Abstract
Background
This study aimed to investigate the effect of over-expressing circular RNA CEP128 (circCEP128) on cell functions and explore the molecular mechanism of which in bladder carcinoma.
Methods
The differentially expressed circRNAs and mRNAs in bladder carcinoma cells and cells in adjacent tissues were screened out using microarray analysis. Expression levels of circRNAs and mRNAs in tissues and cells were determined by qRT-PCR. Expression of SOX11 was detected by western blot. Luciferase reporter assay and RNA pull-down assay were used to investigate the interactions between the specific circRNA, miRNA and mRNA. Cell cycle and apoptosis were measured using flow cytometry after transfection. MTT assay was also performed to detect the cell proliferation.
Results
In present study, circCEP128 and SOX11 were observed significantly up-regulated in bladder cancer tissues, while the expression of miR-145-5p was decreased in cancer samples compared to normal samples. Cytoscape was used to visualize circCEP128-miRNA-target gene interactions based on the TargetScan and circular RNA interactome, which revealed that circCEP128 served as a sponge of miR-145-5p and indirectly regulated SOX11. Knockdown of circCEP128 induced the inhibition of cell proliferation and the increased bladder cancer cell apoptosis rate.
Conclusions
CircCEP128 functions as a ceRNA for miR-145-5p, which could up regulates SOX11 and further promotes cell proliferation and inhibits cell apoptosis of bladder cancer.
Keywords: Bladder cancer, Circle RNA, circCEP128, miR-145-5p, SOX11
Background
Global Cancer Statistics alarms that bladder cancer, a tumor in urinary system, caused 130,000 deaths each year (Li et al., 2017). In China, the mortality and morbidity of bladder cancer ranked first among all the tumors in urinary system (Huang et al., 2016). Especially for the patients suffer from muscle-invasive bladder cancer, the occurrence of metastasis is more frequently and the prognosis is poorer than other kinds of bladder cancers (Zhong et al., 2017). Besides, most clinical trials of chemotherapy for advanced bladder carcinoma displayed limited benefits. Therefore, it is urgent for us to identify novel molecular targets for inhibiting the tumorigenesis of bladder cancer (Zhong et al., 2016).
MicroRNAs (miRNAs) are endogenous small noncoding RNA molecules (19–22 nucleotides in length) that negatively modulate protein-coding genes expression through binding to the specific sequence of genes (Enokida et al., 2016). Increasing evidences have demonstrated that miRNAs usually expressed abnormally in bladder cancer and resulted in multiple alterations during the development of cancer, such as promoting or inhibiting carcinogenesis (Li et al., 2017). MicroRNA-145 (miR-145) has been frequently reported to be down-regulated in various human cancers, including prostate cancer, bladder cancer and colon cancer, as well as in B-cell malignancies (Noguchi et al., 2013). Exogenous miR-145 has also been speculated as tumor inhibitor when being administrated intravesically (Minami et al., 2017).
Circular RNAs (circRNAs), without cap and polyA tails, forms covalently closed continuous loops by non-sequential back-splicing of pre-mRNA transcripts. They generally expressed in eukaryotic cells, but the biological function of them remains unclear (Zhong et al., 2017). CircRNAs are less prone to degrade since they are resistant to exonuclease, the property of which makes circRNAs superior than linear RNAs in terms of biological markers (Huang et al., 2016). Multiple properties of circRNAs have been identified in recent years, among which “miRNA sponges” role was most frequently discussed (Shang et al., 2016). Zhong et al., 2017 investigated competing endogenous RNA mechanism in bladder cancer, among which circRNA MYLK was profoundly discussed but circCEP128 was neglected (Zhong et al., 2017). CircTCF25 and circHIPK3 have been fully illustrated in other studies about ceRNA network and variant pathways (Li et al., 2017; Huang et al., 2016; Zhong et al., 2016). However, this speculation refers to cirCEP128 as a new bio-functional marker which has a close interaction with miR-145-5p in bladder cancer.
SOX11, as an intronless gene regulates cell fate, functions in tumorigenesis and adult neurogenesis. It is known to promote tissue remodeling, progenitor cell expansion and differentiation of a number of cell types, including neural progenitor cells (Oliemuller et al., 2017). SOX11 is a diagnostic and prognostic antigen in B cell lymphomas and has recently been demonstrated to have tumor suppressor functions (Sernbo et al., 2011). It was found that miR-223-3p inhibitor restrains ovarian cancer development by increasing SOX11 expression (Fang et al., 2017). Therefore, we hypothesized that SOX11 might be regulated by miR-145-5p in human bladder cancer.
In present study, the expression profiles of circRNA and miRNA in cells of bladder tumor and adjacent tissues were illustrated clearly. The expression of SOX11 was also detected. The directly interactions among circRNA, miRNA and SOX11 were confirmed by luciferase assay. We hypothesized that circCEP128 might function as a competing endogenous RNA (ceRNA) for miR-145-5p in regulating SOX11, which might further inhibit cell proliferation and promote cell apoptosis of bladder cancer. The study might be of biological and clinical importance.
Methods
Clinical specimens
Ten pairs of bladder tumor tissues and adjacent bladder tissues were obtained from patients who suffered from radical cystectomy at the First Affiliated Hospital of Xiamen University. The normal bladder urothelium samples were collected with a distance of ≥3 cm from the edge of cancer tissues in the resected bladder. All the specimens for histological and pathological detections were snap-frozen in liquid nitrogen and stored at − 80 °C after surgical removal. Samples were obtained from the patients with proper informed consent and approved by the Institutional Review Board of the First Affiliated Hospital of Xiamen University.
Cell culture and treatment
Human bladder cancer cells lines (RT-112, 5637, BIU-87 and TCCSUP) and human embryonic kidney cells lines (HEK293T) were all purchased from the BNCC cell bank (Beijing, China). Cells were inoculated in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS (GIBCO BRL, NY, USA), 1% Glutamax (35,050, Invitrogen, Carlsbad, CA, USA), 1% Non-essential Amino Acids1 (111,401, Invitrogen) and 1% Sodium Pyruvate 100 mM Solution (113,600,070, Invitrogen). Cultured cells were stored at 37 °C in a humidified atmosphere (5% CO2 and 95% O2).
