Abstract
Background and Aims
Turgor-driven plant cell growth depends on cell wall structure and mechanics. Strengthening of cell walls on the basis of an association and interaction with silicon (Si) could lead to improved nutrient uptake and optimized growth and metabolism in rice (Oryza sativa). However, the structural basis and physiological mechanisms of nutrient uptake and metabolism optimization under Si assistance remain obscure.
Methods
Single-cell level biophysical measurements, including in situ non-invasive micro-testing (NMT) of NH4+ ion fluxes, atomic force microscopy (AFM) of cell walls, and electrolyte leakage and membrane potential, as well as whole-cell proteomics using isobaric tags for relative and absolute quantification (iTRAQ), were performed.
Key Results
The altered cell wall structure increases the uptake rate of the main nutrient NH4+ in Si-accumulating cells, whereas the rate is only half in Si-deprived counterparts.
Conclusions
Rigid cell walls enhanced by a wall-bound form of Si as the structural basis stabilize cell membranes. This, in turn, optimizes nutrient uptake of the cells in the same growth phase without any requirement for up-regulation of transmembrane ammonium transporters. Optimization of cellular nutrient acquisition strategies can substantially improve performance in terms of growth, metabolism and stress resistance.
Keywords: Nutrient uptake of NH4+, silicon (Si), cell wall-bound Si, iTRAQ, membrane stability, metabolism, AFM, XPS, Oryza sativa
INTRODUCTION
Silicon (Si) is a beneficial nutrient of rice plants, and its deficiency not only reduces stress resistance but also leads to growth inhibition (Epstein, 1994). Rice has evolved unique strategies to deposit large amounts of intra- or extracellular silica (Savant et al., 1997) by taking up silicic acid using Si transporters (Ma et al., 2006). In addition to silica, recent evidence suggests that the cell wall in rice cells contains Si in a bound form for cross-linking of hemicellulose polysaccharides (He et al., 2013, 2015).
Adequate Si fertilization greatly increases rice yield and improves grain quality, probably by enhancing biotic and abiotic stress resistance (Meharg and Meharg, 2015). Isa et al. (2010) showed that supply of Si to the lsi1 (low-silicon) mutant resulted in increased growth and accumulation of Si with the formation of only a few silica bodies, suggesting that complexation of Si with wall polysaccharides (He et al., 2015) may play an important role in enhancing growth independently of silica deposition. Using the same low-Si rice mutant lsi1, Detmann et al. (2012) showed that Si increased grain yield by an increase in nitrogen (N) use efficiency, and altered primary metabolism by stimulating amino acid remobilization. Moreover, Si may influence enzymes associated with amino acid synthesis and carbon (C) metabolism (Lavinsky et al., 2016; Sanglard et al., 2016). Nwugo and Huerta (2011) investigated proteomic changes of rice leaves and showed that Si plays a more active role in physiological processes than previously proposed.
Irrespective of whether the form is present in inorganic silica or organosilicon or both, no attempt has been made to elucidate the mechanisms underlying such responses, especially N use efficiency, at the single-cell level. The aim of this study is to explore the role of wall-bound Si in enhancing N uptake in individual rice cells. In this context, we hypothesize that such a role is achieved by stabilizing cell membranes to improve N uptake rate without up-regulation of relevant transmembrane transporter proteins. To test this hypothesis, we used a combination of non-invasive micro-test technology (NMT) to determine NH4+ and NO3− ion fluxes, isobaric tags for relative and absolute quantification (iTRAQ) to analyse whole-cell proteomics, atomic force microscopy (AFM) to probe cell wall mechanical responses to deformation, and electrolyte leakage and fluorescent staining to assess the membrane stability/membrane potential of rice suspension cells in the same growth phase. We demonstrate that wall-bound Si alters the rigidity of the cell wall and hence the stability of membranes, promoting individual cells to take up the main nutrient NH4+ and optimizing growth/metabolism in the same growth phases without any requirement for up-regulation of transmembrane ammonium transporters.
MATERIALS AND METHODS
Cell culture and cell wall isolation for AFM and XPS
Rice cells (Oryza sativa L. ‘Zhonghua 11’) were suspension-cultured in the absence (–Si) and presence (+Si) of sodium silicate (Na2SiO3) at 1.0 mm for 3 months as previously described (Liu et al., 2013). The cells were subcultured at 5-d intervals by using newly prepared nutrient solution to replace old liquid medium. Isolations of the cell walls and lyophilized cell walls for AFM and X-ray photoelectron spectroscopy (XPS) analyses were based on previously reported methods (He et al., 2013, 2015). For analyses of Young’s modulus, Origin and SPSS software was used.
Measurements of NH4+, NO3− and K+ ion fluxes
Net NH4+ fluxes were measured using NMT [NMT100 Series, YoungerUSA, Amherst, MA; Xuyue (Beijing) Sci. & Tech. Co., Ltd, Beijing, China]. Prior to flux measurements, the ion-selective microelectrodes were manufactured, pre-pulled and silanized, and micropipettes (Φ1.5 ± 0.5 µm, XY-CGQ-02, YoungerUSA) were first filled with a backfilling solution (NH4+, 100 mm NH4Cl; NO3−, 10 mm KNO3; K+, 100 mm KCl, pH 7.0) to a length of approx. 1.0 cm from the tip, and then were front-filled with columns of selective liquid ion-exchange cocktails (NH4+, XY-SJ-NH4; NO3−, XY-SJ-NO3; K+, XY-SJ-K). An Ag/AgCl wire microsensor holder (YG003-Y11, YoungerUSA) was inserted in the back of the electrode, which makes electrical contact with the electrolyte solution. YG003-Y11 was used as the reference microsensor, and the microelectrodes were then calibrated at the start and end time of each test with cultural media of different concentrations (NH4+, NO3− and K+: 0.05 and 0.5 mm). Only electrodes with Nernstian slopes >50 mm per decade for NH4+ and K+ and <−50 mm per decade for NO3− were used.
Rice suspension cells (200 µL) were put in the middle of glass coverslips pretreated with a poly-l-lysine (Sigma-Aldrich, St Louis, MO, USA) solution in the measuring chamber, and were equilibrated for 15 min and washed twice with corresponding measuring solution. Three millilitres of measuring solution was slowly added to the chamber. Ion fluxes were monitored in the following solutions: (1) NH4+: 0.1 mm NH4Cl, 0.1 mm CaCl2, 0.1 % sucrose, pH 6.0 adjusted with HCl; (2) NO3−: 0.1 mm KNO3, 1 mm KCl, 0.1 mm CaCl2, 0.1 % sucrose, pH 6.0 adjusted with HCl; (3) K+: 0.1 mm KCl, 0.1 mm CaCl2, 0.1 mm MgCl2, 0.5 mm NaCl, 0.3 mm MES, 0.2 mm Na2SO4, 0.1 % sucrose, pH 6.0 adjusted with HCl.
NMT was used to measure the concentration gradient of the target ion by moving the microelectrode repeatedly from one point to another perpendicular to cell surfaces with 10 µm of the preset excursion between two points at a frequency of approx. 0.3 Hz. A continuous flux recording of more than 600 s was conducted for each rice cell in measuring solutions. The calculation of ionic fluxes (pmol cm−2 s−1) was obtained by imFluxes V2.0 (YoungerUSA) software and JCal V3.3 (Xuyue, Beijing, China). SPSS software was used for data statistical analyses.
Proteomics using iTRAQ
Total proteins were extracted according to Ma et al. (2016a, b). A mixture of extracted proteins of +Si and –Si cell samples (2 treatments × 2 biological replicate samples, 100 µg for each) was used as the reference (REF). Protein digestion was performed by a filter aided sample preparation (FASP) method (Wisniewski et al., 2009), and the resulting peptide mixtures were labelled with iTRAQ isobaric tagging reagents according to the manufacturer’s instructions (Applied Biosystems, Carlsbad, CA, USA) (Ma et al., 2016). After labelling and being multiplexed, samples were dried in a vacuum centrifuge and then stored at −20 °C.
