Abstract
Background
The death receptors TRAIL-R1 and TRAIL-R2 are frequently overexpressed in cancer and there is an emerging evidence for their important role in malignant progression, also in the case of pancreatic ductal adenocarcinoma (PDAC). In their canonical localization at the plasma membrane, TRAIL-R1/−R2 may induce cell death and/or pro-inflammatory signaling leading to cell migration, invasion and metastasis. Although, they have repeatedly been found intracellular, in the cytoplasm and in the nucleus, their functions in intracellular locations are still not well understood. Likewise, studies dealing with the prognostic relevance of TRAIL-Rs located in particular cellular compartments are very rare. For PDAC, the correlation of nuclear TRAIL-R2 with worse patients’ prognosis has been shown recently. Corresponding data on TRAIL-R1 are not available so far.
Methods
In the present study we analyzed the expression of TRAIL-R1 in 106 PDACs and 28 adjacent, peritumoral non-malignant pancreatic ducts with special emphasis on its cytoplasmic and nuclear localization and correlated the immunohistochemical findings with clinico-pathological patient characteristics.
Results
TRAIL-R1 was found in 93.4% of all PDAC samples. Cytoplasmic staining was present with very similar intensity in tumor and normal tissue. In contrast, nuclear TRAIL-R1 staining was significantly stronger in tumor compared to normal tissue (p = 0.006). Interestingly, we found that the number of cells with cytoplasmic TRAIL-R1 staining negatively correlates with tumor grading (p = 0.043). No such correlation could be detected for nuclear TRAIL-R1. Neither, cytoplasmic nor nuclear TRAIL-R1 staining showed a correlation with other clinico-pathological parameter such as pTNM categories. However, Kaplan-Meier analyses revealed significantly prolonged median survival of patients with positive cytoplasmic TRAIL-R1 expression in more than 80% of tumor cells compared to patients with tumors containing a smaller quantity of cells positively stained for cytoplasmic TRAIL-R1 (20 vs. 8 months; p = 0.004).
Conclusion
Cytoplasmic TRAIL-R1 is a positive prognostic marker for patients with PDAC. Our findings indicate that loss of cytoplasmic TRAIL-R1 results in recurrent disease with more malignant phenotype thus suggesting anti-tumor activities of cytoplasmic TRAIL-R1 in PDAC.
Keywords: TRAIL-R1, Death receptor, Immunhistology, Pancreatic cancer
Background
Despite tremendous efforts in molecular and clinical oncology, pancreatic ductal adenocarcinoma (PDAC) still remains one of the deadliest cancers with a mortality rate almost equal to its incidence rate [1]. Its dismal prognosis results from the lack of early diagnostic options, its highly aggressive growth and a resistance to current radio- and chemotherapeutic treatments [2]. Thus, identification of new prognostic markers provides a strategy to uncover still unknown players driving PDAC malignancy and potentially to identify novel therapeutic targets.
TNF-related apoptosis inducing ligand (TRAIL) and its death inducing receptors TRAIL-R1 and TRAIL-R2 are promising candidates for the development of such novel targeted strategies. The rationale behind this assumption is the original observations that i) TRAIL induces apoptosis preferentially in tumor cells leaving normal healthy cells alive; ii) tumor cells usually express high levels of either TRAIL-R1 or TRAIL-R2 or both. These facts led to the development of different TRAIL formulations as well as agonistic TRAIL-R1- or TRAIL-R2-specific antibodies for treatment of human malignancies. However, relatively soon it has been recognized that many tumor cells are resistant to TRAIL induced apoptosis, the fact explaining the disappointing results from clinical trials [3]. In addition, it became evident that TRAIL-R1 and TRAIL-R2 may respond to TRAIL - apart from apoptosis induction - with activation of different non-apoptotic signal transduction pathways like NF-kB, ERK1/ERK2, JNK, Src and AKT [4], which can lead to the inhibition of apoptosis as well as to cell proliferation, migration and invasion. Most importantly, TRAIL receptor signaling may enhance cancer cell invasion and metastasis in vivo [5–7]. Thus, therapeutic concepts are needed which combine TRAIL receptor targeting agents with agents sensitizing tumor cells and reducing the unwanted, non-death-inducing signaling of the receptors. The important concern regarding TRAIL-receptor based anti-tumor therapy is also the observed preference for the usage of the particular TRAIL death receptor for the transmission of the TRAIL-mediated signaling in tumor cells. Generalized predictions on main death receptor responsibility for apoptosis induction in given cancer types are difficult. It is widely accepted that the preference for either TRAIL-R1 or TRAIL-R2 is a cell type specific feature. A comprehensive compilation of tumor cell lines together with their preferences for usage of the particular TRAIL death receptor is provided by Roosmalen et al. [8]. In PDAC cells, others and we have shown that regardless of the simultaneous presence of TRAIL-R1 and TRAIL-R2 at the cell surface, these cells use predominantly TRAIL-R1 when treated with recombinant TRAIL [9, 10]. Consequently, TRAIL-R1-targeting variants of TRAIL and agonistic TRAIL-R1 specific antibodies were expected to have higher therapeutic effects in the treatment of PDAC than they in fact do [3]. Interestingly, a recent report revealed that some PDAC cell lines show preference for TRAIL-R2 in inducing cell death [11] pointing to an unexpected high diversity of TRAIL receptor preference even in the same tumor entity.
Of note, under physiological conditions TRAIL has been shown to be an important effector molecule in the tumor immunosurveillance [12, 13]. On the other hand, malignant cells themselves can produce TRAIL and this may lead to an increase of their invasive and migratory properties [7]. Thus, the expression levels of TRAIL receptors as well as the preference for the usage of TRAIL-R1 or TRAIL-R2 in TRAIL-induced apoptotic/non-apoptotic signaling may be an essential factor determining both, the tumor initiation and progression.
The biological responses to TRAIL are attributed to the function of TRAIL receptors at the plasma membrane. Interestingly, although the intracellular presence of TRAIL-R1 and/or TRAIL-R2 has repeatedly been noticed, only recently the question of biological relevance and eventually specific functions of intracellular receptors began to be addressed. Obviously, sequestration of the receptors in the cytoplasm or in the nucleus, frequently observed in cancer, could represent one of the strategies used by these cells to escape TRAIL-induced apoptosis. Indeed, such mechanisms have been proposed for both cytoplasmic [14–16] and nuclear TRAIL receptors [16–18]. More recently, specific function of nuclear TRAIL-R2 has also been uncovered [19]. In the nucleus, TRAIL-R2 interacts with the microprocessor complex and impairs the maturation of the miRNA let-7. This leads to the increased levels of the malignancy promoting factors HMGA2 and Lin28B and enhances tumor cell proliferation in vitro and in vivo [19]. Likewise, specific functions of cytoplasmic TRAIL death receptors have also been proposed lately. Concrete, in response to endoplasmic reticulum (ER) stress, as a part of unfolded protein response (UPR), cytoplasmic TRAIL-R1 and TRAIL-R2 both are able to aggregate and induce cell death [20, 21].
