The replication of influenza A virus is regulated by host factors. However, the mechanisms by which lncRNAs regulate IAV infection are not well understood. We identified that lnc-ISG20 is upregulated during IAV infection and is also an interferon-stimulated gene. We demonstrated that lnc-ISG20 can enhance ISG20 expression, which in turn inhibits IAV replication. Our studies indicate that lnc-ISG20 functions as a competing endogenous RNA that binds miR-326 and reduces its inhibitory effect on ISG20. Taken together, our findings reveal the mechanistic details of lnc-ISG20 negatively regulating IAV replication. These findings indicate that lnc-ISG20 plays an important role during the host antiviral immune response.
KEYWORDS: lnc-ISG20, ISG20, influenza A virus, lnc-RNA, antiviral immunity, miR-326
ABSTRACT
Long noncoding RNAs (lncRNAs) are involved in many aspects of cellular processes, including the antiviral immune response. To identify influenza A virus (IAV)-related lncRNAs, we performed RNA deep sequencing to compare the profiles of lncRNAs in A549 and HEK293T cells with or without IAV infection. We identified an IAV-upregulated lncRNA named lnc-ISG20 because it shares most of its sequence with ISG20. We found that lnc-ISG20 is an interferon-stimulated gene similar to ISG20. Overexpression of lnc-ISG20 inhibited IAV replication, while lnc-ISG20 knockdown favored viral replication, suggesting that lnc-ISG20 is inhibitory to IAV replication. Further study indicated that overexpression of lnc-ISG20 enhances ISG20 protein levels, while knockdown of lnc-ISG20 reduces ISG20 protein levels in A549 cells induced with poly(I·C) and Sendai virus. We demonstrated that lnc-ISG20 inhibits IAV replication in an ISG20-dependent manner. As lnc-ISG20 did not affect the mRNA level of ISG20, we postulated that lnc-ISG20 may function as endogenous RNA competing with ISG20 to enhance its translation. Indeed, we identified that microRNA 326 (miR-326) is a mutual microRNA for both ISG20 and lnc-ISG20 that targets the 3′ untranslated region of ISG20 mRNA to inhibit its translation. We confirmed that lnc-ISG20 can bind miR-326, which in turn decreased the amount of miR-326 bound to ISG20 mRNA. In conclusion, we identified that the IAV-upregulated lnc-ISG20 is a novel interferon-stimulated gene that elicits its inhibitory effect on IAV replication by enhancing ISG20 expression. We demonstrated that lnc-ISG20 functions as a competitive endogenous RNA to bind miR-326 to reduce its inhibition of ISG20 translation. Our results revealed the mechanism by which lnc-ISG20 inhibits IAV replication.
IMPORTANCE The replication of influenza A virus is regulated by host factors. However, the mechanisms by which lncRNAs regulate IAV infection are not well understood. We identified that lnc-ISG20 is upregulated during IAV infection and is also an interferon-stimulated gene. We demonstrated that lnc-ISG20 can enhance ISG20 expression, which in turn inhibits IAV replication. Our studies indicate that lnc-ISG20 functions as a competing endogenous RNA that binds miR-326 and reduces its inhibitory effect on ISG20. Taken together, our findings reveal the mechanistic details of lnc-ISG20 negatively regulating IAV replication. These findings indicate that lnc-ISG20 plays an important role during the host antiviral immune response.
INTRODUCTION
Influenza A virus (IAV) causes a highly contagious respiratory disease in humans, threatening public health and representing a considerable financial burden worldwide (1–3). IAV belongs to the Orthomyxoviridae family. Its genome consists of eight segments of negative-sense single-stranded RNA encoding at least 16 proteins, which are coated by viral nucleoprotein (NP) (4, 5).
IAV infection activates the expression of type I interferons (IFNs) and triggers the host antiviral immune response (6). Type I IFNs have broad effects on the innate immune system against viruses, and their ability to restrict IAV replication is largely dependent on the induction of interferon-stimulated genes (ISGs) (7). For example, Mx1 inhibits IAV replication by interfering with viral RNP complex assembly (8), IFITM1, -2, and -3 restrict the early step in IAV replication (9), oligoadenylate synthetase (OAS) activates RNase L to cleave viral RNA, thereby limiting virus replication (10), and protein kinase R (PKR) inhibits the translation of viral mRNAs by phosphorylating the α subunit of eukaryotic initiation factor 2 (11). Moreover, ISG20, a member of the 3′-to-5′ exonuclease superfamily, displays effective antiviral activity against several RNA viruses, including IAV, vesicular stomatitis virus (VSV), hepatitis C virus (HCV), and encephalomyocarditis virus (EMCV), mostly relying on its RNase activity (12–16). In addition, ISG20 inhibits IAV replication by interacting with viral NP protein and inhibiting viral polymerase activity (17).
Long noncoding RNAs (lncRNAs) are a large class of non-protein-coding transcripts with a minimum length of 200 nucleotides (nt) (18). LncRNAs can bind to DNA, RNA, or proteins to elicit their functions in regulating different processes, such as transcription, mRNA stabilization, and protein translation. It has been reported that the lncRNA HOTAIR acts as a scaffold for recruiting PRC2 to repress transcription (19). In contrast, the lncRNA BACE1-AS increases the stability of BACE1 mRNA by forming a RNA duplex, which is associated with the development of Alzheimer's disease (20). Interestingly, lncRNA BGL3 functions as a competitive endogenous RNA (ceRNA) to bind microRNAs (miRNAs) and regulate the translation of the tumor suppressor PTEN (21).
Recently, lncRNAs have also been found to play important roles during virus infection and the antiviral immune response (22). LncRNA-GAS5 suppresses HCV infection by binding the HCV NS3 protein (23), while lncRNA CMPK2 benefits HCV replication as a negative regulator of protein-coding ISGs (24). The lncRNA NEAT1 can enhance the transcription of interleukin-8 and modulate HIV-1 posttranscriptional expression (25, 26). LncRNA NKILA can inhibit NF-κB signaling by blocking IκB kinase-induced IκB phosphorylation and preventing the degradation of IκB (27).
Recent studies have focused on the differential expression lncRNAs during IAV infection through microarray analysis (28–30). The IAV infection-induced lncRNA NRAV serves as a negative regulator of antiviral innate immunity by suppressing the transcription of multiple ISGs (30). LncRNAs VIN and BISPR are also involved in IAV-host interactions, though the mechanisms are still not clear (28, 31). Nevertheless, the roles of lncRNAs during IAV replication and type I IFN induction are understudied.
Here, we identified that the IAV-upregulated lncRNA named lnc-ISG20 is an ISG. We found that lnc-ISG20 inhibits IAV replication by enhancing ISG20 protein levels. Further studies demonstrated that lnc-ISG20 acts as a competing endogenous RNA that binds to miRNA 326 (miR-326) to reduce its inhibition of ISG20 mRNA translation. Our results revealed the mechanism by which lnc-ISG20 inhibits IAV replication.
