Histone PTMs convey information that regulates chromatin compaction and DNA accessibility. Histone reader proteins bind specific PTMs and translate their effects by modifying chromatin and/or by recruiting effectors that alter chromatin structure or activity. In this study, CaLCuV was used to characterize the activities of two Arabidopsis Agenet domain histone readers, EML1 and EML3. We show that eml1 mutants are hypersusceptible to CaLCuV, whereas eml3 plants are more tolerant of infection than wild-type plants. We also demonstrate that EML1 and EML3 associate with histones and viral chromatin in planta and that both proteins bind peptides containing H3K36, a PTM associated with active gene expression. Consistent with antiviral activity, EML1 suppresses CaLCuV gene expression and reduces Pol II access to viral chromatin. By linking EML1 and EML3 to pathogenesis, these studies have expanded our knowledge of histone reader proteins and uncovered an additional level of viral chromatin regulation.
KEYWORDS: geminivirus, histone reader proteins, EML1, EML3
ABSTRACT
Histone posttranslational modifications (PTMs) impart information that regulates chromatin structure and activity. Their effects are mediated by histone reader proteins that bind specific PTMs to modify chromatin and/or recruit appropriate effectors to alter the chromatin landscape. Despite their crucial juxtaposition between information and functional outcome, relatively few plant histone readers have been identified, and nothing is known about their impact on viral chromatin and pathogenesis. We used the geminivirus Cabbage leaf curl virus (CaLCuV) as a model to functionally characterize two recently identified reader proteins, EMSY-LIKE 1 (EML1) and EML3, which contain Tudor-like Agenet domains predictive of histone PTM binding function. Here, we show that mutant Arabidopsis plants exhibit contrasting hypersusceptible (eml1) and tolerant (eml3) responses to CaLCuV infection and that EML1 deficiency correlates with RNA polymerase II (Pol II) enrichment on viral chromatin and upregulated viral gene expression. Consistent with reader activity, EML1 and EML3 associate with nucleosomes and with CaLCuV chromatin, suggesting a direct impact on pathogenesis. We also demonstrate that EML1 and EML3 bind peptides containing histone H3 lysine 36 (H3K36), a PTM usually associated with active gene expression. The interaction encompasses multiple H3K36 PTMs, including methylation and acetylation, suggesting nuanced regulation. Furthermore, EML1 and EML3 associate with similar regions of viral chromatin, implying possible competition between the two readers. Regions of EML1 and EML3 association correlate with sites of trimethylated H3K36 (H3K36me3) enrichment, consistent with regulation of geminivirus chromatin by direct EML targeting.
IMPORTANCE Histone PTMs convey information that regulates chromatin compaction and DNA accessibility. Histone reader proteins bind specific PTMs and translate their effects by modifying chromatin and/or by recruiting effectors that alter chromatin structure or activity. In this study, CaLCuV was used to characterize the activities of two Arabidopsis Agenet domain histone readers, EML1 and EML3. We show that eml1 mutants are hypersusceptible to CaLCuV, whereas eml3 plants are more tolerant of infection than wild-type plants. We also demonstrate that EML1 and EML3 associate with histones and viral chromatin in planta and that both proteins bind peptides containing H3K36, a PTM associated with active gene expression. Consistent with antiviral activity, EML1 suppresses CaLCuV gene expression and reduces Pol II access to viral chromatin. By linking EML1 and EML3 to pathogenesis, these studies have expanded our knowledge of histone reader proteins and uncovered an additional level of viral chromatin regulation.
INTRODUCTION
Histone posttranslational modifications (PTMs) provide information that regulates the structure and function of chromatin, impacting fundamental processes such as gene expression, RNA splicing, and DNA replication and repair (1–4). Distinct combinations of histone PTMs, indicative of different epigenetic states, have been mapped to precise regions of the Arabidopsis thaliana genome and are associated with particular structural and functional environments (5). Most studies to date have focused on the relationship between epigenetic states, chromatin structure, and transcriptional activity.
Chromatin modulators establish, maintain, or alter chromatin structure. Modulators can be broadly classified according to their ability to deposit PTMs (writers), to remove PTMs (erasers), to bind and interpret PTMs (readers), or to reposition nucleosomes (remodelers). Histone writer proteins generate PTMs by adding chemical moieties to specific amino acid residues, often in the ultraconserved histone tails. Modifications include, but are not limited to, acetylation (ac) and methylation (me) of lysine residues. Lysine acetylation (e.g., histone H3 lysine 9 acetylation [H3K9ac]) reduces positive charge and generally increases the accessibility and transcriptional activity of nucleosome-bound DNA. Lysine methylation does not alter charge and can either promote or repress gene expression, largely depending on the position modified. For example, H3K4 and H3K36 methylation is characteristic of actively transcribed genes, while H3K9 and H3K27 methylation is a hallmark of transcriptional gene silencing (TGS) (1, 6–8). In addition, the extent of methylation at a particular residue, mono- di-, or trimethylation (me1, me2, or me3, respectively), can specify distinct structural and functional chromatin states.
Histone reader proteins interpret histone PTMs by binding specific modifications. Reader proteins sometimes possess multiple PTM binding activities and/or additional chromatin modulator activities (e.g., reader/writer domains). Readers are also frequently components of multiprotein complexes that contain other modulators (9–14). Hence, reader proteins represent a crucial link between histone PTMs and the effectors that determine local chromatin structure and activity. However, considering their importance, relatively few plant histone readers and their binding targets have been identified and characterized, limiting our understanding of these proteins and especially how they might impact the structure and function of viral chromatin related to pathogenesis.
Histone reader domains are grouped into classes or families based on their domain structure and organization (9). A structurally conserved group known as the Royal Family is largely responsible for recognizing methylated lysine and arginine residues (15, 16). In Arabidopsis, there are 32 proteins that collectively contain 71 copies of the Royal Family Agenet domain, a structural homolog of animal Tudor domains (17, 18). Until recently, only two proteins with Agenet or Agenet-like domains have been linked to a specific histone PTM target. The tandem Tudor-like fold of the SAWADEE domain of SAWADEE HOMEODOMAIN HOMOLOGUE 1 (SHH1) recognizes H3K9me2 and recruits RNA polymerase IV, a component of the RNA-directed DNA methylation (RdDM) pathway, to silenced loci (19, 20). The Agenet domain-containing ABAP1 INTERACTING PROTEIN 1 (AIP1), important for flowering, is believed to interact with unmodified histones (17, 21).
Four Arabidopsis EMSY-LIKE (EML) proteins have recently been identified as potential histone readers based on their Agenet domain-containing structure (17). EML proteins are characterized by a central Agenet domain and an EMSY N-terminal (ENT) domain. They are homologs of the chromatin-associated R-INTERACTING FACTOR 1 (RIF1) from maize, critical for the expression of genes regulating anthocyanin biosynthesis in the context of chromatin (22). EML activity is also involved in the expression of RPP7, an Arabidopsis resistance gene that confers race-specific immunity to the fungus Hyaloperonospora arabidopsidis (23). In a very recent report (24), the Agenet domain of EML1 was identified primarily as an H3K4me3 histone reader, with some specificity for the H3K36me3 modification.
Geminiviruses cause disease in major crops around the world, including cassava, tomato, beans, and maize (25, 26). These viruses package small circular, single-stranded DNA (ssDNA) genomes in twin icosahedral particles that give the family its name. In the host cell nucleus, viral ssDNA is converted into a double-stranded DNA (dsDNA) replicative form that associates with histones to form nonintegrating minichromosomes. The viral chromatin serves as the template for replication and transcription, which depends on host polymerases and associated machineries. In a classic example of host defense and pathogen counterdefense, multiple studies have shown that plants employ DNA and H3K9 methylation to compact and silence geminivirus chromatin, while viral pathogenicity factors inhibit repressive chromatin methylation (27–36). Collectively, these studies have established geminiviruses as unique model systems to investigate de novo chromatin formation and epigenetic modification in plant cells.
Here, we used the geminivirus Cabbage leaf curl virus (CaLCuV) to characterize the activities and functions of EML1 and EML3. While Arabidopsis plants lacking EML1 or EML3 do not display obvious phenotypes under standard growth conditions, mutant eml1 plants show hypersusceptibility and eml3 plants show increased tolerance to viral infection. Consistent with its defensive role, EML1 is involved in inhibiting RNA polymerase II (Pol II) association with viral chromatin and suppressing viral gene expression. We also demonstrate that EML1 and EML3 have properties consistent with histone reader function: they interact with nucleosomes, associate with viral chromatin, and bind peptides containing multiple modifications of H3K36. In addition, regions of EML1 and EML3 association with viral chromatin overlap sites of H3K36me3 enrichment, further supporting a direct role for these proteins in the regulation of geminivirus chromatin.
