Abstract
Silicon carbide has been shown to be biocompatible and is used as a coating material for implanted medical devices to prevent biofilms. Silicon carbide nanomaterials are also promising in cell tracking due to their stable and strong luminescence, but more comprehensive studies of this material on the nanoscale are needed. Here, we studied the toxicity of silicon carbide nanomaterials on human mesenchymal stem cells in terms of metabolism, viability, adhesion, proliferation, migration, oxidative stress, and differentiation ability. We compared two different shapes and found that silicon carbide nanowires are toxic to human mesenchymal stem cells but not to cancer cell lines at the concentration of 0.1 mg/mL. Control silicon carbide nanoparticles were biocompatible to human mesenchymal stem cells at 0.1 mg/mL. We studied the potential mechanistic effect of silicon carbide nanowires on human mesenchymal stem cells’ phenotype, cytokines secretion, and gene expressions. These findings suggest that the toxic effect of silicon carbide nanomaterials to human mesenchymal stem cells are dependent on morphology.
Keywords: SiC nanowires, SiC nanoparticles, cytotoxicity, cytocompatibility, human mesenchymal stem cell
Table of Contents:
1. Introduction
Silicon carbide (SiC) is temperature tolerant, radiation resistant, and chemically inert.[1, 2] SiC is a wide bandgap semiconductor with high breakdown electric field strength, high saturated electrons’ drift velocity and a high thermal conductivity.[3] Due to these unique electrical properties, SiC has become a promising bio-sensor for a variety of medical applications[4–6] including glucose sensing,[7] DNA detection,[8] and neural probing.[9] SiC is also used to coat medical devices and implants due to its excellent strength, low density (in comparison with metals), stability, and chemical inertness.[10]
SiC nanomaterials also have highly strong and stable luminescence.[11, 12] Compared to organic dyes and fluorescent proteins, SiC nanomaterials have a higher quantum yield, lower photobleaching, and more stable fluorescence. Cubic symmetric SiC (3C-SiC) nanoparticles have intense photoluminescence in the visible spectral range in aqueous solutions.[13] These SiC nanoparticles maintain their uniform dispersity and luminescent properties in aqueous solutions after storage in air for over 7 months.[14] SiC nanowires with an oxidized layer have recently shown markedly enhanced optical emission from organic photosensitizers resulting in efficient energy transfer.[6] SiC nanomaterials are also more biocompatible than quantum dots because SiC does not release heavy metal ions to the surrounding biological environment.[15]
The strong luminescence and nano-scale size make SiC nanomaterials great candidates for labeling and tracking live cells.[4, 16, 17] Botsoa et al. demonstrated that 3C-SiC nanoparticles can enter 3T3-L1 fibroblasts with no toxicity after one week of incubation.[4] The photoluminescence of 3C-SiC nanoparticles enables cell-tracking using fluorescence imaging. SiC nanowires (SiCNWs) can also enter epithelial cells, breast cancer cells, and normal human dermal fibroblasts.[2] These studies highlight the significant potential of SiC nanoparticles (SiCNPs)[4] and SiCNWs[18, 19] including a role in cell tracking to understand the fate of stem cells and to improve the efficacy of stem cell therapy.[20]
However, it is critical to understand the cytotoxicity of nanomaterials, including SiC, before using them for cell tracking. Previous work showed that macrophages, fibroblasts, and osteoblast-like cells have a dose-dependent toxicity response to both alpha- and 3C-SiC with no measurable toxicity below 0.1 mg/mL.[21] Cacchioli et al. proved the cytocompatibility of the SiCNWs to epithelial cells, breast cancer cells, and normal human dermal fibroblasts by analyzing cell proliferation, cell cycle progression, and oxidative stress.[2] However, Mzyk et al. showed that SiC nanoparticles prepared by plasma activated chemical vapor deposition were toxic to mouse fibroblasts.[22]
These controversies have limited the translation and development of SiC nanomaterials for medical applications. Such controversies arise from the complexity of factors that affect the cytotoxicity of nanomaterials. While some nanoparticles like quantum dots are intrinsically toxic because they contain and release heavy metal ions,[23, 24] SiC nanomaterials could potentially have good biocompatibility because they only contain nontoxic elements—silicon, carbon, and oxygen (due to surface oxidation). However, the cytotoxicity of nanomaterials also depends on dosage, morphology, structure, and surface properties[2, 25–27] besides their elemental compositions.[28]
For example, Napierska et al. found that the cytotoxicity of monodisperse silica nanoparticles in human endothelial cells is size-dependent: the smaller silica nanoparticles showed higher cytotoxicity than larger particles.[29] The shape of nanomaterials is another factor that affects their cytotoxicity. Silver nanowires have significant antimicrobial activity, but low cytotoxicity to human cells compared to silver nanoparticles.[30] Crystallinity also affects cytotoxicity. Zhang et al. found that crystalline silica particles induced apoptosis and generated toxic reactive oxygen species (ROS) while amorphous silica nanoparticles are generally biocompatible.[27] The same group also found that the toxicity of amorphous silica nanoparticles depends on the concentration of hydroxyl groups on their surface and the potential to generate ROS.[27]
Although there have been previous studies on the cytotoxicity of SiC-based nanomaterials,[31] there are very few reports with human mesenchymal stem cells (hMSCs). Mesenchymal stem cells (MSCs) have a great capacity for self-renewal while maintaining their multipotency, therefore, MSCs have been widely investigated in regenerative medicine. To understand the toxicity of nanomaterials to hMSCs, it is important to develop nanomaterials for hMSC tracking and understand the complex toxicity of nanomaterials before transitioning to in vivo applications. We present here a careful study on the differences in cytotoxicity between SiCNWs and SiCNPs to hMSCs in vitro. Our study focuses on the cytocompatibility of different shapes of SiC nanomaterials and how that affects the metabolism, viability, adhesion, proliferation, migration, oxidative stress, and differentiation of hMSCs. We also compared the cytotoxicity of SiCNWs to hMSCs and a breast cancer cell line.
2. Experimental methods
2.1. SiC nanomaterials synthesis and characterization
The SiC nanowires (SiCNWs) were synthesized in a polymer pyrolysis chemical vapor deposition (PPCVD) route similar to our previous report.[18] The polymethylsilane (PMS) used in this work was provided by the National University of Defense Technology (Changsha, China).[32] Briefly, 4.0 g PMS was mixed well with 5.0 g activated carbon fine powder (100 mesh, with a BET surface area of 390 m2/g) in a ceramic boat (30 mm × 60 mm) to make a slurry, followed by pushing the slurry into the center of a tubular corundum furnace (Φ 41 mm, 1200 mm in length). A cleaned graphite substrate (30 mm × 50 mm) was placed at downstream of the slurry. The furnace was heated to 1300° C at 10° C/min with ultra-high-purity argon atmosphere at 5mL/min. The samples were maintained under these conditions for 3 h, and then allowed to cool to room temperature naturally. Finally, a large amount of cotton-like nanowires was found covering the graphite substrate. We collected the nanowires and then grinded them in a ball mill for 0.5h. The SiC nanoparticles were purchased from US Research Nanomaterials, Inc (#US2161 and #US2022). Both SiC nanoparticles and nanowires were sterilized by heating to 600° Cor 2 hours before cell labeling. SEM images of SiC nanomaterials were taken using an FEI Quanta FEG 250 SEM at an accelerating voltage of 5 kV (Zeiss, Germany). EDX spectra were performed at an accelerating voltage of 20 KV. TEM images were performed on a JEOL 1200 EX II with a Gatan Orius 600 camera.
2.2. Surface modification of the SiC nanomaterials
First, we added 5 mg fluorescein isothiocyanate (FITC) to 1 mL ethanol and 100 μL (3-aminopropyl) triethoxysilane (APTES) and then rotated overnight in dark at room temperature. Then, 10 mg SiC nanomaterials were dispersed in 5 mL toluene and 1 mL ethanol, then 1 mL of silanized fluorescein mixture was added and stirred for 1 hour at 110° C. The product was then washed with ethanol thrice to remove unbound fluorescein.