Interestingly, although numbers of immunohistological studies addressed the issue of the impact of differentially expressed TRAIL death receptors in tumor and corresponding normal healthy tissue, the clinical relevance of TRAIL receptors present in particular intracellular compartment, cytoplasm or nucleus was analyzed only sporadically. For PDAC we reported recently that high levels of nuclear TRAIL-R2 correlates with worse prognosis for PDAC patients suffering from early stage PDACs [19]. The level of TRAIL-R2 in the cytoplasm of PDAC cells, although significantly higher than in healthy tissue, did not correlate with any clinico-pathological parameter. To the best of our knowledge, no corresponding data for TRAIL-R1 are available so far. To fill this gap, we evaluated the clinical relevance of high levels of TRAIL-R1 in tumor tissues of PDAC patients with special emphasis to the possible differential impact of its cytoplasmic and nuclear localization.
Methods
We retrieved formalin fixed and paraffin embedded PDAC and adjacent, peritumoral non-malignant tissue samples from the archives of the Institute of Pathology of the University Hospital Schleswig-Holstein and Christian-Albrechts-University Kiel, resected between 1999 and 2010. Follow-up data were retrieved from the Epidemiological Cancer Registry Schleswig–Holstein, Germany, hospital records and general practitioners. Patients were included if histology confirmed an adenocarcinoma of the pancreas. pTNM stage was determined according to the 8th edition of the Union for International Cancer Control (UICC) guidelines [22]. This study was approved by the local institutional review board of the Medical Faculty of the Christian-Albrechts-University of Kiel (A-110/99).
Immunohistochemical staining
For immunohistochemistry 3 μm paraffin sections were deparaffinized in xylol and re-hydrated in a descending alcohol series. Antigen retrieval was achieved by heating for 15 min at 89 °C in citrate-buffer (pH 6.0). Intrinsic biotin and avidin binding sites were blocked with Avidin-Biotin Blocking Kit (Vector Laboratories, Burlingham, CA), endogenous peroxidase-activity with Hydrogen-Peroxide Block (15 min, RT; Thermo Scientific, Fremont, CA) and unspecific background was reduced with Ultra-Vision Block (5 min, RT; Thermo Scientific, Fremont, CA, USA). Slides were incubated with primary antibodies as previously described (clone TR1.02; Ganten et al., 2009 [23]). Bound antibodies were detected by a Super Sensitive IHC Detection System (BioGenex, San Ramon, USA). For color development, a Fast Red system (Sigma, Deisenhofen, Germany) was used. Washing steps were done with Tris-buffered saline supplemented with Tween (TBST). All slides were counterstained with hemalum and cover slipped.
Microscopic evaluations and histopathological scoring
Evaluation of the staining was performed on a Leica DM 1000-Microscope (Leica, Wetzlar, Germany) and a two-dimensional scoring system was applied to semi-quantitatively assess the TRAIL-R1 expression data. The intensity of the staining was judged on an arbitrary scale of 0 to 3 with 0: no staining; 1: weak staining; 2: moderate staining and 3: strong staining. In samples with varying staining intensities, strongest values were stated. In addition, the percentage of stained cells was quantified and scaled from 0 to 4 with 0: no positive cells; 1: 1–10%; 2: 10–50%; 3: 51–80%; and 4: 81–100% positively stained cells. Both values were summarized in a sum score (Table 1) and separately assessed for cytoplasm and nuclei by two independent pathologists.
Table 1.
Histomorphological evaluation score
| Staining intensity | Points | Number of positive cells | Points |
|---|---|---|---|
| Negative | 0 | 0% | 0 |
| Weak positive | 1 | < 10% | 1 |
| Moderate positive | 2 | 10–50% | 2 |
| Strong positive | 3 | 51–80% | 3 |
| – | – | 81–100% | 4 |
Statistical analyses
Statistical analyses were done using SPSS 23.0 (SPSS, IBM Corporation, Armonk, NY, USA). For the correlation of the clinico-pathological patient characteristics and TRAIL-R1 expression, data were dichotomized and Kendall’s Tau (τ) was used. Only patients with existing follow-up data were included. Patients who died within 14 days after surgery and patients who received neoadjuvant treatment were excluded. Consequently, for these analyses 97 out of 106 patients were included. Out of these, 19 patients were censored because they were either alive or lost in follow up. The overall postoperative survival was analyzed. Staining intensities of tumor and normal tissues were compared using the Wilcoxon test as a nonparametric test for paired samples. Survival analyses were performed by Kaplan-Meier estimates and statistical evaluations were done by log-rank tests. P values ≤0.05 were considered significant.
Results
Patient collective
To investigate the significance of TRAIL-R1 for pancreatic cancer biology, we analyzed the staining intensity, the percentage of stained cells and the intracellular distribution of this receptor in sections of 106 tumors and 28 morphologically normal corresponding peritumoral ducts from 106 patients suffering from PDAC. Out of 106 patients, 51 (48.1%) were female and median age was 65 years (range 47–85 years). Cancer of the pancreatic head, corpus and tale were found in 75/106 (70.8%), 7/106 (6.6%) and 8/106 (7.5%) cases, respectively. In 16/106 cases (15.1%), the localization was not specified. The detailed clinico-pathological patient characteristics are summarized in Table 2. Most of the patients have undergone surgery at stage T3 (94/106; 88.7%) and had already developed lymph node metastases (84/106, 79.2%), whereas no patient was operated at stage T1. Resected tumors were well or moderately differentiated in 66.1% of the cases. In 89.6% (95/106), patients were free of distant metastases at the time of resection. Resection without residual tumor load was achieved in 74/106 patients (69.8%).
Table 2.
