Abstract
Background
Infectious diarrhea is a common problem in the developing world, especially among people living with HIV/AIDS. Traditional diagnostic methods such as stool culture and microscopic examination are limited by resources and poor sensitivity. The use of molecular diagnostics for enteropathogen detection in this region of sub-Saharan Africa has not been fully explored. We sought to identify risk factors and characterize enteropathogens from diarrheic stools of HIV-positive patients in Gondar, Ethiopia using multiplex molecular panels targeting key infectious agents.
Methods
A cross-sectional study of 100 stool samples was performed. Samples were collected consecutively from HIV- positive patients presenting with diarrhea at University of Gondar Hospital clinic, a major center in NW Ethiopia. Genomic DNA was extracted from stool and processed using a multiplex molecular panel Allplex™ [Seegene, Canada]. Correlations between patient characteristics, symptoms, public health risk factors, and enteropathogen type (s) were studied. Eighty-six samples were successfully analyzed by molecular methods.
Results
The mean age was 35 with 43% male. Eighty percent lived in an urban area, 18% had access to well water only, and 81% practiced proper hand hygiene. The majority of patients (72%) were receiving HAART with a median CD4 cell count of 362/μL. Multiple pathogens were detected in 94% of specimens, with an average of 5 enteropathogens per sample. Common bacteria, viruses, and parasites detected were Shigella spp./enteroinvasive E. coli (80%), enterotoxigenic E. coli (73%), Norovirus (16%) and B. hominis (62%). CD4 cell count < 500/ μL was associated with the presence of viruses (p = 0.004) and the absence of STEC (p = 0.010). The use of HAART or CD4 levels was not associated with the number of enteropathogens detected.
Conclusions
Diarrheic stool from HIV-positive outpatients in Gondar, Ethiopia had on average 5 enteropathogens present in their stool. Shigellaspp./enteroinvasive E. coli and enterotoxigenic E. coli are the major pathogens, not dissimilar to immunocompetent individuals in low income countries.
Keywords: HIV, Enteropathogens, Molecular, Diarrhea, Diagnostics
Background
Diarrheal illness contributes to significant mortality and morbidity within the HIV-positive population [1]. It accounts for 1 in 9 deaths for children worldwide and the death rate increases 11 -fold in children with HIV [2]. Furthermore, diarrhea is associated with reduced quality of life and can cause psychological and social burden on afflicted patients [3, 4]. This is most notable in low-resource settings such as Ethiopia in which HIV/AIDS occurs in 1.5% of adults [5].
The variety of organisms known to cause diarrhea presents an inherent challenge in treatment to the clinician, and, in diagnosis, to the laboratory due to shortage of reagents, laboratory set up and skilled manpower in developing countries. Clinical laboratories currently utilize an array of different methodologies to test for bacterial, parasitic, and viral causes of diarrhea among HIV/AIDS patients, a strategy that suffers from poor sensitivity, potentially long turnaround times, and complicated ordering practices and workflows [6–14]. Additionally, there are limited or no testing methods routinely available for most diarrheagenic Escherichia coli strains [15] and certain enteric viruses.
Molecular techniques for enteropathogen detection provides a comprehensive, rapid, and streamlined alternative to conventional methods for the microbiological diagnosis of diarrhea in the laboratory setting. The potential advantages include improved performance parameters, a more extensive menu of pathogens, and a turnaround time as short as 1 h [15]. Diagnostic yields in terms of pathogens detected are also approximately 30% higher and multiple enteropathogens are more commonly detected per sample, occurring in up to 35% of positive samples [16].
Prevalence of enteropathogens is regionally dependent, with stark differences between developed and developing countries. A study conducted in the United States on 1556 diarrheal stools using the Biofire FilmArray® platform detected a wide variety of enteropathogens in approximately half (832) of the stool samples collected. In the study, EPEC (41.8%), C. difficile (24.5%), EAEC (13.1%) and norovirus GI/GII (8.4%) were the most prevalent organisms detected in positive samples and 73% had multiple enteropathogens [15]. In contrast, a study on 91 diarrheal stool of symptomatic Peruvians demonstrated higher prevalence of diarrheagenic E. coli strains and mixed enteropathogens. The most common organisms detected in their study was Shigella spp./enteroinvasive E. coli (EIEC) at 93%, G. lamblia (70%), enteroaggregative E. coli (EAEC) (60%), and enterotoxigenic E. coli (ETEC) (48%). In addition, 100% of their samples contained mixed enteropathogens [17]. Our study’s aim was to identify epidemiological risk factors and characterize enteropathogens from diarrheic stools of HIV-positive patients in Gondar, Ethiopia using multiplex molecular panels targeting key infectious agents.