2.3. Cell culture and labeling
The hMSCs were purchased from Lonza and were used between passage 2 and 10. The growth media was also purchased from Lonza. The seeding density was approximately 5,000 cells/cm2 unless specified. Cells were passaged when they reached about 90% confluence with TrypLE Express (Life Technologies) with about 5–8 days between each passage. Labeling with SiC nanoparticles or nanowires was performed without any exogenous transfection agents. The SiC nanomaterials were added to media and allowed to incubate for 4 hours. The adherent cells were washed three times with excessive PBS before detachment or other treatment.
2.4. SEM and EDX sample preparation
Cells were cultured in a 6-well plate until 80% confluence and then incubated with 1 ml of 50 μg/mL SiC nanoparticles or nanowires for 4 hours. Cells without any nanomaterials were used as the control. After 4 hours, cells were washed with sterilized PBS thrice, detached, isolated from the media, and then resuspended and kept in 500 μL 0.1% glutaraldehyde in PBS for 30 minutes. We then centrifuged and resuspended the cells in PBS. Drops of cells were added onto sterile plastic slides. The cells were immersed in 0.1% glutaraldehyde again for 60 minutes right after the drops were dried in the air. We then transferred the plastic slides carefully to 0.5% glutaraldehyde and left them in the solution for 1h. After that, the cells were dehydrated through a series of alcohols and hexamethyldisilazane (Sigma Aldrich) then dried with liquid CO2 (Tousimis AutoSamdri 815A, USA) until the critical point was reached. Samples were then sputter coated with an iridium layer using a coating device (Emitech K575X Sputter Coater). SEM images were taken using an FEI Quanta FEG 250 SEM at an accelerating voltage of 5 kV (Zeiss, Germany). EDX spectra and mapping were performed at an accelerating voltage of 20 KV.
2.5. Differentiation experiments
Low passage number (≤ 6)) MSCs were used for differentiation experiments and done at least in duplicate. Cells were plated and incubated as described above. Stained cells were imaged with a bright field microscopy (Keyence BZ-9000, Germany).
For osteogenic inducted differentiation, standard media was replaced with osteogenic media (hMSC Osteogenic BulletKit, Lonza PT-3002) after the cells were labeled with SiC nanomaterials. Standard media was used for non-induced cells. The media for both induced and non-induced cells was changed every 2–3 days. One week after the non-induced unlabeled cells reached 100% confluence, all cells were fixed with 70% ethanol on ice for one hour and then stained with freshly made 2% Alizarin Red in water for 7 minutes followed by washing with water until no excess stain was removable. Dissolving the colored complex in 10% acetic acid and measuring the optical density at 402 nm quantitated the degree of osteogenesis.
For the adipogenic protocol, standard media was replaced with adipogenic induction media (hMSC Adipogenic BulletKit, Lonza PT-3004) after the cells were labeled with SiC nanomaterials. The cells were then incubated in maintenance media and induction media for 2–3 days alternatively. One week after the non-induced unlabeled cells reached 100% confluence, all cells were fixed in 10% formalin for 45 minutes and washed with water then 60% isopropanol. Oil red O (Sigma Aldrich) was used to stain the adipogenic cells. The stain solution was freshly made by adding 18 mL water to 27 mL of 3 mg/mL Oil Red O in isopropanol followed by filtration after 10 minutes. The cells were allowed to stain for 5 minutes and then washed with water.
2.6. Cell metabolism, viability, and oxidative stress
Cell metabolism was studied after labeling hMSCs with SiC nanomaterials using CellTiter 96® AQueous One Solution cell proliferation assay (MTS, Promega). After co-incubation with SiC nanomaterials for 4 hours, 20 μL of the assay reagent was pipetted into the samples in 100 μL of growth media. This was allowed to incubate under standard conditions for 4 hours. We then transferred 80 μL of the sample solutions to a new plate and read the absorbance at 490 nm on a spectrophotometer (SpectraMax M5, Molecular Devices). Cell viability was studied with the calcein AM cell viability assay kit (Biotium). Labeled cells were incubated with 100 μL of the reagent assay (containing 2 μM calcein AM) for approximately 1 hour and then the fluorescence was read at 517 nm with an excitation at 494 nm on a spectrophotometer (SpectraMax M5, Molecular Devices). Quantitative ROS measurement was performed by staining cells with 2’,7’-dichlorofluorescin diacetate (DCFDA, Sigma Aldrich) assay. H2O2-treated cells and nontreated cells were included as positive and negative controls, respectively. The fluorescence intensity was read at 530 nm with an excitation at 485 nm on a spectrophotometer (SpectraMax M5, Molecular Devices).
2.7. Cell adhesion, proliferation, migration, and cytoskeleton staining
For cell adhesion assessment, we plated non-labeled hMSCs and SiC-labeled hMSCs at 5,000 cells/cm2 in 96-well plate. After incubation for 24 hours, we washed the cells with PBS to remove non-adherent cells and then determined the cell count with a resazurin assay.[33] Adhesion was calculated as the ratio of attached cells to plated cells, and all groups were normalized to the adhesion of non-labeled hMSCs. For cell proliferation, the viability was assessed by a resazurin assay 1, 4, and 7 days after seeding. The fluorescence intensities were normalized to day 1.
For the cell migration ability assessment, cells were labeled in a 6-well plate when they reached approximately 60% confluence. Cells were allowed to grow overnight after removing the free nanomaterials. Then, a gap among cells was created by scraping cells away with a pipette tip. The gap was imaged repeatedly with light microscopy over one week. Cells inside and outside the gap on the microscope images were counted with ImageJ. For cytoskeleton staining assay, the SC-labeled and unlabeled cells were fixed with 4% paraformaldehyde, permeated with 0.5% Triton® X-100 (v/v) in PBS, and then stained with 2 drops of ActinGreen™ 488 ReadyProbes™ Reagent (ThermoFisher) and NucBlue™ Live ReadyProbes™ Reagent (ThermoFisher) at room temperature for 10 minutes. The cells were then washed gently with PBS thrice to remove extra fluorophores.
2.8. Surface marker and cytokine secretion analysis
The cluster of differentiation proteins CD73 (clone: REA804), CD90 (clone: REA897), and CD105 (clone: 43A4E1) on hMSCs were evaluated by flow cytometry (FACSCanto 2, BD Biosciences). The following fluorophore-conjugated monoclonal antibodies and corresponding isotypes were used (Miltenyi Biotec): CD73-PE, CD90-FITC, and CD105-APC. For cytokine secretion analysis, cells were incubated in fresh growth media for 48 hours after labeling. Then the cell media was collected, and cytokines in the media were analyzed with a human inflammation MAP® v.1.0 profile (Myriad RBM).
2.9. RNA isolation and RT-PCR analysis
The total amount of cellular RNA was extracted from cells in two T25 flasks when the confluence reached to 90%, using GeneJet RNA Purification Kit (Thermo Scientific, #K0731). RNA was quantified by using a NanoDrop 2000 (Thermo Scientific). One microgram of total RNA was reverse transcribed and quantitative PCR was performed. Samples were normalized to EF1α, and the ratio change to unlabeled cells was calculated.
3. Results and Discussion
3.1. Characterization and Surface Modification of SiC Nanomaterials
We synthesized SiCNWs by polymer pyrolysis chemical vapor deposition.[18] The SiCNWs were grown on clean graphite. To remove the potential impurities from graphite, the products were calcined at 600° C air for 2 hours. The SiCNPs were made by chemical vapor deposition. We also calcined these SiCNPs at 600° C in air for 2 hours.
The SiCNPs are 78.8 ± 18.0 nm (abbreviated as SiCNP80, n = 200) and 603.2 ± 97.6 nm (abbreviated as SiCNP600, n = 179) by TEM, respectively. The width of the SiCNWs were 82.3 ± 12.4 nm (n=199). Their morphologies and elemental analysis data are shown in Figure 1. The Pt and Ir arise from the coatings used for scanning electron microscopy (SEM). Elemental analysis showed a small peak for oxygen in the SiC nanomaterials, which indicates oxidation of the surface of SiC nanomaterials similar to the literature[2]. The oxidized layer can be modified with functional moieties. In this study, all SiCNPs and SiCNWs were modified with FTIC; green fluorescence overlaps with their intrinsic luminescence (Figure 2). This indicates that we can modify the SiC nanomaterials with targeting molecules for specific cell imaging.
Figure 1.
