Abstract
It is becoming clear that most eukaryotic transposable elements (TEs) owe their evolutionary success in part to horizontal transfer events, which enable them to invade new species. Recent large-scale studies are beginning to unravel the mechanisms and ecological factors underlying this mode of transmission. Viruses are increasingly recognized as vectors in the process but also as a direct source of genetic material horizontally acquired by eukaryotic organisms. Because TEs and endogenous viruses are major catalysts of variation and innovation in genomes, we argue that horizontal inheritance has had a more profound impact in eukaryotic evolution than is commonly appreciated. To support this proposal, we compile a list of examples, including some previously unrecognized, whereby new host functions and phenotypes can be directly attributed to horizontally acquired TE or viral sequences. We predict that the number of examples will rapidly grow in the future as the prevalence of horizontal transfer in the life cycle of TEs becomes even more apparent, firmly establishing this form of non-Mendelian inheritance as a consequential facet of eukaryotic evolution.
Introduction
Transposable elements (TEs) are segments of DNA able to move from one locus to another in a given genome and to replicate themselves in the process [1]. TEs are found in nearly all organisms and frequently constitute the major portion of the genome [2–6]. They can be transmitted from one host to another in two ways: through vertical inheritance from parent to offspring, or through horizontal transfer (HT) between non-mating organisms [7,8]. HT can be viewed as a way for TEs to ensure their long-term persistence, by jumping from hosts able to suppress their transposition to naïve ones in which they can spawn new copies [9]. It is still unclear how much TEs as a whole rely on horizontal versus vertical transmission to propagate in eukaryotes, however it has become apparent that most known TE types have propagated horizontally at some point during their evolutionary history. Given the profound influence that TEs and their associated activities have exerted on eukaryotic genomes, it follows that the horizontal transfer of TEs (HTT) must represent an important facet of eukaryotic evolution [7]. However, cases of eukaryotic TEs for which there is robust evidence of both HT and direct functional consequences are still scarce or have remained unrecognized. Here we provide an update on the trends characterizing HTT in eukaryotes, with an emphasis on plants and animals, and we compile a list of cases whereby a given TE shows evidence of HT and has had direct evolutionary consequences for the host lineage where it was introduced. We argue that HT is likely to be a widespread mechanism ensuring the long-term persistence of TEs and that HTT should be regarded as an important source of genetic variation, whose impact on the host’s biology has been underappreciated [10,11].
HTT is pervasive in plants and animals
An event of HTT is suspected when similarity between TE copies from different host species is anomalously high given the divergence time of the species. For example, copies of the Mariner_Tbel family of transposons from the Northern tree shrew (Tupaia belangeri) display up to 87% nucleotide identity over their entire length (~1,300 bp) with elements found in the harvester ant (Pogonomyrmex barbaratus) [12]. These sequences, like most TE copies, have evolved neutrally after their insertion in the host genome. Thus, such a level of interspecific sequence identity is incompatible with their vertical inheritance since the divergence of mammals and insects, which occurred more than 550 million years ago [12]. The most likely explanation is that this TE family was introduced horizontally in one or both of these species well after they diverged from a common ancestor. Based on this type of analysis, as well as on other criteria reviewed elsewhere [13,14], no less than 2,836 HTT events (retrieved from HTT-db [15] as of October 2017) have been recorded since the first unquestionable case of HTT, that of the P element in Drosophila melanogaster, reported in 1990 [16] (Figure 1). Among the many HTT stories documented over the past few years, one achieved an unprecedented level of precision in the timing of an HTT event, which again involved the P element but this time invading D. simulans, the species sister to D. melanogaster. Population sequencing revealed that this transposon occurs only in D. simulans populations sampled after 2010, and that its presence in this species is best explained by a single, very recent HTT event of one particular P element variant from D. melanogaster [17]. This HTT event, which was nearly “caught in the act” provides an outstanding opportunity, together with experimental evolution studies [18], to characterize the first steps of TE invasion and the host response in natural populations.
Figure 1. Numbers of horizontal transfers of transposable elements and vertebrate endogenous retroviruses.
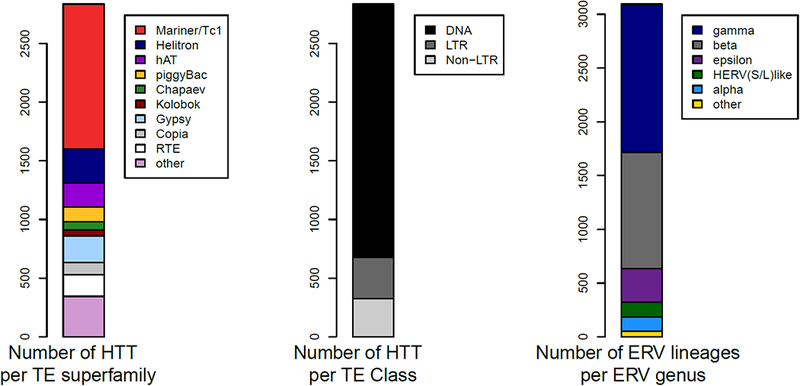
The barplots show the numbers of HTT events per TE superfamily (A) and per TE class (B) as taken from HTT-db in October 2017 (Dotto et al. 2015). The barplot in C) shows the number of clusters of related ERV sequences for each ERV family unearthed from 65 vertebrate genomes by Hayward et al. (2015). The presence of >3000 ERV lineages in vertebrates, each inferred to descend from a discrete endogenisation event [96], suggests that this form of HTT has been pervasive during vertebrate evolution.
Another recent study took advantage of the large number of whole genome sequences available in public databases to perform a systematic survey of HTT among 195 insect species [19]. More than 2,000 HTT events were inferred to explain the distribution of TEs in these insects. This is by far the highest number of HTT inferred in a single study, and it suggests that the actual number of HTT that occurred over the entire evolutionary history of insects is orders of magnitude higher [19]. Comparable projections were made based on a systematic search for HT of long terminal repeat (LTR) retrotransposons among 40 plant species [20]. Non-LTR retrotransposons (notably those of the RTE clade) also appear to have transferred repeatedly during eukaryotic evolution [21]. The pervasiveness of HTT was also apparent from a recent study focusing on the distribution of DNA transposons of the mariner family across 20 Drosophila genomes, in which almost all mariner lineages tested (24 out of 26) were found to have transferred horizontally, often repeatedly [14]. These findings are in line with a seminal study quantifying HTT among three Drosophila species, which estimated an average rate of 0.04 HTT per TE family per million years [22]. Thus, a plethora of HTTs punctuates the evolutionary history of TEs in a variety of eukaryote lineages. To these can be added another common form of HT in eukaryotes, which involves the acquisition of viral sequences also known as endogenous viral elements (Box 1). Some EVEs such as the endogenous retroviruses (ERVs) of vertebrates can be readily affiliated with TEs and, like them, are capable of spreading vertically and horizontally (Box 1; Figure 1).