Methods
Patient population
Diarrheic stool was collected consecutively from all pediatric and adult patients with HIV who attended an antiretroviral therapy (ART) clinic at University of Gondar Hospital, Ethiopia from January 2016, to May 2016. All patients who had diarrhea, defined as 3 or more loose or watery stools during a 24-h period, were included in the study. Acute and chronic diarrhea was defined as diarrhea lasting less than 2 weeks and greater than 4 weeks, respectively. Patients who were on antibiotics or anti-parasitic agents were excluded. Socio-demographic data was collected via patient questionnaires (created in Amharic) and face-to-face patient interviews. The study protocol was reviewed and approved by research ethics committee of School of Biomedical and Laboratory Sciences, College of Medicine and Health Sciences, University of Gondar (SBMLS/641/08). A permission letter was obtained from ART clinic, University of Gondar Hospital (para/1002/08). Informed assent and written consent was obtained from each study participant after explaining the objective, benefit and risk of the study with vernacular language that the study participant could understand.
Specimen handling
Diarrheic stool was collected on site using sterile containers. Wet mount microscopy, using direct saline and iodine smear, and modified acid-fast staining was performed for the detection of Cryptosporidium, Isospora and Cyclospora species. Stool microscopy was performed by two experienced laboratory technologists and discordant results were resolved by a local microbiology expert. After microscopy was performed, specimens were stored at 4 °C for 1 month until further molecular testing was performed.
Molecular testing
Total nucleic acid was extracted from the stool specimens using QIAamp DNA Stool Mini Kit (QIAGEN, Germany) according to the manufacturer’s instructions [18] and subsequently stored at -20 °C. The extracted stool DNA was then shipped to the University of Calgary, in Calgary, Alberta, for molecular analysis using the Allplex™ [Seegene, Canada] kit on the CFX96TM Real-time PCR instrument (Bio-Rad, Canada). The Allplex™ gastrointestinal panel comprises bacteria including Campylobacter, Clostridium difficle toxin A/B, Plesiomonas shigelloides, Salmonella, Vibrio, Yersinia enterocolitica, and diarrheagenic E. coli/shigella including Enteroaggregative E. coli (EAEC), Enteropathogenic E. coli (EPEC), Enterotoxigenic E. coli (ETEC), Shiga-like toxin-producing E. coli (STEC), E. coli 0157 and Shigella/Enteroinvasive E. coli (EIEC). Parasites detected included Cryptosporidum, Cyclospora cayetnensis, Entamoeba histolytica, Giardia lamblia, Blastocystis hominis (BH) and Dientamoeba fragilis (DF). Viruses detected included Adenovirus F 40/41, Astrovirus, Norovirus GI/GII, Rotavirus A and Sapovirus. Negative and positive controls were included in all extraction and amplification procedures. Gene targets are considered proprietary at this time (Seegene, Canada).
Statistical analysis
Correlations between patient characteristics, symptoms, epidemiological risk factors and enteropathogen (s) were explored using STATA (Version 14.1). Categorical variables were analyzed using Pearson chi-squared or Fisher’s exact test. Continuous variables were analyzed using two paired t-test for normally distributed variables, two-sample Wilcoxon rank-sum test for non-normally distributed variables and linear regression or Spearman rank test for two continuous variables.
Results
Diarrheic stool from 100 patients was collected. Fourteen samples were excluded from the molecular testing arm due to insufficient sample volume or sample processing deficiency. Patient demographics are outlined in Table 1. The mean age was 34.9 with 5 patients under the age of 14. 43.0% of patients were male, 80.2% lived in an urban area, 17.4% had access to well water only, 88.4% had access to proper latrines, and 81.4% practiced proper hand hygiene. Patients median duration of diarrhea was 10 days with 53.5% having acute diarrhea and 7.0% having chronic diarrhea. Median bowel movement frequency per day was 5. The median CD4 cell count was 361.5/μL with 36.1% having CD4 cell counts < 200/μL. The majority (72.1%) of patients were receiving highly active antiretroviral therapy (HAART) at the time of collection.
