutralizing anti-Env antibodies in the containment of immunodeficiency virus infection.
IMPORTANCE A better understanding of the immune correlates of protection against HIV might facilitate the development of a prophylactic vaccine. Therefore, we investigated simian immunodeficiency virus (SIV) infection outcomes in rhesus macaques expressing the major histocompatibility complex class I allele Mamu-B*17. Approximately 21% of Mamu-B*17+ macaques spontaneously controlled chronic phase viremia after SIV infection, an effect that may involve CD8+ T cells targeting Mamu-B*17-restricted SIV epitopes. We vaccinated Mamu-B*17+ macaques with genes encoding immunodominant epitopes in Vif and Nef alone (group 1) or together with env (group 2). Although neither vaccine regimen prevented SIV infection, 5/8 group 2 vaccinees controlled viremia to below detection limits shortly after infection. This outcome, which was not observed in group 1, was associated with vaccine-induced, nonneutralizing Env-binding antibodies. Together, these findings suggest a limited contribution of Vif- and Nef-specific CD8+ T cells for virologic control in Mamu-B*17+ macaques and implicate anti-Env antibodies in containment of SIV infection.
INTRODUCTION
Despite improvements in prevention strategies and antiretroviral therapy (ART) coverage, thousands of new human immunodeficiency virus (HIV) infections are still occurring every day, highlighting the need for an effective HIV vaccine (1). Eliciting robust protection against HIV infection has not been straightforward, as seen by the failure of most HIV vaccines tested in humans to date (2–6). Although the RV144 trial remains the only report of vaccine-mediated reduction in HIV infection rates (7), the observed results were modest and short-lived and continue to be contested (8, 9). The unsatisfactory performance of mainstream HIV vaccine regimens underscores the need to better understand the nature of effective anti-lentiviral immune responses.
Elite controllers (ECs) are a small fraction of HIV-infected individuals who spontaneously control chronic-phase viremia in the absence of ART (10). Certain major histocompatibility complex class I (MHC-I) alleles, such as HLA-B*27 and HLA-B*57, are associated with elite control of HIV-1 infection (11), implying an immunological basis for this phenotype. Indeed, CD8+ T cells targeting viral epitopes restricted by “protective” MHC-I molecules and natural killer (NK) cells are widely thought to be important mediators of antiviral activity in ECs (12, 13). The study of ECs thus provides a useful framework to investigate the basis for immune containment of lentivirus replication. Similar to the case with human ECs, certain rhesus macaque (RM) MHC-I alleles are also associated with elite control of simian immunodeficiency virus (SIV) infection. Indeed, approximately 21% of unvaccinated RMs expressing Mamu-B*17 control chronic-phase viral replication after infection with SIVmac239 (14). The incidence of elite control in Mamu-B*08+ RMs is higher, reaching 50% of infected animals (15). Curiously, the peptide binding motifs of the Mamu-B*08 and Mamu-B*17 molecules resemble those of HLA-B*27 and HLA-B*57, respectively (16, 17). This similarity is not explained by sequence homology between the human and RM MHC-I alleles, thereby implicating the presented peptide as an important determinant of elite control.
The immunodominant SIV epitopes restricted by Mamu-B*08 and Mamu-B*17 have a common feature, that is, their location in the accessory proteins Vif and Nef (18–20). While CD8+ T-cell responses targeting these Vif and Nef epitopes are crucial for virologic control in Mamu-B*08+ RMs (21), it is not clear to what extent Vif- and Nef-specific CD8+ T cells contribute to the EC phenotype of Mamu-B*17+ RMs. We set out to clarify this issue by conducting an SIV vaccine trial in Mamu-B*17+ RMs. We hypothesized that vaccine-induced CD8+ T cells targeting the immunodominant Mamu-B*17-restricted Vif HW8 (amino acids 66 to 73) and Nef IW9 (amino acids 165 to 173) epitopes would increase the incidence of elite control in Mamu-B*17+ RMs following infection with SIVmac239. Because vaccine-elicited anti-Env antibodies (Abs) have been linked to delayed acquisition of immunodeficiency virus infection following repeated mucosal challenges (22–24), we also evaluated whether these humoral responses would increase the protective efficacy of Vif HW8- and Nef IW9-specific CD8+ T cells induced by vaccination. To this end, we used a heterologous prime/boost/boost/boost/boost (PBBBB) immunization regimen to vaccinate two groups of Mamu-B*17+ RMs with genes encoding the Vif HW8 and Nef IW9 epitopes. These epitopes were delivered by themselves (group 1) or together with env (group 2). As a result, vaccinees in both groups mounted Vif- and Nef-specific CD8+ T cells but only the ones in group 2 developed Env-specific Abs. We assessed the efficacies of both regimens by repeatedly challenging vaccinees along with sham-vaccinated MHC-I-matched control RMs (group 3) intrarectally (i.r.) with a marginal dose of SIVmac239. The challenge outcomes varied greatly between groups 1 and 2, thereby revealing important aspects of immune containment of lentivirus replication. Here we discuss the relevance of these findings for HIV vaccine development and for understanding the basis of elite control of SIV replication in Mamu-B*17+ RMs.
RESULTS
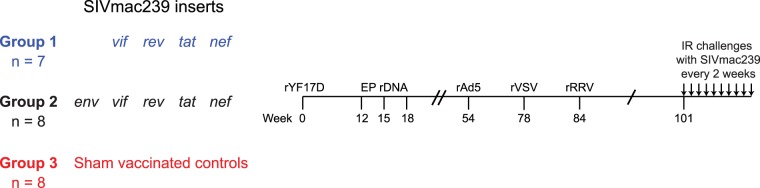
Twenty-three RMs expressing the MHC-I allele Mamu-B*17 were used in this study. These animals were divided among three groups, depending on which immunogens they received. RMs in group 1 (n = 7) were vaccinated with genes encoding the immunodominant Vif HW8 and Nef IW9 epitopes, whereas those in group 2 (n = 8) received the same inserts with the addition of env (Fig. 1). The SIV sequences were delivered by a recombinant yellow fever virus 17D (rYF17D) prime followed by three boosts with recombinant DNA (rDNA) plasmids delivered by intramuscular electroporation (EP rDNA). Subsequently, vaccinees in groups 1 and 2 were boosted once with each of the following recombinant viral vectors: adenovirus type 5 (rAd5), vesicular stomatitis virus (rVSV), and rhesus monkey rhadinovirus (rRRV) (Fig. 1). We used this PBBBB vaccine regimen because recurrent antigen stimulation is thought to facilitate the induction of effective cellular and humoral immune responses against lentiviruses (25, 26). It should be noted that some of the vaccine vectors employed in this study included segments of vif and nef, or full-length vif and nef fused with other genes, such as tat and rev (see Materials and Methods). As a result, RMs in groups 1 and 2 also developed cellular immune responses against Tat and Rev. Additionally, some of the vaccine-encoded immunogens also included the subdominant Mamu-B*17-restricted epitopes Nef MW9 (amino acids 195 to 203) and, in the case of group 2, Env FW9 (amino acids 830 to 838). Finally, eight Mamu-B17+ RMs were immunized with vectors encoding irrelevant antigens or lacking any inserts (“empty” vectors) and served as the controls for this experiment (group 3) (Fig. 1).
FIG 1.
Experimental design. Twenty-three Mamu-B*17+ RMs were vaccinated with an rYF17D/EP rDNA/rAd5/rVSV/rRRV regimen and divided into three groups depending on which vaccine inserts they received. Animals in group 1 (n = 7) were vaccinated with vif and nef sequences. Animals in group 2 (n = 8) were vaccinated with the same SIV inserts plus env. RMs in group 3 were sham vaccinated with vectors lacking SIV genes or expressing irrelevant inserts and served as the control group for this experiment. At study week 101, vaccine efficacy was assessed by subjecting all animals to repeated intrarectal (i.r.) challenges with a marginal dose of SIVmac239 (200 TCID50) every 2 weeks.
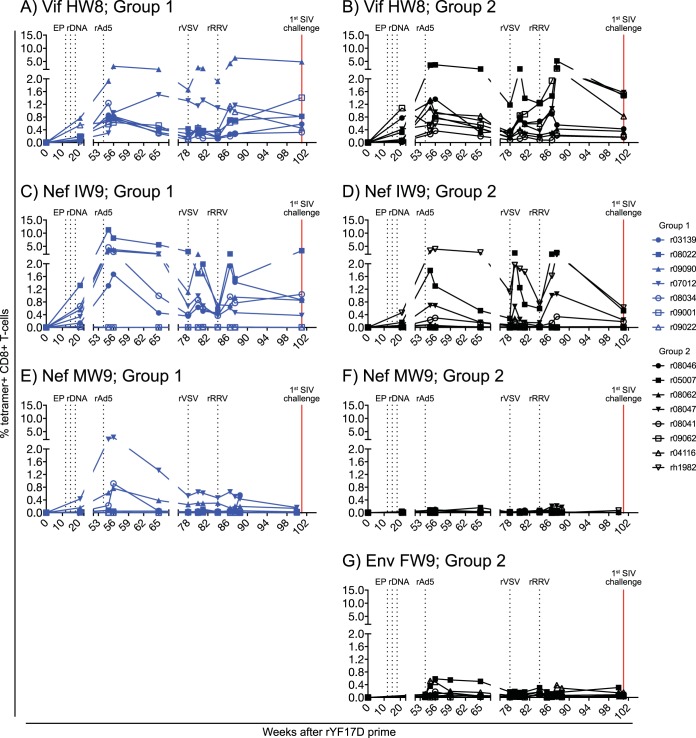
We monitored vaccine-induced CD8+ T-cell responses against Mamu-B*17-restricted epitopes by staining peripheral blood mononuclear cells (PBMC) with fluorochrome-labeled MHC-I tetramers. This analysis revealed that the PBBBB regimen generated high-frequency SIV-specific CD8+ T-cell responses in the majority of the group 1 and group 2 vaccinees (Fig. 2). Consistent with the immunodominance of Vif HW8 and Nef IW9 during SIV infection (20), vaccine-elicited CD8+ T cells in groups 1 and 2 were primarily directed against these two epitopes (Fig. 2A to D). Curiously, while Nef MW9-specific CD8+ T cells were undetectable or at borderline levels in all group 2 vaccinees, low to modest frequencies of these responses were observed in several RMs in group 1 (Fig. 2E and F). A few macaques in group 2 developed Env FW9-specific CD8+ T cells, but these responses remained scarce throughout the vaccine phase (Fig. 2G).
FIG 2.
Development of vaccine-induced CD8+ T-cell responses against Mamu-B*17-restricted SIV epitopes in groups 1 and 2. Fluorochrome-labeled Mamu-B*17 tetramers folded with peptides corresponding to SIV epitopes were used to track vaccine-elicited SIV-specific CD8+ T cells in PBMC from RMs in group 1 (A, C, and E) and group 2 (B, D, F, and G). The percentages of live tetramer+ CD8+ T cells specific for Vif HW8 (A and B), Nef IW9 (C and D), Nef MW9 (E and F), and Env FW9 (G) are shown at multiple time points throughout the vaccine phase. No data for Env FW9-specific CD8+ T cells are available for the group 1 RMs because those animals were not vaccinated with env. The times of each vaccination (black vertical dotted lines) and the day of the first i.r. SIV challenge (red vertical solid line) are indicated in each graph. RMs in groups 1 and 2 are color-coded in blue and black, respectively.
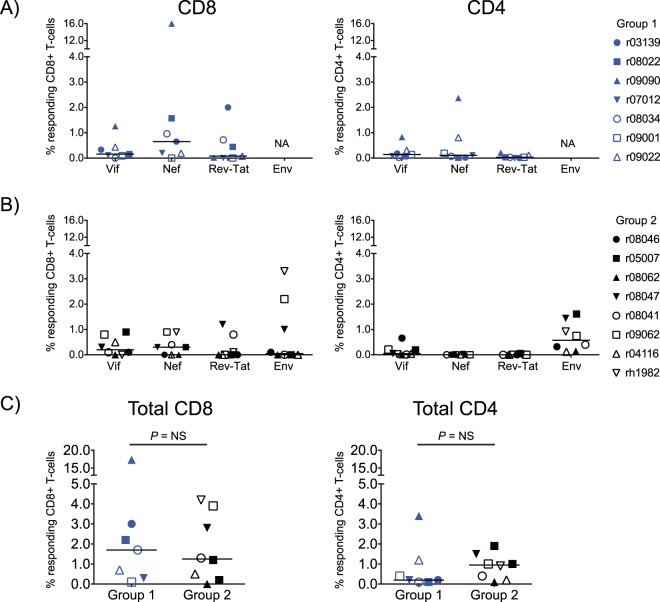
We also determined the magnitude of vaccine-induced SIV-specific T-cell responses in groups 1 and 2 by performing intracellular cytokine staining (ICS) in PBMC at the time of the first i.r. SIV challenge (Fig. 3). In accordance with the MHC-I tetramer analysis, vaccine-induced CD8+ T-cell responses in groups 1 and 2 were mainly directed against Vif and Nef, although a few animals in each group developed responses against Rev, Tat, and Env (group 2 only) as well (Fig. 3A and B). Vaccine-elicited CD4+ T-cell responses in group 2 were detected at higher frequencies than those in group 1 and focused primarily on Env (Fig. 3A to C). There was no significant difference in the total magnitude of vaccine-elicited SIV-specific CD8+ or CD4+ T-cell responses between groups 1 and 2 (Fig. 3C).
FIG 3.
Vaccine-induced SIV-specific CD4+ and CD8+ T-cell responses in groups 1 and 2 at the time of the first SIV challenge. CD8+ and CD4+ T-cell responses were measured in PBMC by ICS using pools of peptides (15-mers overlapping by 11 amino acids) spanning the appropriate SIVmac239 proteins. Peptides covering the Rev and Tat proteins were tested together. The percentages of responding CD4+ or CD8+ T cells displayed in all panels were calculated by adding the frequencies of positive responses producing any combination of three immunological functions (IFN-γ, TNF-α, and CD107a). The magnitude and specificity of vaccine-induced CD8+ (left panels) and CD4+ (right panels) T-cell responses are shown for group 1 (A) and group 2 (B). (C) Comparison of the total magnitude of vaccine-elicited SIV-specific CD8+ (left) and CD4+ (right) between groups 1 and 2. The Mann-Whitney U test was used for these comparisons, and no statistically significant difference was found. RMs in groups 1 and 2 are color-coded in blue and black, respectively, and each symbol corresponds to one vaccinee. Lines represent medians. NA, not applicable; NS, not significant.