RESULTS
Plants lacking EML1 are hypersusceptible to CaLCuV infection.
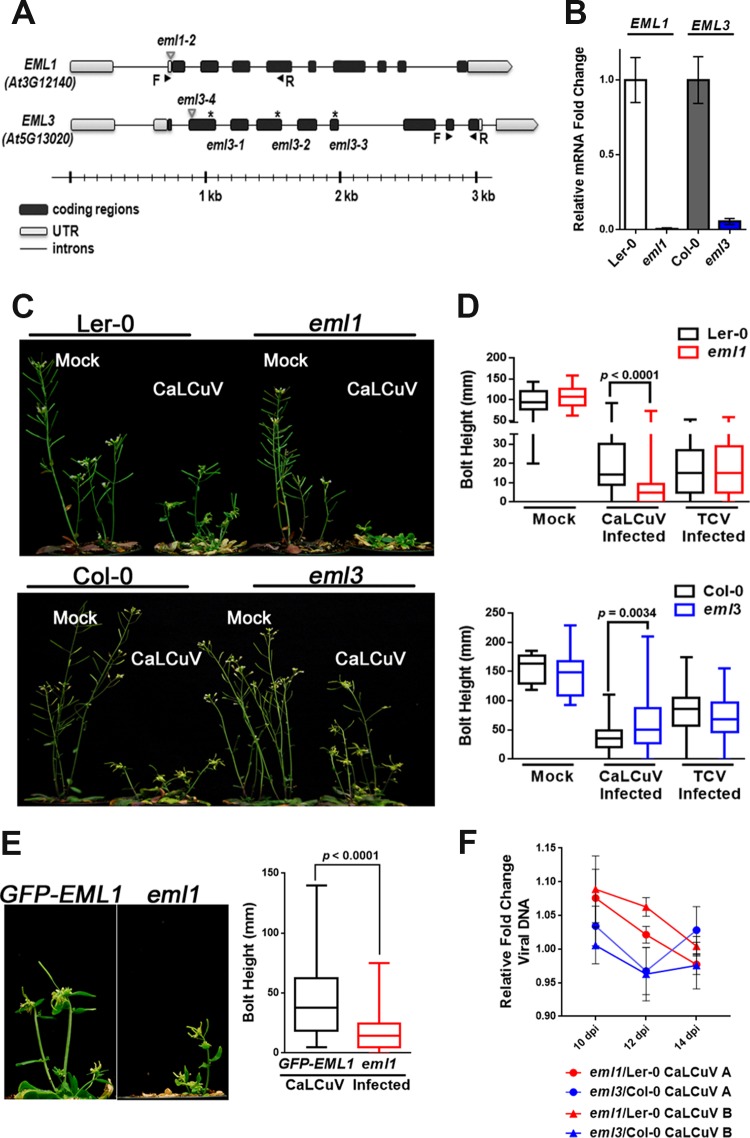
EML1 and EML3 were selected to investigate function and interaction with geminiviruses, as mutant plants exhibited distinct infection phenotypes. Mutant insertion lines (eml1-2 and eml3-4 [here referred to as eml1 and eml3, respectively]) for both of these genes were available although in different Arabidopsis accessions. The eml1 line is in the Landsberg erecta (Ler-0) background, whereas eml3 is in the Columbia (Col-0) background. Sites of Agrobacterium tumefaciens transfer DNA (T-DNA) or transposon insertion within these two genes are illustrated in Fig. 1A.
FIG 1.
Mutant eml1 plants exhibit hypersusceptibility and eml3 plants show increased tolerance to CaLCuV infection. (A) Illustration of transposon/T-DNA insertion sites (triangles) and EMS mutations (asterisks) within EML1 and EML3 genes. Arrows indicate the positions of forward (F) and reverse (R) primers used to amplify transcript cDNAs. UTR, untranslated region. (B) Relative EML1 and EML3 mRNA levels, measured by RT-qPCR, in extracts from silique tissue of eml mutants and cognate wild-type plants using the 2−ΔΔCT method. Values were normalized to values for PP2A (At1g13320). (C) Representative photographs of mock-inoculated and CaLCuV-infected wild-type Ler-0 plants and eml1 plants (top) and wild-type Col-0 and eml3 plants (bottom). (D) Bolt heights were measured from CaLCuV-infected plants (Col-0, n = 96; eml3, n = 46; Ler-0, n = 45; eml1, n = 67), mock-inoculated plants (Col-0, n = 9; eml3, n = 9; Ler-0, n = 11; eml1, n = 19), and TCV-infected plants (Col-0, n = 70; eml3, n = 62; Ler-0, n = 80; eml1, n = 51). The average bolt height per plant was calculated for each treatment, and distributions are depicted in box-and-whisker plots. (E) Representative photographs and bolt measurements of CaLCuV-infected, transgenic GFP-EML1 plants (n = 68) and eml1 plants (n = 34). Bolt heights were measured and plotted as described above for panel D. (F) Levels of CaLCuV DNA A and DNA B in silique and floral tissues of eml1 and eml3 plants, collected at 10, 12, and 14 days postinoculation (dpi), were quantified by qPCR, normalized to 18S ribosomal DNA levels, and compared to viral DNA levels in cognate wild-type plants. Data presented in panels B and D to F were compiled from a minimum of three biological replicates. Significance values were determined using Student's two-tailed t test. Bars in panels B and F indicate standard errors.
To confirm that the eml1 and eml3 mutations compromise the expression of their respective genes, RNA was isolated from wild-type and mutant plants, and quantitative reverse transcriptase PCR (RT-qPCR) was used to assess transcript levels. This analysis verified that EML1 and EML3 transcripts were greatly reduced in mutant compared to cognate wild-type plants (Fig. 1B). Nevertheless, visual inspection revealed no obvious differences in phenotypes between mock-inoculated wild-type and mutant plants (Fig. 1C).
To assess infection phenotypes, plants were inoculated with CaLCuV, and disease symptoms in eml1 and eml3 mutants were compared with those in wild-type plants. Remarkably, under inoculation conditions that normally elicit relatively mild symptoms, infected eml1 plants exhibited very severe stunting, culminating in death or the absence of reproductive shoots (bolts) in 32% of the plants (Fig. 1C, top). In contrast, eml3 mutants were more tolerant of infection than wild-type plants (Fig. 1C, bottom). Similar opposing phenotypes were also noted following infection with the geminivirus Beet curly top virus (BCTV) (not shown). However, as CaLCuV infection phenotypes were more prominent, this virus was used in further studies.
To objectively assess the extent of stunting resulting from CaLCuV infection, the lengths of reproductive shoots (bolt heights) were measured and compared. Measurements confirmed that bolts of surviving eml1 mutants were on average considerably shorter than those of wild-type Ler-0 plants (Fig. 1D, top) and that bolts of infected eml3 mutants were significantly taller than those of infected Col-0 plants (Fig. 1D, bottom).
Infection phenotypes could be attributable to either a direct impact of EML1 and EML3 on CaLCuV or an indirect effect on host defense gene expression. To address this question, eml1 and eml3 plants were inoculated with Turnip crinkle virus (TCV), an RNA virus. No differences in visual symptoms and bolt height were observed between TCV-infected eml1 or eml3 plants and wild-type plants (Fig. 1D), indicating that defenses against TCV are not impaired in the mutants. Thus, CaLCuV infection phenotypes are likely not a general effect of reduced stress tolerance or basal defenses, which are typically effective against a broad spectrum of pathogens.
We concluded that plants deficient for EML1 or EML3 respond to CaLCuV in distinctly different ways and that these proteins specifically impact geminiviruses. Hence, we hypothesized that EML1 provides an antiviral function that suppresses the activity of CaLCuV chromatin, whereas EML3 promotes virulence. Because the eml1 phenotype is more dramatic, we focused mostly on EML1 and used EML3 largely for comparison.
EML1 transgene expression rescues plant responses to CaLCuV.
Insertion lines often have additional mutations in loci other than those annotated (37). To confirm that the phenotype observed in eml1 plants was due specifically to a lack of EML1 expression, a transgene encoding EML1 cDNA with green fluorescent protein (GFP) fused at its N terminus and driven by the native EML1 promoter was introduced into eml1 plants (pEML1::GFP-EML1 [here referred to as GFP-EML1]). Transgene expression was verified by confocal fluorescence microscopy and by Western blotting using GFP antibody as a probe (not shown), and three independent, homozygous lines were selected for analysis. Following CaLCuV inoculation, the GFP-EML1-expressing transgenic lines displayed significantly less stunting than eml1 mutants, as judged by visual inspection and bolt measurements (Fig. 1E), confirming that the eml1 infection phenotype is due to the absence of EML1 protein. Complementation of the eml3 insertion line used in infectivity studies proved technically challenging. Other eml3 mutant lines were pursued (ethyl methanesulfonate [EMS] mutants shown in Fig. 1A), but all of these lines exhibited severe growth defects, suggesting the presence of additional off-target mutations.