Table 1.
Demographics of Included Study Participants (N = 86)
| Age, mean (SD) | 34.9 (12.2) |
|---|---|
| Age, median | 35 |
| Male (%) | 43.0 |
| Urban (%) | 80.2 |
| Tap water (%) | 82.6 |
| Access to toilet (%) | 88.4 |
| Practices proper hand hygiene (%) | 81.4 |
| Receiving HAART (%) | 72.1 |
| CD4 count/μl, mean (SD) | 359.9 (278.2) |
| CD4 count/μl, median | 361.5 |
| CD4 count < 50/μl (%) | 12.8 |
| CD4 count 50–200/μl (%) | 23.3 |
| CD4 count 200–500/μl (%) | 30.2 |
| CD4 count > 500/μl (%) | 34.9 |
| Duration diarrhea, median (days) | 10 |
| Diarrhea duration < 14 days (%) | 53.5 |
| Diarrhea duration 14–30 days (%) | 39.5 |
| Diarrhea duration > 30 days (%) | 7.0 |
| Frequency diarrhea per day, median | 5 |
| Total organisms, mean (SD) | 5.2 (2.3) |
Out of the 100 specimens, 28 were positive for intestinal parasites using wet mounts and modified acid fast staining. C. parvum was the most common parasite at 28.6% followed by E. histolytica (17.9%), G. lamblia at (14.3%), C. cayetanensis (14.3%), S. stercoralis, (7.1%) and hookworm (3.6%).
The enteropathogens detected by the Allplex™ panel are outlined in Fig. 1. No failures were detected among the negative and positive controls. Only one patient had no organisms detected (when excluding B. hominis, D. fragilis and Aeromonas spp). The most common organisms detected were Shigella spp./enteroinvasive E. coli (EIEC) at 80.2%, enterotoxigenic E. coli (ETEC) at 73.3%, Aeromonas spp. at 73.3%, and enteroaggregative E. coli (EAEC) at 59.3%. Parasites detected included Blastocystis hominis (61.6%), Giardia lamblia (17.4%), Cryptosporidium spp. (10.5%), and Dientamoeba fragilis (8.1%). Viruses detected included norovirus GI/GII (16.3%), rotavirus A (4.7%) and adenovirus 40/41 (3.5%). Multiple pathogens were detected in 94.1% of stool specimens, with 64.0% having 5 or more enteropathogens (33.7% if excluding B. hominis, D. fragilis and Aeromonas spp.).
Figure 1.
Enteropathogens Detected (%) in HIV Patient Participants with Diarrhea (N = 86) using the Allplex™ gastrointestinal panels [Seegene, Canada]
Older age was associated with Campylobacter spp., only occurring in patients between over the age of 35 (p = 0.04). D. fragilis was associated with increased frequency of diarrhea (median 4 movements when D. fragilis present compared to median 5, p = 0.02). Use of well water was associated with Cryptosporidium (33.3% with well water vs 6.0%, p = 0.001).
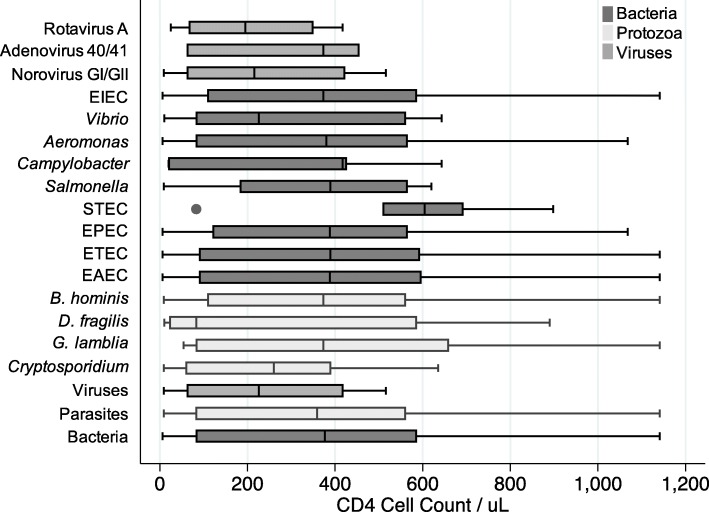
A box plot describing enteropathogen frequencies with CD4 cell count is outlined in Fig. 2. Only viral enteropathogens and STEC were associated with CD4 cell counts. A CD4 cell count < 500/μL was associated with the presence of a viral enteropathogen (p = 0.004) and the absence of STEC (p = 0.018) (Table 2). Lower CD4 cell counts were also associated with a longer duration of diarrhea (p = 0.0015) and older age (p = 0.011). Associations between organisms at CD4 count < 200/uL and < 50/uL were analyzed and none were found to be statistically significant.