Box 1: Endogenization of viral sequences as a source of horizontally-acquired DNA.
It has long been appreciated that viral sequences can become integrated in the genome of their host, either as part of their normal replication cycle or accidently through host-encoded reverse transcription and/or recombination activities. Such integration of viral DNA, when it occurs in germ cell lineage, can lead to the vertical transmission and fixation of viral sequences in the host population. Genome sequence mining over the past decade has revealed that such endogenous viral elements are more widespread and diverse than ever imagined, spanning every major type of virus and a wide range of host organisms and unearthing a precious fossil record of past viral infections [97,98]. Most viruses gave rise to just a few endogenous elements per host genome, but some have spawn massive genomic invasions [99]. In vertebrates, endogenous retroviruses (ERVs) are by far the most common, for two main reasons. First, integration of the viral genome into the host chromosome is a required step in retroviral replication and therefore both reverse transcription and integration activities are encoded by the viral genome itself. Second, much like TEs, ERVs can continue to generate copies of themselves long after their horizontal introduction, either through germline reinfections or retrotransposition events [100,101]. As a result, ERVs can rapidly spread in the host population and occasionally attain high copy numbers. This is most apparent in mammalian genomes where multiple waves of ERV invasions together amount to 5–10% of nuclear genome content [102]. A recent study identified no less than 3,100 clusters of related ERV sequences in 65 vertebrate genomes [96], implying that thousands of independent endogenisation events occurred during vertebrate evolution (Figure 1). Furthermore, the phylogeny of ERV lineages in vertebrates indicates that they have undergone more than 1,000 HT events at the host family level, thus the number of HTs is likely to be extremely high at the species-level across vertebrate diversity [96]. Indeed, there is accumulating evidence that some retroviruses have been quite promiscuous as shown by their ability to infiltrate the germline of widely diverged species, for instance belonging to different mammalian orders (ref. 86). Thus, viruses represent a substantial source of genetic material assimilated by eukaryotic genomes, and the boundary between endogenous viruses and transposable elements is becoming increasingly blurry in favor of the notion of a dynamic continuum of invasive genetic elements [103,104].
In comparison to the multiplicity of reports on HTT, evidence for the long-term vertical persistence of TEs is scarce. The best-documented cases involve non-LTR retrotransposons. For instance, the distribution and evolution of R1 and R2 elements in arthropods is consistent with vertical inheritance being the predominant mode of propagation for these elements [23]. The evolutionary stability of these elements may be linked to their specificity for insertion within the ribosomal DNA locus, a relatively “safe haven” in the genome. The LINE-1 family of elements, while it may have been introduced horizontally in the common ancestor of therian mammals (placentals and marsupials)[21], also appears to be exceptional for its continuous activity throughout more than 160 My [24,25], with no clear evidence of HT events and only rare cases of extinction during therian evolution [24,26–28]. To our knowledge, there is no report of DNA transposon or LTR retrotransposon families with such level of evolutionary stability. Rather, the picture emerging from recent systematic studies is that of an overwhelming reliance of most TEs on HT to ensure their long-term persistence. However, the rapid evolutionary erosion and turnover of TE sequences in some host lineages may hamper our ability to trace their vertical origins. The continuing expansion of genome sequencing for diverse taxa should eventually enable a rigorous assessment of the role of HTT in the evolutionary persistence and diversification of TEs.
Ecological factors facilitating HTT
While the cellular mechanisms by which TEs are transferred and reach the germline of a new organism remain largely obscure [29,30], recent systematic analyses are beginning to unravel ecological factors and global patterns governing HTT. First, in plants and insects, the number of horizontally acquired TEs shared between species negatively correlates with the genetic distance between species [19,20,22]. In fact, this so-called phylogenetic distance effect is not limited to intragenomic parasites as it has been observed previously for most pathogens and parasites [31,32]. It can be explained by the fact that pathogens are more capable to adapt to environments similar to where they come from, which are more likely to be found in closely related hosts. The rate of HTT may also be higher between closely related species than between distant ones, especially if such transfers are facilitated by introgressive hybridization which is more likely to occur between close species [33,34]. Interestingly, in insects, the phylogenetic effect is more pronounced for retrotransposons than DNA transposons [19]. In addition, the latter class accounts for more than 75% of all HTT events reported so far (Figure 1), a trend that is increasingly evident [13,29,35]. The higher propensity of DNA transposons to undergo HT and their greater ability to colonize distant hosts compared to retrotransposons may be due to their simpler genetic organization and lower reliance on host factors. Indeed, transposase and substrate DNA are typically the only components required to mediate transposition of these elements in vitro [36–39]. Furthermore, DNA transposons may have relatively minimal requirements for transposition: transposases can mobilize elements that may be distantly related, and the transposed sequences do not need to be transcribed themselves [40]. Furthermore, the transposase is often encoded by an intronless gene driven by a ‘minimal’ promoter. For instance, the promoter of the Drosophila DNA transposon Bari is able to drive transcription in human, yeast, and E. coli cells, unlike that of the Drosophila copia retrotransposon [41]. Thus DNA transposons have evolved a number of features that likely facilitate HTT.
Another trend emerging from the study of HTT in insects is that the number of HTTs increases significantly as geographical distance decreases between hosts [19]. This effect may be expected because species that are geographically distant from each other are less likely to interact than sympatric species. However, it is remarkable that the signal supporting this trend emerges even at the very global scale of biogeographic realms. Finally, an increasing number of HTT events have been inferred to have occurred between parasites and their hosts, such as between blood-feeding insects and vertebrates, lampreys and teleost fishes, or nematodes and birds [21,42–53]. As the sampling of HTT events increase, it should become possible to test the hypothesis according to which host-parasite interactions facilitate HTT, within a robust statistical framework [54].