RESULTS
Identification and characterization of IAV-induced lnc-ISG20.
To identify IAV-related lncRNAs and their functions during viral infection, we used RNA deep sequencing to analyze the lncRNA profiles in A549 and HEK293T (293T) cells with or without IAV infection. As shown in Fig. 1A and Table S1 in the supplemental material, a number of lncRNAs were upregulated or downregulated after IAV infection. The cluster heat map shows that 139 upregulated lncRNAs and 150 downregulated lncRNAs were found in both cell lines. We noticed that there was an IAV-upregulated lncRNA (NR_130134, NONHSAG017802) located at the same chromosomal locus as the ISG20 gene (Fig. 1B). We named it lnc-ISG20 and confirmed that it is upregulated by IAV infection using reverse transcription-quantitative PCR (RT-qPCR) (Fig. 1C and D). ISG20 is an interferon-stimulated gene. It has been found that ISG20 can inhibit IAV replication (14, 17). As lnc-ISG20 is also upregulated during viral infection, we sought to investigate the function and mechanism of lnc-ISG20 on IAV replication as well as its correlation with ISG20. We performed 5′ and 3′ rapid amplification of cDNA ends (RACE) to determine the sequence of lnc-ISG20 and Northern blotting to examine the size of lnc-ISG20 and ISG20 (Fig. 1E and F). To verify that lnc-ISG20 is indeed a noncoding RNA, we constructed a C-terminally FLAG-tagged lnc-ISG20 in three reading frames and transfected these constructs into 293T cells. Immunoblotting revealed that lnc-ISG20 cannot be translated (Fig. 1G). Next, we examined the subcellular localization of lnc-ISG20. As shown in Fig. 1H, lnc-ISG20 mainly localized in the cytoplasm.
FIG 1.
Identification and characterization of IAV infection-induced lnc-ISG20. (A) A549 or HEK293T cells were infected with IAV (A/WSN/33, A/CA04, or A/PR8) at an MOI of 1 for 8 h. Total RNA was extracted and subjected to RNA-seq analysis. The expression values are shown in shades of red and blue, indicating expression above and below the median expression value across all the samples (log scale 2, from −2 to +2), respectively. (B) Schematic diagram of ISG20 and lnc-ISG20. (C) A549 and HEK293T cells were infected with A/WSN/33, A/CA04, or A/PR/8 at an MOI of 1 for 8 h. Total RNA was extracted and subjected to RT-PCR for lnc-ISG20. (D) A549 and HEK293T cells were infected with A/WSN/33 at the indicated MOI for 0, 4, 8, 12, and 16 h. The amount of lnc-ISG20 was quantified by RT-qPCR. (E) A549 cells were infected with A/WSN/33 for 16 h. Total RNA was prepared for 5′ and 3′ RACE for lnc-ISG20. Images of the RACE PCR products of lnc-ISG20 (left) and sequencing data (right) are presented. (F) A549 cells were infected with A/WSN/33 at an MOI of 0.1 for 16 h. Total RNA was extracted and subjected to Northern blot analysis. The probe used in the assay whose results are presented in the left panel was specific for lnc-ISG20, and the probe used in the assay whose results are presented in the right panel recognizes both the lnc-ISG20 and ISG20 mRNA isoforms. The experiments were repeated twice, and representative data are presented. (G) Full-length lnc-ISG20 was cloned into pcDNA3.1 with an N-terminal start codon and a C-terminal FLAG tag in all three reading frames. The T in lnc-ISG20-T-FLAG and TT in lnc-ISG20-TT-FLAG represent thymidine nucleotide(s). The plasmids were transfected into HEK293T cells for 48 h. Cell lysates were harvested and subjected to immunoblotting with anti-ISG20 or anti-β-actin. The empty vector and FLAG-ISG20 served as the negative and positive controls, respectively. (H) A549 cells were fractionated into cytoplasmic (C) and nuclear (N) fractions. The RNA was extracted from each fraction and subjected to quantitative RT-PCR for lnc-ISG20, and MALAT1, U6, β-actin, and RPS18 mRNAs were used as controls. The fractionated lysates were immunoblotted with lamin B and tubulin. The experiments whose results are presented in panels C, D, and H were repeated twice, and the results were consistent. The data are expressed as the mean for triplicate samples from one independent experiment.
Lnc-ISG20 is an interferon-stimulated gene.
Because lnc-ISG20 is located at the same locus as ISG20, we wondered whether it was also an ISG. Therefore, we examined the levels of lnc-ISG20 and ISG20 in A549 cells infected with Sendai virus (SeV) or transfected with poly(I·C). We found that both lnc-ISG20 and ISG20 were significantly induced upon Sendai virus infection or poly(I·C) transfection (Fig. 2A and B). Then, we treated A549 cells with beta IFN (IFN-β) and measured the levels of lnc-ISG20 and ISG20 by real-time PCR. The data showed that the lnc-ISG20 levels were significantly higher in IFN-β-treated cells than in control cells, which is similar to the pattern of ISG20 (Fig. 2C). These results indicate that lnc-ISG20 is an ISG.
FIG 2.

Lnc-ISG20 is inducible as the IFN-β signaling pathway is activated. (A, B, C) A549 cells were infected with Sendai virus at the indicated MOI for 8 h (A) and transfected with poly(I·C) (0.5 μg/ml) (B) or treated with IFN-β (10 ng/ml) (C) for the indicated times. Total RNA was extracted and subjected to RT-qPCR for lnc-ISG20 and ISG20. Samples were normalized to β-actin. Data are shown as the mean ± SD (n = 3).
lnc-ISG20 inhibits IAV replication.
