Abstract
The recent genomic revolution, characterised by surges in the number of available genetic tests and known genetic associations, calls for improved genetic literacy amongst medical scientists and clinicians. This has been driven by next generation sequencing, a technology allowing multiple genes to be sequenced in parallel, thereby reducing the time and financial costs associated with genetic testing in both research and clinical settings. Endocrinology is an intuitive setting in which to consider the principles of genetic testing because endocrine disorders are due to defects in circumscribed pathways, providing clues to candidate genes. This article discusses genetic testing in contemporary endocrine practice with reference to examples of endocrine genetic disorders or multisystem genetic disorders with endocrine manifestations. Monogenic disorders are prioritised as these form the bulk of endocrine genetic disorders and the associated genetic testing is readily understandable, clinically available and practice-changing. Although it remains true that genetic testing should be embarked upon only if the result will alter management, the clinical utility of genetic testing is often underestimated and there are expanding indications for genetic testing across all areas of endocrinology.
Introduction
Cytogenetic testing in the late 1950s led to the discovery of relatively common chromosomal disorders such as Turner’s1 and Klinefelter’s syndromes.2 In the 1980s and 1990s, a combination of linkage and positional cloning followed by Sanger sequencing identified the causative mutations in many recognised familial disorders, including: tumour disorders such as multiple endocrine neoplasia (MEN) type 1 (MEN1),3 MEN2A,4 MEN2B (also known as MEN3)5 and von Hippel-Lindau (vHL) syndrome;6 hormone biosynthetic defect disorders such as the congenital adrenal hyperplasias (CAH)7 and glucocorticoid suppressible hyperaldosteronism (familial hyperaldosteronism type 1);8 autoimmunity through defective self-recognition by T cells in autoimmune polyendocrinopathy, candidiasis and ectodermal dystrophy (APECED) syndrome;9 and hormone receptor defects such as familial hypocalciuric hypercalcaemia (FHH)10 and thyroid hormone resistance.11 Completion of the human genome project, astute phenotyping of familial disorders and the introduction of next generation sequencing (NGS; also known as massively parallel sequencing) then unravelled numerous less common endocrine disorders, including some where a familial basis was not initially recognised, such as many of the familial phaeochromocytoma-paraganglioma (PPGL) syndromes,12 AIP-related familial isolated pituitary adenomas (FIPA),13 and ARMC5-related bilateral macronodular adrenal hyperplasia (BMAH).14
This review provides a contemporary clinical and laboratory framework for genetic testing in endocrinology. We focus on monogenic disorders as these disorders are suitable for assessment by current genetic testing methodologies; specifically, cytogenetic tests such as karyotype and single nucleotide polymorphism (SNP) array looking for genetic variation at the chromosomal level, and molecular tests including Sanger sequencing of single genes and NGS of multiple or all genes. Although individually uncommon, endocrine genetic disorders together constitute a significant proportion of endocrinology. More common endocrine disorders such as type 2 diabetes and osteoporosis are polyallelic but testing is not feasible for clinical purposes yet.
Clinical Evaluation
Genetic testing should be preceded by comprehensive phenotyping to determine the likelihood of finding a genetic disorder and to identify candidate genes. This staged form of patient evaluation is critical in avoiding variants of unknown significance which may be regarded as the ‘incidentalomas’ of genetic testing.
The decision to pursue genetic testing should be an index of how likely a disorder is to be genetic multiplied by how likely a genetic result would be to alter management of the individual or their family (Table 1). For example, germline RET mutations are found in only 7% of patients with sporadic medullary thyroid cancer and yet testing should be considered in all affected individuals as finding a mutation in the RET proto-oncogene, and thereby diagnosing MEN2, allows for testing of family members and lifesaving prophylactic thyroidectomy in those who have inherited the mutation.15
Table 1.
Examples of indications for genetic testing in endocrinology.
| Indication | Clinical example | Informative result | Benefit of result |
|---|---|---|---|
| Differentiation or confirmation of clinical disorders | Mild, longstanding PTH-dependent hypercalcaemia and intermediate urinary calcium creatinine clearance ratio | CASR mutation | Diagnosis of familial hypocalciuric hypercalcaemia which does not necessitate treatment, rather than primary hyperparathyroidism which is treated surgically |
| Disease monitoring | Sporadic medullary thyroid cancer | RET mutation | Rationale for current and ongoing assessment for other MEN2-related tumours |
| Therapeutic guidance | Lean individual with longstanding diabetes mellitus, glycosuria and a positive family history of diabetes | HNF1A mutation | Confirmation of sulphonylurea-sensitive subtype of monogenic diabetes to facilitate safe and appropriate insulin withdrawal and replacement with a sulphonylurea |
| Prognostication | Lean individual with longstanding mild hyperglycaemia | GCK mutation | Diagnosis of GCK diabetes which is non-progressive and not associated with vascular complications, therefore not requiring treatment |
| Family testing primarily to guide surveillance | Clinical MEN1 syndrome | MEN1 mutation | Possibility of targeted mutation testing in presymptomatic at-risk family members to obviate the need for tumour surveillance in those who test negative |
| Family testing primarily to guide management | Clinical MEN2 syndrome | RET mutation | Possibility of targeted mutation testing in family members to facilitate prophylactic thyroidectomy |
| Family planning | Biochemically diagnosed non-classical congenital adrenal hyperplasia | One classic mutation and one variant allele in CYP21A2 | Risk of classic congenital adrenal hyperplasia where the partner also carries a classic mutation in CYP21A2 may be overcome by IVF and preimplantation genetic diagnosis |
Some endocrine genetic disorders have well known, characteristic patterns, whilst others are exceedingly rare. It is impossible to know all genetic disorders but generic clues may be useful, such as young-onset and/or aggressive disease or the development of multiple rare disorders within an individual, or one or more rare disorders within multiple family members. An autosomal dominant family history of sulphonylurea-responsive diabetes mellitus in a lean individual with glycosuria immediately brings to mind HNF1A-related monogenic diabetes. GATA6-related monogenic diabetes is little known by comparison but the phenotype of pancreatic agenesis, congenital heart disease and other congenital malformations should raise suspicion for a genetic aetiology worthy of evaluation. Skin manifestations may hint at heritable syndromes in patients with apparently sporadic tumours. For instance, the classic interscapular rash of cutaneous lichen amyloidosis in a patient with apparently sporadic medullary thyroid cancer, phaeochromocytoma or primary hyperparathyroidism should prompt consideration of MEN2.16
The primary aim of the family history is to determine whether the disorder in the presenting patient (the ‘proband’) is likely to be genetic. Broad questions may be illuminating. For example, a family history of primary hyperparathyroidism may be represented only by affected members having fractures and renal calculi. Extending to a three-generation pedigree is critical in identifying disorders with reduced penetrance. For example, affected relatives with familial isolated pituitary adenoma syndrome are not uncommonly separated by unaffected relatives.17 Consanguinity between the parents of an affected individual is an important clue to autosomal recessive disorders. If genetic testing has been deemed to be useful, a secondary aim of the family history is to determine which family member should be tested first. Environmental risk factors should be sought in each individual to identify alternative explanations for disease predisposition. In a family suspected to have monogenic diabetes, an obese individual may be the presenting patient and they may ultimately be found to carry the genetic mutation responsible for the family’s diabetes, but the patient may alternatively transpire to be a ‘phenocopy’ with hyperglycaemia due to sporadically occurring type 2 diabetes related to their obesity rather than the familial mutation. Testing a lean family member would be more worthwhile because of their greater likelihood of having the family’s genetic disorder rather than type 2 diabetes.
