Version Changes
Revised. Amendments from Version 1
In Version 2 of this article we have addressed the comments of the two reviewers, and included more detail about integrating datasets, workflows, authentication and privacy considerations. We have also included a second figure (Figure 2), a flowchart showing how the data life cycle considerations might be applied to an example research project.
Abstract
Throughout history, the life sciences have been revolutionised by technological advances; in our era this is manifested by advances in instrumentation for data generation, and consequently researchers now routinely handle large amounts of heterogeneous data in digital formats. The simultaneous transitions towards biology as a data science and towards a ‘life cycle’ view of research data pose new challenges. Researchers face a bewildering landscape of data management requirements, recommendations and regulations, without necessarily being able to access data management training or possessing a clear understanding of practical approaches that can assist in data management in their particular research domain.
Here we provide an overview of best practice data life cycle approaches for researchers in the life sciences/bioinformatics space with a particular focus on ‘omics’ datasets and computer-based data processing and analysis. We discuss the different stages of the data life cycle and provide practical suggestions for useful tools and resources to improve data management practices.
Keywords: data sharing, data management, open science, bioinformatics, reproducibility
Introduction
Technological data production capacity is revolutionising biology 1, but is not necessarily correlated with the ability to efficiently analyse and integrate data, or with enabling long-term data sharing and reuse. There are selfish as well as altruistic benefits to making research data reusable 2: it allows one to find and reuse one’s own previously-generated data easily; it is associated with higher citation rates 3, 4; and it ensures eligibility for funding from and publication in venues that mandate data sharing, an increasingly common requirement (e.g. Final NIH statement on sharing research data, Wellcome Trust policy on data management and sharing, Bill & Melinda Gates Foundation open access policy). Currently we are losing data at a rapid rate, with up to 80% unavailable after 20 years 5. This affects reproducibility - assessing the robustness of scientific conclusions by ensuring experiments and findings can be reproduced - which underpins the scientific method. Once access to the underlying data is lost, replicability, reproducibility and extensibility 6 are reduced.
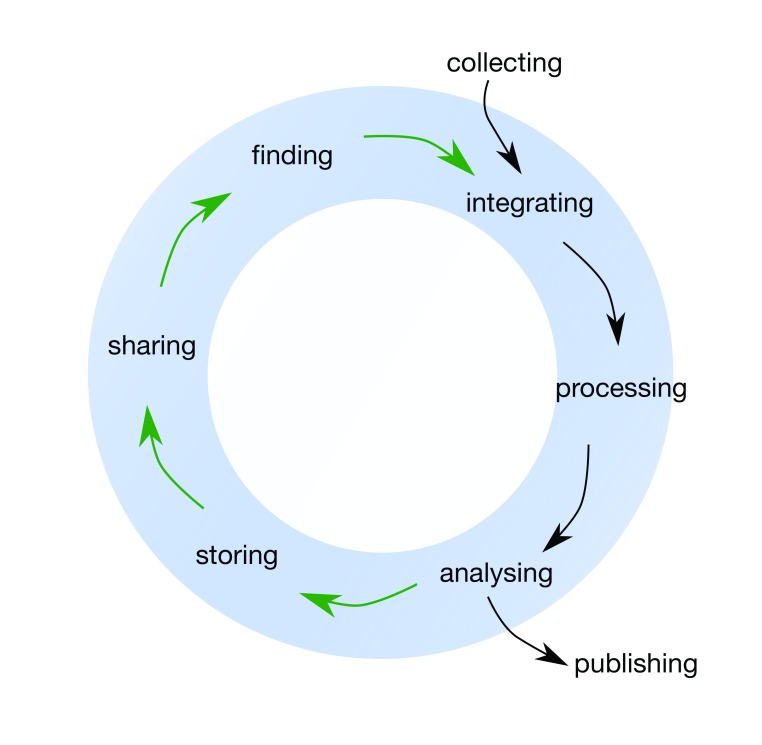
At a broader societal level, the full value of research data may go beyond the initial use case in unforeseen ways 7, 8, so ensuring data quality and reusability is crucial to realising its potential value 9– 12. The recent publication of the FAIR principles 9, 13 identifies four key criteria for high-quality research data: the data should be Findable, Accessible, Interoperable and Reusable. Whereas a traditional view of data focuses on collecting, processing, analysing data and publishing results only, a life cycle view reveals the additional importance of finding, storing and sharing data 11. Throughout this article, we present a researcher-focused data life cycle framework that has commonalities with other published frameworks [e.g. the DataONE Data Life Cycle, the US geological survey science data lifecycle model and 11, 14– 15], but is aimed at life science researchers specifically ( Figure 1).
Figure 1. The Data Life Cycle framework for bioscience, biomedical and bioinformatics data that is discussed throughout this article.
Black arrows indicate the ‘traditional’, linear view of research data; the green arrows show the steps necessary for data reusability. This framework is likely to be a simplified representation of any given research project, and in practice there would be numerous ‘feedback loops’ and revisiting of previous stages. In addition, the publishing stage can occur at several points in the data life cycle.
Learning how to find, store and share research data is not typically an explicit part of undergraduate or postgraduate training in the biological sciences 16– 18, though some subdomains (e.g. ecology) have a history of data management advice 8, 19. The scope, size and complexity of datasets in many fields has increased dramatically over the last 10–20 years, but the knowledge of how to manage this data is currently limited to specific cohorts of ‘information managers’ (e.g. research data managers, research librarians, database curators and IT professionals with expertise in databases and data schemas 18). In response to institutional and funding requirements around data availability, a number of tools and educational programs have been developed to help researchers create Data Management Plans to address elements of the data lifecycle 20; however, even when a plan is mandated, there is often a gap between the plan and the actions of the researcher 10.
This publication targets life science researchers wanting to improve their data management practice but will also be relevant to life science journals, funders, and research infrastructure bodies. It arose from a 2016 workshop series on the data lifecycle for life science researchers run by EMBL Australia Bioinformatics Resource 21, which provided opportunities to (i) map the current approaches to the data life cycle in biology and bioinformatics, and (ii) present and discuss best practice approaches and standards for key international projects with Australian life scientists and bioinformaticians. Throughout the article we highlight some specific data management challenges mentioned by participants. An earlier version of this article can be found on bioRxiv ( https://doi.org/10.1101/167619).
Finding data
In biology, research data is frequently published as supplementary material to articles, on personal or institutional websites, or in non-discipline-specific repositories like Figshare and Dryad 22. In such cases, data may exist behind a paywall, there is no guarantee it will remain extant, and, unless one already knows it exists and its exact location, it may remain undiscovered 23. It is only when a dataset is added to a public data repository, along with accompanying standardized descriptive metadata (see Collecting data), that it can be indexed and made publicly available 24. Data repositories also provide unique identifiers that increase findability by enabling persistent linking from other locations and permanent association between data and its metadata.
In the field of molecular biology, a number of bioinformatics-relevant organisations host public data repositories. National and international-level organisations of this kind include the European Bioinformatics Institute (EMBL-EBI) 25, the National Centre for Biotechnology Information (NCBI) 26, the DNA Data Bank of Japan (DDBJ) 27, the Swiss Institute of Bioinformatics (SIB) 28, and the four data center members of the worldwide Protein Data Bank 29, which mirror their shared data with regular, frequent updates. This shared central infrastructure is hugely valuable to research and development. For example, EMBL-EBI resources have been valued at over £270 million per year and contribute to ~£1 billion in research efficiencies; a 20-fold return on investment 30.