Fig. 2.
Box Plot graphic (median and interquartile range) depicting CD4 cell counts per μl in relation to the enteropathogen detected (N = 86). E. coli 0157 and C. difficle excluded from box plot due to insufficient positive cases. The black solid circle denotes a single outlier case
Table 2.
Proportion of pathogens detected in patients with CD4 cell counts < 500/μL
| Pathogen | CD4 < 500/μL (N = 56) | CD4 > 500/μL (N = 30) | p-valuea |
|---|---|---|---|
| STEC (%) | 1.8 | 16.7 | 0.018 |
| Norovirus GI/GII (%) | 23.2 | 3.3 | 0.029 |
| Rotavirus A (%) | 7.1 | 0 | 0.293 |
| Adenovirus 40/41 (%) | 5.4 | 0 | 0.549 |
| Viruses (%) | 28.6 | 3.3 | 0.004 |
a2-sided Fisher’s exact test
The number of organisms detected in one sample was not associated with any specific demographic, including CD4 cell count, administration of HAART, age, duration of diarrhea or frequency of diarrhea. This was also the case when excluding B. hominis, D. fragilis and Aeromonas spp.
Discussion
Detection of intestinal parasites was overall higher using multiplex PCR compared to microscopy in our study. While Cryptosporidium spp. detection was similar, multiplex PCR yielded more G. lamblia, B. hominis, and D. fragilis but less E. histolytica and C. cayetanensis. Notably, microscopy is unable to distinguish E. histolytica (pathogen) from E. dispar (non-pathogen) whereas molecular detection is specific for E. histolytica. Multiplex PCR is known to be more sensitive than conventional techniques for parasite detection therefore it is not clear why C. cayetanensis was not detected using multiplex PCR in our study [15]. Shigella spp./enteroinvasive enteroinvasive E. coli was the most commonly detected organism (s) in our study at 80.2%, followed by ETEC (73.3%), Aeromonas spp. (73.3%), EAEC (59.3%) and EPEC (54.7%). This is similar to a study among symptomatic Peruvians in which Shigella spp./enteroinvasive E. coli was the most commonly detected organism at 93% followed by G. lamblia (70%), EAEC (60%), ETEC (48%) and EPEC (41%). G.lamblia was of lower prevalence in our study.
In contrast, detection of diarrheagenic E. coli strains are much lower when examining stool from patients in developed countries, highlighting their common frequency in sanitation poor settings [19]. Even in asymptomatic individuals, prevalence of diarrheagenic strains of E. coli are high in developing countries. For instance, one study detected an enteropathogen in 61% of asymptomatic travelers, with EPEC, EAEC and ETEC being most commonly detected at 42, 28, and 9%, respectively [20].
Our study reported a 24.5% prevalence of enteric viruses with the most common being norovirus GI/GII at 16.3%. This finding is higher than that reported in other studies examining diarrheal stool from adults with HIV/AIDS, which showed an enteric virus prevalence of 15.9–17%. However, those studies were done using less sensitive conventional techniques [21, 22]. A study examining microbiological etiologies of diarrhea in adult Peruvians detected enteric viruses, with multiplex PCR, in 18% of samples, the majority being norovirus GI/GII at 12% [17]. To our knowledge, no study has yet examined diarrheic stool on HIV/AIDS patients using multiplex PCR for comparison.
Debate exists in the literature as to whether B. hominis, D. fragilis and Aeromonas spp. are true enteropathogens with virulence potential [23, 24]. Among several studies examining HIV populations, there has been no statistically significant difference between B. hominis found in HIV patients with or without diarrhea [25] and in D. fragilis found in HIV-negative or HIV-positive individuals [26]. Several studies on HIV-positive patients did report higher detection of Aeromonas spp. in those with diarrhea compared to those without [27, 28]. The lack of a control group, unfortunately, makes our study unable to draw any conclusions with respect to the aforementioned organisms and their intrinsic virulence.