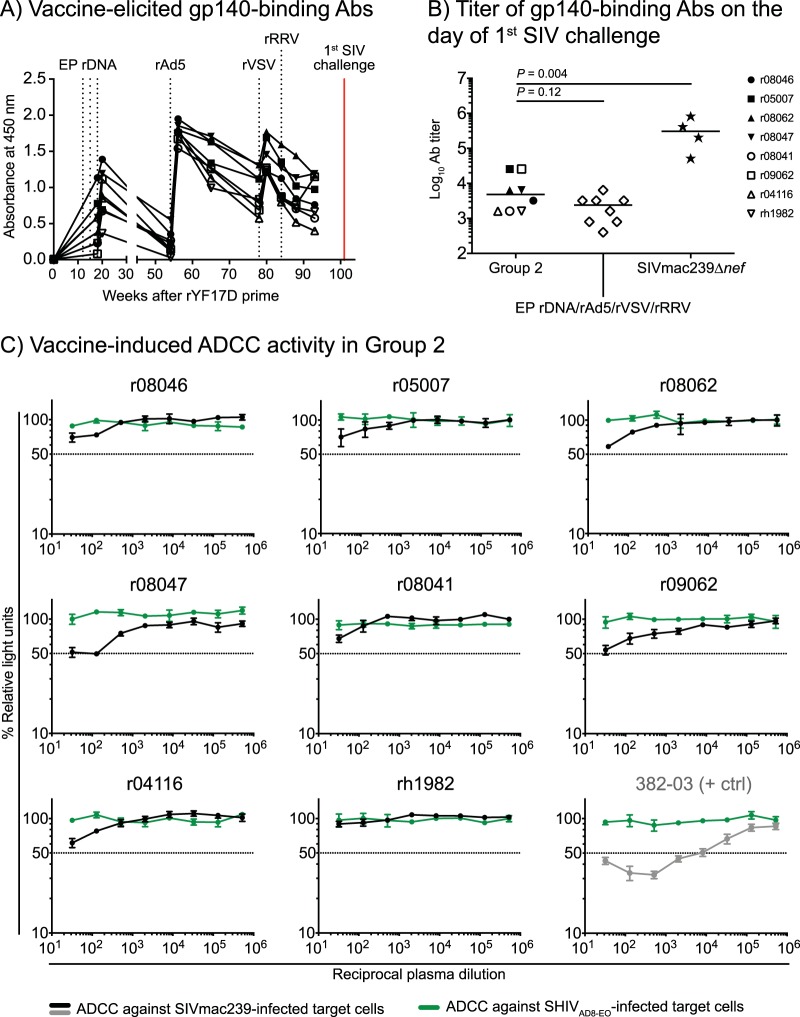
Vaccine-elicited Env-specific humoral responses in group 2 were also evaluated at multiple time points during the vaccine phase. A longitudinal analysis of gp140-binding Abs showed substantial increases in these responses after the EP rDNA and rAd5 vaccinations (Fig. 4A). Subsequent boosting with rVSV, but not with rRRV, resulted in a modest rise in these responses (Fig. 4A). We also quantified vaccine-elicited gp140-binding Abs on the day of the 1st SIV challenge (Fig. 4B). The median endpoint titer of gp140-binding Abs in group 2 was 4,800 (Fig. 4B), a value that was twice as high as that induced by an EP rDNA/rAd5/rVSV/rRRV vaccine regimen recently tested by our group (27). However, these responses were still about 100-fold lower than those generated by live attenuated SIV vaccination (Fig. 4B). Low levels of NK cell-mediated, Ab-dependent cellular cytotoxicity (ADCC) against SIVmac239-infected cells were also detected in plasma from r08047 and r09062 collected on the day of the 1st SIV challenge, but ADCC activity was either absent or at borderline levels in the other animals (Fig. 4C). No neutralizing antibodies (nAbs) against SIVmac239 were detected on the day of the 1st SIV challenge or after SIV infection (because none of the samples resulted in a 50% reduction in SIV infection in vitro at the lowest reciprocal serum dilution [1:20], the 50% inhibitory dose for each animal was considered <20).
FIG 4.
Vaccine-induced Env-specific humoral immune responses in group 2. (A) Env-binding antibodies (Abs) were measured by ELISA using plate-bound gp140 at multiple time points during the vaccine phase. Straight 1:200 dilutions of plasma from each RM in group 2 were used for this analysis. (B) Log-transformed endpoint titers of vaccine-induced gp140-binding Abs in sera from the group 2 vaccinees collected at the time of the first SIV challenge. As a reference, these values were plotted alongside the endpoint titers of gp140-binding Abs in four RMs that had been infected with SIVmac239Δnef for 28 weeks as part of a previous experiment (70). The endpoint titers of gp140-binding Abs in macaques vaccinated with an EP rDNA/rAd5/rVSV/rRRV regimen encoding env, gag, vif, tat, rev, and nef are also shown (27). (C) Ab-dependent cellular cytotoxicity (ADCC) activity was screened against SIVmac239-infected target cells using plasma collected from the group 2 vaccinees at the time of the first i.r. SIV challenge (black lines). SHIVAD8-EO-infected target cells were used as internal controls (green lines) for nonspecific killing. The decrease in relative light units (RLU) indicates the loss of virus-infected cells in the presence of an NK cell line during the duration of the assay. Dotted lines indicate 50% activity. Plasma from an SIV-infected RM (382-03) with a defined ADCC titer against SIVmac239-infected cells was used as a positive control for these measurements.
To assess vaccine efficacy, all RMs in groups 1 to 3 were subjected to repeated i.r. challenges with a marginal dose of SIVmac239 every 2 weeks, starting at study week 101 (Fig. 1). The challenge inoculum consisted of 200 50% tissue culture infective doses (TCID50) of an in vivo-titrated SIVmac239 stock. As a reference, the challenge dose employed in this study typically infects 80% of SIV naive RMs after six i.r. exposures. RMs in groups 1 to 3 became infected at similar rates (Fig. 5), indicating that vaccine-induced immune responses did not block acquisition of SIVmac239 infection.
FIG 5.
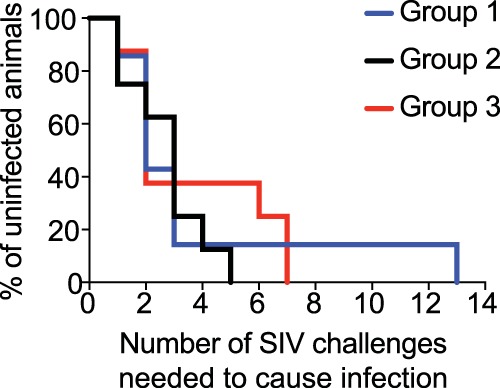
Kaplan-Meier rate of infection after repeated i.r. challenges with SIVmac239. Macaques in groups 1 to 3 were inoculated intrarectally with 200 TCID50 of SIVmac239 every other week. The rate of SIV infection in groups 1 and 2 was not significantly different than that of group 3. The P value for the comparison of group 1 versus group 3 was 0.96. The P value for the comparison of group 2 versus group 3 was 0.28.
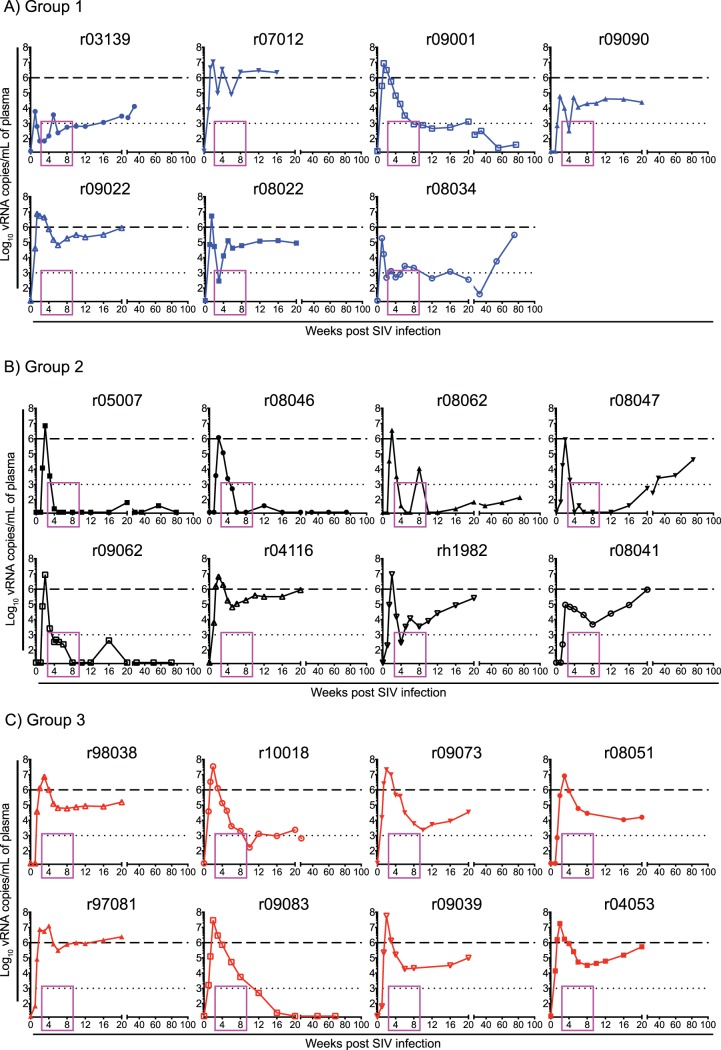
Contrary to our expectations based on control of SIVmac239 replication in Mamu-B*08+ RMs vaccinated with Vif and Nef (21), vaccine-induced CD8+ T-cell responses targeting the immunodominant Mamu-B*17-restricted Vif HW8 and Nef IW9 epitopes resulted in only a modicum of virologic control in group 1 (Fig. 6A). Although two vaccinees (r09001 and r08034) fared well in the chronic phase, none of the group 1 RMs suppressed viremia to <15 viral RNA (vRNA) copies/ml and the latter RM lost control of viral replication after week 32 postinfection (p.i.) (Fig. 6A). The number of infected monkeys with chronic-phase viral loads (VLs) below 1,000 vRNA copies/ml, even if transiently as in the case of r08034, was similar to that in group 3, where r10018 developed a viral set point of approximately 1,000 vRNA copies/ml, while r09083 controlled viral replication to <15 vRNA copies/ml at week 20 p.i. (Fig. 6A and C).
FIG 6.
Plasma viral RNA levels after SIVmac239 infection. Viral load (VL) traces for individual animals in group 1 (A), group 2 (B), and group 3 (C). VLs were log transformed and correspond to the number of vRNA copies/milliliter of plasma. The dotted lines in all the graphs are for reference only and indicate a VL of 103 vRNA copies/ml. The dashed lines are also for reference only and denote a VL of 106 vRNA copies/ml. The pink rectangle in each graph frames the interval during which five group 2 vaccinees controlled viremia to <15 vRNA copies/ml. Groups 1, 2, and 3 are color-coded in blue, black, and red, respectively.
Surprisingly, however, group 2 exhibited an entirely unexpected outcome after infection. Despite experiencing peak VLs in excess of 8.5 × 105 vRNA copies/ml of plasma, five group 2 vaccinees (r05007, r08046, r08062, r08047, and r09062) controlled viral replication to <15 vRNA copies/ml by weeks 4 to 8 p.i. (Fig. 6B). This corresponded to an average reduction in VLs of 5.3 logs, which was effected within only 2 to 6 weeks after peak viremia. Four of the five group 2 vaccinees that showed early viral suppression maintained control of viral replication throughout the chronic phase. RM r08047 was the exception, as its VLs progressively increased after week 20 p.i. (Fig. 6B). Of note, after decreasing viremia to <15 vRNA copies/ml, the group 2 controllers experienced occasional VLs blips in the ensuing weeks, similar to those reported by Hansen et al. (28) and Winstone et al. (29). RM r08062 began to deviate from this pattern at week 16 p.i., after which viremia became persistent, albeit at low levels (140 vRNA copies/ml at week 75 p.i. [Fig. 6B]).
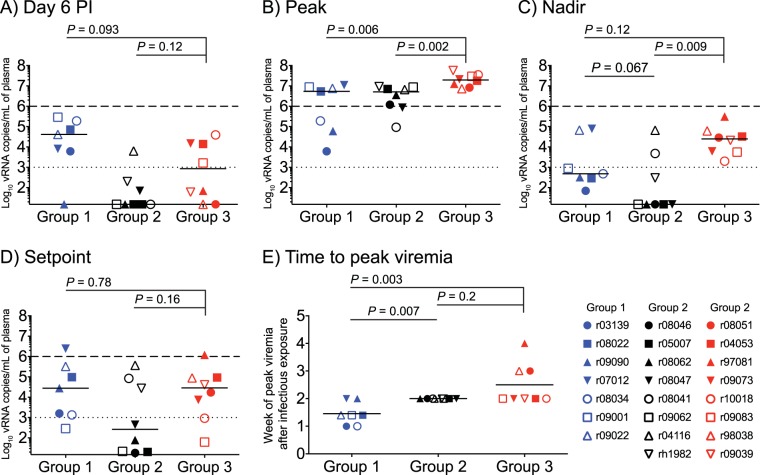
We compared VLs among groups 1 to 3 and made several observations about postinfection viral control in these groups. First, five group 2 vaccinees had VLs that were either at or below the limit of detection (15 vRNA copies/ml) on day 6 p.i. (Fig. 7A), implying early control of viral replication. By comparison, only one vaccinee in group 1 (r09090) and two monkeys in group 3 (r08051 and r09038) had VLs below 15 vRNA copies/ml at this time point (Fig. 7A). These differences were not, however, statistically significant (Fig. 7A). Second, peak VLs were significantly lower in groups 1 and 2 than in group 3, although these reductions were modest (Fig. 7B). Curiously, the two RMs (r03139 and r09090) with the lowest peak VLs were in group 1 (Fig. 7B). Third, although set point VLs were not significantly decreased in either vaccinated group, group 2 (but not group 1) exhibited a significant reduction in nadir VLs, consistent with stringent control of postacute viremia being the most distinctive feature of group 2 (Fig. 7C and D). Fourth, it took a median of 1.46 weeks for the group 1 macaques to experience peak viremia after SIV infection, compared to medians of 2 and 2.5 weeks in groups 2 and 3, respectively (Fig. 7E). This difference between group 1 and the other groups was statistically significant, although it is not clear why viral replication peaked earlier in the group 1 vaccinees. There was no statistically significant difference in the times to peak VLs between groups 2 and 3, despite a few outliers in the latter group (Fig. 7E).
FIG 7.