Consequences of HTT for genome evolution
TEs have shaped eukaryotic genomes deeply and in myriads ways [2,3,55]. Their movement and repetitive nature represent a potent source of genomic variation and genetic disorders [17,56–58]. Because HT is often responsible for the initial colonization of genomes by TEs, HT must be regarded as an important process in eukaryotic evolution [7,11,59,60]. This syllogism is supported by numerous studies in mammalian genomes showing how ERV-derived sequences have been a recurrent source of new coding or regulatory sequences promoting a multitude of cellular functions [61–63] as well as dysfunctions [64–66]. Cases implicating non-viral elements are less commonly recognized, but also exist (Table 1, Figure 2). In insects for example, it was estimated that ~2% of the nuclear genome on average (and up to 24%) derives from horizontally-transferred TEs, a figure that likely remains a gross underestimate [19]. The picture is similar in bats where at least 6% of the Myotis lucifugus genome derives from horizontally-acquired Helitrons [67]. These and other massive inflations of genome size generated through HTT [19,21,43,67,68] suggest that the process must have had a long-lasting impact on genome evolution along these lineages. For instance, several families of Helitrons introduced in vespertilionid bats have caused substantial remodeling and divergence across their genomes, including the capture, duplication, and reshuffling of hundreds of host exonic sequences and promoters [67].
Table 1.
Examples of horizontally acquired transposable elements with documented evolutionary or phenotypic impact
| TE name | Locus | Species | TE size (bp) | TE type | Type of impact | Evidence for HTT* | References |
|---|---|---|---|---|---|---|---|
| P | Multiple copies and P neogenes | Drosophila | 2900 | Class 2; P | Hybrid dysgenesis; P neogenes involved in repression of P activity | see References column | [16,17,79,80] |
| hobo | Multiple copies | Drosophila | 3016 | Class 2; hAT | Hybrid dysgenesis | see References column | [81,82] |
| SPIN | Spider gene (NM_183088.2) | Murid rodents | 2867 | Class 2; hAT | Exaptation of transposase domain inferred from purifying selection on an orthologous copy shared by mouse and rats; unknown function | see References column | [83] |
| Helibat | Multiple | Vespertilionid bats | Class 2; Helitron | Capture, duplication, fusion of genes and regulatory region; remodeling of gene expression | see References column | [67] | |
| Bari1 | Juvenile hormone epoxy hydrolase (Jheh) genes | Drosophila | 1750 | Class 2; Tc1/Mariner | Adaptive insertion in intergenic region between Jheh2 and Jheh3 genes, downregulates expression of both genes | see References column | [14,22,84] |
| Rider | Sun gene; phytoene synthase gene PSY1; FER gene | Tomato (Solanum lycopersicum) | 4867 | Class 1; Copia | TE-mediated gene duplication causing a change in fruit shape; insertion into PSY1 inducing the production of yellow flesh; insertion into FER gene resulting in iron deficiency | Closest copy in spinach (107 My**), 79% identical over 4239 bp; dS TE = 1.1; lowest dS gene = 1.34; Rider was also found to be horizontally transferred in Arabidopsis [85] | [70,71,85–87] |
| Tsc2 | Ruby gene | Blood orange (Citrus sinensis) | 5454 | Class 1; Copia | TE induces cold-dependent modification of Ruby expression, which encodes an activator of anthocyanin production | Closest copy in asparagus (148 My), 82% identical over 4010 bp; dS TE = 0.48; lowest dS gene = 0.78 | [72,88] |
| Tip100 | CHS-D gene | Morning glory (Ipomoea purpurea) | 3873 | Class 2; hAT | TE insertion into CHS-D intron suppresses anthocyanin production | Closest copy in cannabis (110 My), 80% identical over 2426 bp; dS TE = 0.47; lowest dS gene = 0.74 | [69] |
| Hsmarl | SETMAR gene | Anthropoid primates | 1035 | Class 2; Mariner | Exaptation of Transposase domain giving birth to a new gene involved in DNA replication/repair | Closest copy in ant Ooceraea biroi (500 My), 73% identical over 1106 bp; dS TE = 0.6; lowest dS gene = 1.2 | [89,90] |
| SORE-1 | GmphyA2 gene | Soybean (Glycine max) | 6238 | Class 1; Copia | TE insertion inactivates GmphyA2 and induces photoperiod-insensitivity | Closest copy in banana (148 My), 85% identical over 5540 bp; dS TE = 0.57; lowest dS gene = 1.3 | [91] |
| ONSEN (ATCOPIA 78) | Abscisic acid (ABA) responsive gene | Arabidopsis (Arabidopsis thaliana) | 4077 | Class 1; Copia | Insertion of ONSEN into ABA responsive gene induces ABA-insensitive phenotype | Closest copy in asparagus (148 Myrs), 89% identical over 4077 bp; dS TE = 0.52; lowest dS gene= 1.14 | [92] |
| raider | Genes upregulated under UV stress | Maize (Zea mais) | 6527 | Class 1; Copia | Copies of raider are enriched within 1 kb of the TSS of genes up-regulated under UV stress. Stimulate stress-responsive gene expression | Closest copy in rice (40 Myrs), 88% identical over 4356 bp; dS TE = 0.35; lowest dS gene = 42 | [93] |
HTT of TEs for which a function has been demonstrated was searched using each TE copy as a query in blastn (megablast option) on the Whole Genome Sequences Genbank database. Hits from different species were retained when nucleotide identity was equal or higher to 79% for plants diverging from the original species by at least 40 myrs or 73% between insects and primates. The TE copy from the best hit was in each case submitted to further analysis, which consisted in comparing the global and synonymous (dS) TE distance to the dS calculated for five conserved genes evolving under purifying selection. These genes are SMC1, SMC2, MSH1, MLH1 and MCM5, taken from Zhang et al. (2012) [94]for plants and RPSA, RPLP0, RPL7, RPS5, RPL12 for the comparison between primates and insects. In all comparisons the dS calculated between TE copies was lower than the lowest dS calculated between the five conserved genes, strongly supporting HT of these TEs.
Indicates the time since when the two species in which the TE copy was found diverged, according to [95]
Figure 2. Phenotypic consequences of the horizontal introduction of transposable elements in plants.
