Abstract
To investigate the consequences of hybridization between species, we studied three replicate hybrid populations that formed naturally between two swordtail fish species, estimating their fine-scale genetic map and inferring ancestry along the genomes of 690 individuals. In all three populations, ancestry from the “minor” parental species is more common in regions of high recombination and where there is linkage to fewer putative targets of selection. The same patterns are apparent in a reanalysis of human and archaic admixture. These results support models in which ancestry from the minor parental species is more likely to persist when rapidly uncoupled from alleles that are deleterious in hybrids. Our analyses further indicate that selection on the swordtail hybrids stems predominantly from deleterious combinations of epistatically-interacting alleles.
Understanding speciation is central to understanding evolution, but so much about the process still puzzles us. Foundational work in evolutionary biology envisioned speciation as an ordered process in which reproductive barriers, once established, prevent gene flow between species (1). We now realize, however, that speciation is much more dynamic, with evidence of historical and ongoing hybridization visible in the genomes of myriad species (2–5). The ubiquity of hybridization raises the question of how species that interbreed remain distinct.
At least part of the answer lies in widespread selection on hybrid genomes (1). Analyses of hominin and swordtail fish hybrids indicate that ancestry from the “minor” parent species (i.e., the parent that contributed less to the gene pool of hybrids) is decreased near functionally important elements (4, 6, 7), presumably because such regions are enriched for harmful alleles. Aside from these observations, however, little is known about how hybrid genomes evolve. Decades of experimental work have demonstrated that Bateson-Dobzhansky-Muller incompatibilities (BDMIs) are a central mechanism underlying reproductive isolation once species are formed (8–10), but the importance of BDMIs in the evolution of hybrid genomes remains unknown, as does the role of other modes of selection. Notably, when there is introgression from a species with a lower effective population size, hybrids may suffer from increased genetic load (“hybridization load”) due to the introduction of weakly deleterious alleles (6, 11, 12). Depending on the environment in which hybrids find themselves, alleles that underlie ecological adaptations in the parental species may also be deleterious (13, 14). Complicating matters further, the sources of selection on hybrids will likely vary from system to system, depending on the extent of genetic and ecological differentiation between the parental species as well as the differences in their effective population sizes.
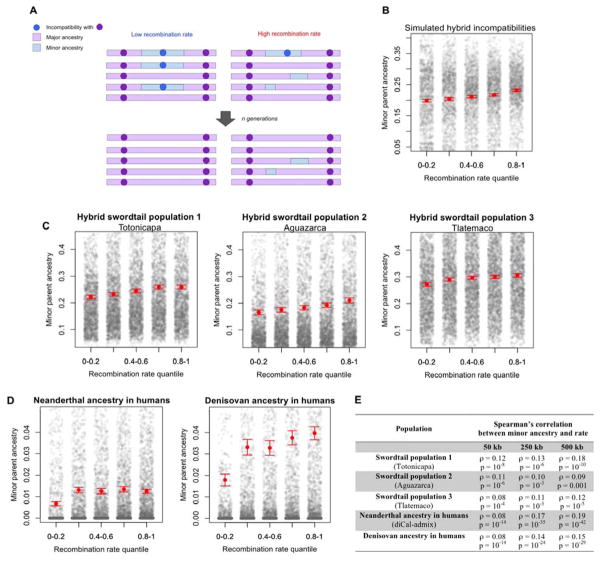
Regardless of the source of selection, however, one feature is expected to play a central role in mediating its effects: variation in recombination rates along the genome (6, 11, 15–17). In models of BDMIs, neutral ancestry from the minor parent is more likely to persist in regions of higher recombination, where it is more rapidly uncoupled from mutations deleterious on the prevalent (i.e., major parent) genetic background (Fig. 1A,B; 17). Similarly, in models of hybridization load, all else being equal, shorter linkage blocks tend to carry fewer weakly deleterious mutations and thus be less rapidly purged by selection (6, 11; Fig. S1). Previous studies have reported patterns consistent with these expectations (18–20), but without investigating ancestry patterns and their relationship to local recombination rates, or distinguishing among sources of selection.
Figure 1. Relationships between minor parent ancestry and recombination rates.