Numerous repositories are available for biological data (see Table 1 for an overview), though repositories are still lacking for some data types and sub-domains 31. Due to privacy regulations, human data is generally not freely available and these repositories typically require access requests on an individual dataset basis 32, 33. Tools like the dbGAP browser 34 and the Beacon Network 35 can assist in identifying relevant limited-access datasets and reduce the burden associated with requesting and downloading data.
Many specialised data repositories exist outside of the shared central infrastructure mentioned, often run voluntarily or with minimal funding. Support for biocuration, hosting and maintenance of these smaller-scale but key resources is a pressing problem 36– 38. The quality of the user-submitted data in public repositories 39, 40 can mean that public datasets require extra curation before reuse. Unfortunately, due to low uptake of established methods (see the EMBL-EBI and NCBI third-party annotation policies; 41) to correct the data 40, the results of extra curation may not find their way back into the repositories. Repositories are often not easily searched by generic web search engines 31. Registries, which form a secondary layer linking multiple, primary repositories, may offer a more convenient way to search across multiple repositories for data relevant to a researcher’s topics of interest 42.
Table 1. Overview of some representative databases, registries and other tools to find life science data.
A more complete list can be found at FAIRsharing.
| Database/
registry |
Name | Description | Datatypes | URL |
|---|---|---|---|---|
| Database | Gene Ontology | Repository of functional roles of gene products,
including: proteins, ncRNAs, and complexes. |
Functional roles as determined experimentally or
through inference. Includes evidence for these roles and links to literature |
http://geneontology.org/ |
| Database | Kyoto
Encyclopedia of Genes and Genomes (KEGG) |
Repository for pathway relationships of
molecules, genes and cells, especially molecular networks |
Protein, gene, cell, and genome pathway
membership data |
http://www.genome.jp/kegg/ |
| Database | OrthoDB | Repository for gene ortholog information | Protein sequences and orthologous group
annotations for evolutionarily related species groups |
http://www.orthodb.org/ |
| Database
with analysis layer |
eggNOG | Repository for gene ortholog information with
functional annotation prediction tool |
Protein sequences, orthologous group
annotations and phylogenetic trees for evolutionarily related species groups |
http://eggnogdb.embl.de/ |
| Database | European
Nucleotide Archive (ENA) |
Repository for nucleotide sequence information | Raw next-generation sequencing data, genome
assembly and annotation data |
http://www.ebi.ac.uk/ena |
| Database | Sequence Read
Archive (SRA) |
Repository for nucleotide sequence information | Raw high-throughput DNA sequencing and
alignment data |
https://www.ncbi.nlm.nih.gov/sra/ |
| Database | GenBank | Repository for nucleotide sequence information | Annotated DNA sequences | https://www.ncbi.nlm.nih.gov/genbank/ |
| Database | ArrayExpress | Repository for genomic expression data | RNA-seq, microarray, CHIP-seq, Bisulfite-seq and
more (see https://www.ebi.ac.uk/arrayexpress/ help/experiment_types.html for full list) |
https://www.ebi.ac.uk/arrayexpress/ |
| Database | Gene
Expression Omnibus (GEO) |
Repository for genetic/genomic expression data | RNA-seq, microarray, real-time PCR data on
gene expression |
https://www.ncbi.nlm.nih.gov/geo/ |
| Database | PRIDE | Repository for proteomics data | Protein and peptide identifications, post-translational
modifications and supporting spectral evidence |
https://www.ebi.ac.uk/pride/archive/ |
| Database | Protein Data
Bank (PDB) |
Repository for protein structure information | 3D structures of proteins, nucleic acids and
complexes |
https://www.wwpdb.org/ |
| Database | MetaboLights | Repository for metabolomics experiments and
derived information |
Metabolite structures, reference spectra and
biological characteristics; raw and processed metabolite profiles |
http://www.ebi.ac.uk/metabolights/ |
| Ontology/
database |
ChEBI | Ontology and repository for chemical entities | Small molecule structures and chemical
properties |
https://www.ebi.ac.uk/chebi/ |
| Database | Taxonomy | Repository of taxonomic classification information | Taxonomic classification and nomenclature data
for organisms in public NCBI databases |
https://www.ncbi.nlm.nih.gov/taxonomy |
| Database | BioStudies | Repository for descriptions of biological studies,
with links to data in other databases and publications |
Study descriptions and supplementary files | https://www.ebi.ac.uk/biostudies/ |
| Database | Biosamples | Repository for information about biological
samples, with links to data generated from these samples located in other databases |
Sample descriptions | https://www.ebi.ac.uk/biosamples/ |
| Database
with analysis layer |
IntAct | Repository for molecular interaction information | Molecular interactions and evidence type | http://www.ebi.ac.uk/intact/ |
| Database | UniProtKB
(SwissProt and TrEMBL) |
Repository for protein sequence and function
data. Combines curated (UniProtKB/SwissProt) and automatically annotated, uncurated (UniProtKB/TrEMBL) databases |
Protein sequences, protein function and
evidence type |
http://www.uniprot.org/ |
| Database | European
Genome- Phenome Archive |
Controlled-access repository for sequence and
genotype experiments from human participants whose consent agreements authorise data release for specific research use |
Raw, processed and/or analysed sequence and
genotype data along with phenotype information |
https://www.ebi.ac.uk/ega/ |
| Database
with analysis layer |
EBI
Metagenomics |
Repository and analysis service for
metagenomics and metatranscriptomics data. Data is archived in ENA |
Next-generation sequencing metagenomic
and metatranscriptomic data; metabarcoding (amplicon-based) data |
https://www.ebi.ac.uk/metagenomics/ |
| Database
with analysis layer |
MG-RAST | Repository and analysis service for
metagenomics data. |
Next-generation sequencing metagenomic and
metabarcoding (amplicon-based) data |
http://metagenomics.anl.gov/ |
| Registry | Omics DI | Registry for dataset discovery that currently
spans 11 data repositories: PRIDE, PeptideAtlas, MassIVE, GPMDB, EGA, Metabolights, Metabolomics Workbench, MetabolomeExpress, GNPS, ArrayExpress, ExpressionAtlas |
Genomic, transcriptomic, proteomic and
metabolomic data |
http://www.omicsdi.org |
| Registry | DataMed | Registry for biomedical dataset discovery that
currently spans 66 data repositories |
Genomic, transcriptomic, proteomic,
metabolomic, morphology, cell signalling, imaging and other data |
https://datamed.org |
| Registry | Biosharing | Curated registry for biological databases, data
standards, and policies |
Information on databases, standards and
policies including fields of research and usage recommendations by key organisations |
https://biosharing.org/ |
| Registry | re3data | Registry for research data repositories across
multiple research disciplines |
Information on research data repositories, terms
of use, research fields |
http://www.re3data.org |
Collecting data
The most useful data has associated information about its creation, its content and its context - called metadata. If metadata is well structured, uses consistent element names and contains element values with specific descriptions from agreed-upon vocabularies, it enables machine readability, aggregation, integration and tracking across datasets: allowing for Findability, Interoperability and Reusability 9, 31. One key approach in best-practice metadata collection is to use controlled vocabularies built from ontology terms. Biological ontologies are tools that provide machine-interpretable representations of some aspect of biological reality 31, 43. They are a way of organising and defining objects (i.e. physical entities or processes), and the relationships between them. Sourcing metadata element values from ontologies ensures that the terms used in metadata are consistent and clearly defined. There are several user-friendly tools available to assist researchers in accessing, using and contributing to ontologies ( Table 2).
