Herpes simplex virus (HSV) establishes a lifelong infection and remains dormant (latent) in our nerve cells. Occasionally HSV reactivates to cause disease, with HSV-1 typically causing cold sores whereas HSV-2 is the most common cause of genital herpes. The details of how HSV reactivates are largely unknown. Most of HSV's genes are silent during latency, with the exception of RNAs made from the latency-associated transcript (LAT) region. While viruses that make less LAT do not reactivate efficiently, these viruses also do not establish latency as efficiently. Here we deliver a ribozyme that can degrade the LAT to the nerve cells of latently infected rabbits using a gene therapy vector. We show that this treatment blocks reactivation in the majority of the rabbits. This work shows that the LAT RNA is important for reactivation and suggests the potential of this treatment as a therapy for treating HSV infections.
KEYWORDS: gene therapy, latency, lncRNA, reactivation, ribozymes
ABSTRACT
During herpes simplex virus (HSV) latency, most viral genes are silenced, with the exception of one region of the genome encoding the latency-associated transcript (LAT). This long noncoding RNA was originally described as having a role in enhancing HSV-1 reactivation. However, subsequent evidence showing that the LAT blocked apoptosis and promoted efficient establishment of latency suggested that its effects on reactivation were secondary to establishment. Here, we utilized an adeno-associated virus (AAV) vector to deliver a LAT-targeting hammerhead ribozyme to HSV-1-infected neurons of rabbits after the establishment of HSV-1 latency. The rabbits were then induced to reactivate latent HSV-1. Using this model, we show that decreasing LAT levels in neurons following the establishment of latency reduced the ability of the virus to reactivate. This demonstrates that the HSV-1 LAT RNA has a role in reactivation that is independent of its function in establishment of latency. In addition, these results suggest the potential of AAV vectors expressing LAT-targeting ribozymes as a potential therapy for recurrent HSV disease such as herpes stromal keratitis, a leading cause of infectious blindness.
IMPORTANCE Herpes simplex virus (HSV) establishes a lifelong infection and remains dormant (latent) in our nerve cells. Occasionally HSV reactivates to cause disease, with HSV-1 typically causing cold sores whereas HSV-2 is the most common cause of genital herpes. The details of how HSV reactivates are largely unknown. Most of HSV's genes are silent during latency, with the exception of RNAs made from the latency-associated transcript (LAT) region. While viruses that make less LAT do not reactivate efficiently, these viruses also do not establish latency as efficiently. Here we deliver a ribozyme that can degrade the LAT to the nerve cells of latently infected rabbits using a gene therapy vector. We show that this treatment blocks reactivation in the majority of the rabbits. This work shows that the LAT RNA is important for reactivation and suggests the potential of this treatment as a therapy for treating HSV infections.
INTRODUCTION
Herpes simplex virus (HSV) establishes a lifelong latent infection within peripheral nerve ganglia following initial infection of the mucus membranes supplied by the neurons. During latency, the viral genomes are maintained as circular episomes within the nucleus of the neuron. Periodically, the virus reactivates from individual neurons, and the newly produced virions are transported via the nerve fibers to the original site of infection. HSV typically infects the mucosal epithelia of the oral cavity or eyes (HSV-1) and also the genitalia (HSV-2), where it can cause cold sores or ocular herpes and genital herpes, respectively. Of special significance, HSV-1 reactivation in the eyes of some individuals is responsible for herpes stromal keratitis (HSK), which is the leading cause of infectious blindness in the United States (1). During latency, most viral genes are silenced, with the exception of one region of the genome encoding the latency-associated transcript (LAT). The HSV-1 LAT is transcribed from the viral long repeats as an 8.3- to 8.5-kb polyadenylated primary transcript (Fig. 1). This long noncoding RNA (lncRNA) is the only viral transcript known to accumulate abundantly during latency (2–4). The primary transcript is spliced to yield a 2.0-kb intron (5). This intron is highly stable, with an observed half-life of 24 h in cell culture (6). It is also stable in vivo and is easily detectable by in situ hybridization within about 25 to 30% of latently infected ganglionic neurons (7, 8). Genetic studies involving deletions and mutations within the LAT promoter and 5′ exon-coding region have yielded a multitude of phenotypes. These pertain to antiapoptosis (9), neuronal tropism (10), reactivation (11–14), leakiness of lytic gene transcription during latency (15), and histone H3 methylation (16, 17). Study of the LAT region is further complicated by the fact that there are at least 8 microRNAs as well as other noncoding RNAs (ncRNAs) transcribed from the LAT locus, whose functions are still being elucidated by mutational analysis (for a review, see reference 18). While LAT promoter mutants display many phenotypes, the mechanism of the LAT RNA in these processes is unknown.
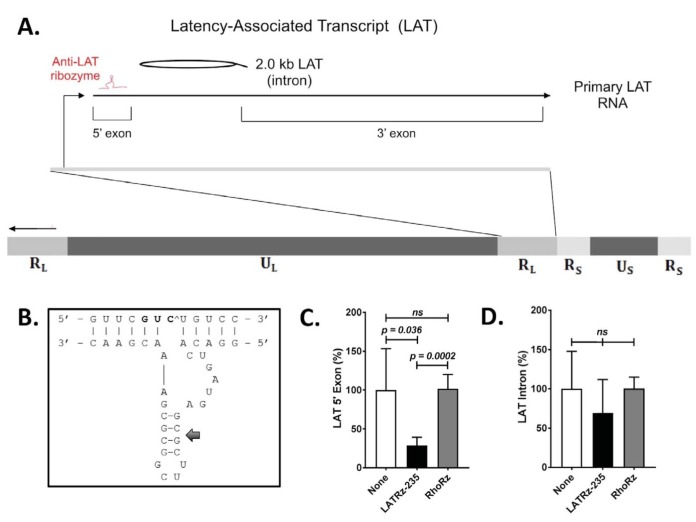
FIG 1.
