Abstract
Background
BCL2 19 kD protein-interacting protein 3 (BNIP3) is a BH3-containing protein of the BCL-2 family; it can regulate cell death, autophagy, and cytoprotection. The upregulation of BNIP3 has been reported to relate to progression and poor prognosis in different cancer types. However, the clinical significance of BNIP3 in uveal melanoma (UM) is still unknown.
Material/Methods
In our study, 47 patients with UM were enrolled; the expression of BNIP3 was detected with immunohistochemistry. According to BNIP3 immunohistochemical scores, the patients were divided into BNIP3 high- and low-expression subgroups. The correlation between the expression of BNIP3 and clinicopathological factors was evaluated with Fisher’s test; the associations with survival rates were analyzed with log-rank test. The independent prognostic factors were identified with the Cox-regression model.
Results
BNIP3 was mainly localized in the cytoplasm, and high expression of BNIP3 accounted for 31.9% (15/47) of the patients in our study. High expression of BNIP3 was demonstrated to be significantly associated with more pigment (P=0.018) and deeper scleral invasion (P=0.013). High expression of BNIP3 was also correlated with lower overall survival rate (P=0.006). Multivariate analysis confirmed positive ciliary body involvement and lymphatic infiltration as independent prognostic factors.
Conclusions
High expression of BNIP3 was significantly associated with poor prognosis of patients with UM, indicating that BNIP3 detection could help stratify high-risk patients and identify new therapies targeting BNIP3 as a promising approach to treat UM.
MeSH Keywords: bcl-2-Associated X Protein; Ciliary Body; Melanoma, Experimental; Uveal Neoplasms
Background
Uveal melanoma (UM) is a primary adult malignancy of the eye and has the highest mortality and morbidity worldwide [1]. It is characterized with early metastasis and unfavorable prognosis. Along with histopathology, several factors have been identified as predictive of the prognosis of patients with UM, such as tumor cell type, size of the nucleoli, mitosis, and activity of proliferation. These factors could be used to stratify high-risk patients and those with poor prognosis, to help inform individual treatment decisions. However, the identification of new prognostic biomarkers in UM is lacking, partially because of the rarity of UM cases and the difficulty of obtain tissue specimens for such evaluations.
BCL2 19 kD protein-interacting protein 3 (BNIP3) is a BH3-containing protein of the BCL-2 family, which has been demonstrated to regulate cell death, autophagy, and cytoprotection. BNIP3 is an unclassical BH3-containing protein because it interacts directly with BCL-2 family members via its C-terminal transmembrane domain instead of BH3 domain. BNIP3 plays dual effects on cell survival. On one hand, BNIP3 activates caspase-independent necrosis-like cell death by opening the mitochondrial permeability transition pore [2]. On the other hand, BNIP3 can initiate the mitochondrial autophagy, which is essential for cell survival in extreme environments [3]. The upregulation of BNIP3 has been reported to be correlated with progression and prognosis in several types of cancers such as non-small cell lung cancer, breast cancer, and salivary adenoid cystic carcinoma [4–6].
In recent years, gene expression profiling provided a breakthrough into UM individual treatment. Onken et al. revealed two molecular classes of UM with different prognosis based on gene expression profiling that provides more detail to stratify high-risk patients for individual treatment [7]. In UM, BNIP3 has been proven to support melanoma cell migration and vasculogenic mimicry by orchestrating the actin cytoskeleton [8]. However, its clinical significance has not yet been revealed. In our study, the expression of BNIP3 was detected with immunohistochemistry (IHC) in 47 Um cases. Moreover, its correlation with clinicopathological factors and prognostic values were analyzed to evaluate the clinical significance of BNIP3.
Material and Methods
Patients
Formalin-fixed paraffin-embedded (FFPE) tissue samples were obtained from the Department of Pathology of Linyi Central Hospital from 71 patients who underwent enucleation and were diagnosed as having primary UM from 2000 to 2013. The specimens were obtained with prior consent of each patient enrolled in the study. Twenty-four patients were excluded because of the absence of follow-up records or due to use of chemotherapy or radiotherapy prior to enucleation. The final validation cohort consisted of 47 patients. The diagnosis was double confirmed by two pathologists and the clinicopathological factors were retrieved from the patients’ medical records and pathological reports. The study was approved by the Ethics Committee of Linyi Central Hospital and conducted in guidance of the principle of the Helsinki Declaration.