Experiments were carried out with an Easy nLC system (Thermo Fisher Scientific, Waltham, MA, USA) that was coupled to Q Exactive mass spectrometer (MS) (Thermo Finnigan, San Jose, CA, USA). The Q Exactive MS output RAW files were analysed via the MASCOT engine (Matrix Science, London, UK; version 2.2) for protein identification, and Proteome Discoverer (Thermo, version 1.3) was used for quantification. Data were searched against a non-redundant International Protein Index Oryza sativa sequence database from uniprotOryzasativa.fasta. The MASCOT search results were further processed using the programs BuildSummary, Isobaric Labeling Multiple File Distiller and Identified Protein iTRAQ Statistic Builder within the Proteomics Tools (version 3.05; details from the Research Center for Proteome Analysis, www.proteomics.ac.cn). The program Build-Summary was used for assembling protein identifications based on a target-decoy search in shotgun proteomics. All reported data were at the 99 % confidence level for protein identification as determined by a false discovery rate (FDR) of 1 % (Sandberg et al., 2012; Ma et al., 2016a, b).
The programs Isobaric Labeling Multiple File Distiller and Identified Protein iTRAQ Statistic Builder were used to calculate protein ratios, in which sample REF was used as reference based on the weighted average of the intensity of report ions in each identified peptide. The final protein ratios were then normalized by the median average protein ratio for unequal mixes of the different labelled samples. Using the Proteome Discoverer software (Thermo Scientific), protein quantification was based on the signature peak areas and corrected according to the Xcalibur Proteome Discoverer (Version 1.3 User Guide). The iTRAQ ratios were selected for further statistical analysis if they match the criteria of (1) being detected in both biological replicates, and (2) having mean expression changes of >1.20 or <0.83 to the relative sample (Ma et al., 2016a, b). Mean expression changes of >1.20, <0.83 and 0.83–1.20 represent up-, down- and un-regulation, respectively, of proteins between +Si and –Si cells in this study.
Cell membrane stability and membrane potential measurements
Electrolyte leakage was used to assess cell membrane stability (Whitlow et al., 1992). Cells were washed with ultrahigh-purity water three times. After drying on filter paper, approx. 0.1 g of cells (f. wt) were collected in each treatment (+Si/–Si) and put into 30 mL water or 1–10 % polyethylene glycol (PEG) solution to record the conductivity at 25 °C in real time. Also, the membrane stability was evaluated by changes of cell membrane potential. Instable membranes, especially in the presence of PEG-induced osmotic stress, can result in a depolarization of the cell membrane potential (Ober and Sharp, 2003), and DiBAC4 (3) fluorescent dye can enter the depolarized cell and bind to intracellular membranes to emit enhanced fluorescence (Konrad and Hedrich, 2008; Sun et al., 2010). The DiBAC4 (3)-dependent fluorescence of rice suspension cells was measured with a Leica SP8 confocal microscope (Leica Microsystems GmbH, Wetzlar, Germany) and fluorescence intensity (average grey value) was measured with image processing software (Adobe Systems; Leica Application Suite Advanced Fluorescence; Leica Microsystems). SPSS software was used for statistical analyses.
RESULTS
Uptake of ammonium (NH4+) and nitrate (NO3−) in rice cells in the absence and presence of Si
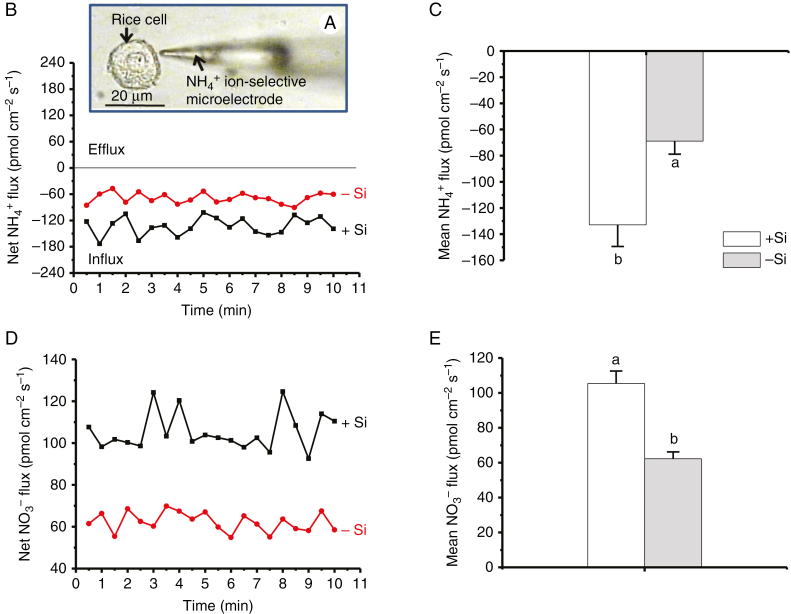
Cells of similar sizes were chosen to avoid different growth phases (Supplementary Data, Fig. S1). Net ammonium (NH4+) fluxes of rice suspension cells were measured using NMT (Fig. 1A). Cells cultivated in solutions with 1 mm silicic acid (+Si) for 3 months exhibited a steady net NH4+ influx of 133 ± 17 pmol cm−2 s−1 (n = 8) during the testing period (Fig. 1B). By contrast, a mean NH4+ influx of 69 ± 10 pmol cm−2 s−1 (n = 8) was observed in rice cells cultivated in solutions without Si (–Si), almost half the value in +Si cells (Fig. 1C). In addition, regardless of the absence or presence of Si, nitrate (NO3−) ions were all in efflux (Fig. 1D, E), suggesting that NH4+ is the main N source in rice (Fan et al., 2016).
Fig. 1.
NH4+ fluxes. (A) Rice (Oryza sativa) suspension cells cultured in the absence (–Si) and presence (+Si) of 1.0 mm silicic acid for 3 months after transplanting to solution medium from solid culture medium prior to NMT measurements. Arrows show a probed rice cell and NH4+ ion-selective microelectrode, respectively. (B, D) Net and (C, E) mean fluxes of NH4+ and NO3− ions (mean ± SD, n = 8), respectively. Different lower-case letters in C and E indicate a significant difference at P < 0.05.
Effects of Si on the protein expression of rice suspension cells
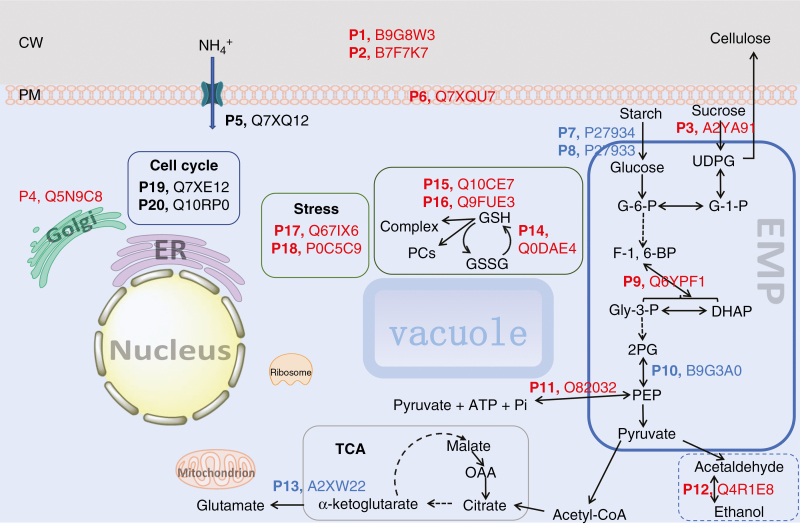
As predicted, the total protein concentration [46 ± 2 mg g−1 f. wt (n = 3)] of the +Si cells was greater than that [38 ± 0.5 mg g−1 (n = 3)] of –Si cells (Fig. S2) due to a significant difference in NH4+ influxes (Fig. 1). To understand the origin of the NH4+ uptake and protein synthesis optimization in the presence of Si in rice suspension cells, we used iTRAQ to detect and identify 20 proteins related to the cell wall (P1–P3), plasma membrane (P4–P6), energy metabolism (P7–P13), stress/stimulus response (P14–P18) and cell cycle (P19 and P20) (Table 1 and Fig. 2). Among these function-known proteins, 13 down-regulated (P1–P4, P6, P9, P11, P12 and P14-–P18), four up-regulated (P7, P8, P10 and P13) and three unregulated (i.e. not affected by Si treatment) (P5, P19 and P20) proteins were identified for +Si cells, whereas 13 up-regulated, four down-regulated and three unregulated proteins were identified for the –Si cells (Table 1 and Fig. 2). Others included 17 unregulated but function-known membrane-related proteins (Table S1), and 57 Si-regulated proteins including 20 function-unknown and 37 unanalysable, function-decentralized proteins (Table S2).
Table 1.