SIV plasma viral load comparisons among groups 1, 2, and 3. (A) Plasma VLs measured on day 6 postinfection (p.i.). (B) Peak VLs. (C) Nadir VLs. (D) Setpoint VLs, calculated as the geometric mean of VLs measured between week 8 p.i. and the last chronic phase time point available. (E) Time to peak viremia, determined as the week p.i. when each animal experienced its peak VL. The dotted lines in panels A to D are for reference only and indicate a VL of 103 vRNA copies/ml. The dashed lines in panels A to D are also for reference only and denote a VL of 106 vRNA copies/ml. Lines represent medians and P values were calculated using the Mann-Whitney U test. Groups 1, 2, and 3 are color-coded in blue, black, and red, respectively, and each symbol corresponds to one vaccinee.
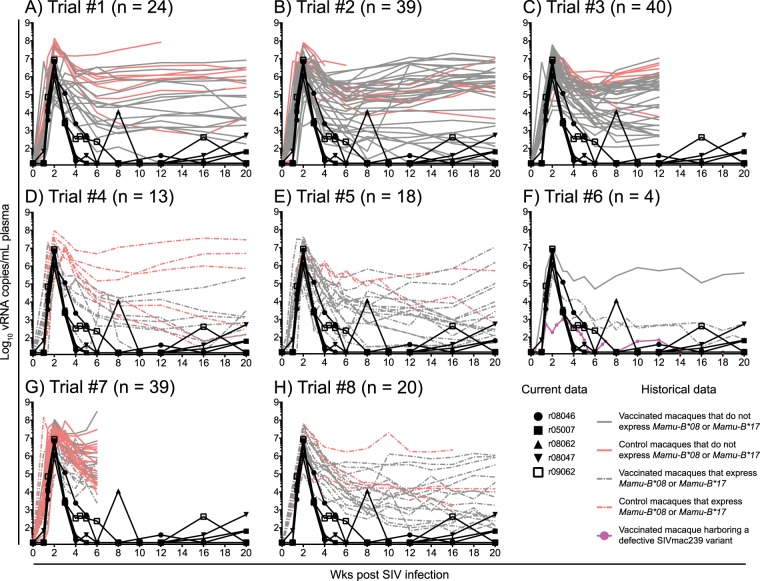
It is important to emphasize that the level of control of chronic-phase SIVmac239 replication manifested by the five controller group 2 vaccinees is exceedingly rare. In fact, of the 197 RMs that have been rectally infected with SIVmac239 as part of eight previous and ongoing SIV vaccine trials conducted by our group, none exhibited the 5.3-log reduction in VLs within 2 to 6 weeks of peak viremia observed in the five group 2 controllers (Fig. 8). Of note, this cumulative past experience also included RMs that expressed MHC-I alleles associated with elite control of SIV infection (Fig. 8D to H). These animals were vaccinated with various heterologous prime boost regimens with vaccine strains carrying vif, nef, rev, and tat (Fig. 8D), nef only (Fig. 8E), rev, tat, and nef (Fig. 8F), and vif only (Fig. 8G). The Mamu-B*08+ vaccinees in Fig. 8H were vaccinated with vif and nef minigenes containing Mamu-B*08-restricted epitopes or with minigenes of other regions of the SIV proteome that lack CD8+ T-cell determinants restricted by Mamu-B*08. As expected, a fraction of these SIVmac239-infected RMs became ECs, but even in these cases, viremia was rarely reduced to <15 vRNA copies/ml by weeks 4 to 8 p.i. (Fig. 8D to H). These different kinetics of virologic control after SIVmac239 infection underscore the uniqueness of the five group 2 controllers.
FIG 8.
Viral loads in the five group 2 controllers compared to those in RMs historically infected with SIVmac239. The outcome of SIVmac239 infection in the five group 2 vaccinees that manifested early control of viral replication was compared to those of 197 RMs that were rectally infected with SIVmac239 as part of eight SIV vaccine trials conducted by our group. The historical VLs for each independent SIV vaccine trial are plotted in panels A to H and include both vaccinated (gray lines) and control (salmon lines) RMs. VLs from RMs that expressed MHC-I alleles associated with elite control of SIV infection (i.e., Mamu-B*08 and Mamu-B*17) are shown in dashed lines. VLs from RMs that did not express these protective MHC-I alleles are shown in solid lines. To better visualize the early control of viral replication manifested by the five group 2 vaccinees, their VLs (black lines) were also plotted in each panel. Additionally, only the first 20 weeks of SIV infection are shown in each graph. (A) In trial 1, 24 RMs were vaccinated with env, gag, vif, rev, tat, and nef delivered by either rRRV alone or rRRV followed by two boosts with rAd5 and rVSV. Eight RMs served as the controls for the challenge phase (M. A. Martins, D. C. Tully, N. Pedreño-Lopez, B. von Bredow, M. G. Pauthner, Y. C. Shin, M. Yuan, N. S. Lima, D. J. Bean, L. Gonzalez-Nieto, A. Domingues, M. J. Gutman, H. S. Maxwell, D. M. Magnani, M. J. Ricciardi, V. K. Bailey, J. D. Altman, D. R. Burton, K. Ejima, D. B. Allison, D. T. Evans, E. G. Rakasz, C. L. Parks, M. C. Bonaldo, S. Capuano III, J. D. Lifson, R. C. Desrosiers, T. M. Allen, D. I. Watkins, unpublished data). (B) The details of this experiment were published recently (27). Briefly, 32 RMs were vaccinated with an EP rDNA/rAd5/rVSV/rRRV regimen encoding four different sets of SIV inserts. Eight RMs served as the controls for this experiment. One vaccinee did not get infected, so VL traces for 39 RMs are shown in the graph. (C) The details of this experiment were published recently (71). Briefly, four different mixed-modality vaccine regimens were used to deliver minigenes of SIV gag, vif, and nef to 32 RMs. Eight RMs served as the controls for this experiment. (D) Ten Mamu-B*08+ RMs were vaccinated with a rAd5/rVSV/rRRV regimen including vif, rev, tat, and nef, and six MHC-I-matched RMs served as the controls for the challenge phase (Martins et al., unpublished). Three vaccinees did not acquire SIV infection in this experiment, so VL traces for 13 RMs are shown in the graph. (E) The details of this experiment were published recently (72). Briefly, 16 Mamu-B*08+ RMs were vaccinated with an EP rDNA/rYF17D/rAd5 regimen containing nef, and two MHC-I-matched macaques served as the controls for the challenge phase. (F) The details of this experiment were published recently (30). Four RMs, two of which were Mamu-B*08+, were vaccinated with an EP rDNA/rYF17D/rRRV regimen containing either gag or nef. For unknown reasons, the animal highlighted in pink harbored a nef deletion SIV variant as early as week 2 p.i. The replicative fitness cost imposed by this nef deletion likely underlies the stringent control of viral replication manifested by this animal. (G) Twenty RMs were vaccinated with an EP rDNA/recombinant vaccinia/rVSV/rAd5/rRRV regimen encoding vif only. Twenty MHC-I-matched RMs served as the controls for the challenge phase. Eight vaccinees and nine control RMs expressed either Mamu-B*08 or Mamu-B*17 (Martins et al., unpublished). One vaccinee did not acquire SIV infection, so VL traces for 39 RMs are shown in the graph. (H) The details of this experiment were published elsewhere (21). Briefly, 16 Mamu-B*08+ RMs were vaccinated with an rYF17D/rAd5 regimen including either vif and nef minigenes containing Mamu-B*08-restricted epitopes or regions of the SIV proteome lacking epitopes restricted by Mamu-B*08. Four MHC-I-matched RMs served as the controls for the challenge phase.
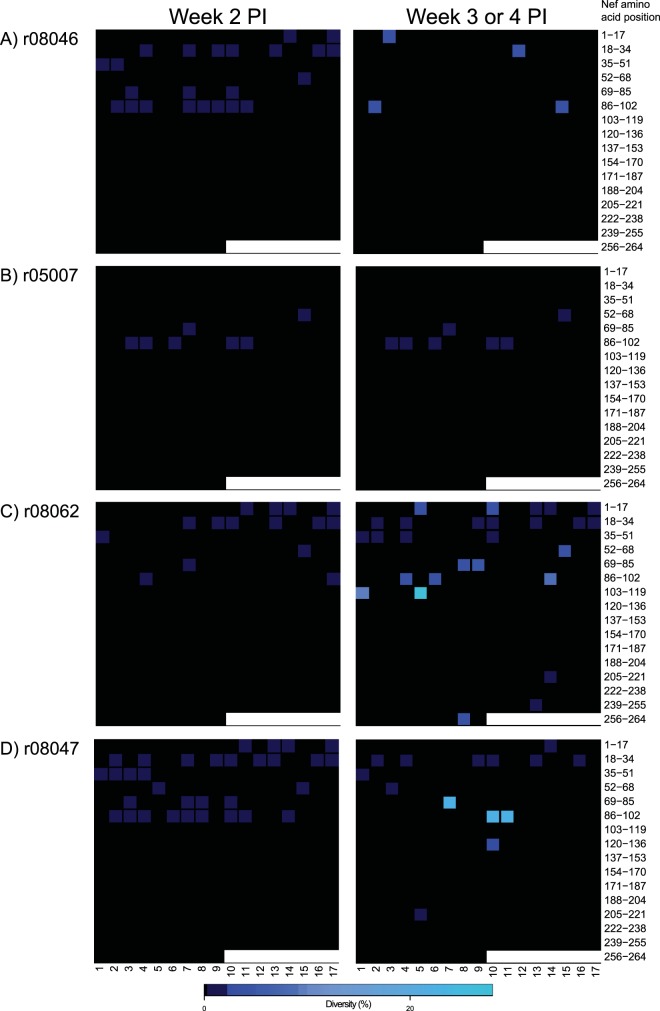
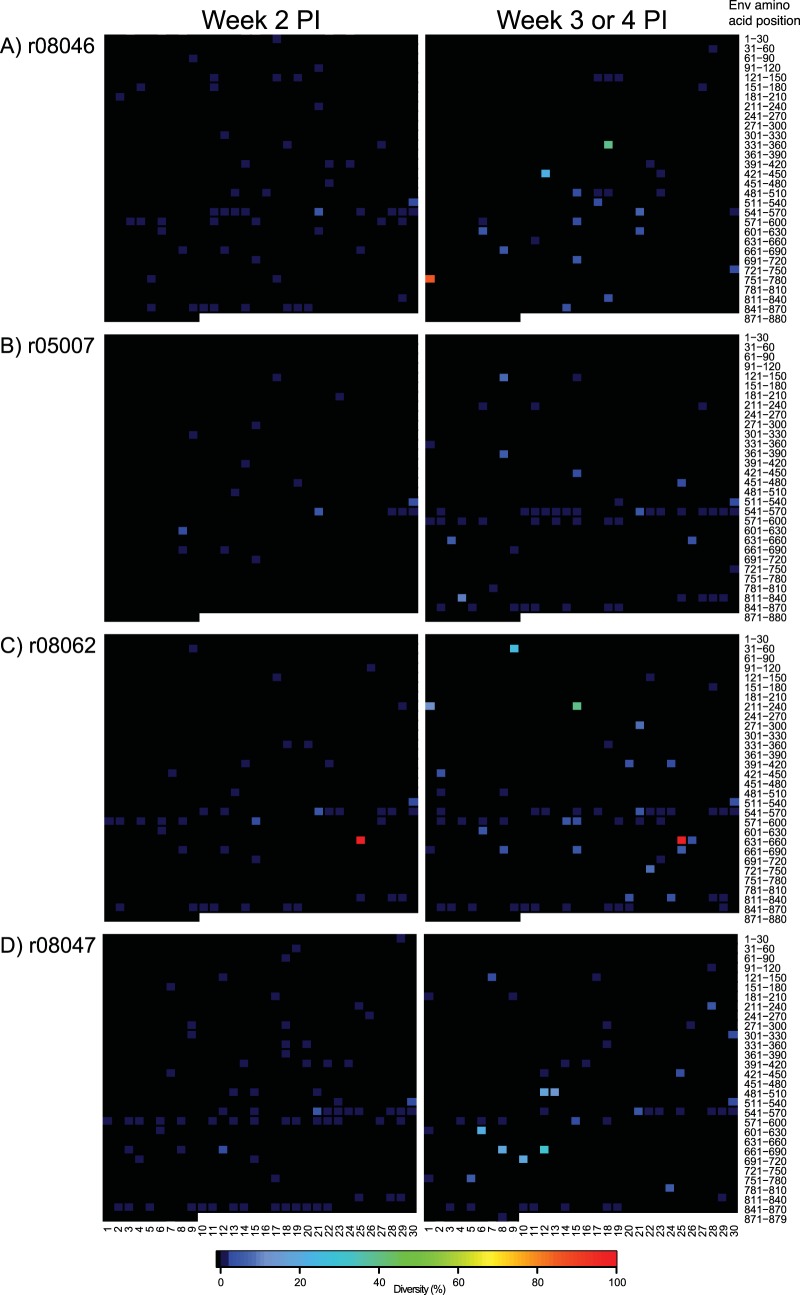
We have recently reported an extraordinary case of control of SIVmac239 infection in a vaccinated RM (Fig. 8E) (30). However, the virus replicating in that animal contained a deletion in nef as early as week 2 p.i., which likely compromised its replicative fitness. Given this precedent, we explored the possibility that the group 2 controllers harbored attenuated viruses, even though we have previously shown that the SIVmac239 challenge stock employed in this study is made almost entirely of wild-type genomes (30). We conducted this analysis in acute-phase (weeks 2 to 4 p.i.) plasma from the four RMs manifesting the earliest control of viral replication (r08046, r05007, r08062, and r08047) and found >99% homogeneity in nef sequences amplified at week 2 p.i. (Fig. 9). Viral sequence variation was more prevalent at the later time points, especially in r08062 and r08047 (Fig. 9C and D), but none of the nef sequences analyzed contained insertions or deletions (Fig. 9). Of note, both the Nef IW9 and Nef MW9 epitopes were essentially intact in these acute-phase samples (Table 1). Thus, gross genetic defects in nef were not detected in the acute-phase virus of 4/5 group 2 controllers, corroborating the interpretation that the outstanding control of SIVmac239 replication manifested by these RMs was vaccine mediated.
FIG 9.
nef sequence diversity in acute-phase plasma from four group 2 controllers. Heat maps illustrate the levels of sequence diversity at each codon in the nef open reading frame. The range of amino acids covered by each row is indicated on the right-hand side of the figure and should be used only as a reference because both synonymous and nonsynonymous mutations are considered in these heat maps. The codon corresponding to the first amino acid in Nef is located in the top left corner of each grid. Each row spans 17 amino acids of the Nef protein, except for the bottom row, which covers 9 amino acids and includes the last codon at position 264. The images on the left correspond to plasma samples collected at week 2 postinfection. The images on the right correspond to plasma samples collected at week 3 p.i., or week 4 p.i. in the case of r08046. Data from r08046, r05007, r08062, and r08047 are shown in panels A, B, C, and D, respectively.