Schematic of the HSV-1 genome showing the regions encoding the HSV-1 LAT and design and in vitro testing results of a ribozyme targeting the LAT. (A) The LAT locus (located in both copies of the RL region of the genome) is expanded to show the 8.5-kb primary LAT, the 5′ and 3′ exons, and the stable 2.0-kb intron. (B) Design of a synthetic hammerhead ribozyme targeting the LAT 5′ exon. The 5′ exon of the HSV-1 LAT was analyzed for hammerhead cleavage sequences. Antisense flanking regions were designed around the conserved catalytic domain to hybridize with the region surrounding the selected triplet. Mfold analysis predicted base pairing in stem II (arrow), but not in the flanking regions, so as to rule out intramolecular base pairing interfering with efficient binding between the ribozyme and the target. (C and D) Quantification of the HSV-1 LAT 5′ exon (C) and the 2.0-kb LAT intron (D) in rabbit skin cells following transfection with a LAT-expressing plasmid and either mock treated or cotransfected with LATRz-235 or RhoRz. RNA was isolated, cDNA was transcribed, and LAT levels were analyzed by TaqMan real-time PCR that was normalized to the untargeted cellular control rabbit GAPDH. Normalized data are graphed as percentages of the 5′ exon or LAT intron in treated cells relative to untreated cells (None). Each bar represents the average expression of four wells of a 24-well plate (n = 4). Error bars represent standard deviations.
LAT promoter mutants exhibit a reduction in genome copy numbers 2- to 3-fold within sensory ganglia compared to wild-type HSV, presumably due to the LAT's ability to protect neurons from apoptosis (9, 19) or cell death (20) during the later stages of the acute infection, as the virus establishes neuronal latency. Given this observation, it has been generally accepted that decreased reactivation of LAT mutants is a consequence of fewer latent genomes being available to reactivate (21), though conclusive evidence is lacking. Therefore, the goal of this study was to determine if the LAT RNA is directly involved in facilitating HSV-1 reactivation. To examine this specific function of the LAT RNA, we performed in vivo knockdown of the LAT after latency had been established. Adeno-associated virus (AAV) vectors were used to deliver a ribozyme to target the HSV-1 LAT being expressed in latently infected sensory neurons. This approach exploits the ability of some serotypes of AAV to be inoculated on the skin or in the cornea, subsequently taken up by nerve termini, and ultimately transported by axonal transport to the neuronal cell bodies, where the HSV-1 latent infection resides (22). We have previously used this approach to deliver small interfering RNAs (siRNAs) to latently infected neurons in the rabbit trigeminal ganglia (TG) to knockdown the cellular insulator protein CTCF (23). Here we demonstrate that using an AAV vector to deliver a ribozyme targeting the LAT after latency has been established blocks induced reactivation in >60% of the rabbit eyes.
RESULTS
A ribozyme targeting the HSV-1 LAT knocks down the LAT RNA in vitro.
For targeting the LAT, we chose a hammerhead ribozyme, a class of catalytic RNA molecules composed of a catalytic loop and three stems that cleave RNAs with high specificity (24). Hammerhead ribozymes can be engineered to act in trans (25, 26) to specifically target virtually any RNA sequence of interest. Our group has designed multiple such ribozymes against HSV-1 transcripts and tested their efficacy both in vitro and in vivo (27). However, to date none of these ribozymes have been directed to the LAT RNA, nor have they been delivered to neurons in vivo. A synthetic hammerhead ribozyme targeting the HSV-1 LAT 5′ exon (Fig. 1A) was designed, synthesized, and cloned into an AAV-green fluorescent protein (GFP) plasmid and was designated LATRz-235, indicating the target cleavage site at the 235th nucleotide of the LAT 5′ exon sequence (Fig. 1B and Table 1). The 5′ exon region of the LAT was targeted initially because this region corresponds to the regions mapped by deletion mutants that resulted in decreased reactivation (28). The efficiency of target knockdown was tested by cotransfecting rabbit skin cells with this plasmid alongside a LAT expression plasmid and measuring the amount of LAT RNA by reverse transcription followed by TaqMan real-time PCR (Fig. 1C). RNA levels were normalized to the nontargeted housekeeping gene transcript for GAPDH (glyceraldehyde-3-phosphate dehydrogenase). Identical transfections were done using a plasmid encoding an antirhodopsin ribozyme that is not predicted to have an effect on LAT RNA expression as a control.
TABLE 1.
Sequence of LATRz-235 and its target in the LAT 5′ exon
| Ribozyme | Sequence | Target sequence |
|---|---|---|
| LATRz-235 | GGACACUGAUGAGCGCUUCGGCGCGAAACGAAC | GUUCGUCUGUCC |
| RhodRz397a | GUACCAAAGCGCGGCUUCGCGAGUAGUCAAGCC | CAUGGUCUUCGG |
Design and characterization described in reference 26.
LAT 5′ exon expression was reduced by 71% in cells cotransfected with the plasmid expressing LATRz-235 compared to LAT levels in cells transfected with the LAT expression plasmid only. In contrast, LAT 5′ exon expression was not affected by cotransfection of the control non-HSV-specific antirhodopsin ribozyme (Fig. 1C). When levels of the LAT intron were examined, there was no significant reduction in level, likely due to its long half-life (Fig. 1D). No amplification was detected in controls lacking reverse transcriptase (not shown). These results demonstrate that LATRz-235 is effective in reducing the levels of the primary LAT transcript. We then used LATRz-235 to knock down LAT in vivo using the strategy and overall design depicted in Fig. 2A and B).