IHC and score evaluation
Streptavidin peroxidase complex method was used for IHC staining according to the literature [9–11]. Briefly, the FFPE tissue specimens were first deparaffinized and rehydrated with xylene and graded alcohol, and then incubated in boiled citrate buffer (pH=6.0) for optimal antigen retrieval. Hydrogen peroxide at 3% was used to incubate the slides for 15 minutes to attenuate the endogenous activity. The unspecific binding was blocked by incubation in 1% bovine serum albumin (Beyotime Biotechnology, Shanghai, China) for 1 hour at 37°C. Primary antibody of BNIP3 (Cat No. 44060, Cell Signaling Technology, USA) at 1: 100 dilution was used to incubate the specimens overnight at 4°C. After rinsing with phosphate buffered solution, the secondary antibody (Beyotime Biotechnology, Shanghai, China) labeled with streptavidin-biotin-peroxidase reagent was used to incubate the slides for 2 hours at room temperature and the 3,3′-diaminobenzidine solution (Beyotime Biotechnology, Shanghai, China) was applied for antigen visualization. Finally, the slides were counterstained with hematoxylin (Beyotime Biotechnology, Shanghai, China).
The results of IHC were first evaluated by 2 senior pathologists to select the densest tumor area. The IHC tumor area was semi-quantified with the software ImageJ-pro (National Institutes of Health, MD, USA) via calculating the intensity and area of cell staining. The ROC curve was applied to set the cutoff of IHC values, which was defined as the point with the highest specificity plus sensitivity [12–14]. The cutoff divided the cohort into BNIP3 high-expression and low-expression subgroups, which was used to evaluate the influence of BNIP3 on tumor progression and prognosis. The cutoff was confirmed as 65.45 by ROC curve.
Statistical analysis
All data were analyzed using the software SPSS 17.0 (IBM Corporation, New York, USA). The overall survival rate (OS) was estimated from the date of operation to the date of death or last follow-up.
The correlation between BNIP3 expression and the clinicopathological factors was analyzed with Fisher’s test. The statistical differences between BNIP3 high- and low-expression subgroups were evaluated with log-rank test and survival curves were displayed with Kaplan-Meier test. Independent prognostic factors were identified using the Cox-regression hazard model. P values <0.05 was considered to be statistically significant.
Results
BNIP3 expression in cytoplasm of UM
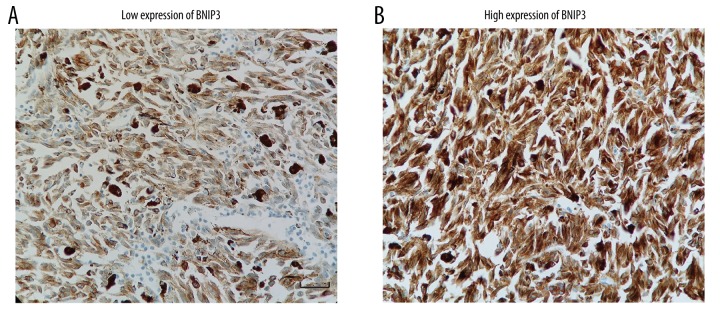
The expression of BNIP3 in UM was detected by IHC on FFPE tissue specimens. BNIP3 was widely expressed in the cytoplasm of UM tissues with variable abundance of expression (Figure 1A, 1B). The cohort of patients was divided into two subgroups: high expression of BNIP3 and low expression of BNIP3. The percentages of BNIP3 low-expression and high-expression cases were 68.1% (32/47) and 31.9% (15/47), respectively.
Figure 1.