TABLE 1.
Percentage of viral sequences encoding intact Mamu-B*17-restricted CD8+ T-cell epitopes in acute-phase samples from four group 2 controllers
| Animal ID | % of viral sequences encoding indicated epitope |
|||||||
|---|---|---|---|---|---|---|---|---|
| Wk 2 p.i. |
Wk 3 or 4 p.i. |
|||||||
| Vif HW8 | Nef IW9 | Nef MW9 | Env FW9 | Vif HW8 | Nef IW9 | Nef MW9 | Env FW9 | |
| r08046a | 100 | 99.7 | 100 | 99.7 | 100 | 100 | 100 | 96.6 |
| r05007 | 100 | 100 | 100 | 100 | 100 | 100 | 98.3 | 98.2 |
| r08062 | 100 | 99.4 | 99.3 | 99.8 | 100 | 99.7 | 99.5 | 94.9 |
| r08047 | NAb | NA | NA | NA | 100 | 100 | 100 | 100 |
No week 3 p.i. plasma was available from r08046, so the sample collected at week 4 p.i. was used instead.
NA, not available.
We also analyzed env sequence evolution in the aforementioned acute-phase samples and found no overlap in the diversity patterns detected in r08046, r05007, r08062, and r08047 (Fig. 10). In spite of many low-frequency mutations scattered throughout env in all four RMs, only a few variants were present at frequencies of ≥10% (Fig. 10). RM r08046 exhibited an R751G substitution at week 4 p.i. that was present in 81% of sequence reads (Fig. 10A). This polymorphism is frequently observed in viruses isolated from SIVmac239-infected RMs and does not by itself significantly alter virus infectivity in vitro (31). One of the env changes in r08062 resulted in a stop codon in gp120 (W225Stop) and comprised 40% of the circulating viral quasispecies at week 3 p.i. (Fig. 10C). However, viral variants harboring this stop codon were no longer detected in week 8 plasma from r08062 (see Fig. S1 in the supplemental material). This RM also exhibited a synonymous mutation in the Leu codon at position 656 (CTA→TTA) in 100% of sequence reads (Fig. 10C). The other mutations in r08062 and in r08047 modified amino acids at various positions in gp120 and gp41 (Fig. 10C and D). Variants of the Env FW9 epitope were detected at weeks 2 to 4 p.i., but these mutants never reached frequencies of >6% (Table 1). A summary of amino acid substitutions in Env detected at frequencies of ≥10% in these acute-phase samples is shown in Fig. 11. Thus, a shared pattern of env sequence evolution did not explain the rapid postacute-phase virologic control manifested by 4/5 group 2 controllers.
FIG 10.
env sequence diversity in acute-phase plasma from four group 2 controllers. Heat maps illustrate the levels of sequence diversity at each codon in the env open reading frame. The range of amino acids covered by each row is indicated on the right-hand side of the figure only as a reference because both synonymous and nonsynonymous mutations are considered in these heat maps. When applicable, the frequency and location of amino acid substitutions are described in the text and in Fig. 11. The codon corresponding to the first amino acid in Env is located in the top left corner of each grid. Each row spans 30 amino acids of the Env protein, except for the bottom row, which covers 9 amino acids and includes the last residue at position 879. The images on the left correspond to plasma samples collected at week 2 postinfection. The images on the right correspond to plasma samples collected at week 3 p.i., or week 4 p.i. in the case of r08046. Data from r08046, r05007, r08062, and r08047 are shown in panels A, B, C, and D, respectively.
FIG 11.
Summary of amino acid substitutions in Env present at ≥10% frequencies in acute phase samples from four group 2 controllers. Each substitution is enclosed by a square, and its details are listed in a text box outside the Env main sequence. The details provided for each amino acid substitution include the animal identifier (ID), time point postinfection, position in the Env protein, and the frequency of sequence reads displaying the wild-type (boldface type) and mutant residues. As a reference, several topological features of the SIV Env protein are shown, including the five variable loops (V) in gp120, the gp120/gp41 cleavage site, the membrane spanning domain, and the highly immunogenic region of gp41. The location of the Mamu-B*17-restricted Env FW9 epitope is also indicated in the carboxyl terminus of gp41.
We then searched for vaccine-induced immune signatures that might explain the distinct virologic outcomes observed in groups 1 and 2. Eight immunological variables determined either at the time of the first i.r. SIV challenge or at week 2.4 p.i. were selected for this analysis (Table 2). These variables were then compared with nadir VLs—the virologic marker most affected by vaccine-induced immune responses (Table 2). The only immunological predictor of virologic control that emerged from this analysis was the endpoint titer of vaccine-induced gp140-binding Abs in group 2 at the time of the first SIV challenge, which inversely correlated with nadir VLs (Table 2; Fig. 12). This association was not, however, statistically significant after correcting for multiple comparisons. Collectively, these data suggest that in the context of potent vaccine-elicited SIV-specific T-cell responses, increasing titers of gp140-binding Abs may result in substantial control of viral replication in Mamu-B*17+ RMs.
TABLE 2.
Correlations between immunological variables measured in vaccinees in groups 1 and 2 and their respective nadir VLs
| Immunological variable | Group(s) included in the analysis | Time point | Correlation coefficient | Unadjusted P value | Bonferroni-corrected P value |
|---|---|---|---|---|---|
| Total frequency of SIV-specific CD8+ T-cell responses | 1 and 2 | Time of 1st SIV challenge | −0.18 | 0.53 | 1.0 |
| Total frequency of SIV-specific CD4+ T-cell responses | 1 and 2 | Time of 1st SIV challenge | −0.25 | 0.37 | 1.0 |
| Total frequency of Mamu-B*17 tetramer+ CD8+ T cells | 1 and 2 | Time of 1st SIV challenge | −0.19 | 0.5 | 1.0 |
| Total frequency of Mamu-B*17 tetramer+ CD8+ T cells | 1 and 2 | Wk 2.4 p.i. | 0.4 | 0.14 | 1.0 |
| Area-under-the-curve values for ADCC | 2 | Time of 1st SIV challenge | −0.49 | 0.22 | 1.0 |
| Area-under-the-curve values for ADCC | 2 | Wk 2.4 p.i. | 0.14 | 0.75 | 1.0 |
| Titer of gp140-binding Abs | 2 | Time of 1st SIV challenge | −0.85 | 0.008 | 0.06 |
| Titer of gp140-binding Abs | 2 | Wk 2.4 p.i. | 0.28 | 0.51 | 1.0 |
FIG 12.
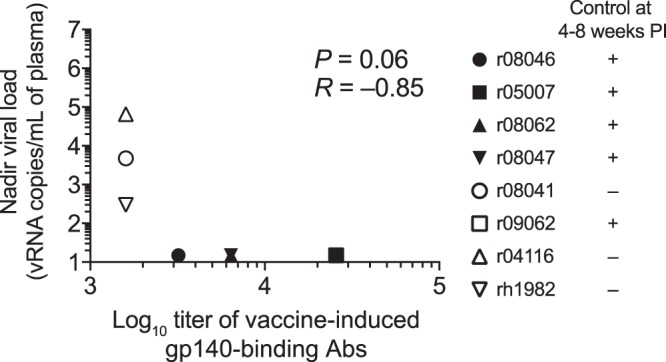
Association between titers of vaccine-induced gp140-binding Abs at the time of the 1st SIV challenge and nadir viral loads in group 2. This is a graphical representation of the comparison between the log10 endpoint titers of vaccine-induced gp140-binding antibodies at the time of the first i.r. SIV challenge and nadir viral loads in group 2 listed in Table 2. The symbol for macaque r09062 (open black square) is masked by the symbol of r05007 (filled black square) because both animals had the same titer of gp140-binding antibodies (25,600) and nadir VL (15 vRNA copies/ml of plasma). The Bonferroni-corrected P value is shown. Each symbol corresponds to one group 2 vaccinee.
DISCUSSION
In this study, we explored whether vaccinating Mamu-B*17+ RMs with inserts encoding Mamu-B*17-restricted SIV epitopes would increase the frequency of elite control after SIVmac239 infection, as is the case with vaccination of Mamu-B*08+ RMs with Mamu-B*08-restricted SIV epitopes (21). We also examined whether the addition of env to the vaccine regimen would confer protection from mucosal acquisition of the challenge virus, as has been reported previously (22–24). To our surprise, none of the vif- and nef-vaccinated group 1 animals controlled viral replication to <15 vRNA copies/ml of plasma, suggesting that the underlying mechanisms of elite control differ between Mamu-B*17+ and Mamu-B*08+ RMs. Even though the inclusion of env in the vaccine did not prevent acquisition of SIVmac239, we identified remarkable levels of virologic control in most of the group 2 vaccinees. Indeed, 5/8 group 2 RMs (vaccinated with vif, nef, and env) suppressed viral replication to <15 vRNA copies/ml of plasma by 4 to 8 weeks p.i. This virologic control did not correlate with vaccine-induced SIV-specific T cells or NK cell-mediated ADCC activity, nor did it depend on anti-SIVmac239 nAbs, because serological neutralizing activity against SIVmac239 was not detected at the time of the 1st SIV challenge or even shortly after infection. Vaccine-elicited gp140-binding Abs at the time of challenge were the only immune predictor of virologic control in group 2.
Approximately 50% of Mamu-B*08+ and 21% of Mamu-B*17+ SIV-naive RMs become ECs after SIVmac239 infection (14, 15, 20). Given the virulence of SIVmac239, elite control in this context is defined as having a set point VL of ≤1,000 vRNA copies/ml of plasma. Of note, expression of these alleles does not affect acute-phase viremia after primary SIVmac239 infection. Indeed, peak VLs are often indistinguishable between unvaccinated SIVmac239-infected RMs expressing Mamu-B*08 or Mamu-B*17 and animals lacking these protective MHC-I alleles (14, 15). Despite the predisposition of Mamu-B*17+ RMs to control SIV replication, the kinetics with which the five group 2 controllers suppressed viremia in the present experiment was completely different from that seen in typical EC RMs. Our historical VL analysis of SIVmac239-infected animals shows that only a few RMs expressing MHC-I alleles associated with elite control of SIV infection controlled viremia to <15 vRNA copies/ml by week 20 p.i. (Fig. 8D to H). Even in those few cases, it took, on average, 12 weeks for their plasma VLs to reach <15 vRNA copies/ml. By comparison, the five group 2 controllers in the present experiment controlled viremia to <15 vRNA copies/ml after an average of 6 weeks of infection. Vaccine-induced Env-specific immune responses appeared to be required for this impressive outcome, considering that they were the only feature that distinguished group 2 from group 1. This conclusion is also supported by the inverse association between vaccine-elicited gp140-binding Ab titers and nadir VLs in group 2. Nevertheless, assuming that Abs were involved in control, it is not clear how these vaccine-elicited gp140-specific Abs contributed to virologic control, considering that they lacked neutralizing activity against SIVmac239 and their ability to recruit NK cell-mediated ADCC activity against SIVmac239-infected cells did not predict postinfection VLs. One caveat to these analyses is the small number of animals in group 2, which limited our ability to accurately identify immune correlates of virologic control. Additionally, given the wide range of nonneutralizing Ab effector functions reported for HIV-specific Abs (32), it is possible that antiviral activities other than NK cell-mediated ADCC might have explained the rapid control of viral replication observed in some of the group 2 vaccinees. Furthermore, the absence of a group of MHC-I-matched RMs vaccinated with env only also precludes any definitive conclusions regarding the role of vaccine-induced Env-specific immune responses in the challenge outcome. Such a group would have revealed whether or not vaccine-induced Env-specific immune responses are sufficient for rapid postacute-phase control of viral replication in Mamu-B*17+ RMs and also to what extent vaccine-elicited T-cell responses against Vif and Nef contributed to virologic containment in the five group 2 controllers. Additional experiments will be needed to elucidate these issues.
The observation that the group 1 vaccine regimen had little effect on viral replication compared to the control group suggests that unlike the case with Mamu-B*08+ RMs (21), preexisting SIV-specific CD8+ T cells alone do not facilitate long-term virologic suppression in Mamu-B*17+ RMs. These discordant outcomes imply a differential dependence on Vif- and Nef-specific CD8+ T cells for elite control of SIV replication in Mamu-B*17+ and Mamu-B*08+ animals. In support of this notion, it is possible to predict which unvaccinated Mamu-B*08+ RMs will become ECs after SIVmac239 infection based on patterns of viral sequence evolution in Mamu-B*08-restricted epitopes shortly after infection (33). In contrast, the emergence of CD8+ T-cell “escape” viral variants does not dictate the ability of certain Mamu-B*17+ animals to contain SIV viremia (20). In this regard, it is important to mention that other host genetic factors have also been associated with control of immunodeficiency virus replication (34). Thus, variability in such factors may explain the different SIV infection outcomes observed in vif- and nef-vaccinated Mamu-B*08+ versus Mamu-B*17+ RMs. Additional studies will be needed to elucidate the differences in elite control between Mamu-B*08+ and Mamu-B*17+ animals.
Importantly, the ability of gp140-binding Ab titers to predict vaccine-mediated control of SIVmac239 replication is not limited to Mamu-B*17+ RMs. We have recently reported that an EP rDNA/rAd5/rVSV/rRRV vaccine regimen including env, gag, vif, rev, tat, and nef resulted in substantial reductions in plasma VLs in SIVmac239-infected RMs lacking MHC-I alleles associated with elite control (27). Similar to the present study, vaccine-elicited Env-specific Abs lacked detectable neutralizing activity against SIVmac239, but their gp140-binding titers correlated with virologic control. Although vaccinees were not protected from SIV infection in either case, the fact that vaccine-induced gp140-binding Abs predicted control of viremia in these two independent settings is instructive for two reasons. First, the observation that high levels of vaccine-induced Env-specific Ab responses might improve control of viral replication in RMs regardless of their MHC-I genotype broadens the applicability of the present findings. Second, any immunological signature associated with containment of SIVmac239 infection warrants further investigation given the stringency of this molecular clone as a challenge virus (35). Indeed, most unvaccinated or drug-naive SIVmac239-infected Indian RMs experience high peak (106 to 109 vRNA copies/ml) and set point (106 vRNA copies/ml) VLs and are euthanized due to AIDS-defining illnesses by 2 years after infection (36). Additionally, the SIVmac239 Env glycoprotein is exceptionally difficult to neutralize (37–41), likely due to its closed conformation. Innumerable vaccine regimens have been evaluated using SIVmac239 challenges, but relatively few have resulted in substantial reductions in viremia after infection (27, 29, 42–45) or prevented virus spread beyond the initial foci of infected cells (28, 46). However, except for live attenuated SIV vaccines (47, 48), no active immunization strategy has consistently afforded sterilizing protection against challenge with SIVmac239. While certain features of the antiviral T-cell response elicited by live attenuated SIV vaccines have been identified as correlates of protection (48–51), vaccine efficacy in this case also relies heavily on Env-specific humoral immunity (52). The fact that only a fraction of SIVmac239Δnef-vaccinated RMs develops detectable anti-SIVmac239 nAb responses implies that Ab-mediated effector functions other than neutralization (e.g., ADCC) can also contribute to the efficacy of live attenuated SIV vaccines (52, 53). In this light, it is noteworthy that the titers of gp140-binding Abs elicited by SIVmac239Δnef vaccination exceeded those generated by our mixed-modality vaccine regimens approximately 100-fold (Fig. 4). Group 2 had greater titers of these responses than the aforementioned EP rDNA/rAd5/rVSV/rRRV group, but this difference was not statistically significant. Although it is impossible at this point to establish a causal link between these anti-Env humoral responses and containment of viral replication, future studies should evaluate if the robust efficacy of live attenuated SIV vaccination can be replicated by matching its titers of gp140-binding Abs.