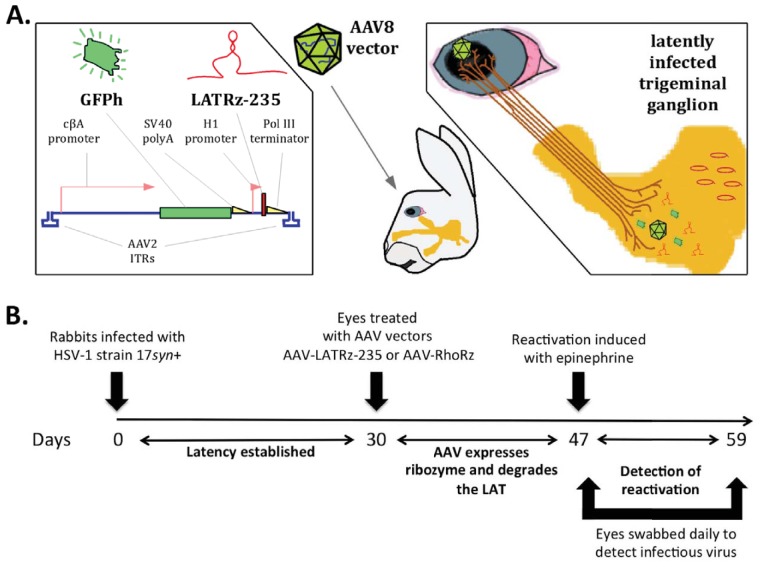
FIG 2.
Delivery of ribozyme targeting the HSV-1 LAT into latently infected rabbits. (A) The LAT-targeting ribozyme was cloned and packaged in an AAV vector (AAV-LATRz-235) that also contains a humanized green fluorescent protein (GFPh) reporter. This vector can be applied to the corneas of rabbits latently infected with HSV-1, allowing the AAV to be taken up by nerve termini under the cornea, and subsequent transport of the AAV to the trigeminal ganglia (TG) neurons. (B) Time line for testing the effect of the LAT ribozyme on epinephrine-induced reactivation.
Delivery of the AAV vector expressing the LAT ribozyme to the eyes of latently infected rabbits blocks HSV-1 reactivation.
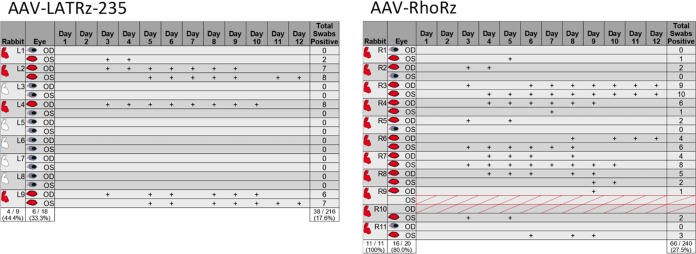
Since the goal of this study was to determine whether in vivo knockdown of the LAT RNA after latency is established affects the ability of HSV-1 to reactivate, the rabbit model was used. The advantage of the rabbit model for the study of HSV-1 reactivation is that the latent virus can be efficiently and synchronously induced to reactivate following transcorneal iontophoresis of β-adrenergic agonists, such as epinephrine (29). In addition, the rabbit model has many features that parallel HSV-1 recurrent disease in humans, including reactivation at the original site of infection. Rabbits are infected in both eyes with HSV-1 by ocular scarification, and after 30 days, the virus is latent within the trigeminal ganglia, and no infectious virus can be detected in the TG or in the tears, except for occasional spontaneous shedding (30, 31). In this study, both eyes of latently infected rabbits were treated with either of two AAV vectors. One set of rabbits was treated with an AAV vector expressing a ribozyme targeting human rhodopsin (AAV-RhoRz) as a control, whereas the other set of rabbits were treated with the HSV-1 LAT-targeting ribozyme (AAV-LATRz-235). We have previously demonstrated that AAV applied to the rabbit eye is efficiently transported to the TG and can efficiently transduce >70% of the total neurons (22). Seventeen days after treatment with AAV, the rabbit eyes were iontophoresed with epinephrine and swabbed daily for 12 days to collect tears. These swabs were then assayed for the presence of infectious virus to measure reactivation. HSV-1 reactivation was detected (at least one positive swab) in only 33% of rabbit eyes treated with AAV-LATRz-235 compared to 80% of those treated with AAV-RhoRz (Fig. 3) (P = 0.04). In addition, the total number of positive swabs was significantly reduced in the AAV-LATRz-235-treated group (P = 0.0026). Therefore, AAV-LATRz-235 vector treatment significantly blocked detectable reactivation in the majority of the rabbits and eyes compared to controls and reduced the overall shedding of virus.
FIG 3.
Treatment with AAV LATRz-235 reduces induced reactivation in vivo. Eyes of latently infected rabbits were treated with either AAV-LAT Rz-235 or AAV-RhoRz. Seventeen days after AAV vector treatment, the rabbits were induced to reactivate. The eyes were then swabbed daily for 12 days to detect infectious HSV-1 as evidence of reactivation. Data are grouped by rabbits (L1 to L9, treated with the AAV LATRz-235, and R1 to R11, treated with the AAV-RhoRz) and by right (OD) and left (OS) eyes of each rabbit. A plus sign (+) indicates that infectious virus was detected in that eye for that day. Rabbits or eyes that exhibited at least one positive swab during the 12-day period are in red. Red crosshatches through OS for R9 and OD for R10 indicate that these eyes were prevented from being iontophoresed due to corneal scarring and were excluded from the reactivation study. Percentages of rabbits, eyes, or swabs that were positive for the two treatment groups are indicated at the bottom of each column. Exact versions of chi-square analysis for all pairwise comparisons with control (AAV-RhoRz) were performed.