Clinico-pathological patient characteristics on the basis of the TNM status (according to the UICC Classification of Malignant Tumors). Given are the total number of patients and the percentage (%)
| Feature | n | % |
|---|---|---|
| T – category | ||
| T1 | 0 | 0.0 |
| T2 | 3 | 2.8 |
| T3 | 94 | 88.7 |
| T4 | 9 | 8.5 |
| N - category | ||
| N0 | 22 | 20.8 |
| N1 | 84 | 79.2 |
| NX | 0 | 0.0 |
| M - category | ||
| M0 | 69 | 65.1 |
| M1 | 11 | 10.4 |
| MX | 26 | 24.5 |
| Venous invasion | ||
| V0 | 79 | 74.5 |
| V1 | 19 | 17.9 |
| V2 | 3 | 2.8 |
| VX | 5 | 4.7 |
| Perineural invasion | ||
| Pn0 | 39 | 36.8 |
| Pn1 | 59 | 55.7 |
| PnX | 8 | 7.5 |
| Lymphatic invasion | ||
| L0 | 30 | 28.3 |
| L1 | 71 | 67.0 |
| LX | 5 | 4.7 |
| R - status | ||
| R0 | 74 | 69.8 |
| R1 | 28 | 26.4 |
| R2 | 2 | 1.9 |
| RX | 2 | 1.9 |
| Histopathological grading | ||
| G1 | 11 | 10.4 |
| G2 | 59 | 55.7 |
| G3 | 35 | 33.0 |
| G4 | 1 | 0.9 |
T1: Tumor < 2 cm within pancreas; T2: > 2 cm, T3 over pancreas without infiltration of A. mes. Sup. or Truncus coeliacus, T4: vessel infiltration
Expression of TRAIL-R1 in PDAC and non-malignant adjacent tissue
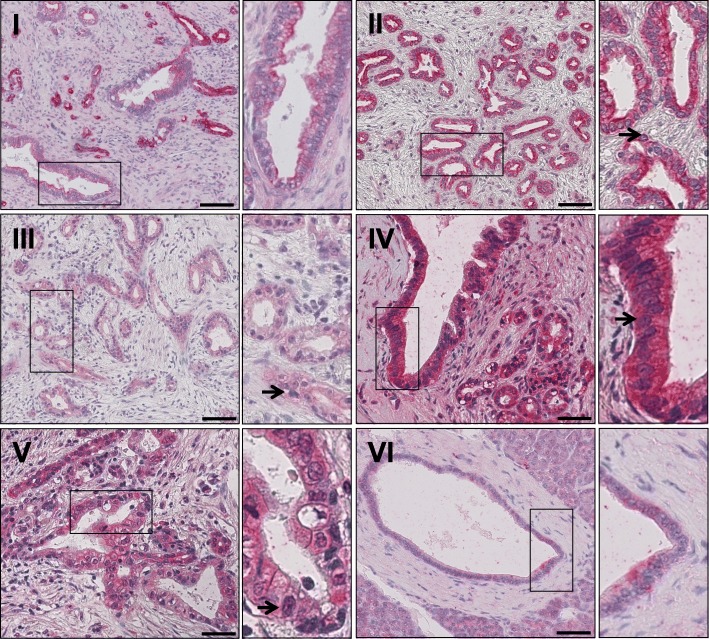
TRAIL-R1 was expressed in 93.4% (99/106) of all PDAC samples (Table 3a). Analysis of its intracellular distribution revealed a cytoplasmic and nuclear localization, whereas plasma membrane staining was not distinct and therefore not evaluable. Representative images showing expression pattern of TRAIL-R1 in tumor tissue and non-malignant, adjacent tissue are shown in Fig. 1.
Table 3.
Cytoplasmic and nuclear TRAIL-R1 expression in malignant and non-malignant ducts
| cytoplasm | nuclei | |||||||
|---|---|---|---|---|---|---|---|---|
| a) | n | % | n | % | ||||
| Positive tumor cells | ||||||||
| 0% | 7 | 6.6 | 70 | 66.0 | ||||
| < 10% | 5 | 4.8 | 9 | 8.5 | ||||
| 11–50% | 11 | 10.4 | 16 | 15.1 | ||||
| 51–80% | 22 | 20.8 | 10 | 9.4 | ||||
| > 80% | 61 | 57.5 | 1 | 0.9 | ||||
| Staining intensity | ||||||||
| Negative | 7 | 6.6 | 70 | 66.0 | ||||
| Weak positive | 55 | 51.9 | 28 | 26.4 | ||||
| Moderate positive | 24 | 22.6 | 8 | 7.5 | ||||
| Strong positive > 80% | 20 | 18.9 | 0 | 0.0 | ||||
| Sum score | ||||||||
| 0 | 7 | 6.6 | 70 | 66.0 | ||||
| 2 | 4 | 3.8 | 9 | 8.5 | ||||
| 3–4 | 25 | 23.6 | 21 | 19.8 | ||||
| 5–6 | 57 | 53.8 | 6 | 5.7 | ||||
| 7 | 13 | 13.0 | 0 | 0.0 | ||||
| b) | tumor | non-malignant | tumor | non-malignant | ||||
| n | % | n | % | n | % | n | % | |
| Staining intensity | ||||||||
| Negative | 7 | 6.6 | 3 | 10.7 | 70 | 66.0 | 26 | 92.9 |
| Weak positive | 55 | 51.9 | 14 | 50.0 | 28 | 26.4 | 2 | 7.1 |
| Moderate positive | 24 | 22.6 | 6 | 21.5 | 8 | 7.5 | 0 | 0.0 |
| Strong positive | 20 | 18.9 | 5 | 17.9 | 0 | 0.0 | 0 | 0.0 |
| Staining pattern: tumor vs. non-malignant | no difference ns, p = 0.698 |
p = 0.006 | ||||||
a) number of positive cells, staining intensity and corresponding sum score are shown for cytoplasm and nucleus separately. Additionally, particular staining intensity in relation to histologic specification (tumor vs. peritumoral non-malignant) and for cytoplasmic vs. nuclear staining are provided (b)
Fig. 1.