Twenty predicted proteins in rice (Oryza sativa L.) suspension cells by iTRAQ
| Protein | Gene | Accession number | Name | Enzyme activity | Fold change (+Si/–Si) |
|---|---|---|---|---|---|
| Cell wall-related proteins | |||||
| P1 | OsJ_34867 | B9G8W3 | Glycosidase | Hydrolysing O-glycosyl compounds | +D |
| P2 | BGLU31 | B7F7K7 | Beta-glucosidase 31 | β-1,4-d-glucanase | +D |
| P3 | OsI_22003 | A2YA91 | Sucrose synthase | Provides UDP-glucose and fructose | +D |
| Cell membrane-related proteins | |||||
| P4 | Os01g0708600 | Q5N9C8 | TRAPP complexes | Golgi vesicle transport | +D |
| P5 | AMT1-1 | Q7XQ12 | Ammonium transporter 1 member 1 | Ammonium transporter activity | – |
| P6 | Os04g0452000 | Q7XQU7 | Probable protein phosphatase 2C 41 | [a protein]-serine/threonine phosphate + H2O = [a protein]-serine/threonine + phosphate. | +D |
| Energy metabolic process-related proteins | |||||
| P7 | AMY1.4 | P27934 | Alpha-amylase isozyme 3E | Endohydrolysis of (1->4)-alpha-d-glucosidic linkages | +U |
| P8 | AMY1.3 | P27933 | Alpha-amylase isozyme 3D | +U | |
| P9 | Os08g0120600 | Q6YPF1 | Fructose-bisphosphate aldolase | d-Fructose 1,6-bisphosphate = glycerone phosphate + d-glyceraldehyde 3-phosphate | +D |
| P10 | OsJ_29149 | B9G3A0 | Enolase | 2-Phospho-d-glycerate = phosphoenolpyruvate + H2O | +U |
| P11 | PPDK | O82032 | Pyruvate, phosphate dikinase | ATP + pyruvate + phosphate = AMP + phosphoenolpyruvate + diphosphate | +D |
| P12 | ADH2 | Q4R1E8 | Alcohol dehydrogenase 2 | An alcohol + NAD+ = an aldehyde or ketone + NADH | +D |
| P13 | GDH2 | A2XW22 | Glutamate dehydrogenase 2, mitochondrial | 2-oxoglutarate + NH3 + NAD(P)H = l-glutamate + H2O + NAD(P)+ | +U |
| Response to stimulus | |||||
| P14 | GRXC8 | Q0DAE4 | Glutaredoxin-C8 | electron carrier activity, protein disulfide oxidoreductase activity | +D |
| P15 | GSTU1 | Q10CE7 | Probable glutathione S-transferase GSTU1 | RX + glutathione = HX + R-S-glutathione | +D |
| P16 | LOC_Os10g38489 | Q9FUE3 | Glutathione S-transferase GSTU6 | transferase activity | +D |
| P17 | PDIL1-4 | Q67IX6 | Protein disulfide isomerase-like 1-4 | Catalyses the rearrangement of –S–S– bonds in proteins | +D |
| P18 | Os07g0638300 | P0C5C9 | 1-Cys peroxiredoxin A | 2 R′-SH + ROOH = R′-S-S-R′ + H2O + ROH | +D |
| Cell cycle | |||||
| P19 | LOC_Os10g30580 | Q7XE16 | Cell division cycle protein 48 | Cell division; cell cycle |
– |
| P20 | LOC_Os03g05730 | Q10RP0 | – | ||
The ‘+’ indicates there is significant difference, and the ‘–‘ indicates there is no statistically significant difference between the +Si and –Si cell. UUp-regulated for +Si cells, Ddown-regulated for +Si cells.
Fig. 2.
Schematic illustrating the influence of Si on intracellular biochemical pathways in suspension cells of rice. Blue, red and black type in numbered proteins (P1–P20) indicates Si-induced up-regulation, down-regulation and unregulation, respectively. Arrows indicate the direction of regulation. CW, cell wall; PM, plasma membrane; ER, endoplasmic reticulum; TCA, tricarboxylic acid cycle; EMP, the Embden–Meyerhof–Parnas pathway, i.e. glycolysis; UDPG, uridine diphosphate glucose; G-6-P, glucose-6-phosphate; G-1-P: glucose-1-phosphate; F-1, 6-BP, fructose 1, 6-bisphosphate; Gly-3-P, d-glyceraldehyde 3-phosphate; DHAP, dihydroxyacetone phosphate; 2PG, 2-phosphoglycerate; PEP, phosphoenolpyruvate; OAA, oxaloacetic acid; GSH, glutathione; GSSG, glutathione disulfide; PCs, phytochelatins.
Morphological and mechanical properties of cell walls in the absence and presence of Si
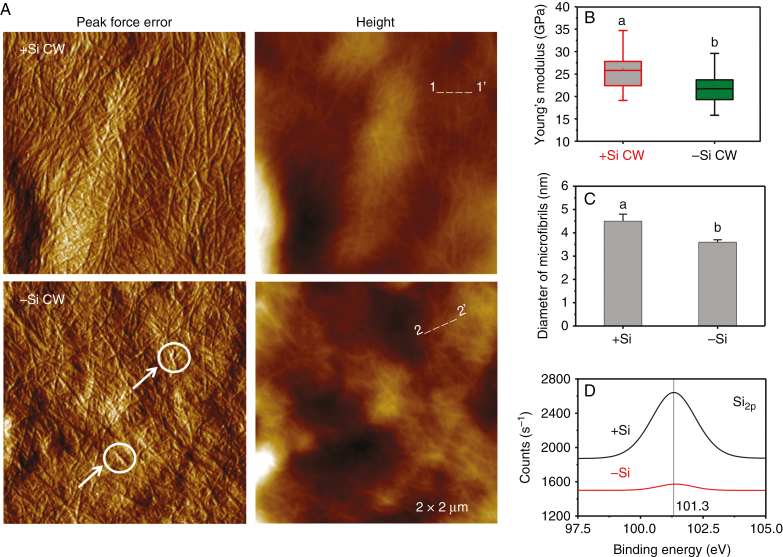
Cell walls isolated from rice suspension cells cultivated in the absence and presence of 1 mm silicic acid for 3 months were analysed using both AFM and XPS (Fig. 3). AFM peak force error images show that the cellulose microfibrils of +Si cell walls are much denser than those of –Si cell walls, which contain many broken and shorter fibrils that are randomly deposited on cell surfaces (Fig. 3A). Regarding Si supply, a clear skeleton of cellulose microfibrils with an increased diameter was seen under AFM based on height measurements (Fig. 3C). Accordingly, the average Young’s moduli of cell walls in the presence and absence of Si were 26.2 ± 3.8 GPa (n = 10) and 22.5 ± 3.4 GPa (n = 10) (Fig. 3B), respectively, measured by AFM force curves. In addition, the Si 2p core-level XPS spectra of +Si cell walls exhibit a clear peak at 101.3 ± 0.3 eV (n = 10), compared to –Si cell walls (Fig. 3D), suggesting the presence of a possible organosilicon component within the cell wall rather than deposition of inorganic silica at cell surfaces (He et al., 2013).
Fig. 3.
Atomic force microscopy (AFM) and X-ray photoelectron spectroscopy (XPS) analysis of the cell wall (CW) of rice suspension cells. (A) AFM PeakForce and height images of the extracted cell wall from +Si and –Si cells. Arrows and circles show the cellulose fibrils are shorter and broken in –Si cells. (B) Average Young’s modulus of the cell wall. The horizontal lines of each box represent the 25th, 50th and 75th percentile values, and the whiskers outside the box extend to the 5th and 95th percentile values. Different lowercase letters indicate a significant difference at P < 0.05. (C) Diameter (averaged height) of cellulose microfibrils or bundles of microfibrils in the +Si and –Si cells measured along lines 1→1’ and 2→2’ in A by AFM. (D) XPS of the cell wall chemical composition for Si 2p. Values are means ± SD (n = 10).