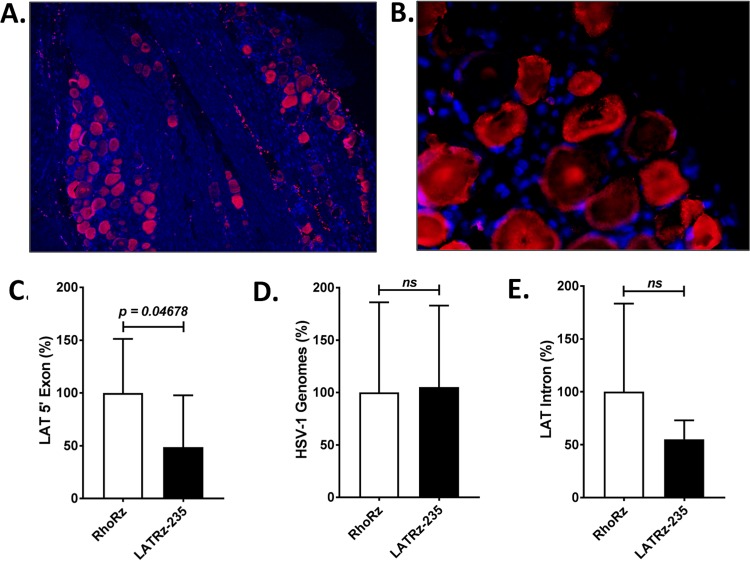
After the reactivation study was complete, rabbit TG were dissected, and one TG per rabbit was processed and analyzed by immunohistochemistry (IHC) to detect GFP expression from AAV-LATRz-235. These analyses revealed that >80% of the neurons in the TG were transduced by AAV and expressed GFP (Fig. 4A and B). The other TG was subjected to DNA/RNA extraction. The RNA was reverse transcribed, and quantitative PCR (qPCR) analysis was performed to quantify the HSV-1 LAT (Fig. 4C). Significantly less LAT was detected in the AAV LATRz-235-treated groups. When the DNA from the same TG was analyzed by qPCR, there were no differences in HSV-1 genomes between the two groups (Fig. 4D). Taken together, these results demonstrate that AAV LATRz-235 decreases LAT levels in the latent ganglia and that this reduction results in decreased reactivation, without destabilizing latent genomes or measurably interfering with their maintenance. Finally, when the levels of the LAT intron RNA were examined by reverse transcriptase quantitative PCR (RT-qPCR), there was no significant difference between the AAV-LATRz-235- and AAV-RhoRz-treated groups (Fig. 4E). This is consistent with the degree of knockdown of the LAT intron in vitro (Fig. 1D) and is likely due to the high stability of the intron.
FIG 4.
Analysis of rabbit TG for AAV transduction, LAT expression, and HSV-1 DNA load. (A and B) Immunofluorescence (IF) demonstrating long-term expression of GFP in a high percentage of latently infected rabbit TG neurons. IF with mouse primary antibody to GFP was performed to identify populations of neurons harboring the AAV-LATRz-235 vector, which expressed GFP from the CBA promoter. Alexa Fluor 548 (Invitrogen) was used as the secondary antibody for immunofluorescence visualization using a Leica deconvolution microscope with Slidebook 5.0. Panel A represents an original magnification of ×10 and panel B the original magnification of ×40 from the same section. DAPI staining in blue represents nuclei of satellite cells present in the sections. RNA was isolated and analyzed by RT-PCR using primers and probes specific for the LAT 5′exon or LAT 2.0-kb intron regions of the genome. Relative quantities were determined for the LAT and then normalized to the relative quantities of rabbit GAPDH RNA to account for variations in RNA recovery between samples. (C) LAT 5′ exon expression significantly decreased following treatment with AAV-LATRz-235 expressing the anti-LAT ribozyme. The average values for each virus are presented along with standard deviations (P < 0.05). (D) HSV-1 genome load was determined by qPCR using primers for the HSV-1 DNA polymerase gene (see Materials and Methods and Table 2). (E) LAT stable 2.0-kb intron RNA levels do not show significant decrease following treatment with AAV-LATRz-235 (n = 7 for AAV-RhoRz [control]; n = 6 for AAV-LATRz-235). All PCR analyses were performed in triplicate, and statistical analyses were performed as one-way analysis of variance (ANOVA) using SigmaPlot 12.5.
DISCUSSION
In this study, we demonstrate that a ribozyme that targets the HSV-1 LAT, when delivered to the eye of a rabbit that has been latently infected with HSV-1, completely blocks detectable reactivation of the virus in >60% of the eyes compared to eyes treated with an AAV vector expressing a ribozyme targeting a heterologous target. This result supports a direct role of the HSV-1 LAT RNA in facilitating efficient reactivation from latency, which is separate from its function in enhancing establishment of latency.
It is important to note that these findings are not in conflict with studies that have provided clear evidence for a role of the HSV-1 LAT in efficient establishment (19, 32) and/or maintenance (33, 34) of latent infections. Studies have shown that HSV-1 recombinants with deletions in the LAT promoter and portions of the 5′ exon coding region have an increase in apoptotic neurons during the acute infection (9, 20, 35), resulting in a 2- to 3-fold decrease in total HSV-1 DNA detected in the ganglia during latency (36). Even though LAT promoter mutants also exhibited a 90% or greater reduction in their ability to reactivate in the rabbit eye (14, 37), it previously was not possible to determine the role of qualitative or quantitative differences in establishment of latency on the ability to reactivate. Our ability in the present study to knock down the LAT RNA after latency is established without altering the integrity of the genome allowed us to separate the LAT's effects on reactivation from effects on establishment. This may open the door for future studies to tease out the specific aspects of the LAT that mediate this role in reactivation.
Given the data showing that the LAT protects against apoptosis, we considered the possibility that knockdown of the LAT as in this study could render neurons that are latently infected with HSV more susceptible to apoptosis (38), especially after subjecting them to epinephrine induction. As shown in Fig. 4D, we saw no evidence of decrease in HSV genomes in the TG from both sets of rabbits after the experiment was complete. It is plausible, however, that our experiment did not have sufficient statistical power to see a subtler decrease. A related point is whether other effects of the LAT in maintaining latency could have affected reactivation (34, 39). Again, while we did not see any decrease in total latent genomes over the 17-day period following ribozyme treatment, we feel this 17-day time frame was probably too short to observe any effects of the depleted LAT on maintaining latency. We plan to look at this in future experiments over a period of several months following LAT ribozyme treatment.