Representative images of TRAIL-R1 staining in PDAC tissue (I-V) and non-neoplastic pancreatic duct (VI). I: Tumors with strong cytoplasmic TRAIL-R1 staining in 51–80% cells and no nuclear TRAIL-R1 staining. II: Tumors with strong positive cytoplasm in > 80% cells with negative nuclear staining. III: Tumors with weak positive cytoplasmic staining intensity in > 80% of the cells. Weak positive staining in 10–50% of the nuclei. IV: Tumors with moderate positive cytoplasmic staining in > 80% cells. Weak positive nuclei in 51–80% cells. V: Tumors with moderate positive nuclei in 51–80% of the nuclei. Weak positive cytoplasmic staining in > 80% of the cells. VI: Non-neoplastic duct with weak to moderate positive cytoplasm staining and without positive nuclei. Magnifications corresponding to the rectangles in the large pictures are shown in the small windows. Arrows indicate exemplary nuclear staining. Scale bar marks 100 μm (I – III + VI) or 50 μm (IV – V)
In the cytoplasm, we found weak, moderate and strong TRAIL-R1 expression in 51.9, 22.6 and 18.9% (55/106; 24/106 and 20/106) of the cases, respectively (Table 3b). In 78.3% of the tumors, over 50% of the carcinoma cells showed positive cytoplasmic expression (Fig. 1). Cytoplasmic staining of TRAIL-R1 was present with very similar intensity in tumor and normal tissue.
In contrast, apparent differences in the nuclear TRAIL-R1 staining frequency and intensity were observed in tumor versus normal tissue (Table 3b, p = 0.006). Whereas overall 34% of tumors showed nuclear presence of TRAIL-R1 with either weak (26.4%; 28/106) or moderate (7.6%; 8/106) staining intensity, only 7.1% (2/28) of normal ducts expressed TRAIL-R1 in the nucleus and this with only weak intensity.
Correlation of TRAIL-R1 expression with clinico-pathological parameters and patient survival
Next, we correlated the expression level of TRAIL-R1 and its intracellular distribution (cytoplasm and nucleus) with diverse clinico-pathological parameters like tumor stage (T), nodal spread (N), distant metastasis (M), grading (G), lymphatic invasion (L), venous invasion (V) and perineural invasion. As shown in Table 4, we found a significant negative correlation of the amount of cells positively stained for TRAIL-R1 in the cytoplasm with a tumor grading (τ = − 0.228; p = 0.043). Apart from that, no other correlation was found for cytoplasmic or nuclear staining with any of the parameters.
Table 4.
Correlation of TRAIL-R1 expression with clinico-pathological parameters
| Staining parameter | Tumor stage | Lymph nodes | Metastasis | Grading | Lymph vessels | Venous invasion | Perineural invasion |
|---|---|---|---|---|---|---|---|
| Intensity cytoplasm | τ = −0.018 p = 0.880 | τ = 0.045 p = 0.703 | τ = −0.096 p = 0.413 | τ = −0.099 p = 0.382 | τ = 0.127 p = 0.277 | τ = 0.096 p = 0.404 | τ = −0.033 p = 0.779 |
| Number pos. cytoplasm | τ = − 0.115 p = 0.327 | τ = − 0.080 p = 0.496 | τ = 0.070 p = 0.548 | τ = − 0.228 p = 0.043 | τ = − 0.018 p = 0.877 | τ = 0.044 p = 0.705 | τ = 0.104 p = 0.375 |
| Sum score cytoplasm | τ = 0.012 p = 0.918 | τ = −0.006 p = 0.960 | τ = −0.062 p = 0.598 | τ = − 0.105 p = 0.351 | τ = 0.071 p = 0.542 | τ = 0.096 p = 0.407 | τ = − 0.015 p = 0.896 |
| Intensity nuclei | τ = − 0.210 p = 0.073 | τ = − 0.006 p = 0.960 | τ = 0.129 p = 0.272 | τ = − 0.079 p = 0.482 | τ = − 0.064 p = 0.584 | τ = 0.096 p = 0.407 | τ = 0.154 p = 0.188 |
| Number pos. nuclei | τ = − 0.100 p = 0.392 | τ = 0.172 p = 0.142 | τ = 0.021 p = 0.858 | τ = −0.057 p = 0.611 | τ = − 0.005 p = 0.963 | τ = 0.100 p = 0.385 | τ = 0.148 p = 0.205 |
| Sum score nuclei | τ = −0.028 p = 0.831 | τ = − 0.010 p = 0.929 | τ = 0.142 p = 0.224 | τ = 0.079 p = 0.483 | τ = 0.125 p = 0.285 | τ = 0.134 p = 0.245 | τ = 0.147 p = 0.210 |
Abbreviations: τ Kendall’s τ, p p value
Shown are correlation coefficients Kendall’s τ as well as the significance of the correlation
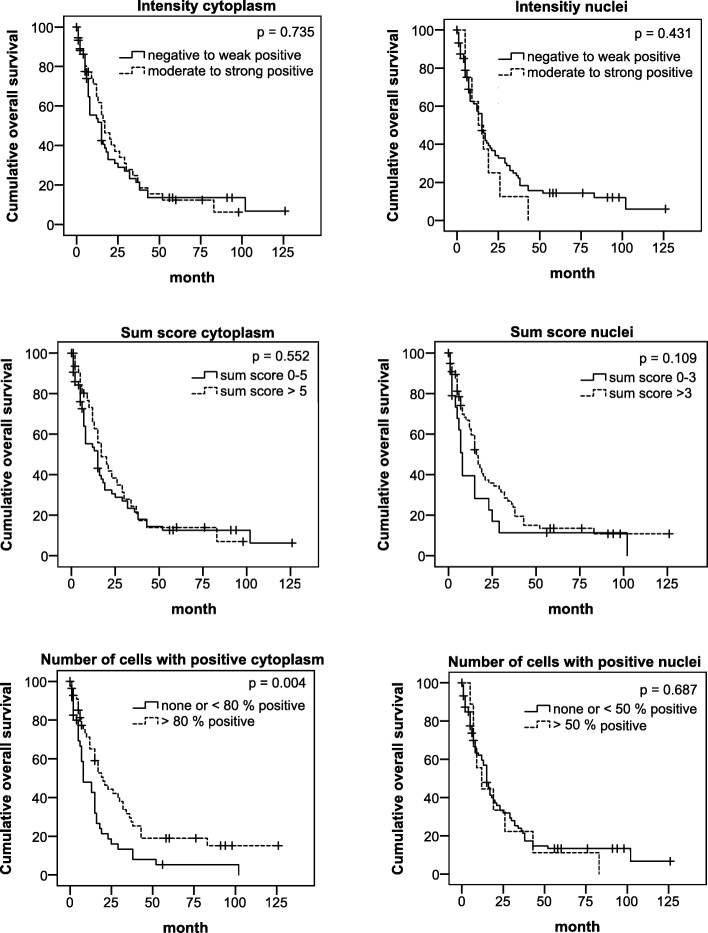
Further we explored whether the TRAIL-R1 expression pattern could be of prognostic value. To address this issue, we dichotomized the results for intensity and amount of positive cells as well as the sum score in a group with strong and in a group with weak expression of TRAIL-R1 (see Table 5) and analyzed these data by Kaplan-Meier analysis. Cumulative survival was compared by log rank test and p values ≤0.05 were considered significant.