Table 2. Useful ontology tools to assist in metadata collection.
| Tool | Task | URL |
|---|---|---|
| Ontology Lookup
Service |
Discover different ontologies and their contents | http://www.ebi.ac.uk/ols/ |
| OBO Foundry | Table of open biomedical ontologies with information
on development status, license and content |
http://obofoundry.org/ |
| Zooma | Assign ontology terms using curated mapping | http://www.ebi.ac.uk/spot/zooma/ |
| Webulous | Create new ontology terms easily | https://www.ebi.ac.uk/efo/webulous/ |
| Ontobee | A linked data server that facilitates ontology data
sharing, visualization, and use. |
http://www.ontobee.org |
Adopting standard data and metadata formats and syntax is critical for compliance with FAIR principles 9, 24, 31, 42, 44. Biological and biomedical research has been considered an especially challenging research field in this regard, as datatypes are extremely heterogeneous and not all have defined data standards 44, 45; many existing data standards are complex and therefore difficult to use 45, or only informally defined, and therefore subject to variation, misrepresentation, and divergence over time 44. Nevertheless, well-established standards exist for a variety of biological data types ( Table 3). FAIRsharing is a useful registry of data standards and policies that also indicates the current status of standards for different data types and those recommended by databases and research organisations 42.
Table 3. Overview of common standard data formats for ‘omics data.
A more complete list can be found at FAIRsharing.
| Data type | Format name | Description | Reference or URL for format specification | URLs for repositories
accepting data in this format |
|---|---|---|---|---|
| Raw DNA/RNA
sequence |
FASTA
FASTQ HDF5 SAM/BAM/ CRAM |
FASTA is a common text format to store DNA/RNA/Protein
sequence and FASTQ combines base quality information with the nucleotide sequence. HDF5 is a newer sequence read formats used by long read sequencers e.g. PacBio and Oxford Nanopore. Raw sequence can also be stored in unaligned SAM/BAM/ CRAM format |
74
75 https://support.hdfgroup.org/HDF5/ https://samtools.github.io/hts-specs/ https://www.ncbi.nlm.nih.gov/ sra/docs/submitformats/ http://www.ebi.ac.uk/ena/ submit/data-formats |
|
| Assembled
DNA sequence |
FASTA
Flat file AGP |
Assemblies without annotation are generally stored in
FASTA format. Annotation can be integrated with assemblies in contig, scaffold or chromosome flat file format. AGP files are used to describe how smaller fragments are placed in an assembly but do not contain the sequence information themselves |
41
http://www.ebi.ac.uk/ena/submit/contig-flat-file http://www.ebi.ac.uk/ena/submit/scaffold-flat-file https://www.ncbi.nlm.nih.gov/assembly/agp/AGP_ Specification/ |
http://www.ebi.ac.uk/ena
/submit/genomes-sequence- submission |
| Aligned DNA
sequence |
SAM/BAM
CRAM |
Sequences aligned to a reference are represented in
sequence alignment and mapping format (SAM). Its binary version is called BAM and further compression can be done using the CRAM format |
https://samtools.github.io/hts-specs/ |
https://www.ncbi.nlm.nih.gov/
sra/docs/submitformats/#bam |
| Gene model or
genomic feature annotation |
GTF/GFF/
GFF3 BED GB/GBK |
General feature format or general transfer format are
commonly used to store genomic features in tab-delimited flat text format. GFF3 is a more advanced version of the basic GFF that allows description of more complex features. BED format is a tab-delimited text format that also allows definition of how a feature should be displayed (e.g. on a genome browser). GenBank flat file Format (GB/GBK) is also commonly used but not well standardised |
https://github.com/The-Sequence-Ontology/
Specifications/blob/master/gff3.md https://genome.ucsc.edu/FAQ/FAQformat.html https://genome.ucsc.edu/FAQ/FAQformat.html https://www.ncbi.nlm.nih.gov/Sitemap/ samplerecord.html |
http://www.ensembl.org/info/
website/upload/gff.html http://www.ensembl.org/info/ website/upload/gff3.html |
| Gene functional
annotation |
GAF
(GPAD and RDF will also be available in 2018) |
A GAF file is a GO Annotation File containing annotations
made to the GO by a contributing resource such as FlyBase or Pombase. However, the GAF standard is applicable outside of GO, e.g. using other ontologies such as PO. GAF (v2) is a simple tab-delimited file format with 17 columns to describe an entity (e.g. a protein), its annotation and some annotation metadata |
http://geneontology.org/page/go-annotation-file-
format-20 |
http://geneontology.org/page/
submitting-go-annotations |
| Genetic/genomic
variants |
VCF | A tab-delimited text format to store meta-information as
header lines followed by information about variants position in the genome. The current version is VCF4.2 |
https://samtools.github.io/hts-specs/VCFv4.2.pdf |
http://www.ensembl.org/info/
website/upload/var.html |
| Interaction data | PSI-MI XML
MITAB |
Data formats developed to exchange molecular interaction
data, related metadata and fully describe molecule constructs |
http://psidev.info/groups/molecular-interactions | http://www.ebi.ac.uk/intact |
| Raw metabolite
profile |
mzML
nmrML |
XML based data formats that define mass spectrometry
and nuclear magnetic resonance raw data in Metabolomics |
http://www.psidev.info/mzml
http://nmrml.org/ |
|
| Protein sequence | FASTA | A text-based format for representing nucleotide sequences
or protein sequences, in which nucleotides or amino acids are represented using single-letter codes |
74 | www.uniprot.org |
| Raw proteome
profile |
mzML | A formally defined XML format for representing mass
spectrometry data. Files typically contain sequences of mass spectra, plus metadata about the experiment |
http://www.psidev.info/mzml | www.ebi.ac.uk/pride |
| Organisms and
specimens |
Darwin Core | The Darwin Core (DwC) standard facilitates the exchange
of information about the geographic location of organisms and associated collection specimens |
http://rs.tdwg.org/dwc/ |
Most public repositories for biological data (see Table 1 and Storing data section) require that minimum metadata be submitted accompanying each dataset ( Table 4). This minimum metadata specification typically has broad community input 46. Minimum metadata standards may not include the crucial metadata fields that give the full context of the particular research project 46, so it is important to gather metadata early, understand how to extend a minimum metadata template to include additional fields in a structured way, and think carefully about all the relevant pieces of metadata information that might be required for reuse.
Table 4. Some community-designed minimum information criteria for metadata specifications in life sciences.
A more complete list can be found at FAIRsharing.
| Name | Description | Examples of projects/databases that
use this specification |
URL |
|---|---|---|---|
| MINSEQE | Minimum Information about a high-
throughput SEQuencing Experiment |
Developed by the Functional Genomics
Data Society. Used in the NCBI Sequence Read Archive, ArrayExpress |
http://fged.org/site_media/pdf/MINSEQE_1.0.pdf |
| MIxS - MIGS/MIMS | Minimum Information about a
(Meta)Genome Sequence. The MIMS extension includes key environmental metadata |
Developed by the Genomic Standards
Consortium. Numerous adopters including NCBI/EBI/DDBJ databases |
http://wiki.gensc.org/index.php?title=MIGS/MIMS |
| MIMARKS | Minimum Information about a
MARKer gene Sequence. This is an extension of MIGS/MIMS for environmental sequences |
Developed by the Genomic Standards
Consortium. Numerous adopters including NCBI/EBI/DDBJ databases |
http://wiki.gensc.org/index.php?title=MIMARKS |
| MIMIx | Minimum Information about a
Molecular Interaction eXperiment |
Developed by the Proteomics Standards
Initiative. Adopted by the IMEx Consortium databases |
http://www.psidev.info/mimix |
| MIAPE | Minimum Information About a
Proteomics Experiment |
Developed by the Proteomics Standards
Initiative. Adopted by PRIDE, World- 2DPAGE and ProteomeXchange databases |
http://www.psidev.info/miape |
| Metabolomics
Standards Initiative (MSI) standards |
Minimal reporting structures that
represent different parts of the metabolomics workflow |
Developed by the Metabolomics
Standards Initiative (MSI) and the Coordination of Standards in Metabolomics (COSMOS) consortium |
http://www.metabolomics-msi.org/ |
| MIRIAM | Minimal Information Required
In the Annotation of Models. For annotation and curation of computational models in biology |
Initiated by the BioModels.net effort.