Total viral DNA accumulation is not significantly altered in eml1 and eml3 plants.
Changes in host susceptibility often, but not always, correlate with altered viral genome levels. To assess this in CaLCuV-infected eml1 and eml3 plants, the amounts of DNA A and DNA B, both of which are required for systemic infection, were determined by using quantitative PCR (qPCR). Primers were chosen to avoid the common region, which contains sequences shared by both viral genome components. Plants were examined at 10, 12, and 14 days postinoculation (dpi), and samples consisted of DNA isolated from pooled tissue of 4 infected plants to minimize plant-to-plant variation.
As measured by qPCR, EML1 or EML3 deficiency did not affect total viral DNA accumulation. No obvious differences were detected in either CaLCuV DNA A or DNA B levels at any time point in mutant compared to cognate wild-type plants (Fig. 1F). This result may seem surprising, especially for hypersusceptible eml1 plants that might be expected to accumulate more viral DNA than wild-type plants. However, the severe stunting in eml1 plants suggests that CaLCuV may be more meristem invasive in the mutants, leaving fewer cells capable of supporting a productive infection. In any case, these results indicate that the hypersusceptibility and tolerance phenotypes of eml1 and eml3 plants, respectively, do not correlate with differences in total viral DNA accumulation at the whole-plant level.
EML1 and EML3 expression is induced by CaLCuV infection.
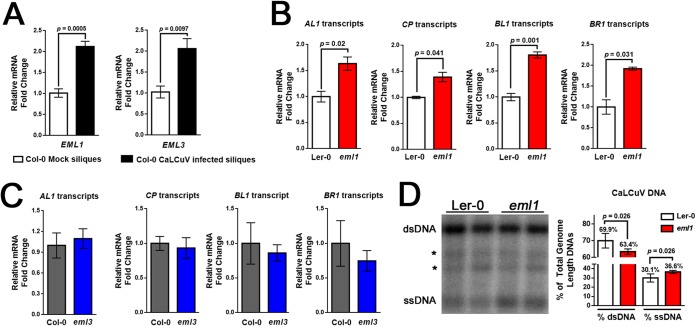
In Arabidopsis, EML1 and EML3 are expressed primarily in siliques and seeds, with low-level expression in other tissues. However, a previous microarray study showed that EML1 expression was significantly upregulated in rosette leaves following CaLCuV infection (38). To further assess the impact of CaLCuV infection on EML1 and EML3 expression, RT-qPCR was employed to measure transcript levels in RNA isolated from the siliques of infected and mock-inoculated wild-type Col-0 plants. This comparative analysis revealed a significant, >2-fold increase in transcript levels for both genes in RNA samples from infected plants (Fig. 2A). Thus, EML1 and EML3 expression levels are increased by CaLCuV infection, suggesting that both genes are responsive to geminiviruses and likely play a role in the infection process.
FIG 2.
Viral gene expression is upregulated in eml1 plants. (A) EML1 and EML3 transcript levels were measured by RT-qPCR in extracts from silique tissue of mock-inoculated and CaLCuV-infected plants. Transcripts were quantified by the 2−ΔΔCT method. (B and C) Viral transcripts from the CaLCuV A genome (AL1 and CP) and B genome (BL1 and BR1) were measured by RT-qPCR and compared in silique and floral tissues of Ler-0 and eml1 mutant plants (B) or Col-0 and eml3 plants (C). (D) The ratio of viral dsDNA to ssDNA was measured by DNA blotting (left). Locations of dsDNA and ssDNA, as well as background signals (asterisks), are shown. The percentage of each form relative to the total (dsDNA plus ssDNA) is shown to the right. Bars indicate standard errors for a minimum of three biological replicates, each with at least two technical replicates. Significance values were determined using Student's two-tailed t test.
Viral gene expression is enhanced in hypersusceptible eml1 plants.
To determine whether phenotypic changes observed in eml1 and eml3 plants were accompanied by changes in viral gene expression, we determined the steady-state levels of selected viral transcripts in extracts from infected wild-type and mutant plants using RT-qPCR. Viral transcripts were further normalized to viral DNA levels determined using qPCR. In all cases, RNA and DNA were obtained from the same extracts. AL1 and CP transcripts from DNA A were selected for analysis, as it was possible to design primers that did not overlap other transcripts (see Fig. 3A for reference). Both of the nonoverlapping DNA B transcripts, BL1 and BR1, were also evaluated.
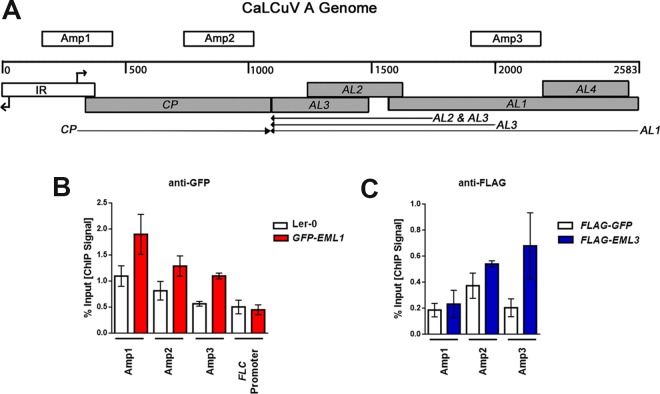
FIG 3.
EML1 and EML3 associate with similar regions of CaLCuV DNA A. (A) Linear representation of the circular dsDNA A genome component (2,583 bp), showing viral open reading frames (shaded boxes), the intergenic region (IR) containing divergent promoters and transcription start sites (right-angle arrows), viral transcripts (arrows), and the amplicons (Amp 1, 2, and 3) tested in ChIP experiments. (B) ChIP-qPCR was performed with nuclear extracts from silique and floral tissues of CaLCuV-infected GFP-EML1 transgenic plants and Ler-0 control plants, using GFP antibody and primers amplifying the indicated amplicons. An amplicon within the FLC promoter served as a negative control. (C) ChIP-qPCR was performed with nuclear extracts from N. benthamiana leaf tissue coinfiltrated with constructs to express CaLCuV DNA A, CaLCuV DNA B, and FLAG-EML3 or FLAG-GFP (control), using FLAG antibody and primers amplifying the indicated amplicons. Bars indicate standard errors for data compiled from at least three biological replicates, each with at least two technical replicates.
Consistent with a hypersusceptible phenotype, we found that levels of AL1 and CP transcripts were moderately but significantly increased ∼1.5-fold in eml1 plants relative to wild-type Ler-0 plants. Similar significant increases approaching 2-fold were also noted for BL1 and BR1 transcript levels (Fig. 2B). In contrast, levels of these same viral transcripts were not significantly different between eml3 and wild-type Col-0 plants, although AL1 transcript levels were slightly increased and BR1 levels were somewhat decreased (Fig. 2C). Thus, the tolerance phenotype of eml3 mutants does not strongly correlate with changes in viral gene expression.
While transcript levels were normalized to total viral DNA levels determined by qPCR, it was possible that the amounts of potential transcription template (i.e., dsDNA) might vary between wild-type Ler-0 and eml1 plants. Because qPCR does not discriminate between the various viral DNA forms, gel blot hybridization was employed to examine the proportions of dsDNA to ssDNA in extracts from wild-type and eml1 plants. DNA was isolated from the pooled tissue of 3 to 4 infected plants and incubated with NcoI to linearize covalently closed and open circular CaLCuV dsDNA. Following hybridization with labeled CP-specific oligonucleotide probes, signals representing full-length CaLCuV dsDNA and ssDNA were quantified from six different samples of pooled plant extracts. The values for the wild type were compared to those for the mutant. Representative samples (2 of each) are shown in Fig. 2D. This analysis revealed a moderate but significant reduction in the proportion of dsDNA to ssDNA in eml1 (63% to 37%) compared to Ler-0 plants (70% to 30%) (Fig. 2D, graph), indicating that mutant plants contain less potential dsDNA transcription template and suggesting that our measurements of steady-state viral transcript levels (per unit template) in this mutant are underestimated (Fig. 2B).
EML1 and EML3 bind CaLCuV chromatin.
Chromatin immunoprecipitation (ChIP) was carried out to determine whether EML proteins directly associate with CaLCuV chromatin. As specific antibodies against the EML proteins are not available, EML1 binding was evaluated using GFP antibody with extracts from CaLCuV-infected, transgenic GFP-EML1 plants. Extracts from infected Ler-0 plants served as the control.