Table 5.
Impact of TRAIL-R1 expression pattern on survival of PDAC patient
| Staining parameter | Total number of patients | Number of deceased patients | Median survival ± SD (95% CI) in months |
p - value |
|---|---|---|---|---|
| Intensitiy cytoplasm | ||||
| Negative to weak positive | 60 | 48 | 15 ± 3.611 (7.922–22.078) | |
| Moderate to strong positive | 37 | 30 | 17 ± 3.513 (10.115–23.885) | 0.735 |
| Number of cells with positively stained cytoplasm | ||||
| ≤ 80% | 40 | 37 | 8 ± 3.041(2.041–13.959) | |
| > 80% | 57 | 41 | 20 ± 3.855 (12.444–27.556) | 0.004 |
| Sum score cytoplasm | ||||
| ≤ 5 | 65 | 52 | 15 ± 3.733 (7.682–22.318) | |
| > 5 | 32 | 26 | 17 ± 3.992 (9.177–24.823) | 0.552 |
| Staining intensity nuclei | ||||
| Negative to weak positive | 89 | 70 | 15 ± 1.573 (11.918–18.082) | |
| Moderate to strong positive | 8 | 8 | 13 ± 4.950 (3.298–22.702) | 0.431 |
| Number of cells with positively stained nucleus | ||||
| ≤ 50% | 88 | 69 | 15 ± 1.430 (12.198–17.802) | |
| > 50% | 9 | 9 | 12 ± 4.472 (3.235–20.765) | 0.687 |
| Sum score nuclei | ||||
| ≤ 3 | 19 | 17 | 8 ± 1.023 (5.995–10.005) | |
| > 3 | 78 | 61 | 16 ± 1.622 (12.821–19.179) | 0.109 |
Abbreviations: SD standard deviation, CI confidence interval
P-values were estimated by log-rank-test with p ≤ 0.05 considered as significant
Neither the intensity of cytoplasmic staining nor the sum score showed a significant correlation with the patient survival. Likewise, nuclear staining showed no prognostic relevance.
Since the immunostaining revealed differences in number of stained cells per tumor, we wondered whether this parameter could be of prognostic relevance for the patients. Importantly, we found that patients with tumors in which > 80% of the cells express TRAIL-R1 in the cytoplasm have significantly prolonged median survival compared to the patients whose tumors show TRAIL-R1 positivity in less than 80% cells (20 vs. 8 months; p = 0.004; Fig. 2).
Fig. 2.
Kaplan-Meier analyses of the cumulative survival of patients with differential expression of TRAIL-R1. P-values were calculated by the log rank test and p ≤ 0.05 was considered significant
Discussion
Identification of the prognostic factors related to survival of cancer patients represents a strategy to understand the molecular mechanisms driving tumor progression and therapy resistance, and may consequently support the development of novel therapeutic strategies. The expression levels of TRAIL-R1 and TRAIL-R2 were shown to be of prognostic relevance for different tumor entities. In addition to their localization at the plasma membrane, TRAIL-R1 and TRAIL-R2 are also found in the cytoplasm and in the nucleus of several cell types. Especially in tumor cells, diminished plasma membrane but enhanced intracellular presence of these receptors was frequently observed. Importantly, despite these observations and emerging evidences for distinct compartment-specific functions of TRAIL-Rs, their cumulative expression levels - regardless of their intracellular distribution - are mainly taken into account when immunohistochemical studies are evaluated. However, recent studies on the TRAIL-R2-expression in PDAC tissues suggest the necessity of considering the intracellular distribution of TRAIL-Rs. Thus, the expression levels of TRAIL-R2 may emerge as either a positive or a negative prognostic marker, depending on the subcellular distribution (plasma membrane vs. nucleus) [19, 24].
Immunohistochemical studies described high cytoplasmic levels of TRAIL receptors in different cancer types, e.g. colorectal cancer [25–27], breast cancer cell lines [23, 28], renal cell carcinoma [29], NSCLC [30, 31], melanoma [32, 33], PDAC [19, 24], hepatocellular carcinoma [14], and glioblastoma multiforme [34]. Notably, the levels of just these intracellular receptors turned out to be of prognostic relevance in different tumor types [16]. Intriguingly, whereas high intracellular levels of TRAIL-R1 mainly correlated with positive patient’s prognosis, increased levels of TRAIL-R2 often correlated with shorter patient’s survival (for review [16]). These observations point to the existence of different, receptor-specific activities of cytoplasmic TRAIL death receptors and, in addition, suggest anti-tumor activities of intracellular TRAIL-R1, at least in some tumor entities.
In our present study, we identified cytoplasmic TRAIL-R1 as a positive prognostic marker for patients with PDAC. Interestingly, whereas overall staining intensity showed no prognostic relevance, the number of TRAIL-R1 positive cells per tumor turned out to be important for patient’s outcome. Noteworthy, we used an antibody for immunochemistry which is able to detect membrane expressed TRAIL-R1 as it has been shown before [23]. Specifically, patients with tumors in which more than 80% of cells showed cytoplasmic TRAIL-R1 staining had significantly prolonged survival compared to patients whose tumors presented with less than 80% cells positively stained for TRAIL-R1. In line with these findings, a significant negative correlation between number of cells with positive stained TRAIL-R1 in the cytoplasm and tumor grading was found. These results indicate that loss of cytoplasmic TRAIL-R1 may support recurrent disease with more malignant phenotype.
Little is known about the origin and sub-cytoplasmic localization of intracellular TRAIL death receptors. Cytoplasmic TRAIL death receptors have been detected in Golgi vesicles [32], endosomes [32] and autophagosomes [35]. In addition, their presence in soluble cytoplasmic fractions was also reported [19].
Likewise, the function(s) of cytoplasmic TRAIL receptors is still not fully understood. Sequestration of these receptors in autophagosomes could act as a strategy by which tumor cells escape TRAIL-induced apoptosis [17, 18]. On the other hand, internalization of TRAIL death receptors in response to TRAIL-treatment has been demonstrated, and may represent a part of TRAIL-induced signal transduction pathway [36]. Recently, the importance of cytoplasmic TRAIL-R1 in inducing cell death as a consequence of unresolved unfolded protein response (UPR) has been demonstrated [20, 21]. Noteworthy, in this case TRAIL-R1 mediated cell death is independent of TRAIL. Efficient UPR activation represents a characteristic feature of many human cancers. It allows the tumor cells to survive and adapt to adverse environmental conditions, promotes dormancy and also tumor growth, progression and therapy resistance. In this context, loss of cytoplasmic TRAIL-R1 would select cancer cells which are resistant to UPR-induced apoptosis and thus cells with more aggressive phenotype.