Non-genetic investigations may guide genetic testing. Hypercalcaemia, suggesting concomitant primary hyperparathyroidism, should prompt consideration of MEN2 in patients with phaeochromocytoma or medullary thyroid cancer, or MEN1 in patients with pituitary adenoma or gastroenteropancreatic neuroendocrine tumours. Negative succinate dehydrogenase subunit B (SDHB) immunohistochemical staining in PPGL specimens suggests a loss-of-function mutation in an SDHx gene (SDHA, SDHB, SDHC or SDHD),18 whilst negative parafibromin immunohistochemical staining in parathyroid adenomas suggests a loss-of-function mutation in CDC73 (formerly HRPT2) which may cause familial hyperparathyroidism or hyperparathyroidism-jaw tumour syndrome. Phenotypic tests may at other times be sufficient for diagnosis. For example, classical or non-classical congenital adrenal hyperplasia due to 21-hydroxylase deficiency may be unequivocally diagnosed using basal or ACTH-stimulated 17-hydroxyprogesterone levels, respectively; however, subsequent sequencing of the CYP21A2 gene may still be indicated to guide reproductive planning.
Genetic Counselling
Genetic disorders may be associated with feelings of fear, despair and guilt which should be addressed in an empathetic, non-directive manner before and after genetic testing. Pretest counselling should address the utility of genetic testing specific to the patient and their disorder. Detecting a ‘premutation’ (55–199 CGG repeats) in FMR1 in a woman with premature ovarian insufficiency not only provides an aetiological answer to her presentation, but also allows screening of at-risk family members and informs reproductive planning. The latter is pertinent as the trinucleotide repeat region is unstable and prone to inter-generation expansion, particularly during maternal inheritance, whereby expansion to a full mutation (>200 CGG repeats) in male offspring produces fragile X syndrome including intellectual disability.19 By contrast, the primary aim of genetic testing in an older patient with medullary thyroid cancer may be facilitation of family testing.
These benefits must be balanced against the potential drawbacks of genetic testing, including the discovery of unexpected health risks or information on parentage. As in the identification of a RET mutation in an individual with medullary thyroid cancer which reclassifies their disease as MEN2, the health risks may be partly related to the genetic disorder, or they may be completely unrelated if a mutation is incidentally found in an unrelated gene such as BRCA1/2. Patients should be informed that results may have implications for family members; however, this is often the desired consequence of genetic testing and family members will not automatically know their own genetic status despite learning the results of their relatives. Exceptions to this include patients being informed of a germline genetic result in their monozygotic twin or in both their affected parent and their offspring in an autosomal dominant disorder. In Australia, genetic results may impact upon certain types of insurance such as life insurance, income protection and mortgage protection. Positive screening for the familial mutation in seemingly unaffected family members may impact their ability to obtain such insurance, but for affected individuals, their history of the disorder in question likely bears greater influence on their ability to obtain insurance. Policies that are already established and not due for renewal are usually unaffected, as is private health cover. Genetic test results may also impose employment restrictions in occupations where maximum physical fitness is required or comprehensive health insurance is integrated with the employment, such as the armed services. Specimen collection, DNA storage protocols, opportunity for consent withdrawal and the protection of confidentiality should be discussed, and patients should be informed that genetic testing may not reveal the genetic cause of even the most clearly inherited disorder.
Laboratories differ but patients are usually required to sign a consent form before genetic testing can proceed. Depending on the consent form, patients may be given the option to know only results immediately relevant to their disorder or all results, including variants of unknown significance and results that are unrelated to the disorder in question. Some consent forms provide the option for patients to know all unrelated results or only medically actionable results where there are options for prevention, early diagnosis or treatment. The distress following notification of a non-actionable mutation with severe health consequences must be considered. Counselling patients for genetic tests involving multiple genes is more complex because of the increasing risk of these incidental findings. In most instances, such counselling should be performed by genetic counsellors and/or clinical geneticists, whereas limited testing of single genes or mutations may be within the scope of practice of non-geneticist clinicians.
Genetic Testing
The choice of genetic testing methodology is dictated by clinical, laboratory and funding circumstances. For example, in the current Medicare-unassisted environment, requesting clinicians may request Sanger sequencing of an individual gene or staged Sanger sequencing of multiple genes based on clinical suspicion because the initial costs are less, even though upfront NGS to analyse multiple genes simultaneously may be more informative and less expensive overall. On the other hand, laboratories may prefer NGS requests as this allows for the testing of multiple patient samples from disparate clinical settings and, potentially, economies of scale. The final decision of which test to pursue is usually made by the requesting clinician with relevant laboratory input.
Testing for monogenic disorders utilises germline DNA extracted from peripheral blood leucocytes, hair follicles or buccal swabs. Which genetic test to then perform depends on the anticipated genetic cause of the disorder in question. Online tools can assist in collating potential genetic causes (e.g. Online Mendelian Inheritance in Man database20 and GeneReviews21) and clinically available tests (e.g. Genetic Testing Registry22 and RCPA Genetic Tests and Laboratories Website23).