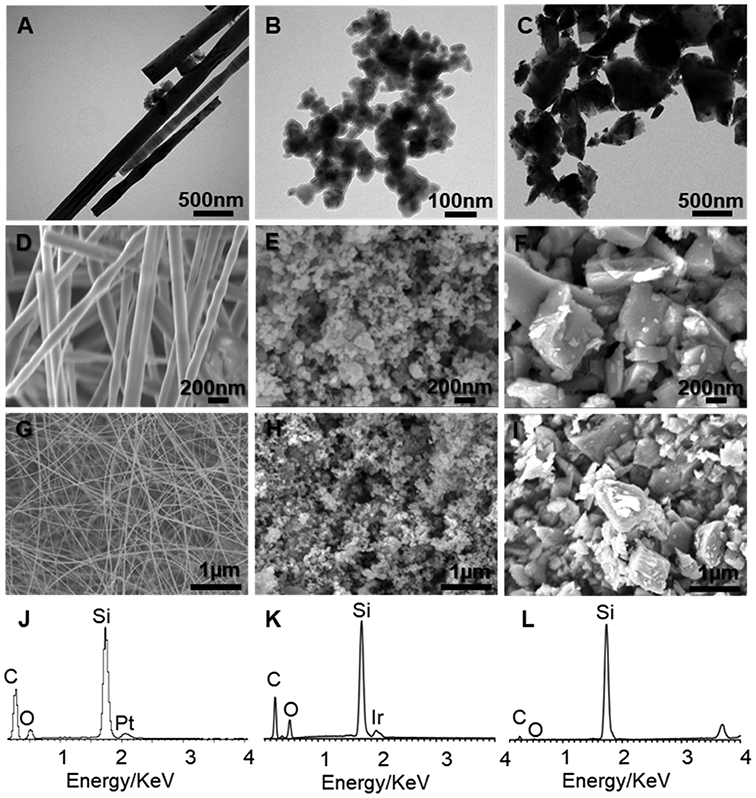
Characterization of SiC nanomaterials. TEM images of (A) SiCNW and SiCNP with an average size of around (B) 80 nm (SiCNP80) and (C) 600 nm (SiCNP600). SEM images of (D, G) SiCNW and (E, H) SiCNP80 and (F, I) SiCNP600. The width of SiCNW is around 80 nm. EDX spectra of (J) SiCNW, (K) SiCNP80, and (L) SiCNP600 show the presence of silicon, carbon, and oxygen. The oxygen in the EDX spectra is due to the oxidation of the SiC nanomaterials surface. The Pt and Ir stem from the coatings for SEM sample preparation.
Figure 2.
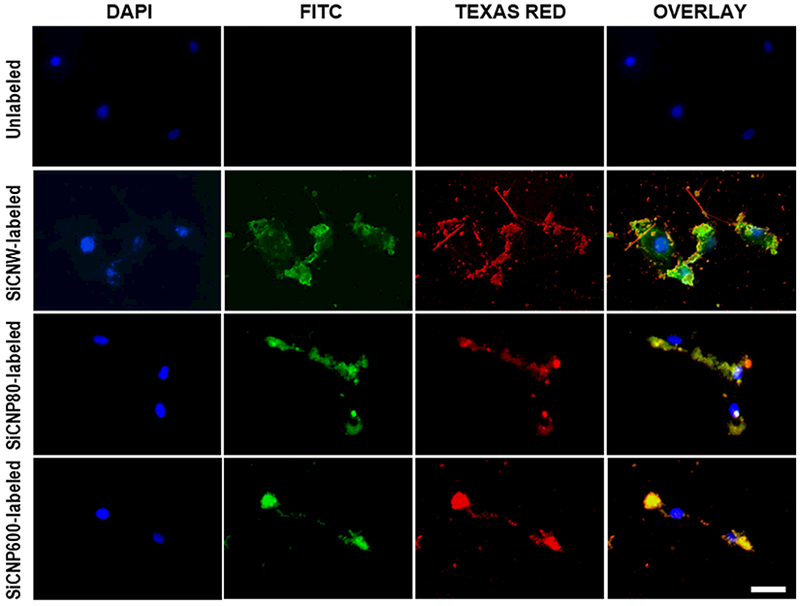
Labeling hMSC with FITC-conjugated SiC nanomaterials. FITC was conjugated onto the oxidized surface of SiC nanomaterials through silane. Fluorescence of FITC (green) overlaps with the photoluminescence of SiC nanomaterials (red) and shows the SiC surface oxidization and modification. Blue fluorescence is from hMSC nucleus stained with NucBlue®. All of the SiC nanomaterials can label hMSCs. The scale bars are 50 μm.
3.2. Labeling hMSCs with SiC Nanomaterials
Next, we used these materials to label stem cells because stem cell labeling with exogenous contrast agents is a common method to track cells and may improve the efficacy of stem cell therapy.[20, 31, 34, 35] We incubated hMSCs with SiCNWs, SiCNP80, and SiCNP600 for four hours. The free nanoparticles were washed away three times with sterile PBS. We also used fluorescein-conjugated SiC to study the distribution of the SiC nanomaterials in the cells. The SiC nanomaterials showed fluorescent signals in both FITC and Texas Red channels. We can use this signal to track the location of hMSCs under a fluorescence microscope (Figure 2).
Next, untreated cells and all three SiC-treated hMSCs were imaged with SEM and analyzed with energy dispersive X-ray spectra (EDX) (Figure 3). Figure 3 A–D show unlabeled hMSCs as well as those labeled with SiCNP80, SiCNP600, and SiCNWs. The area in the red dotted rectangles is enlarged in Figure 3 E–H and provides more details on the cells’ surface morphology. Non-treated hMSCs are covered by microvilli and small vesicles similar to other reported SEM images.[36] A Si peak is seen for the SiCNPs- and SiCNWs-labeled hMSCs but not control hMSCs (Figure 3 I–L). EDX mapping of cells also shows that the SiC-labeled hMSCs had Si all over the cells while control cells had negligible Si signals. (Figure S1). The SiC nanomaterials on the cell surface were removed before sampling, and the Si signal was thus derived from the SiCs that enter the cells at an accelerating voltage of 20 KV. The SiCNWs-labeled hMSCs have a wire-like structure protruding from the membrane (Figure 3H), which could be microvilli for adhesion or tunneling nanotubes.[37] EDX mapping proves that most of these wire-like structures were SiCNWs because they contained silicon as indicated by the green arrows (Figure S1). One wire-like structure was broken into two parts and does not contain silicon (red arrow), and it is very likely to be a microvillus or tunneling nanotube.
Figure 3.
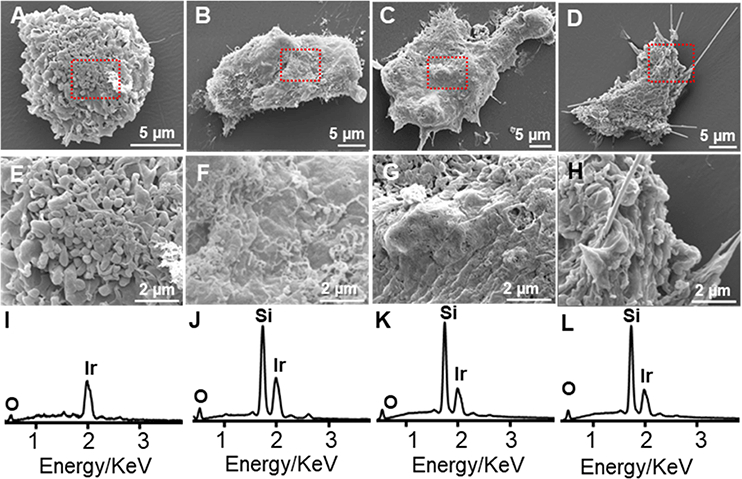
SEM and EDX of hMSCs. SEM images of (A) unlabeled hMSCs, (B) hMSCs labeled with SiCNP80, (C) SiCNP600, and (D) SiCNWs. Images (E-H) are enlarged areas from the rectangles in (A-D) respectively. EDX spectra of (I) unlabeled hMSCs, (J) hMSCs labeled with SiCNP80, (K) SiCNP600, and (L) SiCNWs. Cells labeled with SiC nanomaterials show a Si peak while the unlabeled hMSCs have no Si peak. Iridium peaks are due to the coatings for SEM sample preparation.