Effects of Si on cell membrane stability
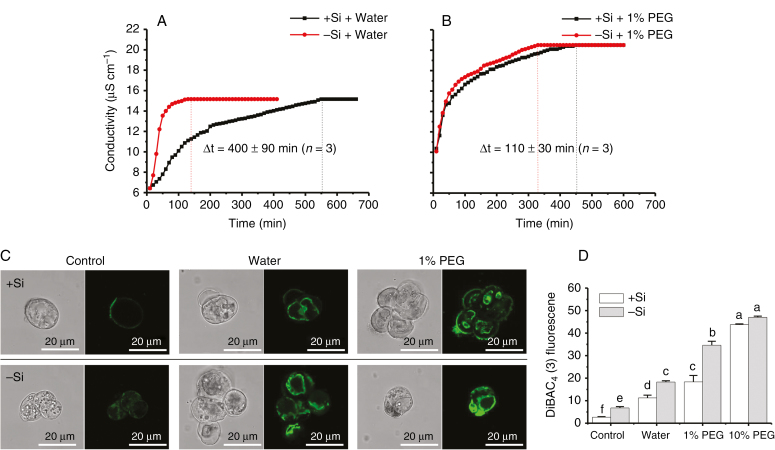
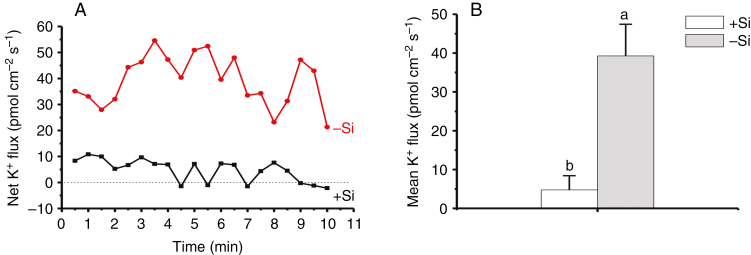
Cell membrane stability was first assessed by measuring the time that is needed for electrolyte leakage to reach equilibrium, by monitoring real-time conductivity (Fig. 4A, B). Time differences (∆t) between +Si and –Si cells were 400 ± 90 min (n = 3) and 110 ± 30 min (n = 3), under the solution conditions of pure water (Fig. 4A) and 1 % PEG solutions (Fig. 4B), respectively. Second, the change in cell membrane potential can be used to evaluate the membrane stability based on DiBAC4 (3)-dependent fluorescence (Fig. 4C). Figure 4D shows that the fluorescence intensity of –Si cells was significantly greater than that of +Si cells in the absence and presence of osmotic stress (hypotonicity, pure water; hypertonicity, 1 % PEG). After shifting to extreme stress of 10 % PEG, the significant difference between +Si and –Si cells vanished (Fig. 4D), either due to complete depolarization of the cells, or because most cells (>70 %) were close to death regardless of the absence and presence of Si (Figs S3 and S4). These results based on two different methods indicate that cell membrane stability can be improved significantly by Si-accumulating cell walls, compared to Si-deprived ones. Consistent with the above results, a mean K+ efflux of 39.3 ± 8.2 pmol cm−2 s−1 (n = 8) was observed in –Si cells, whereas a very low efflux of 4.8 ± 3.7 pmol cm−2 s−1 (n = 8) was exhibited in +Si cells (Fig. 5).
Fig. 4.
Cell membrane stability in the presence of osmotic stress. Plots of conductivity against time in the presence of (A) hypotonicity (in pure water) or (B) hypertonicity (in 1 % PEG solutions). (C) Fluorescence distribution and (D) intensity of –Si and +Si cells using DiBAC4 (3) dye (mean ± SD, n = 3). Different lower-case letters in D show a significant difference at P < 0.05.
Fig. 5.
K+ fluxes of rice suspension cells cultivated in the absence and presence of 1.0 mm silicic acid for 3 months. (A) Net and (B) mean fluxes of K+ ions within 10 min of NMT measurements in the measuring solutions containing 0.1 mm K+ at pH 6.0 (mean ± SD; n = 8). Different lower-case letters in B demonstrate a significant difference at P < 0.05.
DISCUSSION
Mechanisms of ammonium uptake optimization
The possible mechanisms to promote uptake of NH4+ ions by Si bound to the cell wall include accelerated cell division in the cell cycle (i.e. in different growth phases for +Si and –Si cells), enhanced expression/synthesis of ammonium transporters in cell membrane, or wall-stabilized cell membranes with improved ammonium transport efficiency.
Cell division/cell cycle proteins.
We selected cells with almost the same sizes for NMT measurements (Fig. S1) to rule out the effects of cell division/cell cycle, and it has been demonstrated that about 60 % of rice cells through suspension culture were in G1 phase (Lendvai et al., 2002; Lee et al., 2004). Although we did not isolate and sort all cells in the same phase of the cell-cycle dynamics for iTRAQ measurements, two unregulated proteins (P19 and P20) (Table 1) were identified, and they are cell division/cell cycle molecules that are highly conserved from yeast to plants to animals and essential for the survival of any eukaryotic cell (Morreale et al., 2009). In particular, cell division cycle protein 48 (P20, Cdc48p) exhibits important roles in cell cycle processes, such as membrane fusion (Latterich et al., 1995), endoplasmic reticulum-associated degradation (ERAD) (Rabinovich et al., 2002) and spindle disassembly at the end of mitosis (Cao et al., 2003). Other proteins, such as plant cyclins (Lee et al., 2003; Nelissen et al., 2016), are functionally important in the G2/early M phase for CycA1;1 and late M phase for CycB2;1 and CycB2;2 (Umeda et al., 1999), but we did not detect them, probably due to their very low concentrations. Moreover, HAL3 (Sun et al., 2009) proteins are also involved in the control of cell cycle progression, particularly in the G1/S transition. However, they are light-regulated proteins and suspension-cultured cells are in darkness, so it is reasonable that OsHAL3 was not detectable and identified by iTRAQ.
Finally, cell-plate formation is an important phase of plant cell mitosis in M phase. Si could accelerate the regeneration of the rice cell wall by crosslinking of wall components (He et al., 2015), suggesting that this organosilicon component may accelerate cell-cycle progression in rice. In our previous study, after a long-term culture (over 100–150 d), compared to a time period of 90 d in the present study, +Si and –Si cells may have been in different phases of the cell cycle (He et al., 2013). Brzezinski et al. (1990) also showed that Si deprivation halted the progression of cells through the cell cycle in marine diatoms. A very recent study has shown that Si promotes cytokinin biosynthesis and delays senescence in Arabidopsis and Sorghum (Markovich et al., 2017), suggesting that Si may accelerate the cell cycle. However, there was no difference in the cell cycle proteins between the treatments in the present study. Therefore, the results do not support this hypothesis that organosilicon promotes rice cell cycle progression exactly.
Ammonium transporters.
We identified the ammonium transporter protein AMT1;1 (P5), which was unregulated by Si (0.83 < fold change = 1.14 < 1.20) (Table 1 and Fig. 2). Membrane proteins of the AMT1 and AMT2 subfamilies represent the major pathways for ammonium transport in plants (Loqué and von Wirén, 2004; Yuan et al., 2007). Ammonium uptake from solution in rice is mainly regulated by three ammonium transporters, and their corresponding genes, OsAMT1;1, 1;2 and 1;3 (Oryza sativa ammonium transporter), have been isolated and examined (Sonoda et al., 2003): OsAMT1;1 is more constitutively expressed in a concentration-dependent manner and thus may serve as a key ammonium transport in both roots and shoots, whereas OsAMT1;2 and 1;3 are low-affinity transporters (Sonoda et al., 2003) with quite low expression levels (Fig. S5). Ammonium influx studies on a T-DNA insertion line in AMT1;1 of Arabidopsis thaliana revealed that in N-deficient roots, AtAMT1;1 confers approx. 30 % of the total ammonium uptake capacity (Loqué et al., 2006). Therefore, AMT1;1 proteins serve as a major transporter for high-affinity ammonium uptake in rice, and this wall-bound form of Si in rice suspended cells does not promote the expression of AMT1;1 to enhance the uptake of NH4+ ions.
Enhanced cell membrane stability after Si crosslinking.
Well-organized wall polysaccharide networks maintain resistance to the extreme tension (several hundred MPa) on the relatively thin cell wall generated by the turgor pressure within the solute-filled cell (Höfte, 2001; Geitmann and Ortega, 2009). The lack of Si results in wall defects (Fig. 3A) with thinner cellulose microfibrils (Fig. 3C), and decreases the mechanical strength (Fig. 3B) against turgor pressure. However, we could not determine whether Si affects the basic unit of cellulose or probably affects the connection between microfibrils through interfering with the hemicellulose. The wall defects may induce membrane deformation, and thus lead to depolarization. Membrane proteins in the deformed cell membrane, such as transmembrane ammonium transporters, may fuse together to form oligomers and aggregates aided by the instability and tension inherent with the high curvature of membranes (McMahon and Gallop, 2005). In such cases, transport efficiency would decrease. Moreover, the membrane potential plays a key role in the transport efficiency of voltage-gated channels (such as K+ channels). Voltage-gated channels are sensitive to changes in membrane potential (membrane depolarization or hyperpolarization) and the change in membrane potential does not influence the degree of channel protein opening, but does control the opening frequency (Alberts et al., 2010).