The most common cytogenetic test in the endocrine setting is karyotyping to diagnose aneuploid disorders such as Turner’s syndrome (45,X) and Klinefelter’s syndrome (45,XXY). Increasing the number of cells analysed may reveal mosaicism with therapeutic ramifications. The presence of the Y chromosome in all or any cells in phenotypic females poses a risk of gonadoblastoma, whilst monosomy X in mosaicism with a normal female karyotype (45,X/46,XX) is associated with milder ovarian insufficiency, sometimes allowing for menarche and even spontaneous conception. Karyotyping in disorders of sexual development may be complemented by fluorescent in situ hybridisation (FISH), which employs probes to identify and quantify relevant genes such as SRY on the Y chromosome. A positive FISH result in this case indicates the presence of the Y chromosome or translocation of the SRY gene onto the X chromosome, the latter being the most common cause of 46,XX complete gonadal dysgenesis. SNP array is a cytogenetic test with resolution between karyotype and sequencing methodologies, identifying copy number variants (CNVs) to an approximate resolution of 0.2 MB (200,000 bp). Endocrine disorders that are well suited to investigation by SNP array include Prader-Willi syndrome (15q11.2 microdeletion), DiGeorge syndrome (22q11.2 microdeletion) and, more recently, X-linked acrogigantism (XLAG; Xq26.3 microduplication).24
Molecular testing is based on nucleotide variation and typically involves gene sequencing either by Sanger sequencing, which is a sequencing-by-termination method targeting an individual gene, or NGS, usually via a sequencing-by-synthesis platform. NGS begins with DNA fragmentation and amplification of these DNA fragments into thousands of copies which can be simultaneously sequenced. NGS thus enables parallel sequencing of multiple genes in a cost-effective manner, although bioinformatic costs rise incrementally with the number of genes analysed. Sanger sequencing costs are dependent on the number of amplicons (continuous DNA segments) required. Because each exon requires one or more amplicons, sequencing costs rise with gene size and exon number. If a disorder is caused by a few known mutations, targeted mutation analysis is most cost-effective with Sanger sequencing of only the regions immediately flanking each mutation of interest. For example, the activating M918T mutation in the RET proto-oncogene accounts for 95% of cases of MEN2B, where patients have a Marfanoid body habitus and mucosal neuromas in addition to the typical MEN2 predisposition to medullary thyroid cancer and phaeochromocytoma.15 Targeted mutation analysis for M918T is particularly time- and cost-efficient in this scenario as RET is a large gene consisting of 20 exons. MEN1, in contrast, occurs due to loss-of-function mutations which may arise anywhere along the MEN1 gene, mandating sequencing of the entire gene. NGS may be preferred in the evaluation of MEN1 not only because the whole gene must be assessed, but also because differential diagnoses in this phenotype include CDKN1B mutations resulting in MEN4, which is clinically indistinguishable from MEN1, and AIP mutations resulting in FIPA.17 Another use of targeted mutation analysis is in relatives at risk of a known familial mutation. Whilst it may take weeks or months to initially find the culprit mutation in a proband, the turnaround time is closer to a fortnight to determine the status of family members subsequently tested by targeted mutation analysis using Sanger sequencing.
NGS is well suited to the investigation of disorders with multiple potential genetic causes, which now include the majority of endocrine genetic disorders, including most endocrine tumour syndromes, disorders of sexual development, congenital hypopituitarism, monogenic diabetes and hypophosphataemic rickets. Even single gene disorders may prove to have multiple genetic causes. For instance, ARMC5 is presently the only gene implicated in familial Cushing’s syndrome due to bilateral macronodular adrenal hyperplasia (BMAH) but some families do not harbour ARMC5 sequencing variants and research is ongoing to discover other causative genes.25 NGS may also elucidate digenic disorders where genetic mutations in two genes act synergistically, as hypothesised in cases of Kallman’s syndrome, familial pituitary tumour syndrome and pituitary stalk interruption syndrome.17,26,27 NGS may be performed in the form of a panel of genes based on a group of disorders, such as metabolic bone diseases, or a single disorder, such as pituitary adenoma. Such panels are cost- and time-efficient but they do not accommodate ongoing genetic discoveries. A carefully designed and optimised genetic panel immediately becomes outdated following the discovery of a previously unknown causative gene.17 Virtual panels, whereby a much wider panel or all coding regions are sequenced but only the genes of interest are interrogated, may overcome this problem as it is a comparatively simpler task to return to the data and analyse a new gene of interest than to return to the patient’s stored DNA to perform another genetic test.28 An alternative to NGS standard or virtual panels is staged gene Sanger sequencing whereby the most likely culprit gene is sequenced first, followed by genes of decreasing probability, but this may ultimately be more expensive than NGS depending on when the causative mutation is found.17
If all genes are to be sequenced and analysed, this may either be in the form of whole genome sequencing (WGS), which covers all coding and non-coding regions, or whole exome sequencing (WES), which is restricted to coding regions. WGS is more expensive but provides insight into vast lengths of untranslated regions (UTR) and potential enhancer, promoter and repressor sites. WES is often considered more cost-effective as it is limited to the most understandable genetic data in contemporary practice and it typically provides greater depth of coverage of the coding regions of interest, though this varies by platform. These broader sequencing methodologies are advantageous where there is ongoing research into novel candidate genes in addition to multiple known susceptibility genes. WGS additionally provides information about copy number variants when analysed using appropriate bioinformatic pipelines.29
There are various pitfalls in sequencing (Table 2). False negative results may occur in the setting of large insertions/deletions (referred to as ‘indels’). Sanger sequencing may identify indels up to the size of an amplicon (approximately 300 bp), whilst NGS panels and standard WES may identify deletions up to approximately 400 bp and insertions up to approximately 50 bp. The precise thresholds depend on the mapper, variant caller, read length and DNA fragment in question. Larger indels may be identified by multiplex ligation-dependent probe amplification (MLPA) using commercially obtained reagents and probe-mixes to produce electrophoresis peaks which may be smaller or larger than reference samples, indicating deletions or insertions, respectively. This may be performed routinely in disorders where CNVs are a recognised cause, such as Von Hippel-Lindau syndrome where exonic or whole gene VHL deletions are found in 20–30% of cases,30 or it may be done when sequencing variants are not identified despite a highly suspicious phenotype. SNP array and quantitative PCR are other methods of identifying CNVs. False negatives may also arise if a point mutation is located in an area not sequenced – for instance, in another gene when only certain genes are sequenced or in a deep intronic area in any sequencing methodology apart from WGS as Sanger sequencing, NGS panels and WES generally only sequence exons and <10 nucleotides flanking each exon. If suspected, intronic mutations may be detected by mRNA reverse transcription and amplification (RT-PCR). This is valuable in neurofibromatosis type 1, where 2–3% of cases are associated with deep intronic NF1 variants affecting splicing with abnormal mRNA results despite normal sequencing results.31 Both VHL and NF1 are relevant in endocrinology because of their associations with phaeochromocytoma. Other than WGS, sequencing may also fail to detect promoter mutations as found in the PTEN gene in up to 10% of patients with Cowden syndrome, characterised by thyroid, endometrial and breast neoplasia, and hamartomas.32
Table 2.