(A) In the presence of hybrid incompatibilities, minor parent ancestry is more likely to persist in regions of high recombination. (B) Shown is one, randomly-chosen replicate of simulations under plausible parameters for swordtail species (21). (C) Relationship between minor parent ancestry and recombination rate in swordtails and (D) in humans (see Fig. S8). Data are summarized in 50 kb windows in swordtail analyses and 250 kb in humans, so that the number of windows is similar. (E) Spearman’s correlations between average minor parent ancestry and recombination rate at several scales; see Table S2 for complete results and 25 for details of the Denisovan analysis. In B–D, red points and whiskers indicate the mean with two standard errors of the mean determined by bootstrapping; gray points show raw data. Quantile binning is for visualization; statistical tests were performed on the unbinned data.
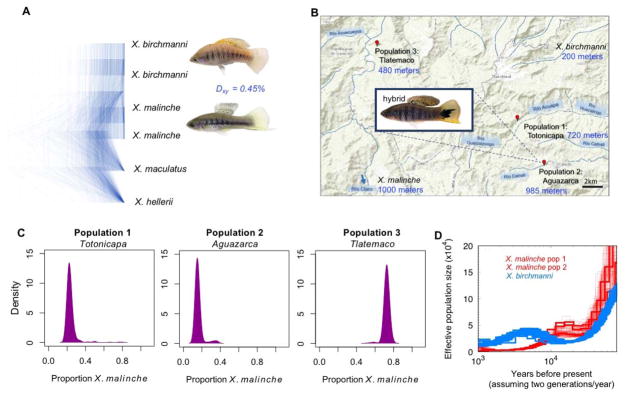
To ask these questions, we took advantage of naturally occurring hybrid populations between sister species of swordtail fish, Xiphophorus birchmanni and X. malinche (Fig. 2A–D; 21). The species are ~0.5% divergent at the nucleotide level and, due to the small effective population size of X. malinche, incomplete lineage sorting between the two is relatively rare (21; Fig. 2A,D). We focused on three hybrid populations that formed independently between the two species fewer than 100 generations ago (22). Previous analyses of hybrids between these species suggested that there are on the order of 100 unlinked BDMI pairs segregating (22, 23), with estimated selection coefficients ~0.03–0.05 (22), in addition to which there could also be linked BDMIs.
Figure 2. Hybridization between X. birchmanni and X. malinche.
(A) Maximum likelihood trees from RAxML for 1,000 alignments of randomly selected 10 kb regions. Dxy refers to the average nucleotide divergence between X. birchmanni and X. malinche. (B) Locations of hybrid populations in river systems in Hidalgo, Mexico; listed in blue are elevations of the hybrid populations and typical elevations for parental populations. (C) Inferred ancestry proportions of individuals from each population. (D) Effective population sizes inferred from three X. malinche genomes (sampled from two populations) and 20 X. birchmanni genomes; 50 bootstraps are shown for one individual from each X. malinche population (21).
To infer local ancestry patterns, we generated ~1X low coverage whole genome data for 690 hybrids sampled from the three hybrid populations, then estimated local ancestry patterns by applying a hidden Markov model to ~1 million sites genome-wide (21, 24). Two of the hybrid populations derive on average 75–80% of their genomes from X. birchmanni, whereas individuals in the third population derive on average 72% of their genomes from X. malinche (Fig. 2C; 21), with median homozygous tract lengths for the minor parent ranging from 84 kb to 225 kb across the three populations (21).
Our previous work (25) indicated that local recombination rates should be conserved between X. birchmanni and X. malinche (21). To consider the relationship between local ancestry and recombination rate, we inferred a fine-scale genetic map for X. birchmanni from patterns of linkage disequilibrium (LD; Table S1; 21). We also generated a crossover map from ancestry switch-points in hybrids, which was concordant with the one obtained for X. birchmanni (Fig. S2; 21).