Adopted by the EBI BioModels database and others |
http://co.mbine.org/standards/miriam |
| MIAPPE | Minimum Information About a Plant
Phenotyping Experiment. Covers study, environment, experimental design, sample management, biosource, treatment and phenotype |
Adopted by the Plant Phenomics and
Genomics Research Data Repository and the Genetic and Genomic Information System (GnpIS) |
http://cropnet.pl/phenotypes/wp-content/uploads/2016/04/MIAPPE.pdf |
| MDM | Minimal Data for Mapping for
sample and experimental metadata for pathogen genome-scale sequence data |
Developed by the Global Microbial
Identifier Initiative and EBI. Complies with EBI ENA database submission requirements |
http://www.ebi.ac.uk/ena/submit/pathogen-data |
| FAANG sample
metadata specification |
Metadata specification for biological
samples derived from animals (animals, tissue samples, cells or other biological materials). Complies with EBI database requirements and BioSamples database formats |
Developed and used by the Functional
Annotation of Animal Genomes Consortium |
https://github.com/FAANG/faang-metadata/blob/master/docs/faang_
sample_metadata.md |
| FAANG experimental
metadata specification |
Metadata specification for
sequencing and array experiments on animal samples |
Developed and used by the Functional
Annotation of Animal Genomes Consortium |
https://github.com/FAANG/faang-metadata/blob/master/docs/faang_
experiment_metadata.md |
| FAANG analysis
metadata specification |
Metadata specification for analysis
results |
Developed and used by the Functional
Annotation of Animal Genomes Consortium. NB no public repository exists for this specific datatype |
https://github.com/FAANG/faang-metadata/blob/master/docs/faang_
analysis_metadata.md |
| SNOMED-CT | Medical terminology and
pharmaceutical product standard |
Commercial but collaboratively-designed
product |
http://www.snomed.org/snomed-ct |
Integrating, processing and analysing data
Where existing and/or newly-collected datasets are to be used in the same experiment, they must first be integrated. This may involve initial processing of one or more datasets so that they share format and granularity, or so that relevant fields map correctly. The researcher also needs to ensure integration at ‘dependency’ level: for example, controlled vocabularies or genome assemblies used in data generation/processing must match or be easily converted. The plethora of autonomous data repositories has created problems with mapping data and annotations among repositories 47, 48. Current large-scale efforts aim to improve interoperability using Linked Data and other Semantic Web tools 48 as well as extensive ontology development (see Collecting data section). The Monarch Initiative is an example of a project that achieves new insights by integrating existing data from multiple sources: in this case, data from animal and human genetic, phenotypic and other repositories is brought together via a custom data flow to help identify unrecognised animal models for human disease 49. In smaller projects, the need for individual researchers to integrate data will often inform the way new data is collected, to ensure it matches existing datasets, creating a feedback loop in the data lifecycle that highlights the need for prior planning ( Figure 2). Seamless solutions are still some way off 50 for all but a handful of applications.
Figure 2. Flowchart of the data life cycle stages applied to an example research project.
Bold text indicates new data, software or workflow objects created during the project. Solid thin arrows indicate movement of objects from creation to storage and sharing. Dashed thin arrows indicate where downstream entities should influence decisions made at a given step. (For example, the choice of format, granularity, metadata content and structure of new data collected may be influenced by existing software requirements, existing data characteristics and requirements of the archive where the data will be deposited). Purple stars indicate objects for which the FAIR principles 9 can provide further guidance. Dotted thin arrows indicate citation of an object using its unique persistent identifier. Brown stars indicate where FAIRsharing can help identify appropriate archives for storing and sharing.
Recording and reporting how research data is processed and analysed computationally is crucial for reproducibility and assessment of research quality 1, 51. This can be aided by scientific workflow approaches that facilitate both recording and reproducing processing and analysis steps 1, though many experiments will require ‘one-off’ workflows that may not function with existing workflow management systems. Full reproducibility requires access to the software, software versions, workflow, dependencies and operating system used as well as the data and software code itself 1, 52. Therefore, although computational work is often seen as enabling reproducibility in the short term, in the long term it is fragile and reproducibility is limited (e.g. discussion by D. Katz, K. Hinsen and C.T. Brown). Best-practice approaches for preserving data processing and analysis code involve hosting source code in a repository where it receives a unique identifier and is under version control; where it is open, accessible, interoperable and reusable - broadly mapping to the FAIR principles for data. Github and Bitbucket, for example, fulfil these criteria, and Zenodo additionally generates Digital Object Identifiers (DOIs) for submissions and guarantees long-term archiving. Workflows can also be preserved in repositories along with relevant annotations (reviewed in 1). A complementary approach is containerised computing (e.g. Docker) which bundles operating system, software, code and potentially workflows and data together. Several recent publications have suggested ways to improve current practice in research software development to aid in reproducibility 15, 53– 55.
The same points hold for wet-lab data production: for full reproducibility within and outside the lab, it is important to capture and enable access to specimen cell lines, tissue samples and/or DNA as well as reagents 56. Wet-lab methods can be captured in electronic laboratory notebooks and reported in the Biosamples database 57, protocols.io or OpenWetWare; specimens can be lodged in biobanks, culture or museum collections 58– 62; but the effort involved in enabling full reproducibility remains extensive. Electronic laboratory notebooks are frequently suggested as a sensible way to make this information openly available and archived 63. Some partial solutions exist (e.g. LabTrove, BlogMyData, Benchling and others 64), including tools for specific domains such as the Scratchpad Virtual Research Environment for natural history research 65. Other tools can act as or be combined to produce notebooks for small standalone code-based projects (see 66 and update), including Jupyter Notebook, Rmarkdown, and Docker. However, it remains a challenge to implement online laboratory notebooks to cover both field/lab work and computer-based work, especially when computer work is extensive, involved and non-modular 51. Currently, no best-practice guidelines or minimum information standards exist for use of electronic laboratory notebooks 6. We suggest that appropriate minimum information to be recorded for most computer-based tasks should include date, task name and brief description, aim, actual command(s) used, software names and versions used, input/output file names and locations, script names and locations, all in a simple text format.
In the authors’ experience, the data processing and analysis stage is one of the most challenging for openness. As reported elsewhere 16– 18, we have observed a gap between modern biological research as a field of data science, and biology as it is still mostly taught in undergraduate courses, with little or no focus on computational analysis, or project or data management. This gap has left researchers lacking key knowledge and skills required to implement best practices in dealing with the life cycle of their data.