Molecular genetic methodologies according to expected ability to detect abnormalities in different genetic settings.
| Next generation sequencing | ||||||
|---|---|---|---|---|---|---|
| Abnormality | Sanger sequencing | MLPA | Panel testing# | Whole exome testing | Whole genome testing | Long-read sequencing |
| Small indels (<50 bp) | Y | Y | Y | Y | Y | Y |
| Medium indels (50–300 bp) | M* | Y | M* | M* | Y | Y |
| Large indels (>300 bp) | N | Y | N | N | Y | Y |
| Mutation in unsuspected gene | N | N | N | Y | Y | Y |
| Intronic mutation | M** | N | M** | M** | Y | Y |
| Promoter mutation | M** | N | M** | M** | Y | Y |
| Pseudogenes | M* | Y | N | N | N | M* |
| GC-rich genes | M* | N | N | N | N | Y |
| Triplet repeat disorders | M* | N | N | N | N | Y |
A selected panel of genes which are sequenced and analysed, does not include ‘virtual panels’ where all genes may be sequenced with only a selected panel of genes being analysed;
depends on length;
depends on location;
Y, abnormality likely to be detected; M, abnormality may be detected; N, abnormality likely to be missed.
Sequencing may produce false positive results if the bioinformatic analysis of variants is not rigorous, leading to misclassification of a benign polymorphism as a disease-causing mutation. False positives may also arise in pseudogenes. These are genes which share a similar sequence to one or more other genes. Such pseudogenes may be mistaken for one another on NGS which has lower specificity for a given nucleotide sequence than Sanger sequencing. Clinically relevant examples include CYP21A2 which is associated with congenital adrenal hyperplasia and PMS2 associated with Lynch syndrome where NGS may incorrectly call variants in related genes as being variants in the genes of interest. Sanger sequencing with amplicons targeting areas of difference between the target gene and the respective pseudogenes usually overcomes this issue. Another indication for Sanger sequencing is guanine-cytosine (GC) rich genes as the three hydrogen bonds between these two nucleotides lead to high melting points, secondary structures and ultimately poor amplification in the automated process of NGS. One such gene is GATA6, which causes pancreatic agenesis/hypoplasia and congenital heart disease and should be interrogated by Sanger sequencing in preference to the usual monogenic diabetes panels.33 False positive results may also occur with NGS of repetitive regions, making it unsuitable for the evaluation of such disorders as fragile X syndrome due to triple repeat expansion in FMR1.34
McCune-Albright syndrome is a noteworthy endocrine disorder as it is a mosaic disorder due to a heterozygous postzygotic mutation. As the mutation is not inherited, parents cannot be affected and there is no risk to offspring as the mutation is incompatible with life in the germline state – any offspring born to affected individuals will have inherited the wild-type GNAS allele. GNAS testing may still be useful in individuals with mild phenotypes like monostotic fibrous dysplasia where securing the diagnosis of McCune-Albright syndrome would prompt surveillance for the various associated endocrinopathies including precocious puberty, hyperthyroidism and growth hormone excess. If no GNAS mutations are found on DNA sequencing using peripheral blood leucocytes, then DNA should be extracted from affected tissues. As the classic café-au-lait skin lesions often fail to demonstrate the GNAS mutation due to the paucity of melanocytes, resected endocrine and other tissues are more suitable for somatic testing.35
Genome-wide association studies (GWAS) are limited to research purposes as they compare SNPs shared between affected versus unaffected individuals, which act as surrogate markers of disease status rather than being causative mutations. GWAS may provide insights into seemingly polygenic endocrine disorders such as osteoporosis, but they add little to risk prediction beyond that achieved by family history and traditional risk factors. Such tests should not be recommended to patients and if results are presented following direct-to-consumer testing, expert advice should be sought from clinical geneticists and genomic pathologists to determine if they contain any meaningful results, the accuracy of such results and whether accredited clinical testing is required.
Result Interpretation
NGS can yield tens to millions of variants, depending on the proportion of the genome sequenced, in an individual’s genetic code compared to the human reference genome. Raw data are therefore filtered by ‘bioinformatic pipelines’ which use measures such as conservation of the nucleotide position across species and biophysicochemical differences in the mutant protein to prioritise variants. As these pipelines differ by software and between laboratories, reviewing raw data in a different bioinformatic laboratory may be enlightening when the initial results are apparently negative.
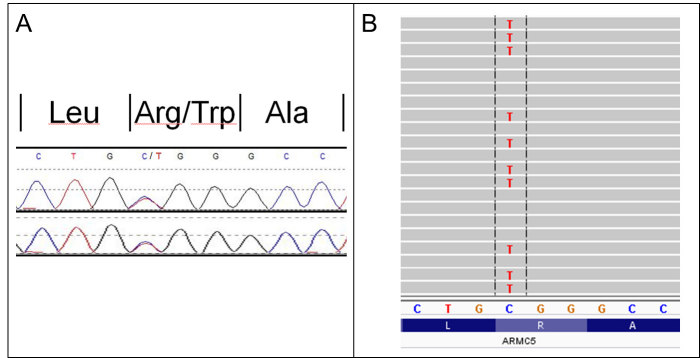
Whether detected by Sanger sequencing or NGS (Figure), variants should be classified according to the American College of Medical Genetics and Genomics (ACMG) categories: 1) benign, 2) likely benign, 3) of uncertain significance, 4) likely pathogenic, and 5) pathogenic.36 Classification is based on features incorporated into most bioinformatic pipelines. For instance, nonsense variants (premature termination codon due to nucleotide substitution) and frameshift variants (altered nucleotide reading frame due to nucleotide insertion/deletion) argue for pathogenicity whilst silent variants (nucleotide substitution with unaltered amino acid sequence) suggest the variant to be benign. Although, a silent variant may be causative if it produces aberrant splicing, which may be predicted by in silico (software) tools that are also incorporated into most bioinformatic pipelines. The ACMG criteria also include more clinical factors such as variant cosegregation in multiple affected family members, and laboratories should liaise with clinicians where appropriate. Another key criterion is the frequency of the variant in control databases. The population frequency threshold to support a variant as being benign versus pathogenic should be considered in comparison to disease frequency. A mutation causing a severe, rare autosomal dominant disorder such as septo-optic dysplasia with midline defects including congenital hypopituitarism should occur extremely infrequently or not at all in population databases. In contrast, a mutation causing a common monogenic disorder such as GCK monogenic diabetes may well be found in population databases as these do not strictly consist of purely healthy individuals. Class 5 (pathogenic) and sometimes class 4 (likely pathogenic) variants are considered to be causative of the associated disorder and may be used in further patient and family management. Class 3 variants, also known as variants of unknown significance (VUS), are particularly prevalent and difficult to elucidate in uncommon, less studied genetic disorders such as familial pituitary tumour syndromes where there is comparatively little genetic literature compared to, say, familial breast and colorectal cancer syndromes.17 Performing a functional assay of the mutated versus wild-type protein products may provide clarification, but this is often limited by access to relevant expertise and sufficient funding. Class 2 (likely benign) and 1 (benign) variants are generally regarded as negative or ‘uninformative’ results.