Following the Si crosslinking of the wall components, the mechanical properties of the cell walls are improved (Fig. 3B). This, in turn, will confer stability of cell membranes on individual cells (Fig. 4) and maintain intracellular ion homeostasis. A significantly lower K+ efflux did exist in +Si cells as compared with –Si cells (Fig. 5). Due to the high conductivity for K+, the membrane potential of resting cells is close to the equilibrium potential of this cation (Konrad and Hedrich, 2008). A negative membrane potential of about –120 mV will suppress K+ efflux in stabilized cell membranes. The results shown in Figs 4 and 5 indicate that cell membranes of +Si cells are more stable than those of –Si cells, and this is direct evidence of an Si-induced enhanced resistance to turgor-related imbalances. In this respect, Luyckx et al. (2017) have demonstrated an effect of silica on bast fibre growth and a mechanical role in the resistance to turgor pressure during elongation. This suggests that both organosilicon and silica may have a similar role but with different strengths in providing enhanced mechanical resistance to turgor-driven expansion (Ma et al., 2016a, b).
Collectively, the rigid cell walls enhanced by a wall-bound form of Si as the structural basis stabilize cell membranes. This, in turn, optimizes NH4+ uptake of the cells via improved transport efficiency in the same growth phase without a requirement for up-regulation of the main transmembrane ammonium transporter.
Over-expression of cell wall-related proteins (P1–P3) to strengthen the cell wall of –Si cells
Walls of growing cells are extremely sophisticated dynamic assembled networks consisting of cellulose, hemicellulose and pectin polysaccharides, as well as protein compounds (Franková and Fry, 2013). The assembly and extension of the wall is in part mediated by wall-associated enzymes, which cleave, rearrange or cross-link polysaccharides (Höfte, 2001; O’Neill et al., 2001).
Splitting and reconnecting chains of (oligo)polysaccharides by enzymes catalysing cross-linking of wall components.
We identified three down-regulated wall proteins (P1, P2 and P3) (Table 1). Glycosidase (P1) belongs to the glycosyl hydrolase 17 family and hydrolyses 1, 3-β-glucan polysaccharides found in the cell wall matrix of plants (Thomas et al., 2000). These isozymes include the endo-1, 3-β-glucanases (Subfamily A) and the endo-1, 3; 1, 4-β-glucanases (Subfamily B) (Thomas et al., 2000). 1, 3-β-Glucan (callose) is formed in vitro when isolated plasma membranes are supplied with UDP-glucose (Amor et al., 1995) and is synthesized in vivo when rapid new cell-wall formation takes place (Verma and Gu, 1996). Callose and other similar carbohydrates have been suggested to serve as possible molecules in silica deposition in horsetail (Equisetum arvense) (Law and Exley, 2011). However, this organosilicon species in rice suspended cells is different from silica templated by callose as proposed by Law and Exley (2011), and thus no direct relationship can be made.
According to iTRAQ results in Table 1, a transport protein particle (TRAPP, P4) complex for Golgi vesicle transport (Rybak et al., 2014) was present and down-regulated in +Si cells. It has been suggested that cell-plate vesicles may contain a membrane-bound sucrose synthase (P3) providing UDP-glucose for the synthesis of 1, 4-β-glucan chains of cellulose (Amor et al., 1995) and subsequent strengthening of the newly formed cell wall (Thomas et al., 2000). Sucrose synthase (P3), a glycosyltransferase (GT), catalyses the reversible reaction (sucrose + UDP = UDPglucose + fructose) (Schmölzer et al., 2016), and pH values of 5.5–7.5 promote the formation of UDP-glucose through high free energy of hydrolysis of the sucrose linkage at sometimes substantial rates in disrupted cells (Amor et al., 1995). In contrast, synthesis of UDP-glucose via UDP-glucose pyrophosphorylase requires two ATP equivalents (Amor et al., 1995), and energy conservation could be especially important for these Si-deprived cells. Moreover, β-glucosidases (P2) are glycosyl hydrolases that hydrolyse the β-O-glycosidic bond and are found to have high exoglucanase activity, consistent with a role in cell wall growth and remodelling (Opassiri et al., 2006).
Insoluble cellulose (1, 4-β-glucan) microfibrils are embedded in soluble polysaccharides, such as xyloglucan and mixed-linkage (1→3, 1→4)-β-d-glucan (MLG). Xyloglucan binds non-covalently to cellulose to form a structural network that can determine the physicochemical and mechanical properties of the cell wall (Carpita and Gibeaut, 1993; Cosgrove, 2005). Rice plants produce 1,3;1,4-β-glucan polysaccharides in the cell wall that should have diverse roles in growth (Thomas et al., 2000): breakdown of these polymers may provide substantial nutrition for wall growth and/or facilitate the access of other hydrolases to their substrates (Fincher, 1989). An example is the xyloglucan endotransglucosylase/hydrolase (XTH) for seamlessly splitting and reconnecting chains of xyloglucan by catalysing covalent cross-linking between cellulose and xyloglucan-oligosaccharide, and between xyloglucan and xyloglucan-oligosaccharide (Shinohara et al., 2017). Our previous observations have shown that Si may control the porosity of the cell wall, as shown by the increased pore number and size of cell walls of Si-starved, protoplast-derived cell cultures (He et al., 2015).
Si crosslinking of wall components.
Our previous results have shown that most of the Si is in hemicellulose in rice cells, where Si may covalently crosslink the hemicellulose polysaccharides to form Si–O–C or Si–C bonds (He et al., 2015) rather than SiO2 (He et al., 2013). This was also confirmed in the present study by XPS (Fig. 3D). The Si 2p peak at 100.1–101.3 eV has been considered to be due to an Si–O–C or O–Si–C chemical environment from simple chemical systems such as Si-organic monolayers (Boukherroub et al., 2000), and silica (fully oxidized silicon Si4+) is at approx. 103.2 eV (He et al., 2013). The hemicellulose-bound form of Si could improve the mechanical properties (Fig. 3B) and regeneration of the cell wall (He et al., 2015). Major hemicelluloses contain the xylans, xyloglucans, mannans and MLGs (Scheller and Ulvskov, 2010). MLG is restricted to Si-accumulating Poales and Equisetum (Fry et al., 2008; Sørensen et al., 2008; Currie and Perry, 2009), suggesting that MLG may serve as a ligand that complexes with Si.
The present AFM results showed that the cellulose microfibrils of –Si cell walls contain many broken and shorter fibrils that are randomly deposited on cell surfaces (Fig. 3A, C). Also, our previous study provided evidence that obvious defects (holes) were present on the protoplast-derived cell surfaces in –Si cells (He et al., 2015). This suggests that the assembly of the cellulose microfibrils with other wall components, such as hemicellulose (which constitutes a hydrated matrix occupying the space between cellulose microfibrils) (Somerville et al., 2004; He et al., 2015), is improved in +Si cells during cell wall formation.
This cell wall-bound form of Si could crosslink cell wall polysaccharides or other matrix-soluble molecules and significantly improves the mechanical properties and regeneration of cell walls. However, up-regulating wall-related proteins/enzymes (P1–P3) in Si-deprived cells may be required for subsequent strengthening of the newly formed cell wall by splitting and reconnecting chains of wall polysaccharides by catalysing cross-linking of the cellulose–hemicellulose and hemicellulose–hemicellulose. This is consistent with AFM results that the diameter of cellulose microfibrils in Si-modified cells is greater than that in Si-limited cells (Fig. 3C), and sucrose synthase (P3, Susy), a key enzyme for cellulose synthesis, was up-regulated in –Si cells. Moreover, Si may not exert a direct role in cross-linking cellulose fibrils and their deposition/crystallization; the presence of Si may only induce over-expression of sucrose synthase to promote new cell wall formation because celluloses are not the ligands for the formation of the Si–wall complexes (He et al., 2015). Thus, Si-accumulating cells may take advantage of the formation of Si–O–C or Si–C bonds that can partially substitute C–O–C bonds in polysaccharide chains, and it would be unnecessary to up-regulate these three proteins.
