Viruses are obligate parasites, and the production of progeny viruses relies strictly on the host translation machinery. Therefore, the efficient modulation of host mRNA translation benefits viral replication, spread, and evolution. In this study, we provide evidence that porcine reproductive and respiratory syndrome virus (PRRSV) infection induces host translation shutoff and that the viral nonstructural protein nsp2 is associated with this process. Many viruses induce host translation shutoff by phosphorylating eukaryotic initiation factor 2α (eIF2α). However, PRRSV nsp2 does not induce eIF2α phosphorylation but attenuates the mTOR signaling pathway, another pathway regulating the host cell translational machinery. We also found that PRRSV-induced host translation shutoff was partly reversed by eliminating the effects of eIF2α phosphorylation or reactivating the mTOR pathway, indicating that PRRSV infection induces both eIF2α phosphorylation-dependent and -independent host translation shutoff.
KEYWORDS: porcine reproductive and respiratory syndrome virus, host translation shutoff, nonstructural protein 2 (nsp2)
ABSTRACT
Porcine reproductive and respiratory syndrome virus (PRRSV) is an Arterivirus that has caused tremendous economic losses in the global swine industry since it was discovered in the late 1980s. Inducing host translation shutoff is a strategy used by many viruses to optimize their replication and spread. Here, we demonstrate that PRRSV infection causes host translation suppression, which is strongly dependent on viral replication. By screening PRRSV-encoded nonstructural proteins (nsps), we found that nsp2 participates in the induction of host translation shutoff and that its transmembrane (TM) domain is required for this process. nsp2-induced translation suppression is independent of protein degradation pathways and the phosphorylation of eukaryotic initiation factor 2α (eIF2α). However, the overexpression of nsp2 or its TM domain significantly attenuated the mammalian target of rapamycin (mTOR) signaling pathway, an alternative pathway for modulating host gene expression. PRRSV infection also attenuated the mTOR signaling pathway, and PRRSV-induced host translation shutoff could be partly reversed when the attenuated mTOR phosphorylation was reactivated by an activator of the mTOR pathway. PRRSV infection still negatively regulated the host translation when the effects of eIF2α phosphorylation were completely reversed. Taken together, our results demonstrate that PRRSV infection induces host translation shutoff and that nsp2 is associated with this process. Both eIF2α phosphorylation and the attenuation of the mTOR signaling pathway contribute to PRRSV-induced host translation arrest.
IMPORTANCE Viruses are obligate parasites, and the production of progeny viruses relies strictly on the host translation machinery. Therefore, the efficient modulation of host mRNA translation benefits viral replication, spread, and evolution. In this study, we provide evidence that porcine reproductive and respiratory syndrome virus (PRRSV) infection induces host translation shutoff and that the viral nonstructural protein nsp2 is associated with this process. Many viruses induce host translation shutoff by phosphorylating eukaryotic initiation factor 2α (eIF2α). However, PRRSV nsp2 does not induce eIF2α phosphorylation but attenuates the mTOR signaling pathway, another pathway regulating the host cell translational machinery. We also found that PRRSV-induced host translation shutoff was partly reversed by eliminating the effects of eIF2α phosphorylation or reactivating the mTOR pathway, indicating that PRRSV infection induces both eIF2α phosphorylation-dependent and -independent host translation shutoff.
INTRODUCTION
Viral infection always increases the expression of the host innate response effector proteins, which may limit viral replication and spread (1). Many viruses can interrupt the cellular translation machinery as a common evasion strategy (2). Initiation is the rate-determining step in translation and is the most frequent target of translational regulation (3). Global translation arrest is usually achieved by the phosphorylation of eukaryotic translation initiation factor 2α (eIF2α), which prevents the recycling of the ternary tRNAMet-GTP-eIF2 complex (4). Eukaryotic translation initiation factor 4E (eIF4E) also plays a critical role in the cap-dependent initiation of translation (3). It is the least-abundant translation initiation factor and is thought to be the rate-limiting protein for translation (5). The activity of eIF4E is regulated by the eIF4E-binding proteins (4E-BPs), which release eIF4E when it is hyperphosphorylated by mechanistic target of rapamycin (mTOR) but sequester eIF4E when it is hypophosphorylated (6). Thus, eIF4E is partly regulated by its phosphorylation (7). The phosphorylation of eIF4E can be activated by the mitogen-activated protein kinase (MAPK)-interacting kinases MNK1/2, two factors downstream of the MEK/ERK and p38 MAPK pathways (8).
Porcine reproductive and respiratory syndrome (PRRS) is an economically important infectious disease typically characterized by severe reproductive failure in sows and respiratory distress in piglets and growing pigs (9). The causative agent of the disease has been identified as PRRS virus (PRRSV), an enveloped, single-stranded positive-sense RNA virus classified in the family Arteriviridae (10). Its approximately 15-kb genome encodes at least 10 open reading frames (ORFs): ORF1a, ORF1b, ORF2a, ORF2b, ORF3 to -7, and ORF5a. ORF1a and ORF1b encode two polyproteins, which can be cleaved into 14 mature nonstructural proteins (11, 12). The other ORFs encode eight structural proteins: glycoprotein 2 (GP2), GP3, GP4, GP5, GP5a, matrix (M) protein, envelope (E) protein, and nucleocapsid (N) protein (13). PRRS is one of the most economically significant diseases affecting pig production worldwide and has caused great economic losses (14). Unfortunately, conventional vaccines and other strategies are insufficient to sustainably control PRRS (15). A better understanding of the virus-host interactions during PRRSV infection should facilitate the development of more-effective control measures.
PRRSV infection modulates normal cellular processes such as autophagy (16, 17), apoptosis (18, 19), and host innate immune responses (20–22). It is well known that PRRSV induces the phosphorylation of eIF2α and the appearance of stress granules (23, 24). PRRSV GP4 and GP5 alter the gene expression profiles of stably transfected host cell lines (25). However, the direct regulation of the translation of the host mRNAs during PRRSV infection is poorly understood. In this study, we report that PRRSV infection clearly inhibits host protein synthesis. We found that PRRSV nonstructural protein 2 (nsp2) plays a crucial role in this process. The possible mechanisms were investigated, and our results suggest that both the attenuation of the mTOR signaling pathway and the phosphorylation eIF2α are involved in PRRSV-induced host translation shutoff.
RESULTS
PRRSV infection induces translation suppression in cells.
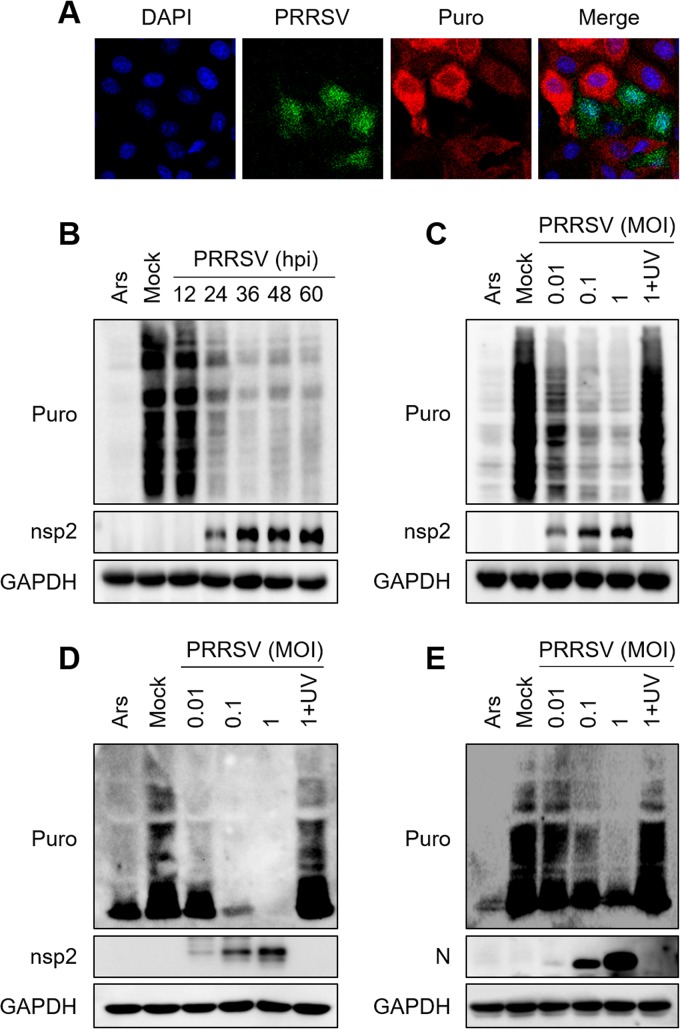
To test whether PRRSV infection induces host translation shutoff, we monitored translation using a short pulse of puromycin (puro), which is incorporated into nascent polypeptides and can be detected by immunofluorescence with an antibody directed against puromycin (ribopuromycylation assay) (26). As shown in Fig. 1A, MARC-145 cells infected with PRRSV strain WUH3 (type 2 PRRSV) showed strong translational repression, whereas the uninfected cells showed ongoing translation. To examine the time at which host translation was shut off after PRRSV infection, cellular polypeptide synthesis was monitored at different time points after PRRSV infection. As a positive control, cells were treated with sodium arsenite (Ars), an efficient inducer of eIF2α phosphorylation that inhibits global protein translation (27). Host translation shutoff was evident at 24 h postinfection (hpi) and gradually became more distinct as infection progressed (Fig. 1B). PRRSV infection also reduced the level of cell translation in a dose-dependent manner (Fig. 1C). It should be noted that UV-inactivated PRRSV lost the ability to suppress translation (Fig. 1C), indicating that the viral replication process is required for host translation shutoff.
FIG 1.