Microarray analysis
Microarray datasets GSE92675 at platform GPL19978 were used. Four pairs of matched bladder tumor tissues and adjacent tissues were prepared for analyzing the expression level and biological functions of circRNAs. Differentially expressed genes were identified using t-test (P < 0.05) combined with fold change (FC > 2). As for mRNA analysis, 19 pairs of tumor tissues and adjacent tissues obtained from TCGA (https://cancergenome.nih.gov/) were screened by R programme (Fold change> 2, P < 0.001).
MiRNA targets prediction of circCEP128
We predicted the miRNA-binding sites of circCEP128 and SOX11 using TargetScan (http://www.targetscan.org/) and circular RNA interactome (https://circinteractome.nia.nih.gov/), respectively.
Cell transfection
TCCSUP and BIU-87 cells were transfected with corresponding plasmids using the Lipofectamine 2000 (Invitrogen) in the light of the manufacturer’s recommendations. And the cells were harvested at 48 h after transfection. Cells were generally assigned to different groups as follows: (1) negative control (NC) group: bladder cancer cells transfected with pCDNA3.1 (GenePharma, Shanghai, China). (2) mimics group: bladder cancer cells transfected with miR-145-5p mimic. (3) inhibitor group: bladder cancer cells transfected with miR-145-5p inhibitor. (4) si-circRNA group: bladder cancer cells transfected with si-circCEP128. (5) si-CEP128 group: bladder cancer cells transfected with si-CEP128. (6) si-SOX11 group: bladder cancer cells transfected with si-SOX11. (7) si-circRNA and inhibitor group: bladder cancer cells transfected with si-circCEP128 and miR-145-5p inhibitor. (8) si-SOX11 and inhibitor group: bladder cancer cells transfected with si-SOX11 and miR-145-5p (Thermo Fisher, Waltham, MA, USA). Si-circCEP128 and siCEP128 were designed on Thermo Fisher (https://rnaidesigner.thermofisher.com/) and produced by Sangon Biotech (Shanghai, China) which were shown in Table 1.
Table 1.
SiRNA sequences
| sense | anti-sense | |
|---|---|---|
| si-circCEP128 | 5’-GAGAGCUUGAACAGGAAUU-3’ | 3’-AAUUCCUGUUCAAGCUCUC-5’ |
| si-CEP128 | 5’-GCGCTACACCAAATACAAA-3’ | 3’-UUUGUAUUUGGUGUAGCGC-5’ |
| si-SOX11 | 5’-GCGAGAAGATCCCGTTCAT-3’ | 3’-AUGAACGGGAUCUUCUCGC-5’ |
qRT-PCR assay
The reverse transcription of RNA was developed using Prime Script RT Master Mix (Takara, Japan). PCR reaction was performed with PCR Master Mix (2×) (ThermoFisher Scientific, Waltham, Ma, USA). For the quantitative determination of circRNA, miRNA and mRNA, real-time PCR analysis was performed using SYBRremix Ex TaqTM kit (Takara, Tokyo, Japan) with GAPDH and U6 as internal controls. All analyses were performed using the StepOnePlus Real-Time PCR System (Applied Biosystems, Carlsbad, CA, USA). The primer sequence was listed in Table 2.
Table 2.
Primer sequences
| Genes | Primers |
|---|---|
| circCEP128 | F: 5’-ACCCACATCGCTGGTTAGC-3’ |
| R: 5’-TCGATCACCTTCTGCTTTCGT-3’ | |
| SOX11 | F: 5’-GCCTCTTTTCTGCTGGGTCT-3’ |
| R: 5’-ACTGAAAACCTCCTCCGCTG-3’ | |
| hsa-miR-155-5p | F: 5’-TGCCTCCAACTGACTCCTAC-3’ |
| R: 5’-GCGAGCACAGAATAATACGAC-3’ | |
| hsa-miR-223-3p | F: 5’-GGGGTGTCAGTTTGTCAAA-3’ |
| R: 5’-CAGTGCGTGTCGCGTGGAGT-3’ | |
| hsa-miR-145-5p | F: 5’-CTCACGGTCCAGTTTTCCCA-3’ |
| R: 5’-ACCTCAAGAACAGTATTTCCAGG-3’ | |
| GAPDH | F: 5’-GGAAAGCTGTGGCGTGAT-3’ |
| R: 5′-AAGGTGGAAGAATGGGAGTT-3’ | |
| U6 | F: 5’-GCTTCGGCAGCACATATACTAAAAT-3’ |
| R: 5’-CGCTTCACGAATTTGCGTGTCAT-3’ |
Immunohistochemistry
Bladder tissues were immunostained by anti-SOX11 (1:200, Abcam, Cambridge, MA, USA) and HRP-conjugated goat anti-rabbit IgG (1:1000, Abcam), respectively. The resultant immunostaining images were captured using the AxioVision Rel.4.6 computerized image analysis system (Carl Zeiss, Oberkochen, Germany). Proteins expression levels were analyzed using Image-Pro Plus version 6.0 (Media Cybernetics, MD) by calculating the integrated optical density in each stained area (IOD/area).
Western blot
Cell lysates were prepared with RIPA buffer (Thermo Scientific). The concentration was determined using a bicinchoninic acid (BCA) protein assay kit (Pierce, Thermo Scientific). Immunoreactive bands were detected by using the Immobilon ECL substrate kit (Millipore, Merck KGaA, Germany). The images were acquired by using BioSpectrum 600 Imaging System (UVP, CA, USA). Antibodies used included primary and secondary antibodies, primary antibodies including anti-SOX11 (1:1000, Abcam), Bcl-2 (1:500, Abcam), Bax (1:500, Abcam), Cleaved-caspase3 (Anti-active Caspase-3, 1:500, Abcam) and anti-GAPDH (1:10000, Abcam); secondary antibody was HRP-conjugated secondary goat anti-rabbit IgG (1:2000, Abcam).
Fluorescence in situ hybridization (FISH)
TCCSUP and BIU-87 cells were performed with cytospin, the collected cells were fixed with Carnoy’s fixative (3:1 methanol (ThermoFisher Scientific, Leicestershire, UK): acetic acid (Sigma-Aldrich, Bornem, Belgium) and then air dried for 5 min. CircCEP128 was localized in bladder cells by FISH using CEP Y SpectrumGreen DNA probe (ACCB Biotech, Beijing, China) under the manufacturer’s instructions. After that, the slides with collected cells were mounted with Fluorescence Mounting Medium (Antifade) (Abace Biology, Beijing, China) to counterstain all nucleic on the slide. Subsequently, the slide was scanned at 20-fold magnification using Carl Zeiss Short Distance Plan- Apochromat® objective.