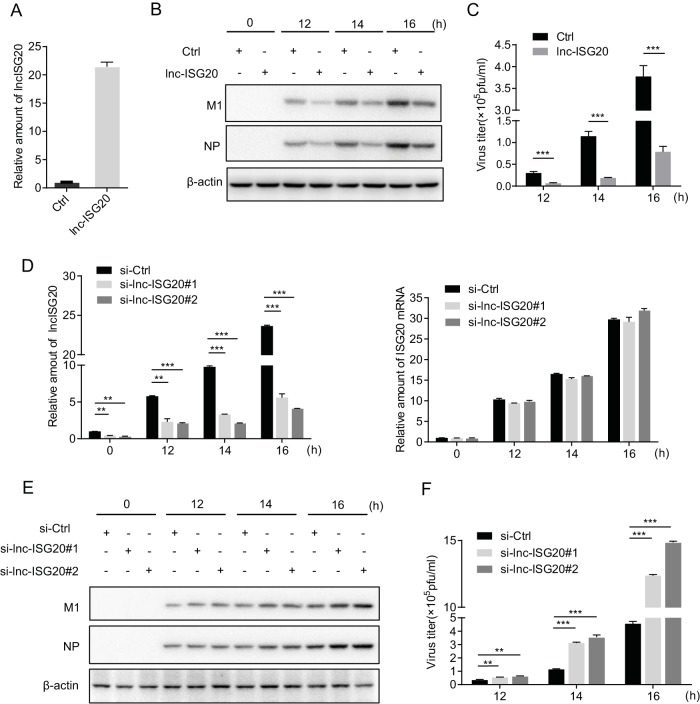
To understand the function of lnc-ISG20 during IAV infection, we generated the A549-lnc-ISG20 cell line, which ectopically expresses lnc-ISG20, and the control cell line A549-ctrl (Fig. 3A). Then, we infected the cells with IAV (A/WSN/33) for the times indicated in Fig. 3. Cell lysates were harvested for immunoblotting, and supernatants were collected for plaque assays. We found that both the protein levels of M1 and NP and viral titers were lower in A549-lnc-ISG20 cells than in control cells (Fig. 3B and C). Next, we knocked down lnc-ISG20 in A549 cells by RNA interference and analyzed the IAV infection as described above. We examined the levels of both lnc-ISG20 and ISG20 by real-time PCR. Treatment with small interfering RNA (siRNA) specific for lnc-ISG20 (si-lnc-ISG20) reduced only the amounts of lnc-ISG20 and not those of ISG20 (Fig. 3D). Immunoblotting indicated that the amounts of M1 and NP protein were increased in cells with reduced amounts of lnc-ISG20 compared to control cells (Fig. 3E), and virus titers were significantly higher in cells with reduced amounts of lnc-ISG20 than in control cells (Fig. 3F). Taken together, these results demonstrate that lnc-ISG20 elicited an inhibitory effect on IAV replication.
FIG 3.
Lnc-ISG20 inhibits IAV replication. (A) A549 cells were infected with lentivirus carrying lnc-ISG20 or control virus. The GFP-positive cells were sorted by fluorescence-activated cell sorting, expanded, and named A549-lnc-ISG20 and A549-ctrl. The expression level of lnc-ISG20 was determined by RT-qPCR, and the data represent the means for triplicate samples from one independent experiment. The experiments were repeated twice. (B, C) A549-lnc-ISG20 and A549-ctrl cells were A549 cells ectopically expressing lnc-ISG20 and A549 control cells, respectively, infected with the A/WSN/33 virus at an MOI of 0.01 for the indicated times. (B) The cell lysates were harvested for immunoprecipitation and immunoblotting assays. (C) The supernatants of infected cells were collected for plaque assays to measure virus titers. (D, E, F) A549 cells were transfected with lnc-ISG20-specific siRNA (si-lnc-ISG20#1 and -#2) or control siRNA (si-Ctrl) and then infected with the A/WSN/33 virus at an MOI of 0.01 for the indicated times. (D) The expression levels of lnc-ISG20 (left) and ISG20 (right) mRNA were measured by RT-qPCR, and the data represent the means for triplicate samples from one independent experiment. The experiments were repeated three times. (E) The cell lysates were harvested for immunoblotting with the indicated antibodies. (F) The supernatants were collected for plaque assays to measure the virus titers. All experiments for the plaque assays were repeated three times, and the results were consistent. The presented data are expressed as the means for triplicate samples from one experiment. **, P < 0.01; ***, P < 0.001 (Student's t test).
Lnc-ISG20 positively regulates the protein level of ISG20.
It has been reported that ISG20 can inhibit IAV replication (14, 17). We sought to determine if lnc-ISG20 suppresses IAV by affecting the level of ISG20. Therefore, we examined the ISG20 protein level in A549-lnc-ISG20 and control cells. We found that the level of ISG20 protein in A549-lnc-ISG20 cells was higher than that in A549-ctrl cells, as cells were transfected with poly(I·C) or infected with Sendai virus (Fig. 4A and B). Next, we transfected A549 cells with si-lnc-ISG20 or control siRNA and performed experiments similar to those described above. We found that the amount of ISG20 protein was lower in si-lnc-ISG20-treated cells than in control cells (Fig. 4C and D). We also performed an experiment to determine whether knockdown of lnc-ISG20 during influenza A virus infection affects ISG20 expression. We found that during IAV infection, the level of ISG20 protein in lnc-ISG20 knockdown cells is lower than that in control cells (Fig. 4E). We then compared the ISG20 mRNA levels in lnc-ISG20-overexpressing cells and control cells and found no significant difference either at the basal level or upon Sendai virus induction in A549-lnc-ISG20 and control cells (Fig. 4F). These results suggest that lnc-ISG20 positively regulates the level of ISG20 protein.
FIG 4.
Overexpression of lnc-ISG20 upregulates ISG20 protein levels but does not affect the level of ISG20 mRNA. (A, B) A549-lnc-ISG20 and A549-ctrl cells were transfected with poly(I·C) (0.5 μg/ml) (A) or infected with Sendai virus at the indicated MOI for 8 h (B). (C, D, E) A549 cells were transfected with si-lnc-ISG20 or control siRNA (si-Ctrl) and then transfected with poly(I·C) (C), infected with Sendai virus (D), or infected with A/WSN/33 virus at an MOI of 0.01 (E). Cell lysates were harvested and subjected to immunoblotting with anti-ISG20 or anti-β-actin antibodies. The above-described experiments were repeated 3 times, and representative data are presented. (F) A549-lnc-ISG20 and A549-ctrl cells were infected with Sendai virus for 8 h. Total RNA was extracted for RT-qPCR to quantify ISG20 mRNA. The experiments were independently performed twice, and consistent results were observed. Data were calculated as the means for triplicate samples. ns, not significant.
Next, we asked whether lnc-ISG20 elicits its inhibitory effect on IAV through ISG20. Therefore, we expressed lnc-ISG20 in A549 cells and then transfected the cells with si-ISG20 or control siRNA, followed by infection with A/WSN/33 (Fig. 5A). The supernatants and cell lysates were then harvested to examine the replication of IAV. Immunoblotting indicated that the levels of viral M1 and NP proteins were lower in lnc-ISG20-expressing cells than in control cells (Fig. 5B, first and second lanes). However, the levels of viral M1 and NP proteins were significantly higher in cells transfected with siRNA specific for ISG20 (si-ISG20), even when lnc-ISG20 was overexpressed (Fig. 5B, third and fourth lanes). Consistently, plaque assays demonstrated that the virus titers were decreased in lnc-ISG20-overexpressing cells, while the virus titers were greatly increased in ISG20 knockdown cells, even when lnc-ISG20 was overexpressed (Fig. 5C). These results demonstrate that lnc-ISG20 inhibits IAV replication in an ISG20-dependent manner.
FIG 5.