In conclusion, here we show that an rYF17D/EP rDNA/rAd5/rVSV/rRRV vaccine regimen, which has env and genes expressing Mamu-B*17-restricted CD8+ T-cell epitopes, resulted in stringent control of viral replication in Mamu-B*17+ RMs shortly after SIVmac239 infection. We present evidence suggesting that vaccine-elicited Env-specific Abs devoid of detectable neutralizing activity against SIVmac239 were critical mediators of virologic control and that similar outcomes may be achieved in RMs that do not express protective MHC-I alleles. However, the exact mechanism by which these humoral responses exerted antiviral activity and to what extent cellular immune responses contributed to viral suppression remain undefined. These findings advance our understanding of vaccine-mediated control of lentivirus infection and establish a new benchmark for evaluating HIV vaccine candidates in the SIVmac239 challenge model.
MATERIALS AND METHODS
Research animals and ethics statement.
Twenty-four RMs expressing the Mamu-B*17 MHC-I allele were originally enrolled in this vaccine trial. Unfortunately, RM r04105 had to be euthanized due to recurrent diarrhea and weight loss, thus reducing the size of group 1 to seven RMs. Because r04105 was euthanized at study week 39—before it was boosted with rAd5—this animal was not included in the immunological comparisons of vaccine-induced T-cell responses between groups 1 and 2.
The details regarding animal welfare described herein are identical to those published for one of our previous experiments (54). “The Indian RMs (Macaca mulatta) utilized in this study were housed at the Wisconsin National Primate Research Center (WNPRC). All animals were cared for in accordance with the guidelines of the Weatherall report and the principles described in the National Research Council's Guide for the Care and Use of Laboratory Animals under a protocol approved by the University of Wisconsin Graduate School Animal Care and Use Committee” (animal welfare assurance no. A3368-01; protocol no. G00696). (Republished from reference 54. For further information on the care of the animals used, see reference 55.) “Furthermore, the RMs in this study were managed according to the animal husbandry program of the WNPRC, which aims at providing consistent and excellent care to nonhuman primates at the center. This program is employed by the Colony Management Unit and is based on the laws, regulations, and guidelines promulgated by the United States Department of Agriculture (e.g., the Animal Welfare Act and its regulations, and the Animal Care Policy Manual), Institute for Laboratory Animal Research (e.g., Guide for the Care and Use of Laboratory Animals, 8th edition), Public Health Service, National Research Council, Centers for Disease Control, and the Association for Assessment and Accreditation of Laboratory Animal Care International. The nutritional plan utilized by the WNPRC is based on recommendations published by the National Research Council. Specifically, RMs were fed twice daily with 2050 Teklad Global 20% Protein Primate Diet and food intake was closely monitored by Animal Research Technicians. This diet was also supplemented with a variety of fruits, vegetables, and other edible objects as part of the environmental enrichment program established by the Behavioral Management Unit. Paired/grouped animals exhibiting stereotypical and/or incompatible behaviors were reported to the Behavioral Management staff and managed accordingly. All primary enclosures (i.e., stationary cages, mobile racks, and pens) and animal rooms were cleaned daily with water and sanitized at least once every 2 weeks. Lights were on a 12:12 diurnal schedule. Vaccinations were performed under anesthesia (Ketamine administered at 5 to 12 mg/kg depending on the animal) and all efforts were made to minimize suffering. Euthanasia was performed at the end of the study or whenever an animal experienced conditions deemed distressful by one of the veterinarians at the WNPRC. All euthanasia were [sic] performed in accordance with the recommendations of the Panel on Euthanasia of the American Veterinary Medical Association and consisted of an IV overdose (greater than or equal to 50 mg/kg or to effect) of sodium pentobarbital or equivalent, as approved by a clinical veterinarian, preceded by ketamine (at least 15 mg/kg body weight) given by the intramuscular (IM) route.” (Republished from reference 54.) Additional animal information, including MHC-I alleles, age at the beginning of study, and sex, is shown in Table 3.
TABLE 3.
Animal characteristics
| Group | Animal ID | Age (yrs) at beginning of study | Sexa | MHC class I allele(s) |
|---|---|---|---|---|
| 1 | r04105b | 8.6 | F | Mamu-B*17 |
| r03139 | 9.6 | F | Mamu-B*17 | |
| r08022 | 4.8 | F | Mamu-B*17 | |
| r09090 | 3.6 | M | Mamu-B*17 | |
| r07012 | 5.6 | F | Mamu-B*17 | |
| r08034 | 4.8 | M | Mamu-B*17 | |
| r09001 | 4.3 | M | Mamu-B*17 | |
| r09022 | 4 | M | Mamu-B*17 | |
| 2 | r08046 | 4.6 | M | Mamu-B*17 |
| r05007 | 8.2 | F | Mamu-B*17 | |
| r08062 | 4.4 | M | Mamu-B*17 | |
| r08047 | 4.6 | M | Mamu-A*02, -B*17 | |
| r08041 | 4.7 | F | Mamu-B*17 | |
| r09062 | 3.8 | F | Mamu-A*02, -B*17 | |
| r04116 | 8.6 | F | Mamu-B*17 | |
| rh1982 | 15 | F | Mamu-B*17 | |
| 3 | r08051 | 4.6 | M | Mamu-B*17 |
| r04053 | 8.9 | F | Mamu-B*17 | |
| r97081 | 15.6 | F | Mamu-B*17 | |
| r09073 | 3.8 | M | Mamu-B*17 | |
| r10018 | 3.1 | M | Mamu-A*02, -B*17 | |
| r09083 | 3.7 | F | Mamu-B*17 | |
| r98038 | 14.9 | M | Mamu-B*17 | |
| r09039 | 3.9 | M | Mamu-B*17 |
F, female; M, male.
Monkey r04105 had to be euthanized at study week 39.
Vaccinations.
The parental live attenuated YF17DD vaccine strain and four rYF17D vectors expressing individual SIVmac239 inserts were used in this study. The SIVmac239 inserts encoded (i) Vif amino acids 1 to 110, (ii) Nef amino acids 45 to 210, and (iii) the amino terminus of gp41 (Env amino acids 526 to 690). In order to increase the stability of the last rYF17D vector, the gp41 fusion peptide was inactivated and the Cys residues at positions 611 and 617 were mutated to Ala. The last SIVmac239 insert (iv) consisted of a fusion of gp120 segments corresponding to amino acids 64 to 109, 209 to 315, and 343 to 502. The codon usage of the aforementioned SIVmac239 inserts matched that of YF17D. RMs in group 2 were vaccinated subcutaneously with 300,000 PFU of each of rYF17D vectors 1 to 4. These constructs were delivered in separate injections, each containing a final volume of 0.5 ml. RMs in group 1 were vaccinated in the same way, except that they received only vectors 1 and 2. RMs in group 3 were sham vaccinated with the same dose of the parental YF17DD vaccine. The construction of some of the rYF17D vectors used here has been published elsewhere (56, 57).
A series of three EP rDNA vaccinations, given at 3-week intervals, started at study week 12. Four pCMVkan plasmids carrying SIVmac239 minigenes were used in the EP rDNA vaccinations (58). These minigenes were identical to the ones delivered by the rYF17D vectors, except that the amino acid substitutions described above were reverted to wild type and the SIV open reading frames were optimized for mammalian expression. RMs in groups 1 and 2 were vaccinated with pCMVkan constructs carrying the aforementioned vif and nef minigenes. In addition to these constructs, RMs in group 2 also received two pCMVkan plasmids carrying env minigenes described above. RMs in group 3 were sham vaccinated with empty pCMCkan plasmid. One milligram of each pCMVkan construct and 0.1 mg of the rhesus interleukin-12-expressing AG157 plasmid were resuspended in 0.5 ml of phosphate-buffered saline (PBS) and loaded into separate injections. These DNA formulations were administered intramuscularly by the TriGrid in vivo electroporation system (Ichor Medical Systems, Inc., San Diego, CA). Muscles in the thighs and forearms were used for these vaccinations, and these anatomical sites were rotated in subsequent immunizations so that each location did not receive vectors bearing the same SIV insert twice.
The rAd5 boost occurred at study week 54. RMs in groups 1 and 2 were boosted with two rAd5 vectors produced by Viraquest, Inc., each bearing the same vif or nef minigenes as described above, which were also optimized for mammalian expression. RMs in group 2 also received an rAd5 vector encoding the full-length SIVmac239 gp160 glycoprotein. RMs in group 3 were sham vaccinated with empty Ad5 vectors lacking any inserts. The two latter Ad5 constructs were produced by the International AIDS Vaccine Initiative. A dose of 1011 viral particles of each rAd5 vector was administered intramuscularly to sites in the forearms and thighs.
The rVSV boost occurred at study week 78. The rVSV vector was based on a modified attenuated virus strain developed by Profectus Biosciences (59). An rVSV vector encoding a fusion of the SIVmac239 Nef, Tat, and Vif proteins was used in RMs in both groups 1 and 2. A separate rVSV vector encoding the SIVmac239 Env protein was administered to the group 2 vaccinees only. SIV Env was modified by replacing the cytoplasmic tail with the corresponding sequence from VSV G (60). RMs in group 3 were sham vaccinated with an rVSV construct encoding the malaria CSP antigen. All three rVSV vectors were provided by Profectus Biosciences. A dose of 107 PFU of each rVSV vector was administered intramuscularly to separate sites in the forearms and thighs.
The rRRV boost occurred at study week 84. The group 1 RMs were vaccinated with a mixture of three rRRV vectors, each expressing SIVmac239 vif, nef, and a fusion of the rev-tat-nef genes. The group 2 RMs received the same rRRV vectors with the addition of an rRRV construct expressing full-length env. The group 3 RMs were sham vaccinated with an rRRV vector encoding enhanced green fluorescent protein. One milliliter of PBS containing 7.1 × 107 genome copies of each appropriate rRRV vector was administered via both the intravenous and i.r. routes. Details about the generation of these rRRV vectors have been described previously (42).
SIVmac239 challenges.
Seventeen weeks after the rRRV boost, RMs in groups 1 to 3 were subjected to repeated i.r. inoculations of 200 TCID50 (4.8 × 105 vRNA copies) of the same SIVmac239 stock described in reference 30. These i.r. challenges occurred every 2 weeks. Plasma VLs were assessed 6 and 10 days after each exposure. Once an animal experienced a positive VL at either one of these time points, it was no longer challenged. Only RMs that remained aviremic at both time points were rechallenged on day 14.
SIV RNA viral load measurements.
The following description of plasma SIV RNA viral loads is identical to that published in one of our recent articles (27). “VLs were measured using 0.5 ml of EDTA-anticoagulated rhesus macaque plasma based on a modification of a previously published [sic]. Total RNA was extracted from plasma samples using QIAgen DSP virus/pathogen Midi kits, on a QIASymphonyXP laboratory automation instrument platform. Six replicate two step RT-PCRs were performed per sample using a random primed reverse transcription reaction, followed by 45 cycles of PCR using the following primers and probe: forward primer: SGAG21: 5′-GTCTGCGTCAT(dP)TGGTGCA TTC-3′; reverse primer SGAG22: 5′-CACTAG(dK)TGTCTCTGCACTAT(dP)TGTTTTG-3′; probe: PSGAG23: 5′-FAM-CTTC(dP)TCAGT(dK)TGTTTCACTTTCTCTTCTGCG-BHQ1-3′. The limit of reliable quantitation on an input volume of 0.5 ml of plasma was 15 vRNA copies/ml.” (Republished from reference 27. For more information on measurement of VLs, see reference 61.)
Quantification of Mamu-B*17 tetramer+ CD8+ T cells in PBMC.
The following description is similar to that published in one of our recent articles (27). Fluorochrome-labeled Mamu-B*17 tetramers produced at the NIH Tetramer Core Facility were used to quantify SIV-specific CD8+ T cells in peripheral blood mononuclear cells (PBMC) according to a recently published protocol (62). Approximately 800,000 PBMC were incubated in R10 medium (RPMI 1640 medium supplemented with GlutaMax [Life Technologies], 10% fetal bovine serum [FBS; VWR], and 1% antibiotic/antimycotic [VWR]) with titrated amounts of each tetramer at room temperature for 45 min. The cells were then stained with fluorochrome-labeled monoclonal antibodies (MAbs) directed against the surface molecules CD3 (clone SP34-2), CD8α (clone RPA-T8), CD14 (clone M5E2), CD16 (clone 3G8), and CD20 (clone 2H7) for 25 min. This step also included an amine-reactive dye (ARD; LIVE/DEAD fixable aqua dead cell stain; Life Technologies). The cells were then washed with wash buffer (Dulbecco's PBS with 0.1% bovine serum albumin and 0.45 g/liter of NaN3) and fixed with PBS containing 2% paraformaldehyde (PFA) for 20 min at 4°C. The cells were washed one more time before they were acquired in a special-order BD LSR II cytometer. The gating strategy used to analyze the data has been described elsewhere (62). Briefly, we used FlowJo 9.6 to determine the percentages of live CD14− CD16− CD20− CD3+ CD8+ tetramer+ lymphocytes in PBMC.
ICS assays.