Flow cytometry (FCM) assay
Transfected cells were subjected to PI staining for detection with Cell Cycle assay Kit (ab112116, Abcam). Then they were subjected to FITC-Annexin V and PI double staining for flow cytometry detection (EPICS, XL-4, Beckman, CA, USA) according to manufacturer’s instructions. Cells were trypsinized, resuspended and incubated with 1.0 μl of PI and 5.0 μl of Annexin V-fluorescein and the apopotosis rate was determined by flow cytometry (FACScan; Becton Dickinson, MountainView, CA, USA) and analyzed with analyzed using Flowjo 7.6 software (BD Bioscience, San Jose, CA, USA).
MTT assay
Well transfected cells were seeded into 96-well plates at 2 × 104 cells/ml in a 5% CO2 atmosphere and incubated overnight. 20 μl MTT reagent (Sigma-Aldrich, Bornem, Belgium) was added to each well respectively at 0, 24, 48 and 72 h. at an absorbance of 490 nm, cell viability was detected with an automatic enzyme-linked immune detector. The experiment was repeated in triplicate.
Luciferase reporter assay
Dual-luciferase reporter assay system (Promega, Madison, WI, USA) was operated for the co-transfection of HEK293T cells. Mutagens were used to mutate the sequence in 3’UTR of circCEP128 and SOX11. The mutant circCEP128 was transfected with miR-145-5p or empty vector as negative control. The mutant SOX11 was also transfected with miR-145-5p and empty control for two groups. The luciferase results were detected by Luc-Pair™ Duo-Luciferase Assay Kit (Yeasen, Shanghai, China) and the luciferase activity in empty vector cells was normalized as 1.
RNA pull-down assay
Cells transfected with biotinylated miR-145-5p or mutant miR-145-5p mimics (50 nM) using Lipofectamine RNAiMax (Life Technologies) were harvested and sonicated 48 h after transfection. Remaining cell lysates were incubated with C-1 magnetic beads (Life Technologies) at 4 °C for 2 h and then purified using RNeasy Mini Kit (QIAGEN, Duesseldorf, Germany) for analysis, following detected RNA enrichment by qRT-PCR.
Tunnel staining
TCCSUP were cultured on coverslips after transfected with miR-145-5p mimics, miR-145-5p inhibitor, si-circRNA, or si-SOX11. 48 h after, cells were harvested and fixed with 4% paraformaldehyde. Cell apoptosis analysis was performed using a tunnel staining kit (Abcam, USA). Cell nuclei were stained with DAPI. All fluorescent images were examined using a Leica DM3000 microscope and photographed using a DFC 420 camera (Leica, Germany).
EdU incorporation
At the last 18 h of cell culture, Edu-labeling reagent (Invitrogen) (1:1000 dilution) was added into medium. EdU was detected using the Click-iT kit (Invitrogen) following the manufacturer’s protocol. The slides were counterstained with DAPI (4′,6-diamidino-2-phenylindole). Images were captured and analyzed using OpenLab software, and cells were quantitated using ImageJ software.
Statistical analysis
All data were shown as mean ± standard error of the mean (SEM). Statistical analyses were performed using Graphpad Prism statistical software (Version 6.0; La Jolla, CA, USA). Statistical significance (P < 0.05) was determined by Student’s t-test (unpaired) for two-group compared and chi-square test was used to assess the RNA correlation.
Results
CircCEP128 (hsa_circ_0102722) and SOX11 are significantly up-regulated in bladder cancer
A total of 433 circRNAs were differentially expressed (Fold change> 2 and P < 0.05) with the analysis of circRNA microarray from GEO database. 169 circRNAs were significantly down-regulated, and 264 circRNAs were up-regulated in 4 bladder cancer samples. The top ten up- and down-regulated circRNAs were chosen by fold change to draw the cluster heat map (Fig. 1a). CircCEP128 was up-regulated in bladder tumor tissues with a fold change value of 5.06 (Table 3). Data of 19 pairs sample (tumor and adjacent tissue of patients) were obtained from TCGA and analyzed by R software, the cluster heat map was drawn (Fig. 1b). SOX11 was up-regulated in bladder tumor tissues with a fold change value of 11.56 (Table 4). Immunochemistry staining showed more SOX11-positive cells in tumor tissues than that of adjacent normal tissues (Fig. 1c, P < 0.01). The highly expressions of circCEP128 and SOX11 were validated in 10 paired of bladder tissues by RT-PCR (Fig. 1d, P < 0.01). Analysis of clinicopathologic features, circCEP128 and SOX11 showed significant higher expressions in bladder cancer tissues compared with adjacent normal tissue (Table 5). Gender and age had no relationships with the expression of circCEP128 or SOX11except for tumor size, TNM stage and lymphatic metastasis. CircCEP128 and SOX11 had a positively related dependency both in cancer tissues and in adjacent normal tissues (Fig. 1e, P < 0.05). Thus we hypothesized that circCEP128 may function as a ceRNA (competing endogenous RNAs) and promote the expression of SOX11.
Fig. 1.
CircCEP128 and SOX11 are up-regulated in bladder cancer. a The cluster heatmap showed some of the differentially expressed circRNAs over 2-fold change between tumor tissues and adjacent normal tissues. Red color indicates high expression level, and green color indicates low expression level. The red arrow indicates hsa_circ_0102722 (circCEP128). b The cluster heat map showed some of the differentially expressed mRNAs over 2-old change in tumor tissues and adjacent normal tissues. The red arrow indicates SOX11. c Immunochemistry staining showed more SOX11-positive cells in tumor tissues than in adjacent normal tissues. Scale bar, 50 μm. **P < 0.01, compared with adjacent tissues. d CircCEP128 and SOX11 were highly expressed in tumor tissues. **P < 0.01, compared with adjacent tissues. e According to the results of qRT-PCR, circCEP128 and SOX11 were positively related in cancer tissues and in cancer adjacent tissues
Table 3.