The expression of BNIP3 in UM tissues is detected by immunohistochemistry staining. (A) Representative image of the low expression of BNIP3. The score calculated by ImageJ-pro is 44.81. Scale bar: 50 um. (B) Representative image of the high expression of BNIP3. The score calculated by ImageJ-pro is 89.27.
The correlation between BNIP3 high-/low-expression and clinicopathological factors was determined by the Fisher’s test (Table 1). The analysis included the clinicopathological factors such as patients’ age, sex, UM histologic type, the largest tumor diameter, pigment, scleral invasion, ciliary body involvement, and lymphatic infiltration. The percentages of groups stratified with these parameters are displayed on Table 1. The high expression of BNIP3 was demonstrated to be significantly associated with more pigment (P=0.018) and deeper scleral invasion (P=0.013).
Table 1.
Correlation between BNIP3 and clinicopatholgic factors.
| Characters | Number | Percentage | BNIP3 | P* | |
|---|---|---|---|---|---|
| Low | High | ||||
| Age | 0.252 | ||||
| <50 | 37 | 78.72% | 27 | 10 | |
| ≥50 | 10 | 21.28% | 5 | 5 | |
| Sex | 0.851 | ||||
| Male | 21 | 44.68% | 14 | 7 | |
| Female | 26 | 55.32% | 18 | 8 | |
| Histologic type | 0.877 | ||||
| Spindle | 32 | 68.09% | 22 | 10 | |
| nonspindle | 15 | 31.91% | 10 | 5 | |
| Largest tumor diameter(mm) | 0.355 | ||||
| <15 | 20 | 42.55% | 12 | 8 | |
| ≥15 | 27 | 57.45% | 20 | 7 | |
| Pigment | 0.018 | ||||
| <1/3 | 13 | 27.66% | 11 | 2 | |
| <1/3–2/3 | 31 | 65.96% | 21 | 10 | |
| >2/3 | 3 | 6.38% | 0 | 3 | |
| Scleral invasion | 0.013 | ||||
| Superfacial | 34 | 72.34% | 27 | 7 | |
| Medial + Deep | 13 | 27.66% | 5 | 8 | |
| Ciliary body involvement | 0.188 | ||||
| No | 40 | 85.11% | 29 | 11 | |
| Yes | 7 | 14.89% | 3 | 4 | |
| Lymphatic infiltration | 0.836 | ||||
| No | 40 | 85.11% | 27 | 13 | |
| Yes | 7 | 14.89% | 5 | 2 | |
Means calculated by Fisher test.
BNIP3 is the abbreviation for BCL2 19 kD protein-interacting protein 3.
Study parameters correlated with poorer prognosis
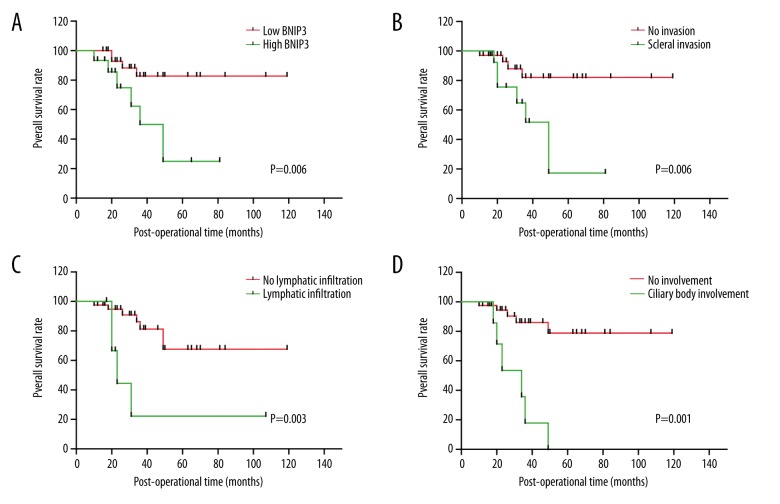
The correlation between BNIP3 subgroups (high-/low-expression) and the 5-year overall survival (OS) rate of UM was analyzed with univariate analysis (Table 2). The statistical significance was evaluated with log-rank test. The survival rates are displayed on Figure 2. Patients with high expression of BNIP3 had poorer prognosis compared with those with low expression of BNIP3 (P=0.006, 5-year OS 25.0% vs. 82.7%) (Figure 2A). Additionally, deeper scleral invasion, positive ciliary body involvement, and lymphatic infiltration were all significantly associated with unfavorable prognosis (P=0.006, 0.001, and 0.003, respectively) (Figure 2B–2D).