Publishing data
Traditionally, scientific publications included raw research data, but in recent times datasets have grown beyond the scope of practical inclusion in a manuscript 11, 51. Selected data outputs are often included without sharing or publishing the underlying raw data 14. Journals increasingly recommend or require deposition of raw data in a public repository [e.g. 67], although exceptions have been made for publications containing commercially-relevant data 68. The current data-sharing mandate is somewhat field-dependent 5, 69 and also varies within fields 70. For example, in the field of bioinformatics, the UPSIDE principle 71 is referred to by some journals (e.g. Bioinformatics), while others have journal- or publisher-specific policies (e.g. BMC Bioinformatics).
The vast majority of scientific journals require inclusion of processing and analysis methods in ‘sufficient detail for reproduction’ (e.g. Public Library of Science submission and data availability guidelines; International Committee of Medical Journal Editors manuscript preparation guidelines; Science instructions for authors; Elsevier Cell Press STAR Methods; and 72), though journal requirements are diverse and complex 73, and the level of detail authors provide can vary greatly in practice 76, 77. More recently, many authors have highlighted that full reproducibility requires sharing data and resources at all stages of the scientific process, from raw data (including biological samples) to full methods and analysis workflows 1, 6, 61, 77. However, this remains a challenge 78, 79, as discussed in the Processing and analysing data section. To our knowledge, strategies for enabling computational reproducibility are currently not mandated by any scientific journal.
A recent development in the field of scientific publishing is the establishment of ‘data journals’: scientific journals that publish papers describing datasets. This gives authors a vehicle to accrue citations (still a dominant metric of academic impact) for data production alone, which can often be labour-intensive and expensive yet is typically not well recognised under the traditional publishing model. Examples of this article type include the Data Descriptor in Scientific Data and the Data Note in GigaScience, which do not include detailed new analysis but rather focus on describing and enabling reuse of datasets.
The movement towards sharing research publications themselves (‘Open Access Publishing’) has been discussed extensively elsewhere [e.g. 23, 80, 81]. Publications have associated metadata (creator, date, title etc.; see Dublin Core Metadata Initiative metadata terms) and unique identifiers (PubMed ID for biomedical and some life science journals, DOIs for the vast majority of journals; see Table 5). The ORCID system enables researchers to claim their own unique identifier, which can be linked to their publications. The use of unique identifiers within publications referring to repository records (e.g. genes, proteins, chemical entities) is not generally mandated by journals, although it would ensure a common vocabulary is used and so make scientific results more interoperable and reusable 82. Some efforts are underway to make this easier for researchers: for example, Genetics and other Genetics Society of America journals assist authors in linking gene names to model organism database entries.
Table 5. Identifiers throughout the data life cycle.
| Name | Relevant stage of
data life cycle |
Description | URL |
|---|---|---|---|
| Digital Object Identifier (DOI) | Publishing, Sharing,
Finding |
A unique identifier for a digital (or physical or
abstract) object |
https://www.doi.org/ |
| Open Researcher and
Contributor ID (ORCID) |
Publishing | An identifier for a specific researcher that
persists across publications and other research outputs |
https://orcid.org/ |
| Repository accession
number |
Finding, Processing/
Analyzing, Publishing, Sharing, Storing |
A unique identifier for a record within a
repository. Format will be repository-specific. Examples include NIH UIDs (unique identifiers) and accession numbers; ENA accession numbers; PDB IDs |
For example,
https://support.ncbi.nlm.nih.gov/link/portal/28045/28049/
Article/499/ http://www.ebi.ac.uk/ena/submit/accession-number-formats |
| Pubmed ID (PMID) | Publishing | An example of a repository-specific unique
identifier: PubMed IDs are used for research publications indexed in the PubMed database |
https://www.ncbi.nlm.nih.gov/pubmed/ |
| International Standard
Serial Number (ISSN) |
Publishing | A unique identifier for a journal, magazine or
periodical |
http://www.issn.org/ |
| International Standard Book
Number (ISBN) |
Publishing | A unique identifier for a book, specific to the
title, edition and format |
https://www.isbn-international.org |
Storing data
While primary data archives are the best location for raw data and some downstream data outputs ( Table 1), researchers also need local data storage solutions during the processing and analysis stages. Data storage requirements vary among research domains, with major challenges often evident for groups working on taxa with large genomes (e.g. crop plants), which require large storage resources, or on human data, where privacy regulations may require local data storage, access controls (e.g. the GA4GH Security Technology Infrastructure document) and conversion to non-identifiable data if data is to be shared (see Sharing data section). For data where privacy is a concern, one approach is separating the data storage from the analysis location and limiting the analysis outputs to ‘nondisclosive’ results 83. An example is DataShield 83, which is mostly used for public health rather than ‘omics’ data. Subdomain-specific practice should be considered when choosing appropriate formats and linking metadata, as outlined in 84. In addition, long-term preservation of research data should consider threats such as storage failure, mistaken erasure, bit rot, outdated media, outdated formats, loss of context and organisational failure 85.
Sharing data
The best-practice approach to sharing biological data is to deposit it (with associated metadata) in a primary archive suitable for that datatype 8 that complies with FAIR principles. As highlighted in the Storing data section, these archives assure both data storage and public sharing as their core mission, making them the most reliable location for long-term data storage. Alternative data sharing venues (e.g. FigShare, Dryad) do not require or implement specific metadata or data standards. This means that while these venues have a low barrier to entry for submitters, the data is not FAIR unless submitters have independently decided to comply with more stringent criteria. If available, an institutional repository may be a good option if there is no suitable archive for that datatype.
Data with privacy concerns (for example, containing human-derived, commercially-important or sensitive environmental information) can require extensive planning and compliance with a range of institutional and regulatory requirements as well as relevant laws 86 (for the Australian context, see the Australian National Data Service Publishing and Sharing Sensitive Data Guide, the National Health and Medical Research Council statement on ethical conduct in human research, and the Australian National Medical Research Storage Facility discussion paper on legal, best practice and security frameworks). In particular, it is often necessary for users of the data to be correctly identified, and to subsequently be authenticated via a mechanism such as OpenID, eduGAIN, or (in the Australian context), AAF, which places the onus on ensuring users are correctly identified with institutions that issue their credentials. Knowing who the users are can be used to restrict access, require compliance with the conditions under which the data is provided, and track user activity as an audit trail. The Data Access Compliance Office of the International Cancer Genome Consortium is an example of how to manage requests for access to controlled data. Large-scale collaborations such as the Global Alliance for Genomics and Health (GA4GH) are leading the way in approaches to sharing sensitive data across institutions and jurisdictions ( 87; also see the GA4GH Privacy and Security Policy). Importantly, plans for data sharing should be made at the start of a research project and reviewed during the project, to ensure ethical approval is in place and that the resources and metadata needed for effective sharing are available at earlier stages of the data life cycle 3.
In our experience, the majority of life science researchers are familiar with at least some public primary data repositories, and many have submitted data to them previously. A common complaint is around usability of current data submission tools and a lack of transparency around metadata requirements and the rationale for them. Some researchers raise specific issues about the potential limitations of public data repositories where their data departs from the assumptions of the repository (e.g. unusual gene models supported by experimental evidence can be rejected by the automated NCBI curation system). In such cases, researchers can provide feedback to the repositories to deal with such situations, but may not be aware of this - it could be made clearer on the repository websites. Again, this points in part to existing limitations in the undergraduate and postgraduate training received by researchers, where the concepts presented in this article are presented as afterthoughts, if at all. On the repository side, while there is a lot of useful information and training material available to guide researchers through the submission process (e.g. the EMBL-EBI Train Online webinars and online training modules), it is not always linked clearly from the database portals or submission pages themselves. Similarly, while there are specifications and standards available for many kinds of metadata [ Table 4; also see FAIRsharing], many do not have example templates available, which would assist researchers in implementing the standards in practice.