Log2 (Fold Change) of ten increased circular RNA and ten decreased circular RNA
| Circular RNA | Log2(Fold Change) |
|---|---|
| hsa_circ_0135888 | 4.88 |
| hsa_circ_0136628 | 4.93 |
| hsa_circ_0135566 | 4.97 |
| hsa_circ_0102722 | 5.06 |
| hsa_circ_0000144 | 5.71 |
| hsa_circ_0115632 | 4.64 |
| hsa_circ_0058063 | 4.85 |
| has_circ_0110572 | 5.30 |
| hsa_circ_0000066 | 5.62 |
| hsa_circ_0096464 | 5.30 |
| hsa_circ_0105820 | − 3.03 |
| hsa_circ_0133859 | −2.63 |
| hsa_circ_0092275 | −2.71 |
| hsa_circ_0050871 | −2.63 |
| hsa_circ_0133491 | −2.89 |
| hsa_circ_0066475 | −4.10 |
| hsa_circ_0073018 | −3.41 |
| hsa_circ_0097271 | −3.44 |
| hsa_circ_0092342 | −3.83 |
| hsa_circ_0111302 | −3.24 |
The positive number means circular RNA was increased in tumor samples, the minus means circular RNA was decreased in tumor samples
Table 4.
Log2 (Fold Change) of ten increased mRNA and ten decreased mRNA
| mRNA | Log2 (Fold Change) |
|---|---|
| MMP27 | −14.42 |
| ANGPTL5 | −12.50 |
| FBXL21 | −13.47 |
| PRAC | −14.15 |
| LGI1 | −12.83 |
| LCN6 | −13.61 |
| ADH1A | −12.92 |
| LRRTM1 | −13.19 |
| LY6G6D | −14.23 |
| GRIA1 | −12.39 |
| CST4 | 10.29 |
| CST1 | 10.75 |
| HIST1H2AH | 10.68 |
| EN1 | 10.96 |
| IBSP | 11.36 |
| CST2 | 12.33 |
| C5orf46 | 13.47 |
| IGFL2 | 11.94 |
| SOX11 | 11.56 |
| CHRNA1 | 10.61 |
The positive number means mRNA was increased in tumor samples, the minus means mRNA was decreased in tumor samples
Table 5.
The relationship between circCEP128/SOX11 expression and clinicopathological characteristics in bladder cancer patients
| Items | Patients (N = 10) | circCEP128 RNA (X ± SD) | P value | SOX11 mRNA (X ± SD) | P value |
|---|---|---|---|---|---|
| N | 10 | 0.00563 ± 0.00235 | 0.0038b | 3.2565 ± 2.0583 | 0.0032b |
| T | 10 | 0.02568 ± 0.01897 | 6.0354 ± 1.5568 | ||
| Gender | 0.7704 | 0.4643 | |||
| Male | 4 | 0.02258 ± 0.01989 | 3.3564 ± 3.0254 | ||
| Female | 6 | 0.01897 ± 0.01765 | 4.8697 ± 3.0658 | ||
| Age (years) | 0.3785 | 0.463 | |||
| < 60 | 2 | 0.00897 ± 0.00458 | 2.7856 ± 1.8975 | ||
| > =60 | 8 | 0.02305 ± 0.02035 | 5.6578 ± 4.9876 | ||
| Tumor size | 0.0291a | 0.0168a | |||
| < 10 | 5 | 0.01756 ± 0.00968 | 1.8756 ± 0.9873 | ||
| > =10 | 5 | 0.03457 ± 0.01057 | 5.5893 ± 4.5760 | ||
| TNM stage | 0.0361a | 0.0263a | |||
| I-II | 3 | 0.01687 ± 0.00952 | 3.8976 ± 1.0687 | ||
| III-IV | 7 | 0.04356 ± 0.01689 | 6.3654 ± 1.3873 | ||
| Lymphatic metastasis | 0.0313a | 0.0343a | |||
| NO | 3 | 0.00897 ± 0.00563 | 1.7056 ± 0.9856 | ||
| Yes | 7 | 0.03056 ± 0.01347 | 6.0325 ± 2.7846 | ||
N: Adjacent normal tissues; T: Bladder cancer tissues; Student-t test
aP < 0.05, bP < 0.01 was recognized as a significant difference
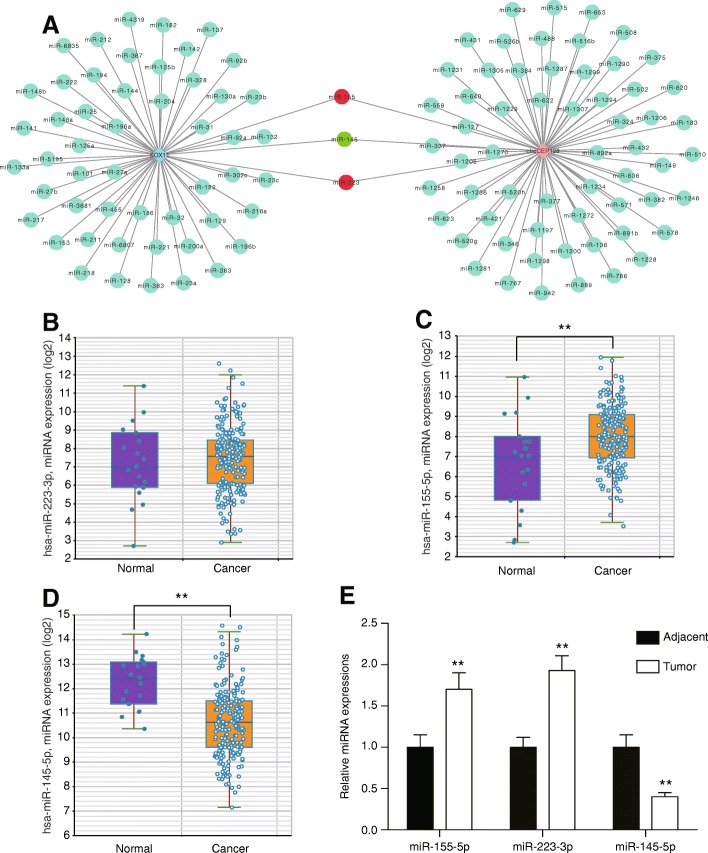
CeRNA analysis for circCEP128 according to the database
In Fig. 2a, miR-155, miR-223 and miR-145 were found to have the same targeting relations with circCEP128 and SOX11 by using circular RNA interactome (https://circinteractome.nia.nih.gov/Circular_RNA/circular_rna.html) and TargetScan (http://www.targetscan.org/). Firstly, the analysis results had been verified in database of starBase v2.0 (http://starbase.sysu.edu.cn/) with TCGA data in bladder cancers, which showed that miR-155-5p obviously up-regulated while miR-145-5p obviously down-regulated in tumor cells (P < 0.01, Fig. 2c and d), miR-223-3p expression had no statistically change between normal and tumor cells (P > 0.05, Fig. 2b). Subsequently, the experiments of qRT-PCR were conducted in 10 bladder cancer tissues (Fig. 2e). It showed that miR-155-5p, miR-233-3p were upregulated and miR-145-5p was downregulated in tumor cells (P < 0.01). The down-regulation of miR-145-5p was speculated as a competitive target of circCEP128 and SOX11 in bladder cancer.