Table 2.
Correlation between exprsssion of BNIP3 and the overall survival rates.
| Characters | 3-year OS rate (%) | 5-year OS rate (%) | P* |
|---|---|---|---|
| Age | 0.861 | ||
| <50 | 65.8 | 65.8 | |
| ≥50 | 58.3 | 58.3 | |
| Sex | 0.750 | ||
| Male | 79.7 | 69.7 | |
| Female | 75.1 | 56.3 | |
| Histologic type | 0.764 | ||
| Spindle | 75.4 | 56.6 | |
| Nonspindle | 65.5 | 65.5 | |
| Largest tumour diameter(mm) | 0.250 | ||
| <15 | 88.0 | 78.2 | |
| ≥15 | 65.3 | 49.5 | |
| Pigment | 0.360 | ||
| <1/3 | 90.0 | 90.0 | |
| <1/3–2/3 | 69.2 | 51.9 | |
| >2/3 | 50.0 | 50.0 | |
| Scleral invasion | 0.006 | ||
| Superfacial | 82.1 | 82.1 | |
| Medial + Deep | 51.8 | 17.3 | |
| Ciliary body involvement | 0.001 | ||
| No | 86.0 | 78.9 | |
| Yes | 17.9 | 0.0 | |
| Lymphatic infiltration | 0.003 | ||
| No | 81.2 | 67.7 | |
| Yes | 22.2 | 22.2 | |
| BNIP3 | 0.006 | ||
| Low | 82.7 | 82.7 | |
| High | 49.9 | 25.0 | |
Means calculated by log-rank test.
BNIP3 is the abbreviation for BCL2 19 kD protein-interacting protein 3. HR means hazard radio and CI means confidence interval.
Figure 2.
Survival curves stratified with BNIP3, scleral invasion, ciliary body involvement, and lymphatic infiltration. (A) High expression of BNIP3 is correlated with lower overall survival rates (P=0.006). (B) Positive scleral invasion is correlated with lower overall survival rates (P=0.006). (C) Positive ciliary body involvement is correlated with lower overall survival rates (P=0.003). (D) Positive lymphatic infiltration is correlated with lower overall survival rates (P=0.001).
Table 3.
Independent prognostic factors by multivariate analysis.
| Characters | HR | 95%CI | P* |
|---|---|---|---|
| Age | 0.05–2.10 | 0.235 | |
| <50 | 1 | ||
| ≥50 | 0.32 | ||
| Sex | 0.13–5.84 | 0.892 | |
| Male | 1 | ||
| Female | 1 | ||
| Scleral invasion | 0.67–14.8 | 0.145 | |
| Superfacial | 1 | ||
| Medial + Deep | 3.15 | ||
| Ciliary body involvement | 1.29–27.1 | 0.022 | |
| No | 1 | ||
| Yes | 5.91 | ||
| Lymphatic infiltration | 1.26–45 | 0.027 | |
| No | 1 | ||
| Yes | 7.54 | ||
| BNIP3 | 0.28–7.47 | 0.651 | |
| Low | 1 | ||
| High | 1.46 | ||
Means calculated by Cox-regression model.
BNIP3 is the abbreviation for BCL2 19 kD protein-interacting protein 3. HR means hazard radio and CI means confidence interval.
Identification of independent prognostic factors in UM
The independent prognostic factors of UM were identified with the Cox-regression hazard model (Table 2). All the significant parameters verified in univariate analysis were enrolled into the hazard model except for patient age and sex. In our cohort, the positive ciliary body involvement and lymphatic infiltration were identified as independent prognostic factors (P=0.022 and 0.027, respectively), suggesting positive ciliary body involvement or lymphatic infiltration could lead to poor prognosis directly. BNIP3 high expression was not proven to be an independent prognostic factor in our study (P=0.651).