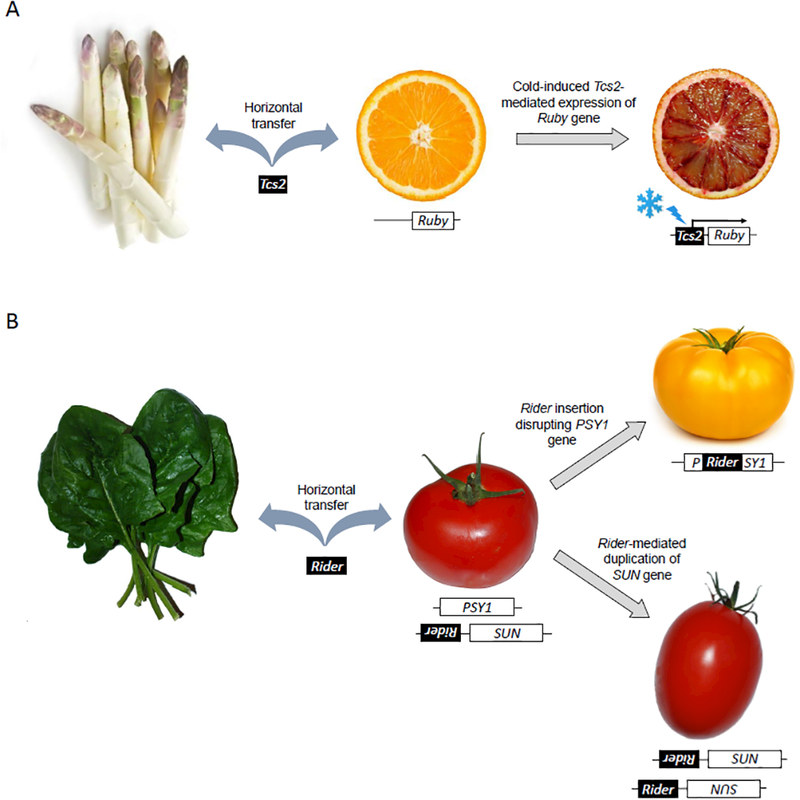
TEs are depicted as black boxes, host genes as white boxes. A. The LTR retrotransposon Tcs2 provided an alternative start site in Jingxian blood orange, inducing cold-dependent overexpression of the Ruby gene, which encodes a MYB transcription factor controlling anthocyanin biosynthesis. This results in the production of orange fruits with more deeply pigmented flesh [72,88]. We found that Tcs2 has been horizontally transferred, either directly or indirectly via intermediate species between orange and asparagus (Table 1). The recent introduction of the Tcs2 element in the orange genome is supported by the fact that it is currently actively transposing [72,88]. B. The LTR retrotransposon Rider triggered the apparition of at least two traits in tomato (yellow flesh and elongated shape) that have been selected by breeders. One copy of Rider is responsible for a duplication of the SUN locus, which leads to increased SUN expression and results in the production of elongated fruits characteristic of the ‘Roma’ variety of tomato [71]. Another copy of Rider disrupts the phytoene synthase (PSY1) gene. The resulting lack of carotenoid leads to the production of yellow tomato flesh [70]. We found that Rider has been horizontally transferred, either directly or indirectly via intermediate species between tomato and spinach (Table 1). The recent introduction of the Rider element in the tomato genome is supported by its absence from the potato (Solanum tuberosum) genome, which diverged only 8 Mya from tomato [85]. Importantly, Cheng et al. (2009) also reported that Rider was horizontally transferred between tomato and the Arabidopsis lineage [85].
To provide further evidence for the role of HTT in the evolution of host genetic novelty and new traits, we revisited the origin of a sample of TEs for which there is direct experimental evidence of their involvement in host physiology or development. We compiled a collection of 28 TEs reported in the literature to have phenotypic consequences (Supplementary Table 1; Table 1) and used their sequence as queries in blastn searches against Genbank’s whole genome sequence database with default parameters. Our non-exhaustive list is purposely biased towards plant TEs with well-known phenotypic consequences. We reasoned that the recent emergence of these plant TE alleles selected by breeders for agronomic traits would facilitate our ability to trace their origins and detect closely related elements in other plant genomes, which are also well represented in current databases. For six of them, we obtained strong evidence supporting the notion that the element implicated derives from a TE family introduced horizontally in that species lineage (Table 1). Among these are textbook examples of TEs acting as drivers of phenotypic changes, including a Tip100 transposon that induces changes in flower color patterns in morning glories [69], the Rider retrotransposon responsible for the ‘yellow flesh’ phenotype [70] and independently for the oval shape of ‘Roma’ tomatoes [71] and, a Tcs2 element that triggers the production of red flesh in blood oranges [72] (Figure 2). These examples also illustrate the various ways by which TEs can influence the expression of host genes, including insertional gene inactivation [70], gene rearrangement [71] and cis-regulatory effects [72] (Figure 2). Searches for adaptive mutations caused by TEs in Drosophila are also inherently biased for young insertions [73], which in turn can be more readily traced to HTT events (see example of Bari1 in Table 1). Nonetheless, there is evidence for relatively ancient cooption events of transposase genes derived from TE families clearly introduced via HT in various mammals (e.g. Spider and SETMAR genes in Table 1). Overall, our preliminary screen highlights a number of cases whereby a direct link between HTT and phenotypic change can be established, which bodes well for future studies aiming at systematically characterizing the consequences of HTT.
Conclusions and perspectives
Recent large-scale analyses suggest that HTT is pervasive in plants and animals and instrumental to the long-term evolutionary persistence of most types of TEs. Despite the growing recognition of this phenomenon, there is still limited understanding of the factors that influence the likelihood or success of HTT, including factors acting at the level of individuals and species (e.g. physiology, population genetics) or their interactions as well as the role of the environment (e.g. ecology, geography). A few systematic studies of HTT conducted for large sets of species have begun to reveal some statistically robust patterns governing HTT, such as phylogenetic and geographic distance effects, but await validation with broader sampling and more diverse sets of organisms. The influence of many other factors linked to the environment (e.g. aquatic versus terrestrial) or the life history (e.g. diet, host-parasite interactions) of the species, while tantalizing, have remained circumstantial and await to be tested more formally [21,42–53,74]. A robust conceptual framework combining functional ecology and network theory would be an attractive way to conduct such analysis [54]. Superimposing HTT over ecological networks may also yield new insights on the molecular and cellular routes underlying these transfers in eukaryotes, which so far have remained broadly elusive [29,30].