Because transgenic EML3 plants are not available, EML3 association was examined using a transient expression system in Nicotiana benthamiana. Leaves were coinfiltrated with cultures of A. tumefaciens containing plasmids capable of expressing CaLCuV DNA A, DNA B, and FLAG-EML3 or FLAG-GFP as control. The DNA A and DNA B plasmids each contain 1.5 copies of their respective viral DNAs, from which circular monomeric genome components are released by replication and amplified when introduced into plant cells (39). The replication of monomeric DNA A and DNA B following infiltration was verified by DNA gel blot analysis (not shown). FLAG-EML3 and FLAG-GFP were expressed from TRBO, a replicating Tobacco mosaic virus (TMV)-based vector that is released from the Agrobacterium plasmid and amplified when delivered to plant cells (40). ChIP was performed using FLAG antibody with nuclear extracts obtained from infiltration zones. In these and all ChIP experiments, extracts were sonicated under conditions that shear DNA into ∼500-bp fragments.
Three regions of CaLCuV DNA A were selected for qPCR analysis of DNA precipitated by GFP antibody (to analyze binding of EML1) or FLAG antibody (to analyze binding of EML3). The locations of these amplicons (Amp 1, Amp 2, and Amp 3), each ∼290 bp long, are illustrated in Fig. 3A. Amp 1 extends over the rightward half of the CaLCuV intergenic region (IR), which contains divergent Pol II promoters flanking the origin of replication. It includes the CP promoter and transcription start site (TSS) as well as the 5′-terminal portion of the CP coding sequence (CDS). Amp 2 covers the central and 3′-terminal portions of the CP CDS. Finally, Amp 3 spans the central region of the AL1 CDS, which also contains sequences that comprise secondary promoters responsible for AL2/AL3 transcription (41). An amplicon corresponding to the promoter of FLOWERING LOCUS C (FLC) was used as a negative control for experiments using transgenic GFP-EML1 plants based on previous, preliminary results demonstrating a lack of EML1 binding to this genomic region.
Remarkably, we found that both GFP-EML1 and FLAG-EML3 associated with the region encompassed by Amp 3 (Fig. 3B and C). At Amp 3, the EML1 signal was ∼2-fold higher than the background, while the EML3 signal was more than 3-fold over the background. Both EML1 and EML3 were also associated with Amp 2, showing a moderate ∼1.5-fold enrichment over the background in each case. However, only EML1 appeared to bind the region covered by Amp 1 (∼2-fold over the background). We also confirmed the absence of EML1 binding at the FLC promoter (Fig. 3B). FLC could not be evaluated in EML3 ChIP studies, as annotation for the N. benthamiana genome is not available. However, the absence of a signal at Amp 1 confirmed the specificity of FLAG precipitation. From these experiments, we concluded that EML1 and EML3 bind CaLCuV chromatin and, with the exception of Amp 1, interact with similar regions of the viral genome.
EML1 and EML3 bind nucleosomes.
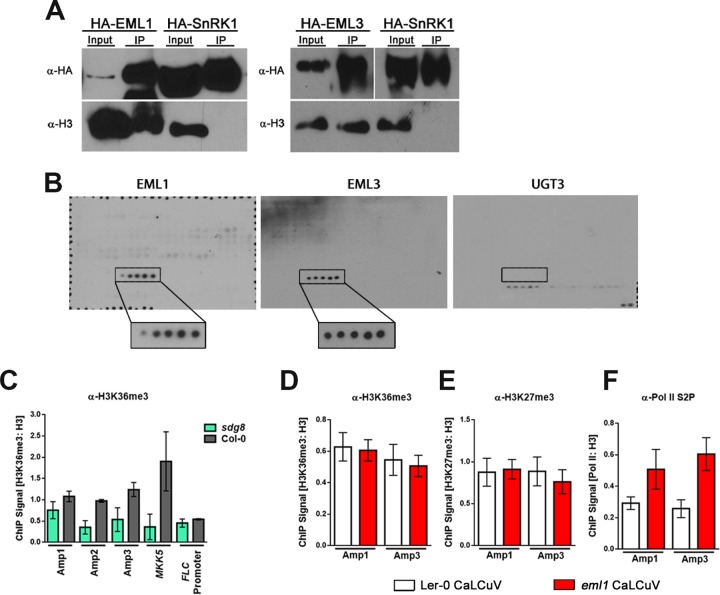
Although EML1 and EML3 were shown to interact with CaLCuV chromatin, it was still unclear whether this interaction was due to the association of EML proteins with nucleosomes. To address this question, coimmunoprecipitation (co-IP) experiments were performed with transiently expressed proteins in N. benthamiana. Leaves were infiltrated with Agrobacterium cultures harboring TRBO plasmids capable of expressing N-terminal hemagglutinin (HA)-tagged EML1 or EML3. The HA-tagged kinase domain of SNF1-RELATED KINASE 1 (SnRK1) was used as a negative control. Extracts were obtained from infiltration zones, and proteins were precipitated with HA antibody. As histone proteins are tightly associated within a nucleosome, histone H3 antibody was used as a proxy for nucleosomes in precipitates.
As shown in Fig. 4A, endogenous plant nucleosomes (as detected by H3 antibody) coimmunoprecipitated with EML1 and EML3 but not with the kinase domain of the control protein SnRK1. These results confirm that EML1 and EML3 associate with nucleosomes, supporting their roles as histone readers in planta.
FIG 4.
EML1 and EML3 are histone readers that recognize H3K36. (A) HA-tagged EML proteins and the kinase domain of SnRK1 (control) were transiently expressed in N. benthamiana leaves. Immunoprecipitation (IP) was performed with HA antibody (α-HA), and immunodetection was performed with HA antibody or histone H3 antibody (α-H3) as a proxy for nucleosomes. (B) Purified His-tagged EML1, EML3, and UGT3 (control) protein association with the MODified histone peptide array (Active Motif). Positive signals detected with His-EML1 and His-EML3 are boxed, with boxes enlarged for increased visibility. Positive signals represent H3 peptide fragments (amino acids 26 to 45) containing, from left to right, unmodified H3K36, H3K36me1, H3K36me2, H3K36me3, and H3K36ac. The box on the UGT3 blot marks the position of the same H3K36-containing peptide fragments for which no binding was detected. Detected signals on this blot are a result of nonspecific binding, based on the presence of signals representing background control peptides (bottom right) after a 3-fold-longer exposure. Probing was performed 3 times for each protein, and the representative blots are shown. (C) ChIP-qPCR was performed with nuclear extracts from silique and floral tissues of wild-type (Col-0) or sdg8 plants infected with CaLCuV using H3K36me3 antibody (α-H3K36me3) and primers amplifying the indicated regions of CaLCuV DNA A (Fig. 3A) or positive-control (MKK5) and negative-control (FLC promoter) loci. Values were normalized to data from ChIP-qPCRs performed with histone H3 antibody using the same extracts. (D to F) ChIP-qPCR was performed with nuclear extracts from silique and floral tissues of Ler-0 (wild-type) and eml1 plants infected with CaLCuV, using H3K36me3 antibody (D), H3K27me3 antibody (E), or elongating Pol II antibody (Pol II phosphorylated at C-terminal domain serine 2 [Pol II S2P]) (F). Primers employed amplified the indicated regions of CaLCuV DNA A. Values were normalized to data from ChIP-qPCRs performed with H3 antibody using the same extracts. Bars indicate standard errors for a minimum of three biological replicates, each with at least two technical replicates.
EML1 and EML3 recognize H3K36.
To determine the specificity of EML protein binding to nucleosomes, EML1 and EML3 containing an N-terminal histidine tag (6×His) were expressed and purified from Escherichia coli. The purified proteins were incubated with MODified histone peptide array slides (Active Motif) printed with 19-mer peptide sequences that span the N-terminal tails of all four canonical histones. Peptide sequences also cover a total of 384 unique histone PTM combinations. EML1 and EML3 binding was detected using a His tag-specific antibody. His-tagged maize UDP GLYCOSYLTRANSFERASE 3 (UGT3) was used as a negative control.