Corresponding to known literature, lower CD4 cell counts in our study were associated with a longer duration of diarrhea [29]. Lower CD4 cell counts were not associated with known opportunistic enteropathogens (Cryptosporidium and Isospora) but our sample size was small (N = 9). We detected an association between lower CD4 cell counts and the presence of enteric viruses (norovirus GI/II, rotavirus A, adenovirus 40/41) in this study. Only one virus was detected when CD4 cell count was > 500/μL. Several studies have associated enteric viruses with more advanced HIV infection, suggesting that CD4 deficiency helps mediate enteric viral replication and infection [22, 30, 31]. Astrovirus has been implicated as a potential opportunistic pathogen but none was detected in this study [32]. A study examining stool from symptomatic HIV patients with diarrhea in London, UK found that adenovirus and rotavirus were associated with significantly lower CD4 cell counts compared to coronavirus or norovirus [21]. Our study found that rotavirus and norovirus were independently associated with lower CD4 cell counts. Adenovirus occurred more frequently at lower CD4 cell counts but was not statistically significant, likely due to small sample size in our study (N = 5).
Ninety-four percent of specimens in our study contained a mixture of enteropathogens, with an average of 5 organisms per sample. When excluding B. hominis, D. fragilis and Aeromonas spp., prevalence of mixed infection was still high at 90% with a mean of 4 organisms per sample. High prevalence of mixed enteropathogens has been demonstrated in stool from residents and travelers in developing countries. For instance, 25% and 53% of stool specimens from asymptomatic travelers and travelers with diarrhea, respectively, had two or more bacterial pathogens present when using molecular techniques [20]. All stool samples from symptomatic Peruvians had mixed enteropathogens present with a mean of 4.4 pathogens per specimen when tested using the FilmArray® panel [17]. In a longitudinal cohort study of 147 infants in Dhaka, Bangladesh, multiple enteropathogens were also observed in both asymptomatic surveillance and diarrheal stools with the average number of organisms being 4.3 and 5.6, respectively [33]. In contrast, no enteropathogens were detected in non-diarrheal stool samples taken from infants within their first year of life in Virginia, US [33]. Our study has several limitations. Notably, there was no control group for comparison and our overall sample size was not large enough to provide any reliable conclusions on less commonly detected enteropathogens (e.g. Cryptosporidium spp., Campylobacter spp.).
Conventional approaches, including culture, microscopy, and antigen-based tests have significant limitations such as the limit of detection and the need for labour-intensive procedures. Molecular diagnostics, especially PCR based tests, are rapidly changing research and practice in infectious diseases. A syndromic approach to the diagnosis of diarrheal disease, with its broad range of potential infectious etiologies, may benefit from the use of multiplex molecular formats. However, their applicability due to cost and capital equipment requirements in developing countries is unclear.
In our study, we found a high number of mixed enteropathogens in diarrheal stools taken from symptomatic HIV patients from Ethiopia when using multiplex PCR. To our knowledge, this is the first study to demonstrate this finding in people living with HIV/AIDS in a developing setting using multiplex molecular methods. Most specimens were mixed with diarrheagenic E. coli strains which were similar in prevalence to those seen among diarrheal stools in symptomatic Peruvians [17]. This finding highlights the difficulties in determining colonization versus infection when using extremely sensitive diagnostic methods such as multiplex PCR, and the specific etiological role of each organism in an individual patient. The use of quantitative cutoffs for enteropathogen detection using multiplex PCR has been explored but further clinical studies are needed, especially in developing countries, where high rates of colonization with diarrheagenic strains of E. coli exists [33, 34]. One concern is that inappropriate antimicrobial use may escalate with this technology without a fuller understanding of infection versus colonization. Nevertheless, simplification of these technologies, reduction in cost, and better clinical understanding will undoubtedly enhance patient management.
Conclusion
In conclusion, multiplex PCR panels are elaborate far greater complexity of infection in HIV patients than traditional methods in resource-limited settings. Establishing the true etiology of diarrhea and treatment approach remains challenging. CD4 cell counts < 500/μL were associated with increased detection of gastrointestinal viruses; an interesting finding that requires further investigation.