Figure.
Pictorial examples of genetic results yielded by different sequencing methods. The same germline ARMC5 mutation (GRCh37/hg19, Chr16:g.31476121C>T; NM_001105247.1, c.1777C>T, p.Arg593Trp) is shown following Sanger sequencing (A) and next generation sequencing (B).
Laboratory reports should be written cautiously in a way that allows incorporation of the patient’s clinical context. MEN1 genetic testing is negative in up to 20% of patients with clinical MEN1, defined as two MEN1-related tumours in an individual or one MEN1-related tumour in a first-degree relative of an individual with MEN1.37 A negative MEN1 genetic result therefore does not negate a clinical diagnosis of MEN1 and tumour surveillance should be continued. By contrast, a patient with apparently sporadic medullary thyroid cancer is diagnosed with MEN2 if an activating RET mutation is detected in their germline DNA and the patient and their family should be managed accordingly.38
Management
Result disclosure closes the loop of genetic counselling and should be performed by the requesting clinician. The category of genetic variant(s) found and implications for the patient and their family should be addressed. In all cases, genetic results should be conveyed in terms of current knowledge but genotype-phenotype relationships are often uncertain. The risk of disease associated with a particular mutated gene may fall with time as initial estimates are often based on large families with high penetrance which were amenable for discovery studies. In addition, some disorders where gene testing is negative may later be suitable for repeat testing as further causative genes are identified, as has occurred in PPGL (now associated with at least 26 genes influencing mitochondrial oxidative metabolism or tumour growth regulation), FHH (classically due to CASR mutations but now also due to GNA11 and AP2S1 mutations) and MEN4 (an MEN1-like disorder due to CDKN1B mutations).
In the case of a VUS, the affected patient should be advised that the variant does not explain their disorder currently, that testing family members for the VUS will not clarify whether they have inherited the disorder, and that only further research can discern whether the VUS is a benign polymorphism or a causative mutation. Such research includes segregation studies, but a larger body of evidence is required to upgrade a VUS to a class 5 (pathogenic) or even a class 4 (likely pathogenic) variant. It is critical that family members understand that any such testing is for the purpose of segregation analysis rather than predictive testing. Failing to convey this message may lead family members to incorrectly think they are no longer at risk of having the disorder simply because they have tested negative for the VUS and they may thus abandon potentially lifesaving surveillance. Another challenge is incidental findings in genes unrelated to the phenotype in question, which is a risk that increases with the number of genes tested. An informed pretest decision by the patient as to whether they wish to know such results is imperative.
Finding the culprit mutation provides the opportunities of family testing and reproductive planning. ‘Cascade testing’ refers to testing for a known familial mutation in at-risk family members, starting with first-degree relatives and proceeding to second-degree relatives if the linking relative has the mutation. Alternatively, family testing may be ad hoc if linking relatives are deceased, unavailable or uninterested in genetic testing, or if expedited testing is required because of emerging clinical features or for family planning. Because of issues surrounding consent, confidentiality and contact tracing, clinical genetics services remain the hub for family testing. The chance of offspring inheriting genetic disorders depends on inheritance pattern. In couples where each partner is heterozygous for a mutation associated with an autosomal recessive disorder such as APECED due to AIRE mutations, 25% of children will be homozygous for the mutation and develop the disorder, 50% of children will be heterozygous carriers and 25% will be neither affected nor carriers. In autosomal dominant disorders such as AIP-related FIPA, children of an affected individual with a heterozygous mutation will have a 50% chance of inheriting the mutation, although not all individuals with the mutation will be affected due to variable penetrance (the chance of a disease manifesting or not in an individual with the mutation). X-linked dominant disorders such as X-linked hypophosphataemic rickets due to either hemizygous or heterozygous PHEX mutations will be inherited by all daughters and no sons of an affected man and by half of all daughters and sons of an affected female. Imprinting refers to silencing of an allele depending on the parent of origin. An example of maternal imprinting is SDHD-related PPGL where 50% of offspring will be affected if born to an affected father, or unaffected carriers if born to an affected mother. Mitochondrial DNA mutations – such as m.3243A>G which may cause maternally inherited diabetes and deafness – are exclusively inherited from the maternal side and disease manifestation depends on overall and tissue-specific mutant load. However, mitochondrial disorders may also arise due to nuclear gene defects with either autosomal recessive or dominant inheritance. The health consequences for an individual carrying the causative mutation depends on penetrance as well as expressivity (the degree of disease manifestation in an individual with the mutation).
An alternative to genetic testing of family members in familial endocrine disorders is phenotypic screening. This is ideal where the test is simple and highly sensitive and the disorder highly penetrant – for instance, serum calcium screening in first-degree relatives of a person with FHH. In contrast, cascade testing for a known familial ARMC5 mutation has superseded the multistep dynamic testing that was previously used in familial BMAH. Detecting subclinical ACTH-independent cortisol rises in response to varying stimuli was proposed to predict which family members would develop overt BMAH-related Cushing’s syndrome; however, these cumbersome tests were later found to correlate poorly with mutational status.39 Whether to perform genotypic or phenotypic screening also depends on family structure. If an older patient with no children is suspected to have HNF1A or HNF4A monogenic diabetes, phenotypic screening by substitution of insulin with a sulphonylurea with close monitoring by an experienced endocrinologist may be a quick, inexpensive and safe option. If this older patient had several lean children and grandchildren with diabetes, then a monogenic diabetes gene panel in one individual followed by targeted sequencing of the familial mutation (if found) would be more efficient in determining the aetiology of diabetes in each affected family member.