Surprisingly, despite the strikingly different phenotypes in mutant plants, both EML1 and EML3 bound peptides containing H3K36. Even more unexpectedly, all five peptide sequences containing the H3K36 residue were bound by EML1 and EML3, regardless of its PTM. As indicated by the boxed signals in Fig. 4B, EML1 and EML3 bound peptides containing (from left to right) unmodified H3K36, H3K36me1, H3K36me2, H3K36me3, and H3K36ac. The control UGT3 protein showed binding for the slide only at the background level, based on the presence of signals representing background control peptides in the bottom right corner, and only after much longer exposure (Fig. 4B). None of the weak nonspecific signals coincided with the H3K36-containing peptide fragments. We concluded that UGT3 did not bind to the array. We can infer from these results that EML1 and EML3 recognize histone peptides containing H3K36, as only these five peptides on the slide include this residue. The five bound peptides span H3 amino acids 26 to 45 and thus also encompass H3K27, a target of repressive methylation. However, EML1 and EML3 did not bind any of the multiple peptides spanning H3 amino acids 16 to 35 and containing unmodified; mono-, di-, or trimethylated; or acetylated H3K27 either alone or in combination with other PTMs, indicating that binding involves the H3K36 residue. The recent identification of the EML1 Agenet domain as an H3K4me3 histone reader that also recognizes H3K36me3 additionally supports the contribution of the H3K36 residue to the binding specificity of EML proteins (24).
CaLCuV chromatin contains H3K36me3.
Although EML1 and EML3 bound to H3 peptides containing unmodified K36 as well as methylated and acetylated forms, both proteins exhibited a slight preference for H3K36me3. Thus, ChIP was used to determine if this modification is present on the CaLCuV genome. Viral chromatin in extracts from infected wild-type Col-0 plants and from plants deficient for SET DOMAIN GROUP 8 (sdg8), the predominant H3K36me2 and H3K36me3 writer protein in Arabidopsis, was precipitated with H3K36me3 antibody. To account for the decreased association of nucleosomal H3 on chromatin in sdg8 mutants, H3K36me3 levels were normalized to H3 levels, also determined by ChIP.
Using the amplicons shown in Fig. 3A, we observed similar levels of H3K36me3 association with Amp 2 and Amp 3, with signals at both amplicons being ∼3-fold higher than the sdg8 background. Somewhat less H3K36me3 was detected at Amp 1 (Fig. 4C). Considerable H3K36me3 signal (nearly 5-fold over the sdg8 level) was also observed at a positive-control locus, MITOGEN-ACTIVATED PROTEIN KINASE KINASE 5 (MKK5), but not at FLC, which served as a negative control. These results indicate that H3K36me3 is present on viral chromatin and that regions of H3K36me3 enrichment for the most part correspond to sites of EML1 and EML3 association.
EML1 decreases Pol II association with viral chromatin.
The recognition of a histone modification by a reader protein can result in downstream changes in the chromatin landscape, including erasure, deposition, or spread of histone PTMs. Factors that impact transcription might also be recruited. Because mutant plants exhibit elevated viral gene expression levels, we investigated whether the eml1 mutation was associated with changes in viral chromatin (histone) modifications. Modifications such as H3K36me3 are typically associated with chromatin environments that are permissive for transcription, while H3K27me3 is generally inhibitory. Furthermore, the removal of one of these marks can lead to the ectopic introduction of the other, accompanied by changes in gene expression. Thus, we asked whether the absence of EML1 reader activity alters the relative levels of H3K36me3 and H3K27me3.
ChIP was performed with extracts from CaLCuV-infected eml1 and wild-type Ler-0 plants using H3K36me3 or H3K27me3 antibodies, and levels of these marks were normalized to the level of histone H3 and compared at Amp 1 and Amp 3. We found that at both amplicons, H3K36me3 and H3K27me3 levels were similar in wild-type and mutant plants (Fig. 4D and E), suggesting that EML1 does not act to recruit proteins that alter the amounts and locations of these PTMs.
Because viral gene expression is upregulated in eml1 mutants, we also compared levels of elongating Pol II on viral chromatin between mutant and wild-type plants. Elongating Pol II, characterized by phosphorylated serine 2 in the C-terminal domain (Pol II S2P), was detected by ChIP using an antibody that specifically recognizes this modification, and levels were again monitored at Amp 1 and Amp 3. In agreement with increased viral gene expression (Fig. 2B), the association of elongating Pol II with viral chromatin was increased nearly 2-fold at Amp 1 and almost 2.5-fold at Amp 3 in eml1 compared to wild-type plants (Fig. 4F). These results indicate that EML1 likely promotes changes in viral chromatin that inhibit Pol II association and activity.
DISCUSSION
While the number of identified histone PTMs continues to increase, much less is known about their corresponding histone reader proteins and especially the effects elicited by reader association with chromatin (14, 18). This is particularly true in plants where only a few reader proteins have been identified. In this study, we determined the targets of two novel reader proteins, EML1 and EML3, and characterized their interaction with geminivirus chromatin.
We found that eml1 plants were hypersusceptible to CaLCuV, indicating that EML1 plays an important role in defense against this virus. Consistent with defensive function, the absence of EML1 resulted in increased viral gene expression, which correlated with increased amounts of elongating Pol II on viral chromatin. In contrast, eml3 plants displayed tolerance to CaLCuV that was not accompanied by changes in viral gene expression, suggesting that EML3 and EML1 play distinctly different roles during infection. These opposing infection phenotypes highlight the utility of geminiviruses for studying epigenetic events, especially in cases where mutations otherwise have modest or no obvious effects on the plant phenotype.
The severity of disease symptoms observed in eml1 mutants is consistent with increased CP transcript levels and the increased proportion of ssDNA to dsDNA detected in these plants. Our previous work demonstrated that a lack of CP leads to a marked decrease in geminivirus ssDNA levels, highlighting the importance of CP for ssDNA accumulation (42). In addition, while still able to systemically spread through host plants, geminiviruses carrying mutations in CP elicit delayed and attenuated disease symptoms (43). Thus, we speculate that increased CP transcript and ssDNA levels in eml1 mutants are at least partly responsible for accelerating systemic spread and symptom development. The elevated amounts of BR1 and BL1 movement gene transcripts observed in eml1 plants also likely contribute to enhanced cell-to-cell and systemic virus spread.
Several lines of evidence support the idea that EML1 and EML3 are histone reader proteins. First, both proteins contain Agenet domains, which are related to Tudor domains that typically bind methylated lysine and arginine residues (10, 12, 15). We further demonstrated that EML1 and EML3 bind nucleosomes and that both proteins associate with CaLCuV chromatin. We also showed that, despite opposite impacts on infection, both EML1 and EML3 bind peptides containing H3K36, with a slight preference for H3K36me3, a mark typically associated with active chromatin. Consistent with this, we found that H3K36me3 is present on CaLCuV chromatin and that regions of EML1 and EML3 association largely coincide with H3K36me3 enrichment. A very recent publication that characterized a number of putative Arabidopsis histone readers corroborates our findings that EML1 is a histone reader, although those authors report that EML1 binds both H3K36me3 and H3K4me3, with a higher affinity for H3K4me3 (24). However, in our histone peptide binding study (Fig. 4B), while 37 peptides containing the H3K4me3 histone mark were present on the array, no association with this mark was observed in any of three independent replications of this experiment. The observed difference in binding affinities might be explained by our use of the full-length EML1 protein to test the association with histone marks, while Zhao et al. employed the isolated Agenet domain. It is possible that the presence of the ENT domain at the N terminus of EML proteins alters specificity in favor of the H3K36me3 mark.
At present, only one protein family has been assigned an in planta H3K36-reading function in plants. Bu and coauthors have shown that MORF RELATED PROTEINS GENE 1 (MRG1) and MRG2 are dual readers that recognize both H3K4me3 and H3K36me3 to regulate the expression of genes involved in the photoperiodic control of flowering time (44). These proteins utilize chromodomains to bind these specific marks. A similar dual reading preference was observed for rice MRG702, a close homolog of MRG1/2 (45). EML1 and EML3 may represent a second family that also binds both H3K4me3 and H3K36 (24; this study). Although a preference for H3K36me3 was noted in our study, EML1 and EML3 bound peptides containing mono-, di-, and trimethylated H3K36 as well as H3K36ac and unmodified H3K36 residues. This rather broad recognition profile could be a consequence of the in vitro binding conditions employed in our experiments. Alternatively, promiscuous binding might have a structural basis, and we therefore analyzed the EML Agenet domain, as it is the most likely candidate for histone binding. The multiple alignment shown in Fig. 5 demonstrates a high level of sequence conservation when Agenet domains of EML1 and EML3 are compared with Agenet domains of the closely related EML2 and EML4 and also with the uncharacterized plant Tudor/PWWP/MBT superfamily domain-containing proteins AT1G80810, AT4G31880, and AT5G10950. Upon comparing these Arabidopsis Agenet domain-containing proteins with the Tudor domains of the human H3K36 readers PHD Finger Protein 1 (PHF1) and PHF19, we observed that only three of the four residues comprising the aromatic cage involved in binding H3K36me3 are present in the EML proteins (12) (Fig. 5). Secondary-structure modeling studies suggest that the Agenet domains of EML proteins contain four β sheets that form a Tudor-like β barrel fold (17). Thus, while EML1 and EML3 Agenet domains contain the four β strands needed to make the characteristic β barrel of Royal Family domains, the aromatic cage may be incomplete, allowing promiscuous target recognition. Alternatively, the three conserved residues might be sufficient to form the aromatic cage required for H3K36 binding, much like a demonstrated requirement of the aromatic cage of EML1 for H3K4me3 binding (24).