PRRSV infection reduces the translation level in cells. (A) MARC-145 cells were infected with PRRSV strain WUH3 (multiplicity of infection [MOI] = 0.5). At 24 h postinfection (hpi), puromycin (5 μg/ml) was added to the medium and the cells were incubated at 37°C for 25 min. The cells were washed twice with PBS and then fixed, permeabilized, and processed for IFA. A PRRSV-specific polyclonal antibody was used to detect PRRSV infection (green). Levels of cellular translation were measured with an antipuromycin antibody (red). Nuclei were stained with DAPI (blue). (B, C) MARC-145 cells were infected with PRRSV (MOI = 0.5) for different times (B) or with different doses of PRRSV (C) for 36 h. As a positive control, uninfected cells were treated with 0.5 mM Ars for 1 h. The cells were lysed and analyzed with Western blotting. Nascent polypeptides exclusively labeled with puromycin were detected with an antipuromycin monoclonal antibody. An anti-nsp2 antibody was used to confirm PRRSV replication, and an anti-GAPDH antibody was used as the protein loading control. (D, E) PAMs were infected with different doses of PRRSV strain WUH3 (D) or GZ11-G1 (E) for 24 h. Cells were treated, and the levels of cellular translation were detected by Western blotting as described for panel C. An anti-N antibody was used to confirm the replication of PRRSV strain GZ11-G1 because the monoclonal antibody against nsp2 of PRRSV strain WUH3 does not react with PRRSV strain GZ11-G1.
PRRSV preferentially infects porcine alveolar macrophages (PAMs) in vivo, and the corresponding results on PAMs are more convincing than those obtained with other cells. To this end, PAMs were infected with different doses of PRRSV strain WUH3. The results showed that PRRSV infection reduced the translation level of PAMs in a dose-dependent manner (Fig. 1D). Based on the genetic and antigenic differences, PRRSV are divided into two genotypic classes, European type 1 PRRSV and North American type 2 PRRSV (10). To test whether type 1 PRRSV also inhibits host translation, PAMs were infected with PRRSV strain GZ11-G1 (28), a type 1 PRRSV. As shown in Fig. 1E, the cellular translation levels obviously decreased in a dose-dependent manner after infection. Taken together, these results demonstrate that the two types of PRRSV infection can induce host translation shutoff.
PRRSV nsp2 and nsp5 induce host translation shutoff.
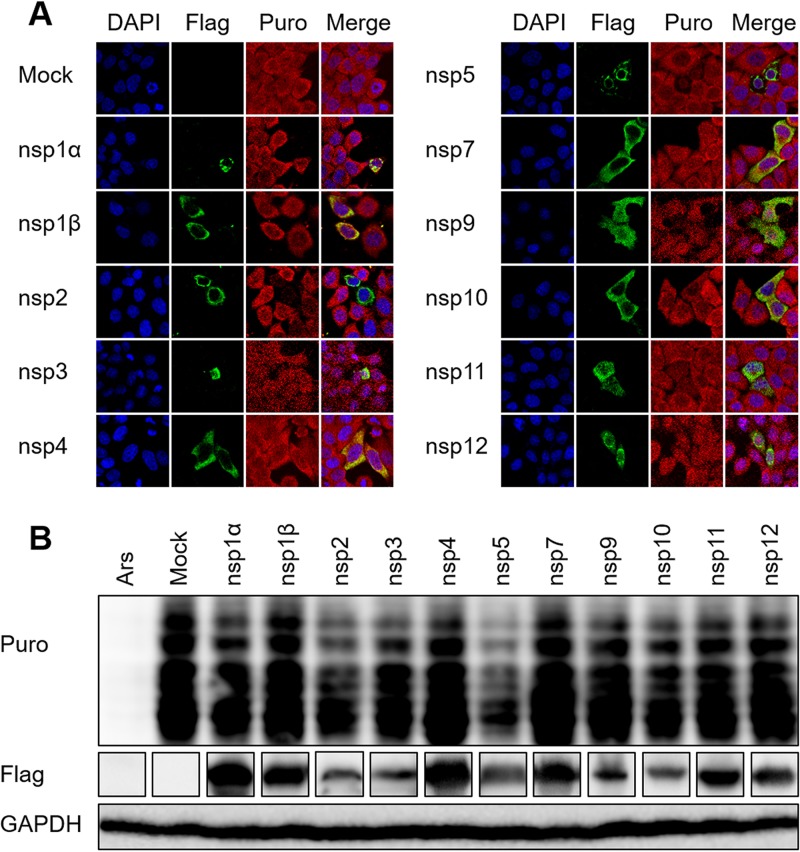
Because PRRSV-induced host translation shutoff is strictly dependent on viral multiplication, we speculated that viral nonstructural proteins participate in this process. Therefore, we investigated the effects of the nonstructural proteins on host translation. HeLa cells were transfected with eukaryotic expression constructs encoding individual PRRSV nonstructural proteins and treated with puromycin. The puromycylated native peptide chains were detected with an antipuromycin antibody, and an anti-Flag-tag antibody was used to verify the expression of viral proteins. As shown in Fig. 2A, the ectopic expression of nsp2 or nsp5 induced a strong reduction of the translation rate, whereas the other nonstructural proteins had little effect. To confirm these results, HEK293T cells were transfected with each of these constructs. Western blotting was performed 30 h after transfection to measure the translation levels with an antibody directed against puromycin. Consistent with the results of the indirect immunofluorescence assay (IFA), nsp2 and nsp5 strongly induced host translation shutoff (Fig. 2B). Because a previous study demonstrated that PRRSV nsp5 induces autophagic cell death in cultured cells (29), we selected nsp2 to explore the exact mechanism of host translation shutoff.
FIG 2.
PRRSV nsp2 and nsp5 induce host translation shutoff. (A) HeLa cells grown in 24-well plates were transfected with plasmid (1 μg) encoding the individual PRRSV proteins or the empty vector (pCAGGS-Flag). At 30 h posttransfection, puromycin was added to the medium and the cells were incubated at 37°C for 25 min. A mouse monoclonal antibody specific for puromycin was used to detect the nascent polypeptides with an immunofluorescence assay (red). Expression of PRRSV proteins was confirmed by immunoblotting with an anti-Flag antibody. (B) HEK293T cells were treated as described for panel A, and the levels of cellular translation were detected with Western blotting.
The C-terminal TM domain of nsp2 is associated with its ability to block translation.
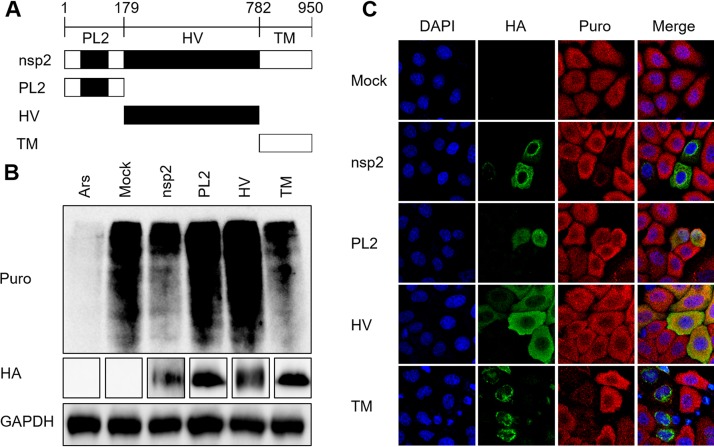
To identify which domain of nsp2 is related to its suppression of host translation, three truncation mutants of nsp2 were generated based on its functional domains: the PL2 (N-terminal papain-like cysteine protease), HV (functionally unspecified middle hypervariable region), and TM (transmembrane) domains (Fig. 3A) (30). HEK293T cells were transfected with constructs encoding nsp2 or one of its three truncation mutants. A Western blot assay was performed 30 h after transfection to determine the translation levels. Cells treated with Ars were used as the positive control. As shown in Fig. 3B, cells expressing nsp2 or TM showed dramatic translation suppression, which was consistent with the IFA results in HeLa cells (Fig. 3C). These results demonstrate that the C-terminal TM domain, rather than the PL2 or HV domain, of nsp2 is involved in host translation shutoff.
FIG 3.
The C-terminal TM domain of PRRSV nsp2 blocks translation. (A) Schematic representation of truncation mutants of PRRSV nsp2 used in this study. PL2, N-terminal papain-like cysteine protease domain; HV, functionally unspecified middle hypervariable region; TM, transmembrane domain. (B) HEK293T cells grown in six-well plates were transfected with pCAGGS-HA-nsp2, pCAGGS-HA-PL2, pCAGGS-HA-HV, or pCAGGS-HA-TM (4 μg). At 30 h posttransfection, puromycin was added to the medium, and the cells were incubated at 37°C for 25 min. Cells treated with 0.5 mM Ars were used as a positive control. Mouse monoclonal antibody specific for puromycin was used to detect the nascent polypeptides with Western blotting. An anti-HA-tag antibody was used to confirm the expression of nsp2 and its mutants. (C) HeLa cells grown in 24-well plates were transfected with plasmid (1 μg) encoding nsp2, PL2, HV, or TM or the empty vector. At 30 h posttransfection, the cells were treated with puromycin, fixed, and analyzed with confocal microscopy.