Regarding the consequences of HTT, it will be important to connect genome-wide assessments of the putative function of TEs to systematic searches for evidence supporting HT of these TEs. We believe that TEs that rely more heavily on HT to persist are also more likely to be involved in conflicts with their hosts. Such conflicts may induce arms-race dynamics, or repeated cycles of adaptation/counter-adaptation that foster new interactions between TEs and cellular host factors [75]. The resulting diversity of these interactions may in turn increase the likelihood of TE co-option for particular cellular pathways [76]. One testable prediction of this model is that TEs that transfer frequently would also undergo co-option more often than those that transfer less frequently. Finally, it has long been hypothesized that TEs could drive host diversification, reinforce population divergence, and promote reproductive isolation (for instance through processes akin to hybrid dysgenesis), ultimately leading to speciation [10,11,77,78]. Given that TEs regularly invade new genomes via HT, one prediction of this hypothesis is that high rates of speciation may be linked to high rates of HTT. This is one of many avenues for future investigation in this burgeoning area.
Supplementary Material
Acknowledgements
We apologize to our colleagues whose relevant work and original articles could not be cited owing to constraint in the number of references. We thank Alex Hayward for discussions on vertebrate endogenous retroviruses. C.G. acknowledges funding from Agence Nationale de la Recherche, project ANR-15-CE32-0011-01 TransVir. C.F. acknowledges funding from the National Institutes of Health, including projects GM122550, GM112972 and GM077582.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Clément Gilbert, Laboratoire Evolution, Génomes, Comportement, Ecologie; CNRS Université Paris-Sud UMR 9191; IRD UMR 247; Avenue de la Terrasse, Bâtiment 13, Boite Postale 1, 91198 Gif sur Yvette, France. clement.gilbert@egce.cnrs-gif.fr.
Cédric Feschotte, Department of Molecular Biology and Genetics, Cornell University, Ithaca, NY, USA. cf458@cornell.edu.
References
- 1.Craig NL C R: Mobile DNA II. Washington (DC): American Society for Microbiology Press.; 2002. [Google Scholar]
- 2.Feschotte C, Pritham EJ: DNA transposons and the evolution of eukaryotic genomes. Annu Rev Genet 2007, 41:331–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cordaux R, Batzer MA: The impact of retrotransposons on human genome evolution. Nat Rev Genet 2009, 10:691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hua-Van A, Le Rouzic A, Boutin TS, Filee J, Capy P: The struggle for life of the genome’s selfish architects. Biol Direct 2011, 6:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chenais B, Caruso A, Hiard S, Casse N: The impact of transposable elements on eukaryotic genomes: from genome size increase to genetic adaptation to stressful environments. Gene 2012, 509:7–15. [DOI] [PubMed] [Google Scholar]
- 6.Pritham EJ: Transposable Elements and Factors Influencing their Success in Eukaryotes. J Hered 2009, 100:648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schaack S, Gilbert C, Feschotte C: Promiscuous DNA: horizontal transfer of transposable elements and why it matters for eukaryotic evolution. Trends Ecol Evol 2010, 25:537–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wallau GL, Vieira C, Loreto ÉLS: Genetic exchange in eukaryotes through horizontal transfer: connected by the mobilome. Mob DNA 2018, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hartl DL, Lohe AR, Lozovskaya ER: Modern thoughts on an ancyent marinere: function, evolution, regulation. Annu Rev Genet 1997, 31:337–58. [DOI] [PubMed] [Google Scholar]
- 10.Jurka J, Bao W, Kojima KK, Kohany O, Yurka MG: Distinct groups of repetitive families preserved in mammals correspond to different periods of regulatory innovations in vertebrates. Biol Direct 2012, 7:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oliver KR, Greene WK: Transposable elements and viruses as factors in adaptation and evolution: an expansion and strengthening of the TE-Thrust hypothesis. Ecol Evol 2012, 2:2912–2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oliveira SG, Bao W, Martins C, Jurka J: Horizontal transfers of Mariner transposons between mammals and insects. Mob DNA 2012, 3:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Silva JC, Loreto EL, Clark JB: Factors that affect the horizontal transfer of transposable elements. Curr Issues Mol Biol 2004, 6:57–71. [PubMed] [Google Scholar]
- 14.Wallau GL, Capy P, Loreto E, Le Rouzic A, Hua-Van A: VHICA, a New Method to Discriminate between Vertical and Horizontal Transposon Transfer: Application to the Mariner Family within Drosophila. Mol Biol Evol 2016, 33:1094–109.* This paper provides a detailed pipeline to detect HTT along a known host phylogeny and shows that most (24 out of 26) of the mariner elements found in 20 Drosophila genomes have been horizontally transferred, often multiple times.
- 15.Dotto BR, Carvalho EL, Silva AF, Duarte Silva LF, Pinto PM, Ortiz MF, Wallau GL: HTT-DB: horizontally transferred transposable elements database. Bioinformatics 2015, 31:2915–7.* A comprehensive database compiling all known cases of horizontal transfer of TEs and viruses in eukaryotic genomes.
- 16.Daniels SB, Peterson KR, Strausbaugh LD, Kidwell MG, Chovnick A: Evidence for horizontal transmission of the P transposable element between Drosophila species. Genetics 1990, 124:339–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kofler R, Hill T, Nolte V, Betancourt AJ, Schlötterer C: The recent invasion of natural Drosophila simulans populations by the P-element. Proc Natl Acad Sci 2015, 112:6659–6663.** This study reports a near-contemporary case of HTT in nature; the P element appears to have invaded Drosophila simulans during the second half of the 20th century.