Lnc-ISG20 inhibits IAV replication in an ISG20-dependent manner. (A, B, C) A549 cells were transduced with lentivirus expressing lnc-ISG20 or control lentivirus and then transfected with si-ISG20 or control siRNA, as indicated (A). Then, the cells were infected with A/WSN/33 virus (MOI = 0.01, 14 h). (B) Cell lysates were harvested for immunoblotting with the indicated antibodies. (C) The supernatants from infected cells were collected for plaque assays to measure the virus titer. The data in panels A and C are expressed as the means for triplicate samples from one independent experiment. The experiments were repeated three times. **, P < 0.01; ***, P < 0.001 (Student's t test).
Lnc-ISG20 acts as a ceRNA to bind miR-326 and enhances the expression of ISG20.
Because lnc-ISG20 did not affect the level of ISG20 mRNA, we hypothesized that lnc-ISG20 may regulate the translation of ISG20. Lnc-ISG20 and ISG20 share the same 3′ untranslated region (UTR), so it is possible that lnc-ISG20 affects the translation of ISG20 through miRNA. Therefore, we first predicted the miRNAs that might target the ISG20 3′ UTR using the TargetScan database. Among them, seven miRNAs were more abundant in A549 and 293T cells than in HeLa cells according to the database, as previously reported (32). Because it is known that IAV replicates more efficiently in A549 and 293T cells than in HeLa cells, we chose these seven miRNAs to further analyze whether they can target the ISG20 3′ UTR using 3′ UTR luciferase reporter assays. As shown in Fig. 6A, miR-326 reduced the activity of the ISG20 3′ UTR luciferase reporter. To confirm that miR-326 can target the ISG20 3′ UTR, we generated the shorter form of the ISG20 3′ UTR (ISG20 3′ UTR-S) and ISG20 3′ UTR mutant reporters, as shown in Fig. 6B, and performed luciferase reporter assays. We found that miR-326 can inhibit the luciferase activity of the ISG20 3′ UTR-S reporter but not the ISG20 3′ UTR-S mutant reporter (Fig. 6C). Next, we generated A549 cell lines ectopically expressing S1 aptamer-tagged lnc-ISG20 or the mutant variant in which the miR-326-targeted region was mutated. We collected cell lysates and performed S1-pulldown assays. The results indicated that lnc-ISG20 can enrich for miR-326, while the lnc-ISG20 mutant cannot (Fig. 6D).
FIG 6.
miR-326 can target lnc-ISG20 and the ISG20 3′ UTR. (A) HEK293T cells were transfected with the pGLO-ISG20 3′ UTR and the indicated miRNA mimics or scrambled miRNA as a negative control. Cell lysates were harvested for dual-luciferase assays. The relative luciferase activity was quantified. The presented data are expressed as the means from one experiment. The experiments were repeated three times. *, P < 0.05 (Student's t test). (B) The pGLO-ISG20 3′ UTR-S (ISG20 3′ UTR) and pGLO-ISG20 3′ UTR-S mutant (ISG20 3′ UTR-mut) were generated by making mutations in the predicted miR-326-binding region. The mutated nucleotides are shown in red. (C) 293T cells were transfected with the miR-326 mimic and reporter plasmids, as indicated. Cell lysates were harvested for dual-luciferase assays. The relative luciferase activity was quantified, and the data represent the means for triplicate samples from one independent experiment. The experiments were repeated twice. **, P < 0.01 (Student's t test). (D) A549 cell lines ectopically expressing S1 aptamer-tagged lnc-ISG20 or lnc-ISG20-mut were generated. Cell lysates were harvested and incubated with streptavidin-coupled beads at 4°C for 3 h. The amount of miR-326 bound to the beads was assessed by RT-qPCR, and the data represent the means for triplicate samples. The experiments were repeated twice. ***, P < 0.01 (Student's t test). (E, F, G) A549 cells were transfected with a control mimic (Ctrl), miR-326 mimic (miR-326), control inhibitor (Ctrl-inhibitor), or miR-326 inhibitor and then transfected with poly(I·C) (0.5 μg/ml, 14 h) (E), infected with SeV at an MOI of 0.2 for 8 h (F), or infected with the A/WSN/33 virus at an MOI of 0.01 for 12 h (G). Cell lysates were harvested and subjected to immunoblotting with anti-ISG20 or anti-β-actin as an internal control. (H) A549 cells were infected with the A/WSN/33 virus at an MOI of 0.01 for the indicated times. The relative amount of miR-326 was measured by RT-qPCR, and the data represent the means for triplicate samples from one independent experiment. The experiments were repeated twice. *, P < 0.05 (Student's t test).
Subsequently, we examined whether miR-326 can inhibit the expression of the ISG20 protein. We transfected A549 cells with miR-326 mimics or an miR-326 inhibitor and then transfected the cells with poly(I·C) or infected the cells with Sendai virus. As shown in Fig. 6E and F, the level of ISG20 protein in the miR-326 mimic-transfected cells was lower than that in control cells, while the level of ISG20 protein in miR-326 inhibitor-transfected cells was higher than that in control cells. These results demonstrate that miR-326 interferes with ISG20 expression. We then performed experiments with the miR-326 mimic and miR-326 inhibitor to examine the effect of miR-326 on influenza A viral replication. We found that miR-326 favors viral replication, while miR-326 inhibition inhibits IAV replication (Fig. 6G). We also examined miR-326 expression during influenza A viral infection. As shown in Fig. 6H, the amount of miR326 was reduced 20 to 25% during viral infection.
We hypothesized that lnc-ISG20 acts as a ceRNA for ISG20 mRNA, meaning that lnc-ISG20 binds to miR-326 to reduce free miR-326, which in turn enhances the expression of ISG20. To test this, we first analyzed whether lnc-ISG20 can reduce the amount of miRNA-bound ISG20 mRNA using the AGO2 radioimmunoprecipitation assay (RIPA). We collected cell lysates from A549-ctrl cells, A549-lnc-ISG20 cells, and A549 cells expressing the lnc-ISG20 mutant (A549-lnc-ISG20-mut cells) and subjected them to immunoprecipitation with anti-AGO2 antibody (Fig. 7A, top) followed by RT-qPCR to quantify the AGO2-bound ISG20 mRNA. We found that that the amount of AGO2-bound ISG20 mRNA in lnc-ISG20-expressing A549 cells was significantly reduced compared to that in control cells or lnc-ISG20 mutant-expressing cells (Fig. 7A, bottom). Next, we examined whether miR-326 can reverse the effect of lnc-ISG20 on the enhancement of ISG20 expression. We transfected A549 cells with lnc-ISG20 or lnc-ISG20 mutant expression plasmids and cotransfected the cells with different amounts of miR-326 or control miRNA, followed by transfection with poly(I·C). Then, we collected cell lysates to examine ISG20 expression by immunoblotting. We found that lnc-ISG20 enhanced ISG20 expression, similar to the results shown in Fig. 4. However, ISG20 expression was decreased in cells cotransfected with miR-326 and lnc-ISG20 compared to that in cells transfected with lnc-ISG20 alone (Fig. 7B), indicating that miR-326 antagonized the upregulation of ISG20 expression enhanced by lnc-ISG20.