The TRAIL-TRAIL-R system has been shown to be of crucial importance in the tumor immune surveillance [12, 13]. Cytoplasmic TRAIL death receptors may represent a reservoir of receptors, which upon stimulation localize to the plasma membrane and boost the primary response of cells to TRAIL. Since PDAC cells preferentially utilize TRAIL-R1 to induce apoptosis in response to TRAIL, loss of TRAIL-R1 could lead to an escape of immune surveillance and accelerate the recurrent tumor growth.
Alternatively, it is also possible that cytoplasmic TRAIL-R1, via direct protein-protein interaction, sequesters TRAIL-R2 in the cytoplasm thus inhibiting its malignancy-promoting nuclear functions. According to this scenario, cells, which have lost cytoplasmic TRAIL-R1, would also present with a more aggressive phenotype.
Which, if any of these potential cytoplasmic TRAIL-R1 functions, accounts for the obviously anti-tumoral functions of cytoplasmic TRAIL-R1 remains to be elucidated. The existence of further, still unknown functions of intracellular TRAIL receptors is very likely.
Likewise, the cellular pathways leading to the observed loss of TRAIL-R1 in a subset of PDAC cells are not known yet. Recently, several mechanisms negatively regulating the cellular levels of TRAIL-R1, but not TRAIL-R2, have been registered in cancer cells. Thus, hypermethylation of TRAIL-R1 promotor leading to an epigenetic silencing of TRAIL-R1 gene was detected in ovarian cancer cells [37]. Furthermore, at the transcriptional level, negative regulation of TRAIL-R1 promotor by GLI3 as well as miR-25-dependent decrease of TRAIL-R1 levels were reported for cholangiocarcinoma cells [38, 39]. At the post-translational level, specific degradation of TRAIL-R1 protein was described in breast cancer and melanoma cells. Here, membrane-associated RING-CH (MARCH)-8 ubiquitin ligase targeted TRAIL-R1, but not TRAIL-R2, for lysosomal degradation. Interestingly, plasma levels of miR-25 are significantly elevated in PDAC and evaluation of the levels of miR-25 together with MIC-1 and CA19–9 was shown to be able to distinguish between PDAC, benign pancreatic disorders and other GI cancers [40].
Conclusion
We show that cytoplasmic TRAIL-R1 may serve as a novel prognostic marker for PDAC patients. In addition, our data point to the necessity to investigate the evidently underestimated biological functions of intracellular TRAIL receptors.
Acknowledgements
We thank Gökhan Alp and Sandra Krüger for excellent technical assistance.
Funding
The study was financially supported by Erich und Gertrud Roggenbuck-Stiftung and intramural funding given to CH.
Availability of data and materials
The clinical datasets supporting the conclusions of this study were derived from the patient files (paper and electronic form). Therefore, restrictions to availability apply due to data protection regulations. Anonymized data are, however, available from the corresponding author on reasonable request and with permission of the University Hospital Schleswig-Holstein and the local review board.
Abbreviations
- CI
Confidence interval
- GI
Gastrointestinal
- IHC
Immunohistochemistry
- PDAC
Pancreatic ductal adenocarcinoma
- SD
Standard deviation
- TRAIL
TNF-related apoptosis inducing ligand
- UPR
Unfolded protein response
Authors’ contributions
JPG and CH wrote the manuscript, prepared the figures and evaluated the data. FS and CB performed the histological examination and where together with JPG and C Röder major contributor to the data analyses. CR, TB, JHE and HK supported infrastructure and organizational issues. AT designed the study, analyzed the data, designed and wrote the manuscript. All authors read and approved the final manuscript. Some of the data are part of the doctoral thesis of FS.
Ethics approval and consent to participate
All patients have agreed to participate in the study and have signed informed consent before collecting material. This study was approved by the local institutional review board of the Medical Faculty of the Christian-Albrechts-University of Kiel (A-110/99).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jan-Paul Gundlach, Email: Jan-Paul.Gundlach@uksh.de.
Charlotte Hauser, Email: Charlotte.Hauser@uksh.de.
Franka Maria Schlegel, Email: Franka.Schlegel@outlook.de.
Christine Böger, Email: Christine.Boeger@uksh.de.
Christian Röder, c.roeder@email.uni-kiel.de.
Christoph Röcken, Email: Christoph.Roecken@uksh.de.
Thomas Becker, Email: Thomas.Becker@uksh.de.
Jan-Hendrik Egberts, Email: Jan-Hendrik.Egberts@uksh.de.
Holger Kalthoff, hkalthoff@email.uni-kiel.de.