In all three hybrid populations, the probability of carrying ancestry from the minor parent increases with the local recombination rate (Fig. 1C, Table S2). The relationship remains irrespective of the choice of scale (Fig. S3) and after thinning the SNP and ancestry data to control for possible differences in the reliability of estimated recombination rates or the power to call ancestry across windows (21). This pattern is not expected under neutrality (Fig. S1) but can readily be generated under several models of selection, including selection against BDMIs, hybridization load, or widespread ecological selection against loci from the minor parent (Fig. 1B, Fig. S1). Thus, our finding supports models in which minor parent ancestry persists where it was more likely to have been rapidly uncoupled from the deleterious alleles with which it was originally linked (21).
In principle, the chance of minor parent ancestry persisting should be a function of the exact number of deleterious alleles to which it was linked since hybridization occurred. Local recombination rates are one proxy for this (unknown) parameter, as are the number of coding or conserved base pairs nearby. Both features predict average minor parent ancestry (Fig. S4; Fig. S5; 21), but in our data recombination is the stronger predictor and remains similarly strong after controlling for the number of coding (or conserved) base pairs (Table S2,S3).
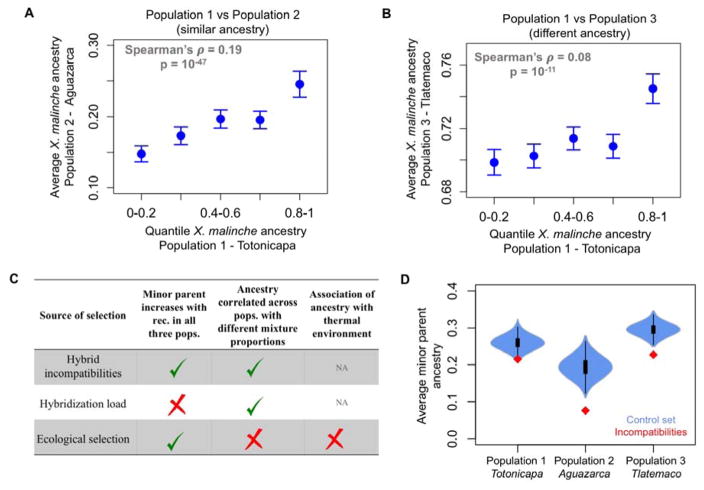
To investigate the source of section on hybrids, we considered correlations in local ancestry between pairs of hybrid populations: though weaker between populations with different major parent ancestries, it was in all cases significantly positive (controlling for the recombination rate; Fig. 3A,B). These correlation patterns should not arise from ecological selection, but are expected from selection against hybridization load, as well as, less intuitively, from selection on the same BDMIs (see Fig. S6; 21).
Figure 3. Evidence for BDMIs being the major source of selection on hybrids.
(A, B) Correlations in ancestry between independently formed swordtail hybrid populations (in 0.1 cM windows; Fig. S9). Points show the mean and whiskers indicate two standard errors of the mean; correlations are calculated on unbinned data. (C) Predictions for different sources of selection on hybrids. (D) Average minor parent ancestry is unusually depleted in 50 kb windows containing putative unlinked BDMIs (red points, from 23) compared to 1,000 null datasets (blue; see 21). Importantly, low minor parent ancestry at putative BDMIs is not expected as a result of how they were originally identified (21).
Further evidence about the source of selection comes from an analysis of genome sequences from X. malinche (3, 22) and X. birchmanni, which indicates that X. malinche has had a lower long-term effective population size over the last ~20,000 generations (Fig. 2D; 21), as reflected in its four-fold lower heterozygosity (0.03% vs 0.12% per base pair). Accordingly, the X. malinche genome carries significantly more derived putatively deleterious alleles than does X. birchmanni (a 2.5% excess, following 33; 21, 26). As a result of this difference, the three hybrid populations of swordtail fish provide an informative contrast: whereas BDMIs should lead to selection against minor parent ancestry in all three populations, hybridization load should favor the major parent in the first two populations (Totonicapa and Aguazarca) and the minor parent in the third (Tlatemaco; Fig. 2C, Fig. 3C). The fact then that minor parent ancestry also increases with recombination in the Tlatemaco population (Fig. 2C) indicates that hybrid incompatibilities are the dominant source of selection, rather than hybridization load (Fig. 3C, Fig. S7; 21). In principle, ecological selection favoring the major parent could also produce a positive correlation between recombination and minor parent ancestry (but not positive correlations in ancestry between populations; Fig. 3A,B). However, this explanation would require two of the hybrid populations to occur in more birchmanni-like environments and one in a more malinche-like environment, when available evidence suggests otherwise (Fig. 2B; (21)).