The ribozyme used in this study targets the 5′ exon region of the LAT primary transcript for cleavage. Cleaving the RNA in this region would be predicted to decrease levels of the primary LAT, spliced LAT, and the LAT intron. In both our in vitro and in vivo analyses, we observed an approximately 70% decrease in the 5′ exon RNA as determined by qRT-PCR analyses. In contrast, however, we saw only a 35 to 40% decrease in the LAT intron. This is likely due to the high stability and long half-life of the LAT intron compared to the LAT primary transcript. Previous work suggesting that the reactivation phenotype of HSV is localized to the promoter/5′ exon coding region of the LAT (28, 37) may be consistent with the degree of decreased reactivation observed in this study. It will be useful to determine if using multiple ribozymes to target different regions of the LAT, which could lead to more-efficient transcript knockdown, could further decrease reactivation. It should be noted that we initially chose to use a ribozyme as opposed to an siRNA or short hairpin RNA (shRNA) because ribozymes are active in the nucleus, which is important since the LAT is a nuclear RNA. Recent studies have shown that transfected siRNAs can efficiently target nuclear transcripts, so this is an option for future studies. In either instance, the ability of AAV to deliver RNA cleaving or silencing RNAs to neurons in vivo offers the potential to dissect the roles of viral or cellular RNAs without altering other functions on the genomes through the use of gene editing or deletion. This offers a powerful and complementary approach to mutation/deletion strategies, especially for genomes of viruses like the herpesviruses that possess a high density of cis-acting genetic elements that cannot easily be untangled. This point is well illustrated in the repeat regions of HSV-1 (18).
Finally, the approach of delivering LAT-targeting ribozymes that block reactivation opens the door to using this approach to treat recurrent HSV-1 disease. AAV vectors are maintained as extrachromosomal episomes (for a review, see reference 40) and express their transgenes for long periods of time in nondividing cells such as neurons (41). AAV can be delivered to human eyes using only minor modifications of clinical practices currently used to treat corneal infections. Therefore, this approach may embody a new treatment for herpes stromal keratitis, the leading infectious cause of blindness in the United States, and for treating recurrent oral and genital herpes as well.
MATERIALS AND METHODS
Viruses and cells.
A low-passage-number stock of HSV-1 strain 17syn+ was obtained from J. Stevens. Viruses were propagated on rabbit skin (RS) cells as described previously (42). Briefly, RS cells were propagated in minimal essential medium (MEM; Gibco Life Technologies, Gaithersburg, MD) with 5% fetal bovine serum (FBS) and antibiotics at 37°C in a humidified 5% carbon dioxide atmosphere. To assess cytopathic effect (CPE) of tear film swabs in the reactivation studies, primary rabbit kidney (PRK) cells were propagated in MEM with 5% FBS under the same conditions.
Target selection and design of synthetic hammerhead ribozymes.
The LAT 5′ exon and intron regions of the HSV-1 genome (NCBI database accession number NC_001806) were scanned for hammerhead ribozyme cleavage sites. The nucleotide triplets GUC, CUC, and UUC were specifically selected as potential target sites. Hybridizing arms were designed complementary to the sequence surrounding the cleavage site. Mfold software (43) was used to predict the secondary structure of each ribozyme. Ribozymes predicted to have a correctly folded catalytic core and open hybridizing arms were carried forward to in vitro testing. An example of sequence selection, design, and predicted folding is shown in Fig. 1.
Cloning synthetic ribozymes into AAV plasmids.
Selected synthetic hammerhead ribozymes targeting the LAT were cloned into scAAV2-GFP plasmids for in vitro testing and subsequent vector production. In this vector, humanized GFP (GFPh) is expressed from the CBA promoter consisting of the chimeric chicken beta actin proximal promoter plus the immediate early enhancer from cytomegalovirus. The self-complementary AAV (scAAV) plasmid contains a modified terminal repeat (TR) that results in the generation of a self-complementary copy of the transgene during packaging to improve transduction efficiency (44). The ribozyme sequence was placed downstream of an H1 promoter and terminated by a PolIII terminator sequence. These minigenes were synthesized by GenScript with SalI restriction endonuclease sites at each end. AAV plasmids containing the AAV2 TRs and the insert were digested with SalI (New England BioLabs) and ligated. Ligations were transformed into SURE electroporation-competent cells (Stratagene) and grown in LB medium. Plasmids were isolated and purified by CsCl centrifugation. Insertion was confirmed by PCR and Sanger sequencing. The construction and characterization of the AAV-RhoRz (also referred to as RhodRz397) were described previously (45). The AAV plasmids were pseudotyped into AAV8 Y733F modified capsids.
In vitro testing of synthetic hammerhead ribozymes.
Lipofectamine 2000 (Invitrogen) was used to cotransfect a plasmid encoding synthetic hammerhead ribozymes and a plasmid encoding the target LAT transcript into rabbit skin cells. Each transfection was performed in triplicate in a 24-well plate format. A plasmid expressing an antirhodopsin ribozyme was cotransfected in control wells to account for nonspecific effects. Twenty-four hours posttransfection, RNA was isolated using TRIzol (Ambion) and used for subsequent reverse transcription and qPCR analysis as described below.
Rabbit inoculation with HSV-1 and establishment of latency.
New Zealand White rabbits (2 to 3 kg) were inoculated with 1 × 105 to 2 × 105 PFU of virus per eye in a suspension of 25 μl MEM with 5% FBS. The viral suspension was placed in the cul-de-sac of the scarified eye, and the eye was closed and massaged for 30 s with care to prevent virus loss (46). On postinoculation (PI) days 3, 5, 7, 10, and 14, all corneas were examined by slit lamp microscopy (SLE) with 0.1% fluorescein for evidence of acute herpetic keratitis. On PI day 30, all eyes were examined to ensure that the corneal epithelium was free of lesions. Only eyes that were normal by SLE were included in the reactivation study. Rabbits that exhibited acute lesions with subsequent recovery were considered latently infected. All animal care and procedures described conducted for the studies in this paper were approved by the Louisiana State University Health Sciences Center Institutional Animal Care and Use Committee.