FIG 7.

lnc-ISG20 functions as a ceRNA to reduce miR-326 binding to ISG20 mRNA and enhance the expression of ISG20 protein. (A) A549 cells were transduced with lentivirus expressing lnc-ISG20, lnc-ISG20-mut, or control lentivirus. The cell lysates were harvested for immunoprecipitation (IP) with anti-AGO2 antibody by RT-qPCR to quantify AGO2-bound ISG mRNA. The experiment was repeated twice, and the results were consistent. The data are the means for triplicate samples from one experiment. **, P < 0.01 (Student's t test). (B) A549 cells were transduced with lentivirus expressing lnc-ISG20, lnc-ISG20-mut, or control lentivirus, as well as with different concentrations of miR-326 mimics (50, 100, or 200 pmol), followed by transfection with poly(I·C) (0.5 μg/ml). Cell lysates were harvested for immunoblotting with anti-ISG20 antibody and anti-β-actin antibody as an internal control.
In summary, we determined that the IAV-upregulated lnc-ISG20 is an ISG. We found that lnc-ISG20 inhibits IAV replication by upregulating the protein levels of ISG20. Further studies demonstrated that lnc-ISG20 acts as ceRNA by reducing miR-326 binding to the ISG20 3′ UTR (Fig. 8). We revealed that lnc-ISG20 is an inhibitory host factor for IAV replication. Thus, our study provides new evidence that lncRNAs play important roles in the antiviral immune response.
FIG 8.
Schematic map of lnc-ISG20 regulating IAV replication. IAV infection triggers the transcription of type I interferons, such as IFN-β. IFN-β stimulates the transcription of downstream genes (ISG20 and lnc-ISG20). Lnc-ISG20 acts as a ceRNA by reducing miR-326 binding to target ISG20 mRNA and enhances the translation of ISG20, which in turn inhibits IAV replication. IFN-βR, IFN-β receptor.
DISCUSSION
As a large number of lncRNAs have been identified, an increasing number of functions for them have been explored. Currently, it has been shown that lncRNAs are involved in antiviral responses by regulating ISG expression. For example, lncRNA#32 binds to activating transcription factor 2 (ATF2) and positively regulates the host antiviral response, which in turn inhibits EMCV replication and hepatitis B virus and HCV infection (33). In addition, lncRNAs may act as inhibitory factors in antiviral innate immunity. Chen and colleagues demonstrated that the lncRNA NRAV modulates antiviral responses through suppression of ISG transcription (30). They found that ectopically expressed NRAV significantly promotes IAV replication and virulence, while silencing of NRAV suppresses IAV replication, suggesting that the downregulation of NRAV is a host antiviral immune response (30). Here, we determined that lnc-ISG20 acts as a ceRNA to enhance ISG20 expression, which inhibits IAV replication. We propose that this may be a general regulatory mechanism for a group of lncRNAs in antiviral immunity. If so, it will be worth identifying these lncRNAs for further investigation to expand our knowledge of the function of lncRNAs and the network of the innate immune response.
As an IFN-induced gene, ISG20 encodes a 3′-to-5′ exonuclease with specificity for single-stranded RNA. A previous report demonstrated that ISG20 is involved in the antiviral functions of IFN (14). ISG20-overexpressing cells display resistance to infections by vesicular stomatitis virus, IAV, and encephalomyocarditis virus. ISG20 specifically interferes with the expression of VSV mRNA and protein but does not affect the expression of cellular control genes. Interestingly, the expression of ISG20 in cells triply deficient in PKR, RNase L, and Mx proteins confers resistance to VSV infection, suggesting that ISG20 may represent an alternative antiviral pathway against VSV infection. In addition, ISG20 is upregulated in macrophages induced by a variety of bacteria, implying that ISG20 may have a more general role against different kinds of pathogens (34). Therefore, it is reasonable for host cells to evolutionarily generate an effective way to ensure expression of ISG20 during infection. In this study, we revealed that upregulation of lnc-ISG20 in virus-infected cells is a strategy for host cells to sequester miR-326 away from ISG20 mRNA, which enhances the translation of the ISG20 protein. This may represent a unique method for host cells to regulate the antiviral immune response. We propose that lnc-ISG20 is an ISG because it can be induced in cells shortly after treatment with IFN-β. In recent years, a number of lncRNAs have been identified as ISGs as deep sequencing data have become available (24, 35, 36). However, their precise roles are not fully determined. It may be worth investigating whether some of them elicit their functions in a similar way as lnc-ISG20.
Most studies of miR-326 focus on its function in tumorigenesis. Indeed, miR-326 serves as a tumor suppressor in melanoma by targeting KRAS and suppressing the AKT and extracellular signal-regulated kinase signaling pathways. miR-326 can inhibit gastric cancer cell proliferation by downregulating NOB1 (37). However, Houzet and colleagues demonstrated that miR-326 is a predicted microRNA targeting the HIV genome (38). Overexpression of miR-326 restricts HIV replication, while knockdown of miR-326 increases HIV-1 replication, implying that miR-326 possesses miRNA-mediated restriction of HIV replication (38). Recently, it was also reported that miR-326 can target NOS2 mRNA and inhibit macrophage activation in chronic liver injury (39). Here, we identified that miR-326 serves as an inhibitory factor in the antiviral immune response by targeting ISG20. These findings indicate that miR-326 can play various roles by targeting different mRNAs.
We observed that the lncRNA profile varied in cells infected with different strains of IAV (Fig. 1A). This implies that the abundance of lncRNAs may determine the host susceptibility to different strains of IAV. However, our current knowledge of the identity and complexity of lncRNAs is still limited. Therefore, it is critical to explore the function of lncRNAs to elucidate the precise mechanisms of the interaction between the host and pathogen.
MATERIALS AND METHODS
Cell lines and virus strains.
Human lung adenocarcinoma epithelial (A549) cells, human embryonic kidney HEK293T (293T) cells, and Madin-Darby canine kidney (MDCK) cells were obtained from the China Infrastructure of Cell Line Resources. To generate stable cell lines expressing lnc-ISG20 or mutant lnc-ISG20 (lnc-ISG20-mut), A549 cells were infected with lentivirus carrying lnc-ISG20 or lnc-ISG20-mut genes, and green fluorescent protein (GFP)-positive cells were sorted and expanded. All cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum (Pan Biotech).
The A/WSN/33 (A/WSN; H1N1), A/Puerto Rico/8/34 (A/PR8; H1N1), and A/California/2009/04 (A/CA04; H1N1) viruses were propagated in MDCK cells. Sendai virus (SeV) was propagated in 9-day-old, specific-pathogen-free embryonated eggs (Merial) at 37°C for 3 days.
Antibodies and reagents.