The following description is similar to that published in one of our recent articles (27). Freshly isolated PBMC were cultured in R10 medium containing unlabeled costimulatory MAbs against CD28 and CD49d and a phycoerythrin-conjugated MAb specific for CD107a. The tubes were kept in a 5.0% CO2 incubator for 9 h at 37°C. One hour into the incubation period, brefeldin A (Biolegend, Inc.) and GolgiStop (BD Biosciences) were added to all tubes in order to inhibit protein transport. The antigen stimuli consisted of three or five pools of SIVmac239 peptides (15-mers overlapping by 11 amino acids) corresponding to (i) the entire Vif protein (amino acids 1 to 214), (ii) the entire Nef protein (amino acids 1 to 263), (iii) both Rev (amino acids 1 to 107) and Tat (amino acids 1 to 130) proteins, (iv) Env gp120 (amino acids 1 to 531), and (v) Env gp41 (amino acids 516 to 879). The final concentration of each 15-mer in the intracellular cytokine staining (ICS) tubes was 1.0 μM. The same steps as outlined above were used to stain the cells with MAbs against surface markers and subsequently fix them. The surface staining master mix included MAbs against CD4 (clone OKT4; Biolegend, Inc.) and CD8 (clone RPA-T8; Biolegend, Inc.), in addition to the same MAbs against CD14, CD16, and CD20 and the ARD reagent described above. For 10 min, cells were permeabilized by homogenization in permeabilization buffer (1× BD FACS lysing solution 2 [Becton Dickinson] and 0.05% Tween 20 [Sigma-Aldrich]) and then were washed with wash buffer. Lastly, cells were incubated for 1 h in the dark at room temperature with MAbs against CD3 (clone SP34-2; BD Biosciences), gamma interferon (IFN-γ; clone 4S. B3; Biolegend, Inc.), tumor necrosis factor alpha (TNF-α; clone Mab11; BD Biosciences), and CD69 (clone FN50; Biolegend, Inc.). Once this incubation was completed, cells were washed and stored at 4°C until acquisition.
After gating on live CD14− CD16− CD20− CD3+ lymphocytes, we selected either CD4+ or CD8+ cells for subsequent analyses. Cells were considered positive for IFN-γ, TNF-α, or CD107a only if these molecules were coexpressed with CD69, a marker of recent activation. Leukocyte activation cocktail (LAC; BD Pharmingen)-stimulated cells stained with fluorochrome-labeled control MAbs of the same isotypes as those against IFN-γ, TNF-α, and CD107a guided the identification of positive populations. Two criteria were used to determine a positive response. First, the frequency of events had to be ≥2-fold higher than their corresponding values in background-subtracted negative-control tests. Second, the gates for each response had to contain ≥10 events. These calculations were performed with Microsoft Excel, and results were presented as the percentage of responding CD4+ or CD8+ T cells, that is, live CD14− CD16− CD20− CD3+ lymphocytes of either subset producing any combination of IFN-γ, TNF-α, and CD107a.
Anti-Env antibody measurements by enzyme-linked immunosorbent assay (ELISA).
The following description is identical to that reported by our group in a recent publication (27). “Vaccine induced anti-Env responses were measured by ELISA. To begin, the ELISA plate was coated with 100 μl of purified SIVmac239 gp140 protein (Immune Technology Corp. IT-001-140p) at a concentration of 0.5 μg/ml and incubated overnight at RT. On the following day, the plate was washed with 1× PBS-Tween20 and wells were blocked with 300 μl of 5% powdered milk in PBS for 1 h at 37°C. Subsequently, the plate was washed and 100 μl of diluted plasma samples were added to the corresponding wells. After a 1-hr incubation at RT, the plate was washed and 100 μl of a 1:2,000 dilution of Goat Anti-Monkey IgG-HRP antibody (Santa Cruz Biotechnology, sc-2458) were added to all wells for 1 h at 37°C. Finally, the plate was washed before being developed with 100 μl of 3,3′,5,5′-Tetramethylbenzidine (EMD Millipore, 613544-100ML). After a short incubation, the reaction was stopped with TMB Stop Solution (Southern Biotech, 0412-01) and the plate was read (Biotek Synergy 2) at 450 nm. The endpoint antibody titers of vaccine-induced anti-Env antibody responses were measured in serum collected at the time of SIV challenge. These titers were determined as the greatest dilution at which the absorbance in experimental wells was at least 2-fold higher than that measured in pooled prevaccination serum from all animals in the experiment.” (Republished from reference 27.)
Antibody-dependent cellular cytotoxicity assay.
The following description is identical to that reported by our group in a recent publication (27). “The SIVmac239 and SHIVAD8-EO stocks used in ADCC assays were produced by transfection of infectious molecular clones into HEK293T cells using GenJet transfection reagent (SignaGen). Virus-containing supernatants were collected 48 and 72 h posttransfection and stored at −80°C. The SHIVAD8-EO clone was provided by Malcom Martin (NIAID, Bethesda, MD). After heat inactivation for 30 min at 56°C, RM plasma samples were tested for nonspecific ADCC due to the presence of antibodies to human cellular antigens by coincubating uninfected CEM.NKR-CCR5-sLTR-Luc target cells (AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH) with an NK cell line (KHYG-1 cells) expressing RM CD16 at a 10:1 effector-to-target ratio in the presence of serial dilutions of plasma. This NK cell line was developed in-house, as described previously. Nonspecific lysis was detected as a reduction in background luciferase activity (% RLU) for target cells incubated with NK cells in the presence compared to the absence of plasma. Plasma samples that directed ADCC against uninfected cells were depleted of anti-human antibodies by repeated cycles of incubation with CEM.NKR-CCR5-sLTR-Luc cells, followed by centrifugation and plasma transfer, until ADCC responses to uninfected cells were no longer detectable.” (Republished from reference 27. For additional information regarding testing for nonspecific ADCC, see reference 63; for additional information regarding development of NK cell line KHYG-1, see reference 60.)
“To measure ADCC activity in plasma of vaccinated animals, CEM.NKR-CCR5-sLTR-Luc target cells were infected with SIVmac239 or SHIVAD8-EO (internal negative control) by spinoculation for 3 h at 1200 × g in the presence of 40 μg/ml Polybrene (EMD Millipore). Four days postinfection, target cells were incubated with the NK cell line KHYG-1 at a 10:1 effector-to-target ratio in the presence of serial plasma dilutions. Luciferase activity was measured after 8 h using the britelite plus luciferase assay system (PerkinElmer). Triplicate wells were tested at each plasma dilution, and wells containing effector cells incubated with uninfected or infected target cells in the absence of plasma were used to determine background and maximal luciferase activity, respectively. ADCC responses were calculated from the dose-dependent loss of luciferase activity in the presence of plasma relative to background and maximal luciferase control wells.” (Republished from reference 27.)
Pseudovirus neutralization assays.
The following description is identical to that reported by our group in a recent publication (27). “Replication incompetent SIVmac239 pseudovirus was produced by cotransfecting env plasmids with an env-deficient backbone plasmid (pSG3Δenv) in HEK293T cells in a 1:2 ratio, using the X-tremeGENE 9 transfection reagent (Roche). Pseudovirus was harvested after 72 h by sterile-filtration (0.22 μm) of cell culture supernatants, and neutralization was tested by incubating pseudovirus and serum for 1 h at 37°C before transferring them onto TZM-bl cells (AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH) as previously described. Neutralization was measured in duplicate wells within each experiment. Neutralization was tested starting at 1:20 serum dilutions followed by nine serial 3-fold dilutions to ensure highest sensitivity and range of detection. Neutralization IC50 titers were calculated using the ‘One site—Fit logIC50′ regression in Graphpad Prism v7.0. We could not detect vaccine-induced nAb titers against SIVmac239 pseudovirus in any of the RMs in group 2 at the time of the first SIV challenge.” (Republished from reference 27. For additional information on testing neutralization of pseudovirus, see reference 64.)
Amplicon-based sequencing of SIV nef and env.
Viral RNA was isolated from plasma using the QIAamp viral RNA minikit (Qiagen) according to the manufacturer's protocol and eluted in 60 μl of AVE buffer, aliquoted, and stored at −80°C for future use. Viral RNA was reverse transcribed and amplified by using the SuperScript III one-step reverse transcription-PCR (RT-PCR) system with high-fidelity Platinum Taq polymerase (Invitrogen). The 3′ half of the genome was amplified in two amplicons using primer pair SIVmac239-Amp3-5660-F (5′-GGCATAGCCTCATAAAATATCTG-3′) and SIVmac239-Amp3-8487-R (5′-ATTGCAGAACCTGCCGTTG-3′) and primer pair SIVmac239-Amp4-7821-F (5′-CAGTCACCATTATGTCTGGATTG-3′) and SIVmac239-Amp4-10235-R (5′-GAATACAGAGCGAAATGCAGTG3′). All SIVmac239 primer positions are based on published SIVmac239 sequence present in GenBank (accession number M33262). The RT-PCR conditions were as follows: 50°C for 30 min; 94°C for 2 min; 40 cycles of 94°C for 15 s, 55°C for 30 s, and 68°C for 3 min; and 68°C for 5 min.
Amplicons were visualized on a 1.0% agarose gel and purified using the Purelink quick gel extraction kit (Invitrogen). RT-PCR products were quantified using a QuantiFluor-ST fluorometer (Promega) and analyzed for quality using an Agilent 2100 bioanalyzer with high-sensitivity DNA chips. PCR amplicons were fragmented and barcoded using N exteraXT DNA library prep kit, as per the manufacturer's protocol. Samples were pooled and sequenced on an Illumina MiSeq platform, using a 2- by 250-bp V2 reagent kit. Paired-end reads obtained from Illumina MiSeq were then processed using our internal sequencing analysis pipeline, which consists of removing PCR duplicate reads using FastUniq (65), quality trimming with trimmomatic (66), and de novo assembly using VICUNA (67), with finishing and annotation completed with V-FAT (https://www.broadinstitute.org/viral-genomics/v-fat). Reads were then aligned to the consensus assembly using Mosaik v2.1.73 with V-Phaser v2.0 used to call intrahost variants as described elsewhere (67–69).
Statistics.
Peak VLs were determined as the highest VL measurement within the first 4 weeks after infection. Nadir VLs were considered the lowest VL measurement between peak and week 8 p.i. Setpoint VLs were calculated as the geometric mean of all VLs measured between week 8 p.i. and the last chronic-phase time point available. The Mann-Whitney U test was used to compare the total magnitudes of vaccine-induced SIV-specific T-cell responses between groups 1 and 2. The Kaplan-Meier method and log rank test were used to determine whether the group 1 and group 2 vaccine regimens affected acquisition of SIV infection. For this analysis, the time to productive infection was analyzed using the Kaplan-Meier method and the differences between groups 1 and 3 and groups 2 and 3 were evaluated using log rank tests. The Mann-Whitney U test was also used to determine the efficacy of each vaccine regimen in reducing viral replication. Viral loads at multiple time points were compared between each of groups 1 and 2 and the control group 3. Lastly, the Spearman rank correlation was used to indicate immune correlates of protection. All significance tests were two tailed.
Accession number(s).
All raw sequence reads have been deposited in the NCBI Sequence Read Archive under the study accession number SRP016012, with experimental accession numbers SRX3797244 to SRX3797252.
Supplementary Material
ACKNOWLEDGMENTS
We thank Teresa Maidana Giret for confirming the MHC-I genotype of the monkeys in this study; Leydi Guzman for administrative assistance; all members of the Immunology Services Unit at the WNPRC; Kelli Oswald, Rebecca Shoemaker, Randy Fast, Mary Lopez, and Marina Kemelman for excellent technical support; and Eric Peterson and Kristin Crosno for providing excellent care of the rhesus macaques used in the experiment. We thank John Eldridge and Profectus Biosciences for providing rVSV-SIV vaccines based on their attenuated rVSV vector platform.
This work was funded by Public Health Service (PHS) grants R56 AI049120 (D.I.W.), R37 AI052056 (D.I.W.), and P01 AI104715 (T.M.A.) from the National Institute of Allergy and Infectious Diseases. Partial support came from PHS grant R01 AI121135 (D.T.E.), federal funds from the Office of Research Infrastructure Programs (P51 OD011106), and the National Cancer Institute, National Institutes of Health, under contract no. HHSN261200800001E (J.D.L.). IAVI's work is made possible by generous support from many donors, including The Bill & Melinda Gates Foundation, the Ministry of Foreign Affairs of Denmark, Irish Aid, the Ministry of Finance of Japan, the Ministry of Foreign Affairs of the Netherlands, the Norwegian Agency for Development Cooperation (NORAD), the United Kingdom Department for International Development (DFID), and the United States Agency for International Development (USAID). The full list of IAVI donors is available at http://www.iavi.org. The contents are the responsibility of the International AIDS Vaccine Initiative and do not necessarily reflect the views of USAID or the U.S. government.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
J.D.L. is employed by Leidos Biomedical Research, Inc., the Prime Contractor for the Operations and Technical Support Contract for the Frederick National Laboratory for Cancer Research (FNLCR), which exists solely to operate the FNLCR on behalf of the National Cancer Institute and National Institutes of Health. J.D.L.'s laboratory is supported by this contract (contract HHSN261200800001E) from the National Cancer Institute, National Institutes of Health. There are no competing interests or relevant declarations related to employment, consultancy, patents, products in development, or marketed products. T.M.A.'s spouse was an employee of Bristol-Myers Squibb (BMS), which has a focus in virology, specifically treatments for hepatitis B and C and HIV/AIDS. T.M.A.'s spouse no longer works for BMS and retained only a small stock interest in the public company. T.M.A.'s interests were reviewed and managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict-of-interest policies.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.00690-18.