FIG 5.

Agenet domains have an incomplete aromatic cage. Multiple-sequence alignments comparing the Agenet domains of EML1-4, three Arabidopsis Tudor/PWWP/MBT superfamily domain proteins (AT1G80810, AT4G31880, and AT5G10950) sharing a high percentage of amino acid sequence similarity with EML Agenet domains, and the Tudor domains of the structural animal homologs PHF1 and PHF19 were constructed using ClustalW2. Highlighted residues in PHF1 and PHF19 are characterized, conserved aromatic residues necessary for the formation of the aromatic cage for H3K36me3 binding. Asterisks mark identical residues. Dots mark similar residues with various degrees of conservation.
Further in vitro and in vivo studies will be required to determine whether EML1 and EML3 have similar or different binding preferences for H3K36 PTMs. Different binding preferences could explain their strikingly different impacts on geminivirus pathogenicity. Alternatively, EML1 and EML3 may have very similar binding activities but are components of distinct protein complexes that condition different functional states. Our results showing that both EML1 and EML3 associate with Amp 2 and Amp 3, while EML1 additionally associates with Amp 1, lend some support to this speculation. These association patterns are also interesting in light of the positional and functional effects of H3K36me3 localization within genomes. In animals and yeast, this histone PTM is often localized to the 3′ end of gene coding regions, where it aids in Pol II elongation by recruiting histone deacetylases that reduce histone turnover/exchange on chromatin (46–48). This in turn decreases Pol II access, limiting aberrant initiation and cryptic transcription downstream of genuine start sites and/or from the antisense strand. In plants, H3K36me3 is localized to the gene body with a slight 5′-end bias and does not appear to play a role in inhibiting cryptic transcription (5, 49). Instead, this PTM is responsible for accelerating Pol II initiation and elongation rates, thus increasing transcript yields (50). As viral transcription is upregulated in eml1 mutant plants, it is interesting to speculate that the antiviral function of EML1 involves binding H3K36 on viral chromatin previously marked as active, rendering it less accessible to Pol II.
In summary, experiments presented in this report demonstrate that EML1 and EML3 have dramatically different effects on the outcome of geminivirus infection. We also verify histone reader activity for Arabidopsis EML1 and EML3 proteins via their association with nucleosomes, interaction with viral chromatin, and binding preference for H3K36 residues. Finally, we provide evidence that EML1 reader activity regulates viral gene expression and Pol II access to viral chromatin.
MATERIALS AND METHODS
Plant materials, inoculation, and bolt height measurement.
Arabidopsis thaliana accessions Col-0 (CS70000) and Ler-0 (CS20) were obtained from the Arabidopsis Biological Resource Center (ABRC) at The Ohio State University. The transposon or T-DNA insertion alleles eml1-2, eml3-4, and sdg8-2 were identified from ABRC stocks CS101744, SALK_106147, and SALK_026442, respectively. The homozygosity of all mutants was verified by genotyping. Arabidopsis plants were grown in a long-day photoperiod (18 h/6 h day/night) at 22°C. Nicotiana benthamiana plants were grown at 27°C with 12 h of light. Plants were agroinoculated with Cabbage leaf curl virus (CaLCuV) or Turnip crinkle virus (TCV), and tissue was collected at ∼2 weeks postinoculation as previously described (27). Average bolt heights were calculated per plant, and a box-and-whisker plot was used to represent their distribution.
Transient expression constructs.
For HA- or FLAG-tagged EML1 and EML3, cDNA was generated from plant RNA using SuperScript III RT (ThermoFisher). Primers containing the restriction sites PacI and AscI were used to amplify EML1 or EML3 coding sequences (CDSs) using high-fidelity Pfx polymerase (Invitrogen). Primers will be provided upon request. Purified PCR products and Ti plasmid-based TRBO destination vectors bearing N-terminal HA or FLAG tags (40) were digested with PacI and AscI and ligated using T4 DNA ligase (Invitrogen). Constructs were transformed into Agrobacterium tumefaciens strain C58C1. The SnRK1 construct was previously described (51).
GFP-EML1 transgenic lines.
A GFP-tagged EML1 construct driven by the EML1 promoter was generated using multisite Gateway technology (Invitrogen). The EML1 promoter region (1,349 bp upstream from the start codon) was amplified from Ler-0 genomic DNA. The EML1 CDS was amplified from Ler-0 cDNA, and the GFP sequence was amplified from pGWB6 (52). All PCRs used primers with suitable attachment site (attB) adapters. Primers will be provided upon request. PCR fragments were cloned into the following pDONR vectors using BP Clonase II (Invitrogen): GFP into pDONR221, the promoter into pDONRP4P1R, and the EML1 CDS into pDONRP2RP3. The three constructs were combined in an LR reaction into pH7m34GW (VIB-UGent Center for Plant Systems Biology) and used to transform Agrobacterium strain GV3101. Agrobacterium cells were used to transform the eml1-2 mutant by floral dip, and transgenic plants were selected on hygromycin. Reporter expression and transgene complementation were verified in 3 independent homozygous T3 lines.
Agroinfiltration and transient infection.
N. benthamiana plants were grown until 3 to 4 weeks postgermination. For coimmunoprecipitation experiments, younger leaves were infiltrated with Agrobacterium (strain C58C1) containing TRBO vectors designed to express HA-tagged EML1, EML3, or SnRK1. For transient infection and ChIP experiments, leaves were infiltrated with a mixture of Agrobacterium cells carrying CaLCuV DNA A, CaLCuV DNA B (GV3111), and a TRBO vector expressing FLAG-tagged EML3 (C58C1) or GFP (GV3101). Cultures were combined in a 1:1:1 ratio. Infiltration zones were collected at 3 to 5 days postinfiltration.
Nucleic acid extraction, qPCR, RT-qPCR, and DNA gel blot analysis.
Total RNA was extracted from uninfected silique tissue using PureLink RNA reagent (ThermoFisher Scientific) according to the manufacturer's instructions. Isolation of DNA and RNA from symptomatic floral heads and siliques of CaLCuV-infected plants was performed as described previously (36). RNA was reverse transcribed using SuperScript III with oligo(dT) primers, and cDNAs were used to perform quantitative reverse transcriptase PCR (RT-qPCR) with Bio-Rad iQ SYBR green supermix and the Bio-Rad CFX96 real-time PCR detection system (34). PP2A (At1G13320) was used as a reference gene, and comparisons of steady-state host and viral transcript levels were performed using the 2−ΔΔCT method (53). Viral transcript levels were normalized to viral DNA levels determined using qPCR. DNA gel blotting and analysis were carried out essentially as previously reported (36).
Coimmunoprecipitation.
Infiltrated N. benthamiana leaf tissue was ground into a fine powder and lysed in solution (100 mM Tris-HCl [pH 8.0], 1 mM EDTA, 10% glycerol, 0.5% Triton X-100, 1 mM dithiothreitol [DTT], 10 mM NaCl, Sigma protease inhibitor cocktail). Two units of Benzonase (Sigma) were added to each lysate. Samples were rotated at 4°C for 1 h and sonicated using a Diagenode Bioruptor (at 200 W) as follows: 3 rounds of 10 min of sonication (30 s on and 30 s off per min). Lysates were then rotated for an hour at 4°C and centrifuged at ∼13,000 rpm for 10 min at 4°C to pellet insoluble cellular debris. Supernatants were incubated with HA-conjugated beads (catalog number sc805-AC; Santa-Cruz) to immunoprecipitate HA-tagged EML1, EML3, and SnRK1 overnight at 4°C. Beads were washed the next day three times in lysis buffer containing 75 mM NaCl. Immunoprecipitated samples were separated by 12% SDS-PAGE (at 100 V), transferred to polyvinylidene difluoride (PVDF) membranes (at 120 mA), and probed with HA-horseradish peroxidase (HRP) antibody (catalog number H6533; Sigma) (1:2,000) or with histone H3 antibody (catalog number A01502-40; GenScript) (1:2,000). A rabbit IgG-HRP antibody (catalog number A1949; Sigma) was used to detect the H3 antibody.
Purification of EML proteins and histone peptide array probing.
EML3 and EML1 coding sequences were cloned as His6 fusion proteins. The EML3 CDS was inserted into the Gateway vector pDEST17 (Invitrogen) for expression in E. coli BL21-Al cells (Invitrogen). The EML1 CDS was inserted into pRSETA (Invitrogen) for expression in E. coli BL21(DE3)/pLysS. A His-tagged UGT construct in pET-28a(+) was obtained as an E. coli Rosetta/pLysS transformant (Novagen).