Treatment of rabbit eyes with AAV LATRz-235 and AAV RhoRz vectors.
Rabbit corneas were abraded using a 27 gauge needle and forming an 8 × 8 lattice crosshatch pattern across the entire surface of the cornea. Following abrasion, 1 1011 × 1011 genome copies of either LATRz-235 or AAV RhoRz in a total volume of no less than 25 μl and no greater than 40 μl of saline was applied directly to the surface of the cornea. The eyelids were gently massaged over the eye.
Induction of HSV-1 reactivation in rabbits.
Transcorneal iontophoresis (0.8 mA for 8 min) of 0.01% epinephrine was performed on each eye once daily for 3 consecutive days (PI days 31, 32, and 33). To assess the frequency of reactivation, tears were collected daily on each of the three reactivation stimulus days (prior to each iontophoresis procedure) and for an additional 9 days thereafter (total of 12 consecutive days following iontophoresis initiation). Tear film collection consisted of inserting a Dacron swab (Puritan, Hardwood Products Company, Guilford, ME) into the cul-de-sac of the eye and passed over the corneal surface and conjunctiva. Each eye was swabbed independently, and the swab was placed in a tube containing 1 ml DMEM (1% FBS). Tubes were placed at −80°C overnight and then thawed at 37°C the following day. The Dacron swabs were removed aseptically, and the medium was placed into a 24-well plate containing a monolayer of CV-1 cells. The plates were then monitored for 7 days for evidence of CPE.
Reverse transcription.
To remove any DNA contamination, RNA samples from transfection experiments were DNase treated with Turbo DNA-free (Ambion). cDNA was synthesized using OmniScript (Qiagen) with 10 μM random decamer primers (Ambion). To control for any residual genomic DNA in cDNA samples, no-RT control reactions were performed. Mixtures for these reactions included all components except the OmniScript reverse transcription enzyme.
Real-time PCR.
cDNA samples from transfection experiments were analyzed by TaqMan quantitative real-time PCR on a StepOnePlus thermocycler (Applied Biosystems). Quantities were normalized to a cellular housekeeping gene, rabbit GAPDH gene. Custom TaqMan assay sequences are given in Table 2.
TABLE 2.
Custom TaqMan assay sequences
| Assay name | Forward primer | Reverse primer | Probe |
|---|---|---|---|
| LAT 5′ exon | GGCTCCATCGCCTTTCCT | AAGGGAGGGAGGAGGGTACTG | TCTCGCTTCTCCCC |
| LAT intron | CGCCCCAGAGGCTAAGG | GGGCTGGTGTGCTGTAACA | CCACGCCACTCGCG |
| HSV DNA Pol | AGAGGGACATCCAGGACTTTGT | CAGGCGCTTGTTGGTGTAC | ACCGCCGAACTGAGCA |
| Rabbit GAPDH | GCACCACCAACTGCTTAGC | CCTCCACAATGCCGAAGTG | CTGGCCAAGGTCATCC |
Immunofluorescence.
Slides containing sections of rabbit TG were deparaffinized, hydrated, and peroxidase blocked through a series of washes that included (i) two xylene washes for 5 min each, (ii) two 100% ethanol washes for 2 min each, (iii) one 3% hydrogen peroxide wash (30% hydrogen peroxide was diluted 1:10 in 100% methanol) for 10 min, (iv) one 95% ethanol wash for 3 min, (v) one 70% ethanol wash for 1 min, and (vi) one Tris-buffered saline with Tween 20 (TBST) wash for 1 min. Epitope retrieval was done using the IHC-Tek epitope retrieval steamer set. The retrieval jar was filled with retrieval solution, slides were placed inside, and the jar was placed in the steamer for 1 h. The lid was removed from the jar, and the solution was cooled to room temperature. Slides were washed with TBST and placed in a humidified chamber. Slides were blocked using 1.5% normal serum (goat) for 30 min and washed with TBST. Sections were then incubated with primary antibody (1:200 of mouse anti-GFP) for 24 h at 4°C. Secondary antibody incubation was then performed using 1:1,000 goat anti-mouse Alexa Fluor 548 (Invitrogen) for 2 h at room temperature, followed by a TBST wash. The slides were mounted using ProLong gold mounting medium (Invitrogen) and imaged using a Leica deconvolution microscope equipped with Slidebook 5.0.
ACKNOWLEDGMENTS
This work was supported in part by grants R01 AI48633 and R56 AI101174 from the NIH and an unrestricted grant from Research to Prevent Blindness to A.S.L. and S.S.T. D.M.P. was supported by a Clinical Research Training Award from the NIH (TL1 TR000066). We also thank William Hauswirth and the Center for Vision Research Vector Core lab for providing some of the AAV vectors used in these experiments. This core was supported by a core grant from the National Eye Institute (P30 EY02172).