- 18.Robillard É, Le Rouzic A, Zhang Z, Capy P, Hua-Van A: Experimental evolution reveals hyperparasitic interactions among transposable elements. Proc Natl Acad Sci 2016, 113:14763–14768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peccoud J, Loiseau V, Cordaux R, Gilbert C: Massive horizontal transfer of transposable elements in insects. Proc Natl Acad Sci U S A 2017, 114:4721–4726.** See reference 96
- 20.El Baidouri M, Carpentier MC, Cooke R, Gao D, Lasserre E, Llauro C, Mirouze M, Picault N, Jackson SA, Panaud O: Widespread and frequent horizontal transfers of transposable elements in plants. Genome Res 2014, 24:831–8.** See reference 96
- 21.Ivancevic A, Kortschak D, Bertozzi T, Adelson D: Re-evaluating inheritance in genome evolution: widespread transfer of LINEs between species. 2017, doi: 10.1101/106914.** See reference 96
- 22.Bartolome C, Bello X, Maside X: Widespread evidence for horizontal transfer of transposable elements across Drosophila genomes. Genome Biol 2009, 10:R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eickbush TH, Eickbush DG: Integration, Regulation, and Long-Term Stability of R2 Retrotransposons. Microbiol Spectr 2015, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ivancevic AM, Kortschak RD, Bertozzi T, Adelson DL: LINEs between Species: Evolutionary Dynamics of LINE-1 Retrotransposons across the Eukaryotic Tree of Life. Genome Biol Evol 2016, 8:3301–3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smit AFA, Tóth G, Riggs AD, Jurka J: Ancestral, Mammalian-wide Subfamilies of LINE-1 Repetitive Sequences. J Mol Biol 1995, 246:401–417. [DOI] [PubMed] [Google Scholar]
- 26.Rinehart TA, Grahn RA, Wichman HA: SINE extinction preceded LINE extinction in sigmodontine rodents: implications for retrotranspositional dynamics and mechanisms. Cytogenet Genome Res 2005, 110:416–425. [DOI] [PubMed] [Google Scholar]
- 27.Cantrell MA, Scott L, Brown CJ, Martinez AR, Wichman HA: Loss of LINE-1 Activity in the Megabats. Genetics 2008, 178:393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Platt RN II, Ray DA: A non-LTR retroelement extinction in Spermophilus tridecemlineatus. Gene 2012, 500:47–53. [DOI] [PubMed] [Google Scholar]
- 29.Loreto EL, Carareto CM, Capy P: Revisiting horizontal transfer of transposable elements in Drosophila. Hered Edinb 2008, 100:545–54. [DOI] [PubMed] [Google Scholar]
- 30.Gilbert C, Cordaux R: Viruses as vectors of horizontal transfer of genetic material in eukaryotes. Curr Opin Virol 2017, 25:16–22. [DOI] [PubMed] [Google Scholar]
- 31.Longdon B, Brockhurst MA, Russell CA, Welch JJ, Jiggins FM: The Evolution and Genetics of Virus Host Shifts. PLoS Pathog 2014, 10:e1004395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perlman SJ, Jaenike J: Infection success in novel hosts: an experimental and phylogenetic study of Drosophila-parasitic nematodes. Evol Int J Org Evol 2003, 57:544–557. [DOI] [PubMed] [Google Scholar]
- 33.Mallet J, Besansky N, Hahn MW: How reticulated are species? BioEssays 2016, 38:140–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coyne JA, Orr HA: PATTERNS OF SPECIATION IN DROSOPHILA. Evolution 1989, 43:362–381. [DOI] [PubMed] [Google Scholar]
- 35.Malik HS, Burke WD, Eickbush TH: The age and evolution of non-LTR retrotransposable elements. Mol Biol Evol 1999, 16:793–805. [DOI] [PubMed] [Google Scholar]
- 36.Lampe DJ, Churchill ME, Robertson HM: A purified mariner transposase is sufficient to mediate transposition in vitro. EMBO J 1997, 15:5470–5479. [PMC free article] [PubMed] [Google Scholar]
- 37.Kaufman PD, Rio DC: P element transposition in vitro proceeds by a cut-and-paste mechanism and uses GTP as a cofactor. Cell 1992, 69:27–39. [DOI] [PubMed] [Google Scholar]
- 38.Hencken CG, Li X, Craig NL: Functional characterization of an active Rag-like transposase. Nat Struct Mol Biol 2012, 19:834–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou L, Mitra R, Atkinson PW, Burgess Hickman A, Dyda F, Craig NL: Transposition of hAT elements links transposable elements and V(D)J recombination. Nature 2004, 432:995–1001. [DOI] [PubMed] [Google Scholar]
- 40.Yang G, Nagel DH, Feschotte C, Hancock CN, Wessler SR: Tuned for transposition: molecular determinants underlying the hyperactivity of a Stowaway MITE. Science 2009, 325:1391–4. [DOI] [PubMed] [Google Scholar]
- 41.Palazzo A, Caizzi R, Viggiano L, Marsano RM: Does the Promoter Constitute a Barrier in the Horizontal Transposon Transfer Process? Insight from Bari Transposons. Genome Biol Evol 2017, 9:1637–1645.* An elegant study showing that the permissive promoter of Class 2 transposons may explain, at least in part, why these TEs have a higher propensity to transfer horizontally than Class 1 retrotransposons.
- 42.Yoshiyama M, Tu Z, Kainoh Y, Honda H, Shono T, Kimura K: Possible horizontal transfer of a transposable element from host to parasitoid. Mol Biol Evol 2001, 18:1952–1958. [DOI] [PubMed] [Google Scholar]
- 43.Gilbert C, Schaack S, Pace JK, Brindley PJ, Feschotte C: A role for host-parasite interactions in the horizontal transfer of transposons across phyla. Nature 2010, 464:1347–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuraku S, Qiu H, Meyer A: Horizontal transfers of Tc1 elements between teleost fishes and their vertebrate parasites, lampreys. Genome Biol Evol 2012, 4:929–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walsh AM, Kortschak RD, Gardner MG, Bertozzi T, Adelson DL: Widespread horizontal transfer of retrotransposons. Proc Natl Acad Sci U S A 2013, 110:1012–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang HH, Feschotte C, Han MJ, Zhang Z: Recurrent horizontal transfers of Chapaev transposons in diverse invertebrate and vertebrate animals. Genome Biol Evol 2014, 6:1375–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guo X, Gao J, Li F, Wang J: Evidence of horizontal transfer of non-autonomous Lep1 Helitrons facilitated by host-parasite interactions. Sci Rep 2014, 4:5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Filée J, Rouault J-D, Harry M, Hua-Van A: Mariner transposons are sailing in the genome of the blood-sucking bug Rhodnius prolixus. BMC Genomics 2015, 16:1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang H-H, Shen Y-H, Xu H-E, Liang H-Y, Han M-J, Zhang Z: A novel hAT element in B ombyx mori and R hodnius prolixus: its relationship with miniature inverted repeat transposable elements (MITEs) and horizontal transfer: Evolution of a particular hAT transposon. Insect Mol Biol 2013, 22:584–596. [DOI] [PubMed] [Google Scholar]
- 50.Tang Z, Zhang H-H, Huang K, Zhang X-G, Han M-J, Zhang Z: Repeated horizontal transfers of four DNA transposons in invertebrates and bats. Mob DNA 2015, 6:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang H-H, Shen Y-H, Xiong X-M, Han M-J, Qi D-W, Zhang X-G: Evidence for horizontal transfer of a recently active Academ transposon: Repeated horizontal transfer of Academ transposons. Insect Mol Biol 2016, 25:338–346. [DOI] [PubMed] [Google Scholar]
- 52.Suh A, Witt CC, Menger J, Sadanandan KR, Podsiadlowski L, Gerth M, Weigert A, McGuire JA, Mudge J, Edwards SV, et al. : Ancient horizontal transfers of retrotransposons between birds and ancestors of human pathogenic nematodes. Nat Commun 2016, 7:11396.* In addition to reporting new cases of HTT between host and parasite lineages, this study illustrates the potential of using HTT as a paleontological record of past interactions between species.