Reproductive options include expectant management, adoption, prenatal testing and preimplantation genetic diagnosis (PGD). In autosomal recessive disorders, PGD may be used to select and transfer unaffected embryos: these may be either homozygous for the wild-type allele (25% of embryos, preferable as they are neither affected nor carriers) or heterozygous for the mutation (50% of embryos, especially in couples with a low yield of viable embryos). In autosomal dominant disorders, only homozygous wild-type embryos (50% of embryos) can be selected. Although used less frequently, preimplantation sex selection can be sufficient for patients with X-linked disorders, either selecting male embryos of men with an X-linked dominant disorder or female embryos of women with an X-linked recessive mutation. Three-parent IVF using the affected woman’s oocyte nucleus, an unaffected donor oocyte and the partner’s sperm is an emerging option for women with a pathogenic mitochondrial DNA mutation. These options are especially pertinent in endocrinology as the underlying disorder may cause infertility and thus independently necessitate assisted reproductive technologies. For instance, secondary hypogonadism may be caused by MEN1 or FIPA, and premature ovarian insufficiency may relate to Turner’s syndrome (45,X), an FMR1 premutation, or an AIRE mutation resulting in APECED.
Future Directions
Ongoing technological advancements are reducing the cost of sequencing, which should increase the uptake of genetic testing in endocrine disorders. In our laboratory, the cost of WES has already decreased from A$4500 in 2015 to A$2000 in current practice. This is different to the consumables cost which is closer to A$300 and sometimes, misleadingly, advertised to the public. Regardless, these costs should not be considered prohibitive if the results would influence patient management or guide family testing. It is useful to compare sequencing costs against other typical costs in the evaluation of a patient – for example, A$600 for a brain MRI or A$1000 for an overnight inpatient admission. Unlike these latter costs, the cost of NGS is distributed across a family because, if it reveals the causative mutation in the proband, other members may be evaluated by merely sequencing one amplicon. Genetic testing may ultimately be subsidised by the Medicare Benefits Scheme (MBS) or private health cover as it is in the United States.40 As of 1 November 2017, germline BRCA1/2 genetic testing is MBS-funded in select women with ovarian, fallopian tube or primary peritoneal cancers. This was instigated by the availability of olaparib, a poly ADP ribose polymerase (PARP) inhibitor which is funded by the Pharmaceutical Benefits Scheme for women with tumour recurrence and a known germline BRCA mutation as BRCA-mutated tumours are highly dependent on PARP-mediated DNA repair pathways. The increasing functionality of genetic testing should provide similar funding avenues.
Less than one in five (3,280/19,580) genes are associated with a Mendelian phenotype and 25% (798/3,280) of these genes can cause two or more phenotypes, some with mixed inheritance patterns.41 These figures are expected to evolve due to emerging phenotypic extensions whereby genetic disorders are shown to have a spectrum of clinical manifestations, and allelic series, which is the molecular corollary referring to different mutant alleles within a gene giving rise to a range of phenotypes. International collaborative databases should enhance our understanding of genotype-phenotype correlations and genotyping may ultimately be performed early in clinical evaluation to save patients from the time-consuming, costly and sometimes invasive diagnostic odyssey that complete phenotyping entails. In the paediatric setting, WES has already been shown to reduce clinical costs, increase diagnostic yield and alter patient management compared to traditional diagnostic methodologies.42
As mentioned, even NGS may fail to demonstrate the molecular cause of a clearly genetic disorder, which may reflect intronic, polygenic or epigenetic changes that are frequently missed by NGS.28 NGS may also miss indels and mistake pseudogenes for one another due to the employment of ‘short reads’ of DNA pieced together. Long-read sequencing (Table 2) is the latest sequencing technology whereby reads of up to 1 MB in length allow elucidation of CNV, pseudogenes and triplet repeats and determination of whether variants are in cis (on the same allele) or in trans (on alternate alleles, as required for an autosomal recessive disease to manifest). Long-read sequencing also performs reasonably well in GC-rich genes as DNA amplification is unnecessary and it utilises a more durable polymerase enzyme compared to standard NGS platforms. In addition, the real-time data of long-read sequencing may reveal delays in nucleotide binding as an indicator of methylation or other epigenetic changes.43
Somatic genetic testing of tumour DNA extracted from affected tissues may guide the management of some malignancies – such as the use of BRAF and MEK inhibitors in patients with melanomas with a somatic BRAF mutation – but it is yet to determine the management of endocrine tumours. Although, the recent discovery of somatic CTNNB1 and BRAF mutations in adamantinomatous and papillary craniopharyngiomas, respectively, heralds the use of tumour genetic testing in endocrinology to guide pharmacotherapy with novel beta-catenin inhibitors and vemurafenib.44,45
Conclusions
There is a genetic basis to many endocrinopathies, including both neoplastic and metabolic disorders. Close liaison between endocrinologists, biochemists, clinical geneticists and genomic pathologists is required in the identification of endocrine genetic disorders. If a genetic aetiology is considered possible, the plausible genetic causes and the utility of genetic testing should be determined. If the results of genetic testing would inform the assessment or management of the patient or their family, genetic testing should be arranged and tailored to the type of genetic abnormality in question. As we discover the genetic basis of more endocrine disorders, the utility of genetic testing will rise and scientists and clinicians will increasingly be faced with the issue of genetic testing in endocrinology.
Acknowledgements
We are grateful to Dr Andreas Schreiber and Dr Jinghua Frank Feng for their technical input and Dr Lucia Gagliardi for providing next generation sequencing data for the Figure. SMCD is supported by a Royal Adelaide Hospital A.R. Clarkson Scholarship. HSS is supported by a Cancer Council SA Breast Cancer Research Fellowship.
Footnotes
Competing Interests: None declared.