3.3. SiC Nanomaterials Cytocompatibility
Knowing that these SiC nanomaterials can enter hMSCs and that the stem cell location can be tracked under fluorescence microscopy, we next investigated the impact of SiC nanomaterials on the hMSCs’ metabolism, viability, oxidative stress, adhesion and proliferation, migration, and pluripotency.
We found that SiCNW-labeled hMSCs have similar effects on the metabolism, viability, and oxidative stress as SiCNP-labeled and unlabeled hMSCs. The metabolic activity was studied with an MTS assay [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium]. SiC nanomaterials up to 400 μg/mL did not cause a significant decrease in the cellular metabolism of hMSCs versus unlabeled cells (Figure 4A). The cell viability was also studied with a calcein AM assay. The results showed no decrease in cell viability for all three SiC nanomaterials from 0 to 400 μg/mL after hMSCs were incubated with the nanomaterials for 4 hours (Figure 4B).
Figure 4.
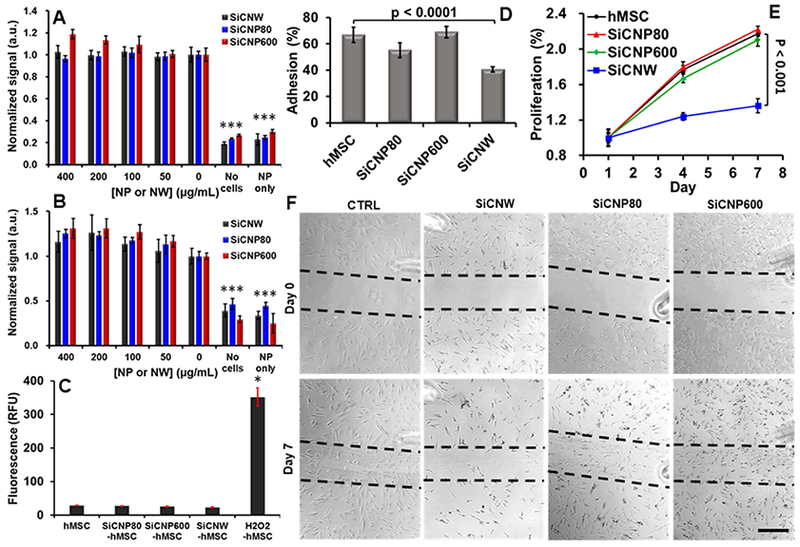
Influence of SiC on hMSCs’ viability, oxidative stress, adhesion, proliferation, and migration ability. (A) The MTS assay showed no significant decrease in cellular metabolism at any combinations of materials and concentrations (up to 400 μg/mL) (p > 0.05). (B) A calcein AM cell viability assay shows no significant decrease in viability at any concentration from 0–400 ug/mL (p > 0.05). (C) SiC labeling did not generate intracellular reactive oxygen species. (D) SiCNWs decreased the adhesion ability of hMSCs by more than 50%. (E) SiCNWs greatly decreased proliferation of hMSCs. (F) A migration assay shows hMSCs can migrate after labeling with SiCNWs and SiCNPs. However, there are fewer cells in the scratched area on the seventh day in hMSCs labeled with SiCNWs compared to the hMSCs labeled with SiC nanoparticles. Scale bar is 500 μm. Dashed black lines were added as guides. *** represents p < 0.005 compared to hMSC groups, unpaired Student’s t-test. Error bars represent standard deviations at least four replicates.
The oxidative stress was measured with a 2’,7’-dichlorofluorescin diacetate ROS assay. Cells exposed to 100 μg/mL of SiC nanomaterials for 4 hours and produced low levels of ROS regardless of the morphology of SiC nanomaterials (Figure 4C). All three assays discussed above were performed within 24 hours after the 4 hours of labeling with SiC nanomaterials. Therefore, we may conclude that SiCNWs have a similar short-term effect on hMSCs as both SiCNP80 and SiCNP600. The SiC nanomaterials have no acute cytotoxicity to hMSCs within 0 to 400 μg/mL. However, that was not the case for long-term viability studies.
For adhesion and proliferation assays (n=8 replicated wells), hMSCs were exposed to SiC nanomaterials and then detached and re-plated, similar to procedures as stem cell therapy. Approximate 67%, 55%, 69%, and 41% of unlabeled, SiCNP80-, SiCNP600-, and SiCNW-labeled hMSCs adhered to the flask 24 hours after the seeding (Figure 4D). This implies that the SiCNW decreased adhesion by 39% (p<0.0001), the SiCNP80 decreased the adhesion by 18%, and the SiCNP600 increased adhesion by 3% (p=0.45) relative to unlabeled cells. Long-term proliferation studies were performed 4 and 7 days after re-plating. The proliferation of SiCNW-labeled hMSCs on days 4 and 7 relative to day 1 were 124% and 136% respectively, which were 30% and 37% lower than the unlabeled hMSCs. The SiCNP600-labeled cells had a proliferation rate that was only 5.7% and 3.4% lower than day unlabeled. The SiCNP80-labeled cells had a slightly higher proliferation rate on days 4 and 7 of 1.7% and 1.0%, respectively. The differences in proliferation between these groups are statistically insignificant except for the SiCNW group (p<0.001). (Figure 4E).
We also performed a migration assay to evaluate the wound healing ability of SiC-labeled hMSCs. Right after treatment with SiC nanomaterials, hMSCs were washed with PBS to remove free nanoparticles. A gap in the cells was created by removing part of cells with a pipet tip. The initial cell density for unlabeled and SiC-labeled in the unscratched area was approximately 3,000 cells/cm2. We monitored the progress daily with microscopy. We counted the cells in scratched area and non-scratched areas in 3 regions of interests.
Microscope images showed that almost no hMSCs, whether labeled or unlabeled, migrated within 24 hours after creation of the gap (Figure S2). On day 3, cells appeared in the gap randomly instead of gradually filling up the gap from the scratched edges, which indicated the migration of cells (Figure S2). In addition, on day 3, the cell density in the gap of unlabeled, SiCNP80-, SiCNP600-, and SiCNW-labeled hMSCs were 49.6%±6.6%, 86.8%±44.2%, 124.2%±41.4%, and 75.5%±29.5% of their initial cell density in the unscratched area, respectively. The standard deviations are calculated based on three different regions of interests. This result indicated that the SiCNWs and SiCNPs had a positive effect on the cells migration ability. To further confirm the effect of SiC nanomaterials on hMSCs, we performed a cytoskeleton staining assay (Figure S3). The result showed that none of the SiC nanomaterials dysregulated the α-actin of hMSCs.
However, SiCNW-labeled hMSCs had the worst wound healing efficacy on day 7, when the cell density in the scratched area of SiCNW-labeled hMSCs was only 29.7%±1.0% of the initial cell density, which was very low when compared to 92.0%±33.4% for the unlabeled, 97.6%±13.0% for the SiCNP80-labeled, and 102.3%±7.5% for the SiCNP600-labeled hMSCs (Figure 4F). This result also indicated the negative effect of SiCNWs on the hMSCs proliferation.