Finally, in all populations, minor parent ancestry is unusually low near previously mapped putative BDMIs (22, 23). Lower minor parent ancestry does not result from the approach used to identify BDMIs (21), but is expected from selection on epistatically-interacting alleles (Fig. 3D; 21). Together, these lines of evidence indicate that BDMIs are the predominant—though not necessarily sole—source of selection filtering minor parent ancestry in these three swordtail hybrid populations (Fig. 3C).
To explore the generality of these relationships, we considered admixture between humans and archaic hominins. Several studies have reported that Neanderthal ancestry tends to decrease with the number of linked coding base pairs and with a measure of purifying selection at linked sites (4, 6, 11), patterns for which both BDMIs and hybridization load—due to the smaller effective population size of Neanderthals (27)—have been proposed as explanations (4, 6, 11). Reanalyzing the data, we found that the proportion of Neanderthal ancestry decreases in regions of the human genome with lower recombination rates (Fig. 1D; Table S2; Table S4). This relationship is seen using three different approaches to infer Neanderthal ancestry (Table S2) and is not explained by variation in power to identify introgression or the number of coding base pairs nearby (Table S2; 21). Repeating these analyses for Denisovan ancestry, we obtained the same pattern (Fig. 1D; Table S2; 21).
As in swordtails, the persistence of archaic hominin ancestry in regions of higher recombination is not expected under neutrality (Fig. S1). However, our conclusion about the source of selection reached for swordtails need not hold for hominins: a priori, because modern humans were less diverged from Neanderthals and Denisovans when they interbred (28), and because plausible models of hybridization load have been shown to provide a good fit to the distribution of Neanderthal ancestry in the human genome (6).
In summary, minor parent ancestry is predicted by the local recombination rate across three replicate admixture events in swordtails, as well as in two cases of admixture in hominins. Together with earlier indications in other species (18–20), our findings show the distribution of minor parent ancestry to be at least in part predictable from genomic features.
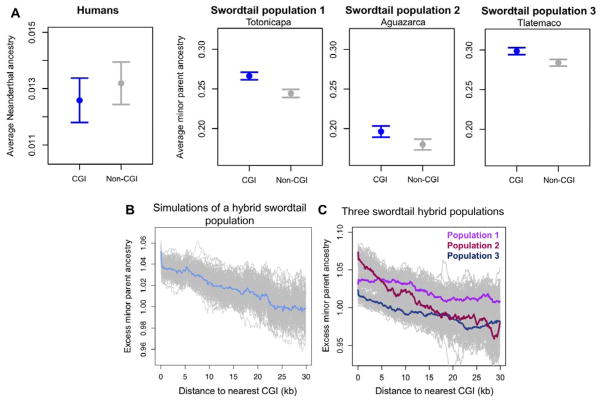
Knowledge of local recombination should therefore provide a guide as to where in the genome minor parent ancestry is expected to be highest. In hominins, meiotic recombination is directed to the genome by binding of the PRDM9 gene; in swordtails, it is not and tends to occur near promoter-like features (21, 25). Accordingly, minor parent ancestry is higher around such features in swordtails but not in humans (Fig. 4A,B; 21). Thus, the mechanism by which recombination is directed to the genome impacts the distribution of minor parent ancestry.
Figure 4. The recombination mechanism shapes the distribution of minor parent ancestry.
(A) Neanderthal ancestry is not elevated in 50 kb windows that overlap with CpG islands (CGIs), when compared to windows that do not, but have similar GC content. The fold difference λ is 0.95 (p=0.91; see 21). The same analysis in swordtail hybrids reveals that minor parent ancestry is higher in windows that overlap CGIs (pop. 1, λ=1.09, p<0.005; pop. 2, λ=1.09, p<0.005; pop. 3, λ=1.02, p<0.005). Points show the mean and whiskers indicate two standard errors of the mean obtained by 1,000 joint bootstraps. (B) Simulations of incompatibility selection in swordtails predict higher minor parent ancestry near CGIs. (C) This prediction is met for all hybrid populations. In B and C, gray lines show results of 500 replicate simulations bootstrapping 5 kb windows; colored lines indicate the mean of all replicates in sliding 5 kb windows.