Influence of Si on intracellular metabolism
Up-regulated, energy metabolic process-related proteins (P7, P8, P10 and P13) in +Si cells.
When the growth, organization and remodelling of the cell wall are improved by wall-bound Si, energy formation and utilization could be enhanced accordingly inside +Si cells. Expectedly, two α-amylase proteins (P7 and P8) were up-regulated in +Si cells (Table 1). α-Amylase is an endo-hydrolase and is considered to be one of the primary enzymes responsible for starch degradation by hydrolysing the 1, 4-α-d-glucosidic linkages in plant cells (Majzlova et al., 2013) to generate glucose for glycolysis, i.e. the Embden–Meyerhof–Parnas (EMP) pathway (Fig. 2). Inside this pathway, a key enzyme, enolase (P10) that catalyses the conversion of 2-phospho-d-glycerate to phosphoenolpyruvate (PEP), was up-regulated in +Si cells (Table 1). At the whole-plant level, Zhu et al. (2016) showed that Si-enhanced assimilate transport provides more energy storage in the root cells, which is beneficial for stress tolerance.
Improved energy generation/utilization in +Si cells should also be of benefit to synthesis of amino acids and proteins by releasing stored energy through the oxidation of acetyl-CoA derived from starch/glucose and chemical energy in the form of ATP in the tricarboxylic acid (TCA) cycle. This cycle provides precursors of certain amino acids, such as α-ketoglutarate; an up-regulated enzyme, glutamate dehydrogenase (P13) that mediates the reductive amination of α-ketoglutarate to yield glutamate (Tsai et al., 2016) was detected in +Si cells, consistent with the results of both increases in uptake of NH4+ ions (Fig. 1A, B) and the total protein concentration in +Si cells (Fig. S2) because glutamate is used by almost all plants in the biosynthesis of proteins.
Compared with +Si cells, glycolytic energy generation is limited in –Si cells, and thus up-regulation of proteins/enzymes in fermentative pathways can compensate for energy insufficiency. In –Si cells, we detected an up-regulated alcohol dehydrogenase (P12, ADH) that is an essential enzyme in the alcohol fermentation pathway for the sustained production of ATP under stress conditions (Umeda and Uchimiya, 1994) by catalysing the reversible reaction from ethanol to acetaldehyde (Table 1 and Fig. 2). Another up-regulated, energy-related protein, pyruvate orthophosphate dikinase (P11, PPDK) (catalysing the reaction: pyruvate + ATP + Pi ↔ PEP + AMP + PPi + 2H+), was present in –Si cells (Table 1). Indeed, cytosolic PPDK is involved in a metabolic response to various environmental stresses and has been shown to function in the synthesis of ATP from PEP (Moons et al., 1998).
Finally, an up-regulated protein, fructose-bisphosphate aldolase (P9), was present in the glycolytic pathway in –Si cells (Table 1 and Fig. 2), and it physically associates with vacuolar H+-ATPase (Lu et al., 2004) that is a primary-active proton pump present in the endomembrane system and is responsible for providing energy for transport of ions and metabolites as a ‘house-keeping’ and as a stress response enzyme (Sze et al., 1999; Ratajczak, 2000). The electrochemical gradient generated by these H+ pumps provides the driving force/energy for the secondary transport of ions and metabolites (Sze et al., 1992) as glycolytic energy generation is limited in –Si cells.
Down-regulated proteins for stimulus/stress responses (P14–P18) in +Si cells.
We detected five down-regulated proteins for stress responses in +Si cells (Table 1). Of the three detected proteins involved in the glutathione (GSH) system, one is glutaredoxin (P14, GRX) that catalyses the reduction of disulfides or glutathione mixed disulfides (Lemaire, 2004) and scavenge reactive oxygen species (Lin et al., 2014), and two (P15 and P16, GSTs) are glutathione transferases involved in cellular detoxification by conjugating the tripeptide (γ-Glu-Cys-Gly) glutathione (GSH) to a wide variety of substrates (Jain et al., 2010). Two additional proteins are protein disulfide isomerase (P17, PDI) as a catalyst of disulfide bond formation and rearrangement (Satoh-Cruz et al., 2010), and 1-cys peroxiredoxin (P18, 1-cysPrx) as a novel antioxidant enzyme able to reduce phospholipid hydroperoxides in vitro by using glutathione as a reductant (Manevich et al., 2002). 1-Cys Prx has the peculiarity of using its unique cysteine residue to accomplish the functions of both peroxidatic and resolving cysteines when GSH is not available (Pedrajas et al., 2016).
These analyses suggest that Si-deprived cells are likely to confront local stress that induces the production of endobiotic toxicants and reactive oxygen species, resulting in the up-regulation of these five proteins (P14–P18) to sequester them as a stress response. This is consistent with an up-regulated protein Ser/Thr phosphatase 2C (P6, PP2C), which is shown to be involved in sophisticated stress signalling (Schweighofer et al., 2004; Xue et al., 2008), such as hyperosmotic stress, to cause cell dehydration due to decreased turgor pressure and water loss (Boudsocq and Laurière, 2005). Indeed, Si-deprived cells are more sensitive to both hyperosmotic and hypo-osmotic stress than Si-accumulating cells (Figs S4 and S5), and Si crosslinking of wall components withstands the effects of water loss and provides cells with a mechanical defence against cell-shape changes (Fig. S4B, C) and the formation of defects (Fig. 3A) exacerbated by osmotic pressure changes.
CONCLUSIONS
The combination of iTRAQ results (Table 1), wall mechanical strength (Fig. 3) and membrane stability (Figs 4 and 5) supports a role of Si bound to the cell wall in stabilizing cell membranes with improved ammonium transport efficiency (Fig. 1) under the same level of expression of AMT1;1 (P5). We demonstrate that the rigid cell walls as the structural basis are able to withstand the enormous turgor pressure exerted by the solute inside each rice cell to stabilize cell membranes and optimize nutrient uptake of rice cells in the same growth phase, thereby supporting survival, growth and metabolism, especially when cells are under osmotic stresses. Further analyses coupling proteomics results with targeted quantitative PCR may provide an additional level of understanding of the actions of Si.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Table S1: Unregulated but function-known membrane-related proteins identified by iTRAQ. Table S2: Si-regulated, including function-unknown, and unanalysable, function-decentralized proteins identified by iTRAQ. Table S3: The primers of the genes used for qPCR analysis. Fig. S1: Rice suspension cells. Fig. S2: Total protein concentrations in rice cells. Fig. S3: Shape changes in rice cells under hyperosmotic stress. Fig. S4: Viability of rice suspension cells cultured in the presence of PEG. Fig. S5: The expression pattern of Oryza sativa ammonium transporters 1 (OsAMT1) in rice.
ACKNOWLEDGEMENTS
This work was supported by the National Natural Science Foundation of China (31672222 and 31172027) and the Fundamental Research Funds for the Central Universities (2662017PY061 and 2662015PY206).