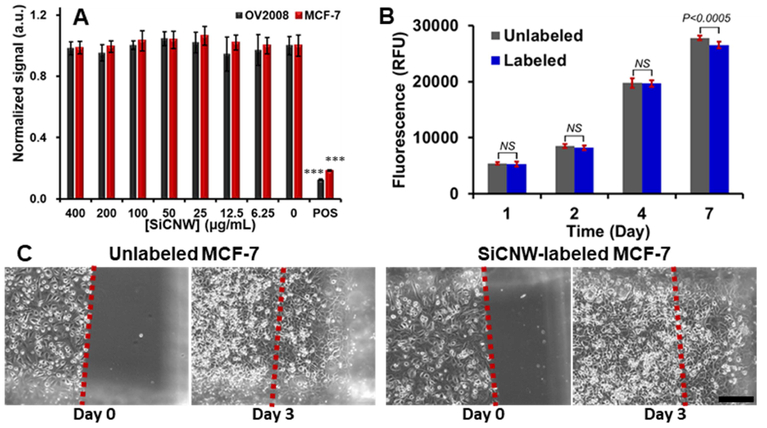
These data showed that SiCNWs were more toxic to hMSCs than SiCNPs at identical labeling concentrations. To confirm that the toxic effect is not because of impurities or endotoxins in the SiCNWs, we performed a control experiment by checking the impact of SiCNWs on cancer cell lines. Similar to prior reports,[2] SiCNWs do not affect the viability, proliferation, and migration ability of cancer cells. A resazurin assay showed the cell viabilities of SiCNW-labeled OV2008 and MCF-7 were over 95% even when the incubation concentration of SiCNWs was up to 0.4 mg/mL (Figure 5A). The proliferation rate of SiCNW-labeled MCF-7 was approximately 97.7%, 96.3%, 99.6%, and 95.6% compared to unlabeled MCF-7 (Figure 5B). Unlabeled and SiCNWs-labeled MCF-7 were plated at a density around 100,000 cells/cm2 on the first day for the migration assay. Three days later, the cell density in the scratched area for unlabeled and SiCNWs-labeled MCF-7 increased from 0 to 286,207 and 289,796 cells/cm2 respectively (Figure 5C). Additionally, the SiCNWs could label breast cancer cells (MCF-7) and track these cells under fluorescence microscopy (Figure S4).
Figure 5.
Influence of SiC on Cancer cells. (A) Viability assay shows that the SiCNWs do not decrease cell viability in both OV2008 and MCF-7 at concentrations up to 400 μg/mL. (B) Proliferation study shows that SiCNW has a negligible effect on the proliferation of MCF-7. (C) Migration assay shows the MCF-7 can migrate after labeled with SiCNW. Scale bar represents 200 μm.
3.4. Effects of SiC Nanomaterials on hMSC Functionality
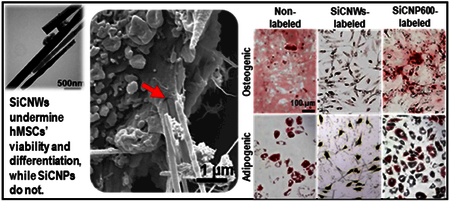
In regenerative medicine, stem cells are often expected to differentiate and replace cells in damaged tissues,[38] which makes it critical that nanomaterial labels do not interfere with hMSC multipotency. Thus, we studied the impact that these SiC nanomaterials had on hMSC multipotency—we found that the effect is morphology-dependent. The SiCNP80 and SiCNP600 did not affect the potency of hMSC, while SiCNWs prevent hMSCs from differentiating into osteogenic and adipogenic cell types (Figure 6, Figure S5A). The hMSCs were first exposed to SiCNPs or SiCNWs at 100 μg/mL for 4 hours and then treated with normal media, osteogenic induction media, or adipogenic induction media. Calcium deposits were detected by Alizarin Red S staining and fatty lipid vacuoles were stained red by Oil Red O staining three weeks after incubation with induction media. Microscopy images showed the presence of SiC nanomaterials in hMSCs even after three weeks (Figure 6).
Figure 6.
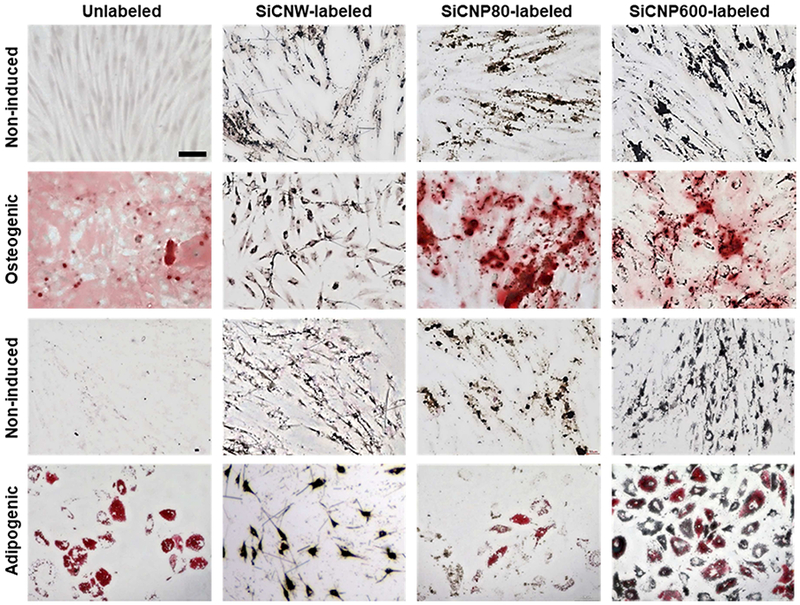
hMSCs’ pluripotency is retained after labeling with SiCNPs but not SiCNWs. Photomicrographs were obtained three weeks after labeling with SiC nanomaterials, and these images clearly showed the presence of SiC nanomaterials in hMSCs even after three weeks. The first two rows were labeled with Alizarin Red S to detect osteogenic cells; the bottom two rows were labeled with Oil Red O to detect adiopenic cells. Non-induced cells did not have adipogenesis or osteogenesis. Unlabeled cells could be induced to either osteocytes or adipocytes. Cells labeled with SiC nanoparticles could also differentiate, but the SiCNWs-labeled cells did not differentiate. All images have the same magnification and the scale bar in the upper-left image presents 100 μm.
The SiCNP600- and SiCNP80-labeled cells differentiated into both osteogenic and adipogenic lineages similar to control (unlabeled) hMSCs (Figure 6). However, cells labeled with SiCNWs failed to differentiate into osteocytes or adipocytes even after treatment with differentiation induction media (Figure 6). Noticeably, 75% more calcium compounds were seen in the SiCNP80-labeled than non-labeled hMSC. Previous work has shown that silicon-based structures [39] can modulate the osteogenic differentiation of cells and that the surface roughness of these materials impacts the differentiation [40] (Figure S5C). However, no obvious calcification process was found in SiCNPs-labeled hMSCs without the osteogenic induction media. The SiCNW-labeled hMSCs do not form vacuoles (red arrows) when treated with adipogenic induction media nor do they form a hollow areas (black arrows) [41] when treated with osteogenic induction media (Figure S5A), which indicates that only SiCNW prohibited the adipogenic and osteogenic differentiation of hMSCs. We also quantified the degree of osteogenesis by dissolving the color compounds and measuring optical density. The SiCNWs-labeled hMSCs could not be differentiated into osteogenic lineages (Figure S5B&C).
3.5. Effects of SiC Nanomaterials on hMSC phenotypes
To understand the mechanism by which SiCNWs dysregulate hMSCs, we studied the phenotypes, cytokine secretion, and gene expression levels of SiCNWs-labeled hMSCs versus unlabeled and SiCNPs-labeled hMSCs. Surface marker proteins (or cluster of differentiation) of hMSCs CD73, CD90, and CD105—were analyzed with flow cytometry (Figure 7A). Approximately 70.7%±7.8%, 52.7%±2.1%, and 65.2%±5.2% of SiCNWs-labeled hMSCs remained positive for CD73, CD90, and CD105 respectively, while over 96% of SiCNP80- and SiCNP600-labeled hMSCs retained these three markers. It is well-accepted that hMSCs display characteristic markers CD73, CD90, and CD105.[42–44] CD73 is a membrane-bound enzyme that catalyzes the conversion of adenosine monophosphate to bioactive adenosine at neutral pH.[45] CD90 is an activation-associated cell adhesion molecule.[46] The CD105 antigen, also known as endoglin, serves as a receptor for the growth and differentiation factors.[47] The SiCNWs-labeled hMSCs have a similar morphology to neurons under microscope (Figure S6). In conclusion, the SiCNWs decreased immunophenotype of hMSCs as well as stemness, while SiCNP80 and SiCNP600 have less impact on the hMSCs.
Figure 7.