What can the research community do to encourage best-practice?
We believe that the biological/biomedical community and individual researchers have a responsibility to the public to help advance knowledge by making research data FAIR for reuse 9, especially if the data were generated using public funding. There are several steps that can assist in this mission:
-
1.
Researchers reusing any data should openly acknowledge this fact and fully cite the dataset, using unique identifiers 8, 10, 31.
-
2.
Researchers should endeavour to improve their own data management practices in line with best practice in their subdomain – even incremental improvement is better than none!
-
3.
Researchers should provide feedback to their institution, data repositories and bodies responsible for community resources (data standards, controlled vocabularies etc.) where they identify roadblocks to good data management.
-
4.
Senior scientists should lead by example and ensure all the data generated by their laboratories is well-managed, fully annotated with the appropriate metadata and made publicly available in an appropriate repository.
-
5.
The importance of data management and benefits of data reuse should be taught at the undergraduate and postgraduate levels 18. Computational biology and bioinformatics courses in particular should include material about data repositories, data and metadata standards, data discovery and access strategies. Material should be domain-specific enough for students to attain learning outcomes directly relevant to their research field.
-
6.
Funding bodies are already taking a lead role in this area by requiring the incorporation of a data management plan into grant applications. A next step would be for a formal check, at the end of the grant period, that this plan has been adhered to and data is available in an appropriate format for reuse 10.
-
7.
Funding bodies and research institutions should judge quality dataset generation as a valued metric when evaluating grant or promotion applications.
-
8.
Similarly, leadership and participation in community efforts in data and metadata standards, and open software and workflow development should be recognised as academic outputs.
-
9.
Data repositories should ensure that the data deposition and third-party annotation processes are as FAIR and painless as possible to the naive researcher, without the need for extensive bioinformatics support 40.
-
10.
Journals should require editors and reviewers to check manuscripts to ensure that all data, including research software code and samples where appropriate, have been made publicly available in an appropriate repository, and that methods have been described in enough detail to allow re-use and meaningful reanalysis 8.
Conclusions
While the concept of a life cycle for research data is appealing from an Open Science perspective, challenges remain for life science researchers to put this into practice. Among attendees of the workshop series that gave rise to this publication, we noted limited awareness among attendees of the resources available to researchers that assist in finding, collecting, processing, analysis, publishing, storing and sharing FAIR data. We believe this article provides a useful overview of the relevant concepts and an introduction to key organisations, resources and guidelines to help researchers improve their data management practices.
Furthermore, we note that data management in the era of biology as a data science is a complex and evolving topic and both best practices and challenges are highly domain-specific, even within the life sciences. This factor may not always be appreciated at the organisational level, but has major practical implications for the quality and interoperability of shared life science data. Finally, domain-specific education and training in data management would be of great value to the life science research workforce, and we note an existing gap at the undergraduate, postgraduate and short course level in this area.
Acknowledgements
The authors thank Dan Bolser for his involvement in the EMBL-ABR Data Life Cycle workshops, and all workshop participants for sharing their experiences and useful discussions.
Funding Statement
This publication was possible thanks to funding support from the University of Melbourne and Bioplatforms Australia (BPA) via an Australian Government NCRIS investment (to EMBL-ABR).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 2; referees: 2 approved]
References
- 1. Cohen-Boulakia S, Belhajjame K, Collin O, et al. : Scientific workflows for computational reproducibility in the life sciences: status, challenges and opportunities. Future Gener Comput Syst. 2017;75:284–298. 10.1016/j.future.2017.01.012 [DOI] [Google Scholar]
- 2. Hampton SE, Anderson SS, Bagby SC, et al. : The Tao of open science for ecology. Ecosphere. 2015;6(7):1–13. 10.1890/ES14-00402.1 [DOI] [Google Scholar]
- 3. Lord P, Macdonald A, Sinnot R, et al. : Large-scale data sharing in the life sciences: Data standards, incentives, barriers and funding models. UK e-Science;2005; Report No.: UKeS-2006-02. Reference Source [Google Scholar]
- 4. Piwowar HA, Vision TJ: Data reuse and the open data citation advantage. PeerJ. 2013;1:e175. 10.7717/peerj.175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vines TH, Albert AY, Andrew RL, et al. : The availability of research data declines rapidly with article age. Curr Biol. 2014;24(1):94–97. 10.1016/j.cub.2013.11.014 [DOI] [PubMed] [Google Scholar]
- 6. Lewis J, Breeze CE, Charlesworth J, et al. : Where next for the reproducibility agenda in computational biology? BMC Syst Biol. 2016;10(1):52. 10.1186/s12918-016-0288-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Voytek B: The Virtuous Cycle of a Data Ecosystem. PLoS Comput Biol. 2016;12(8):e1005037. 10.1371/journal.pcbi.1005037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Whitlock MC: Data archiving in ecology and evolution: best practices. Trends Ecol Evol. 2011;26(2):61–65. 10.1016/j.tree.2010.11.006 [DOI] [PubMed] [Google Scholar]
- 9. Wilkinson MD, Dumontier M, Aalbersberg IJ, et al. : The FAIR Guiding Principles for scientific data management and stewardship. Sci Data. 2016;3:160018. 10.1038/sdata.2016.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Van Tuyl S, Whitmire AL: Water, Water, Everywhere: Defining and Assessing Data Sharing in Academia. PLoS One. 2016;11(2):e0147942. 10.1371/journal.pone.0147942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rüegg J, Gries C, Bond-Lamberty B, et al. : Completing the data life cycle: using information management in macrosystems ecology research. Front Ecol Environ.Ecological Society of America.2014;12(1):24–30. 10.1890/120375 [DOI] [Google Scholar]
- 12. Moody D, Walsh P: Measuring the value of information: an asset valuation approach. European Conference on Information Systems1999;17 Reference Source [Google Scholar]
- 13. Mons B, Neylon C, Velterop J, et al. : Cloudy, increasingly FAIR; revisiting the FAIR Data guiding principles for the European Open Science Cloud. Inf Serv Use.IOS Press;2017;37(1):49–56. 10.3233/ISU-170824 [DOI] [Google Scholar]
- 14. Michener WK, Jones MB: Ecoinformatics: supporting ecology as a data-intensive science. Trends Ecol Evol. 2012;27(2):85–93. 10.1016/j.tree.2011.11.016 [DOI] [PubMed] [Google Scholar]
- 15. Lenhardt WC, Ahalt S, Blanton B, et al. : Data management lifecycle and software lifecycle management in the context of conducting science. J Open Res Softw. 2014;2(1):e15 10.5334/jors.ax [DOI] [Google Scholar]
- 16. Data’s shameful neglect. Nature. 2009;461(7261):145. 10.1038/461145a [DOI] [PubMed] [Google Scholar]