The expression of recombinant His-EML1 and His-UGT3 proteins was induced by adding isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 1 mM. His-EML3 was induced by adding l-arabinose to a final concentration of 0.2%. Recombinant proteins were purified using Ni-nitrilotriacetic acid (NTA) agarose according to the manufacturer's instructions (Qiagen). Eluates were collected into a Pierce 20,000-molecular-weight-cutoff (MWCO) concentrator and subjected to diafiltration to exchange the elution buffer with the interaction buffer (MODified protein domain binding kit; Active Motif). The MODified histone peptide array (Active Motif) was blocked, washed, and prepared according to the manufacturer's instructions. Slides were incubated with purified His-tagged proteins at 4°C overnight, washed, and incubated with primary mouse His6 antibody (1:3,000) (Active Motif) and secondary anti-mouse-HRP antibody (1:2,500) (Active Motif) according to kit instructions. Signals were detected by using the enhanced chemiluminescence (ECL) detection system and analyzed with Active Motif Array Analyze software.
Chromatin immunoprecipitation.
Chromatin immunoprecipitation (ChIP) was performed as described previously, with minor modifications (27). Assays were performed with antibodies against FLAG (catalog number F1804-200UG; Sigma), GFP (catalog number ab290; Abcam), histone H3 (catalog number A01502-40; GenScript), H3K36me3 (catalog number ab9050; Abcam), H3K27me3 (catalog number ab6002; Abcam), or Pol II S2P (catalog number A01633; GenScript), as appropriate. Immunoprecipitated DNA was quantified using qPCR.
ACKNOWLEDGMENTS
We are grateful to Erich Grotewold for the suggestion to use geminiviruses as a tool to test the reader function of EML proteins and his laboratory for providing resources, especially Asela Wijeratne for eml1-2 and eml3-4 mutants and Maria I. Casas for the His-UGT3 construct. We also thank Feng Qu for providing the TCV inoculum and members of the Bisaro laboratory and the laboratories of Keith Slotkin and Michael Poirier for advice and technical support. Finally, we thank Jamie Jackel and Virginia L. Fernandez for performing preliminary experiments related to this work.
Work in the Bisaro laboratory was supported by grants from the National Science Foundation (NSF MCB-1158262 and IOS-1354636) and the U.S. Department of Agriculture, National Institute of Food and Agriculture (USDA/NIFA 2015-6703-22999). Funding for this project was also provided by a grant from the Ohio Plant Biotechnology Consortium (OPBC2011-003) through the Ohio Agricultural Research and Development Center (OARDC) to J.B. Support for T.C. was provided by the Cellular, Molecular, and Biochemical Sciences Training Program (NIH T32-GM-086252 from the National Institute for General Medical Sciences, National Institutes of Health). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Bannister AJ, Kouzarides T. 2011. Regulation of chromatin by histone modifications. Cell Res 21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumar R, Horikoshi N, Singh M, Gupta A, Misra HS, Albuquerque K, Hunt CR, Pandita TK. 2013. Chromatin modifications and the DNA damage response to ionizing radiation. Front Oncol 2:214. doi: 10.3389/fonc.2012.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou H-L, Luo G, Wise JA, Lou H. 2014. Regulation of alternative splicing by local histone modifications: potential roles for RNA-guided mechanisms. Nucleic Acids Res 42:701–713. doi: 10.1093/nar/gkt875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiao J, Lee U-S, Wagner D. 2016. Tug of war: adding and removing histone lysine methylation in Arabidopsis. Curr Opin Plant Biol 34:41–53. doi: 10.1016/j.pbi.2016.08.002. [DOI] [PubMed] [Google Scholar]
- 5.Roudier F, Ahmed I, Berard C, Sarazin A, Mary-Huard T, Cortijo S, Bouyer D, Caillieux E, Duvernois-Berthet E, Al-Shikhley L, Giraut L, Despres B, Drevensek S, Barneche F, Derozier S, Brunaud V, Aubourg S, Schnittger A, Bowler C, Martin-Magniette M-L, Robin S, Caboche M, Colot V. 2011. Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J 30:1928–1938. doi: 10.1038/emboj.2011.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ebbs ML, Bender J. 2006. Locus-specific control of DNA methylation by the Arabidopsis SUVH5 histone methyltransferase. Plant Cell 18:1166–1176. doi: 10.1105/tpc.106.041400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernatavichute YV, Zhang X, Cokus S, Pelligrini M, Jacobsen SE. 2008. Genome-wide association of histone H3 lysine nine methylation with CHG DNA methylation in Arabidopsis thaliana. PLoS One 3:e3156. doi: 10.1371/journal.pone.0003156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang H, Howard M, Dean C. 2014. Antagonistic roles for H3K36me3 and H3K27me3 in the cold-induced epigenetic switch at Arabidopsis FLC. Curr Biol 24:1793–1797. doi: 10.1016/j.cub.2014.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Musselman CA, Lalonde M-E, Cote J, Kutateladze TG. 2012. Perceiving the epigenetic landscape through histone readers. Nat Struct Mol Biol 19:1218–1227. doi: 10.1038/nsmb.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ballare C, Lange M, Lapinaite A, Martin GM, Morey L, Pascual G, Liefke R, Simon B, Shi Y, Gozani O, Carlomagno T, Benitah SA, Di Croce L. 2012. Phf19 links methylated Lys36 of histone H3 to regulation of Polycomb activity. Nat Struct Mol Biol 19:1257–1265. doi: 10.1038/nsmb.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maltby VE, Martin BJE, Schulze JM, Johnson I, Hentrich T, Sharma A, Kobor MS, Howe L. 2012. Histone H3 lysine 36 methylation targets the Isw1b remodeling complex to chromatin. Mol Cell Biol 32:3479–3485. doi: 10.1128/MCB.00389-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Musselman CA, Avvakumov N, Watanabe R, Abraham CG, Lalonde M-E, Hong Z, Allen C, Roy S, Nunez JK, Nickoloff J, Kulesza CA, Yasui A, Cote J, Kutateladze TG. 2012. Molecular basis for H3K36me3 recognition by the Tudor domain of PHF1. Nat Struct Mol Biol 19:1266–1272. doi: 10.1038/nsmb.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du J, Johnson LM, Jacobsen SE, Patel DJ. 2015. DNA methylation pathways and their crosstalk with histone methylation. Nat Rev Mol Cell Biol 16:519–532. doi: 10.1038/nrm4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andrews FH, Strahl BD, Kutateladze TG. 2016. Insights into newly discovered marks and readers of epigenetic information. Nat Chem Biol 12:662–668. doi: 10.1038/nchembio.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maurer-Stroh S, Dickens NJ, Hughes-Davies L, Kouzarides T, Eisenhaber F, Ponting C. 2003. The Tudor domain “Royal Family”: Tudor, Agenet, Chromo, PWWP, and MBT domains. Trends Biochem Sci 28:69–74. doi: 10.1016/S0968-0004(03)00004-5. [DOI] [PubMed] [Google Scholar]
- 16.Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. 2007. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol 14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brasil JN, Cabral LM, Eloy NB, Primo LMF, Barroso-Neto IL, Grangeiro LPP, Gonzalez N, Inze D, Ferreira PCG, Hemerly AS. 2015. AIP1 is a novel Agenet/Tudor domain protein from Arabidopsis that interacts with regulators of DNA replication. BMC Plant Biol 15:270. doi: 10.1186/s12870-015-0641-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Y, Min J. 2016. Structure and function of histone methylation-binding proteins in plants. Biochem J 473:1663–1680. doi: 10.1042/BCJ20160123. [DOI] [PubMed] [Google Scholar]
- 19.Law JA, Vashisht AA, Wohlschlegel JA, Jacobsen SE. 2011. SHH1, a homeodomain protein required for DNA methylation, as well as RDR2, RDM4, and chromatin remodeling factors, associate with RNA polymerase IV. PLoS Genet 7:e1002195. doi: 10.1371/journal.pgen.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Law JA, Du J, Hale CJ, Feng S, Krajewski K, Palanca AMS, Strahl BD, Patel DJ, Jacobsen SE. 2013. Polymerase IV occupancy at RNA-directed DNA methylation sites requires SHH1. Nature 498:385–389. doi: 10.1038/nature12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masuda HP, Cabral LM, De Veylder L, Tanurdzic M, de Almeida EJ, Geelen D, Inze D, Martienssen RA, Ferreira PCG, Hemerly AS. 2008. ABAP1 is a novel plant Armadillo BTB protein involved in DNA replication and transcription. EMBO J 27:2746–2756. doi: 10.1038/emboj.2008.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hernandez JM, Feller A, Morohashi K, Frame K, Grotewold E. 2007. The basic helix-loop-helix domain of maize R links transcriptional regulation and histone modifications by recruitment of an EMSY-related factor. Proc Natl Acad Sci U S A 104:17222–17227. doi: 10.1073/pnas.0705629104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsuchiya T, Eulgem T. 2011. EMSY-Like genes are required for full RPP7-mediated race-specific immunity and basal defense in Arabidopsis. Mol Plant Microbe Interact 24:1573–1581. doi: 10.1094/MPMI-05-11-0123. [DOI] [PubMed] [Google Scholar]