REFERENCES
- 1.UNAIDS. 2016. Global AIDS update. UNAIDS, Geneva, Switzerland. [Google Scholar]
- 2.Buchbinder SP, Mehrotra DV, Duerr A, Fitzgerald DW, Mogg R, Li D, Gilbert PB, Lama JR, Marmor M, Del Rio C, McElrath MJ, Casimiro DR, Gottesdiener KM, Chodakewitz JA, Corey L, Robertson MN. 2008. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): a double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet 372:1881–1893. doi: 10.1016/S0140-6736(08)61591-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flynn NM, Forthal DN, Harro CD, Judson FN, Mayer KH, Para MF. 2005. Placebo-controlled phase 3 trial of a recombinant glycoprotein 120 vaccine to prevent HIV-1 infection. J Infect Dis 191:654–665. doi: 10.1086/428404. [DOI] [PubMed] [Google Scholar]
- 4.Gray GE, Allen M, Moodie Z, Churchyard G, Bekker LG, Nchabeleng M, Mlisana K, Metch B, de Bruyn G, Latka MH, Roux S, Mathebula M, Naicker N, Ducar C, Carter DK, Puren A, Eaton N, McElrath MJ, Robertson M, Corey L, Kublin JG. 2011. Safety and efficacy of the HVTN 503/Phambili study of a clade-B-based HIV-1 vaccine in South Africa: a double-blind, randomised, placebo-controlled test-of-concept phase 2b study. Lancet Infect Dis 11:507–515. doi: 10.1016/S1473-3099(11)70098-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hammer SM, Sobieszczyk ME, Janes H, Karuna ST, Mulligan MJ, Grove D, Koblin BA, Buchbinder SP, Keefer MC, Tomaras GD, Frahm N, Hural J, Anude C, Graham BS, Enama ME, Adams E, DeJesus E, Novak RM, Frank I, Bentley C, Ramirez S, Fu R, Koup RA, Mascola JR, Nabel GJ, Montefiori DC, Kublin J, McElrath MJ, Corey L, Gilbert PB. 2013. Efficacy trial of a DNA/rAd5 HIV-1 preventive vaccine. N Engl J Med 369:2083–2092. doi: 10.1056/NEJMoa1310566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pitisuttithum P, Gilbert P, Gurwith M, Heyward W, Martin M, van Griensven F, Hu D, Tappero JW, Choopanya K. 2006. Randomized, double-blind, placebo-controlled efficacy trial of a bivalent recombinant glycoprotein 120 HIV-1 vaccine among injection drug users in Bangkok, Thailand. J Infect Dis 194:1661–1671. doi: 10.1086/508748. [DOI] [PubMed] [Google Scholar]
- 7.Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, Benenson M, Gurunathan S, Tartaglia J, McNeil JG, Francis DP, Stablein D, Birx DL, Chunsuttiwat S, Khamboonruang C, Thongcharoen P, Robb ML, Michael NL, Kunasol P, Kim JH. 2009. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med 361:2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 8.Desrosiers RC. 2017. Protection against HIV acquisition in the RV144 trial. J Virol 91:e00905-. doi: 10.1128/JVI.00905-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilbert PB, Berger JO, Stablein D, Becker S, Essex M, Hammer SM, Kim JH, Degruttola VG. 2011. Statistical interpretation of the RV144 HIV vaccine efficacy trial in Thailand: a case study for statistical issues in efficacy trials. J Infect Dis 203:969–975. doi: 10.1093/infdis/jiq152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Migueles SA, Connors M. 2010. Long-term nonprogressive disease among untreated HIV-infected individuals: clinical implications of understanding immune control of HIV. JAMA 304:194–201. doi: 10.1001/jama.2010.925. [DOI] [PubMed] [Google Scholar]
- 11.Goulder PJ, Walker BD. 2012. HIV and HLA class I: an evolving relationship. Immunity 37:426–440. doi: 10.1016/j.immuni.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kiepiela P, Ngumbela K, Thobakgale C, Ramduth D, Honeyborne I, Moodley E, Reddy S, de Pierres C, Mncube Z, Mkhwanazi N, Bishop K, van der Stok M, Nair K, Khan N, Crawford H, Payne R, Leslie A, Prado J, Prendergast A, Frater J, McCarthy N, Brander C, Learn GH, Nickle D, Rousseau C, Coovadia H, Mullins JI, Heckerman D, Walker BD, Goulder P. 2007. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat Med 13:46–53. doi: 10.1038/nm1520. [DOI] [PubMed] [Google Scholar]
- 13.Martin MP, Qi Y, Gao X, Yamada E, Martin JN, Pereyra F, Colombo S, Brown EE, Shupert WL, Phair J, Goedert JJ, Buchbinder S, Kirk GD, Telenti A, Connors M, O'Brien SJ, Walker BD, Parham P, Deeks SG, McVicar DW, Carrington M. 2007. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet 39:733–740. doi: 10.1038/ng2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yant LJ, Friedrich TC, Johnson RC, May GE, Maness NJ, Enz AM, Lifson JD, O'Connor DH, Carrington M, Watkins DI. 2006. The high-frequency major histocompatibility complex class I allele Mamu-B*17 is associated with control of simian immunodeficiency virus SIVmac239 replication. J Virol 80:5074–5077. doi: 10.1128/JVI.80.10.5074-5077.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loffredo JT, Maxwell J, Qi Y, Glidden CE, Borchardt GJ, Soma T, Bean AT, Beal DR, Wilson NA, Rehrauer WM, Lifson JD, Carrington M, Watkins DI. 2007. Mamu-B*08-positive macaques control simian immunodeficiency virus replication. J Virol 81:8827–8832. doi: 10.1128/JVI.00895-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loffredo JT, Sidney J, Bean AT, Beal DR, Bardet W, Wahl A, Hawkins OE, Piaskowski S, Wilson NA, Hildebrand WH, Watkins DI, Sette A. 2009. Two MHC class I molecules associated with elite control of immunodeficiency virus replication, Mamu-B*08 and HLA-B*2705, bind peptides with sequence similarity. J Immunol 182:7763–7775. doi: 10.4049/jimmunol.0900111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mothé BR, Sidney J, Dzuris JL, Liebl ME, Fuenger S, Watkins DI, Sette A. 2002. Characterization of the peptide-binding specificity of Mamu-B*17 and identification of Mamu-B*17-restricted epitopes derived from simian immunodeficiency virus proteins. J Immunol 169:210–219. doi: 10.4049/jimmunol.169.1.210. [DOI] [PubMed] [Google Scholar]
- 18.Loffredo JT, Friedrich TC, Leon EJ, Stephany JJ, Rodrigues DS, Spencer SP, Bean AT, Beal DR, Burwitz BJ, Rudersdorf RA, Wallace LT, Piaskowski SM, May GE, Sidney J, Gostick E, Wilson NA, Price DA, Kallas EG, Piontkivska H, Hughes AL, Sette A, Watkins DI. 2007. CD8+ T cells from SIV elite controller macaques recognize Mamu-B*08-bound epitopes and select for widespread viral variation. PLoS One 2:e1152. doi: 10.1371/journal.pone.0001152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loffredo JT, Bean AT, Beal DR, Leon EJ, May GE, Piaskowski SM, Furlott JR, Reed J, Musani SK, Rakasz EG, Friedrich TC, Wilson NA, Allison DB, Watkins DI. 2008. Patterns of CD8+ immunodominance may influence the ability of Mamu-B*08-positive macaques to naturally control simian immunodeficiency virus SIVmac239 replication. J Virol 82:1723–1738. doi: 10.1128/JVI.02084-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maness NJ, Yant LJ, Chung C, Loffredo JT, Friedrich TC, Piaskowski SM, Furlott J, May GE, Soma T, Leon EJ, Wilson NA, Piontkivska H, Hughes AL, Sidney J, Sette A, Watkins DI. 2008. Comprehensive immunological evaluation reveals surprisingly few differences between elite controller and progressor Mamu-B*17-positive simian immunodeficiency virus-infected rhesus macaques. J Virol 82:5245–5254. doi: 10.1128/JVI.00292-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mudd PA, Martins MA, Ericsen AJ, Tully DC, Power KA, Bean AT, Piaskowski SM, Duan L, Seese A, Gladden AD, Weisgrau KL, Furlott JR, Kim YI, Veloso de Santana MG, Rakasz E, Capuano S, Wilson NA, Bonaldo MC, Galler R, Allison DB, Piatak MJ, Haase AT, Lifson JD, Allen TM, Watkins DI. 2012. Vaccine-induced CD8+ T cells control AIDS virus replication. Nature 491:129–133. doi: 10.1038/nature11443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barouch DH, Liu J, Li H, Maxfield LF, Abbink P, Lynch DM, Iampietro MJ, SanMiguel A, Seaman MS, Ferrari G, Forthal DN, Ourmanov I, Hirsch VM, Carville A, Mansfield KG, Stablein D, Pau MG, Schuitemaker H, Sadoff JC, Billings EA, Rao M, Robb ML, Kim JH, Marovich MA, Goudsmit J, Michael NL. 2012. Vaccine protection against acquisition of neutralization-resistant SIV challenges in rhesus monkeys. Nature 482:89–93. doi: 10.1038/nature10766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barouch DH, Alter G, Broge T, Linde C, Ackerman ME, Brown EP, Borducchi EN, Smith KM, Nkolola JP, Liu J, Shields J, Parenteau L, Whitney JB, Abbink P, Ng'ang'a DM, Seaman MS, Lavine CL, Perry JR, Li W, Colantonio AD, Lewis MG, Chen B, Wenschuh H, Reimer U, Piatak M, Lifson JD, Handley SA, Virgin HW, Koutsoukos M, Lorin C, Voss G, Weijtens M, Pau MG, Schuitemaker H. 2015. Protective efficacy of adenovirus/protein vaccines against SIV challenges in rhesus monkeys. Science 349:320–324. doi: 10.1126/science.aab3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vaccari M, Gordon SN, Fourati S, Schifanella L, Liyanage NP, Cameron M, Keele BF, Shen X, Tomaras GD, Billings E, Rao M, Chung AW, Dowell KG, Bailey-Kellogg C, Brown EP, Ackerman ME, Vargas-Inchaustegui DA, Whitney S, Doster MN, Binello N, Pegu P, Montefiori DC, Foulds K, Quinn DS, Donaldson M, Liang F, Lore K, Roederer M, Koup RA, McDermott A, Ma ZM, Miller CJ, Phan TB, Forthal DN, Blackburn M, Caccuri F, Bissa M, Ferrari G, Kalyanaraman V, Ferrari MG, Thompson D, Robert-Guroff M, Ratto-Kim S, Kim JH, Michael NL, Phogat S, Barnett SW, Tartaglia J, Venzon D, Stablein DM, Alter G, Sekaly RP, Franchini G. 2016. Adjuvant-dependent innate and adaptive immune signatures of risk of SIVmac251 acquisition. Nat Med 22:762–770. doi: 10.1038/nm.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cirelli KM, Crotty S. 2017. Germinal center enhancement by extended antigen availability. Curr Opin Immunol 47:64–69. doi: 10.1016/j.coi.2017.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Picker LJ, Hansen SG, Lifson JD. 2012. New paradigms for HIV/AIDS vaccine development. Annu Rev Med 63:95–111. doi: 10.1146/annurev-med-042010-085643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martins MA, Shin YC, Gonzalez-Nieto L, Domingues A, Gutman MJ, Maxwell HS, Castro I, Magnani DM, Ricciardi M, Pedreño-Lopez N, Bailey V, Betancourt D, Altman JD, Pauthner M, Burton DR, von Bredow B, Evans DT, Yuan M, Parks CL, Ejima K, Allison DB, Rakasz E, Barber GN, Capuano S, Lifson JD, Desrosiers RC, Watkins DI. 2017. Vaccine-induced immune responses against both Gag and Env improve control of simian immunodeficiency virus replication in rectally challenged rhesus macaques. PLoS Pathog 13:e1006529. doi: 10.1371/journal.ppat.1006529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hansen SG, Ford JC, Lewis MS, Ventura AB, Hughes CM, Coyne-Johnson L, Whizin N, Oswald K, Shoemaker R, Swanson T, Legasse AW, Chiuchiolo MJ, Parks CL, Axthelm MK, Nelson JA, Jarvis MA, Piatak MJ, Lifson JD, Picker LJ. 2011. Profound early control of highly pathogenic SIV by an effector memory T-cell vaccine. Nature 473:523–527. doi: 10.1038/nature10003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winstone N, Wilson AJ, Morrow G, Boggiano C, Chiuchiolo MJ, Lopez M, Kemelman M, Ginsberg AA, Mullen K, Coleman JW, Wu CD, Narpala S, Ouellette I, Dean HJ, Lin F, Sardesai NY, Cassamasa H, McBride D, Felber BK, Pavlakis GN, Schultz A, Hudgens MG, King CR, Zamb TJ, Parks CL, McDermott AB. 2011. Enhanced control of pathogenic simian immunodeficiency virus SIVmac239 replication in macaques immunized with an interleukin-12 plasmid and a DNA prime-viral vector boost vaccine regimen. J Virol 85:9578–9587. doi: 10.1128/JVI.05060-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martins MA, Tully DC, Shin YC, Gonzalez-Nieto L, Weisgrau KL, Bean DJ, Gadgil R, Gutman MJ, Domingues A, Maxwell HS, Magnani DM, Ricciardi M, Pedreño-Lopez N, Bailey V, Cruz MA, Lima NS, Bonaldo MC, Altman JD, Rakasz E, Capuano S, Reimann KA, Piatak M, Lifson JD, Desrosiers RC, Allen TM, Watkins DI. 2017. Rare control of SIVmac239 infection in a vaccinated rhesus macaque. AIDS Res Hum Retroviruses 33:843–858. doi: 10.1089/aid.2017.0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sato S, Yuste E, Lauer WA, Chang EH, Morgan JS, Bixby JG, Lifson JD, Desrosiers RC, Johnson WE. 2008. Potent antibody-mediated neutralization and evolution of antigenic escape variants of simian immunodeficiency virus strain SIVmac239 in vivo. J Virol 82:9739–9752. doi: 10.1128/JVI.00871-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chung AW, Kumar MP, Arnold KB, Yu WH, Schoen MK, Dunphy LJ, Suscovich TJ, Frahm N, Linde C, Mahan AE, Hoffner M, Streeck H, Ackerman ME, McElrath MJ, Schuitemaker H, Pau MG, Baden LR, Kim JH, Michael NL, Barouch DH, Lauffenburger DA, Alter G. 2015. Dissecting polyclonal vaccine-induced humoral immunity against HIV using systems serology. Cell 163:988–998. doi: 10.1016/j.cell.2015.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mudd PA, Ericsen AJ, Burwitz BJ, Wilson NA, O'Connor DH, Hughes AL, Watkins DI. 2012. Escape from CD8(+) T cell responses in Mamu-B*00801(+) macaques differentiates progressors from elite controllers. J Immunol 188:3364–3370. doi: 10.4049/jimmunol.1102470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walker BD, Yu XG. 2013. Unravelling the mechanisms of durable control of HIV-1. Nat Rev Immunol 13:487–498. doi: 10.1038/nri3478. [DOI] [PubMed] [Google Scholar]
- 35.Martins MA, Watkins DI. 27 March 2017. What is the predictive value of animal models for vaccine efficacy in humans? Rigorous simian immunodeficiency virus vaccine trials can be instructive. Cold Spring Harb Perspect Biol doi: 10.1101/cshperspect.a029504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dang Q, Hirsch VM. 2008. Rapid disease progression to AIDS due to simian immunodeficiency virus infection of macaques: host and viral factors. Adv Pharmacol 56:369–398. doi: 10.1016/S1054-3589(07)56012-6. [DOI] [PubMed] [Google Scholar]