- 17. Strasser CA, Hampton SE: The fractured lab notebook: undergraduates and ecological data management training in the United States. Ecosphere.Ecological Society of America.2012;3(12):1–18. 10.1890/ES12-00139.1 [DOI] [Google Scholar]
- 18. Tenopir C, Allard S, Sinha P, et al. : Data Management Education from the Perspective of Science Educators. International Journal of Digital Curation. 2016;11(1):232–251. 10.2218/ijdc.v11i1.389 [DOI] [Google Scholar]
- 19. Alidina HM, Fisher D, Stienback C, et al. : Assessing and managing data.In: Ardron J, Possingham H, Klein C, editors. Marxan Good Practices Handbook; Vancouver, Canada;2008;14–20. Reference Source [Google Scholar]
- 20. Simms S, Strong M, Jones S, et al. : The future of data management planning: tools, policies, and players. International Journal of Digital Curation. 2016;11(1):208–217. 10.2218/ijdc.v11i1.413 [DOI] [Google Scholar]
- 21. Schneider MV, Griffin PC, Tyagi S, et al. : Establishing a distributed national research infrastructure providing bioinformatics support to life science researchers in Australia. Brief Bioinform. 2017. 10.1093/bib/bbx071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Womack RP: Research Data in Core Journals in Biology, Chemistry, Mathematics, and Physics. PLoS One. 2015;10(12):e0143460. 10.1371/journal.pone.0143460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. McKiernan EC, Bourne PE, Brown CT, et al. : How open science helps researchers succeed. eLife. 2016;5: pii: e16800. 10.7554/eLife.16800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sansone SA, Rocca-Serra P, Field D, et al. : Toward interoperable bioscience data. Nat Genet. 2012;44(2):121–126. 10.1038/ng.1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cook CE, Bergman MT, Finn RD, et al. : The European Bioinformatics Institute in 2016: Data growth and integration. Nucleic Acids Res. 2016;44(D1):D20–6. 10.1093/nar/gkv1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. NCBI Resource Coordinators: Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2017;45(D1):D12–D17. 10.1093/nar/gkw1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mashima J, Kodama Y, Fujisawa T, et al. : DNA Data Bank of Japan. Nucleic Acids Res. 2017;45(D1):D25–D31. 10.1093/nar/gkw1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. SIB Swiss Institute of Bioinformatics Members: The SIB Swiss Institute of Bioinformatics’ resources: focus on curated databases. Nucleic Acids Res. 2016;44(D1):D27–37. 10.1093/nar/gkv1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Burley SK, Berman HM, Kleywegt GJ, et al. : Protein Data Bank (PDB): The Single Global Macromolecular Structure Archive. Methods Mol Biol. 2017;1607:627–641. 10.1007/978-1-4939-7000-1_26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Beagrie N, Houghton J: The value and impact of the European Bioinformatics Institute: executive summary. Charles Beagrie Ltd.;2016. Reference Source [Google Scholar]
- 31. Thessen AE, Patterson DJ: Data issues in the life sciences. Zookeys. 2011; (150):15–51. 10.3897/zookeys.150.1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Brookes AJ, Robinson PN: Human genotype-phenotype databases: aims, challenges and opportunities. Nat Rev Genet. 2015;16(12):702–715. 10.1038/nrg3932 [DOI] [PubMed] [Google Scholar]
- 33. Joly Y, Dove ES, Knoppers BM, et al. : Data sharing in the post-genomic world: the experience of the International Cancer Genome Consortium (ICGC) Data Access Compliance Office (DACO). PLoS Comput Biol. 2012;8(7):e1002549. 10.1371/journal.pcbi.1002549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wong KM, Langlais K, Tobias GS, et al. : The dbGaP data browser: a new tool for browsing dbGaP controlled-access genomic data. Nucleic Acids Res. 2017;45(D1):D819–D826. 10.1093/nar/gkw1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Global Alliance for Genomics and Health: GENOMICS. A federated ecosystem for sharing genomic, clinical data. Science. 2016;352(6291):1278–80. 10.1126/science.aaf6162 [DOI] [PubMed] [Google Scholar]
- 36. Costello MJ, Appeltans W, Bailly N, et al. : Strategies for the sustainability of online open-access biodiversity databases. Biol Conserv. 2014;173:155–165. 10.1016/j.biocon.2013.07.042 [DOI] [Google Scholar]
- 37. Oliver SG, Lock A, Harris MA, et al. : Model organism databases: essential resources that need the support of both funders and users. BMC Biol. 2016;14:49. 10.1186/s12915-016-0276-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kaiser J: BIOMEDICAL RESOURCES. Funding for key data resources in jeopardy. Science. 2016;351(6268):14. 10.1126/science.351.6268.14 [DOI] [PubMed] [Google Scholar]
- 39. Schnoes AM, Brown SD, Dodevski I, et al. : Annotation error in public databases: misannotation of molecular function in enzyme superfamilies. PLoS Comput Biol. 2009;5(12):e1000605. 10.1371/journal.pcbi.1000605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bengtsson-Palme J, Boulund F, Edström R, et al. : Strategies to improve usability and preserve accuracy in biological sequence databases. Proteomics. 2016;16(18):2454–2460. 10.1002/pmic.201600034 [DOI] [PubMed] [Google Scholar]
- 41. ten Hoopen P, Amid C, Buttigieg PL, et al. : Value, but high costs in post-deposition data curation. Database (Oxford). 2016;2016: pii: bav126. 10.1093/database/bav126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. McQuilton P, Gonzalez-Beltran A, Rocca-Serra P, et al. : BioSharing: curated and crowd-sourced metadata standards, databases and data policies in the life sciences. Database (Oxford). 2016;2016: pii: baw075. 10.1093/database/baw075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Malone J, Stevens R, Jupp S, et al. : Ten Simple Rules for Selecting a Bio-ontology. PLoS Comput Biol. 2016;12(2):e1004743. 10.1371/journal.pcbi.1004743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rocca-Serra P, Salek RM, Arita M, et al. : Data standards can boost metabolomics research, and if there is a will, there is a way. Metabolomics. 2016;12:14. 10.1007/s11306-015-0879-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tenenbaum JD, Sansone SA, Haendel M: A sea of standards for omics data: sink or swim? J Am Med Inform Assoc. 2014;21(2):200–203. 10.1136/amiajnl-2013-002066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Taylor CF, Field D, Sansone SA, et al. : Promoting coherent minimum reporting guidelines for biological and biomedical investigations: the MIBBI project. Nat Biotechnol. 2008;26(8):889–896. 10.1038/nbt.1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gomez-Cabrero D, Abugessaisa I, Maier D, et al. : Data integration in the era of omics: current and future challenges. BMC Syst Biol. 2014;8 Suppl 2:I1. 10.1186/1752-0509-8-S2-I1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Goble C, Stevens R: State of the nation in data integration for bioinformatics. J Biomed Inform. 2008;41(5):687–693. 10.1016/j.jbi.2008.01.008 [DOI] [PubMed] [Google Scholar]