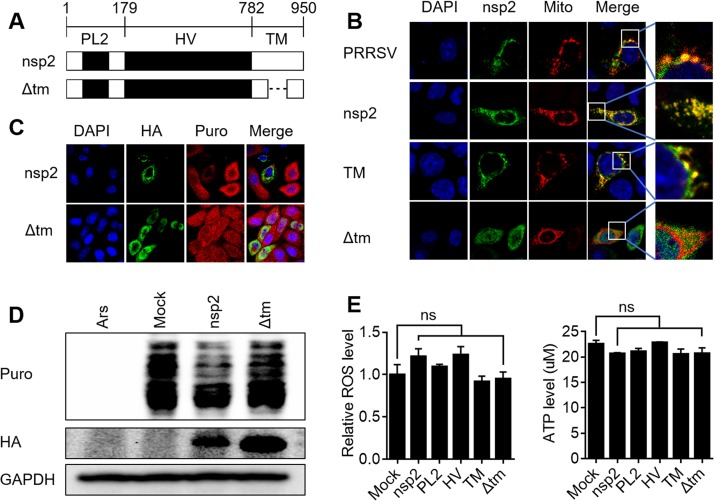
The TM domain contains a predicted transmembrane helix, which may determine the subcellular localization and function of nsp2. Therefore, we generated a deletion mutant, designated nsp2-Δtm, lacking the transmembrane sequence of nsp2 (Fig. 4A). Previous studies have demonstrated that PRRSV nsp2 interacts with many mitochondrial proteins (31–33), and our laboratory has also observed that nsp2 localizes to the mitochondria (unpublished data), so we tested the exact intracellular localizations of nsp2 and nsp2-Δtm. To this end, HeLa cells were transfected with plasmids encoding nsp2 or nsp2-Δtm together with the pDsRed2-Mito plasmid, which expresses the Mito-DsRed2 fusion protein and targets it to the mitochondria. As shown in Fig. 4B, most of the cells transfected with nsp2 or the TM mutant showed some degree of punctate spotting on their mitochondria, similar to those observed in MARC-145 cells after PRRSV infection. In contrast, nsp2-Δtm displayed a more-diffuse pattern of staining throughout the cells (Fig. 4B). At the same time, nsp2-Δtm lost its ability to inhibit translation, as was demonstrated with the IFA (Fig. 4C) and Western blot analyses (Fig. 4D). We speculated that nsp2 may directly impair the mitochondria, so we measured the production of reactive oxygen species (ROS) and the generation of ATP in HEK293T cells transfected with a plasmid expressing nsp2 or its mutants. However, neither ROS production nor ATP generation was visibly impaired after the ectopic expression of nsp2 or its mutants (Fig. 4E). These results indicate that PRRSV nsp2 induces host translation shutoff depending on its membrane localization. Despite its colocalization with the mitochondria, nsp2 had no effects on the main mitochondrial functions.
FIG 4.
PRRSV nsp2 induces host translation shutoff depending on its membrane location. (A) Schematic representation of PRRSV nsp2-Δtm, which lacks amino acids 851 to 873, a region predicted to be a transmembrane helix by the TMHMM server v2.0 (http://www.cbs.dtu.dk/services/TMHMM/). (B) MARC-145 cells grown in 24-well plates were transfected with 0.5 μg of pDsRed2-Mito and then infected with PRRSV for 36 h. HeLa cells were cotransfected with 0.5 μg of pDsRed2-Mito and 0.5 μg of pCAGGS-HA-nsp2, pCAGGS-HA-TM, or pCAGGS-HA-Δtm plasmid for 30 h. The cells were fixed and analyzed with confocal microscopy. (C, D) HeLa cells (C) or HEK293T cells (D) were transfected with pCAGGS-HA-nsp2 or pCAGGS-HA-Δtm. After treatment with puromycin, the effects of nsp2 and Δtm on translation were measured with an immunofluorescence assay (C) or Western blotting (D). (E) HEK293T cells grown in 24-well plates were transfected with 1 μg of pCAGGS-HA-nsp2, pCAGGS-HA-PL2, pCAGGS-HA-HV, pCAGGS-HA-TM, or pCAGGS-HA-Δtm. At 30 h posttransfection, ROS and ATP levels were measured with a reactive oxygen species assay kit or ATP assay kit. The results shown are the means ± SD from three independent experiments. ns, not significant.
nsp2-induced host translation shutoff is not dependent on proteasomal, autophagic, apoptotic, or transcriptional regulation.
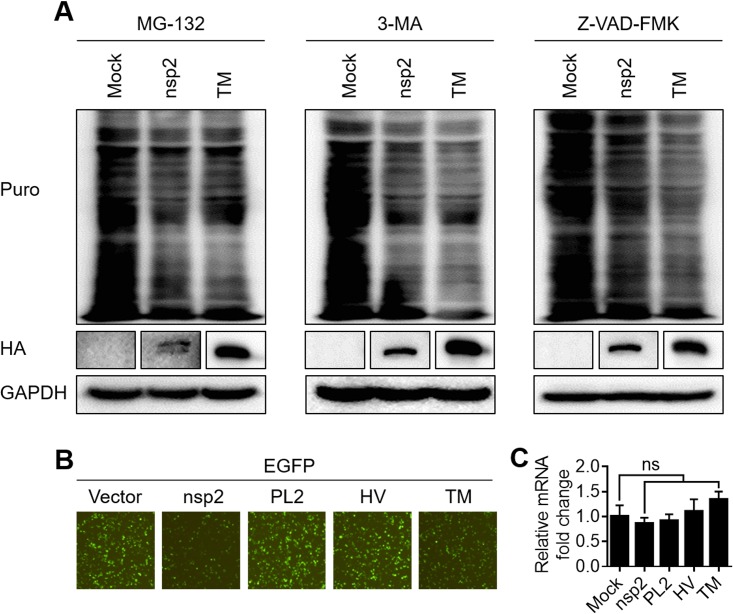
The ubiquitin-proteasome system, the autophagy-lysosome system, and apoptosis are the three major intracellular protein degradation pathways in eukaryotic cells (34, 35). To test whether these pathways are associated with nsp2-induced host translation shutoff, the proteasomal inhibitor MG132, the autophagy inhibitor 3-MA, or the apoptosis inhibitor Z-VAD-FMK was added to HEK293T cells transfected with the nsp2 or TM expression constructs. The cellular translation levels did not recover markedly after treatment with any of the three inhibitors (Fig. 5A). Based on these results, we conclude that neither proteasomes, nor autophagy, nor apoptosis is involved in the translation suppression induced by nsp2.
FIG 5.
PRRSV nsp2 and its TM domain suppress host gene expression specifically at the translation level. (A) HEK293T cells grown in six-well plates were transfected with 4 μg of pCAGGS-HA-nsp2 or pCAGGS-HA-TM or empty vector. At 16 h after transfection, the cells were treated with MG-132 (20 μM), 3-MA (5 mM), or Z-VAD-FMK (10 μM) for 14 h. After treatment with puromycin, translation was measured as described above. (B) HEK293T cells grown in 24-well plates were cotransfected with 0.5 μg of pEGFP-C1 and pCAGGS-HA-nsp2, pCAGGS-HA-PL2, pCAGGS-HA-HV, or pCAGGS-HA-TM. At 30 h posttransfection, the expression of EGFP was analyzed with an inverted fluorescence microscope. (C) RNA was extracted from the cells shown in panel B, and real-time PCR was used to measure the content of EGFP mRNA. The individual transcripts were normalized to the expression level of GAPDH housekeeping gene transcripts. The results shown are the means ± SD from three independent experiments.
To investigate whether nsp2 and TM reduce the transcription levels of host cells, HEK293T cells were cotransfected with plasmids expressing the individual nsp2 mutants and a plasmid expressing enhanced green fluorescent protein (EGFP). As shown in Fig. 5B, EGFP expression was clearly reduced in the presence of nsp2 or the TM mutant, whereas the PL2 and HV mutants had no effect on EGFP expression. Total RNA was isolated from the samples, and the EGFP mRNA was quantified with quantitative real-time PCR (qPCR) analysis. The results in Fig. 5C show that there were no significant differences in the EGFP mRNA levels in the cells transfected with nsp2, the truncation mutants, or the empty vector. Taken together, these data demonstrate that nsp2 and TM cause host translation shutoff specifically at the level of translation.
PRRSV nsp2 does not induce the phosphorylation of eIF2α.
The phosphorylation of eIF2α is a key mechanism of translational control (36, 37). This event is vital to the regulation of global protein levels and is essential for the maintenance of cellular homeostasis (38). To test whether eIF2α plays a part in nsp2-induced translational arrest, we measured the eIF2α phosphorylation levels in cells overexpressing nsp2 or its mutants. The results showed that whereas Ars induced the marked upregulation of eIF2α phosphorylation, nsp2 and its truncation mutants did not (Fig. 6A). To confirm that eIF2α phosphorylation is not involved in the nsp2-induced inhibition of translation, we measured the mRNA levels of ATF4 and GADD34, which are induced in response to eIF2α phosphorylation (39). As shown in Fig. 6B and C, the expression of ATF4 and GADD34 mRNAs did not change significantly with time in cells overexpressing nsp2 compared with their expression in mock-transfected cells. These results support the conclusion that PRRSV nsp2 does not trigger eIF2α phosphorylation, suggesting that there is little connection between eIF2α phosphorylation and nsp2-induced translation arrest.
FIG 6.
PRRSV nsp2 has no effect on eIF2α phosphorylation. (A) HEK293T cells grown in six-well plates were transfected with 4 μg of pCAGGS-HA-nsp2, pCAGGS-HA-PL2, pCAGGS-HA-HV, pCAGGS-HA-TM, or empty vector. At 30 h posttransfection, the cells were harvested and analyzed with Western blotting and the indicated antibodies. (B, C) HEK293T cells grown in 24-well plates were transfected with the pCAGGS-HA-nsp2 plasmid (1 μg) or empty vector. At 30 h posttransfection, the total RNA was extracted from the cells and analyzed for the abundance of endogenous ATF4 (B) and GADD34 mRNAs (C) using real-time PCR. The results are the means ± SD from three independent experiments.
PRRSV induces both eIF2α phosphorylation-dependent and -independent host translation shutoff.