- 53.Wang X, Liu X: Close ecological relationship among species facilitated horizontal transfer of retrotransposons. BMC Evol Biol 2016, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Venner S, Miele V, Terzian C, Biémont C, Daubin V, Feschotte C, Pontier D: Ecological networks to unravel the routes to horizontal transposon transfers. PLOS Biol 2017, 15:e2001536.* This study proposes important guidelines to uncover the factors facilitating HTT in eukaryotes by combining functional ecology and network theory.
- 55.Levin HL, Moran JV: Dynamic interactions between transposable elements and their hosts. Nat Rev Genet 2011, 12:615–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.O’Donnell KA, Burns KH: Mobilizing diversity: transposable element insertions in genetic variation and disease. Mob DNA 2010, 1:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hancks DC, Kazazian HH: Roles for retrotransposon insertions in human disease. Mob DNA 2016, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lai X, Schnable JC, Liao Z, Xu J, Zhang G, Li C, Hu E, Rong T, Xu Y, Lu Y: Genome-wide characterization of non-reference transposable element insertion polymorphisms reveals genetic diversity in tropical and temperate maize. BMC Genomics 2017, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Casacuberta E, González J: The impact of transposable elements in environmental adaptation. Mol Ecol 2013, 22:1503–1517. [DOI] [PubMed] [Google Scholar]
- 60.Drezen J-M, Josse T, Bézier A, Gauthier J, Huguet E, Herniou E: Impact of Lateral Transfers on the Genomes of Lepidoptera. Genes 2017, 8:315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Frank JA, Feschotte C: Co-option of endogenous viral sequences for host cell function. Curr Opin Virol 2017, 25:81–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Römer C, Singh M, Hurst LD, Izsvák Z: How to tame an endogenous retrovirus: HERVH and the evolution of human pluripotency. Curr Opin Virol 2017, 25:49–58. [DOI] [PubMed] [Google Scholar]
- 63.Horie M: The biological significance of bornavirus-derived genes in mammals. Curr Opin Virol 2017, 25:1–6. [DOI] [PubMed] [Google Scholar]
- 64.Kassiotis G, Stoye JP: Immune responses to endogenous retroelements: taking the bad with the good. Nat Rev Immunol 2016, 16:207–219. [DOI] [PubMed] [Google Scholar]
- 65.Babaian A, Mager DL: Endogenous retroviral promoter exaptation in human cancer. Mob DNA 2016, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Perron H, Lang A: The Human Endogenous Retrovirus Link between Genes and Environment in Multiple Sclerosis and in Multifactorial Diseases Associating Neuroinflammation. Clin Rev Allergy Immunol 2010, 39:51–61. [DOI] [PubMed] [Google Scholar]
- 67.Thomas J, Phillips CD, Baker RJ, Pritham EJ: Rolling-Circle Transposons Catalyze Genomic Innovation in a Mammalian Lineage. Genome Biol Evol 2014, 6:2595–2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thomas J, Schaack S, Pritham EJ: Pervasive Horizontal Transfer of Rolling-Circle Transposons among Animals. Genome Biol Evol 2010, 2:656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Habu Y, Hisatomi Y, Iida S: Molecular characterization of the mutable flaked allele for flower variegation in the common morning glory. Plant J Cell Mol Biol 1998, 16:371–376. [DOI] [PubMed] [Google Scholar]
- 70.Fray RG, Grierson D: Identification and genetic analysis of normal and mutant phytoene synthase genes of tomato by sequencing, complementation and co-suppression. Plant Mol Biol 1993, 22:589–602. [DOI] [PubMed] [Google Scholar]
- 71.Xiao H, Jiang N, Schaffner E, Stockinger EJ, van der Knaap E: A retrotransposon-mediated gene duplication underlies morphological variation of tomato fruit. Science 2008, 319:1527–1530. [DOI] [PubMed] [Google Scholar]
- 72.Butelli E, Garcia-Lor A, Licciardello C, Las Casas G, Hill L, Recupero GR, Keremane ML, Ramadugu C, Krueger R, Xu Q, et al. : Changes in Anthocyanin Production during Domestication of Citrus. Plant Physiol 2017, 173:2225–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Barrón MG, Fiston-Lavier A-S, Petrov DA, González J: Population Genomics of Transposable Elements in Drosophila. Annu Rev Genet 2014, 48:561–581. [DOI] [PubMed] [Google Scholar]
- 74.Wang X, Liu X: Close ecological relationship among species facilitated horizontal transfer of retrotransposons. BMC Evol Biol 2016, 16:201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McLaughlin RN, Malik HS: Genetic conflicts: the usual suspects and beyond. J Exp Biol 2017, 220:6–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jangam D, Feschotte C, Betrán E: Transposable Element Domestication As an Adaptation to Evolutionary Conflicts. Trends Genet TIG 2017, 33:817–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dion-Côté A-M, Barbash DA: Beyond speciation genes: an overview of genome stability in evolution and speciation. Curr Opin Genet Dev 2017, 47:17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Parhad SS, Tu S, Weng Z, Theurkauf WE: Adaptive Evolution Leads to Cross-Species Incompatibility in the piRNA Transposon Silencing Machinery. Dev Cell 2017, 43:60–70.e5.* This study shows how the rapid evolution of Drosophila genes involved in TE silencing, likely driven by an arms race with TEs introduced horizontally, leads to their inability to function across species, which may contribute to reproductive isolation
- 79.Ronsseray S, Anxolabéhère D, Périquet G: Hybrid dysgenesis in Drosophila melanogaster: influence of temperature on cytotype determination in the P-M system. Mol Gen Genet MGG 1984, 196:17–23. [DOI] [PubMed] [Google Scholar]