Mouse anti-M1 monoclonal antibody and rabbit anti-NP polyclonal antibody were gifts from Wenjun Liu. Rabbit anti-ISG20 monoclonal antibody (Proteintech), rabbit anti-Argonaute-2 antibody (Abcam), mouse anti-β-actin antibody (Santa Cruz), mouse anti-FLAG antibody (M2) (Sigma), and horseradish peroxidase HRP-conjugated secondary antibodies (The Jackson Laboratory) were purchased from the indicated companies.
Actinomycin D (Sigma), poly(I·C) sodium salt (Sigma), and human IFN-β (PeproTech) were purchased from the indicated companies.
Plasmids, siRNAs, and miRNAs.
The lnc-ISG20 and lnc-ISG20-S1-tagged genes were cloned into the lentiviral pLentiLox3.7 (pLL3.7) plasmid. The lnc-ISG20, lnc-ISG20-T, lnc-ISG20-TT, and ISG20 genes were cloned into the eukaryotic expression vector pcDNA3.1 with an N-terminal ATG start codon and a C-terminal FLAG tag. The full-length 3′ UTR of ISG20 (nt 1 to 813) and the 3′ UTR-ISG20-S (nt 300 to 412) were cloned into the luciferase reporter plasmid pmirGLO (Promega).
Lnc-ISG20-specific siRNAs, ISG20-specific siRNAs, and negative-control siRNAs were transfected into A549 cells using siRNA-Mate (GenePharma) according to the manufacturer's instructions. The sequences of the siRNAs were as follows: si-lnc-ISG20#1, 5′-CUGUAAGUAGAUCCUGCAGTT-3′; si-lnc-ISG20#2, 5′-CUCCUCUGUAAGUAGAUCCTT-3′; si-ISG20#1, 5′-GCUGUGCUGUACGACAAGUTT-3′; and si-ISG20#2, 5′-GAGAUCACCGAUUACAGAATT-3′.
The miRNA mimics or inhibitors were transfected into A549 or 293T cells using the Lipofectamine 2000 reagent (Invitrogen).
Transcriptome sequencing (RNA-seq) analysis.
A549 or 293T cells were infected with influenza A virus (A/WSN/33, A/CA04, A/PR8) at a multiplicity of infection (MOI) of 1 for 8 h or not infected (mock infected). Total RNA was isolated with the TRIzol reagent (Invitrogen). RNA samples with an RNA Integrity Number (RIN) greater than 8, as assessed by an Agilent 2100 bioanalyzer (Agilent, Santa Clara, CA, USA), were used for RNA library construction. The cDNA libraries were prepared using an Illumina TruSeq RNA sample preparation kit (catalog number FC-122-1001; Illumina, San Diego, CA, USA). In brief, 1 μg of total RNA was purified and fragmented. Then, the fragmented RNAs were subjected to reverse transcription for first-strand cDNA synthesis with random primers (Invitrogen), followed by second-strand cDNA synthesis. After end repair and 3′-adenylation, a single A base was added to the 3′ end of the cDNA fragments for subsequent adapter ligation with paired-end (PE) adapters. Then, the cDNAs fragments were purified and enriched by 15 PCR cycles to create the final cDNA libraries. The cDNA libraries were quantified using an Agilent bioanalyzer and a DNA 1000 kit. Each library was sequenced using the HiSeq 2000 platform and generated 100-bp paired-end (PE) reads. After filtering out reads containing sequencing adapters and reads of low quality, the remaining reads were aligned to human long noncoding RNAs in the NONCODE database (version 4.0) (http://www.noncode.org) using the TopHat program. For identification of differentially expressed genes and functional annotation analysis, data from both mock-infected cells and IAV-infected cells were used for differential gene expression analysis. We applied the cufflinks tool to identify differentially expressed genes and measured transcript abundances in fragments per kilobase of exon per million fragments mapped (FPKM).
Quantitative PCR.
cDNA was synthesized from total RNA using an oligo(dT) primer (TransGen) and subjected to quantitative PCR using SYBR green PCR master mix (Toyobo). The relative amounts of the indicated RNAs were calculated using β-actin as an internal control. The primer pairs used were as follows: primer pair lnc-ISG20-F (5′-GCATCCCGACATTGGTTTA-3′) and lnc-ISG20-R (5′-AGCTGCAGGATCTACTTACAGAC-3′) and primer pair ISG20-F (5′-TGGACTGCGAGATGGTGG-3′) and ISG20-R (5′-GGGTTCTGTAATCGGTGAT-3′).
Northern blotting and RACE.
The total RNA from A549 cells infected with IAV (A/WSN/33; MOI, 0.01; 8 h) or mock-infected cells was isolated with the TRIzol reagent (Invitrogen) and subjected to Northern blotting with a NorthernMax-Gly kit (Invitrogen). The probe was labeled with digoxigenin (DIG) using a PCR DIG probe synthesis kit (Roche). The probe specific for lnc-ISG20 was designed to recognize lnc-ISG20 (nt 1 to 155). The probe that recognized both lnc-ISG20 and ISG20 mRNA was designed to target lnc-ISG20 (nt 266 to 572) and ISG20 (nt 788 to 1094) mRNA.
The 5′ and 3′ rapid amplification of cDNA ends (RACE) of lnc-ISG20 was performed using a SMARTer RACE cDNA amplification kit (Clontech). The primers for lnc-ISG20 were as follows: 5′ primer, 5′-GTCCTCTTTCAGTGCCTGGAAGTCGT-3′, and 3′ primer, 5′-CCACGTGTGGTCTGTAAGTAGATCCTGC-3′.
Immunoblotting and immunoprecipitation.
Cells were lysed with ice-cold RIPA buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% NP-40, 1 mM EDTA) supplemented with protease inhibitors (Roche). Proteins were resolved by 10% SDS-PAGE and transferred to polyvinylidene difluoride membranes, followed by immunoblotting with the indicated antibodies.
For AGO2 immunoprecipitation, A549-lnc-ISG20, A549-lnc-ISG20-mut, or A549-ctrl cells were UV cross-linked and lysed with ice-cold lysis buffer (25 mM Tris-HCl, pH 7.5, 150 mM KCl, 2 mM EDTA, 0.5% NP-40, 0.5 mM dithiothreitol [DTT]) supplemented with protease inhibitors and RNase inhibitor on ice for 20 min. The lysates were immunoprecipitated with rabbit anti-AGO2 antibody or rabbit IgG (as a control), followed by the addition of protein A agarose beads. The beads were washed, and bound RNA was eluted and subjected to RT-qPCR with primers specific for ISG20 mRNA. The efficiency of AGO2 immunoprecipitation was detected by immunoblotting of AGO2.
Plaque assay.
MDCK cells were seeded in 12-well plates and infected with serial dilutions of virus in serum-free DMEM supplemented with 4 μg/ml of tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-trypsin for 2 h. Then, the cells were washed with phosphate-buffered saline and overlaid with modified Eagle's medium (MEM) containing 1% agarose (Amresco) and 2 μg/ml of TPCK-trypsin. The contents of the plates were allowed to settle at 4°C for 5 min, and the plates were incubated upside down at 37°C. After 72 h, visible plaques were counted, and viral titers were determined.