- 37.Johnson WE, Morgan J, Reitter J, Puffer BA, Czajak S, Doms RW, Desrosiers RC. 2002. A replication-competent, neutralization-sensitive variant of simian immunodeficiency virus lacking 100 amino acids of envelope. J Virol 76:2075–2086. doi: 10.1128/jvi.76.5.2075-2086.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson WE, Sanford H, Schwall L, Burton DR, Parren PW, Robinson JE, Desrosiers RC. 2003. Assorted mutations in the envelope gene of simian immunodeficiency virus lead to loss of neutralization resistance against antibodies representing a broad spectrum of specificities. J Virol 77:9993–10003. doi: 10.1128/JVI.77.18.9993-10003.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson WE, Lifson JD, Lang SM, Johnson RP, Desrosiers RC. 2003. Importance of B-cell responses for immunological control of variant strains of simian immunodeficiency virus. J Virol 77:375–381. doi: 10.1128/JVI.77.1.375-381.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kilgore KM, Murphy MK, Burton SL, Wetzel KS, Smith SA, Xiao P, Reddy S, Francella N, Sodora DL, Silvestri G, Cole KS, Villinger F, Robinson JE, Pulendran B, Hunter E, Collman RG, Amara RR, Derdeyn CA. 2015. Characterization and implementation of a diverse simian immunodeficiency virus SIVsm envelope panel in the assessment of neutralizing antibody breadth elicited in rhesus macaques by multimodal vaccines expressing the SIVmac239 envelope. J Virol 89:8130–8151. doi: 10.1128/JVI.01221-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Means RE, Greenough T, Desrosiers RC. 1997. Neutralization sensitivity of cell culture-passaged simian immunodeficiency virus. J Virol 71:7895–7902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bilello JP, Manrique JM, Shin YC, Lauer W, Li W, Lifson JD, Mansfield KG, Johnson RP, Desrosiers RC. 2011. Vaccine protection against simian immunodeficiency virus in monkeys using recombinant gamma-2 herpesvirus. J Virol 85:12708–12720. doi: 10.1128/JVI.00865-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Evans DT, Bricker JE, Sanford HB, Lang S, Carville A, Richardson BA, Piatak M, Lifson JD, Mansfield KG, Desrosiers RC. 2005. Immunization of macaques with single-cycle simian immunodeficiency virus (SIV) stimulates diverse virus-specific immune responses and reduces viral loads after challenge with SIVmac239. J Virol 79:7707–7720. doi: 10.1128/JVI.79.12.7707-7720.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iwamoto N, Takahashi N, Seki S, Nomura T, Yamamoto H, Inoue M, Shu T, Naruse TK, Kimura A, Matano T. 2014. Control of simian immunodeficiency virus replication by vaccine-induced Gag- and Vif-specific CD8+ T cells. J Virol 88:425–433. doi: 10.1128/JVI.02634-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matano T, Kobayashi M, Igarashi H, Takeda A, Nakamura H, Kano M, Sugimoto C, Mori K, Iida A, Hirata T, Hasegawa M, Yuasa T, Miyazawa M, Takahashi Y, Yasunami M, Kimura A, O'Connor DH, Watkins DI, Nagai Y. 2004. Cytotoxic T lymphocyte-based control of simian immunodeficiency virus replication in a preclinical AIDS vaccine trial. J Exp Med 199:1709–1718. doi: 10.1084/jem.20040432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hansen SG, Piatak MJ, Ventura AB, Hughes CM, Gilbride RM, Ford JC, Oswald K, Shoemaker R, Li Y, Lewis MS, Gilliam AN, Xu G, Whizin N, Burwitz BJ, Planer SL, Turner JM, Legasse AW, Axthelm MK, Nelson JA, Fruh K, Sacha JB, Estes JD, Keele BF, Edlefsen PT, Lifson JD, Picker LJ. 2013. Immune clearance of highly pathogenic SIV infection. Nature 502:100–104. doi: 10.1038/nature12519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Daniel MD, Kirchhoff F, Czajak SC, Sehgal PK, Desrosiers RC. 1992. Protective effects of a live attenuated SIV vaccine with a deletion in the nef gene. Science 258:1938–1941. doi: 10.1126/science.1470917. [DOI] [PubMed] [Google Scholar]
- 48.Fukazawa Y, Park H, Cameron MJ, Lefebvre F, Lum R, Coombes N, Mahyari E, Hagen SI, Bae JY, Reyes MD, Swanson T, Legasse AW, Sylwester A, Hansen SG, Smith AT, Stafova P, Shoemaker R, Li Y, Oswald K, Axthelm MK, McDermott A, Ferrari G, Montefiori DC, Edlefsen PT, Piatak MJ, Lifson JD, Sekaly RP, Picker LJ. 2012. Lymph node T cell responses predict the efficacy of live attenuated SIV vaccines. Nat Med 18:1673–1681. doi: 10.1038/nm.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adnan S, Colantonio AD, Yu Y, Gillis J, Wong FE, Becker EA, Piatak M, Reeves RK, Lifson JD, O'Connor SL, Johnson RP. 2015. CD8 T cell response maturation defined by anentropic specificity and repertoire depth correlates with SIVΔnef-induced protection. PLoS Pathog 11:e1004633. doi: 10.1371/journal.ppat.1004633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adnan S, Reeves RK, Gillis J, Wong FE, Yu Y, Camp JV, Li Q, Connole M, Li Y, Piatak M, Lifson JD, Li W, Keele BF, Kozlowski PA, Desrosiers RC, Haase AT, Johnson RP. 2016. Persistent low-level replication of SIVΔnef drives maturation of antibody and CD8 T cell responses to induce protective immunity against vaginal SIV infection. PLoS Pathog 12:e1006104. doi: 10.1371/journal.ppat.1006104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lu W, Wan Y, Ma F, Johnson RP, Li Q. 2017. Distinct transcriptome profiles of Gag-specific CD8+ T cells temporally correlated with the protection elicited by SIVΔnef live attenuated vaccine. PLoS One 12:e0173929. doi: 10.1371/journal.pone.0173929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Manrique J, Piatak M, Lauer W, Johnson W, Mansfield K, Lifson J, Desrosiers R. 2013. Influence of mismatch of Env sequences on vaccine protection by live attenuated simian immunodeficiency virus. J Virol 87:7246–7254. doi: 10.1128/JVI.00798-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alpert MD, Harvey JD, Lauer WA, Reeves RK, Piatak MJ, Carville A, Mansfield KG, Lifson JD, Li W, Desrosiers RC, Johnson RP, Evans DT. 2012. ADCC develops over time during persistent infection with live-attenuated SIV and is associated with complete protection against SIV(mac)251 challenge. PLoS Pathog 8:e1002890. doi: 10.1371/journal.ppat.1002890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weatherall D. 2006. The use of non-human primates in research. Wellcome Trust, London, United Kingdom. [Google Scholar]
- 55.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]
- 56.Bonaldo MC, Mello SM, Trindade GF, Rangel AA, Duarte AS, Oliveira PJ, Freire MS, Kubelka CF, Galler R. 2007. Construction and characterization of recombinant flaviviruses bearing insertions between E and NS1 genes. Virol J 4:115. doi: 10.1186/1743-422X-4-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martins MA, Bonaldo MC, Rudersdorf RA, Piaskowski SM, Rakasz EG, Weisgrau KL, Furlott JR, Eernisse CM, Veloso de Santana MG, Hidalgo B, Friedrich TC, Chiuchiolo MJ, Parks CL, Wilson NA, Allison DB, Galler R, Watkins DI. 2013. Immunogenicity of seven new recombinant yellow fever viruses 17D expressing fragments of SIVmac239 Gag, Nef, and Vif in Indian rhesus macaques. PLoS One 8:e54434. doi: 10.1371/journal.pone.0054434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rosati M, von Gegerfelt A, Roth P, Alicea C, Valentin A, Robert-Guroff M, Venzon D, Montefiori DC, Markham P, Felber BK, Pavlakis GN. 2005. DNA vaccines expressing different forms of simian immunodeficiency virus antigens decrease viremia upon SIVmac251 challenge. J Virol 79:8480–8492. doi: 10.1128/JVI.79.13.8480-8492.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fuchs JD, Frank I, Elizaga ML, Allen M, Frahm N, Kochar N, Li S, Edupuganti S, Kalams SA, Tomaras GD, Sheets R, Pensiero M, Tremblay MA, Higgins TJ, Latham T, Egan MA, Clarke DK, Eldridge JH, HVTN 090 Study Group and the National Institutes of Allergy and Infectious Diseases. HIV Vaccine Trials Network, Mulligan M, Rouphael N, Estep S, Rybczyk K, Dunbar D, Buchbinder S, Wagner T, Isbell R, Chinnell V, Bae J, Escamilla G, Tseng J, Fair R, Ramirez S, Broder G, Briesemeister L, Ferrara A. 2015. First-in-human evaluation of the safety and immunogenicity of a recombinant vesicular stomatitis virus human immunodeficiency virus-1 gag vaccine (HVTN 090). Open Forum Infect Dis 2:ofv082. doi: 10.1093/ofid/ofv082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Johnson JE, Schnell MJ, Buonocore L, Rose JK. 1997. Specific targeting to CD4+ cells of recombinant vesicular stomatitis viruses encoding human immunodeficiency virus envelope proteins. J Virol 71:5060–5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li H, Wang S, Kong R, Ding W, Lee FH, Parker Z, Kim E, Learn GH, Hahn P, Policicchio B, Brocca-Cofano E, Deleage C, Hao X, Chuang GY, Gorman J, Gardner M, Lewis MG, Hatziioannou T, Santra S, Apetrei C, Pandrea I, Alam SM, Liao HX, Shen X, Tomaras GD, Farzan M, Chertova E, Keele BF, Estes JD, Lifson JD, Doms RW, Montefiori DC, Haynes BF, Sodroski JG, Kwong PD, Hahn BH, Shaw GM. 2016. Envelope residue 375 substitutions in simian-human immunodeficiency viruses enhance CD4 binding and replication in rhesus macaques. Proc Natl Acad Sci U S A 113:E3413–E3422. doi: 10.1073/pnas.1606636113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gonzalez-Nieto L, Domingues A, Ricciardi M, Gutman MJ, Maxwell HS, Pedreño-Lopez N, Bailey V, Magnani DM, Martins MA. 2016. Analysis of simian immunodeficiency virus-specific CD8+ T-cells in rhesus macaques by peptide-MHC-I tetramer staining. J Vis Exp 2016:e54881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alpert MD, Heyer LN, Williams DE, Harvey JD, Greenough T, Allhorn M, Evans DT. 2012. A novel assay for antibody-dependent cell-mediated cytotoxicity against HIV-1- or SIV-infected cells reveals incomplete overlap with antibodies measured by neutralization and binding assays. J Virol 86:12039–12052. doi: 10.1128/JVI.01650-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sok D, Pauthner M, Briney B, Lee JH, Saye-Francisco KL, Hsueh J, Ramos A, Le KM, Jones M, Jardine JG, Bastidas R, Sarkar A, Liang CH, Shivatare SS, Wu CY, Schief WR, Wong CH, Wilson IA, Ward AB, Zhu J, Poignard P, Burton DR. 2016. A prominent site of antibody vulnerability on HIV envelope incorporates a motif associated with CCR5 binding and its camouflaging glycans. Immunity 45:31–45. doi: 10.1016/j.immuni.2016.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xu H, Luo X, Qian J, Pang X, Song J, Qian G, Chen J, Chen S. 2012. FastUniq: a fast de novo duplicates removal tool for paired short reads. PLoS One 7:e52249. doi: 10.1371/journal.pone.0052249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang X, Charlebois P, Gnerre S, Coole MG, Lennon NJ, Levin JZ, Qu J, Ryan EM, Zody MC, Henn MR. 2012. De novo assembly of highly diverse viral populations. BMC Genomics 13:475. doi: 10.1186/1471-2164-13-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Henn MR, Boutwell CL, Charlebois P, Lennon NJ, Power KA, Macalalad AR, Berlin AM, Malboeuf CM, Ryan EM, Gnerre S, Zody MC, Erlich RL, Green LM, Berical A, Wang Y, Casali M, Streeck H, Bloom AK, Dudek T, Tully D, Newman R, Axten KL, Gladden AD, Battis L, Kemper M, Zeng Q, Shea TP, Gujja S, Zedlack C, Gasser O, Brander C, Hess C, Gunthard HF, Brumme ZL, Brumme CJ, Bazner S, Rychert J, Tinsley JP, Mayer KH, Rosenberg E, Pereyra F, Levin JZ, Young SK, Jessen H, Altfeld M, Birren BW, Walker BD, Allen TM. 2012. Whole genome deep sequencing of HIV-1 reveals the impact of early minor variants upon immune recognition during acute infection. PLoS Pathog 8:e1002529. doi: 10.1371/journal.ppat.1002529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tully DC, Ogilvie CB, Batorsky RE, Bean DJ, Power KA, Ghebremichael M, Bedard HE, Gladden AD, Seese AM, Amero MA, Lane K, McGrath G, Bazner SB, Tinsley J, Lennon NJ, Henn MR, Brumme ZL, Norris PJ, Rosenberg ES, Mayer KH, Jessen H, Kosakovsky Pond SL, Walker BD, Altfeld M, Carlson JM, Allen TM. 2016. Differences in the selection bottleneck between modes of sexual transmission influence the genetic composition of the HIV-1 founder virus. PLoS Pathog 12:e1005619. doi: 10.1371/journal.ppat.1005619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Reynolds MR, Weiler AM, Piaskowski SM, Kolar HL, Hessell AJ, Weiker M, Weisgrau KL, Leon EJ, Rogers WE, Makowsky R, McDermott AB, Boyle R, Wilson NA, Allison DB, Burton DR, Koff WC, Watkins DI. 2010. Macaques vaccinated with simian immunodeficiency virus SIVmac239Delta nef delay acquisition and control replication after repeated low-dose heterologous SIV challenge. J Virol 84:9190–9199. doi: 10.1128/JVI.00041-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Martins MA, Wilson NA, Piaskowski SM, Weisgrau KL, Furlott JR, Bonaldo MC, Veloso de Santana MG, Rudersdorf RA, Rakasz EG, Keating KD, Chiuchiolo MJ, Piatak MJ, Allison DB, Parks CL, Galler R, Lifson JD, Watkins DI. 2014. Vaccination with Gag, Vif, and Nef gene fragments affords partial control of viral replication after mucosal challenge with SIVmac239. J Virol 88:7493–7516. doi: 10.1128/JVI.00601-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Martins MA, Tully DC, Cruz MA, Power KA, Veloso de Santana MG, Bean DJ, Ogilvie CB, Gadgil R, Lima NS, Magnani DM, Ejima K, Allison DB, Piatak MJ, Altman JD, Parks CL, Rakasz EG, Capuano S, Galler R, Bonaldo MC, Lifson JD, Allen TM, Watkins DI. 2015. Vaccine-induced simian immunodeficiency virus-specific CD8+ T-cell responses focused on a single Nef epitope select for escape variants shortly after infection. J Virol 89:10802–10820. doi: 10.1128/JVI.01440-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.