Acknowledgments
We recognize the health clinic staff in University of Gondar Hospital who assisted in the study. We also thank Ms. Tracie Lloyd for expert technical assistance, and Drs. John Gill and David Goldfarb for reading the manuscript and providing valuable feedback.
Funding
Funding for this work was provided in part by Calgary Laboratory Services.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request. Data presented here was previously reported in poster format at ID week in San Diego, CA on4 October, 2017 [35].
Abbreviations
- AIDS
Acquired immunodeficiency syndrome
- ART
Antiretroviral therapy
- BH
Blastocystis hominis
- DF
Dientamoeba fragilis
- DNA
Deoxyribonucleic acid
- EAEC
Enteroaggregative E. coli
- EIEC
Shigella spp./enteroinvasive E. coli
- EPEC
Enteropathogenic E. coli
- ETEC
Enterotoxigenic E. coli
- HIV
Human immunodeficiency virus
- PCR
Polymerase chain reaction
- STEC
Shiga-like toxin-producing E. coli
Authors’ contributions
LS, AA, SG performed the actual experimental work. LS, SG, HT and DRP contributed to the experimental design. LS, HT, WS, DRP, and AG performed data analysis. All authors contributed to the writing of the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The study protocol was reviewed and approved by research ethics committee of School of Biomedical and Laboratory Sciences, College of Medicine and Health Sciences, University of Gondar (SBMLS/641/08). A permission letter was obtained from ART clinic, University of Gondar Hospital (para/1002/08). Informed assent and written consent was obtained from each study participant after explaining the objective, benefit and risk of the study with vernacular language that the study participant could understand.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Feasey NA, Healey P, Gordon MA. Review article: the aetiology, investigation and management of diarrhoea in the HIV-positive patient. Aliment Pharmacol Ther. 2011;34(6):587–603. doi: 10.1111/j.1365-2036.2011.04781.x. [DOI] [PubMed] [Google Scholar]
- 2.Hutton G, Haller L, Bartram J. Global cost-benefit analysis of water supply and sanitation interventions. J Water Health. 2007;5(4):481–502. doi: 10.2166/wh.2007.009. [DOI] [PubMed] [Google Scholar]
- 3.Siegel K, Schrimshaw EW, Brown-Bradley CJ, Lekas H-M. Sources of emotional distress associated with diarrhea among late middle-age and older HIV-infected adults. J Pain Symptom Manag. 2010;40(3):353–369. doi: 10.1016/j.jpainsymman.2010.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tramarin A, Parise N, Campostrini S, Yin DD, Postma MJ, Lyu R, et al. Association between diarrhea and quality of life in HIV-infected patients receiving highly active antiretroviral therapy. Qual Life Res Int J Qual Life Asp Treat Care Rehabil. 2004;13(1):243–250. doi: 10.1023/B:QURE.0000015282.24774.36. [DOI] [PubMed] [Google Scholar]
- 5.Ethiopia Demographic and Health Survey 2011 [Internet]. Central Statistical Agency and ICF International, Addis Ababa and Calverton; 2011 [cited 2017 Oct 26]. Available from: https://dhsprogram.com/pubs/pdf/FR255/FR255.pdf
- 6.Van Lint P, Rossen JW, Vermeiren S, Ver Elst K, Weekx S, Van Schaeren J, et al. Detection of Giardia lamblia, cryptosporidium spp. and Entamoeba histolytica in clinical stool samples by using multiplex real-time PCR after automated DNA isolation. Acta Clin Belg. 2013;68(3):188–192. doi: 10.2143/ACB.3170. [DOI] [PubMed] [Google Scholar]
- 7.Svraka-Latifovic S, Bouter S, Naus H, Bakker LJ, Timmerman CP, Dorigo-Zetsma JW. Impact of transition from microscopy to molecular screening for detection of intestinal protozoa in Dutch patients. Clin Microbiol Infect Off Publ Eur Soc Clin Microbiol Infect Dis. 2014;20(11):O969–O971. doi: 10.1111/1469-0691.12623. [DOI] [PubMed] [Google Scholar]