Anna Trauzold, Phone: +49 431 500 30580, atrauzold@email.uni-kiel.de.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Ansari D, Gustafsson A, Andersson R. Update on the management of pancreatic cancer: surgery is not enough. World J Gastroenterol. 2015;21(11):3157–3165. doi: 10.3748/wjg.v21.i11.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lemke J, von Karstedt S, Zinngrebe J, Walczak H. Getting TRAIL back on track for cancer therapy. Cell Death Differ. 2014;21(9):1350–1364. doi: 10.1038/cdd.2014.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Azijli K, Weyhenmeyer B, Peters GJ, de Jong S, Kruyt FAE. Non-canonical kinase signaling by the death ligand TRAIL in cancer cells: discord in the death receptor family. Cell Death Differ. 2013;20(7):858–868. doi: 10.1038/cdd.2013.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trauzold A, Siegmund D, Schniewind B. TRAIL promotes metastasis of human pancreatic ductal adenocarcinoma. Oncogene. 2006;25(56):7434–7439. doi: 10.1038/sj.onc.1209719. [DOI] [PubMed] [Google Scholar]
- 6.Hoogwater FJH, Nijkamp MW, Smakman N, EJa S, Emmink BL, Westendorp BF, DaE R, Sprick MR, Schaefer U, Van Houdt WJ, et al. Oncogenic K-Ras turns death receptors into metastasis-promoting receptors in human and mouse colorectal cancer cells. Gastroenterology. 2010;138(7):2357–2367. doi: 10.1053/j.gastro.2010.02.046. [DOI] [PubMed] [Google Scholar]
- 7.Karstedt SV, Conti A, Nobis M, Montinaro A, Hartwig T, Lemke J, Legler K, Annewanter F, Campbell A, Taraborrelli L, et al. Cancer-cell-autonomous TRAIL-R signaling promotes KRAS-driven cancer progression, invasion and metastasis. Cancer Cell. 2015;27(4):561–573. doi: 10.1016/j.ccell.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Roosmalen IA, Quax WJ, Kruyt FA. Two death-inducing human TRAIL receptors to target in cancer: similar or distinct regulation and function? Biochem Pharmacol. 2014;91(4):447–456. doi: 10.1016/j.bcp.2014.08.010. [DOI] [PubMed] [Google Scholar]
- 9.Stadel D, Mohr A, Ref C, MacFarlane M, Zhou S, Humphreys R, Bachem M, Cohen G, Moller P, Zwacka RM, et al. TRAIL-induced apoptosis is preferentially mediated via TRAIL receptor 1 in pancreatic carcinoma cells and profoundly enhanced by XIAP inhibitors. Clin Cancer Res. 2010;16(23):5734–5749. doi: 10.1158/1078-0432.CCR-10-0985. [DOI] [PubMed] [Google Scholar]
- 10.Lemke J, Noack A, Adam D, Tchikov V, Bertsch U, Roder C, Schutze S, Wajant H, Kalthoff H, Trauzold A. TRAIL signaling is mediated by DR4 in pancreatic tumor cells despite the expression of functional DR5. J Mol Med (Berl) 2010;88(7):729–740. doi: 10.1007/s00109-010-0619-0. [DOI] [PubMed] [Google Scholar]
- 11.Mohr A, Yu R, Zwacka RM. TRAIL-receptor preferences in pancreatic cancer cells revisited: both TRAIL-R1 and TRAIL-R2 have a licence to kill. BMC Cancer. 2015;15:494. doi: 10.1186/s12885-015-1508-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takeda K, Smyth MJ, Cretney E, Hayakawa Y, Kayagaki N, Yagita H, Okumura K. Critical role for tumor necrosis factor-related apoptosis-inducing ligand in immune surveillance against tumor development. J Exp Med. 2002;195(2):161–169. doi: 10.1084/jem.20011171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cretney E, Takeda K, Yagita H, Glaccum M, Peschon JJ, Smyth MJ. Increased susceptibility to tumor initiation and metastasis in TNF-related apoptosis-inducing ligand-deficient mice. J Immunol. 2002;168(3):1356–1361. doi: 10.4049/jimmunol.168.3.1356. [DOI] [PubMed] [Google Scholar]
- 14.Kriegl L, Jung A, Engel J, Jackstadt R, Gerbes AL, Gallmeier E, Reiche JA, Hermeking H, Rizzani A, Bruns CJ, et al. Expression, cellular distribution, and prognostic relevance of TRAIL receptors in hepatocellular carcinoma. Clin Cancer Res. 2010;16(22):5529–5538. doi: 10.1158/1078-0432.CCR-09-3403. [DOI] [PubMed] [Google Scholar]
- 15.Chen JJ, Shen HC, Rivera Rosado LA, Zhang Y, Di X, Zhang B. Mislocalization of death receptors correlates with cellular resistance to their cognate ligands in human breast cancer cells. Oncotarget. 2012;3(8):833–842. doi: 10.18632/oncotarget.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bertsch U, Röder C, Kalthoff H, Trauzold A. Compartmentalization of TNF-related apoptosis-inducing ligand (TRAIL) death receptor functions: emerging role of nuclear TRAIL-R2. Cell Death Dis. 2014;5:–e1390. [DOI] [PMC free article] [PubMed]
- 17.Kojima Y, Nakayama M, Nishina T, Nakano H, Koyanagi M, Takeda K, Okumura K, Yagita H. Importin beta1 protein-mediated nuclear localization of death receptor 5 (DR5) limits DR5/tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL)-induced cell death of human tumor cells. J Biol Chem. 2011;286(50):43383–43393. doi: 10.1074/jbc.M111.309377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bai X, Williams JL, Greenwood SL, Baker PN, Aplin JD, Crocker IP. A placental protective role for trophoblast-derived TNF-related apoptosis-inducing ligand (TRAIL) Placenta. 2009;30(10):855–860. doi: 10.1016/j.placenta.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 19.Haselmann V, Kurz A, Bertsch U, Hübner S, Olempska-Müller M, Fritsch J, Häsler R, Pickl A, Fritsche H, Annewanter F, et al. Nuclear death receptor TRAIL-R2 inhibits maturation of let-7 and promotes proliferation of pancreatic and other tumor cells. Gastroenterology. 2014;146(1):278–290. doi: 10.1053/j.gastro.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 20.Lu M, Lawrence DA, Marsters S, Acosta-Alvear D, Kimmig P, Mendez AS, Paton AW, Paton JC, Walter P, Ashkenazi A. Opposing unfolded-protein-response signals converge on death receptor 5 to control apoptosis. Science. 2014;345(6192):98–101. doi: 10.1126/science.1254312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dufour F, Rattier T, Constantinescu AA, Zischler L, Morle A, Ben Mabrouk H, Humblin E, Jacquemin G, Szegezdi E, Delacote F, et al. TRAIL receptor gene editing unveils TRAIL-R1 as a master player of apoptosis induced by TRAIL and ER stress. Oncotarget. 2017;8(6):9974–9985. doi: 10.18632/oncotarget.14285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.GM BJD, Wittekind C, editors. TNM Classification of Malignant Tumours, 8th Edition. 2016. [Google Scholar]