Fig. 2.
CeRNA analysis for circCEP128. a Cytoscope was used to visualize circCEP128-miRNA-target gene interactions based on the TargetScan and circular RNA interactome. Red color indicates high expression level in bladder cancer and green color indicates low expression level in bladder cancer. b-d TCGA bladder cancer patients’ data analysis was that miR-223-3p expression had no significant difference, miR-155-5p expression increased but miR-145-5p expression decreased in tumor samples. **P < 0.01, compared with adjacent samples. e QRT-PCR results showed that miR-155 and miR-223 was up-regulated in tumor tissues while miR-145 was down-regulated in tumor tissues. **P < 0.01, compared with adjacent tissues
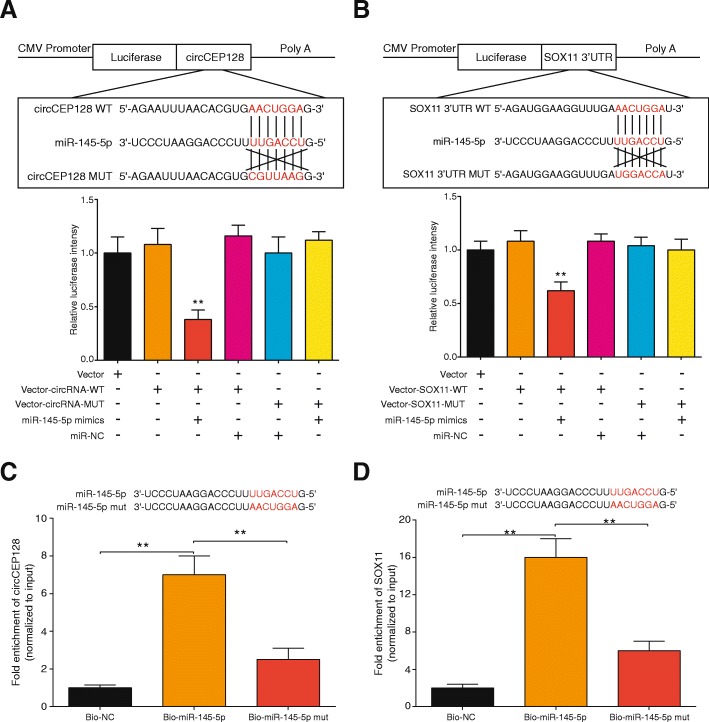
CircCEP128 serves as a sponge of miR-145-5p which directly regulates SOX11
StarBase v2.0 predicted the binding relationship between CircCEP128 and miR-145-5p. Luciferase reporter assay showed that the luciferase intensity of HEK293T cells transfected with circCEP128 wild type or miR-145-5p mimics were significantly reduced, but the luciferase intensity of cells transfected with circCEP128 muted type or miR-145-5p mimics were hardly changed (P > 0.05, Fig. 3a). Similarly, with the targetScan prediction of the binding sequences of miR-145-5p and SOX11, it showed that the luciferase activity was significantly reduced in SOX11 wild type + miR-145-5p mimics co-transfection group (P < 0.01, Fig. 3b). After RNA pull-down assay, the circCEP128 level of the miR-145-5p group was 6.73 times as much as NC group and 2.28 times as much as miR-145-5p-mut group (P < 0.01, Fig. 3c) and the miR-145-5p group was found high SOX11 level which was 16.69 times as much as NC group (P < 0.01, Fig. 3d).
Fig. 3.
CircCEP128 serves as a sponge of miR-145-5p and indirectly targets at SOX11. a and b Binding sites of miR-145-5p in circCEP128/SOX11 3’UTR (Up). Luciferase reporter assay revealed that miR-145-5p was able to reduce the luciferase intensity more than 40% by targeting at wild type RNAs (WT, Down). **P < 0.01, compared with the group with only vector. c and d RNA pull-down assays indicated the physical interactions between circCEP128 and miR-145-5p or between miR-145-5p and SOX11
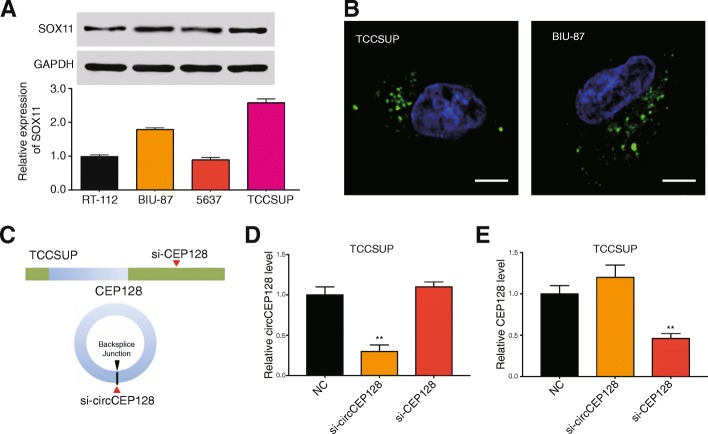
Knockdown of circCEP128 inhibits expression of SOX11
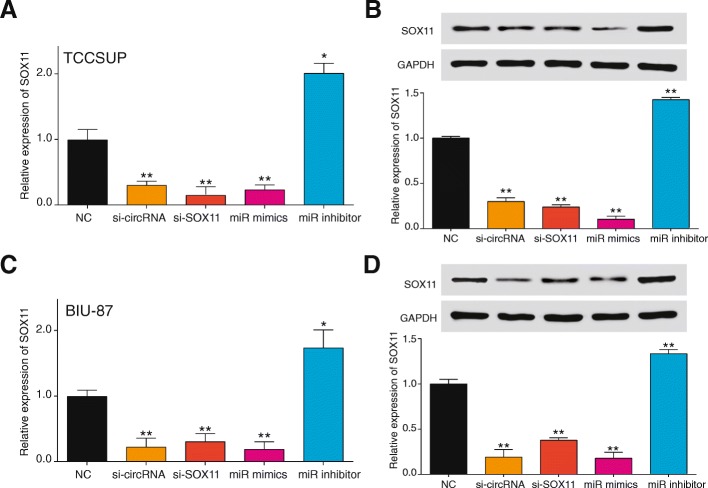
In bladder cancer cell lines, SOX11 expressed higher in BIU-87 and TCCSUP compared with RT-112 and 5637 (Fig. 4a). TSSCP showed the highest expression level of SOX11. Experiments of fluorescent probe localized circCEP128 in the plasma of bladder cell lines (Fig. 4b). Si-circCEP128 (Fig. 4c) could knockdown the expression of circCEP128 while si-CEP128 had no effect on altering the expression of circCEP128 (P > 0.05, Fig. 4d). Similarly, Si-circCEP128 had no effect on regulating the expression of CEP128, while si-CEP128 could knockdown the expression of CEP128 (P < 0.01, Fig. 4e). Expression of SOX11 was detected by qRT-PCR (P < 0.01, Fig. 5a and c) and western blot (P < 0.01, Fig. 5b and d) in TCCSUP and BIU-87 cells. Expression of SOX11 was increased greatly in miRNA inhibitor group while decreased dramatically in si-SOX11 group. The results verified that knockdown of circCEP128 inhibited expression of SOX11 and it could be guessed that knockdown of circCEP128 promotes expression of miR-145-5p to inhibit expression of SOX11.