- 8.Vanniasinkam T, Lanser JA, Barton MD. PCR for the detection of campylobacter spp. in clinical specimens. Lett Appl Microbiol. 1999;28(1):52–56. doi: 10.1046/j.1365-2672.1999.00474.x. [DOI] [PubMed] [Google Scholar]
- 9.de Boer RF, Ott A, Kesztyüs B, Kooistra-Smid AMD. Improved detection of five major gastrointestinal pathogens by use of a molecular screening approach. J Clin Microbiol. 2010;48(11):4140–4146. doi: 10.1128/JCM.01124-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kele B, Lengyel G, Deak J. Comparison of an ELISA and two reverse transcription polymerase chain reaction methods for norovirus detection. Diagn Microbiol Infect Dis. 2011;70(4):475–478. doi: 10.1016/j.diagmicrobio.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 11.Castriciano S, Luinstra K, Petrich A, Smieja M, Lee C, Jang D, et al. Comparison of the RIDASCREEN norovirus enzyme immunoassay to IDEIA NLV GI/GII by testing stools also assayed by RT-PCR and electron microscopy. J Virol Methods. 2007;141(2):216–219. doi: 10.1016/j.jviromet.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 12.Siqueira JAM, Júnior ECS, da C LA, Gabbay YB. Molecular analysis of norovirus in specimens from children enrolled in a 1982-1986 study in Belém, Brazil: a community-based longitudinal study. J Med Virol. 2017;89(11):1894–1903. doi: 10.1002/jmv.24812. [DOI] [PubMed] [Google Scholar]
- 13.Gautam R, Lyde F, Esona MD, Quaye O, Bowen MD. Comparison of premier™ Rotaclone®, ProSpecT™, and RIDASCREEN® rotavirus enzyme immunoassay kits for detection of rotavirus antigen in stool specimens. J Clin Virol Off Publ Pan Am Soc Clin Virol. 2013;58(1):292–294. doi: 10.1016/j.jcv.2013.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wolffs PFG, Bruggeman CA, van Well GTJ, van Loo IHM. Replacing traditional diagnostics of fecal viral pathogens by a comprehensive panel of real-time PCRs. J Clin Microbiol. 2011;49(5):1926–1931. doi: 10.1128/JCM.01925-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buss SN, Leber A, Chapin K, Fey PD, Bankowski MJ, Jones MK, et al. Multicenter evaluation of the BioFire FilmArray gastrointestinal panel for etiologic diagnosis of infectious gastroenteritis. J Clin Microbiol. 2015;53(3):915–925. doi: 10.1128/JCM.02674-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Byrne S. Towards routine diagnosis of gastrointestinal infections by molecular technology in Australian laboratories. Department of microbiology and molecular pathology [dissertation] [Internet]. Sullivan Nicolaides Pathology; 2017 [cited 2017 Oct 26]. Available from: http://city.asnevents.com.au/assets/Towards-the-Routine-Diagnosis-of-Gastrointestinal-Infections-by-Molecular-Technology-in-Australian-Laboratories-Shane-Byrne-Resubmission.pdf
- 17.Gregory M. Evaluation of infectious diarrhea in travelers to Peru. In Denver, Colorado: American Society for Microbiology Conference; 2013 [cited 2017 Oct 26]. Available from: http://biofiredefense.com/media/Evaluation-of-Infectious-Diarrheas-in-Travelers-to-Peru-Using-the-FilmArray-GI-Panel.pdf
- 18.Vandenberg N, van Oorschot RAH. Extraction of human nuclear DNA from feces samples using the QIAamp DNA stool mini kit. J Forensic Sci. 2002;47(5):993–995. doi: 10.1520/JFS15502J. [DOI] [PubMed] [Google Scholar]
- 19.Otto CC, Chen LH, He T, Tang Y-W, Babady NE. Detection of gastrointestinal pathogens in oncology patients by highly multiplexed molecular panels. Eur J Clin Microbiol Infect Dis Off Publ Eur Soc Clin Microbiol. 2017;36(9):1665-72. [DOI] [PMC free article] [PubMed]
- 20.Lääveri T, Antikainen J, Pakkanen SH, Kirveskari J, Kantele A. Prospective study of pathogens in asymptomatic travellers and those with diarrhoea: aetiological agents revisited. Clin Microbiol Infect Off Publ Eur Soc Clin Microbiol Infect Dis. 2016;22(6):535–541. doi: 10.1016/j.cmi.2016.02.011. [DOI] [PubMed] [Google Scholar]