Dual-luciferase assay.
For the dual-luciferase assays, 293T cells were seeded in 24-well plates and transfected with pGLO-ISG20 3′ UTR, pGLO-ISG20 3′ UTR-S, or pGLO-ISG20 3′ UTR-S-mut together with an miRNA mimic using the Lipofectamine 2000 reagent (Invitrogen). After 48 h, the cell lysates were harvested for luciferase assays using a dual-luciferase assay kit (Promega). Relative luciferase activity was calculated (firefly luciferase/Renilla luciferase). The experiment was performed in triplicate and independently repeated three times.
RNA pulldown assay.
A549 cell lines that ectopically expressed S1-lnc-ISG20, S1-lnc-ISG20-mut, or control cells were harvested for pulldown assays as previously described (40). Briefly, cells were UV cross-linked and lysed with lysis buffer (10 mM Tris-HCl, pH 7.5, 10 mM NaCl, 10 mM EDTA, 0.5% Triton X-100, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride) with RNase inhibitor for 20 min. The lysates were incubated with streptavidin-coupled beads at 4°C for 3 h. RNA bound to beads was extracted using the TRIzol reagent and subjected to reverse transcription-PCR (RT-PCR) for miR-326.
Statistical analysis.
Statistical comparisons were performed using GraphPad Prism (version 5.0) software (GraphPad Software Inc.). Student's t test was used to analyze the data. A P value of <0.05 was considered statistically significant. Error bars represent the standard error (±SE).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Ministry of Science and Technology of China (2016YFC1200304, 2015CB910502, 2016YFC1200803, and 2016YFD0500206), the National Natural Science Foundation of China (NSFC; 81772989, 31630079, and 31572526), and the Strategic Priority Research Program of the Chinese Academy of Sciences (XDPB0301). Xin Ye and Wenjun Liu are the principal investigators of the Innovative Research Group of the National Natural Science Foundation of China (81621091).
W.C. designed and performed experiments, analyzed data, and wrote the manuscript; J.L. performed RNA sequencing analysis and wrote the manuscript; Q.S. and Q.L. participated in the experiments and bioinformatics analysis; X.L., D.Q., and X.T. participated in the experiments; and W.L. participated in the design of RNA deep sequencing and provided suggestions. X.Y. designed, conceived, and supervised the study and wrote the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.00539-18.
REFERENCES
- 1.Krug RM. 2014. Influenza: an RNA-synthesizing machine. Nature 516:338–339. doi: 10.1038/516338a. [DOI] [PubMed] [Google Scholar]
- 2.Smith GJ, Vijaykrishna D, Bahl J, Lycett SJ, Worobey M, Pybus OG, Ma SK, Cheung CL, Raghwani J, Bhatt S, Peiris JS, Guan Y, Rambaut A. 2009. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 459:1122–1125. doi: 10.1038/nature08182. [DOI] [PubMed] [Google Scholar]
- 3.Paules CI, Marston HD, Eisinger RW, Baltimore D, Fauci AS. 2017. The pathway to a universal influenza vaccine. Immunity 47:599–603. doi: 10.1016/j.immuni.2017.09.007. [DOI] [PubMed] [Google Scholar]
- 4.Eisfeld AJ, Neumann G, Kawaoka Y. 2015. At the centre: influenza A virus ribonucleoproteins. Nat Rev Microbiol 13:28–41. doi: 10.1038/nrmicro3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arranz R, Coloma R, Chichon FJ, Conesa JJ, Carrascosa JL, Valpuesta JM, Ortin J, Martin-Benito J. 2012. The structure of native influenza virion ribonucleoproteins. Science 338:1634–1637. doi: 10.1126/science.1228172. [DOI] [PubMed] [Google Scholar]
- 6.McNab F, Mayer-Barber K, Sher A, Wack A, O'Garra A. 2015. Type I interferons in infectious disease. Nat Rev Immunol 15:87–103. doi: 10.1038/nri3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. 2011. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Verhelst J, Parthoens E, Schepens B, Fiers W, Saelens X. 2012. Interferon-inducible protein Mx1 inhibits influenza virus by interfering with functional viral ribonucleoprotein complex assembly. J Virol 86:13445–13455. doi: 10.1128/JVI.01682-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, Feeley EM, Ryan BJ, Weyer JL, van der Weyden L, Fikrig E, Adams DJ, Xavier RJ, Farzan M, Elledge SJ. 2009. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139:1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y, Banerjee S, Wang Y, Goldstein SA, Dong B, Gaughan C, Silverman RH, Weiss SR. 2016. Activation of RNase L is dependent on OAS3 expression during infection with diverse human viruses. Proc Natl Acad Sci U S A 113:2241–2246. doi: 10.1073/pnas.1519657113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balachandran S, Roberts PC, Brown LE, Truong H, Pattnaik AK, Archer DR, Barber GN. 2000. Essential role for the dsRNA-dependent protein kinase PKR in innate immunity to viral infection. Immunity 13:129–141. doi: 10.1016/S1074-7613(00)00014-5. [DOI] [PubMed] [Google Scholar]
- 12.Gongora C, David G, Pintard L, Tissot C, Hua TD, Dejean A, Mechti N. 1997. Molecular cloning of a new interferon-induced PML nuclear body-associated protein. J Biol Chem 272:19457–19463. doi: 10.1074/jbc.272.31.19457. [DOI] [PubMed] [Google Scholar]
- 13.Dalgaard JZ, Klar AJ, Moser MJ, Holley WR, Chatterjee A, Mian IS. 1997. Statistical modeling and analysis of the LAGLIDADG family of site-specific endonucleases and identification of an intein that encodes a site-specific endonuclease of the HNH family. Nucleic Acids Res 25:4626–4638. doi: 10.1093/nar/25.22.4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Espert L, Degols G, Gongora C, Blondel D, Williams BR, Silverman RH, Mechti N. 2003. ISG20, a new interferon-induced RNase specific for single-stranded RNA, defines an alternative antiviral pathway against RNA genomic viruses. J Biol Chem 278:16151–16158. doi: 10.1074/jbc.M209628200. [DOI] [PubMed] [Google Scholar]
- 15.Jiang D, Guo H, Xu C, Chang J, Gu B, Wang L, Block TM, Guo JT. 2008. Identification of three interferon-inducible cellular enzymes that inhibit the replication of hepatitis C virus. J Virol 82:1665–1678. doi: 10.1128/JVI.02113-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng Z, Wang L, Pan J. 2017. Interferon-stimulated gene 20-kDa protein (ISG20) in infection and disease: review and outlook. Intractable Rare Dis Res 6:35–40. doi: 10.5582/irdr.2017.01004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qu H, Li J, Yang L, Sun L, Liu W, He H. 2016. Influenza A virus-induced expression of ISG20 inhibits viral replication by interacting with nucleoprotein. Virus Genes 52:759–767. doi: 10.1007/s11262-016-1366-2. [DOI] [PubMed] [Google Scholar]