REFERENCES
- 1.Liesegang TJ. 2001. Herpes simplex virus epidemiology and ocular importance. Cornea 20:1–13. doi: 10.1097/00003226-200101000-00001. [DOI] [PubMed] [Google Scholar]
- 2.Stevens JG, Wagner EK, Devi RGB, Cook ML, Feldman LT. 1987. RNA complementary to a herpesvirus alpha gene mRNA is prominent in latently infected neurons. Science 235:1056–1059. doi: 10.1126/science.2434993. [DOI] [PubMed] [Google Scholar]
- 3.Deatly AM, Spivack JG, Lavi E, Fraser NW. 1987. RNA from an immediate early region of the type 1 herpes simplex virus genome is present in the trigeminal ganglia of latently infected mice. Proc Natl Acad Sci U S A 84:3204–3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rock DL, Nesburn AB, Ghaisi H, Ong J, Lewis TL, Lokensgard JR, Wechsler SL. 1987. Detection of latency-related viral RNAs in trigeminal ganglia of rabbits latently infected with herpes simplex virus. J Virol 61:3820–3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farrell MJ, Dobson AT, Feldman LT. 1991. Herpes simplex virus latency-associated transcript is a stable intron. Proc Natl Acad Sci U S A 88:790–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomas DL, Lock M, Zabolotny JM, Mohan BR, Fraser NW. 2002. The 2-kilobase intron of the herpes simplex virus type 1 latency-associated transcript has a half-life of approximately 24 hours in SY5Y and COS-1 cells. J Virol 76:532–540. doi: 10.1128/JVI.76.2.532-540.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mehta A, Maggioncalda J, Bagasra O, Thikkavarapu S, Saikumari P, Valyi-Nagy T, Fraser NW, Block TM. 1995. In situ DNA PCR and RNA hybridization of herpes simplex virus sequences in trigeminal ganglia of latently infected mice. Virology 206:633–640. doi: 10.1016/S0042-6822(95)80080-8. [DOI] [PubMed] [Google Scholar]
- 8.Chen XP, Mata M, Kelley M, Glorioso JC, Fink DJ. 2002. The relationship of herpes simplex virus latency associated transcript expression to genome copy number: a quantitative study using laser capture microdissection. J Neurovirol 8:204–210. doi: 10.1080/13550280290049642. [DOI] [PubMed] [Google Scholar]
- 9.Perng GC, Jones C, Ciacci-Zanella J, Stone M, Henderson G, Yukht A, Slanina SM, Hofman FM, Ghiasi H, Nesburn AB, Wechsler SL. 2000. Virus-induced neuronal apoptosis blocked by the herpes simplex virus latency-associated transcript. Science 287:1500–1503. doi: 10.1126/science.287.5457.1500. [DOI] [PubMed] [Google Scholar]
- 10.Margolis TP, Imai Y, Yang L, Vallas V, Krause PR. 2007. Herpes simplex virus type 2 (HSV-2) establishes latent infection in a different population of ganglionic neurons than HSV-1: role of latency-associated transcripts. J Virol 81:1872–1878. doi: 10.1128/JVI.02110-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leib DA, Bogard CL, Kosz VM, Hicks KA, Coen DM, Knipe DM, Schaffer PA. 1989. A deletion mutant of the latency-associated transcript of herpes simplex virus type 1 reactivates from the latent state with reduced frequency. J Virol 63:2893–2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hill JM, Sedarati F, Javier RT, Wagner EK, Stevens JG. 1990. Herpes simplex virus latent phase transcription facilitates in vivo reactivation. Virology 174:117–125. doi: 10.1016/0042-6822(90)90060-5. [DOI] [PubMed] [Google Scholar]
- 13.Perng G-C, Dunkel EC, Geary PA, Slanina SM, Ghiasi H, Kaiwar R, Nesburn AB, Wechsler SL. 1994. The latency-associated transcript gene of herpes simplex virus type 1 (HSV-1) is required for efficient in vivo spontaneous reactivation of HSV-1 from latency. J Virol 68:8045–8055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bloom DC, Devi-Rao GB, Hill JM, Stevens JG, Wagner EK. 1994. Molecular analysis of herpes simplex virus type 1 during epinephrine induced reactivation of latently infected rabbits in vivo. J Virol 68:1283–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giordani NV, Neumann DM, Kwiatkowski DL, Bhattacharjee PS, McAnany PK, Hill JM, Bloom DC. 2008. During HSV-1 infection of rabbits, the ability to express the LAT increases latent-phase transcription of lytic genes. J Virol 82:6056–6060. doi: 10.1128/JVI.02661-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cliffe AR, Garber DA, Knipe DM. 2009. Transcription of the herpes simplex virus latency-associated transcript promotes the formation of facultative heterochromatin on lytic promoters. J Virol 83:8182–8190. doi: 10.1128/JVI.00712-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwiatkowski DL, Thompson HW, Bloom DC. 2009. The polycomb group protein Bmi1 binds to the herpes simplex virus 1 latent genome and maintains repressive histone marks during latency. J Virol 83:8173–8181. doi: 10.1128/JVI.00686-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Phelan D, Barrozo ER, Bloom DC. 2017. HSV1 latent transcription and non-coding RNA: a critical retrospective. J Neuroimmunol 308:65–101. doi: 10.1016/j.jneuroim.2017.03.002. [DOI] [PubMed] [Google Scholar]
- 19.Perng GC, Slanina SM, Yukht A, Ghiasi H, Nesburn AB, Wechsler SL. 2000. The latency-associated transcript gene enhances establishment of herpes simplex virus type 1 latency in rabbits. J Virol 74:1885–1891. doi: 10.1128/JVI.74.4.1885-1891.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson RL, Sawtell NM. 2001. Herpes simplex virus type 1 latency-associated transcript gene promotes neuronal survival. J Virol 75:6660–6675. doi: 10.1128/JVI.75.14.6660-6675.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sawtell NM, Poon DK, Tansky CS, Thompson RL. 1998. The latent herpes simplex virus type 1 genome copy number in individual neurons is virus strain specific and correlates with reactivation. J Virol 72:5343–5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watson ZL, Ertel MK, Lewin AS, Tuli SS, Schultz GS, Neumann DM, Bloom DC. 2016. Adeno-associated virus vectors efficiently transduce mouse and rabbit sensory neurons coinfected with herpes simplex virus 1 following peripheral inoculation. J Virol 90:7894–7901. doi: 10.1128/JVI.01028-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Washington SD, Edenfield SI, Lieux C, Watson ZL, Taasan SM, Dhummakupt A, Bloom DC, Neumann DM. 2018. Depletion of the insulator protein CTCF results in herpes simplex virus 1 reactivation in vivo. J Virol 92:e00173-. doi: 10.1128/JVI.00173-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Citti L, Rainaldi G. 2005. Synthetic hammerhead ribozymes as therapeutic tools to control disease genes. Curr Gene Ther 5:11–24. doi: 10.2174/1566523052997541. [DOI] [PubMed] [Google Scholar]