- 80.Pinsker W, Haring E, Hagemann S, Miller WJ: The evolutionary life history of P transposons: from horizontal invaders to domesticated neogenes. Chromosoma 2001, 110:148–158. [DOI] [PubMed] [Google Scholar]
- 81.Yannopoulos G, Stamatis N, Monastirioti M, Hatzopoulos P, Louis C: hobo is responsible for the induction of hybrid dysgenesis by strains of Drosophila melanogaster bearing the male recombination factor 23.5MRF. Cell 1987, 49:487–495. [DOI] [PubMed] [Google Scholar]
- 82.Simmons GM: Horizontal transfer of hobo transposable elements within the Drosophila melanogaster species complex: evidence from DNA sequencing. Mol Biol Evol 1992, 9:1050–1060. [DOI] [PubMed] [Google Scholar]
- 83.Pace JK, Gilbert C, Clark MS, Feschotte C: Repeated horizontal transfer of a DNA transposon in mammals and other tetrapods. Proc Natl Acad Sci U A 2008, 105:17023–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gonzalez J, Macpherson JM, Petrov DA: A Recent Adaptive Transposable Element Insertion Near Highly Conserved Developmental Loci in Drosophila melanogaster. Mol Biol Evol 2009, 26:1949–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cheng X, Zhang D, Cheng Z, Keller B, Ling H-Q: A New Family of Ty1-copia-Like Retrotransposons Originated in the Tomato Genome by a Recent Horizontal Transfer Event. Genetics 2009, 181:1183–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ling H-Q, Bauer P, Bereczky Z, Keller B, Ganal M: The tomato fer gene encoding a bHLH protein controls iron-uptake responses in roots. Proc Natl Acad Sci 2002, 99:13938–13943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jiang N, Gao D, Xiao H, van der Knaap E: Genome organization of the tomato sun locus and characterization of the unusual retrotransposon Rider. Plant J 2009, 60:181–193. [DOI] [PubMed] [Google Scholar]
- 88.Butelli E, Licciardello C, Zhang Y, Liu J, Mackay S, Bailey P, Reforgiato-Recupero G, Martin C: Retrotransposons control fruit-specific, cold-dependent accumulation of anthocyanins in blood oranges. Plant Cell 2012, 24:1242–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kim H-S, Williamson EA, Nickoloff JA, Hromas RA, Lee S-H: Metnase Mediates Loading of Exonuclease 1 onto Single Strand Overhang DNA for End Resection at Stalled Replication Forks. J Biol Chem 2017, 292:1414–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cordaux R, Udit S, Batzer MA, Feschotte C: Birth of a chimeric primate gene by capture of the transposase gene from a mobile element. Proc Natl Acad Sci U A 2006, 103:8101–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kanazawa A, Liu B, Kong F, Arase S, Abe J: Adaptive evolution involving gene duplication and insertion of a novel Ty1/copia-like retrotransposon in soybean. J Mol Evol 2009, 69:164–175. [DOI] [PubMed] [Google Scholar]
- 92.Ito H, Gaubert H, Bucher E, Mirouze M, Vaillant I, Paszkowski J: An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature 2011, 472:115–119. [DOI] [PubMed] [Google Scholar]
- 93.Makarevitch I, Waters AJ, West PT, Stitzer M, Hirsch CN, Ross-Ibarra J, Springer NM: Transposable Elements Contribute to Activation of Maize Genes in Response to Abiotic Stress. PLoS Genet 2015, 11:e1004915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang N, Zeng L, Shan H, Ma H: Highly conserved low-copy nuclear genes as effective markers for phylogenetic analyses in angiosperms. New Phytol 2012, 195:923–937. [DOI] [PubMed] [Google Scholar]
- 95.Hedges SB, Marin J, Suleski M, Paymer M, Kumar S: Tree of Life Reveals Clock-Like Speciation and Diversification. Mol Biol Evol 2015, 32:835–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hayward A, Cornwallis CK, Jern P: Pan-vertebrate comparative genomics unmasks retrovirus macroevolution. Proc Natl Acad Sci 2015, 112:464–469.** References 19, 20, 21 and 96 are the first systematic studies of HTT mining dozens of complete genomes; they revealed that HTT is pervasive in plant and animal genome evolution.
- 97.Feschotte C, Gilbert C: Endogenous viruses: insights into viral evolution and impact on host biology. Nat Rev Genet 2012, 13:283–96. [DOI] [PubMed] [Google Scholar]
- 98.Katzourakis A: Editorial overview: Paleovirology: the genomic fossil record, and consequences of ancient viral infections. Curr Opin Virol 2017, 25:ix–xi. [DOI] [PubMed] [Google Scholar]
- 99.Pritham EJ, Putliwala T, Feschotte C: Mavericks, a novel class of giant transposable elements widespread in eukaryotes and related to DNA viruses. Gene 2007, 390:3–17. [DOI] [PubMed] [Google Scholar]
- 100.Belshaw R, Pereira V, Katzourakis A, Talbot G, Paces J, Burt A, Tristem M: Long-term reinfection of the human genome by endogenous retroviruses. Proc Natl Acad Sci U A 2004, 101:4894–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhuo X, Feschotte C: Cross-Species Transmission and Differential Fate of an Endogenous Retrovirus in Three Mammal Lineages. PLoS Pathog 2015, 11:e1005279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhuo X, Rho M, Feschotte C: Genome-wide characterization of endogenous retroviruses in the bat Myotis lucifugus reveals recent and diverse infections. J Virol 2013, 87:8493–501.* A detailed case investigation of the cross-ordinal transmission of a retrovirus and its differential genomic success after horizontal introduction.
- 103.Koonin EV: Viruses and mobile elements as drivers of evolutionary transitions. Philos Trans R Soc B Biol Sci 2016, 371:20150442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Koonin EV, Krupovic M: Polintons, virophages and transpovirons: a tangled web linking viruses, transposons and immunity. Curr Opin Virol 2017, 25:7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