One implication is that the reliance on PRDM9 to direct recombination may not only impact reproductive isolation between species directly (as in mice, 29), but also indirectly. For example, if epistatic interactions often occur between regulatory and coding regions, hybrids between species with recombination concentrated in promoter-like regions may experience greater negative selection due to BDMIs but more opportunities for adaptive introgression. As genomic data accumulate for hybridizing species across the tree of life, the impact of recombination mechanisms on the fate of hybrids can soon be evaluated systematically.
Supplementary Material
Acknowledgments
We thank Y. Brandvain, E. Calfee, G. Coop, J. Pickrell, J. Pritchard, D. Reich, G. Sella, S. Singhal, M. Steinrücken and members of the Przeworski and Sella labs for helpful discussions and/or comments. We thank the Federal Government of Mexico for permission to collect fish. Funding: This project was supported by R01 GM83098 grant to MP, NSF DEB-1405232, HHMI Hanna H. Gray Fellowship, and Milton grant to MS, and grant from the Cancer Prevention Research Institute of Texas to GGR. Author contributions: MS and MP designed the project and wrote the manuscript; MS, DP, CH, LS, JCB collected data; MS, CX, AD, SS performed analyses; PA and GGR provided expertise and technical support. Competing interests: The authors declare no competing interests. Data and Materials Availability: Data are available through NCBI (SRA-SRP130891; SRP018918) and Dryad (doi: XXX).
Footnotes
References and Notes
- 1.Coyne JA, Orr HA. Speciation. Sinaeur Associates; Sunderland, MA: 2004. [Google Scholar]
- 2.Rieseberg LH, Whitton J, Gardner K. Genetics. 1999;152:713–727. doi: 10.1093/genetics/152.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cui R, et al. Evolution. 2013;67:2166–2179. doi: 10.1111/evo.12099. [DOI] [PubMed] [Google Scholar]
- 4.Sankararaman S, et al. Nature. 2014;507:354–357. doi: 10.1038/nature12961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jacobsen F, Omland KE. Mol Ecol. 2011;20:2236–2239. doi: 10.1111/j.1365-294x.2011.05120.x. [DOI] [PubMed] [Google Scholar]
- 6.Juric I, Aeschbacher S, Coop G. Plos Genet. 2016;12 doi: 10.1371/journal.pgen.1006340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schumer M, Cui R, Powell DL, Rosenthal GG, Andolfatto P. Mol Ecol. 2016;25:2661–2679. doi: 10.1111/mec.13602. [DOI] [PubMed] [Google Scholar]
- 8.Masly JP, Presgraves DC. PLoS Biol. 2007;5:1890–1898. doi: 10.1371/journal.pbio.0050243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bomblies K, et al. PLoS Biol. 2007;5:1962–1972. doi: 10.1371/journal.pbio.0050236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee HY, et al. Cell. 2008;135:1065–1073. doi: 10.1016/j.cell.2008.10.047. [DOI] [PubMed] [Google Scholar]
- 11.Harris K, Nielsen R. Genetics. 2016;203:881–891. doi: 10.1534/genetics.116.186890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bierne N, Lenormand T, Bonhomme F, David P. Genet Res. 2002;80:197–204. doi: 10.1017/s001667230200592x. [DOI] [PubMed] [Google Scholar]
- 13.Rieseberg LH, et al. Science. 2003;301:1211–1216. doi: 10.1126/science.1086949. [DOI] [PubMed] [Google Scholar]
- 14.Arnegard ME, et al. Nature. 2014;511:307. doi: 10.1038/nature13301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu CI. J Evol Biol. 2001;14:851–865. [Google Scholar]
- 16.Noor MAF, Grams KL, Bertucci LA, Reiland J. PNAS. 2001;98:12084–12088. doi: 10.1073/pnas.221274498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nachman MW, Payseur BA. Phil Trans Royal Soc B. 2012;367:409–421. doi: 10.1098/rstb.2011.0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brandvain Y, Kenney AM, Flagel L, Coop G, Sweigart AL. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carneiro M, Ferrand N, Nachman MW. Genetics. 2009;181:593–606. doi: 10.1534/genetics.108.096826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geraldes A, Basset P, Smith KL, Nachman MW. Mol Ecol. 2011;20:4722–4736. doi: 10.1111/j.1365-294X.2011.05285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Supporting Online Materials