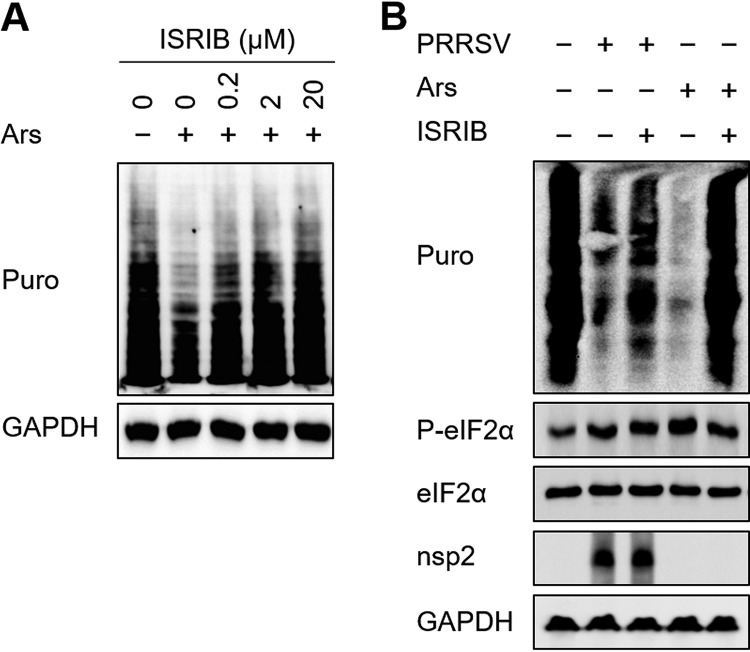
Previous studies have indicated that PRRSV increases the phosphorylation of eIF2α (23, 24). It is well known that the phosphorylation of eIF2α causes translation to stall (40). To evaluate the role of eIF2α phosphorylation on PRRSV-induced translation suppression, we used the drug ISRIB (an integrated stress response inhibitor), which can substantially and comprehensively reverse the downstream effects of eIF2α phosphorylation on translation (41). To determine the appropriate concentration of ISRIB, the levels of cell translation were measured after treatment with Ars and then with different concentrations of ISRIB. As shown in Fig. 7A, 20 μM ISRIB completely reversed the translation arrest caused by Ars and was used as the action concentration. This concentration (20 μM) of ISRIB was then used to test whether it reversed PRRSV-induced host translation shutoff. PRRSV infection reduced cellular translation to quite a low level, and ISRIB reversed the effect to some extent, but not completely (Fig. 7B). These data demonstrate that while PRRSV interrupts host cell translation via eIF2α phosphorylation, it also inhibits cellular mRNA translation in an eIF2α phosphorylation-independent manner.
FIG 7.
PRRSV induces both eIF2α phosphorylation-dependent and -independent host translation shutoff. (A) MARC-145 cells grown in six-well plates were treated with 0.5 mM Ars at 37°C for 1 h. The medium was changed to medium containing different concentrations of ISRIB, and after 25 min, the cells were treated with puromycin, incubated at 37°C for 25 min, and lysed for Western blotting. (B) MARC-145 cells grown in six-well plates were infected with PRRSV (MOI = 0.5). At 36 h postinfection, the cells were treated with 20 μM ISRIB for 25 min and with puromycin for 25 min. Western blotting was performed to analyze the levels of translation and eIF2α phosphorylation.
PRRSV infection and nsp2 overexpression both negatively regulate the mTOR signaling pathway.
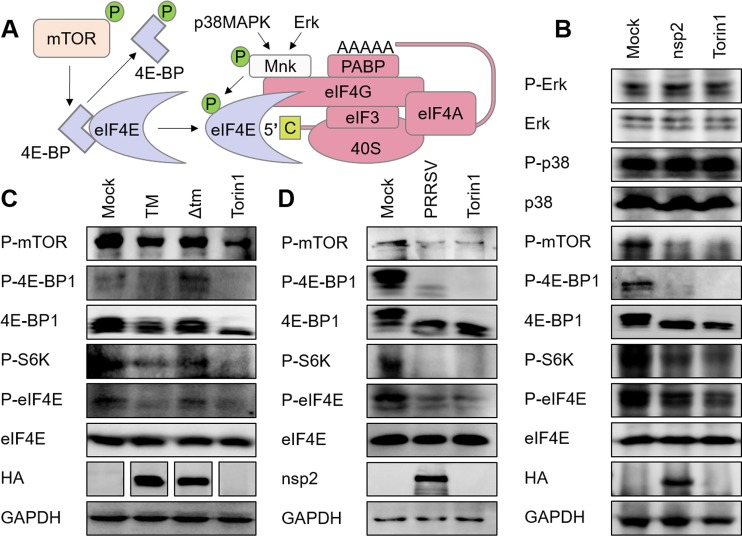
Eukaryotic mRNAs contain a cap structure at their 5′ ends. Ongoing cap-dependent translation requires eIF4E to bind the caps of the mRNAs (3), and eIF4E activity is regulated by both the mTOR and MAPK signaling pathways (Fig. 8A). Therefore, we next investigated whether PRRSV nsp2 impairs eIF4E phosphorylation or the regulators of eIF4E. To this end, HEK293T cells were transfected with an nsp2-expressing plasmid, and the expression of total eIF4E, 4E-BP1, p38 MAPK, ERK, and the phosphorylation levels of eIF4E, p70-S6K (S6K), 4E-BP1, mTOR, p38 MAPK, and ERK were determined with Western blotting. As a positive control, HEK293T cells were treated with Torin 1, an mTOR inhibitor that blocks mTOR phosphorylation and thereby lowers the activation of downstream factors (42). As shown in Fig. 8B, nsp2 significantly reduced the phosphorylation of mTOR, 4E-BP1, S6K, and eIF4E, but not that of ERK or p38 MAPK. We also tested the effects of TM and the Δtm mutant on the mTOR signaling pathway. The results were consistent with our prediction that TM negatively regulated the mTOR signaling pathway, whereas Δtm had only a limited effect (Fig. 8C). PRRSV infection also inhibited the cellular mTOR signaling pathway (Fig. 8D). All of these results demonstrate that PRRSV infection and the ectopic expression of nsp2 or TM inactivate the mTOR signaling pathway, offering a possible explanation for the block of translation caused by PRRSV.
FIG 8.
mTOR signaling pathway is attenuated by PRRSV infection and nsp2 expression. (A) Schematic representation of the regulation of eIF4E activity. (B, C) HEK293T cells grown in six-well plates were transfected with 4 μg of pCAGGS-HA-nsp2 (B) or pCAGGS-HA-TM or pCAGGS-HA-Δtm (C). Cells treated with Torin 1 (250 nM, 6 h) were used as the positive control. The cells were harvested at 30 h posttransfection, and the lysates were analyzed with immunoblotting using antibodies specific for the indicated proteins. (D) MARC-145 cells were infected with PRRSV (MOI = 0.5), harvested at 36 h postinfection, and analyzed with Western blotting.
Attenuation of the mTOR signaling pathway contributes to PRRSV-induced host translation shutoff.
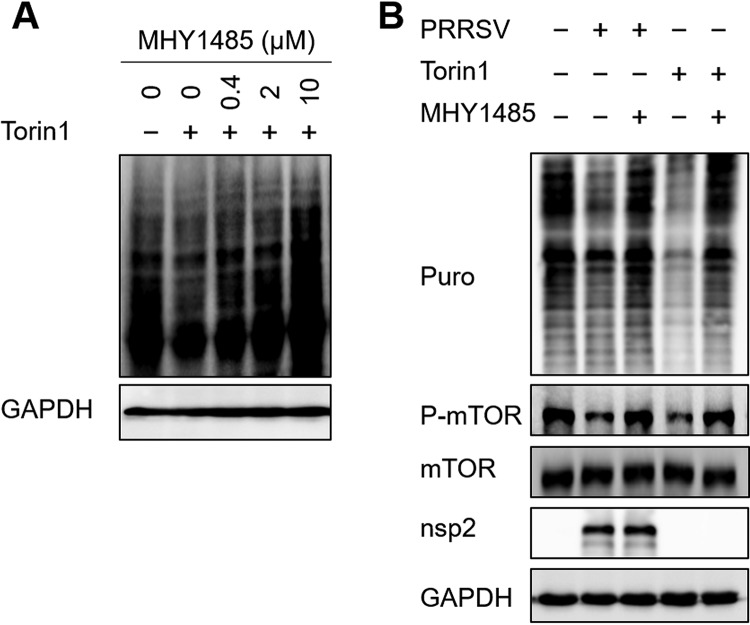
mTOR is thought to modulate protein synthesis, and mTOR inhibition causes the almost-complete arrest of protein synthesis (43, 44). To determine how the attenuation of the mTOR signaling pathway affects translation in PRRSV-infected cells, we used the drug MHY1485, a powerful activator of the mTOR pathway (45). As shown in Fig. 9A, 2 μM MHY1485 was sufficient to completely restore the translation shutoff caused by Torin 1, an inducer of host translation shutoff. The same concentration of MHY1485 rescued most mTOR phosphorylation and the cellular translation level in PRRSV-infected cells (Fig. 9B). Based on these observations, we conclude that PRRSV suppresses host cell translation by inhibiting the mTOR signaling pathway.
FIG 9.
PRRSV-induced host translation shutoff is associated with the attenuation of the mTOR signaling pathway. (A) MARC-145 cells grown in six-well plates were treated with 250 nM Torin 1 at 37°C for 6 h. The medium was changed to medium containing different concentrations of MHY1485, and after 1 h, the cells were treated with puromycin at 37°C for 25 min and lysed for analysis with Western blotting. (B) MARC-145 cells grown in six-well plates were infected with PRRSV (MOI = 0.5). At 36 h postinfection, the cells were treated with 2 μM MHY1485 for 1 h and then with puromycin for 25 min. Western blotting was used to analyze the levels of translation and mTOR phosphorylation.