- 18.Ponting CP, Oliver PL, Reik W. 2009. Evolution and functions of long noncoding RNAs. Cell 136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 19.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. 2010. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G III, Kenny PJ, Wahlestedt C. 2008. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of beta-secretase. Nat Med 14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo G, Kang Q, Zhu X, Chen Q, Wang X, Chen Y, Ouyang J, Zhang L, Tan H, Chen R, Huang S, Chen JL. 2015. A long noncoding RNA critically regulates Bcr-Abl-mediated cellular transformation by acting as a competitive endogenous RNA. Oncogene 34:1768–1779. doi: 10.1038/onc.2014.131. [DOI] [PubMed] [Google Scholar]
- 22.Landeras-Bueno S, Ortin J. 2016. Regulation of influenza virus infection by long non-coding RNAs. Virus Res 212:78–84. doi: 10.1016/j.virusres.2015.08.008. [DOI] [PubMed] [Google Scholar]
- 23.Qian X, Xu C, Zhao P, Qi Z. 2016. Long non-coding RNA GAS5 inhibited hepatitis C virus replication by binding viral NS3 protein. Virology 492:155–165. doi: 10.1016/j.virol.2016.02.020. [DOI] [PubMed] [Google Scholar]
- 24.Kambara H, Niazi F, Kostadinova L, Moonka DK, Siegel CT, Post AB, Carnero E, Barriocanal M, Fortes P, Anthony DD, Valadkhan S. 2014. Negative regulation of the interferon response by an interferon-induced long non-coding RNA. Nucleic Acids Res 42:10668–10680. doi: 10.1093/nar/gku713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imamura K, Imamachi N, Akizuki G, Kumakura M, Kawaguchi A, Nagata K, Kato A, Kawaguchi Y, Sato H, Yoneda M, Kai C, Yada T, Suzuki Y, Yamada T, Ozawa T, Kaneki K, Inoue T, Kobayashi M, Kodama T, Wada Y, Sekimizu K, Akimitsu N. 2014. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol Cell 53:393–406. doi: 10.1016/j.molcel.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Q, Chen CY, Yedavalli VS, Jeang KT. 2013. NEAT1 long noncoding RNA and paraspeckle bodies modulate HIV-1 posttranscriptional expression. mBio 4:e00596-12. doi: 10.1128/mBio.00596-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu B, Sun L, Liu Q, Gong C, Yao Y, Lv X, Lin L, Yao H, Su F, Li D, Zeng M, Song E. 2015. A cytoplasmic NF-kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Cancer Cell 27:370–381. doi: 10.1016/j.ccell.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 28.Winterling C, Koch M, Koeppel M, Garcia-Alcalde F, Karlas A, Meyer TF. 2014. Evidence for a crucial role of a host non-coding RNA in influenza A virus replication. RNA Biol 11:66–75. doi: 10.4161/rna.27504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li F, Chen Y, Zhang Z, Ouyang J, Wang Y, Yan R, Huang S, Gao GF, Guo G, Chen JL. 2015. Robust expression of vault RNAs induced by influenza A virus plays a critical role in suppression of PKR-mediated innate immunity. Nucleic Acids Res 43:10321–10337. doi: 10.1093/nar/gkv1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ouyang J, Zhu X, Chen Y, Wei H, Chen Q, Chi X, Qi B, Zhang L, Zhao Y, Gao GF, Wang G, Chen JL. 2014. NRAV, a long noncoding RNA, modulates antiviral responses through suppression of interferon-stimulated gene transcription. Cell Host Microbe 16:616–626. doi: 10.1016/j.chom.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barriocanal M, Carnero E, Segura V, Fortes P. 2014. Long non-coding RNA BST2/BISPR is induced by IFN and regulates the expression of the antiviral factor tetherin. Front Immunol 5:655. doi: 10.3389/fimmu.2014.00655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruike Y, Ichimura A, Tsuchiya S, Shimizu K, Kunimoto R, Okuno Y, Tsujimoto G. 2008. Global correlation analysis for micro-RNA and mRNA expression profiles in human cell lines. J Hum Genet 53:515–523. doi: 10.1007/s10038-008-0279-x. [DOI] [PubMed] [Google Scholar]
- 33.Nishitsuji H, Ujino S, Yoshio S, Sugiyama M, Mizokami M, Kanto T, Shimotohno K. 2016. Long noncoding RNA #32 contributes to antiviral responses by controlling interferon-stimulated gene expression. Proc Natl Acad Sci U S A 113:10388–10393. doi: 10.1073/pnas.1525022113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nau GJ, Richmond JF, Schlesinger A, Jennings EG, Lander ES, Young RA. 2002. Human macrophage activation programs induced by bacterial pathogens. Proc Natl Acad Sci U S A 99:1503–1508. doi: 10.1073/pnas.022649799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Valadkhan S, Gunawardane LS. 2016. LncRNA-mediated regulation of the interferon response. Virus Res 212:127–136. doi: 10.1016/j.virusres.2015.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Josset L, Tchitchek N, Gralinski LE, Ferris MT, Eisfeld AJ, Green RR, Thomas MJ, Tisoncik-Go J, Schroth GP, Kawaoka Y, Manuel de Villena FP, Baric RS, Heise MT, Peng X, Katze MG. 2014. Annotation of long non-coding RNAs expressed in collaborative cross founder mice in response to respiratory virus infection reveals a new class of interferon-stimulated transcripts. RNA Biol 11:875–890. doi: 10.4161/rna.29442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ji S, Zhang B, Kong Y, Ma F, Hua Y. 2017. miR-326 inhibits gastric cancer cell growth through downregulating NOB1. Oncol Res 25:853–861. doi: 10.3727/096504016X14759582767486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Houzet L, Klase Z, Yeung ML, Wu A, Le SY, Quinones M, Jeang KT. 2012. The extent of sequence complementarity correlates with the potency of cellular miRNA-mediated restriction of HIV-1. Nucleic Acids Res 40:11684–11696. doi: 10.1093/nar/gks912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li W, Chang N, Tian L, Yang J, Ji X, Xie J, Yang L, Li L. 2017. miR-27b-3p, miR-181a-1-3p, and miR-326-5p are involved in the inhibition of macrophage activation in chronic liver injury. J Mol Med 95:1091–1105. doi: 10.1007/s00109-017-1570-0. [DOI] [PubMed] [Google Scholar]
- 40.Dong Y, Yang J, Ye W, Wang Y, Ye C, Weng D, Gao H, Zhang F, Xu Z, Lei Y. 2015. Isolation of endogenously assembled RNA-protein complexes using affinity purification based on streptavidin aptamer S1. Int J Mol Sci 16:22456–22472. doi: 10.3390/ijms160922456. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.