Discussion
UM has dramatically high mortality and is a major health burden worldwide [15]. Radical resection of the tumor at an early stage is still the only curative treatment because of its characteristic of early metastasis. Achievements in new therapeutic approaches for the treatment of UM have been made in recent years. One of these striking breakthroughs was therapy targeting immune-checkpoints. Up to now, there have been three immune-checkpoint inhibitors approved by the US Food and Drug Administration (FDA) for the treatment of unresectable or metastatic melanoma. Those are ipilimumab, nivolumab, and pembrolizumab. The discovery of all these inhibitors was based on the identification of new biomarkers and drug targets like PD-1 receptor. However, exploring new biomarkers for UM is challenging because of its rare frequency as well as its early metastasis rate, which results in lost resect surgery opportunity for many patients, and thus no chance to collect tissue samples. For all these reasons, studies targeting the identification of prognostic biomarkers are relatively rare. In our study, we showed BNIP3 expression was predictive of unfavorable prognosis of UM with a very significant statistical difference (P=0.006). This may provide new insight into the treatment of UM and may help select high-risk UM patients.
In our study, the BNIP3 overexpression was not identified as an independent prognostic factor, indicating that BNIP3 expression may mainly affect tumor progression related to scleral invasion and indirectly influence prognosis. The underlying mechanism of how BNIP3 correlates to scleral invasion and poor prognosis is essential to elucidating the function of BNIP3 in UM and confirming whether BNIP3 is an independent prognostic factor, however, unfortunately, it was not involved in our study. However, based on previous literature, some of our hypothesis could be verified. BNIP3 is essential to two fundamental processes: apoptosis and mitochondrial autophagy, which play opposite functions on cell survival [16]. The ability of autophagy induction of BNIP3 protects tumor cells from dying in environments like starvation or hypoxia [17]. The starvation or hypoxia of tumor cells is normal in all types of cancer, including UM. The role of BNIP3 as a gatekeeper in mitochondrial autophagy has been widely accepted. Mitochondrial autophagy, also known as mitophagy, is usually considered to suppress oncogenesis in tumor initiation and promote tumor progression via supporting tumor cell survival by removing damaged organelles and providing energy by catabolizing cellular macromolecules [18]. During tumor progression, the enhancement of mitophagy is usually regarded as one reason for rapid progression. However, this conclusion is still controversial, and some studies have reported that upregulation of BNIP3 could promote apoptosis of cancer cells [19,20]. BNIP3 plays an important role in the induction of apoptosis as well as autophagy. The former is responsible for cell death while the latter is responsible for survival protection. The function of BNIP3 in cancer may be different for different cancer types and in different cancer phases.
Conclusions
We, for the first time, detected the expression of BNIP3 in 47 cases of UM and demonstrated that BNIP3 is significantly correlated with scleral invasion and poor prognosis, suggesting therapies targeting BNIP3 may be promising for successful treatment of UM. Our study could also help stratify UM patients with high risk and determine the selection of the optimal therapy for each patient.
Footnotes
Conflicts of interest
None.