- 22.Schumer M, et al. eLife. 2014;3 doi: 10.7554/eLife.02535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schumer M, Brandvain Y. Mol Ecol. 2016;25:2577–2591. doi: 10.1111/mec.13641. [DOI] [PubMed] [Google Scholar]
- 24.Andolfatto P, et al. Genome Res. 2011;21:610–617. doi: 10.1101/gr.115402.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baker Z, et al. eLife. 2017;6 doi: 10.7554/eLife.24133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Do R, et al. Nature Genetics. 2015;47:126–131. doi: 10.1038/ng.3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prufer K, et al. Nature. 2014;505:43. doi: 10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orr HA, Turelli M. Evolution. 2001;55:1085–1094. doi: 10.1111/j.0014-3820.2001.tb00628.x. [DOI] [PubMed] [Google Scholar]
- 29.Davies B, et al. Nature. 2016;530:171. doi: 10.1038/nature16931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Picelli S, et al. Genome Res. 2014;24:2033–2040. doi: 10.1101/gr.177881.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schumer M, Cui R, Rosenthal G, Andolfatto P. Mol Ecol Res. 2015 doi: 10.1111/1755-0998.12434. [DOI] [PubMed] [Google Scholar]
- 32.Schumer M, et al. PNAS. 2017;114:10936–10941. doi: 10.1073/pnas.1711238114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pickrell JK, Pritchard JK. PLoS Genet. 2012;8:e1002967–e1002967. doi: 10.1371/journal.pgen.1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schartl M, et al. Nat Genet. 2013;45:567–U150. doi: 10.1038/ng.2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quail MA, Swerdlow H, Turner DJ. In: Current protocols in human genetics. Haines JL, et al., editors. Vol. 18. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li JW, et al. Nucleic Acids Res. 2012;40:D1313–D1317. doi: 10.1093/nar/gkr1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Amores A, et al. Genetics. 2014;197:625–U307. doi: 10.1534/genetics.114.164293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H, Durbin R. Bioinformatics. 2009;25 doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McKenna A, et al. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Purcell S, et al. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Altshuler D, et al. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chan AH, Jenkins PA, Song YS. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1003090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stamatakis A. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 44.Siepel A, Haussler D. Mol Biol Evol. 2004;21:468–488. doi: 10.1093/molbev/msh039. [DOI] [PubMed] [Google Scholar]
- 45.Chen GK, Marjoram P, Wall JD. Genome Res. 2009;19:136–142. doi: 10.1101/gr.083634.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schumer M, et al. Evolution. 2013;67:1155–1168. doi: 10.1111/evo.12009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rambaut A, Grassly NC. Comput Appl Biosci. 1997;13 doi: 10.1093/bioinformatics/13.3.235. [DOI] [PubMed] [Google Scholar]
- 48.O’Connell J, et al. PLoS Genet. 2014;10 [Google Scholar]
- 49.Howie BN, Donnelly P, Marchini J. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Williams AL, Housman DE, Rinard MC, Gifford DK. Genome Biol. 2010;11 doi: 10.1186/gb-2010-11-10-r108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Auton A, McVean G. Genome Res. 2007;17:1219–1227. doi: 10.1101/gr.6386707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Feng CG, et al. eLife. 2017;6 [Google Scholar]
- 53.Wu CI, Li WH. PNAS. 1985;82:1741–1745. doi: 10.1073/pnas.82.6.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li H, Durbin R. Nature. 2011;475:493–U484. doi: 10.1038/nature10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Walling CA, Royle NJ, Metcalfe NB, Lindstrom J. Biol Lett. 2007;3:144–146. doi: 10.1098/rsbl.2006.0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kamm JA, Spence JP, Chan J, Song YS. Genetics. 2016;203:1381–1399. doi: 10.1534/genetics.115.184820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Simons YB, Sella G. Curr Opin Genetics Dev. 2016;41:150–158. doi: 10.1016/j.gde.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cui RF, Schumer M, Rosenthal GG. Bioinformatics. 2016;32:1103–1105. doi: 10.1093/bioinformatics/btv700. [DOI] [PubMed] [Google Scholar]