Fig. 4.
Si-circCEP128 inhibits the expression of circCEP128 not CEP128. a SOX11detection of 4 bladder cancer cells was conducted by qRT-PCR. b Cellar locations of circCEP128 were in TCCUSP and BIU-87 cytoplasm. Scale bar, 2 μm. c Schematic model of the si-circRNAs. si-CEP128 targets the CEP128 linear transcript, si-circCEP128 targets the back-splice junction of circCEP128. d and e Si-circCEP128 knocked down only the circular transcript and did not affect the expression of linear species. Si-CEP128 knocked down only the CEP128 linear transcript but not the circular transcript. **P < 0.01, compared with NC
Fig. 5.
Knockdown of circCEP128 inhibits expression of SOX11. The expression levels of SOX11 were detected following knockdown of circCEP127 using si-circCEP128, miR-145-5p inhibitor, co-transfection with si-circCEP128 and the miR-145-5p inhibitor or si-SOX11 (a) in TCCSUP and (c) in BIU-87. **P < 0.01, compared with NC. SOX11 protein relative expression levels were analyzed by western blotting (b) in TCCSUP and (d) in BIU-87. **P < 0.01, compared with NC
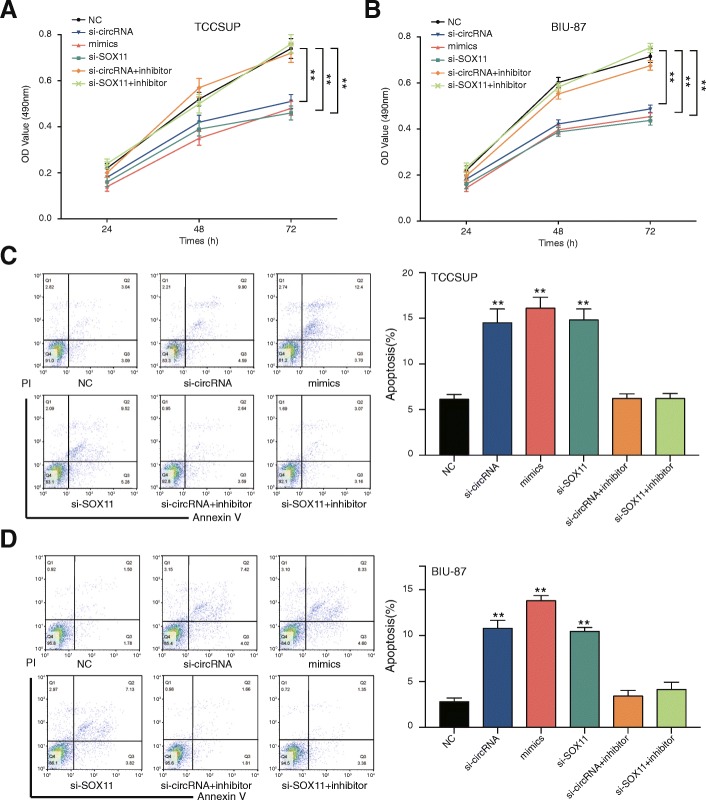
The effect of circCEP128 on bladder tumor cell proliferation, apoptosis and cell cycle
Knockdown of circCEP128 inhibited cell proliferation (P < 0.05, Fig. 6a and b) but promoted cell apoptosis (P < 0.05, Fig. 6c and d). Cell cycle induced bladder cancer cell G1/S arrest in si-cirRNA, miR-145-5p mimics and si-SOX11 group (P < 0.01, Fig. 7a and b). Similar results were generated after improving expression of miR-145-5p or knocking down SOX11 (P < 0.01, Fig. 6a-Fig. 7b). Cell cycle, cell apoptosis rate and cell proliferation were recovered with the addition of miR-145-5p inhibitor after knocking down the circCEP128 (P > 0.05, Fig. 6a-Fig. 7b). Similarly, Cell cycle, cell apoptosis rate and cell proliferation were recovered with si-SOX11 after the inhibition of miR-145-5p (P > 0.05, Fig. 6a-Fig. 7b). As a result, knockdown of circCEP128 induced the inhibition of cell proliferation and increased bladder cancer cell apoptosis rate.
Fig. 6.
Effects of circCEP128 on cell proliferation. MTT assay was also performed to assess cell proliferation after transfection with si-circRNA of circCEP128, miR-145-5p mimics, si-SOX11, co-transfection with si-circRNA and the miR-145-5p inhibitor and co-transfection with si-SOX11 and the miR-145-5p inhibitor (a) in TCCSUP and (b) in BIU-87. Knockdown of circCEP128 inhibited cell proliferation. *P < 0.05, compared with NC. Cell apoptosis was analyzed using flow cytometry (c) in TCCSUP and (d) in BIU-87 with double staining of PI and Annexin V-FITC. Knockdown of circCEP128 induced the increased cell apoptosis rate. **P < 0.05, compared with NC
Fig. 7.