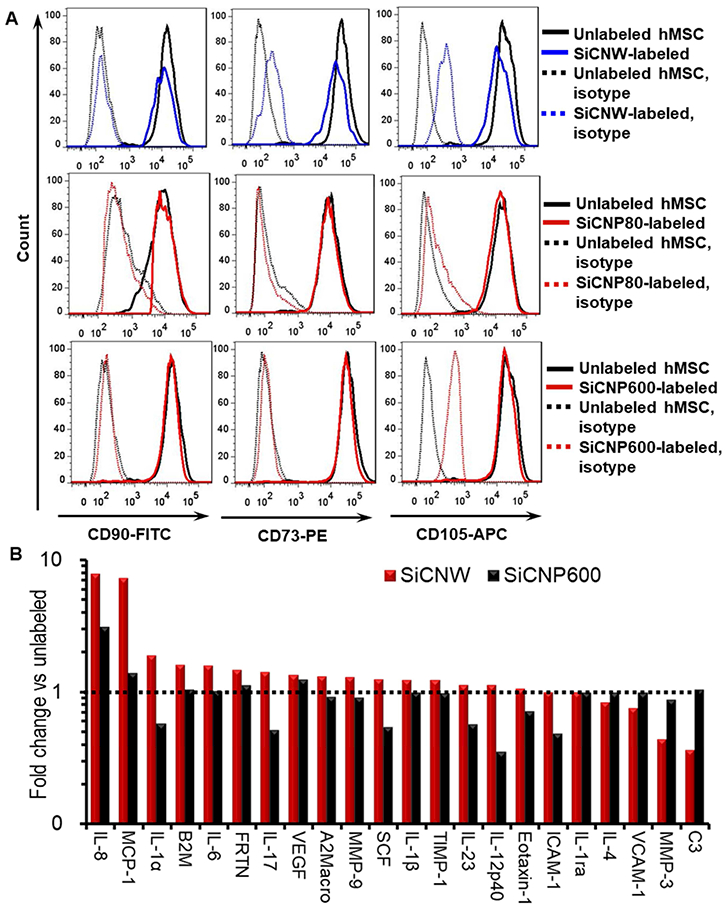
Molecular changes of hMSC labeled with SiC nanomaterials. (A) Phenotypes on the surface of SiC-labeled hMSCs. Fewer SiCNWs-labeled hMSCs remained positive for the CD73, CD90, and CD105 when compared to SiCNP80- and SiCNP600-labeled hMSCs. Unlabeled cells were used as a control and treated with the same antibodies. IgG conjugated with FITC, PE, and APC were used as isotypes. (B) Cytokines secreted by hMSC after labeling with different SiC nanomaterials. SiCNWs-labeled hMSCs secreted 7.2 and 7.8 times more of MCP-1 and IL-8 than unlabeled hMSCs, and they secreted MMP-3 and C3 less than half of unlabeled cells. MCP-1 and IL-8 are pro-inflammatory cytokines. C3 is important to activate the complement system and MMPs play an important role in promoting the differentiation, angiogenesis, proliferation, and migration of hMSCs. SiCNWs have more impact on MCP-1, IL-8, MMP-3, and C3 secretion than SiCNPs600, which may cause the differences in cytotoxicity to hMSCs between the SiCNW and SiCNPs.
3.6. Effects of SiC Nanomaterials on hMSC Gene Expression
We next investigated the gene expression levels in SiCNWs- and SiCNP600-labeled and unlabeled hMSCs via a real-time polymerase chain reaction (RT-PCR). The genes of interest included: 1) senescence-associated genes: cyclin-dependent kinase inhibitors CDKN1A (P21), CDKN2A (p16) and CDKN2B (p15), P53; 2) proliferation-related genes: cyclin-dependent kinase 4 (CDK4), MKi67, and CDK1; 3) adipose-specific genes: lipoprotein lipase (LPL), peroxisome proliferator-activator receptor-gamma2 (PPARG); 4) bone-specific genes: alkaline phosphatase (ALP), bone sialoprotein (BSP); and 5) cartilage-specific genes: aggrecan (AGN), and collagen type II alpha 1 (COL2A).[48–50] The housekeeping gene elongation factor-1α (EF1α) monitored RNA loading. The fold changes of gene expression in SiCNWs-labeled hMSCs versus unlabeled cells for all studied genes are within 200%, and the changes are similar to SiCNP600-labeled hMSCs (Figure S7). Therefore, the cytotoxicity of SiCNWs to hMSCs is unlikely due to the changes of gene expression in hMSCs.
3.7. Effects of SiC Nanomaterials on hMSC Cytokines Secretion
We evaluated cytokine secretion from unlabeled and labeled hMSCs because the cytokines can explain nanoparticle immunotoxicity.[51] Secretome analysis of SiCNW- and SiCNP600-labeled and unlabeled hMSCs showed that 23 of the 46 analyzed proteins were detectable in cell culture media. Cytokine concentrations from the SiCNWs-labeled cells were within one-fold (50%−200%) of unlabeled hMSC except for complement protein 3 (C3), matrix metalloproteinase-3 (MMP-3), monocyte chemotactic protein 1 (MCP-1), and interleukin-8 (IL-8) (Figure 7B). SiCNWs-labeled hMSCs secreted 7.2 and 7.8 times more of MCP-1 and IL-8 than unlabeled hMSCs. In comparison, the SiCNP600-labeled hMSCs secreted 1.4 times more MCP-1 and 3.1 times more IL-8 compared to unlabeled hMSCs. Endocytosis of SiCNWs disrupts the cell wall, which likely leads to the inflammatory reactions and upregulated IL-8 and MCP-1 in hMSCs[52] (Figure S8). Both MCP-1 and IL-8 are pro-inflammatory cytokines. MCP-1 is a key chemokine that regulates migration and infiltration of monocytes/macrophages,[53] and IL-8 is a chemokine that plays a key role in the activation of neutrophils and their recruitment to the site of inflammation.[51] Gerszten et. al. found that both MCP-a and IL-8 can trigger the adhesion of monocytes onto monolayers.[54] We have shown that SiCNW-labeled hMSCs have low levels of adhesion and migration. As a result, the increased secretion of IL-8 and MCP-1 could be an attempt to to compensate for this.[51] These increased IL-8 and MCP-1 levels might prevent hMSCs from differentiating because IL-8 and MCP-1 could inhibit adipocyte differentiation.[55].”
SiCNWs-labeled hMSCs secreted MMP-3 and C3 levels at less than half that of unlabeled hMSCs (Figure 7B). C3 plays a central role in the activation of the complement system, and C3 deficiency causes susceptibility to bacterial infection.[56, 57] MMPs play an important role in promoting the differentiation, angiogenesis, proliferation, and migration of hMSCs.[58] Specifically, MMP-3 is responsible for remodeling the extracellular matrix, which is necessary for wound repair, organismal growth and development, and mediation of immune response.[59] SiCNWs can be internalized by cells through direct penetration,[2] and the SEM images of SiCNWs-labeled hMSCs (Figure 3D&H, Figure S1) also show SiCNWs protruding from the hMSCs’ membranes. Moreover, clusters of SiCNWs can penetrate hMSCs membrane and puncture the cell membrane, which may lead to undesired exchange between hMSCs and the environment (Figure S8). Thus, a deficiency in MMP-3 could hinder wound repair mechanisms. The SiCNP600-labeled hMSCs had MMP-3 and C3 levels that were 88% and 105% that of unlabeled hMSCs, respectively.
4. Conclusion
Here, we report the effects of two different SiC nanostructures on hMSCs. We also show that SiCNWs are biocompatible with MCF-7 and OV2008 immortalized cancer cell lines. Additionally, the SiCNWs can be surface modified with potential utility in molecular imaging or drug delivery.[60, 61] Also, we provided a comprehensive analysis on the impacts of SiC nanomaterials to hMSCs in terms of metabolic activity, viability, oxidative stress, adhesion, proliferation, migration, pluripotency, phenotype, gene expression levels, and cytokine secretion. Our results indicate that the cytotoxicity of SiC nanomaterials to hMSCs is shape-dependent SiCNWs is toxic to hMSCs but SiCNPs (both 80 nm and 600 nm) show no toxicity at the same labeling concentration. Therefore, SiCNPs have great potential as optical contrast agents for hMSCs tracking—they have a stable and strong luminescence[11, 12] but also good cytocompatibility to hMSCs. However, the SiCNWs were shown to be toxic to hMSCs 24 hours after labeling. The SiCNWs adversely affect hMSCs’ adhesion, proliferation, migration ability, multipotency, phenotypes, and cytokine secretion. Importantly, SiCNWs should not be used with hMSCs because of the negative impact they have on the hMSCs’ ability to differentiate into the osteocytic and adipocytic lineages.[62, 63] The toxic effect of SiCNWs to hMSCs may be due to the excessive production of pro-inflammatory cytokines MCP-1 and IL-8 and deficiency in MMP-3 and C3.