- 24.Zhao S, Zhang B, Yang M, Zhu J, Li H. 2018. Systematic profiling of histone readers in Arabidopsis thaliana. Cell Rep 22:1090–1102. doi: 10.1016/j.celrep.2017.12.099. [DOI] [PubMed] [Google Scholar]
- 25.Jeske H. 2009. Geminiviruses. Curr Top Microbiol Immunol 331:185–226. [DOI] [PubMed] [Google Scholar]
- 26.Hanley-Bowdoin L, Bejarano ER, Robertson D, Mansoor S. 2013. Geminiviruses: masters at redirecting and reprogramming plant processes. Nat Rev Microbiol 11:777–788. doi: 10.1038/nrmicro3117. [DOI] [PubMed] [Google Scholar]
- 27.Raja P, Sanville BC, Buchmann RC, Bisaro DM. 2008. Viral genome methylation as an epigenetic defense against geminiviruses. J Virol 82:8997–9007. doi: 10.1128/JVI.00719-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buchmann RC, Asad S, Wolf JN, Mohannath G, Bisaro DM. 2009. Geminivirus AL2 and L2 proteins suppress transcriptional gene silencing and cause genome-wide reductions in cytosine methylation. J Virol 83:5005–5013. doi: 10.1128/JVI.01771-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang X, Xie Y, Raja P, Li S, Wolf JN, Shen Q, Bisaro DM, Zhou X. 2011. Suppression of methylation-mediated transcriptional gene silencing by βC1-SAHH protein interaction during geminivirus-betasatellite infection. PLoS Pathog 7:e1002329. doi: 10.1371/journal.ppat.1002329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Z, Chen H, Huang X, Xia R, Zhao Q, Lai J, Teng K, Li Y, Liang L, Du Q, Zhou X, Guo H, Xie Q. 2011. BSCTV C2 attenuates the degradation of SAMDC1 to suppress DNA methylation-mediated gene silencing in Arabidopsis. Plant Cell 23:273–288. doi: 10.1105/tpc.110.081695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raja P, Jackel JN, Li S, Heard IM, Bisaro DM. 2014. Arabidopsis double-stranded RNA binding protein DRB3 participates in methylation-mediated defense against geminiviruses. J Virol 88:2611–2622. doi: 10.1128/JVI.02305-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Castillo-Gonzalez C, Liu X, Huang C, Zhao C, Ma Z, Hu T, Sun F, Zhou Y, Zhou X, Wang X-J, Zhang X. 2015. Geminivirus-encoded TrAP suppressor inhibits the histone methyltransferase SUVH4/KYP to counter host defense. Elife 4:e06671. doi: 10.7554/eLife.06671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jackel JN, Buchmann RC, Singhal U, Bisaro DM. 2015. Analysis of geminivirus AL2 and L2 proteins reveals a novel AL2 silencing suppressor activity. J Virol 89:3176–3187. doi: 10.1128/JVI.02625-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jackel JN, Storer JM, Coursey T, Bisaro DM. 2016. Arabidopsis RNA polymerases IV and V are required to establish H3K9 methylation, but not cytosine methylation, on geminivirus chromatin. J Virol 90:7529–7540. doi: 10.1128/JVI.00656-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ceniceros-Ojeda EA, Rodriguez-Negrete EA, Rivera-Bustamante RF. 2016. Two populations of viral minichromosomes are present in a geminivirus-infected plant showing symptom remission (recovery). J Virol 90:3828–3838. doi: 10.1128/JVI.02385-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coursey T, Regedanz E, Bisaro DM. 2018. Arabidopsis RNA polymerase V mediates enhanced compaction and silencing of geminivirus and transposon chromatin during host recovery from infection. J Virol 92:e01320-. doi: 10.1128/JVI.01320-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Malley RC, Ecker JR. 2010. Linking genotype to phenotype using the Arabidopsis unimutant collection. Plant J 61:928–940. doi: 10.1111/j.1365-313X.2010.04119.x. [DOI] [PubMed] [Google Scholar]
- 38.Ascencio-Ibanez JT, Sozzani R, Lee TJ, Chu TM, Wolfinger RD, Cella R, Hanley-Bowdoin L. 2008. Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol 148:436–454. doi: 10.1104/pp.108.121038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stenger DC, Revington GN, Stevenson MC, Bisaro DM. 1991. Replicational release of geminivirus genomes from tandemly repeated copies: evidence for rolling circle replication of a plant viral DNA. Proc Natl Acad Sci U S A 88:8029–8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lindbo JA. 2007. TRBO: a high-efficiency Tobacco mosaic virus RNA-based overexpression vector. Plant Physiol 145:1232–1240. doi: 10.1104/pp.107.106377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shung CY, Sunter J, Sirasanagandla SS, Sunter G. 2006. Distinct viral sequence elements are necessary for expression of Tomato golden mosaic virus complementary sense transcripts that direct AL2 and AL3 gene expression. Mol Plant Microbe Interact 19:1394–1405. doi: 10.1094/MPMI-19-1394. [DOI] [PubMed] [Google Scholar]
- 42.Sunter G, Hartitz MD, Hormuzdi SG, Brough CL, Bisaro DM. 1990. Genetic analysis of tomato golden mosaic virus. ORF AL2 is required for coat protein accumulation while ORF AL3 is necessary for efficient DNA replication. Virology 179:69–77. doi: 10.1016/0042-6822(90)90275-V. [DOI] [PubMed] [Google Scholar]
- 43.Gardiner WE, Sunter G, Brand L, Elmer JS, Rogers SG, Bisaro DM. 1988. Genetic analysis of tomato golden mosaic virus: the coat protein is not required for systemic spread or symptom development. EMBO J 7:899–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bu Z, Yu Y, Li Z, Liu Y, Jiang W, Huang Y, Dong A-W. 2014. Regulation of Arabidopsis flowering locus by the histone mark readers MRG1/2 via interaction with CONSTANS to modulate FT expression. PLoS Genet 10:e1004617. doi: 10.1371/journal.pgen.1004617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jin J, Shi J, Liu B, Liu Y, Huang Y, Dong A. 2015. MORF-RELATED GENE702, a reader protein of trimethylated histone H3 lysine 4 and histone H3 lysine 36, is involved in brassionosteroid-regulated growth and flowering time control in rice. Plant Physiol 168:1275–1285. doi: 10.1104/pp.114.255737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL. 2005. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 47.Carvalho S, Raposo AC, Martins FB, Grosso AR, Sridhara SC, Rino J, Carmo-Fonseca M, de Almeida SF. 2013. Histone methyltransferase SETD2 coordinates FACT recruitment with nucleosome dynamics during transcription. Nucleic Acids Res 41:2881–2893. doi: 10.1093/nar/gks1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Venkatesh S, Smolle M, Li H, Gogol MM, Saint M, Kumar S, Natarajan K, Workman JL. 2012. Set2 methylation of histone H3 lysine 36 suppresses histone exchange on transcribed genes. Nature 489:452–455. doi: 10.1038/nature11326. [DOI] [PubMed] [Google Scholar]
- 49.Mahrez W, Arellano MS, Moreno-Romero J, Nakamura M, Shu H, Nanni P, Kohler C, Gruissem W, Hennig L. 2016. H3K36ac is an evolutionarily conserved plant histone modification that marks active genes. Plant Physiol 170:1566–1577. doi: 10.1104/pp.15.01744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu Z, Ietswaart R, Liu F, Yang H, Howard M, Dean C. 2016. Quantitative regulation of FLC via coordinated transcriptional initiation and elongation. Proc Natl Acad Sci U S A 113:218–223. doi: 10.1073/pnas.1518369112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mohannath G, Jackel JN, Lee YH, Buchmann RC, Wang H, Patil V, Adams AK, Bisaro DM. 2014. A complex containing SNF1-related kinase (SnRK1) and adenosine kinase in Arabidopsis. PLoS One 9:e87592. doi: 10.1371/journal.pone.0087592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T. 2007. Development of a series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104:34–41. doi: 10.1263/jbb.104.34. [DOI] [PubMed] [Google Scholar]
- 53.Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]