- 59.Schumer M, Cui R, Rosenthal GG, Andolfatto P. PLoS Genet. 2015;11:5041–5041. doi: 10.1371/journal.pgen.1005041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Presgraves DC. Genetics. 2003;163:955–972. doi: 10.1093/genetics/163.3.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sweigart AL. Genetics. 2010;184:779–U225. doi: 10.1534/genetics.109.112896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maheshwari S, Barbash DA. Ann Rev Genet. 2011;45:331–355. doi: 10.1146/annurev-genet-110410-132514. [DOI] [PubMed] [Google Scholar]
- 63.Culumber ZW, et al. Mol Ecol. 2011;20:342–356. doi: 10.1111/j.1365-294X.2010.04949.x. [DOI] [PubMed] [Google Scholar]
- 64.Culumber ZW, Shepard DB, Coleman SW, Rosenthal GG, Tobler M. J Evol Biol. 2012;25:1800–1814. doi: 10.1111/j.1420-9101.2012.02562.x. [DOI] [PubMed] [Google Scholar]
- 65.Yang Y, Cao W, Wu S, Qian W. Mol Biol Evol. 2017;34:3254–3266. doi: 10.1093/molbev/msx264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hurst LD, Pal C, Lercher MJ. Nat Rev Genet. 2004;5:299–310. doi: 10.1038/nrg1319. [DOI] [PubMed] [Google Scholar]
- 67.Quinlan AR, Hall IM. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Neph S, et al. Bioinformatics. 2012;28:1919–1920. doi: 10.1093/bioinformatics/bts277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Singhal S, et al. Science. 2015;350:928–932. doi: 10.1126/science.aad0843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lam I, Keeney S. Science. 2015;350:932–937. doi: 10.1126/science.aad0814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Persikov AV, Singh M. Nucleic Acids Res. 2014;42:97–108. doi: 10.1093/nar/gkt890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bailey TL, et al. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gravel S. Genetics. 2012;191:607–619. doi: 10.1534/genetics.112.139808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sankararaman S, Patterson N, Li H, Paeaebo S, Reich D. Plos Genetics. 2012;8 doi: 10.1371/journal.pgen.1002947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sankararaman S, Mallick S, Patterson N, Reich D. Curr Biol. 2016;26:1241–1247. doi: 10.1016/j.cub.2016.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McVicker G, Gordon D, Davis C, Green P. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dannemann M, Andres AM, Kelso J. Am J Hum Genet. 2016;98:399–399. doi: 10.1016/j.ajhg.2016.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Enard D, Petrov D. 2017 bioRxiv. [Google Scholar]
- 79.Steinrücken M, Kamm J, Song Y. 2017 bioRxiv. [Google Scholar]
- 80.Steinrücken M, Spence J, Kamm J, Wieczorek E, Song Y. 2017 bioRxiv. [Google Scholar]
- 81.Hinch AG, et al. Nature. 2011;476:170–U167. doi: 10.1038/nature10336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stevison LS, et al. Mol Biol Evol. 2016;33:928–945. doi: 10.1093/molbev/msv331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Auton A, et al. Science. 2012;336:193–198. doi: 10.1126/science.1216872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Racimo F, Sankararaman S, Nielsen R, Huerta-Sanchez E. Nat Rev Genet. 2015;16:359–371. doi: 10.1038/nrg3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vernot B, Akey JM. Science. 2014;343:1017–1021. doi: 10.1126/science.1245938. [DOI] [PubMed] [Google Scholar]
- 86.Vernot B, et al. Science. 2016;352:235–239. doi: 10.1126/science.aad9416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Green RE, et al. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gavrilets S. Evolution. 1997;51:1027–1035. doi: 10.1111/j.1558-5646.1997.tb03949.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.