DISCUSSION
Cap-dependent viral mRNAs are structurally similar to host mRNAs and usually share the same translation system mechanisms. However, when some viruses induce host translational shutoff, they maintain the synthesis of their own gene products. For example, vesicular stomatitis virus (VSV) inhibits the translation of host mRNAs in infected cells but allows the translation of its own capped mRNAs (46). The 5′ untranslated regions (UTRs) of VSV mRNAs are shorter than those of the host mRNAs and probably lack secondary structure. This structural characteristic may allow the efficient translation of VSV mRNAs by the limited eIF4F complexes (47). The translation of the dengue virus (DENV) genome benefits from the optimized use of codons that occur with low prevalence in the host cells, whereas the translation of the host mRNAs is strongly reduced (48, 49). The severe acute respiratory syndrome coronavirus (SARS-CoV) nsp1-induced degradation of host mRNAs inhibits host gene expression in infected cells, but the viral mRNAs appear to be resistant to the cleavage mechanism (50). These examples demonstrate that during their coevolution with their hosts, viruses have evolved different strategies to inhibit host translation while protecting their own protein synthesis.
In this study, we have demonstrated that PRRSV infection induces host translation shutoff and that nsp2 is involved in this process. The PRRSV genome also has a putative type I cap structure at its 5′ end, and the translation of viral mRNAs is cap dependent (51, 52). Although the nsp2 protein band gradually increases in intensity (Fig. 1B), this may be attributed to its accumulation rather than newly synthesized protein. We cannot conclude that viral translation is unaffected during host translation shutoff, but it seems that viral multiplication remains robust in the late stage of PRRSV infection (53). There are several possible explanations for the exemption of viral translation from host translation shutoff. First, virus-induced host translation shutoff is thought to inhibit the host antiviral response (54, 55). Therefore, the interruption of host mRNA translation is a frequent evasion strategy evolved by viruses to enhance their replication. Second, the abundance of viral mRNAs increases exponentially during infection and perhaps competes successfully for the limited translation machinery. This may contribute to the preferential translation of viral mRNAs and, at least in part, counterbalance the effects of global translational suppression on viral proteins. Third, PRRSV imprisons host cellular mRNAs in the nucleus and blocks their nuclear export, thus hindering their translation (56, 57). This is a novel mechanism that guarantees the efficient translation of viral genomic RNAs, with less competition from host mRNAs. It has also been reported that the modification of adenosine at the N6 position (m6A) in the 5′ UTRs of mammalian mRNAs stimulates cap-independent translation (58, 59). The transcripts of HIV-1, influenza A virus, and simian virus 40 contain multiple m6A sites that enhance the expression of viral mRNAs and proteins (60). If the 5′ UTRs of PRRSV mRNAs undergo m6A modification in infected cells and these mRNAs use a cap-independent mechanism for translation, their translation will not be affected by host translation shutoff.
eIF2α is a key factor in the innate immune response to viral infection. Imperceptible changes in eIF2α phosphorylation can dramatically suppress ongoing protein synthesis (2). However, under some conditions, eIF2α phosphorylation has only a limited effect on the translation level. For example, the phosphorylation of PKR and probably eIF2α are not the primary drivers to block the translation of herpes simplex virus true late mRNAs in the absence of virion host shutoff (61). DENV also activates the eIF2α-dependent stress response, but it is uncoupled from the host translation repression induced by DENV infection (48). Consistent with this, our data demonstrate that eIF2α phosphorylation plays only a partial role in PRRSV-induced translation suppression. These data confirm the complexity of the strategies that PRRSV uses to defend against the host's antiviral innate immune response, to guarantee viral multiplication.
eIF4E plays a decisive role in the cap-dependent initiation of translation. Its activity is regulated by the AKT/mTOR, MEK/ERK, and p38 MAPK pathways (3). mTOR phosphorylates 4E-BPs and terminates the arrest of eIF4E, and then Erk and p38 MAPK phosphorylate eIF4E by Mnk1 (62). Many viruses reportedly inhibit the phosphorylation of eIF4E. For example, adenovirus infection inhibits the translation of cellular mRNAs during the late phase of infection, which correlates with the displacement of MNK1 from eIF4G by the viral 100K protein and the dephosphorylation of eIF4E (63). VSV infection results in the dephosphorylation of 4E-BP1 and eIF4E, which is associated with the inhibition of host protein synthesis (47). 4E-BP1 is dephosphorylated during encephalomyocarditis virus (EMCV) infection, and this event is almost simultaneous with the shutoff of protein synthesis caused by EMCV (64). A previous study reported that the phosphorylation of mTOR, 4E-BP, and S6K is reduced 48 h after PRRSV infection (65). Consistent with this finding, we have demonstrated that PRRSV infection inhibits the phosphorylation of eIF4E by inactivating the mTOR signaling pathway, and we speculate that this contributes to the host translation shutoff caused by PRRSV. The exact mechanism by which PRRSV inhibits the mTOR pathway must be established in a future study.
Viral protein expression, followed by the consequent increase in the endoplasmic reticulum (ER) burden, the formation of double-membrane vesicles, and the loss of ER lipids, may contribute to ER stress, which inhibits cellular mRNA expression in virus-infected cells (66). During chronic ER stress, the decline in protein synthesis occurs in an eIF2α phosphorylation-independent manner (67). This process correlates with reduced mTOR activity and the phosphorylation of 4E-BP1, which negatively regulate cap-dependent translation (68). Equine arteritis virus (EAV), an arterivirus similar to PRRSV, reshapes the ER to accommodate viral RNA synthesis (69). PRRSV infection also induces ER stress (16, 70), while blocking protein phosphorylation in the mTOR signaling pathway (Fig. 7D). It is possible that during the short-term phase of infection, ER stress is induced by PRRSV infection, and a certain amount of global translational suppression occurs as a result of eIF2α phosphorylation. With time, chronic ER stress inactivates the mTOR signaling pathway, which may play a dominant role in translation arrest. It is noteworthy that EAV nsp2 modifies the host cell membranes during the formation of the arterivirus replication complex (71). Because the TM domain of PRRSV nsp2 contains a transmembrane structure, it is possible that PRRSV nsp2 induces ER stress via its TM domain, thus inactivating the mTOR signaling pathway and host translation activity. Although attempts were made to construct a recombinant PRRSV with a deletion of the TM region, no viable virus could be rescued, indicating that the TM domain is essential for PRRSV replication (72). Therefore, we could not use infectious clones to evaluate the role of the TM region in host translation shutoff under physiological conditions. In addition, the nsp2-coding region is highly variable in the PRRSV genome, and the HV domain of nsp2 is the most variable region (73). In this study, we demonstrated that the TM domain, rather than the HV domain, is the critical domain for nsp2-mediated host translation shutoff. Relative to the HV domain, the TM domain is conservative among different PRRSV isolates or lineages. Furthermore, both type 1 and type 2 PRRSV can induce host translation shutoff, indicating that the variation of nsp2 HV domain does not affect nsp2's function to induce host translation shutoff.
Viral mRNA translation depends strictly on translation initiation factors, so therapies targeting these factors should provide effective tools for combating viral infections, especially when other therapies fail. Silvestrol, a highly efficient, nontoxic, specific inhibitor of eIF4A, is a potent inhibitor of Ebola virus, coronaviruses, and picornaviruses (74, 75). The drug 4E2RCat reduces human coronavirus 229E replication and reduces both the intra- and extracellular infectious viral titers by preventing the interaction between eIF4E and eIF4G and cap-dependent translation (76). Two specific aptamers that recognize PABP1 reduce the expression of the viral genome and the production of infective influenza virus particles (77). At present, vaccination is the principal measure used to control and treat PRRSV infection. Unfortunately, the current commercially available vaccines failed to provide the desirable protection against challenge with heterologous PRRSV strains (78–80). Therefore, alternative measures for the prevention and control of PRRSV are required. Pharmacological interventions that target the translation initiation factors and their regulators may be an ideal choice.
In conclusion, our study provides evidence that PRRSV infection induces host translation shutoff and that nsp2 is associated with this process. For the two main pathways to modulate host translation arrest, nsp2 attenuates the mTOR signaling pathway but does not induce eIF2α phosphorylation. In the context of virus infection, both eIF2α phosphorylation and the attenuation of the mTOR signaling pathway contribute to PRRSV-induced host translation arrest.
MATERIALS AND METHODS
Cells and viruses.
MARC-145 cells and HEK293T cells were maintained in Dulbecco's modified Eagle's medium (Invitrogen, Asheville, NC, USA) supplemented with 10% heated-inactivated fetal bovine serum (FBS), 10 μg/ml streptomycin sulfate, and 100 U/ml penicillin at 37°C in a humidified 5% CO2 incubator. PAMs were cultured in RPMI 1640 (Sigma-Aldrich, St. Louis, MO, USA) supplemented with 10% heated-inactivated FBS at 37°C in a humidified 5% CO2 incubator. PRRSV strain WUH3 (GenBank accession no. HM853673), a highly pathogenic type 2 PRRSV, was previously isolated from the brains of pigs suffering from a high fever syndrome in China at the end of 2006 (81). PRRSV strain GZ11-G1, a type 1 PRRSV (28), was a gift from Hanchun Yang at China Agricultural University.
Antibodies.