- 21.Thomas PD, Pollok RC, Gazzard BG. Enteric viral infections as a cause of diarrhoea in the acquired immunodeficiency syndrome. HIV Med. 1999;1(1):19–24. doi: 10.1046/j.1468-1293.1999.00004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thea DM, Glass R, Grohmann GS, Perriens J, Ngoy B, Kapita B, et al. Prevalence of enteric viruses among hospital patients with AIDS in Kinshasa, Zaire. Trans R Soc Trop Med Hyg. 1993;87(3):263–266. doi: 10.1016/0035-9203(93)90119-B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stark D, Barratt JLN, van Hal S, Marriott D, Harkness J, Ellis JT. Clinical significance of enteric protozoa in the immunosuppressed human population. Clin Microbiol Rev. 2009;22(4):634–650. doi: 10.1128/CMR.00017-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Janda JM, Abbott SL. The genus Aeromonas: taxonomy, pathogenicity, and infection. Clin Microbiol Rev. 2010;23(1):35–73. doi: 10.1128/CMR.00039-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Albrecht H, Stellbrink HJ, Koperski K, Greten H. Blastocystis hominis in human immunodeficiency virus-related diarrhea. Scand J Gastroenterol. 1995;30(9):909–914. doi: 10.3109/00365529509101600. [DOI] [PubMed] [Google Scholar]
- 26.Stark D, Fotedar R, van Hal S, Beebe N, Marriott D, Ellis JT, et al. Prevalence of enteric protozoa in human immunodeficiency virus (HIV)-positive and HIV-negative men who have sex with men from Sydney Australia. Am J Trop Med Hyg. 2007;76(3):549–552. [PubMed] [Google Scholar]
- 27.Lule JR, Mermin J, Awor A, Hughes P, Kigozi A, Wafula W, et al. Aetiology of diarrhoea among persons with HIV and their family members in rural Uganda: a community-based study. East Afr Med J. 2009;86(9):422–429. doi: 10.4314/eamj.v86i9.54164. [DOI] [PubMed] [Google Scholar]
- 28.Ramalivhana C, Momba M, Igumbor J. Scope and frequency of enteric bacterial pathogens isolated from HIV/AIDS patients and their household drinking water in Limpopo province. Water SA. 1989;33(4):539–548. [Google Scholar]
- 29.Brink A-K, Mahé C, Watera C, Lugada E, Gilks C, Whitworth J, et al. Diarrhea, CD4 counts and enteric infections in a community-based cohort of HIV-infected adults in Uganda. J Inf Secur. 2002;45(2):99–106. doi: 10.1053/jinf.2002.1002. [DOI] [PubMed] [Google Scholar]
- 30.Cunningham AL, Grohman GS, Harkness J, Law C, Marriott D, Tindall B, et al. Gastrointestinal viral infections in homosexual men who were symptomatic and seropositive for human immunodeficiency virus. J Infect Dis. 1988;158(2):386–391. doi: 10.1093/infdis/158.2.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaljot KT, Ling JP, Gold JW, Laughon BE, Bartlett JG, Kotler DP, et al. Prevalence of acute enteric viral pathogens in acquired immunodeficiency syndrome patients with diarrhea. Gastroenterology. 1989;97(4):1031–1032. doi: 10.1016/0016-5085(89)91516-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mendez E, Arias C. Fields Virology. 5. 2007. Astroviruses; pp. 982–1000. [Google Scholar]
- 33.Taniuchi M, Sobuz SU, Begum S, Platts-Mills JA, Liu J, Yang Z, et al. Etiology of diarrhea in Bangladeshi infants in the first year of life analyzed using molecular methods. J Infect Dis. 2013;208(11):1794–1802. doi: 10.1093/infdis/jit507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu J, Platts-Mills JA, Juma J, Kabir F, Nkeze J, Okoi C, et al. Use of quantitative molecular diagnostic methods to identify causes of diarrhoea in children: a reanalysis of the GEMS case-control study. Lancet Lond Engl. 2016;388(10051):1291–1301. doi: 10.1016/S0140-6736(16)31529-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.ID week 2017 [cited 4 October 2017]. Available from https://academic.oup.com/ofid/article/4/suppl_1/S608/4295101.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request. Data presented here was previously reported in poster format at ID week in San Diego, CA on4 October, 2017 [35].