LITERATURE CITED
- Amor Y, Haigler CH, Johnson S, Wainscott M, Delmer DP. 1995. A membrane-associated form of sucrose synthase and its potential role in synthesis of cellulose and callose in plants. Proceedings of the National Academy of Sciences of the United States of America 92: 9353–9357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts B, Bray D, Johnson A et al. . 2010. Essential cell biology, 3rd edn. New York: Garland Publishing. [Google Scholar]
- Boudsocq M, Laurière C. 2005. Osmotic signaling in plants. Multiple pathways mediated by emerging kinase families. Plant Physiology 138: 1185–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukherroub R, Morin S, Sharpe P, Wayner DD, Allongue P. 2000. Insights into the formation mechanisms of Si− OR monolayers from the thermal reactions of alcohols and aldehydes with Si (111)− H. Langmuir 16: 7429–7434. [Google Scholar]
- Brzezinski MA, Olson RJ, Chisholm SW. 1990. Silicon availability and cell-cycle progression in marine diatoms. Marine Ecology Progress Series 67: 83–96. [Google Scholar]
- Cao K, Nakajima R, Meyer HH, Zheng Y. 2003. The AAA-ATPase Cdc48/p97 regulates spindle disassembly at the end of mitosis. Cell 115: 355–367. [DOI] [PubMed] [Google Scholar]
- Carpita NC, Gibeaut DM. 1993. Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. The Plant Journal 3: 1–30. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. 2005. Growth of the plant cell wall. Nature Review Molecular and Cell Biology 6: 850–861. [DOI] [PubMed] [Google Scholar]
- Currie HA, Perry CC. 2009. Chemical evidence for intrinsic ‘Si’ within Equisetum cell walls. Phytochemistry 70: 2089–2095. [DOI] [PubMed] [Google Scholar]
- Detmann KC, Araújo L, Martins SC et al. . 2012. Silicon nutrition increases grain yield, which, in turn, exerts a feedforward stimulation of photosynthetic rates via enhanced mesophyll conductance and alters primary metabolism in rice. New Phytologist 196: 752–762. [DOI] [PubMed] [Google Scholar]
- Epstein E. 1994. The anomaly of silicon in plant biology. Proceedings of the National Academy of Sciences of the United States of America 91: 11–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan XR, Tang Z, Tan YW et al. . 2016. Overexpression of a pH-sensitive nitrate transporter in rice increases crop yields. Proceedings of the National Academy of Sciences of the United States of America 113: 7118–7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fincher GB. 1989. Molecular and cellular biology associated with endosperm mobilization in germinating cereal grains. Annual Review of Plant Biology 40: 305–346. [Google Scholar]
- Franková L, Fry SC. 2013. Biochemistry and physiological roles of enzymes that ‘cut and paste’ plant cell-wall polysaccharides. Journal of Experimental Botany 64: 3519–3550. [DOI] [PubMed] [Google Scholar]
- Fry SC, Nesselrode BH, Miller JG, Mewburn BR. 2008. Mixed-linkage (1→3, 1→4)-β-D-glucan is a major hemicellulose of Equisetum (horsetail) cell walls. New Phytologist 179: 104–115. [DOI] [PubMed] [Google Scholar]
- Geitmann A, Ortega JKE. 2009. Mechanics and modeling of plant cell growth. Trends in Plant Science 14: 467–478. [DOI] [PubMed] [Google Scholar]
- He CW, Wang LJ, Liu J et al. . 2013. Evidence for ‘silicon’ within the cell walls of suspension-cultured rice cells. New Phytologist 200: 700–709. [DOI] [PubMed] [Google Scholar]
- He CW, Ma J, Wang LJ. 2015. A hemicellulose-bound form of silicon with potential to improve the mechanical properties and regeneration of the cell wall of rice. New Phytologist 206: 1051–1062. [DOI] [PubMed] [Google Scholar]
- Höfte H. 2001. A baroque residue in red wine. Science 294: 795–797. [DOI] [PubMed] [Google Scholar]
- Isa M, Bai S, Yokoyama T et al. . 2010. Silicon enhances growth independent of silica deposition in a low-silica rice mutant, lsi1. Plant and Soil 331: 361–375. [Google Scholar]
- Jain M, Ghanashyam C, Bhattacharjee A. 2010. Comprehensive expression analysis suggests overlapping and specific roles of rice glutathione S-transferase genes during development and stress responses. BMC Genomics 11: 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konrad KR, Hedrich R. 2008. The use of voltage-sensitive dyes to monitor signal-induced changes in membrane potential-ABA triggered membrane depolarization in guard cell. The Plant Journal 55: 161–173. [DOI] [PubMed] [Google Scholar]
- Latterich M, Frohlich KU, Schekman R. 1995. Membrane fusion and the cell cycle: Cdc48p participates in the fusion of ER membranes. Cell 82: 885–893. [DOI] [PubMed] [Google Scholar]
- Lavinsky AO, Detmann KC, Reis JV et al. . 2016. Silicon improves rice grain yield and photosynthesis specifically when supplied during the reproductive growth stage. Journal of Plant Physiology 206: 125–132. [DOI] [PubMed] [Google Scholar]
- Law C, Exley C. 2011. New insight into silica deposition in horsetail (Equisetum arvense). BMC Plant Biology 11: 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Das A, Yamaguchi M et al. . 2003. Cell cycle function of a rice B2-type cyclin interacting with a B-type cyclin-dependent kinase. The Plant Journal 34: 417–425. [DOI] [PubMed] [Google Scholar]
- Lee T, Shultz RW, Hanley-Bowdoin L, Thompson WF. 2004. Establishment of rapidly proliferating rice cell suspension culture and its characterization by fluorescence-activated cell sorting analysis. Plant Molecular Biology Reporter 22: 259–267. [Google Scholar]
- Lemaire SD. 2004. The glutaredoxin family in oxygenic photosynthetic organisms. Photosynthesis Research 79: 305–318. [DOI] [PubMed] [Google Scholar]
- Lendvai A, Nikovics K, Bako L, Dudits D, Gyorgyey J. 2002. Synchronization of Oryza sativa L. cv. Taipei-309 cell suspension culture. Acta Biologica Szegediensis 46: 39–41. [Google Scholar]
- Lin Z, Zhang X, Yang X et al. . 2014. Proteomic analysis of proteins related to rice grain chalkiness using iTRAQ and a novel comparison system based on a notched-belly mutant with white-belly. BMC Plant Biology 14: 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Ma J, He C et al. . 2013. Inhibition of cadmium ion uptake in rice (Oryza sativa) cells by a wall-bound form of silicon. New Phytologist 200: 691–699. [DOI] [PubMed] [Google Scholar]
- Loqué D, von Wirén N. 2004. Regulatory levels for the transport of ammonium in plant roots. Journal of Experimental Botany 55: 1293–1305. [DOI] [PubMed] [Google Scholar]
- Loqué D, Yuan L, Kojima S et al. . 2006. Additive contribution of AtAMT1;1 and AtAMT1;3 to high-affinity ammonium uptake across the plasma membrane of nitrogen-deficient Arabidopsis roots. The Plant Journal 48: 522–534. [DOI] [PubMed] [Google Scholar]
- Lu M, Sautin YY, Holliday S, Gluck SL. 2004. The glycolytic enzyme aldolase mediates assembly, expression, and activity of vacuolar H+-ATPase. The Journal of Biological Chemistry 279: 8732–8739. [DOI] [PubMed] [Google Scholar]
- Luyckx M, Hausman JF, Lutts S, Guerriero G. 2017. Impact of silicon in plant biomass production: focus on bast fibres, hypotheses, and perspectives. Plants 6: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JF, Tamai K, Yamaji N et al. . 2006. A silicon transporter in rice. Nature 440: 688–691. [DOI] [PubMed] [Google Scholar]
- Ma J, Sheng HC, Li XL, Wang LJ. 2016a iTRAQ-based proteomic analysis reveals the mechanisms of silicon-mediated cadmium tolerance in rice (Oryza sativa) cells. Plant Physiology and Biochemistry 104: 71–80. [DOI] [PubMed] [Google Scholar]
- Ma J, Zhang XQ, Zhang WJ, Wang LJ. 2016b Multifunctionality of silicified nanoshells at cell surfaces of Oryza sativa. ACS Sustainable Chemistry & Engineering 4: 6792–6799. [Google Scholar]
- Manevich Y, Sweitzer T, Pak JH, Feinstein SI, Muzykantov V, Fisher AB. 2002. 1-Cys peroxiredoxin overexpression protects cells against phospholipid peroxidation-mediated membrane damage. Proceedings of the National Academy of Sciences of the United States of America 99: 11599–11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majzlova K, Pukajova Z, Janecek S. 2013. Tracing the evolution of the alpha-amylase subfamily GH13_36 covering the amylolytic enzymes intermediate between oligo-1,6-glucosidases and neopullulanases. Carbohydrate Research 367: 48–57. [DOI] [PubMed] [Google Scholar]