Effects of circCEP128 on cell cycle. Cell cycle was analyzed using flow cytometry after transfection with si-circRNA of circCEP128, miR-145-5p mimics, si-SOX11, co-transfection with si-circRNA and the miR-145-5p inhibitor and co-transfection with si-SOX11 and the miR-145-5p inhibitor (a) in TCCSUP and (b) in BIU-87. Knockdown of circCEP128 induced G1/S arrest. *P < 0.05, compared with NC
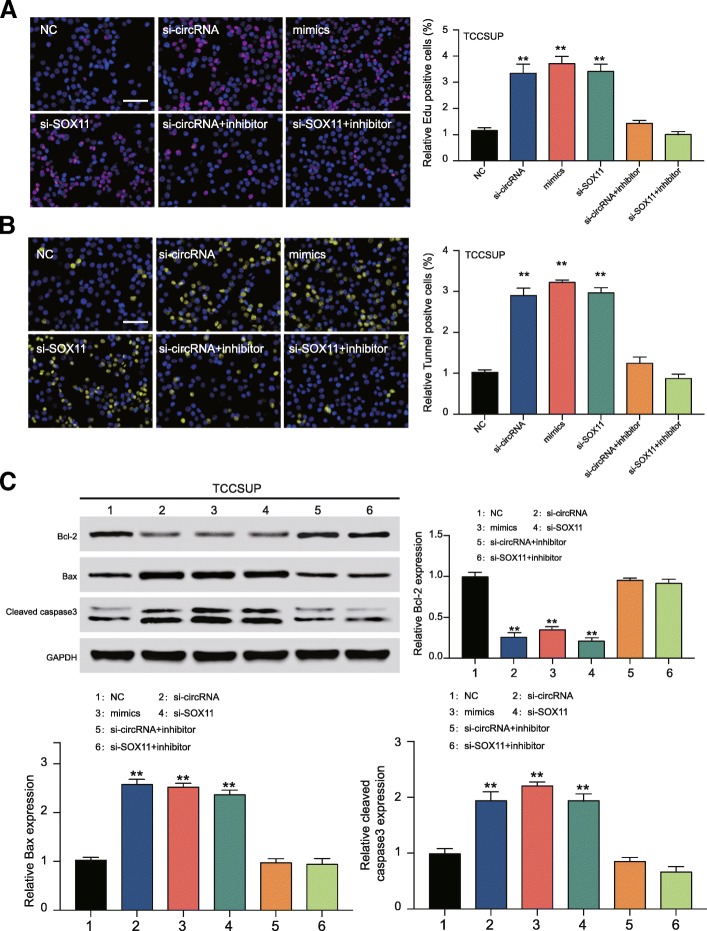
Knockdown of circCEP128 promoted TCCSUP cells damage and apoptosis
Damaged TCCSUP cells marked Edu and Tunnel staining, a strong positive cells presented in si-circRNA, miR-145-5p mimics and si-SOX11 group (P < 0.01, Fig. 8a and b). The effects of si-circRNA and si-SOX11 on TCCSUP had been recovered by miR-145-5p inhibitor. The expression levels of three apoptosis relative proteins were detected (Fig. 8c), the results showed that the expression of Bcl-2 was significantly decreased in si-circRNA group, mimics group as well as si-SOX11 group (P < 0.01). However, the expression levels of bax and cleaved caspase3 were increased obviously in si-circRNA group, mimics group and si-SOX11 group (P < 0.01).All in all, inhibition of circCEP128 induced TCCSUP cell damage and apoptosis through the regulation of miR-145-5p/SOX11.
Fig. 8.
Effects of circCEP128 on TCCSUP cell damage and apoptosis. After transfected with corresponding siRNA or mimics/inhibitor, higher DNA damage happened in si-circRNA, miR-145-5p mimics and si-SOX11 with the experiments of a Edu staining b Tunnel staining. **P < 0.01, compared with NC. Scale bar, 20 μm. c Western blot showed the result of apoptosis related proteins bax and caspase3 were higher but bcl-2 was lower in si-circRNA, miR-145-5p mimics and si-SOX11 groups compared with NC group. **P < 0.01, compared with NC
Discussion
In this study, circCEP128 and SOX11 were found highly expression and positively related in bladder cancer tissues. The relation could be regulated by miR-145-5p, a downregulated miRNA in bladder cancer samples. CircCEP128 served as a sponge of miR-145-5p and indirectly regulated SOX11. Knockdown of circCEP128 could induce the inhibition of cell proliferation and increase bladder cancer cell apoptosis rate.
Many studies have confirmed that stable transcripts with a host of miRNA-binding sites or miRNA response elements (MREs) could function as miRNA sponges (Memczak et al., 2013) and circular RNAs are found to be enriched in functional miRNA binding sites (Huang et al., 2016). For example, Li et al. found that circHIPK3 inhibited migration, invasion and angiogenesis of human invasive bladder cancer by targeting miR-558 (Markopoulos et al., 2017). In addition, other studies indicated that circRNA-MYLK could function as an endogenous sponge for miR-29a in bladder cancer (Zhong et al., 2017). What is more, circRNA CDR1as was reported to negatively regulate miR-7 and increase the levels of miR-7 targets (Xu et al., 2015; Hansen et al., 2013). These conclusions coincide with the results of our study, which indicate that circCEP128 is able to function as a ceRNA for miR-145-5p.
Recently, studies about circular RNA mainly focus on its interactions with miRNA and miRNA targets. For example, it is revealed that circRNA-MYLK functions as a ceRNA for miR-29a, thus boosting VEGF/VEGFR2 expressions and activating downstream Ras/ERK signaling pathway in bladder cancer progression (Zhong et al., 2017). Li et al. demonstrated that cir-ITCH increased the expression of the miRNA target gene ITCH in esophageal squamous cell carcinoma (Li et al., 2015). Other studies showed that the over-expression of circHIPK3 induced efficiently interaction with miR-558 and then down-regulated the expression of HPSE and its downstream targeted MMP-9 and VEGF to attenuate the promoting effect of miR-558 on bladder cancer cell migration, invasion, and angiogenesis (Li et al., 2017). Also, it was speculated that circTCF25 may competitively bind with miR-103a-3p and miR-107 and relieve their suppression on associated target genes (Zhong et al., 2016). Similarly, in our study, we found that circCEP128 served as a sponge of miR-145-5p and indirectly regulated SOX11.