The monoclonal antibodies against PRRSV nsp2, N, and PRRSV-specific immunoglobulins (IgGs) were prepared in our laboratory (32). Rabbit monoclonal antibodies directed against eIF2α, P-eIF2α, p38 MAPK, P-p38 MAPK, ERK1/2, P-ERK1/2, P-mTOR, P-S6K, 4E-BP1, P-4E-BP1, and P-eIF4E were purchased from Cell Signaling Technology (Beverly, MA, USA). Antibodies directed against TIA1 and eIF4E were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA). Horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG antibody, HRP-conjugated goat anti-mouse IgG antibody, and monoclonal antibodies directed against Flag, HA, and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were purchased from MBL Beijing Biotech (Beijing, China). An antibody directed against puromycin was purchased from Millipore (Billerica, MA, USA). Fluorescently labeled antimouse and antirabbit secondary antibodies were purchased from Thermo Fisher Scientific (Waltham, MA, USA). Rabbit anti-pig IgG/Alexa Fluor 488 antibody was purchased from Bioss (Woburn, MA, USA).
Plasmids.
The pCAGGS-MCS vector was modified by inserting the Flag or HA epitope tag at the N terminus to generate the pCAGGS-Flag or pCAGGS-HA vector, respectively, as previously described (82). Expression constructs encoding the nonstructural proteins of PRRSV strain WUH3 have been described previously (83). PRRSV nsp2 and its truncation mutants were inserted into the pCAGGS-HA vector. The pEGFP-C1 vector, which expresses EGFP, and the pDsRed2-Mito vector, which expresses the Mito-DsRed2 fusion protein, were purchased from Clontech (Mountain View, CA, USA).
Chemicals and reagents.
Puromycin, Z-VAD-FMK, and MG132 were from Beyotime Co. (Jiangsu, China). Torin 1 and ISRIB were purchased from Medchem Express (Monmouth Junction, NJ, USA). Sodium arsenite (Ars) and 3-methyladenine (3- MA) were from Sigma-Aldrich.
Quantitative real-time PCR.
Total cellular RNA was extracted with TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions, and 1 μg of each sample was subsequently reverse transcribed to cDNA with the Transcriptor First Strand cDNA synthesis kit (Roche, Mannheim, Germany) according to the manufacturer's instructions. The resulting cDNA was then used as the template in a SYBR green qPCR assay (Applied Biosystems, Foster City, CA, USA). The abundance of individual mRNA transcripts in each sample was assayed three times and normalized to that of GAPDH mRNA (internal control). The specific primer sequences used in this study are listed in Table 1.
TABLE 1.
Sequences of primers used for real-time PCR
| Gene | Primer sequence (5′–3′) |
|
|---|---|---|
| Forward | Reverse | |
| eGFP | AAGGGCATCGACTTCAAGGA | CTTCTCGTTGGGGTCTTTGC |
| GAPDH | TCATGACCACAGTCCATGCC | GGATGACCTTGCCCACAGCC |
| ATF4 | ATGACCGAAATGAGCTTCCTG | GCTGGAGAACCCATGAGGT |
| GADD34 | ATGATGGCATGTATGGTGAGC | AACCTTGCAGTGTCCTTATCAG |
Indirect immunofluorescence assay.
Cells were seeded on circular glass coverslips in 24-well plates and grown to 80% to 90% confluence. At the indicated time points after treatment, the cells were incubated in 4% paraformaldehyde for 15 min and immediately permeabilized with precooled methanol for 10 min. The cells were blocked with 5% bovine serum albumin (BSA) in phosphate-buffered saline (PBS) for 45 min and then incubated with the indicated antibodies for 1 h. The cells were treated with secondary antibodies for 1 h and then with 4′,6-diamidino-2-phenylindole (DAPI; Beyotime, Nantong, China) in PBS (1/200 dilution) for 15 min. The fluorescent images were acquired with an Olympus FV10 laser scanning confocal microscope (Olympus, Tokyo, Japan).
Western blotting.
The total cellular samples were washed twice with ice-cold PBS, lysed in sample buffer (Beyotime) with 1% phosphatase inhibitor cocktail (Sigma-Aldrich), and then resolved with sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride membranes (Millipore, Billerica, MA, USA). The membranes were blocked with 5% BSA in Tris-buffered saline containing Tween 20 and incubated with the primary antibodies.
ROS measurements.
The levels of ROS were determined with a ROS detection kit (Beyotime) according to the manufacturer's protocol. Briefly, cells grown in 24-well plates were washed twice with cold PBS and incubated with 200 μl of 2′,7′-dichlorodihydrofluorescein diacetate (DCFH-DA) solution at room temperature for 30 min in the dark. The fluorescence intensity was recorded at 488 nm with a microplate reader (BioTek, Winooski, VT, USA). The results given are representative of data from three independent experiments.
ATP measurement.
The amounts of cellular ATP were measured with a firefly luciferase-based ATP detection kit (Beyotime) according to the manufacturer's protocol. Briefly, cells grown in 24-well plates were lysed with 200 μl of lysis buffer and centrifuged at 12,000 × g for 5 min at 4°C. One-half of the supernatant was mixed with 100 μl of ATP detection working solution. Luminance was assayed in a luminometer (Promega). Standard curves were generated to calculate the contents of ATP. All the experiments were performed independently in triplicate. Values reported are the means from three replicates ± standard deviations (SD).
ACKNOWLEDGMENTS
We thank Hanchun Yang for providing type 1 PRRSV.
This work was supported by the National Basic Research Program (973) of China (2014CB542700), the Major Project of National Natural Science Foundation of China (31490602), the National Natural Sciences Foundation of China (31372467, 31225027), and the Key Technology R&D Programme of China (2015BAD12B02).
REFERENCES
- 1.Schoggins JW, MacDuff DA, Imanaka N, Gainey MD, Shrestha B, Eitson JL, Mar KB, Richardson RB, Ratushny AV, Litvak V, Dabelic R, Manicassamy B, Aitchison JD, Aderem A, Elliott RM, Garcia-Sastre A, Racaniello V, Snijder EJ, Yokoyama WM, Diamond MS, Virgin HW, Rice CM. 2014. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature 505:691–695. doi: 10.1038/nature12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walsh D, Mathews MB, Mohr I. 2013. Tinkering with translation: protein synthesis in virus-infected cells. Cold Spring Harb Perspect Biol 5:a012351. doi: 10.1101/cshperspect.a012351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Richter JD, Sonenberg N. 2005. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature 433:477–480. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- 4.Kimball SR. 1999. Eukaryotic initiation factor eIF2. Int J Biochem Cell Biol 31:25–29. doi: 10.1016/S1357-2725(98)00128-9. [DOI] [PubMed] [Google Scholar]
- 5.Duncan R, Milburn SC, Hershey JW. 1987. Regulated phosphorylation and low abundance of HeLa cell initiation factor eIF-4F suggest a role in translational control. Heat shock effects on eIF-4F. J Biol Chem 262:380–388. [PubMed] [Google Scholar]
- 6.Gingras AC, Raught B, Gygi SP, Niedzwiecka A, Miron M, Burley SK, Polakiewicz RD, Wyslouch-Cieszynska A, Aebersold R, Sonenberg N. 2001. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev 15:2852–2864. doi: 10.1101/gad.887201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Furic L, Rong L, Larsson O, Koumakpayi IH, Yoshida K, Brueschke A, Petroulakis E, Robichaud N, Pollak M, Gaboury LA, Pandolfi PP, Saad F, Sonenberg N. 2010. eIF4E phosphorylation promotes tumorigenesis and is associated with prostate cancer progression. Proc Natl Acad Sci U S A 107:14134–14139. doi: 10.1073/pnas.1005320107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X, Flynn A, Waskiewicz AJ, Webb BL, Vries RG, Baines IA, Cooper JA, Proud CG. 1998. The phosphorylation of eukaryotic initiation factor eIF4E in response to phorbol esters, cell stresses, and cytokines is mediated by distinct MAP kinase pathways. J Biol Chem 273:9373–9377. doi: 10.1074/jbc.273.16.9373. [DOI] [PubMed] [Google Scholar]
- 9.Rahe MC, Murtaugh MP. 2017. Effector mechanisms of humoral immunity to porcine reproductive and respiratory syndrome virus. Vet Immunol Immunopathol 186:15–18. doi: 10.1016/j.vetimm.2017.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nelsen CJ, Murtaugh MP, Faaberg KS. 1999. Porcine reproductive and respiratory syndrome virus comparison: divergent evolution on two continents. J Virol 73:270–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wootton S, Yoo D, Rogan D. 2000. Full-length sequence of a Canadian porcine reproductive and respiratory syndrome virus (PRRSV) isolate. Arch Virol 145:2297–2323. doi: 10.1007/s007050070022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y, Tas A, Snijder EJ, Fang Y. 2012. Identification of porcine reproductive and respiratory syndrome virus ORF1a-encoded non-structural proteins in virus-infected cells. J Gen Virol 93:829–839. doi: 10.1099/vir.0.039289-0. [DOI] [PubMed] [Google Scholar]
- 13.Sun Y, Han M, Kim C, Calvert JG, Yoo D. 2012. Interplay between interferon-mediated innate immunity and porcine reproductive and respiratory syndrome virus. Viruses 4:424–446. doi: 10.3390/v4040424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chand RJ, Trible BR, Rowland RR. 2012. Pathogenesis of porcine reproductive and respiratory syndrome virus. Curr Opin Virol 2:256–263. doi: 10.1016/j.coviro.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 15.Lager KM, Schlink SN, Brockmeier SL, Miller LC, Henningson JN, Kappes MA, Kehrli ME, Loving CL, Guo B, Swenson SL, Yang HC, Faaberg KS. 2014. Efficacy of type 2 PRRSV vaccine against Chinese and Vietnamese HP-PRRSV challenge in pigs. Vaccine 32:6457–6462. doi: 10.1016/j.vaccine.2014.09.046. [DOI] [PubMed] [Google Scholar]