References
- 1.Ford CE, Jones KW, Polani PE, De Almeida JC, Briggs JH. A sex-chromosome anomaly in a case of gonadal dysgenesis (Turners syndrome) Lancet. 1959;1:711–3. doi: 10.1016/s0140-6736(59)91893-8. [DOI] [PubMed] [Google Scholar]
- 2.Jacobs PA, Strong JA. A case of human intersexuality having a possible XXY sex-determining mechanism. Nature. 1959;183:302–3. doi: 10.1038/183302a0. [DOI] [PubMed] [Google Scholar]
- 3.Chandrasekharappa SC, Guru SC, Manickam P, Olufemi SE, Collins FS, Emmert-Buck MR, et al. Positional cloning of the gene for multiple endocrine neoplasia-type 1. Science. 1997;276:404–7. doi: 10.1126/science.276.5311.404. [DOI] [PubMed] [Google Scholar]
- 4.Donis-Keller H, Dou S, Chi D, Carlson KM, Toshima K, Lairmore TC, et al. Mutations in the RET proto-oncogene are associated with MEN 2A and FMTC. Hum Mol Genet. 1993;2:851–6. doi: 10.1093/hmg/2.7.851. [DOI] [PubMed] [Google Scholar]
- 5.Carlson KM, Dou S, Chi D, Scavarda N, Toshima K, Jackson CE, et al. Single missense mutation in the tyrosine kinase catalytic domain of the RET protooncogene is associated with multiple endocrine neoplasia type 2B. Proc Natl Acad Sci U S A. 1994;91:1579–83. doi: 10.1073/pnas.91.4.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Latif F, Tory K, Gnarra J, Yao M, Duh FM, Orcutt ML, et al. Identification of the von Hippel-Lindau disease tumor suppressor gene. Science. 1993;260:1317–20. doi: 10.1126/science.8493574. [DOI] [PubMed] [Google Scholar]
- 7.White PC, Grossberger D, Onufer BJ, Chaplin DD, New MI, Dupont B, et al. Two genes encoding steroid 21-hydroxylase are located near the genes encoding the fourth component of complement in man. Proc Natl Acad Sci U S A. 1985;82:1089–93. doi: 10.1073/pnas.82.4.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lifton RP, Dluhy RG, Powers M, Rich GM, Cook S, Ulick S, et al. A chimaeric 11 beta-hydroxylase/aldosterone synthase gene causes glucocorticoid-remediable aldosteronism and human hypertension. Nature. 1992;355:262–5. doi: 10.1038/355262a0. [DOI] [PubMed] [Google Scholar]
- 9.Nagamine K, Peterson P, Scott HS, Kudoh J, Minoshima S, Heino M, et al. Positional cloning of the APECED gene. Nat Genet. 1997;17:393–8. doi: 10.1038/ng1297-393. [DOI] [PubMed] [Google Scholar]
- 10.Pollak MR, Brown EM, Chou YH, Hebert SC, Marx SJ, Steinmann B, et al. Mutations in the human Ca(2+)-sensing receptor gene cause familial hypocalciuric hypercalcemia and neonatal severe hyperparathyroidism. Cell. 1993;75:1297–303. doi: 10.1016/0092-8674(93)90617-y. [DOI] [PubMed] [Google Scholar]
- 11.Sakurai A, Takeda K, Ain K, Ceccarelli P, Nakai A, Seino S, et al. Generalized resistance to thyroid hormone associated with a mutation in the ligand-binding domain of the human thyroid hormone receptor beta. Proc Natl Acad Sci U S A. 1989;86:8977–81. doi: 10.1073/pnas.86.22.8977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Toledo RA, Burnichon N, Cascon A, Benn DE, Bayley JP, Welander J, et al. NGS in PPGL (NGSnPPGL) Study Group. Consensus Statement on next-generation-sequencing-based diagnostic testing of hereditary phaeochromocytomas and paragangliomas. Nat Rev Endocrinol. 2017;13:233–47. doi: 10.1038/nrendo.2016.185. [DOI] [PubMed] [Google Scholar]
- 13.Vierimaa O, Georgitsi M, Lehtonen R, Vahteristo P, Kokko A, Raitila A, et al. Pituitary adenoma predisposition caused by germline mutations in the AIP gene. Science. 2006;312:1228–30. doi: 10.1126/science.1126100. [DOI] [PubMed] [Google Scholar]
- 14.Assié G, Libé R, Espiard S, Rizk-Rabin M, Guimier A, Luscap W, et al. ARMC5 mutations in macronodular adrenal hyperplasia with Cushings syndrome. N Engl J Med. 2013;369:2105–14. doi: 10.1056/NEJMoa1304603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wells SA, Jr, Asa SL, Dralle H, Elisei R, Evans DB, Gagel RF, et al. American Thyroid Association Guidelines Task Force on Medullary Thyroid Carcinoma. Revised American Thyroid Association guidelines for the management of medullary thyroid carcinoma. Thyroid. 2015;25:567–610. doi: 10.1089/thy.2014.0335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Sousa SM, McCormack AI. Cutaneous lichen amyloidosis in multiple endocrine neoplasia. Intern Med J. 2016;46:116–7. doi: 10.1111/imj.12925. [DOI] [PubMed] [Google Scholar]
- 17.De Sousa SM, McCabe MJ, Wu K, Roscioli T, Gayevskiy V, Brook K, et al. Germline variants in familial pituitary tumour syndrome genes are common in young patients and families with additional endocrine tumours. Eur J Endocrinol. 2017;176:635–44. doi: 10.1530/EJE-16-0944. [DOI] [PubMed] [Google Scholar]
- 18.Gill AJ. Succinate dehydrogenase (SDH) and mitochondrial driven neoplasia. Pathology. 2012;44:285–92. doi: 10.1097/PAT.0b013e3283539932. [DOI] [PubMed] [Google Scholar]
- 19.Nolin SL, Brown WT, Glicksman A, Houck GE, Jr, Gargano AD, Sullivan A, et al. Expansion of the fragile X CGG repeat in females with premutation or intermediate alleles. Am J Hum Genet. 2003;72:454–64. doi: 10.1086/367713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Online Mendelian Inheritance in Man. [Accessed 11 April 2018]. https://www.ncbi.nlm.nih.gov/omim.
- 21.GeneReviews. [Accessed 11 April 2018]. https://www.ncbi.nlm.nih.gov/books/NBK1116/
- 22.Genetic Testing Registry. [Accessed 11 April 2018]. https://www.ncbi.nlm.nih.gov/gtr/
- 23.RCPA Genetic Tests and Laboratories Website. [Accessed 11 April 2018]. https://www.rcpa.edu.au/Library/Practising-Pathology/RCPA-Genetic-Testing/RGTL/Home.