- 49. Mungall CJ, McMurry JA, Köhler S, et al. : The Monarch Initiative: an integrative data and analytic platform connecting phenotypes to genotypes across species. Nucleic Acids Res. 2017;45(D1):D712–D722. 10.1093/nar/gkw1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Barone L, Williams J, Micklos D: Unmet needs for analyzing biological big data: A survey of 704 NSF principal investigators. PLoS Comput Biol. 2017;13(10):e1005755. 10.1371/journal.pcbi.1005755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hinsen K: ActivePapers: a platform for publishing and archiving computer-aided research [version 3; referees: 3 approved]. F1000Res. 2015;3:289. 10.12688/f1000research.5773.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Piccolo SR, Frampton MB: Tools and techniques for computational reproducibility. Gigascience. 2016;5(1):30. 10.1186/s13742-016-0135-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jiménez RC, Kuzak M, Alhamdoosh M, et al. : Four simple recommendations to encourage best practices in research software [version 1; referees: 3 approved]. F1000Res. 2017;6: pii: ELIXIR-876. 10.12688/f1000research.11407.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Artaza H, Chue Hong N, Corpas M, et al. : Top 10 metrics for life science software good practices [version 1; referees: 2 approved]. F1000Res. 2016;5: pii: ELIXIR-2000. 10.12688/f1000research.9206.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wilson G, Bryan J, Cranston K, et al. : Good enough practices in scientific computing. PLoS Comput Biol. 2017;13(6):e1005510. 10.1371/journal.pcbi.1005510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kazic T: Ten Simple Rules for Experiments' Provenance. PLoS Comput Biol. 2015;11(10):e1004384. 10.1371/journal.pcbi.1004384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Faulconbridge A, Burdett T, Brandizi M, et al. : Updates to BioSamples database at European Bioinformatics Institute. Nucleic Acids Res. 2014;42(Database issue):D50–2. 10.1093/nar/gkt1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Schilthuizen M, Vairappan CS, Slade EM, et al. : Specimens as primary data: museums and 'open science'. Trends Ecol Evol. 2015;30(5):237–238. 10.1016/j.tree.2015.03.002 [DOI] [PubMed] [Google Scholar]
- 59. Turney S, Cameron ER, Cloutier CA, et al. : Non-repeatable science: assessing the frequency of voucher specimen deposition reveals that most arthropod research cannot be verified. PeerJ. 2015;3:e1168. 10.7717/peerj.1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Walters C, Volk GM, Richards CM: Genebanks in the post-genomic age: emerging roles and anticipated uses. Biodiversity.Taylor & Francis;2008;9(1–2):68–71. 10.1080/14888386.2008.9712887 [DOI] [Google Scholar]
- 61. Lloyd K, Franklin C, Lutz C, et al. : Reproducibility: use mouse biobanks or lose them. Nature. 2015;522(7555):151–153. 10.1038/522151a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Watson PH: Biospecimen Complexity-the Next Challenge for Cancer Research Biobanks? Clin Cancer Res. 2017;23(4):894–898. 10.1158/1078-0432.CCR-16-1406 [DOI] [PubMed] [Google Scholar]
- 63. Schnell S: Ten Simple Rules for a Computational Biologist’s Laboratory Notebook. PLoS Comput Biol. 2015;11(9):e1004385. 10.1371/journal.pcbi.1004385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Walsh E, Cho I: Using Evernote as an electronic lab notebook in a translational science laboratory. J Lab Autom. 2013;18(3):229–234. 10.1177/2211068212471834 [DOI] [PubMed] [Google Scholar]
- 65. Smith VS, Rycroft SD, Brake I, et al. : Scratchpads 2.0: a Virtual Research Environment supporting scholarly collaboration, communication and data publication in biodiversity science. Zookeys. 2011; (150):53–70. 10.3897/zookeys.150.2193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Boettiger C: A reproducible R notebook using Docker. In: Kitzes J, Turek D, Deniz F, editors. The practice of reproducible research: case studies and lessons from the data-intensive sciences Oakland, CA: University of California Press;2017. Reference Source [Google Scholar]
- 67. Koshland DE, Jr: The price of progress. Science. 1988;241(4866):637. 10.1126/science.241.4866.637 [DOI] [PubMed] [Google Scholar]
- 68. Jasny BR: Realities of data sharing using the genome wars as case study - an historical perspective and commentary. EPJ Data Sci. 2013;2:1 10.1140/epjds13 [DOI] [Google Scholar]
- 69. Caetano DS, Aisenberg A: Forgotten treasures: the fate of data in animal behaviour studies. Anim Behav. 2014;98:1–5. 10.1016/j.anbehav.2014.09.025 [DOI] [Google Scholar]
- 70. Piwowar HA, Chapman WW: A review of journal policies for sharing research data. Open scholarship: authority, community, and sustainability in the age of Web 20 Proceedings of the 12th International Conference on Electronic Publishing (ELPUB) 2008 Toronto, Canada;2008. Reference Source [Google Scholar]
- 71. National Research Council, Division on Earth and Life Studies, Board on Life Sciences, et al. : Sharing Publication-Related Data and Materials: Responsibilities of Authorship in the Life Sciences. National Academies Press;2003. 10.17226/10613 [DOI] [PubMed] [Google Scholar]
- 72. Kilkenny C, Browne WJ, Cuthill IC, et al. : Improving bioscience research reporting: the ARRIVE guidelines for reporting animal research. PLoS Biol. 2010;8(6):e1000412. 10.1371/journal.pbio.1000412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Naughton L, Kernohan D: Making sense of journal research data policies. Insights.UKSG in association with Ubiquity Press;2016;29(1):84–89. 10.1629/uksg.284 [DOI] [Google Scholar]
- 74. Pearson WR, Lipman DJ: Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988;85(8):2444–2448. 10.1073/pnas.85.8.2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Cock PJ, Fields CJ, Goto N, et al. : The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res. 2010;38(6):1767–1771. 10.1093/nar/gkp1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Iqbal SA, Wallach JD, Khoury MJ, et al. : Reproducible Research Practices and Transparency across the Biomedical Literature. PLoS Biol. 2016;14(1):e1002333. 10.1371/journal.pbio.1002333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Nekrutenko A, Taylor J: Next-generation sequencing data interpretation: enhancing reproducibility and accessibility. Nat Rev Genet. 2012;13(9):667–672. 10.1038/nrg3305 [DOI] [PubMed] [Google Scholar]
- 78. Ioannidis JP, Khoury MJ: Improving validation practices in “omics” research. Science. 2011;334(6060):1230–1232. 10.1126/science.1211811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Errington TM, Iorns E, Gunn W, et al. : An open investigation of the reproducibility of cancer biology research. eLife. 2014;3:e04333. 10.7554/eLife.04333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wolpert AJ: For the sake of inquiry and knowledge--the inevitability of open access. N Engl J Med.Mass Medical Soc;2013;368(9):785–787. 10.1056/NEJMp1211410 [DOI] [PubMed] [Google Scholar]
- 81. Laakso M, Welling P, Bukvova H, et al. : The development of open access journal publishing from 1993 to 2009. PLoS One. 2011;6(6):e20961. 10.1371/journal.pone.0020961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. McMurry JA, Juty N, Blomberg N, et al. : Identifiers for the 21st century: How to design, provision, and reuse persistent identifiers to maximize utility and impact of life science data. PLoS Biol. 2017;15(6):e2001414. 10.1371/journal.pbio.2001414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Wilson RC, Butters OW, Avraam D, et al. : DataSHIELD – new directions and dimensions. Data Science Journal. 2017;16:21 10.5334/dsj-2017-021 [DOI] [Google Scholar]
- 84. Hart EM, Barmby P, LeBauer D, et al. : Ten Simple Rules for Digital Data Storage. PLoS Comput Biol. 2016;12(10):e1005097. 10.1371/journal.pcbi.1005097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Baker M, Keeton K, Martin S: Why traditional storage systems don’t help us save stuff forever. Proc 1st IEEE Workshop on Hot Topics in System Dependability2005;2005–2120. Reference Source [Google Scholar]
- 86. Kahn SD: On the future of genomic data. Science. 2011;331(6018):728–729. 10.1126/science.1197891 [DOI] [PubMed] [Google Scholar]
- 87. Siu LL, Lawler M, Haussler D, et al. : Facilitating a culture of responsible and effective sharing of cancer genome data. Nat Med. 2016;22(5):464–471. 10.1038/nm.4089 [DOI] [PMC free article] [PubMed] [Google Scholar]