- 16.Chen Q, Fang L, Wang D, Wang S, Li P, Li M, Luo R, Chen H, Xiao S. 2012. Induction of autophagy enhances porcine reproductive and respiratory syndrome virus replication. Virus Res 163:650–655. doi: 10.1016/j.virusres.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun MX, Huang L, Wang R, Yu YL, Li C, Li PP, Hu XC, Hao HP, Ishag HA, Mao X. 2012. Porcine reproductive and respiratory syndrome virus induces autophagy to promote virus replication. Autophagy 8:1434–1447. doi: 10.4161/auto.21159. [DOI] [PubMed] [Google Scholar]
- 18.Sur JH, Doster AR, Osorio FA. 1998. Apoptosis induced in vivo during acute infection by porcine reproductive and respiratory syndrome virus. Vet Pathol 35:506–514. doi: 10.1177/030098589803500605. [DOI] [PubMed] [Google Scholar]
- 19.Sur JH, Doster AR, Christian JS, Galeota JA, Wills RW, Zimmerman JJ, Osorio FA. 1997. Porcine reproductive and respiratory syndrome virus replicates in testicular germ cells, alters spermatogenesis, and induces germ cell death by apoptosis. J Virol 71:9170–9179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sang Y, Rowland RR, Blecha F. 2014. Antiviral regulation in porcine monocytic cells at different activation states. J Virol 88:11395–11410. doi: 10.1128/JVI.01714-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loving CL, Osorio FA, Murtaugh MP, Zuckermann FA. 2015. Innate and adaptive immunity against porcine reproductive and respiratory syndrome virus. Vet Immunol Immunopathol 167:1–14. doi: 10.1016/j.vetimm.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoo D, Song C, Sun Y, Du Y, Kim O, Liu HC. 2010. Modulation of host cell responses and evasion strategies for porcine reproductive and respiratory syndrome virus. Virus Res 154:48–60. doi: 10.1016/j.virusres.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou Y, Fang L, Wang D, Cai K, Chen H, Xiao S. 2017. Porcine reproductive and respiratory syndrome virus infection induces stress granule formation depending on protein kinase R-like endoplasmic reticulum kinase (PERK) in MARC-145 cells. Front Cell Infect Microbiol 7:111. doi: 10.3389/fcimb.2017.00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen WY, Schniztlein WM, Calzada-Nova G, Zuckermann FA. 2017. Genotype 2 strains of porcine reproductive and respiratory syndrome virus dysregulate alveolar macrophage cytokine production via the unfolded protein response. J Virol 92:e01251-17. doi: 10.1128/JVI.01251-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee C, Bachand A, Murtaugh MP, Yoo D. 2004. Differential host cell gene expression regulated by the porcine reproductive and respiratory syndrome virus GP4 and GP5 glycoproteins. Vet Immunol Immunopathol 102:189–198. doi: 10.1016/j.vetimm.2004.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmidt EK, Clavarino G, Ceppi M, Pierre P. 2009. SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods 6:275–277. doi: 10.1038/nmeth.1314. [DOI] [PubMed] [Google Scholar]
- 27.McEwen E, Kedersha N, Song B, Scheuner D, Gilks N, Han A, Chen JJ, Anderson P, Kaufman RJ. 2005. Heme-regulated inhibitor kinase-mediated phosphorylation of eukaryotic translation initiation factor 2 inhibits translation, induces stress granule formation, and mediates survival upon arsenite exposure. J Biol Chem 280:16925–16933. doi: 10.1074/jbc.M412882200. [DOI] [PubMed] [Google Scholar]
- 28.Wang X, Yang X, Zhou R, Zhou L, Ge X, Guo X, Yang H. 2016. Genomic characterization and pathogenicity of a strain of type 1 porcine reproductive and respiratory syndrome virus. Virus Res 225:40–49. doi: 10.1016/j.virusres.2016.09.006. [DOI] [PubMed] [Google Scholar]
- 29.Yang L, Wang R, Ma Z, Wang Y, Zhang Y. 2015. Inducing autophagic cell death by Nsp5 of porcine reproductive and respiratory syndrome virus. Austin Virol Retrovirol 2:1014. [Google Scholar]
- 30.Fang Y, Fang L, Wang Y, Lei Y, Luo R, Wang D, Chen H, Xiao S. 2012. Porcine reproductive and respiratory syndrome virus nonstructural protein 2 contributes to NF-kappaB activation. Virol J 9:83. doi: 10.1186/1743-422X-9-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiao Y, Wu W, Gao J, Smith N, Burkard C, Xia D, Zhang M, Wang C, Archibald A, Digard P, Zhou EM, Hiscox JA. 2016. Characterization of the interactome of the porcine reproductive and respiratory syndrome virus nonstructural protein 2 reveals the hyper variable region as a binding platform for association with 14-3-3 proteins. J Proteome Res 15:1388–1401. doi: 10.1021/acs.jproteome.5b00396. [DOI] [PubMed] [Google Scholar]
- 32.Song T, Fang L, Wang D, Zhang R, Zeng S, An K, Chen H, Xiao S. 2016. Quantitative interactome reveals that porcine reproductive and respiratory syndrome virus nonstructural protein 2 forms a complex with viral nucleocapsid protein and cellular vimentin. J Proteomics 142:70–81. doi: 10.1016/j.jprot.2016.05.009. [DOI] [PubMed] [Google Scholar]
- 33.Wang L, Zhou L, Zhang H, Li Y, Ge X, Guo X, Yu K, Yang H. 2014. Interactome profile of the host cellular proteins and the nonstructural protein 2 of porcine reproductive and respiratory syndrome virus. PLoS One 9:e99176. doi: 10.1371/journal.pone.0099176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hershko A, Ciechanover A. 1986. The ubiquitin pathway for the degradation of intracellular proteins. Prog Nucleic Acid Res Mol Biol 33:19–56, 301. [DOI] [PubMed] [Google Scholar]
- 35.Mizushima N, Komatsu M. 2011. Autophagy: renovation of cells and tissues. Cell 147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 36.Hershey JW. 1991. Translational control in mammalian cells. Annu Rev Biochem 60:717–755. doi: 10.1146/annurev.bi.60.070191.003441. [DOI] [PubMed] [Google Scholar]
- 37.Merrick WC. 1992. Mechanism and regulation of eukaryotic protein synthesis. Microbiol Rev 56:291–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gil J, Esteban M, Roth D. 2000. In vivo regulation of protein synthesis by phosphorylation of the alpha subunit of wheat eukaryotic initiation factor 2. Biochemistry 39:7521–7530. doi: 10.1021/bi992868b. [DOI] [PubMed] [Google Scholar]
- 39.Farook JM, Shields J, Tawfik A, Markand S, Sen T, Smith SB, Brann D, Dhandapani KM, Sen N. 2013. GADD34 induces cell death through inactivation of Akt following traumatic brain injury. Cell Death Dis 4:e754. doi: 10.1038/cddis.2013.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sonenberg N, Hinnebusch AG. 2009. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sidrauski C, McGeachy AM, Ingolia NT, Walter P. 2015. The small molecule ISRIB reverses the effects of eIF2alpha phosphorylation on translation and stress granule assembly. Elife 4:e05033. doi: 10.7554/eLife.05033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, Gray NS. 2009. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem 284:8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Daveri E, Maellaro E, Valacchi G, Ietta F, Muscettola M, Maioli E. 2016. Inhibitions of mTORC1 and 4EBP-1 are key events orchestrated by Rottlerin in SK-Mel-28 cell killing. Cancer Lett 380:106–113. doi: 10.1016/j.canlet.2016.06.018. [DOI] [PubMed] [Google Scholar]
- 44.Roux PP, Topisirovic I. 2012. Regulation of mRNA translation by signaling pathways. Cold Spring Harb Perspect Biol 4(11):a012252. doi: 10.1101/cshperspect.a012252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao S, Chen C, Wang S, Ji F, Xie Y. 2016. MHY1485 activates mTOR and protects osteoblasts from dexamethasone. Biochem Biophys Res Commun 481:212–218. doi: 10.1016/j.bbrc.2016.10.104. [DOI] [PubMed] [Google Scholar]
- 46.Wertz GW, Youngner JS. 1970. Interferon production and inhibition of host synthesis in cells infected with vesicular stomatitis virus. J Virol 6:476–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Connor JH, Lyles DS. 2002. Vesicular stomatitis virus infection alters the eIF4F translation initiation complex and causes dephosphorylation of the eIF4E binding protein 4E-BP1. J Virol 76:10177–10187. doi: 10.1128/JVI.76.20.10177-10187.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roth H, Magg V, Uch F, Mutz P, Klein P, Haneke K, Lohmann V, Bartenschlager R, Fackler OT, Locker N, Stoecklin G, Ruggieri A. 2017. Flavivirus infection uncouples translation suppression from cellular stress responses. mBio 8:e02150-16. doi: 10.1128/mBio.02150-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou JH, Zhang J, Sun DJ, Ma Q, Chen HT, Ma LN, Ding YZ, Liu YS. 2013. The distribution of synonymous codon choice in the translation initiation region of dengue virus. PLoS One 8:e77239. doi: 10.1371/journal.pone.0077239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Narayanan K, Huang C, Lokugamage K, Kamitani W, Ikegami T, Tseng CT, Makino S. 2008. Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J Virol 82:4471–4479. doi: 10.1128/JVI.02472-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kappes MA, Faaberg KS. 2015. PRRSV structure, replication and recombination: origin of phenotype and genotype diversity. Virology 479-480:475–486. doi: 10.1016/j.virol.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Snijder EJ, Kikkert M, Fang Y. 2013. Arterivirus molecular biology and pathogenesis. J Gen Virol 94:2141–2163. doi: 10.1099/vir.0.056341-0. [DOI] [PubMed] [Google Scholar]