Source of support: Departmental sources
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. Cancer J Clin. 2014;64(1):9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Imazu T, Shimizu S, Tagami S, et al. Bcl-2/E1B 19 kDa-interacting protein 3-like protein (Bnip3L) interacts with bcl-2/Bcl-xL and induces apoptosis by altering mitochondrial membrane permeability. Oncogene. 1999;18(32):4523–29. doi: 10.1038/sj.onc.1202722. [DOI] [PubMed] [Google Scholar]
- 3.Zhang J, Ney PA. Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell Death Differ. 2009;16(7):939–46. doi: 10.1038/cdd.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giatromanolaki A, Koukourakis MI, Sowter HM, et al. BNIP3 expression is linked with hypoxia-regulated protein expression and with poor prognosis in non-small cell lung cancer. Clin Cancer Res. 2004;10(16):5566–71. doi: 10.1158/1078-0432.CCR-04-0076. [DOI] [PubMed] [Google Scholar]
- 5.Chourasia AH, Tracy K, Frankenberger C, et al. Mitophagy defects arising from BNip3 loss promote mammary tumor progression to metastasis. EMBO Rep. 2015;16(9):1145–63. doi: 10.15252/embr.201540759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Z, Wu H, Huang S, et al. Expression of BNIP3 and its correlations to hypoxia-induced autophagy and clinicopathological features in salivary adenoid cystic carcinoma. Cancer Biomark. 2015;15(4):467–75. doi: 10.3233/CBM-150474. [DOI] [PubMed] [Google Scholar]
- 7.Onken MD, Worley LA, Ehlers JP, Harbour JW. Gene expression profiling in uveal melanoma reveals two molecular classes and predicts metastatic death. Cancer Res. 2004;64(20):7205–9. doi: 10.1158/0008-5472.CAN-04-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maes H, Van Eygen S, Krysko DV, et al. BNIP3 supports melanoma cell migration and vasculogenic mimicry by orchestrating the actin cytoskeleton. Cell Death Dis. 2014;5:e1127. doi: 10.1038/cddis.2014.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu YF, Ge FJ, Han B, et al. High-mobility group box 1 expression and lymph node metastasis in intrahepatic cholangiocarcinoma. World J Gastroenterol. 2015;21(11):3256–65. doi: 10.3748/wjg.v21.i11.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu H, Xu Y, Zhang Q, et al. Correlations between TBL1XR1 and recurrence of colorectal cancer. Sci Rep. 2017;7:44275. doi: 10.1038/srep44275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu YF, Yang XQ, Lu XF, et al. Fibroblast growth factor receptor 4 promotes progression and correlates to poor prognosis in cholangiocarcinoma. Biochem Biophys Res Commun. 2014;446(1):54–60. doi: 10.1016/j.bbrc.2014.02.050. [DOI] [PubMed] [Google Scholar]
- 12.Liu H, Xu Y, Zhang Q, et al. Prognostic significance of TBL1XR1 in predicting liver metastasis for early stage colorectal cancer. Surg Oncol. 2017;26(1):13–20. doi: 10.1016/j.suronc.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 13.Xu Y, Yang X, Li Z, et al. Sprouty2 correlates with favorable prognosis of gastric adenocarcinoma via suppressing FGFR2-induced ERK phosphorylation and cancer progression. Oncotarget. 2017;8(3):4888–900. doi: 10.18632/oncotarget.13982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu H, Liu Z, Li K, et al. TBL1XR1 predicts isolated tumor cells and micrometastasis in patients with TNM stage I/II colorectal cancer. J Gastroenterol Hepatol. 2017;32(9):1570–80. doi: 10.1111/jgh.13749. [DOI] [PubMed] [Google Scholar]
- 15.Lozano R, Naghavi M, Foreman K, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: A systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380(9859):2095–28. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drake LE, Springer MZ, Poole LP, et al. Expanding perspectives on the significance of mitophagy in cancer. Semin Cancer Biol. 2017;47:110–24. doi: 10.1016/j.semcancer.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Novak I. Mitophagy: A complex mechanism of mitochondrial removal. Antioxid Redox Signal. 2012;17(5):794–802. doi: 10.1089/ars.2011.4407. [DOI] [PubMed] [Google Scholar]
- 18.Klionsky DJ. Autophagy revisited: A conversation with Christian de Duve. Autophagy. 2008;4(6):740–43. doi: 10.4161/auto.6398. [DOI] [PubMed] [Google Scholar]
- 19.Chourasia AH, Macleod KF. Tumor suppressor functions of BNIP3 and mitophagy. Autophagy. 2015;11(10):1937–38. doi: 10.1080/15548627.2015.1085136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan J, Yun H, Yang Y, et al. Upregulation of BNIP3 promotes apoptosis of lung cancer cells that were induced by p53. Biochem Biophys Res Commun. 2006;346(2):501–7. doi: 10.1016/j.bbrc.2006.05.160. [DOI] [PubMed] [Google Scholar]