SOX11 have been proved to promote invasive growth and progression of DCIS cells (Oliemuller et al., 2017) and prevent cell differentiation in mantle cell lymphoma (Meggendorfer et al., 2013). However, Sernbo et al. demonstrated that the overexpression of SOX11 could induce growth arrest in ovarian cancer cells (Sernbo et al., 2011). Varied findings may result from differential populations and cancer types (Fang et al., 2017). In this study, SOX11 was a contributor to bladder cancer in terms of proliferation and apoptosis. Therefore, it was verified that circCEP128 acted as a ceRNA for miR-145-5p to regulate SOX11, which further promoted cell proliferation and suppressed cell apoptosis of bladder cancer.
In this study, siRNA could only specifically target the circular form but not the linear form of CEP128; thus the off-target effect have to be considered. Therefore, more effective, accurate and specific methods of RNA interference remain to be exploited. Additionally, further research on functions and mechanisms underlying circCEP128 are required.
Conclusion
In conclusion, the results of our study demonstrate that circCEP128 is up-regulated in human bladder cancer, and is able to sponge miR-145-5p for promoting SOX11 expression with high efficiency. We also demonstrate that knockdown of circCEP128 can effectively inhibit cell proliferation and promote cell apoptosis rate of bladder cancer cells through targeting miR-145-5p/SOX11 axis. Our findings provide novel evidences that circRNAs might act as “microRNA sponges” and provide a new therapeutic target for the treatment of bladder cancer.
Acknowledgments
Funding
This project was supported by Natural Science Foundation of Fujian Province of China (Grant No.2016D009, Grant No.2017 J01355, Grant No.2011–2-60).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- BCA
Bicinchoninic acid
- ceRNA
Competing endogenous RNA
- circCEP128
Circular RNA CEP128
- circRNAs
Circular RNAs
- FCM
Flow cytometry
- FISH
Fluorescence in situ hybridization
- miR-145
MicroRNAs
- miRNAs
MicroRNAs
- MREs
MiRNA response elements
- NC
Negative control
- SEM
Standard error of the mean
Authors’ contributions
Research conception and design: ZW & WH; Data analysis and interpretation: XW, TW, YC & BC; Statistical analysis: ZW, RL & PB; Drafting of the manuscript: ZW; Critical revision of the manuscript: JX; Receiving grant: JX; Approval of final manuscript: all authors.
Ethics approval and consent to participate
Samples were obtained from the patients with proper informed consent and approved by the Institutional Review Board of the First Affiliated Hospital of Xiamen University.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Enokida H, Yoshino H, Matsushita R, Nakagawa M. The role of microRNAs in bladder cancer. Investigative and Clin Urology. 2016;57(Suppl 1):S60–S76. doi: 10.4111/icu.2016.57.S1.S60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang G, Liu J, Wang Q, Huang X, Yang R, Pang Y, et al. MicroRNA-223-3p regulates ovarian Cancer cell proliferation and invasion by targeting SOX11 expression. Int J Mol Sci. 2017;18(6):1208. doi: 10.3390/ijms18061208. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- Huang M, Zhong Z, Lv M, Shu J, Tian Q, Chen J. Comprehensive analysis of differentially expressed profiles of lncRNAs and circRNAs with associated co-expression and ceRNA networks in bladder carcinoma. Oncotarget. 2016;7:47186–47200. doi: 10.18632/oncotarget.9706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Zhang L, Li W, Deng J, Zheng J, An M, et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/beta-catenin pathway. Oncotarget. 2015;6:6001–6013. doi: 10.18632/oncotarget.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zheng F, Xiao X, Xie F, Tao D, Huang C, et al. CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep. 2017;18:1646–1659. doi: 10.15252/embr.201643581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markopoulos GS, Roupakia E, Tokamani M, Chavdoula E, Hatziapostolou M, Polytarchou C, et al. A step-by-step microRNA guide to cancer development and metastasis. Cell Oncol (Dordr) 2017;40:303–339. doi: 10.1007/s13402-017-0341-9. [DOI] [PubMed] [Google Scholar]
- Meggendorfer M, Kern W, Haferlach C, Haferlach T, Schnittger S. SOX11 overexpression is a specific marker for mantle cell lymphoma and correlates with t (11;14) translocation, CCND1 expression and an adverse prognosis. Leukemia. 2013;27:2388–2391. doi: 10.1038/leu.2013.141. [DOI] [PubMed] [Google Scholar]
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- Minami K, Taniguchi K, Sugito N, Kuranaga Y, Inamoto T, Takahara K, et al. MiR-145 negatively regulates Warburg effect by silencing KLF4 and PTBP1 in bladder cancer cells. Oncotarget. 2017;8:33064–33077. doi: 10.18632/oncotarget.16524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi S, Yamada N, Kumazaki M, Yasui Y, Iwasaki J, Naito S, et al. socs7, a target gene of microRNA-145, regulates interferon-beta induction through STAT3 nuclear translocation in bladder cancer cells. Cell Death Dis. 2013;4:e482. doi: 10.1038/cddis.2013.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliemuller E, Kogata N, Bland P, Kriplani D, Daley F, Haider S, et al. SOX11 promotes invasive growth and ductal carcinoma in situ progression. J Pathol. 2017;243:193–207. doi: 10.1002/path.4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sernbo S, Gustavsson E, Brennan DJ, Gallagher WM, Rexhepaj E, Rydnert F, et al. The tumour suppressor SOX11 is associated with improved survival among high grade epithelial ovarian cancers and is regulated by reversible promoter methylation. BMC Cancer. 2011;11:405. doi: 10.1186/1471-2407-11-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang X, Li G, Liu H, Li T, Liu J, Zhao Q, et al. Comprehensive circular RNA profiling reveals that hsa_circ_0005075, a new circular RNA biomarker, is involved in hepatocellular Crcinoma development. Medicine. 2016;95:e3811. doi: 10.1097/MD.0000000000003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Guo S, Li W, Yu P. The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci Rep. 2015;5:12453. doi: 10.1038/srep12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Z, et al. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett. 2017;403:305–17. [DOI] [PubMed]
- Zhong Z, Lv M, Chen J. Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in bladder carcinoma. Sci Rep. 2016;6:30919. doi: 10.1038/srep30919. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.