- 53.Duan E, Wang D, Fang L, Ma J, Luo J, Chen H, Li K, Xiao S. 2015. Suppression of porcine reproductive and respiratory syndrome virus proliferation by glycyrrhizin. Antiviral Res 120:122–125. doi: 10.1016/j.antiviral.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jagger BW, Wise HM, Kash JC, Walters KA, Wills NM, Xiao YL, Dunfee RL, Schwartzman LM, Ozinsky A, Bell GL, Dalton RM, Lo A, Efstathiou S, Atkins JF, Firth AE, Taubenberger JK, Digard P. 2012. An overlapping protein-coding region in influenza A virus segment 3 modulates the host response. Science 337:199–204. doi: 10.1126/science.1222213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kint J, Langereis MA, Maier HJ, Britton P, van Kuppeveld FJ, Koumans J, Wiegertjes GF, Forlenza M. 2016. Infectious bronchitis coronavirus limits interferon production by inducing a host shutoff that requires accessory protein 5b. J Virol 90:7519–7528. doi: 10.1128/JVI.00627-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Han M, Ke H, Zhang Q, Yoo D. 2017. Nuclear imprisonment of host cellular mRNA by nsp1beta protein of porcine reproductive and respiratory syndrome virus. Virology 505:42–55. doi: 10.1016/j.virol.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ke H, Han M, Zhang Q, Rowland R, Kerrigan M, Yoo D. 2018. Type I interferon suppression-negative and host mRNA nuclear retention-negative mutation in nsp1beta confers attenuation of porcine reproductive and respiratory syndrome virus in pigs. Virology 517:177–187. doi: 10.1016/j.virol.2018.01.016. [DOI] [PubMed] [Google Scholar]
- 58.Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, Pestova TV, Qian SB, Jaffrey SR. 2015. 5′ UTR m(6)A promotes cap-independent translation. Cell 163:999–1010. doi: 10.1016/j.cell.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian SB. 2015. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature 526:591–594. doi: 10.1038/nature15377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tsai K, Courtney DG, Cullen BR. 2018. Addition of m6A to SV40 late mRNAs enhances viral structural gene expression and replication. PLoS Pathog 14:e1006919. doi: 10.1371/journal.ppat.1006919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dauber B, Poon D, Dos Santos T, Duguay BA, Mehta N, Saffran HA, Smiley JR. 2016. The herpes simplex virus virion host shutoff protein enhances translation of viral true late mRNAs independently of suppressing protein kinase R and stress granule formation. J Virol 90:6049–6057. doi: 10.1128/JVI.03180-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pyronnet S. 2000. Phosphorylation of the cap-binding protein eIF4E by the MAPK-activated protein kinase Mnk1. Biochem Pharmacol 60:1237–1243. doi: 10.1016/S0006-2952(00)00429-9. [DOI] [PubMed] [Google Scholar]
- 63.Cuesta R, Xi Q, Schneider RJ. 2004. Structural basis for competitive inhibition of eIF4G-Mnk1 interaction by the adenovirus 100-kilodalton protein. J Virol 78:7707–7716. doi: 10.1128/JVI.78.14.7707-7716.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gingras AC, Svitkin Y, Belsham GJ, Pause A, Sonenberg N. 1996. Activation of the translational suppressor 4E-BP1 following infection with encephalomyocarditis virus and poliovirus. Proc Natl Acad Sci U S A 93:5578–5583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pujhari S, Kryworuchko M, Zakhartchouk AN. 2014. Role of phosphatidylinositol-3-kinase (PI3K) and the mammalian target of rapamycin (mTOR) signalling pathways in porcine reproductive and respiratory syndrome virus (PRRSV) replication. Virus Res 194:138–144. doi: 10.1016/j.virusres.2014.09.017. [DOI] [PubMed] [Google Scholar]
- 66.Nakagawa K, Lokugamage KG, Makino S. 2016. Viral and cellular mRNA translation in coronavirus-infected cells. Adv Virus Res 96:165–192. doi: 10.1016/bs.aivir.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guan BJ, Krokowski D, Majumder M, Schmotzer CL, Kimball SR, Merrick WC, Koromilas AE, Hatzoglou M. 2014. Translational control during endoplasmic reticulum stress beyond phosphorylation of the translation initiation factor eIF2alpha. J Biol Chem 289:12593–12611. doi: 10.1074/jbc.M113.543215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Preston AM, Hendershot LM. 2013. Examination of a second node of translational control in the unfolded protein response. J Cell Sci 126:4253–4261. doi: 10.1242/jcs.130336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Knoops K, Barcena M, Limpens RW, Koster AJ, Mommaas AM, Snijder EJ. 2012. Ultrastructural characterization of arterivirus replication structures: reshaping the endoplasmic reticulum to accommodate viral RNA synthesis. J Virol 86:2474–2487. doi: 10.1128/JVI.06677-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huo Y, Fan L, Yin S, Dong Y, Guo X, Yang H, Hu H. 2013. Involvement of unfolded protein response, p53 and Akt in modulation of porcine reproductive and respiratory syndrome virus-mediated JNK activation. Virology 444:233–240. doi: 10.1016/j.virol.2013.06.015. [DOI] [PubMed] [Google Scholar]
- 71.Snijder EJ, van Tol H, Roos N, Pedersen KW. 2001. Non-structural proteins 2 and 3 interact to modify host cell membranes during the formation of the arterivirus replication complex. J Gen Virol 82:985–994. doi: 10.1099/0022-1317-82-5-985. [DOI] [PubMed] [Google Scholar]
- 72.Han J, Liu G, Wang Y, Faaberg KS. 2007. Identification of nonessential regions of the nsp2 replicase protein of porcine reproductive and respiratory syndrome virus strain VR-2332 for replication in cell culture. J Virol 81:9878–9890. doi: 10.1128/JVI.00562-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li B, Fang L, Guo X, Gao J, Song T, Bi J, He K, Chen H, Xiao S. 2011. Epidemiology and evolutionary characteristics of the porcine reproductive and respiratory syndrome virus in China between 2006 and 2010. J Clin Microbiol 49:3175–3183. doi: 10.1128/JCM.00234-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Biedenkopf N, Lange-Grunweller K, Schulte FW, Weisser A, Muller C, Becker D, Becker S, Hartmann RK, Grunweller A. 2017. The natural compound silvestrol is a potent inhibitor of Ebola virus replication. Antiviral Res 137:76–81. doi: 10.1016/j.antiviral.2016.11.011. [DOI] [PubMed] [Google Scholar]
- 75.Muller C, Schulte FW, Lange-Grunweller K, Obermann W, Madhugiri R, Pleschka S, Ziebuhr J, Hartmann RK, Grunweller A. 2018. Broad-spectrum antiviral activity of the eIF4A inhibitor silvestrol against corona- and picornaviruses. Antiviral Res 150:123–129. doi: 10.1016/j.antiviral.2017.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cencic R, Desforges M, Hall DR, Kozakov D, Du Y, Min J, Dingledine R, Fu H, Vajda S, Talbot PJ, Pelletier J. 2011. Blocking eIF4E-eIF4G interaction as a strategy to impair coronavirus replication. J Virol 85:6381–6389. doi: 10.1128/JVI.00078-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rodriguez P, Perez-Morgado MI, Gonzalez VM, Martin ME, Nieto A. 2016. Inhibition of influenza virus replication by DNA aptamers targeting a cellular component of translation initiation. Mol Ther Nucleic Acids 5:e308. doi: 10.1038/mtna.2016.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kristensen CS, Kvisgaard LK, Pawlowski M, Holmgaard Carlsen S, Hjulsager CK, Heegaard PMH, Botner A, Stadejek T, Haugegaard S, Larsen LE. 2018. Efficacy and safety of simultaneous vaccination with two modified live virus vaccines against porcine reproductive and respiratory syndrome virus types 1 and 2 in pigs. Vaccine 36:227–236. doi: 10.1016/j.vaccine.2017.11.059. [DOI] [PubMed] [Google Scholar]
- 79.Murtaugh MP, Genzow M. 2011. Immunological solutions for treatment and prevention of porcine reproductive and respiratory syndrome (PRRS). Vaccine 29:8192–8204. doi: 10.1016/j.vaccine.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 80.Zuckermann FA, Garcia EA, Luque ID, Christopher-Hennings J, Doster A, Brito M, Osorio F. 2007. Assessment of the efficacy of commercial porcine reproductive and respiratory syndrome virus (PRRSV) vaccines based on measurement of serologic response, frequency of gamma-IFN-producing cells and virological parameters of protection upon challenge. Vet Microbiol 123:69–85. doi: 10.1016/j.vetmic.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 81.Li B, Fang L, Liu S, Zhao F, Jiang Y, He K, Chen H, Xiao S. 2010. The genomic diversity of Chinese porcine reproductive and respiratory syndrome virus isolates from 1996 to 2009. Vet Microbiol 146:226–237. doi: 10.1016/j.vetmic.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 82.Zhong H, Wang D, Fang L, Zhang H, Luo R, Shang M, Ouyang C, Ouyang H, Chen H, Xiao S. 2013. Ubiquitin-specific proteases 25 negatively regulates virus-induced type I interferon signaling. PLoS One 8:e80976. doi: 10.1371/journal.pone.0080976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jing H, Fang L, Ding Z, Wang D, Hao W, Gao L, Ke W, Chen H, Xiao S. 2017. Porcine reproductive and respiratory syndrome virus nsp1alpha Inhibits NF-kappaB activation by targeting the linear ubiquitin chain assembly complex. J Virol 91:e01911-16. doi: 10.1128/JVI.01911-16. [DOI] [PMC free article] [PubMed] [Google Scholar]