Supplementary Material
Acknowledgment
F.C. acknowledges Dr. C. Liu for his help in the RT-PCR.
Funding Sources
We acknowledge funding from the NIH including DP2 HL137187 and R00 HL 117048.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest
The authors declare no competing financial interest.
Supporting Information Available: Figure S1—EDX mapping of labeled hMSC, Figure S2—Short-term migration of non-labeled and SiC-labeled hMSCs, Figure S3—Effect of SiC nanomaterials on hMSCs’ cytoskeleton, Figure S4—Labeling MCF-7 with SiC nanomaterials. Figure S5—Quantification of osteogenesis degree, Figure S6—Bright field image of SiCNWs-labeled hMSC, Figure S7—Gene expression of hMSCs labeled with SiC nanomaterials, Figure S8—Magnified SEM image of SiCNWs-labeled hMSCs.
Data availability
The raw/processed data required to reproduce these findings cannot be shared at this time due to technical or time limitations.
REFERENCES
- [1].Yakimova R, Petoral RM, Yazdi GR, Vahlberg C, Spetz AL, Uvdal K. Surface functionalization and biomedical applications based on SiC. J Phys D-Appl Phys. 2007;40:6435–42. [Google Scholar]
- [2].Cacchioli A, Ravanetti F, Alinovi R, Pinelli S, Rossi F, Negri M, et al. Cytocompatibility and Cellular Internalization Mechanisms of SiC/SiO2 Nanowires. Nano Letters. 2014;14:4368–75. [DOI] [PubMed] [Google Scholar]
- [3].Saddow SE, Agarwal A. Advances in silicon carbide processing and applications: Artech House; 2004.
- [4].Botsoa J, Lysenko V, Géloën A, Marty O, Bluet JM, Guillot G. Application of 3C-SiC quantum dots for living cell imaging. Applied Physics Letters. 2008;92:173902. [Google Scholar]
- [5].Seong HK, Choi HJ, Lee SK, Lee JI, Choi DJ. Optical and electrical transport properties in silicon carbide nanowires. Applied Physics Letters. 2004;85:1256–8. [Google Scholar]
- [6].Tatti R, Timpel M, Nardi MV, Fabbri F, Rossi R, Pasquardini L, et al. Functionalization of SiC/SiOx nanowires with a porphyrin derivative: a hybrid nanosystem for X-ray induced singlet oxygen generation. Molecular Systems Design & Engineering. 2017;2:165–72. [Google Scholar]
- [7].Rossi F, Lagonegro P, Negri M, Fabbri F, Salviati G, Alinovi R, et al. Silicon Carbide-Based Nanowires for Biomedical Applications. 2016:311–42. [Google Scholar]
- [8].Bano E, Fradetal L, Ollivier M, Choi J-H, Stambouli V. SiC Nanowire-Based Transistors for Electrical DNA Detection. 2016:261–310. [Google Scholar]
- [9].Knaack GL, Charkhkar H, Cogan SF, Pancrazio JJ. Amorphous Silicon Carbide for Neural Interface Applications. 2016:249–60. [Google Scholar]
- [10].Iannotta S, Romeo A, D’Angelo P, Tarabella G. SiC Biosensing and Electrochemical Sensing: State of the Art and Perspectives. 2016:143–77. [Google Scholar]
- [11].Zhang LG, Yang WY, Jin H, Zheng ZH, Xie ZP, Miao HZ, et al. Ultraviolet photoluminescence from 3C-SiC nanorods. Applied Physics Letters. 2006;89. [Google Scholar]
- [12].Fan JY, Wu XL, Chu PK. Low-dimensional SiC nanostructures: Fabrication, luminescence, and electrical properties. Progress in Materials Science. 2006;51:983–1031. [Google Scholar]
- [13].Wu XL, Fan JY, Qiu T, Yang X, Siu GG, Chu PK. Experimental evidence for the quantum confinement effect in 3C-SiC nanocrystallites. Phys Rev Lett. 2005;94:026102. [DOI] [PubMed] [Google Scholar]
- [14].Fan J, Wu X, Chu PK. Low-dimensional SiC nanostructures: fabrication, luminescence, and electrical properties. Progress in Materials Science. 2006;51:983–1031. [Google Scholar]
- [15].Seleverstov O, Zabirnyk O, Zscharnack M, Bulavina L, Nowicki M, Heinrich J-M, et al. Quantum Dots for Human Mesenchymal Stem Cells Labeling. A Size-Dependent Autophagy Activation. Nano Letters. 2006;6:2826–32. [DOI] [PubMed] [Google Scholar]
- [16].McNeil SE. Nanotechnology for the biologist. Journal of Leukocyte Biology. 2005;78:585–94. [DOI] [PubMed] [Google Scholar]
- [17].Jokerst JV, Gambhir SS. Molecular imaging with theranostic nanoparticles. Accounts Of Chemical Research. 2011;44:1050–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Li GY, Li XD, Chen ZD, Wang J, Wang H, Cho RC. Large Areas of Centimeters-Long SiC Nanowires Synthesized by Pyrolysis of a Polymer Precursor by a CVD Route. J Phys Chem C. 2009;113:17655–60. [Google Scholar]
- [19].Li GY, Ma J, Peng G, Chen W, Chu ZY, Li YH, et al. Room-Temperature Humidity-Sensing Performance of SiC Nanopaper. ACS Appl Mater Interfaces. 2014;6:22673–9. [DOI] [PubMed] [Google Scholar]
- [20].Wang J, Jokerst JV. Stem Cell Imaging: Tools to Improve Cell Delivery and Viability. Stem Cells Int. 2016;2016:9240652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Allen M, Butter R, Chandra L, Lettington A, Rushton N. Toxicity of particulate silicon carbide for macrophages, fibroblasts and osteoblast-like cells in vitro. Bio-medical materials and engineering. 1995;5:151–9. [PubMed] [Google Scholar]
- [22].Mzyk A, Major R, Lackner JM, Bruckert F, Major B. Cytotoxicity control of SiC nanoparticles introduced into polyelectrolyte multilayer films. RSC Adv. 2014;4:31948–54. [Google Scholar]
- [23].Derfus AM, Chan WCW, Bhatia SN. Probing the Cytotoxicity of Semiconductor Quantum Dots. Nano Letters. 2004;4:11–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Bilberg K, Hovgaard MB, Besenbacher F, Baatrup E. In Vivo Toxicity of Silver Nanoparticles and Silver Ions in Zebrafish (Danio rerio). Journal of toxicology. 2012;2012:293784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Rimal B, Greenberg AK, Rom WN. Basic pathogenetic mechanisms in silicosis: current understanding. Curr Opin Pulm Med. 2005;11:169–73. [DOI] [PubMed] [Google Scholar]
- [26].Ekkapongpisit M, Giovia A, Follo C, Caputo G, Isidoro C. Biocompatibility, endocytosis, and intracellular trafficking of mesoporous silica and polystyrene nanoparticles in ovarian cancer cells: effects of size and surface charge groups. International journal of nanomedicine. 2012;7:4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zhang HY, Dunphy DR, Jiang XM, Meng H, Sun BB, Tarn D, et al. Processing Pathway Dependence of Amorphous Silica Nanoparticle Toxicity: Colloidal vs Pyrolytic. Journal of the American Chemical Society. 2012;134:15790–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Bhirde A, Xie J, Swierczewska M, Chen X. Nanoparticles for cell labeling. Nanoscale. 2011;3:142–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Napierska D, Thomassen LCJ, Rabolli V, Liston D, Gonzalez L, Kirsch-Volders M, et al. Size-Dependent Cytotoxicity of Monodisperse Silica Nanoparticles in Human Endothelial Cells. Small. 2009;5:846–53. [DOI] [PubMed] [Google Scholar]
- [30].El-Gamel NEA. Silver(i) complexes as precursors to produce silver nanowires: structure characterization, antimicrobial activity and cell viability. Dalton Transactions. 2013;42:9884–92. [DOI] [PubMed] [Google Scholar]
- [31].Thakor AS, Jokerst JV, Ghanouni P, Campbell JL, Mittra E, Gambhir SS. Clinically Approved Nanoparticle Imaging Agents. J Nucl Med. 2016;57:1833–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Peng P, Lu Y, Liu XY, Feng CX. Study of preceramic polymers. Microchemical Journal. 1996;53:273–81. [Google Scholar]