- 25.Uhlenbeck OC. 1987. A small catalytic oligoribonucleotide. Nature 328:596–600. doi: 10.1038/328596a0. [DOI] [PubMed] [Google Scholar]
- 26.Haseloff J, Gerlach WL. 1992. Simple RNA enzymes with new and highly specific endoribonuclease activities. 1988. Biotechnology 24:264–269. [PubMed] [Google Scholar]
- 27.Liu J, Lewin AS, Tuli SS, Ghivizzani SC, Schultz GS, Bloom DC. 2008. Reduction in severity of a herpes simplex virus type 1 murine infection by treatment with a ribozyme targeting the UL20 gene RNA. J Virol 82:7467–7474. doi: 10.1128/JVI.02720-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bloom DC, Hill JT, Wagner EK, Feldman LF, Stevens JG. 1996. A 348-bp region in the latency associated transcript facilitates herpes simplex virus type 1 reactivation. J Virol 70:2449–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hill JM, Haruta Y, Rootman DS. 1987. Adrenergically induced recurrent HSV-1 corneal epithelial lessions. Curr Eye Res 6:1065–1071. doi: 10.3109/02713688709034878. [DOI] [PubMed] [Google Scholar]
- 30.Nesburn AB, Elliot JH, Leibowitz HM. 1967. Spontaneous reactivation of experimental herpes simplex keratitis in rabbits. Arch Ophthalmol 78:523–529. doi: 10.1001/archopht.1967.00980030525021. [DOI] [PubMed] [Google Scholar]
- 31.Berman EJ, Hill JM. 1985. Spontaneous ocular shedding of herpes simplex virus 1 in latently infected rabbits. Invest Ophthalmol Vis Sci 26:587–590. [PubMed] [Google Scholar]
- 32.Sawtell NM, Thompson RL. 1992. Herpes simplex virus type 1 latency associated transcription unit promotes anatomical site-dependent establishment and reactivation from latency. J Virol 66:2157–2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nicoll MP, Proenca JT, Connor V, Efstathiou S. 2012. Influence of herpes simplex virus 1 latency-associated transcripts on the establishment and maintenance of latency in the ROSA26R reporter mouse model. J Virol 86:8848–8858. doi: 10.1128/JVI.00652-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nicoll MP, Hann W, Shivkumar M, Harman LE, Connor V, Coleman HM, Proenca JT, Efstathiou S. 2016. The HSV-1 latency-associated transcript functions to repress latent phase lytic gene expression and suppress virus reactivation from latently infected neurons. PLoS Pathog 12:e1005539. doi: 10.1371/journal.ppat.1005539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henderson G, Peng W, Jin L, Perng GC, Nesburn AB, Wechsler SL, Jones C. 2002. Regulation of caspase 8- and caspase 9-induced apoptosis by the herpes simplex virus type 1 latency-associated transcript. J Neurovirol 8(Suppl 2):S103–S111. [DOI] [PubMed] [Google Scholar]
- 36.Thompson RL, Sawtell NM. 1997. The herpes simplex virus type 1 latency-associated transcript gene regulates the establishment of latency. J Virol 71:5432–5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perng G-C, Ghiasi H, Slanina SM, Nesburn AB, Wechsler SL. 1996. The spontaneous reactivation function of the herpes simplex virus type 1 LAT gene resides completely within the first 1.5 kilobases of the 8.3-kilobase primary transcript. J Virol 70:976–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ahmed M, Lock M, Miller CG, Fraser NW. 2002. Regions of the herpes simplex virus type 1 latency-associated transcript that protect cells from apoptosis in vitro and protect neural cells in vivo. J Virol 76:717–729. doi: 10.1128/JVI.76.2.717-729.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson RL, Sawtell NM. 2011. The herpes simplex virus type 1 latency associated transcript locus is required for the maintenance of reactivation competent latent infections. J Neurovirol 17:552–558. doi: 10.1007/s13365-011-0071-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grimm D, Buning H. 2017. Small but increasingly mighty: latest advances in AAV vector research, design, and evolution. Hum Gene Ther 28:1075–1086. doi: 10.1089/hum.2017.172. [DOI] [PubMed] [Google Scholar]
- 41.Sehara Y, Fujimoto KI, Ikeguchi K, Katakai Y, Ono F, Takino N, Ito M, Ozawa K, Muramatsu SI. 2017. Persistent expression of dopamine-synthesizing enzymes 15 years after gene transfer in a primate model of Parkinson's disease. Hum Gene Ther Clin Dev 28:74–79. doi: 10.1089/humc.2017.010. [DOI] [PubMed] [Google Scholar]
- 42.Bloom DC, Stevens JG. 1994. Neuronal specific restriction of an HSV recombinant maps to the UL5 gene. J Virol 68:3761–3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zuker M. 2003. Mfold web server for Nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCarty DM, Fu H, Monahan PE, Toulson CE, Naik P, Samulski RJ. 2003. Adeno-associated virus terminal repeat (TR) mutant generates self-complementary vectors to overcome the rate-limiting step to transduction in vivo. Gene Ther 10:2112–2118. doi: 10.1038/sj.gt.3302134. [DOI] [PubMed] [Google Scholar]
- 45.Gorbatyuk MS, Pang JJ, Thomas J Jr, Hauswirth WW, Lewin AS. 2005. Knockdown of wild-type mouse rhodopsin using an AAV vectored ribozyme as part of an RNA replacement approach. Mol Vis 11:648–656. [PubMed] [Google Scholar]
- 46.Creech CC, Neumann DM. 2010. Changes to euchromatin on LAT and ICP4 following reactivation are more prevalent in an efficiently reactivating strain of HSV-1. PLoS One 5:e15416. doi: 10.1371/journal.pone.0015416. [DOI] [PMC free article] [PubMed] [Google Scholar]