- 23.Ganten TM, Sykora J, Koschny R, Batke E, Aulmann S, Mansmann U, Stremmel W, Sinn H-P, Walczak H. Prognostic significance of tumour necrosis factor-related apoptosis-inducing ligand (TRAIL) receptor expression in patients with breast cancer. J Mol Med (Berl) 2009;87(10):995–1007. doi: 10.1007/s00109-009-0510-z. [DOI] [PubMed] [Google Scholar]
- 24.Gallmeier E, Bader DC, Kriegl L, Berezowska S, Seeliger H, Goke B, Kirchner T, Bruns C, De Toni EN. Loss of TRAIL-receptors is a recurrent feature in pancreatic cancer and determines the prognosis of patients with no nodal metastasis after surgery. PLoS One. 2013;8(2):e56760. doi: 10.1371/journal.pone.0056760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strater J, Hinz U, Walczak H, Mechtersheimer G, Koretz K, Herfarth C, Moller P, Lehnert T. Expression of TRAIL and TRAIL receptors in colon carcinoma: TRAIL-R1 is an independent prognostic parameter. Clin Cancer Res. 2002;8(12):3734–3740. [PubMed] [Google Scholar]
- 26.Granci V, Bibeau F, Kramar A, Boissiere-Michot F, Thezenas S, Thirion A, Gongora C, Martineau P, Del Rio M, Ychou M. Prognostic significance of TRAIL-R1 and TRAIL-R3 expression in metastatic colorectal carcinomas. Eur J Cancer. 2008;44(15):2312–2318. doi: 10.1016/j.ejca.2008.06.042. [DOI] [PubMed] [Google Scholar]
- 27.Bavi P, Prabhakaran SE, Abubaker J, Qadri Z, George T, Al-Sanea N, Abduljabbar A, Ashari LH, Alhomoud S, Al-Dayel F, et al. Prognostic significance of TRAIL death receptors in middle eastern colorectal carcinomas and their correlation to oncogenic KRAS alterations. Mol Cancer. 2010;9:203. doi: 10.1186/1476-4598-9-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCarthy MM, Sznol M, DiVito KA, Camp RL, Rimm DL, Kluger HM. Evaluating the expression and prognostic value of TRAIL-R1 and TRAIL-R2 in breast cancer. Clin Cancer Res. 2005;11(14):5188–5194. doi: 10.1158/1078-0432.CCR-05-0158. [DOI] [PubMed] [Google Scholar]
- 29.Macher-Goeppinger S, Aulmann S, Tagscherer KE, Wagener N, Haferkamp A, Penzel R, Brauckhoff A, Hohenfellner M, Sykora J, Walczak H, et al. Prognostic value of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) and TRAIL receptors in renal cell cancer. Clin Cancer Res. 2009;15(2):650–659. doi: 10.1158/1078-0432.CCR-08-0284. [DOI] [PubMed] [Google Scholar]
- 30.Spierings DC, de Vries EG, Timens W, Groen HJ, Boezen HM, de Jong S. Expression of TRAIL and TRAIL death receptors in stage III non-small cell lung cancer tumors. Clin Cancer Res. 2003;9(9):3397–3405. [PubMed] [Google Scholar]
- 31.Cooper WA, Kohonen-Corish MR, Zhuang L, McCaughan B, Kennedy C, Screaton G, Sutherland RL, Lee CS. Role and prognostic significance of tumor necrosis factor-related apoptosis-inducing ligand death receptor DR5 in nonsmall-cell lung cancer and precursor lesions. Cancer. 2008;113(1):135–142. doi: 10.1002/cncr.23528. [DOI] [PubMed] [Google Scholar]
- 32.Zhang XD, Franco AV, Nguyen T, Gray CP, Hersey P. Differential localization and regulation of death and decoy receptors for TNF-related apoptosis-inducing ligand (TRAIL) in human melanoma cells. J Immunol. 2000;164(8):3961–3970. doi: 10.4049/jimmunol.164.8.3961. [DOI] [PubMed] [Google Scholar]
- 33.Zhuang L, Lee CS, Scolyer RA, McCarthy SW, Zhang XD, Thompson JF, Screaton G, Hersey P. Progression in melanoma is associated with decreased expression of death receptors for tumor necrosis factor-related apoptosis-inducing ligand. Hum Pathol. 2006;37(10):1286–1294. doi: 10.1016/j.humpath.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 34.Kuijlen JM, Mooij JJ, Platteel I, Hoving EW, van der Graaf WT, Span MM, Hollema H, den Dunnen WF. TRAIL-receptor expression is an independent prognostic factor for survival in patients with a primary glioblastoma multiforme. J Neuro-Oncol. 2006;78(2):161–171. doi: 10.1007/s11060-005-9081-1. [DOI] [PubMed] [Google Scholar]
- 35.Di X, Zhang G, Zhang Y, Takeda K, Rivera Rosado LA, Zhang B. Accumulation of autophagosomes in breast cancer cells induces TRAIL resistance through downregulation of surface expression of death receptors 4 and 5. Oncotarget. 2013;4(9):1349–1364. doi: 10.18632/oncotarget.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akazawa Y, Mott JL, Bronk SF, Werneburg NW, Kahraman A, Guicciardi ME, Meng XW, Kohno S, Shah VH, Kaufmann SH et al: Death receptor 5 internalization is required for lysosomal permeabilization by TRAIL in malignant liver cell lines. Gastroenterology 2009, 136(7):2365–2376 e2361–2367. [DOI] [PMC free article] [PubMed]
- 37.Horak P, Pils D, Haller G, Pribill I, Roessler M, Tomek S, Horvat R, Zeillinger R, Zielinski C, Krainer M. Contribution of epigenetic silencing of tumor necrosis factor-related apoptosis inducing ligand receptor 1 (DR4) to TRAIL resistance and ovarian cancer. Mol Cancer Res. 2005;3(6):335–343. doi: 10.1158/1541-7786.MCR-04-0136. [DOI] [PubMed] [Google Scholar]
- 38.Kurita S, Mott JL, Almada LL, Bronk SF, Werneburg NW, Sun SY, Roberts LR, Fernandez-Zapico ME, Gores GJ. GLI3-dependent repression of DR4 mediates hedgehog antagonism of TRAIL-induced apoptosis. Oncogene. 2010;29(34):4848–4858. doi: 10.1038/onc.2010.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Razumilava N, Bronk SF, Smoot RL, Fingas CD, Werneburg NW, Roberts LR, Mott JL. miR-25 targets TNF-related apoptosis inducing ligand (TRAIL) death receptor-4 and promotes apoptosis resistance in cholangiocarcinoma. Hepatology. 2012;55(2):465–475. doi: 10.1002/hep.24698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yuan W, Tang W, Xie Y, Wang S, Chen Y, Qi J, Qiao Y, Ma J. New combined microRNA and protein plasmatic biomarker panel for pancreatic cancer. Oncotarget. 2016;7(48):80033–80045. doi: 10.18632/oncotarget.12406. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The clinical datasets supporting the conclusions of this study were derived from the patient files (paper and electronic form). Therefore, restrictions to availability apply due to data protection regulations. Anonymized data are, however, available from the corresponding author on reasonable request and with permission of the University Hospital Schleswig-Holstein and the local review board.