- [33].Cheng K, Li T-S, Malliaras K, Davis DR, Zhang Y, Marbán E. Magnetic Targeting Enhances Engraftment and Functional Benefit of Iron-Labeled Cardiosphere-Derived Cells in Myocardial Infarction. Circulation Research. 2010;106:1570–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Jokerst JV, Khademi C, Gambhir SS. Intracellular aggregation of multimodal silica nanoparticles for ultrasound-guided stem cell implantation. Sci Transl Med. 2013;5:177ra35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Jokerst JV, Thangaraj M, Kempen PJ, Sinclair R, Gambhir SS. Photoacoustic Imaging of Mesenchymal Stem Cells in Living Mice via Silica-Coated Gold Nanorods. ACS Nano. 2012;6:5920–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Katsen-Globa A, Meiser I, Petrenko YA, Ivanov RV, Lozinsky VI, Zimmermann H, et al. Towards ready-to-use 3-D scaffolds for regenerative medicine: adhesion-based cryopreservation of human mesenchymal stem cells attached and spread within alginate-gelatin cryogel scaffolds. Journal of materials science Materials in medicine. 2014;25:857–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Tylek T, Schlegelmilch K, Ewald A, Rudert M, Jakob F, Groll J. Cell communication modes and bidirectional mitochondrial exchange in direct and indirect macrophage/hMSC co-culture models. BioNanoMaterials 2017. [Google Scholar]
- [38].Kim T, Lemaster JE, Chen F, Li J, Jokerst JV. Photoacoustic Imaging of Human Mesenchymal Stem Cells Labeled with Prussian Blue-Poly(l-lysine) Nanocomplexes. ACS Nano. 2017;11:9022–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Amaral M, Costa MA, Lopes MA, Silva RF, Santos JD, Fernandes MH. Si3N4-bioglass composites stimulate the proliferation of MG63 osteoblast-like cells and support the osteogenic differentiation of human bone marrow cells. Biomaterials. 2002;23:4897–906. [DOI] [PubMed] [Google Scholar]
- [40].Peng-Yuan W,CL R, Helmut T, Andrew J, Wei-Bor T, VN H. Screening Mesenchymal Stem Cell Attachment and Differentiation on Porous Silicon Gradients. Advanced Functional Materials. 2012;22:3414–23. [Google Scholar]
- [41].Matsuoka F, Takeuchi I, Agata H, Kagami H, Shiono H, Kiyota Y, et al. Morphology-Based Prediction of Osteogenic Differentiation Potential of Human Mesenchymal Stem Cells. PLoS One. 2013;8:e55082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Gong Z, Niklason LE. Small-diameter human vessel wall engineered from bone marrowderived mesenchymal stem cells (hMSCs). The FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2008;22:1635–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Ramos LT, Sánchez-Abarca LI, Muntión S, Preciado S, Puig N, López-Ruano G, et al. MSC surface markers (CD44, CD73, and CD90) can identify human MSC-derived extracellular vesicles by conventional flow cytometry. Cell Communication and Signaling : CCS. 2016;14:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini F, Krause D, et al. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315–7. [DOI] [PubMed] [Google Scholar]
- [45].Regateiro FS, Cobbold SP, Waldmann H. CD73 and adenosine generation in the creation of regulatory microenvironments. Clinical and Experimental Immunology. 2013;171:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Wetzel A, Chavakis T, Preissner KT, Sticherling M, Haustein U-F, Anderegg U, et al. Human Thy-1 (CD90) on Activated Endothelial Cells Is a Counterreceptor for the Leukocyte Integrin Mac-1 (CD11b/CD18). The Journal of Immunology. 2004;172:3850–9. [DOI] [PubMed] [Google Scholar]
- [47].Pierelli L, Bonanno G, Rutella S, Marone M, Scambia G, Leone G. CD105 (endoglin) expression on hematopoietic stem/progenitor cells. Leukemia & lymphoma. 2001;42:1195–206. [DOI] [PubMed] [Google Scholar]
- [48].Li J, Pei M. Cell senescence: a challenge in cartilage engineering and regeneration. Tissue Engineering Part B: Reviews. 2012;18:270–87. [DOI] [PubMed] [Google Scholar]
- [49].Heymer A, Haddad D, Weber M, Gbureck U, Jakob PM, Eulert J, et al. Iron oxide labelling of human mesenchymal stem cells in collagen hydrogels for articular cartilage repair. Biomaterials. 2008;29:1473–83. [DOI] [PubMed] [Google Scholar]
- [50].Batsali AK, Pontikoglou C, Koutroulakis D, Pavlaki KI, Damianaki A, Mavroudi I, et al. Differential expression of cell cycle and WNT pathway-related genes accounts for differences in the growth and differentiation potential of Wharton’s jelly and bone marrow-derived mesenchymal stem cells. Stem Cell Research & Therapy. 2017;8:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Elsabahy M, Wooley KL. Cytokines as biomarkers of nanoparticle immunotoxicity. Chemical Society Reviews. 2013;42:5552–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Padmanabhan J, Kyriakides TR. Nanomaterials, Inflammation and Tissue Engineering. Wiley interdisciplinary reviews Nanomedicine and nanobiotechnology. 2015;7:355–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Deshmane SL, Kremlev S, Amini S, Sawaya BE. Monocyte Chemoattractant Protein-1 (MCP-1): An Overview. Journal of Interferon & Cytokine Research. 2009;29:313–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Gerszten RE, Garcia-Zepeda EA, Lim Y-C, Yoshida M, Ding HA, Gimbrone MA Jr, et al. MCP-1 and IL-8 trigger firm adhesion of monocytes to vascular endothelium under flow conditions. Nature. 1999;398:718. [DOI] [PubMed] [Google Scholar]
- [55].Gerhardt CC, Romero IA, Cancello R, Camoin L, Strosberg AD. Chemokines control fat accumulation and leptin secretion by cultured human adipocytes. Molecular and Cellular Endocrinology. 2001;175:81–92. [DOI] [PubMed] [Google Scholar]
- [56].Sahu A, Lambris JD. Structure and biology of complement protein C3, a connecting link between innate and acquired immunity. Immunological Reviews. 2001;180:35–48. [DOI] [PubMed] [Google Scholar]
- [57].Matsuyama W, Nakagawa M, Takashima H, Muranaga F, Sano Y, Osame M. Molecular Analysis of Hereditary Deficiency of the Third Component of Complement (C3) in Two Sisters. Internal Medicine. 2001;40:1254–8. [DOI] [PubMed] [Google Scholar]
- [58].Almalki SG, Agrawal DK. Effects of matrix metalloproteinases on the fate of mesenchymal stem cells. Stem Cell Research & Therapy. 2016;7:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Cathcart J, Pulkoski-Gross A, Cao J. Targeting matrix metalloproteinases in cancer: Bringing new life to old ideas. Genes & Diseases. 2015;2:26–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Sun G, Xing W, Xing R, Cong L, Tong S, Yu S. Targeting breast cancer cells with a CuInS2/ZnS quantum dot-labeled Ki-67 bioprobe. Oncology letters. 2018;15:2471–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Fang M, Peng C-w, Pang D-W, Li Y. Quantum Dots for Cancer Research: Current Status, Remaining Issues, and Future Perspectives. Cancer Biology & Medicine. 2012;9:151–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Laflamme MA, Chen KY, Naumova AV, Muskheli V, Fugate JA, Dupras SK, et al. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat Biotech. 2007;25:1015–24. [DOI] [PubMed] [Google Scholar]
- [63].Bolli R, Chugh AR, D’Amario D, Loughran JH, Stoddard MF, Ikram S, et al. Cardiac stem cells in patients with ischaemic cardiomyopathy (SCIPIO): initial results of a randomised phase 1 trial. Lancet. 2011;378:1847–57. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.