- 24.Rostomyan L, Daly AF, Petrossians P, Nachev E, Lila AR, Lecoq AL, et al. Clinical and genetic characterization of pituitary gigantism: an international collaborative study in 208 patients. Endocr Relat Cancer. 2015;22:745–57. doi: 10.1530/ERC-15-0320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gagliardi L, Schreiber AW, Hahn CN, Feng J, Cranston T, Boon H, et al. ARMC5 mutations are common in familial bilateral macronodular adrenal hyperplasia. J Clin Endocrinol Metab. 2014;99:E1784–92. doi: 10.1210/jc.2014-1265. [DOI] [PubMed] [Google Scholar]
- 26.McCormack SE, Li D, Kim YJ, Lee JY, Kim SH, Rapaport R, et al. Digenic Inheritance of PROKR2 and WDR11 Mutations in Pituitary Stalk Interruption Syndrome. J Clin Endocrinol Metab. 2017;102:2501–7. doi: 10.1210/jc.2017-00332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sarfati J, Guiochon-Mantel A, Rondard P, Arnulf I, Garcia-Piñero A, Wolczynski S, et al. A comparative phenotypic study of kallmann syndrome patients carrying monoallelic and biallelic mutations in the prokineticin 2 or prokineticin receptor 2 genes. J Clin Endocrinol Metab. 2010;95:659–69. doi: 10.1210/jc.2009-0843. [DOI] [PubMed] [Google Scholar]
- 28.De Sousa SM, Stowasser M, Feng J, Schreiber AW, Wang P, Hahn CN, et al. ARMC5 is not implicated in familial hyperaldosteronism type II (FH-II) J Hum Hypertens. 2017;31:857–9. doi: 10.1038/jhh.2017.71. [DOI] [PubMed] [Google Scholar]
- 29.De Sousa SM, Kassahn KS, McIntyre LC, Chong CE, Scott HS, Torpy DJ. Case report of whole genome sequencing in the XY female: identification of a novel SRY mutation and revision of a misdiagnosis of androgen insensitivity syndrome. BMC Endocr Disord. 2016;16:58. doi: 10.1186/s12902-016-0141-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoebeeck J, van der Luijt R, Poppe B, De Smet E, Yigit N, Claes K, et al. Rapid detection of VHL exon deletions using real-time quantitative PCR. Lab Invest. 2005;85:24–33. doi: 10.1038/labinvest.3700209. [DOI] [PubMed] [Google Scholar]
- 31.Evans DG, Bowers N, Burkitt-Wright E, Miles E, Garg S, Scott-Kitching V, et al. Northern UK NF1 Research Network. Comprehensive RNA Analysis of the NF1 Gene in Classically Affected NF1 Affected Individuals Meeting NIH Criteria has High Sensitivity and Mutation Negative Testing is Reassuring in Isolated Cases With Pigmentary Features Only. EBioMedicine. 2016;7:212–20. doi: 10.1016/j.ebiom.2016.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou XP, Waite KA, Pilarski R, Hampel H, Fernandez MJ, Bos C, et al. Germline PTEN promoter mutations and deletions in Cowden/Bannayan-Riley-Ruvalcaba syndrome result in aberrant PTEN protein and dysregulation of the phosphoinositol-3-kinase/Akt pathway. Am J Hum Genet. 2003;73:404–11. doi: 10.1086/377109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ellard S, Lango Allen H, De Franco E, Flanagan SE, Hysenaj G, Colclough K, et al. Improved genetic testing for monogenic diabetes using targeted next-generation sequencing. Diabetologia. 2013;56:1958–63. doi: 10.1007/s00125-013-2962-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Forlenza GP, Calhoun A, Beckman KB, Halvorsen T, Hamdoun E, Zierhut H, et al. Next generation sequencing in endocrine practice. Mol Genet Metab. 2015;115:61–71. doi: 10.1016/j.ymgme.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lumbroso S, Paris F, Sultan C European Collaborative Study. Activating Gsalpha mutations: analysis of 113 patients with signs of McCune-Albright syndromea European Collaborative Study. J Clin Endocrinol Metab. 2004;89:2107–13. doi: 10.1210/jc.2003-031225. [DOI] [PubMed] [Google Scholar]
- 36.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thakker RV, Newey PJ, Walls GV, Bilezikian J, Dralle H, Ebeling PR, et al. Endocrine Society. Clinical practice guidelines for multiple endocrine neoplasia type 1 (MEN1) J Clin Endocrinol Metab. 2012;97:2990–3011. doi: 10.1210/jc.2012-1230. [DOI] [PubMed] [Google Scholar]
- 38.Brandi ML, Gagel RF, Angeli A, Bilezikian JP, Beck-Peccoz P, Bordi C, et al. Guidelines for diagnosis and therapy of MEN type 1 and type 2. J Clin Endocrinol Metab. 2001;86:5658–71. doi: 10.1210/jcem.86.12.8070. [DOI] [PubMed] [Google Scholar]
- 39.Gagliardi L, Hotu C, Casey G, Braund WJ, Ling KH, Dodd T, et al. Familial vasopressin-sensitive ACTH-independent macronodular adrenal hyperplasia (VPs-AIMAH): clinical studies of three kindreds. Clin Endocrinol (Oxf) 2009;70:883–91. doi: 10.1111/j.1365-2265.2008.03471.x. [DOI] [PubMed] [Google Scholar]
- 40.Kirmani S. Molecular genetic testing in endocrinology - a practical guide. Endocr Pract. 2012;18:85–9. doi: 10.4158/EP11364.RA. [DOI] [PubMed] [Google Scholar]
- 41.Amberger JS, Bocchini CA, Schiettecatte F, Scott AF, Hamosh A. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015;43:D789–98. doi: 10.1093/nar/gku1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stark Z, Tan TY, Chong B, Brett GR, Yap P, Walsh M, et al. Melbourne Genomics Health Alliance. A prospective evaluation of whole-exome sequencing as a first-tier molecular test in infants with suspected monogenic disorders. Genet Med. 2016;18:1090–6. doi: 10.1038/gim.2016.1. [DOI] [PubMed] [Google Scholar]
- 43.English AC, Richards S, Han Y, Wang M, Vee V, Qu J, et al. Mind the gap: upgrading genomes with Pacific Biosciences RS long-read sequencing technology. PLoS One. 2012;7:e47768. doi: 10.1371/journal.pone.0047768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brastianos PK, Taylor-Weiner A, Manley PE, Jones RT, Dias-Santagata D, Thorner AR, et al. Exome sequencing identifies BRAF mutations in papillary craniopharyngiomas. Nat Genet. 2014;46:161–5. doi: 10.1038/ng.2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Larkin SJ, Preda V, Karavitaki N, Grossman A, Ansorge O. BRAF V600E mutations are characteristic for papillary craniopharyngioma and may coexist with CTNNB1-mutated adamantinomatous craniopharyngioma. Acta Neuropathol. 2014;127:927–9. doi: 10.1007/s00401-014-1270-6. [DOI] [PMC free article] [PubMed] [Google Scholar]