- Markovich O, Steiner E, Kouril S, Tarkowski P, Aharoni A, Elbaum R. 2017. Silicon promotes cytokinin biosynthesis and delays senescence in Arabidopsis and Sorghum. Plant, Cell and Environment 40: 1189–1196. [DOI] [PubMed] [Google Scholar]
- McMahon HT, Gallop JL. 2005. Membrane curvature and mechanisms of dynamic cell membrane remodeling. Nature 438: 590–596. [DOI] [PubMed] [Google Scholar]
- Meharg C, Meharg AA. 2015. Silicon, the silver bullet for mitigating biotic and abiotic stress, and improving grain quality, in rice?Environmental and Experimental Botany 120: 8–17. [Google Scholar]
- Moons A, Valcke R, Van Mondtagu M. 1998. Low‐oxygen stress and water deficit induce cytosolic pyruvate orthophosphate dikinase (PPDK) expression in roots of rice, a C3 plant. The Plant Journal 15: 89–98. [DOI] [PubMed] [Google Scholar]
- Morreale G, Conforti L, Coadwell J, Wilbrey AL, Coleman MP. 2009. Evolutionary divergence of valosin-containing protein/cell division cycle protein 48 binding interactions among endoplasmic reticulum-associated degradation proteins. FEBS Journal 276: 1208–1220. [DOI] [PubMed] [Google Scholar]
- Nelissen H, Gonzalez N, Dirk I. 2016. Leaf growth in dicots and monocots: so different yet so alike. Current Opinion in Plant Biology 33: 72–76. [DOI] [PubMed] [Google Scholar]
- Nwugo CC., Huerta AJ. 2011. The effect of silicon on the leaf proteome of rice (Oryza sativa L.) plants under cadmium-stress. Journal of Proteome Research 10: 518–528. [DOI] [PubMed] [Google Scholar]
- Ober ES, Sharp RE. 2003. Electrophysiological responses of maize roots to low water potentials: relationship to growth and ABA accumulation. Journal of Experimental Botany 54: 813–824. [DOI] [PubMed] [Google Scholar]
- O’Neill M, Eberhard S, Albersheim P, Darvill AG. 2001. Requirement of borate cross-linking of cell wall rhamnogalacturonan II for Arabidopsis growth. Science 294: 846–849. [DOI] [PubMed] [Google Scholar]
- Opassiri R, Pomthong B, Onkoksoong T, Akiyama T, Esen A, Ketudat Cairns JR. 2006. Analysis of rice glycosyl hydrolase family I and expression of Os4bglu 1 2 β-glucosidase. BMC Plant Biology 6: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrajas JR, McDonagh B, Hernandez-Torres F et al. . 2016. Glutathione is the resolving thiol for thioredoxin peroxidase activity of 1-cys peroxiredoxin without being consumed during the catalytic cycle. Antioxidants & Redox Signaling 24: 115–128. [DOI] [PubMed] [Google Scholar]
- Rabinovich E, Kerem A, Frohlich KU, Diamant N, Bar-Nun S. 2002. AAA-ATPase p97⁄Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation. Molecular and Cell Biology 22: 626–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratajczak R. 2000. Structure, function and regulation of the plant vacuolar H+-translocating ATPase. Biochimica et Biophysica Acta 1465: 17–36. [DOI] [PubMed] [Google Scholar]
- Rybak K, Steiner A, Synek L et al. . 2014. Plant cytokinesis is orchestrated by the sequential action of the TRAPPII and exocyst tethering complexes. Developmental Cell 29: 607–620. [DOI] [PubMed] [Google Scholar]
- Sandberg A, Lindell G, Källström BN et al. . 2012. Tumor proteomics by multivariate analysis on individual pathway data for characterization of vulvar cancer phenotypes. Molecular & Cellular Proteomics 11: 3929–3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanglard LMVP, Detmann KC, Martins SCV et al. . 2016. The role of silicon in metabolic acclimation of rice plants challenged with arsenic. Environmental and Experimental Botany 123: 22–36. [Google Scholar]
- Satoh-Cruz M, Crofts AJ, Takemoto-Kuno Y et al. . 2010. Protein disulfide isomerase like 1-1 participates in the maturation of proglutelin within the endoplasmic reticulum in rice endosperm. Plant Cell and Physiology 51: 1581–1593. [DOI] [PubMed] [Google Scholar]
- Savant NK, Snyder GH, Datnoff LE. 1997. Silicon management and sustainable rice production. Advances in Agronomy 58: 151–199. [Google Scholar]
- Scheller HV, Ulvskov P. 2010. Hemicelluloses. Plant Biology 61: 263–289. [DOI] [PubMed] [Google Scholar]
- Schmölzer K, Gutmann A, Diricks M, Desmet T, Nidetzky B. 2016. Sucrose synthase: a unique glycosyltransferase for biocatalytic glycosylation process development. Biotechnology Advances 34: 88–111. [DOI] [PubMed] [Google Scholar]
- Schweighofer A, Hirt H, Meskiene I. 2004. Plant PP2C phosphatases: emerging functions in stress signaling. Trends in Plant Science 9: 236–243. [DOI] [PubMed] [Google Scholar]
- Shinohara N, Sunagawa N, Tamura S, Yokoyama R, Ueda M, Igarashi K, Nishitani K. 2017. The plant cell-wall enzyme AtXTH3 catalyses covalent cross-linking between cellulose and cello-oligosaccharide. Scientific Reports 7: 46099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville C, Bauer S, Brininstool G et al. . 2004. Toward a systems approach to understanding plant cell walls. Science 306: 2206–2211. [DOI] [PubMed] [Google Scholar]
- Sonoda Y, Ikeda A, Saiki S, von Wirén N, Yamaya T, Yamaguchi J. 2003. Distinct expression and function of three ammonium transporter genes (OsAMT1;1-1;3) in rice. Plant and Cell Physiology 44: 726–734. [DOI] [PubMed] [Google Scholar]
- Sørensen I, Pettolino FA, Wilson SM et al. . 2008. Mixed-linkage (1→3, 1→4)-β-D-glucan is not unique to the Poales and is an abundant component of Equisetum arvense cell walls. The Plant Journal 54: 510–521. [DOI] [PubMed] [Google Scholar]
- Sun J, Wang MJ, Ding MQ et al. . 2010. H2O2 and cytosolic Ca2+ signals triggered by the PM H+-coupled transport system mediate K+/Na+ homeostasis in NaCl-stressed Populus euphratica cells. Plant, Cell & Environment 33: 943–958. [DOI] [PubMed] [Google Scholar]
- Sun SY, Chao DY, Li XM et al. . 2009. OsHAL3 mediates a new pathway in the light-regulated growth of rice. Nature Cell Biology 11: 845–851. [DOI] [PubMed] [Google Scholar]
- Sze H, Ward JM, Lai S, Perera I. 1992. Vacuolar-type H+-translocating ATPases in plant endomembranes: subunit organization and multigene families. Journal of Experimental Biology 172: 123–135. [DOI] [PubMed] [Google Scholar]
- Sze H, Li X, Palmgren MG. 1999. Energization of plant cell membranes by H+-pumping ATPases: regulation and biosynthesis. The Plant Cell 11: 677–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas BR, Romero GO, Nevins DJ, Rodriguez RL. 2000. New perspectives on the endo-beta-glucanases of glycosyl hydrolase Family 17. International Journal of Biological Macromolecules 27: 139–144. [DOI] [PubMed] [Google Scholar]
- Tsai KJ, Lin CY, Ting CY, Shih MC. 2016. Ethylene-regulated glutamate dehydrogenase fine-tunes metabolism during anoxia-reoxygenation. Plant Physiology 172: 1548–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeda M, Uchimiya H. 1994. Differential transcript levels of genes associated with glycolysis and alcohol fermentation in rice plants (Oryza sativa L.) under submergence stress, Plant Physiology 106: 1015–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeda M, Iwamoto N, Umeda-Hara C, Yamaguchi M, Hashimoto J, Uchimiya H. 1999. Molecular characterization of mitotic cyclins in rice plants. Molecular and General Genetics 262: 230–238. [DOI] [PubMed] [Google Scholar]
- Verma DPS, Gu X. 1996. Vesicle dynamics during cell-plate formation in plants. Trends in Plant Science 1: 145–148. [Google Scholar]
- Whitlow TH, Bassuk NL, Ranney TG, Reichert DL. 1992. An improved method for using electrolyte leakage to assess membrane competence in plant tissues. Plant Physiology 98: 198–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski JR, Zougman A, Nagaraj N, Mann M. 2009. Universal sample preparation method for proteome analysis. Nature Methods 6: 359–362. [DOI] [PubMed] [Google Scholar]
- Xue T, Wang D, Zhang S et al. . 2008. Genome-wide and expression analysis of protein phosphatase 2C in rice and Arabidopsis. BMC Genomics 9: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L, Loqué D, Ye F, Frommer WB, von Wirén N. 2007. Nitrogen-dependent posttranscriptional regulation of the ammonium transporter AtAMT1;1. Plant Physiology 143: 732–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu YX, Guo J, Feng R, Jia JH, Han WH, Gong HJ. 2016. The regulatory role of silicon on carbohydrate metabolism in Cucumis sativus L. under salt stress. Plant and Soil 406: 231–249. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.